Abstract
The aim of our study was to determine the impact of genetic polymorphisms in the caspase (CASP) genes on prognosis of hepatocellular carcinoma (HCC). We genotyped 7 potentially functional polymorphisms in CASP3, CASP7, CASP8, CASP9, CASP10 genes in 362 HCC patients of receiving surgical resection of HCC tumor. The associations of genotype and haplotype with overall survival (OS) and disease free survival (DFS) were analyzed by using the Cox proportional hazards model. We found that the CASP9 rs4645981 C allele was significantly associated with positive effect on DFS (P = 0.011 and 0.016 for CT+CC vs. TT in univariate and multivariate analysis, respectively), CT genotype was associated with a better OS of HCC than the TT genotype both in univariate and multivariate analysis (P = 0.048 and 0.041, respectively). Moreover, the CASP3 rs2705897 GT genotype showed marginally significant association with decreased OS and DFS, compared with the GG genotype. One haplotype TT/TG in CASP3 (constructed by rs12108497 T>C and rs2705897 T>G) was significantly associated with decreased OS and DFS, compared to the common haplotype TT/TT both in univariate analysis (P = 0.021 and 0.026, respectively) and multivariate analysis (P = 0.025 and 0.030, respectively). The haplotype GT/GT in CASP9 (constructed by rs4645978 A>G and rs4645981 C>T) was significantly associated with decreased DFS both in univariate and multivariate analysis (P = 0.012 and 0.010, respectively). In conclusion, the CASP9 rs4645981 polymorphism, CASP3 and CASP9 haplotypes may be useful prognosis markers for HCC patients with surgical resection of tumor.
Introduction
Hepatocellular carcinoma (HCC), mainly caused by chronic hepatitis B virus (HBV) and hepatitis C virus (HCV) infections, is one of the most commonly diagnosed cancers and the third leading cause of cancer-related death worldwide [1]. In 2012, about 782,500 new HCC cases and 745,500 deaths occurred in the world, making the incidence and mortality rates almost equal [2]. Although attempts have been made to predict recurrence and prognosis in HCC patients using clinical factors, such as positive portal vein thrombosis, large tumor size, increased serum alpha-fetoprotein (AFP), vascular invasion and advanced tumor node metastasis (TNM) stage, the long-term prognosis remains poor with reported 5-year survival rates ranging from 17% to 53% [3]. Therefore it is essential to better understand the mechanism of cancer progression and development in HCC and identify potential biomarkers for prognosis prediction.
Apoptosis is a genetically controlled process of cell suicide, which plays an important role in multicellular organisms [4]. As we all know, inappropriate regulation of apoptosis mechanism facilitates the accumulation of somatic mutations and thereby contributes to tumor initiation, progression as well as metastasis [5–6]. Caspases (CASPs), members of a conserved family involved in signaling and execution in apoptosis pathways, are cysteine-aspartic acid proteases, which can be broadly divided into initiator (upstream) and effector (downstream) CASPs based on their functions [7]. To date, 14 family members have been identified [8]. CASP3, CASP6, and CASP7 belong to effector CASPs and they execute cell death process; CASP8, CASP9, and CASP10 transmit apoptotic signals and they belong to initiator CASPs [9]. All known CASPs possess an active-site cysteine and they cleave their substrates after the aspartic acid residue [10].
Many studies have shown that genetic polymorphisms in CASP genes are associated with risk of various human cancers, including HCC [9, 11–15]. However, the influence of the CASP genes-related polymorphisms on the prognosis of HCC have not been investigated extensively. Therefore, we selected 7 potentially functional SNPs in CASP3, CASP7, CASP8, CASP9, and CASP10 genes and aimed to determine whether polymorphisms in these genes are associated with prognosis of HCC.
Materials and methods
Patients
From April 1996 to September 2009, a total of 362 Chinese Han patients with primary HCC newly diagnosed and received surgical resection of HCC tumor were recruited by the Qidong Liver Cancer institute in Qidong, Jiangsu province, China. The clinical outcomes of HCC were recorded until October 2014, with a median follow-up time of 53.0 months, which range from 2 to 110 months. The clinical diagnosis of HCC was based on the National Comprehensive Cancer Network (NCCN) clinical practice guidelines in oncology and histopathological examination. Patients with secondary liver cancers were excluded from our study. Patients with no other cancers were determined at the initial screening examination and were followed-up every 3months by researchers from the time of enrollment, stopping until death or the last time of follow-up.
There were no restrictions on gender, age and tumor stage for recruitment and 5 ml whole blood was extracted for each subject. Clinical characteristics such as tumor size, differentiation, venous invasion, and son on were collected via medical records with approval of patients. The clinical typing of tumors were determined by the TNM classification system of International Union Against Cancer (edition 6) and the histologic grade of tumor differentiation was assigned by the Edmondson grading system. Overall survival (OS) and disease free survival (DFS) were used as endpoints for the study. OS was calculated from the date of pathologic diagnosis/recruitment to death or the end of available follow-up.
Disease free survival (DFS) was defined as the time from pathologic diagnosis/recruitment to disease recurrence, metastasis, disease specific death or last follow-up.
Written informed consent was obtained from each patient before enrollment, and this study was approved by the Department of Scientific Research of Fudan University as well as the Qidong Liver Cancer Institute.
SNP selection
To select potentially functional SNPs in CASP3, CASP7, CASP8, CASP9, and CASP10 genes, we utilized the International HapMap Project database (http://hapmap.ncbi.nlm.nih.gov/), and the dbSNP database (https://www.ncbi.nlm.nih.gov/projects/SNP/). Finally, a total of 7 SNPs were selected for genotyping (Table 1).
Table 1. SNPs selected in CASP genes and their allele frequencies.
| Gene | Chromosome | Location | Position | SNP | Allele | MAF (CHB)a | MAF (observed)b |
|---|---|---|---|---|---|---|---|
| CASP3 | 4q34 | 5' flank | 185571557 | rs12108497 | T>C | 0.282 | 0.256 |
| 5' flank | 185553098 | rs2705897 | A>C | 0.209 | 0.162 | ||
| CASP7 | 10q25 | T244S | 115489152 | rs2227310 | C>G | 0.427 | 0.391 |
| CASP8 | 2q33-q34 | Intron | 202151163 | rs3769818 | G>A | 0.291 | 0.265 |
| CASP9 | 1p36.21 | 5' flank | 15852034 | rs4645978 | A>G | 0.379 | 0.368 |
| 5' flank | 15851483 | rs4645981 | C>T | 0.214 | 0.136 | ||
| CASP10 | 2q33-q34 | L522I | 202082459 | rs13006529 | T>A | 0.185 | 0.184 |
MAF, minor allele frequency; CHB, Chinese Han in Beijing; SNP, single nucleotide polymorphism.
a MAF in Chinese Han population in Hapmap database.
b MAF in our studied population.
DNA extraction and genotyping
Genomic DNA was extracted from blood samples using the QIAamp DNA Mini Kit (GIAGEN GmbH, Hilden, Germany). Genotyping was performed with Sequenom MassARRAY iPLEX platform by use of allele-specific MALDI-TOF mass spectrometry assay. Polymerase chain reaction (PCR) and extension primers for these 7 SNPs were designed using the MassARRAY Assay Design 3.0 software (Sequenom). Duplicate test samples and two water samples (PCR negative controls) were included in each 96-well plate. Genotyping quality was examined by a detailed QC procedure consisting of >95% successful call rate, duplicate calling of genotypes, internal positive control samples.
Statistical analysis
The haplotypes were constructed for the genes with at least two SNPs using Bayesian algorithm by PHASE software. Survival curves were estimated using the Kaplan-Meier method. Hazard ratios (HRs) and 95% confidence intervals (CIs) were estimated for each analysis by using the Cox proportional hazards regression model. The effects of clinical variables, single SNP and haplotype on OS and DFS were assessed using the Cox proportional hazards regression model and log-rank test. All analyses were performed with SPSS software version 22 (SPSS, Chicago, IL). All tests were two-sided and a P<0.05 was considered statistically significant.
Results
Patient characteristics and clinical predictors
The clinical pathologic characteristics of the 362 HCC patients and their associations with OS are summarized in Table 2. There were 225 (62.2%) deaths at the time of analysis and the overall median survival time (MST) was 34.0 (95%CI, 27.4–40.6) months. In univariate analysis, tumor size and venous invasion were significantly associated with OS (P = 0.028 and 0.029, respectively) and DFS (P = 0.042 and 0.026, respectively). However, none of other clinical characteristics was significantly associated with OS or DFS.
Table 2. Clinical characteristics and their prediction of overall survival and disease free survival in HCC patients.
| Characteristics | No of patients | No of events | 5-y-survival (%) | MST (95%CI) | Overall survival (OS) | Disease free survival (DFS) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Log-rank P |
Hazard ratio (95% CI) | P | Log-rank P | Hazard ratio (95% CI) |
P | |||||
| Number | 362 | 225 | 30 | 34.0 (27.4–40.6) | ||||||
| Age (year) | 0.478 | 0.410 | ||||||||
| ≤50 | 186 | 113 | 30 | 35.0 (23.3–46.7) | 1.00 | |||||
| >50 | 176 | 112 | 29 | 33.0 (24.2–41.8) | 1.10 (0.85–1.43) | 0.483 | 1.11 (0.86–1.45) | 0.417 | ||
| Sex | 0.476 | 0.665 | ||||||||
| female | 63 | 41 | 27 | 31.0 (24.2–37.8) | 1.00 | |||||
| male | 299 | 184 | 30 | 37.0 (27.5–46.5) | 0.89 (0.63–1.24) | 0.481 | 0.93 (0.66–1.30) | 0.670 | ||
| Smoking | 0.265 | 0.200 | ||||||||
| never | 224 | 144 | 26 | 31.0 (23.2–38.8) | 1.000 | |||||
| ever | 138 | 81 | 37 | 39.0 (27.3–50.7) | 0.86 (0.65–1.13) | 0.270 | 0.84 (0.64–1.10) | 0.207 | ||
| Drinking | 0.615 | 0.728 | ||||||||
| never | 142 | 86 | 28 | 35.0 (19.6–50.4) | 1.00 | |||||
| ever | 220 | 139 | 31 | 33.0 (25.0–41.0) | 1.07 (0.82–1.40) | 0.619 | 1.05 (0.80–1.37) | 0.731 | ||
| Family history | 0.257 | 0.299 | ||||||||
| absent | 263 | 158 | 31 | 37.0 (27.3–46.7) | 1.00 | |||||
| present | 81 | 55 | 25 | 29.0 (17.4–40.6) | 1.19 (0.88–1.62) | 0.262 | 1.17 (0.86–1.60) | 0.306 | ||
| unkown | 18 | 18 | ||||||||
| HbsAg | 0.599 | 0.478 | ||||||||
| negative | 59 | 40 | 35 | 22.0 (6.7–37.3) | 1.00 | |||||
| positive | 303 | 185 | 28 | 37.0 (30.2–43.8) | 0.91 (0.65–1.29) | 0.603 | 0.89 (0.63–1.25) | 0.484 | ||
| AFP | 0.395 | 0.266 | ||||||||
| negative | 142 | 95 | 26 | 33.0 (25.7–40.3) | 1.00 | |||||
| positive | 214 | 127 | 32 | 35.0 (26.1–43.9) | 0.89 (0.68–1.16) | 0.400 | 0.86 (0.66–1.12) | 0.273 | ||
| unkown | 6 | 3 | ||||||||
| Tumor size (cm) | 0.026 | 0.039 | ||||||||
| ≤5 | 183 | 107 | 35 | 39.0 (28.1–49.9) | 1.00 | |||||
| >5 | 179 | 118 | 24 | 30.0 (21.0–39.0) | 1.34 (1.03–1.75) | 0.028 | 1.31 (1.01–1.71) | 0.042 | ||
| Differentiation | 0.568 | 0.390 | ||||||||
| Ⅰ+Ⅱ | 196 | 122 | 28 | 37.0 (27.5–46.5) | 1.00 | |||||
| Ⅲ+Ⅳ | 155 | 96 | 32 | 34.0 (26.0–42.0) | 0.93 (0.71–1.21) | 0.572 | 0.89 (0.68–1.16) | 0.397 | ||
| unkown | 11 | 7 | ||||||||
| Tumor capsule | 0.495 | 0.432 | ||||||||
| absent | 177 | 113 | 28 | 31.0 (22.7–39.3) | 1.00 | |||||
| present | 181 | 110 | 31 | 37.0 (26.1–47.9) | 0.91 (0.70–1.19) | 0.499 | 0.90 (0.69–1.17) | 0.439 | ||
| unkown | 4 | 2 | ||||||||
| Venous invasion | 0.026 | 0.023 | ||||||||
| absent | 257 | 150 | 33 | 39.0 (29.3–48.7) | 1.00 | |||||
| present | 102 | 73 | 22 | 26.0 (20.1–31.9) | 1.37 (1.03–1.81) | 0.029 | 1.38 (1.04–1.82) | 0.026 | ||
| unkown | 3 | 2 | ||||||||
| Cirrhosis | 0.706 | 0.705 | ||||||||
| absent | 121 | 79 | 30 | 27.0 (13.6–40.4) | 1.00 | |||||
| present | 239 | 145 | 30 | 36.0 (29.6–42.4) | 0.95 (0.72–1.25) | 0.708 | 0.95 (0.72–1.25) | 0.709 | ||
| unkown | 2 | 1 | ||||||||
| Tumor number | 0.701 | 0.644 | ||||||||
| solitary | 279 | 172 | 30 | 34.0 (26.0–42.0) | 1.00 | |||||
| multiple | 83 | 53 | 27 | 35.0 (24.5–45.5) | 1.06 (0.78–1.45) | 0.704 | 1.07 (0.79–1.46_ | 0.649 | ||
| pTNM stage | 0.225 | 0.339 | ||||||||
| Ⅰ+Ⅱ | 309 | 188 | 31 | 37.0 (30.7–43.3) | 1.00 | |||||
| Ⅲ+Ⅳ | 39 | 27 | 24 | 22.0 (13.0–31.0) | 1.28 (0.86–1.92) | 0.231 | 1.21 (0.81–1.82) | 0.346 | ||
| unkown | 14 | 10 | ||||||||
MST, median survival time; CI, confidence interval; AFP, serum α-fetoprotein.
Association analysis of SNPs with OS and DFS of HCC patients
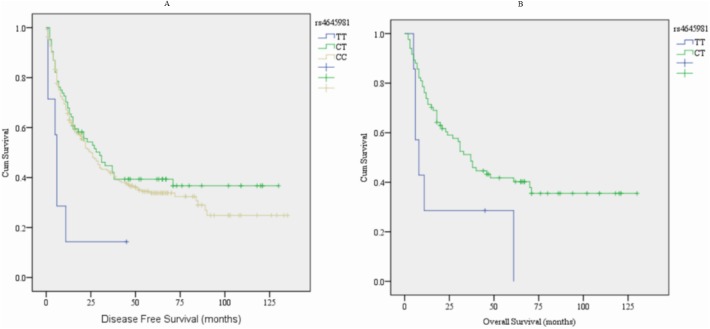
There are 7 SNPs among CASP genes (CASP3, CASP7, CASP8, CASP9, CASP10) in the present study. The associations of these genetic polymorphisms with OS and DFS are detailed in Table 3. In univariate analysis, the CASP3 rs2705897 GT genotype possessed a marginally significant association with decreased OS and DFS, compared with the GG genotype (P = 0.072 and 0.078, respectively, Table 3). The CASP9 rs4645981 CT, CC and CT+CC genotypes were associated with significantly increased DFS, compared with the TT genotype (P = 0.012, 0.013 and 0.011, respectively, Table 3, Fig 1A). However, the CASP9 rs4645981 CC and CT+CC genotypes showed a marginally significant association with positive effect on OS, compared with the TT genotype (P = 0.066 and 0.057, respectively). Furthermore, the rs4645981 CT genotype showed a statistically significant association with OS, compared with the TT genotype (P = 0.048, Table 3, Fig 1B).
Table 3. Univariate and multivariate Cox regression analysis of genotypes in HCC patients.
| Genotype | No of patients | No of events | 5-y-survival | MST (95%CI) |
OS | DFS | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | |||||||||
| Hazard ratio (95% CI) | P | Hazard ratio (95% CI) |
pa |
Hazard ratio (95% CI) | P | Hazard ratio (95% CI) |
pa |
|||||
| CASP3_rs12108497 | ||||||||||||
| CC | 30 | 18 | 46 | 50.0 (8.0–92.0) | 1.0 | 1.0 | ||||||
| CT | 125 | 81 | 36 | 38.0 (22.5–53.5) | 1.32 (0.79–2.20) | 0.288 | 1.35 (0.81–2.25) | 0.254 | 1.45 (0.87–2.43) | 0.155 | 1.49 (0.89–2.49) | 0.129 |
| TT | 206 | 126 | 36 | 31.0 (22.3–39.7) | 1.23 (0.75–2.02) | 0.404 | 1.24 (0.76–2.04) | 0.388 | 1.30 (0.79–2.13) | 0.302 | 1.32 (0.80–2.16) | 0.274 |
| CT+TT | 331 | 207 | 36 | 33.0 (26.8–39.2) | 1.26 (0.78–2.04) | 0.344 | 1.28 (0.79–2.07) | 0.315 | 1.34 (0.83–2.17) | 0.236 | 1.37 (0.84–2.22) | 0.204 |
| CASP3_rs2705897 | ||||||||||||
| GG | 10 | 5 | 47 | 42.0 | 1.0 | 1.0 | 1.0 | 1.0 | ||||
| GT | 96 | 69 | 32 | 27.0 (17.7–36.3) | 2.35 (0.93–5.96) | 0.072 | 2.32 (0.92–5.87) | 0.075 | 2.30 (0.91–5.82) | 0.078 | 2.35 (0.93–5.94) | 0.070 |
| TT | 253 | 151 | 38 | 37.0 (30.3–43.7) | 1.49 (0.61–3.64) | 0.380 | 1.63 (0.67–3.99) | 0.283 | 1.52 (0.62–3.71) | 0.357 | 1.68 (0.68–4.10) | 0.259 |
| GT+TT | 349 | 220 | 36 | 33.0 (26.8–39.2) | 1.67 (0.69–4.06) | 0.259 | 1.77 (0.73–4.31) | 0.207 | 1.69 (0.70–4.12) | 0.245 | 1.83 (0.75–4.46) | 0.182 |
| CASP7_rs2227310 | ||||||||||||
| CC | 139 | 87 | 36 | 33.0 (21.0–45.0) | 1.0 | 1.0 | 1.0 | 1.0 | ||||
| CG | 158 | 95 | 39 | 35.0 (27.8–42.2) | 0.97 (0.72–1.30) | 0.832 | 0.91 (0.68–1.22) | 0.524 | 0.98 (0.73–1.31) | 0.868 | 0.93 (0.69–1.25) | 0.616 |
| GG | 61 | 40 | 33 | 37.0 (17.5–56.5) | 0.98 (0.68–1.43) | 0.929 | 1.01 (0.69–1.48) | 0.954 | 1.03 (0.71–1.50) | 0.876 | 1.06 (0.73–1.55) | 0.751 |
| CG+GG | 219 | 135 | 37 | 37.0 (30.5–43.5) | 0.97 (0.74–1.27) | 0.844 | 0.94 (0.72–1.24) | 0.669 | 1.00 (0.76–1.30) | 0.960 | 0.97 (0.74–1.28) | 0.828 |
| CASP8_rs3769818 | ||||||||||||
| CC | 198 | 122 | 38 | 38.0 (25.7–50.3) | 1.0 | 1.0 | 1.0 | 1.0 | ||||
| CT | 123 | 76 | 34 | 29.0 (18.5–39.5) | 1.08 (0.81–1.43) | 0.619 | 1.08 (0.81–1.44) | 0.620 | 1.05 (0.79–1.40) | 0.740 | 1.05 (0.79–1.40) | 0.734 |
| TT | 32 | 23 | 33 | 33.0 (26.5–39.5) | 1.18 (0.76–1.84) | 0.466 | 1.16 (0.74–1.81) | 0.525 | 1.24 (0.80–1.94) | 0.340 | 1.22 (0.78–1.92) | 0.383 |
| CT+TT | 155 | 99 | 32 | 31.0 (22.2–39.8) | 1.10 (0.84–1.43) | 0.495 | 1.10 (0.84–1.43) | 0.514 | 1.09 (0.84–1.42) | 0.528 | 1.09 (0.83–1.42) | 0.546 |
| CASP9_rs4645978 | ||||||||||||
| AA | 145 | 93 | 35 | 37.0 (29.0–45.0) | 1.0 | 1.0 | 1.0 | 1.0 | ||||
| AG | 165 | 99 | 39 | 31.0 (18.5–43.5) | 0.98 (0.74–1.30) | 0.885 | 0.98 (0.74–1.30) | 0.890 | 1.00 (0.75–0.33) | 0.995 | 1.00 (0.75–1.33) | 0.982 |
| GG | 50 | 32 | 34 | 24.0 (8.0–40.0) | 1.11 (0.74–1.66) | 0.620 | 1.11 (0.74–1.66) | 0.609 | 1.12 (0.75–1.67) | 0.584 | 1.11 (0.74–1.65) | 0.626 |
| AG+GG | 215 | 131 | 36 | 31.0 (22.3–39.7) | 1.01 (0.77–1.32) | 0.952 | 1.01 (0.77–1.32) | 0.953 | 1.03 (0.79–1.34) | 0.854 | 1.02 (0.78–1.33) | 0.883 |
| CASP9_rs4645981 | ||||||||||||
| TT | 7 | 6 | 23 | 8.0 (2.9–13.1) | 1.0 | 1.0 | 1.0 | 1.0 | ||||
| CT | 84 | 50 | 41 | 37.0 (27.7–46.3) | 0.42 (0.18–0.99) | 0.048 | 0.38 (0.15–0.96) | 0.041 | 0.33 (0.14–0.79) | 0.012 | 0.34 (0.14–0.83) | 0.018 |
| CC | 270 | 168 | 36 | 35.0 (27.0–43.0) | 0.47 (0.21–1.05) | 0.066 | 0.51 (0.22–1.17) | 0.112 | 0.35 (0.16–0.80) | 0.013 | 0.37 (0.16–0.86) | 0.021 |
| CT+CC | 354 | 218 | 37 | 35.0 (28.5–41.5) | 0.45 (0.20–1.02) | 0.057 | 0.47 (0.21–1.08) | 0.077 | 0.35 (0.15–0.78) | 0.011 | 0.36 (0.16–0.83) | 0.016 |
| CASP10_rs13006529 | ||||||||||||
| AA | 12 | 8 | 27 | 42.0 (3.5–80.5) | 1.0 | 1.0 | 1.0 | 1.0 | ||||
| AT | 109 | 62 | 40 | 31.0 (17.8–44.2) | 0.89 (0.43–1.87) | 0.764 | 0.95 (0.45–2.01) | 0.895 | 0.96 (0.46–2.00) | 0.912 | 1.02 (0.49–2.16) | 0.951 |
| TT | 236 | 150 | 37 | 34.0 (27.1–40.9) | 0.92 (0.45–1.87) | 0.813 | 1.02 (0.50–2.09) | 0.960 | 1.04 (0.51–2.12) | 0.918 | 1.20 (0.58–2.47) | 0.625 |
| AT+TT | 345 | 212 | 36 | 34.0 (27.4–40.6) | 0.91 (0.45–1.84) | 0.789 | 0.99 (0.49–2.03) | 0.987 | 1.01 (0.50–2.05) | 0.977 | 1.13 (0.56–2.31) | 0.733 |
MST, median survival time; CI, confidence interval.
a Adjusted by tumor size and venous invasion.
Fig 1. Kaplan-Meier survival curves of CASP9 rs4645981 with clinical outcomes of 362 HCC patients.
(A) disease free survival (DFS) (B) overall survival (OS).
A multivariate analysis of genotype association with OS and DFS of HCC patients was conducted by using Cox proportional hazards model, adjusted for the two significant clinical predictors (tumor size and venous invasion), and the results were similar to the univariate analysis. The GT genotype of CASP3 rs2705897 showed a suggestively negative effect on OS and DFS of HCC patients, compared with the GG genotype (P = 0.075 and 0.070, respectively, Table 3). The CASP9 rs4645981 CT, CC and CT+CC genotypes were still significantly associated with increased DFS, compared with the TT genotype (P = 0.018, 0.021 and 0.016, respectively, Table 3). However, the CT+CC genotype of CASP9 rs4645981 presented a suggestively positive effect on OS, and the CT genotype of rs4645981 showed a positive effect on OS of HCC patients, compared with the TT genotype (P = 0.041, Table 3).
Association analysis of haplotypes with OS and DFS of HCC patients
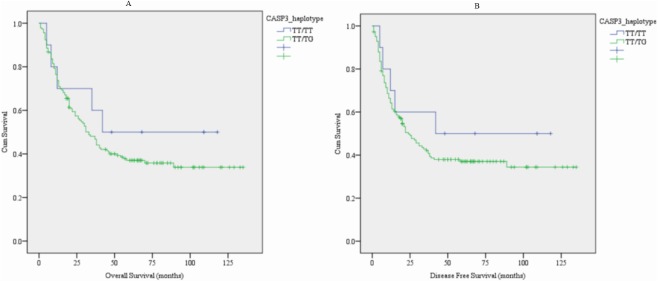
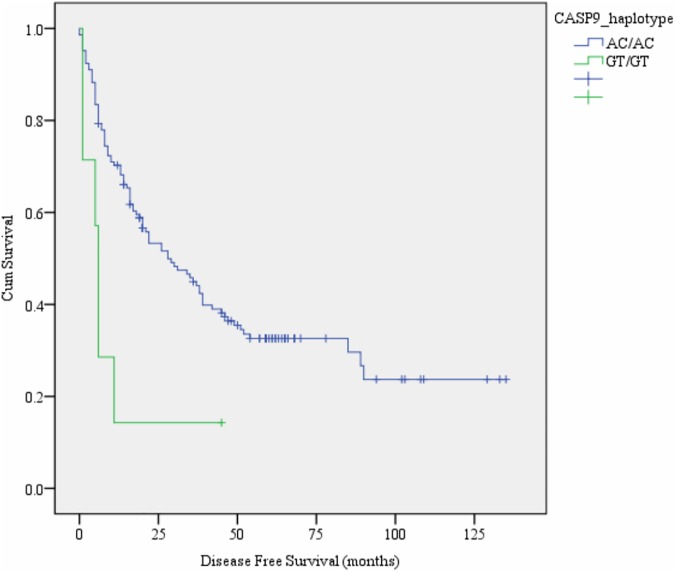
Since two SNPs in both CASP3 (rs12108497 T>C and rs2705897 T>G) and CASP9 (rs4645978 A>G and rs4645981 C>T) were selected in the present study, we constructed haplotypes for each of the two genes. We examined the associations of these haplotypes with OS and DFS of HCC patients. The detailed information is shown in Table 4. We attained 7 haplotypes in CASP3 and 6 haplotypes in CASP9. In univariate analysis, CASP3 haplotype TT/TG was significantly associated with OS (P = 0.021, Fig 2A) and DFS (P = 0.026, Fig 2B), compared to the common haplotype TT/TT (Table 4). CASP9 haplotype GT/GT was significantly associated with decreased DFS and showed marginally associated with OS, compared to the common haplotype AC/AC (P = 0.012, Fig 3, Table 4).
Table 4. Univariate and multivariate Cox regression analysis of haplotypes in HCC patients.
| Haplotype | No of patients | No of events | 5-y-survival | MST (95%CI) |
OS | DFS | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | |||||||||
| Hazard ratio (95% CI) | P | Hazard ratio (95% CI) |
pa |
Hazard ratio (95% CI) | P | Hazard ratio (95% CI) |
pa |
|||||
| CASP3_haplotype | ||||||||||||
| TT/TT | 183 | 108 | 37 | 33.0 (25.7–40.4) | 1.0 | 1.0 | 1.0 | 1.0 | ||||
| CG/CG | 10 | 5 | 47 | 42.0 | 0.69 (0.28–1.69) | 0.416 | 0.64 (0.26–1.58) | 0.333 | 0.68 (0.28–1.66) | 0.395 | 0.62 (0.25–1.53) | 0.299 |
| CG/TT | 62 | 43 | 34 | 36.0 (14.3–57.7) | 1.19 (0.84–1.70) | 0.325 | 1.19 (0.83–1.69) | 0.350 | 1.26 (0.88–1.79) | 0.202 | 1.26 (0.88–1.80) | 0.200 |
| CT/CG | 12 | 8 | 36 | 26.0 (10.7–41.7) | 1.08 (0.53–2.21) | 0.840 | 0.98 (0.47–2.02) | 0.954 | 1.00 (0.49–2.05) | 0.996 | 0.90 (0.44–1.86) | 0.782 |
| CT/CT | 8 | 5 | 60 | 72.0(27.4–116.6) | 0.80 (0.33–1.96) | 0.622 | 0.97 (0.39–2.41) | 0.939 | 0.78 (0.32–1.92) | 0.592 | 0.97 (0.39–2.42) | 0.939 |
| TT/CT | 61 | 38 | 36 | 38.0 (23.4–52.6) | 1.07 (0.74–1.54) | 0.735 | 1.12 (0.77–1.62) | 0.562 | 1.10 (0.76–1.60) | 0.607 | 1.13 (0.78–1.64) | 0.512 |
| TT/TG | 22 | 18 | 24 | 17.0 (8.0–26.0) | 1.81 (1.09–2.98) | 0.021 | 1.77 (1.08–2.93) | 0.025 | 1.77 (1.07–2.91) | 0.026 | 1.74 (1.05–2.87) | 0.030 |
| CASP9_haplotype | ||||||||||||
| AC/AC | 145 | 93 | 35 | 37.0 (19.0–45.0) | 1.0 | 1.0 | 1.0 | 1.0 | ||||
| AC/GC | 100 | 61 | 36 | 30.0 (15.4–44.6) | 1.06 (0.76–1.46) | 0.742 | 1.10 (0.79–1.52) | 0.574 | 1.09 (0.79–1.51) | 0.590 | 1.12 (0.81–1.55) | 0.504 |
| GC/GC | 24 | 13 | 42 | 26.0 (2.9–49.1) | 0.89 (0.50–1.60) | 0.704 | 0.87 (0.49–1.57) | 0.653 | 0.88 (0.49–1.58) | 0.678 | 0.83 (0.46–1.51) | 0.549 |
| GC/GT | 19 | 13 | 28 | 33.0 (11.4–54.6) | 1.14 (0.64–2.03) | 0.666 | 1.23 (0.68–2.21) | 0.498 | 1.11 (0.62–1.99) | 0.717 | 1.18 (0.66–2.13) | 0.571 |
| GT/AC | 64 | 37 | 43 | 38.0 (19.7–56.3) | 0.87 (0.59–1.27) | 0.462 | 0.83 (0.56–1.21) | 0.328 | 0.87 (0.59–1.27) | 0.464 | 0.83 (0.57–1.22) | 0.340 |
| GT/GT | 7 | 6 | 23 | 8.0 (2.9–13.1) | 2.19 (0.95–5.02) | 0.064 | 2.35 (0.98–5.64) | 0.055 | 2.91 (1.26–6.71) | 0.012 | 3.16 (1.32–7.57) | 0.010 |
MST, median survival time; CI, confidence interval.
a Adjusted by tumor size and venous invasion.
Fig 2. Kaplan-Meier survival curves of CASP3_haplotype with clinical outcomes of HCC patients.
(A) overall survival (OS), and (B) disease free survival (DFS).
Fig 3. Kaplan-Meier survival curves of CASP9_haplotype with disease free survival of HCC patients.
Similar results were found in multivariate analysis adjusted for tumor size and venous invasion. The CASP3 haplotype TT/TG presented a negative effect on OS (P = 0.025) and DFS (P = 0.030) of HCC patients, compared to the common haplotype TT/TT (Table 4). Meanwhile, CASP9 haplotype GT/GT presented a negative effect on DFS (P = 0.010) and suggestively negative effect on OS (P = 0.055) of HCC patients, compared to the common haplotype AC/AC (Table 4).
Discussion
Though many investigations have reported associations of SNPs in CASP genes with several types of cancer, studies of genetic polymorphisms in CASP genes on susceptibility to HCC is few, not to mention relationship between CASP polymorphisms and prognosis of HCC. The aim of our study was to evaluate genetic variants of CASP genes in relation to survival outcomes of HCC patients. To the best of our knowledge, this is the first evidence showing the relationship between genetic polymorphisms of CASP genes and prognosis of HCC patients. Our results revealed that CASP9 rs4645981 C allele was significantly increased DFS compared with the T allele and only the CT genotype was significantly associated with positive effect on OS, compared with the TT genotype. Moreover, the haplotype TT/TG (constructed by rs12108497 T>C, rs2705897 T>G) in CASP3 gene was significantly associated with decreased OS and DFS. The haplotype GT/GT in CASP9 was only significantly associated with decreased DFS. These findings suggest that the CASP9 rs4645981 and the haplotype TT/TG in CASP3 and GT/GT in CASP9 may be useful markers for predicting prognosis of HCC patients.
Failure of apoptosis is a hallmark of human cancers. As an effector CASP, CASP3 plays an important role in the execution phase of apoptosis, also in the development and progression of cancers [16–17]. Several previous studies have shown associations of CASP3 polymorphisms on the risk of different types of cancer, including HCC [13, 18–20]. Moreover, studies in several tumor types indicated that the expression levels of CASP3 have effects on the development and survival of cancers [8, 17, 21]. In our study, the haplotype TT/TG (constructed by rs12108497 T>C, rs2705897 T>G) was significantly associated with decreased OS and DFS in patients with HCC. The findings of previous studies and ours suggest that polymorphisms in CASP3 may increase risk of development of HCC and lead to poor survival outcome in patients with HCC, through reducing the apoptotic capacity.
CASPs have two distinct but converging pathways for activation, including extrinsic pathway and intrinsic pathway. CASP9, an important initiator CASP of the intrinsic pathway, is activated by the release of cytochrome c from mitochondria, activates downstream the effector CASP3 and CASP7 [22–23]. Many previous studies have shown that polymorphisms in CASP9 were associated with various cancer types, especially in the promoter region. For example, Theodoropoulos GE et al. [22] evaluated the association between two SNPs (rs4645978, rs4645981) in the promoter region of CASP9 and the risk of breast cancer. They found that the rs4645978 G allele was at high risk for breast cancer development and similar results for the rs4645981 T allele, which was significantly associated with increased risk of breast cancer, compared with those harboring the CC genotype. However, Park JY et al. [24] found that CASP9 rs4645978 polymorphism played a protective role in susceptibility to lung cancer risk and the rs4645981 T allele was at a significantly increased risk of lung cancer compared with those harboring the CC genotype. Moreover, previous studies demonstrated that CASP9 polymorphisms and expression were associated with prognosis of cancers [25–26]. To our best knowledge, the present study showed the first evidence of association between polymorphisms in CASP9 and the prognosis of HCC patients.
We acknowledge that there were several limitations in our study. First, the sample size of the present study was relatively small. Therefore, larger sample size and follow-up studies are warranted to confirm our findings. Second, determination of the exact functional influence was not performed in our study. Functional studies on biological mechanisms are needed to investigate in further studies. Third, other treatment information such as whether or not receiving targeted therapy was not collected in our study, except for surgery which is the most important factor for prognosis of patients. Finally, though two clinical and pathologic characteristics showed significant associations with OS and DFS, including tumor size and venous invasion, it is regretful that we failed to collect accurate information of these factors in our study. We only performed multivariate analysis by adjusting these potential prognostic factors. Further studies are essential to evaluate the role of genetic polymorphisms in HCC patients with more complete and comprehensive clinical pathologic characteristics.
In conclusion, our results provide suggestive evidence that CASP9 and CASP3 genetic polymorphisms may be independent prognosis markers for HCC patients with surgical resection of tumor. This study is the first evidence showing the relationship between genetic polymorphisms of CASP genes and survival outcomes in HCC patients, more comprehensive studies are needed to confirm our findings and investigate the associations between CASP genetic polymorphisms and prognosis of HCC patients.
Supporting information
(ZIP)
Acknowledgments
We thank all the participants in this study.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This project was funded by the National Natural Science Foundation of China (Grant # 31100895/81472618/81670535) to De-Ke Jiang. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Waziry R, Grebely J, Amin J, Alavi M, Hajarizadeh B, George J, et al. Trends in hepatocellular carcinoma among people with HBV or HCV notification in Australia (2000–2014). J Hepatol. 2016; 65: 1086–1093. doi: 10.1016/j.jhep.2016.08.010 [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015; 65: 87–108. doi: 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 3.Singhal A, Jayaraman M, Dhanasekaran DN, Kohli V. Molecular and serum markers in hepatocellular carcinoma: predictive tools for prognosis and recurrence. Crit Rev Oncol Hematol. 2012; 82: 116–40. doi: 10.1016/j.critrevonc.2011.05.005 [DOI] [PubMed] [Google Scholar]
- 4.Raff M. Cell suicide for beginners. Nature. 1998; 396: 119–122. doi: 10.1038/24055 [DOI] [PubMed] [Google Scholar]
- 5.Lowe SW, Lin AW. Apoptosis in cancer. Carcinogenesis. 2000; 21: 485–495. [DOI] [PubMed] [Google Scholar]
- 6.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001; 411: 342–348. doi: 10.1038/35077213 [DOI] [PubMed] [Google Scholar]
- 7.Shivapurkar N, Reddy J, Chaudhary PM, Gazdar AF. Apoptosis and lung cancer: a review. J Cell Biochem. 2003; 88: 885–98. doi: 10.1002/jcb.10440 [DOI] [PubMed] [Google Scholar]
- 8.Pu X, Storr SJ, Zhang Y, Rakha EA, Green AR, Ellis IO, et al. Caspase-3 and caspase-8 expression in breast cancer: caspase-3 is associated with survival. Apoptosis. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoo SS, Choi JE, Lee WK, Choi YY, Kam S, Kim MJ, et al. Polymorphisms in the CASPASE genes and survival in patients with early-stage non-small-cell lung cancer. J Clin Oncol. 2009; 27: 5823–9. doi: 10.1200/JCO.2009.23.1738 [DOI] [PubMed] [Google Scholar]
- 10.Hengartner MO. The biochemistry of apoptosis. Nature. 2000; 407: 770–6. doi: 10.1038/35037710 [DOI] [PubMed] [Google Scholar]
- 11.Dabrowska C, Li M, Fan Y. Apoptotic Caspases in Promoting Cancer: Implications from Their Roles in Development and Tissue Homeostasis. Adv Exp Med Biol. 2016; 930: 89–112. doi: 10.1007/978-3-319-39406-0_4 [DOI] [PubMed] [Google Scholar]
- 12.Sun T, Gao Y, Tan W, Ma S, Shi Y, Yao J, et al. A six-nucleotide insertion-deletion polymorphism in the CASP8 promoter is associated with susceptibility to multiple cancers. Nat Genet. 2007; 39: 605–13. doi: 10.1038/ng2030 [DOI] [PubMed] [Google Scholar]
- 13.Deng B, Liu F, Luo L, Wei Y, Li B, Yang H. CASP3 genetic polymorphisms and risk of Hepatocellular carcinoma: a case-control study in a Chinese population. Tumour Biol. 2016; 37: 8985–91. doi: 10.1007/s13277-015-4779-y [DOI] [PubMed] [Google Scholar]
- 14.Frank B, Hemminki K, Wappenschmidt B, Meindl A, Klaes R, Schmutzler RK, et al. Association of the CASP10 V410I variant with reduced familial breast cancer risk and interaction with CASP8 D302H variant. Carcinogenesis. 2006; 27: 606–609. doi: 10.1093/carcin/bgi248 [DOI] [PubMed] [Google Scholar]
- 15.Cho S, Lee JH, Cho SB, Yoon KW, Park SY, Lee WS, et al. Epigenetic methylation and expression of caspase 8 and survivin in hepatocellular carcinoma. Pathol Int. 2010; 60: 203–11. doi: 10.1111/j.1440-1827.2009.02507.x [DOI] [PubMed] [Google Scholar]
- 16.Soung YH, Lee JW, Kim SY, Park WS, Nam SW, Lee JY, et al. Somatic mutations of CASP3 gene in human cancers. Hum Genet. 2004; 115: 112–5. doi: 10.1007/s00439-004-1129-3 [DOI] [PubMed] [Google Scholar]
- 17.Persad R, Liu C, Wu TT, Houlihan PS, Hamilton SR, Diehl AM, et al. Overexpression of caspase-3 in hepatocellular carcinomas. Mod Pathol. 2004; 17: 861–7. doi: 10.1038/modpathol.3800146 [DOI] [PubMed] [Google Scholar]
- 18.Jang JS, Kim KM, Choi JE, Cha SI, Kim CH, Lee WK, et al. Identification of polymorphisms in the Caspase-3 gene and their association with lung cancer risk. Mol Carcinog. 2008; 47: 383–90. doi: 10.1002/mc.20397 [DOI] [PubMed] [Google Scholar]
- 19.Lin J, Zhang Y, Wang H, Chang J, Wei L, Cao L, et al. Genetic Polymorphisms in the Apoptosis-Associated Gene CASP3 and the Risk of Lung Cancer in Chinese Population. PLoS One. 2016; 11: e0164358 doi: 10.1371/journal.pone.0164358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan S, Li YZ, Zhu XW, Liu CL, Wang P, Liu YL. HuGE systematic review and meta-analysis demonstrate association of CASP-3 and CASP-7 genetic polymorphisms with cancerrisk. Genet Mol Res. 2013; 12: 1561–73. doi: 10.4238/2013.May.13.10 [DOI] [PubMed] [Google Scholar]
- 21.Huang H, Zhang XF, Zhou HJ, Xue YH, Dong QZ, Ye QH, et al. Expression and prognostic significance of osteopontin and caspase-3 in hepatocellular carcinoma patients after curative resection. Cancer Sci. 2010; 101: 1314–9. doi: 10.1111/j.1349-7006.2010.01524.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Theodoropoulos GE, Michalopoulos NV, Pantou MP, Kontogianni P, Gazouli M, Karantanos T, et al. Caspase 9 promoter polymorphisms confer increased susceptibility to breast cancer. Cancer Genet. 2012; 205: 508–12. doi: 10.1016/j.cancergen.2012.08.001 [DOI] [PubMed] [Google Scholar]
- 23.Cingeetham A, Vuree S, Dunna NR, Gorre M, Nanchari SR, Edathara PM, et al. Association of caspase9 promoter polymorphisms with the susceptibility of AML in south Indian subjects. Tumour Biol. 2014; 35: 8813–22. doi: 10.1007/s13277-014-2096-5 [DOI] [PubMed] [Google Scholar]
- 24.Park JY, Park JM, Jang JS, Choi JE, Kim KM, Cha SI, et al. Caspase 9 promoter polymorphisms and risk of primary lung cancer. Hum Mol Genet. 2006; 15: 1963–71. doi: 10.1093/hmg/ddl119 [DOI] [PubMed] [Google Scholar]
- 25.Theodoropoulos GE, Gazouli M, Vaiopoulou A, Leandrou M, Nikouli S, Vassou E, et al. Polymorphisms of caspase 8 and caspase 9 gene and colorectal cancer susceptibility and prognosis. Int J Colorectal Dis. 2011; 26: 1113–8. doi: 10.1007/s00384-011-1217-5 [DOI] [PubMed] [Google Scholar]
- 26.Sträter J, Herter I, Merkel G, Hinz U, Weitz J, Möller P. Expression and prognostic significance of APAF-1, caspase-8 and caspase-9 in stage II/III colon carcinoma: caspase-8 and caspase-9 is associated with poor prognosis. Int J Cancer. 2010; 127: 80. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(ZIP)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.