Abstract
Programmed death-ligand 1 (PD-L1) is a promising target of cancer immune therapy. It not only expressed in tumor cells (TCs) but also up regulated in tumor infiltrating immune cells (TIICs). Although the previous meta-analysis have shown that PD-L1 expression in TCs was a valuable biomarker in predicting cancer prognosis, but few researches systematic evaluated the association between its expression in TIICs and survival of cancer patients. Thus, we performed this meta-analysis to evaluate the prognostic value of PD-L1 expression in TIICs in different types of cancers. Our results are valuable supplements when using PD-L1 expression to predict the survival of cancer patients and to select the beneficial patients from PD-L1 target therapy. PubMed, Embase, Web of Science and the Cochrane Central Search Library were used to perform our systematic literature search. Overall survival (OS) at 5th years and hazard ratios (HRs) were calculated using random effects models. Eighteen studies involving 3674 patients were included. The median positive rate of PD-L1 staining in TIICs was 36.37%. PD-L1 positive expression in TIICs related to a lower risk of death (HR = 0.784, 95%CI: 0.616–0.997, P = 0.047). Subgroup analyses found that PD-L1 positive expression in TIICs indicated a better prognosis especially in breast cancer patients (HR = 0.359, P = 0.041). When using whole tissue section slides, or using ‘any expression in TIICs’ as a cutoff value to assessing the results of IHC staining, PD-L1 expression in TIICs had a good prognostic value in cancer prognosis (HR = 0.587, P = 0.001 and HR = 0.549, P = 0.002). Our findings suggested that PD-L1 expression in TIICs was related to a better survival of cancer. The comprehensive evaluation of tumor cells and tumor infiltrating immune cells are required when evaluating the effect of PD-L1 expression on prognosis of cancer in future research.
Introduction
Cancer remains the most prominent global health-related threat[1, 2]. Traditional therapies such as tumorectomy, radiotherapy and chemotherapy are still the main treatments, but their efficacies are unsatisfactory in most cancers, especially in advanced cancers[3]. Recently, variable new cancer treatments have emerged, with immune checkpoint inhibition being one of the most promising therapies[4, 5].
Among the immune checkpoint molecules, programmed death 1 (PD-1) and its ligand, programmed death-ligand 1 (PD-L1), constitute a pair of negative co-stimulatory molecules that can suppress the functions of T cells and mediate the immune escape of cancers[6, 7]. PD-1 and PD-L1 inhibitors were developed by numerous pharmaceuticals companies and well studied in several clinical trials[8, 9]. A meta-analysis including 20 trials reported that patients with positive PD-L1 expression might have a decreased risk of mortality compared to negative cases when treated with anti PD-1/PD-L1 antibodies[10]. And the expression of PD-L1 not only linked to the response of immune checkpoint therapy but also associated with the prognosis of several types of cancer, such as non-small-cell lung cancer[11], gastric cancer[12], and breast cancer[13].
Although there has been already a lot of literatures published investigated the associations between PD-L1 expression and cancer prognosis using the method of meta-analysis[14]. However, all of them focused only on the PD-L1 expression in tumor cells. As we know, PD-1/PD-L1 pathway plays an important role in the cancer-specific immune response. PD-L1 is not only expressed in tumor cells but also up-regulated in tumor infiltrating immune cells (TIICs), including tumor infiltrating lymphocytes, mononuclear cells and other immune cells[15–17]. Current clinical trials have shown that the expression of PD-L1 in TIICs is also indicative of a higher response rate to PD-L1/PD-1 targeted therapy[18, 19]. Therefore, not only tumor cell-based but also immune cell-based PD-L1 expression appears to be clinically valuable. Recently, a number of studies have reported that the expression of PD-L1 in TIICs which was correlated with the survival of patients with tumors, but have failed to reach consistent conclusions [20–26]. In addition, there has been no research systematic evaluates the predicted value of PD-L1 positive expression in TIICs in cancer prognosis.
Thus, we performed this meta-analysis to evaluate the prognostic value of PD-L1 expression in TIICs in different types of cancers. Our results are valuable supplements when using PD-L1 expression to predict the survival of cancer patients and to select the beneficial patients from PD-L1 target therapy.
Materials and methods
Search strategy
PubMed, Embase, Web of Science and the Cochrane Central Search Library were used to perform our systematic literature search (until December 2016). Key words used included “programmed death-ligand 1 or PD-L1 or B7-H1 or CD274” and “tumor infiltrating lymphocyte or TIL or tumor infiltrating immune cells or TIIC or tumor infiltrating mononuclear cells or TIMC or tumor stroma” and “cancer or carcinoma or tumor” and “prognosis or survival”; the results were limited to human studies. In addition, we searched the reference lists of the reviews on related topics by hand to identify additional studies.
Inclusion and exclusion criteria
The eligible studies were included in this meta-analysis based on the following criteria: (1) PD-L1 expression has been measured by immunohistochemistry (IHC) stain in tumor infiltrating immune cells rather than in tumor cells; (2) studies reported 5-year OS, HR with 95% confidence interval (95% CIs), or reported original survival curves; (3) studies were published in English, and their full texts were available. Exclusion criteria for this study were as follows: (1) conference abstracts, letters, reviews and unpublished studies; and (2) insufficient data to report the hazard ratios and 95% CI, or could not extract the data from Kaplan-Meier curves. If duplicate data presented in more than one study, the largest or most recent study was included.
Data extraction and quality assessment
Two reviewers (Zhao TC and Wu YH) identified relevant articles independently. The details of these surveys included the author’s name, date of publication, type of cancer, type of pathological section, number of patients, tumor stage, age of patients, duration of follow up, antibody, staining location, and cut-off value in assessing the positive expression of PD-L1 in tumor infiltrating immune cells. Newcastle-Ottawa Scale (NOS) was used for quality assessment[27]. NOS scores no less than 6 were defined as high-quality studies.
Statistical analysis
Data were analyzed using STATA version 12.0 (STATA Corporation, College Station, TX, USA). The 3-year OS, 5-year OS, HR and 95%CI were extracted from the original studies, tables or recalculated from Kaplan—Meier curves using the program of Engauge Digitizer (http://sourceforge.net/projects/digitizer/), and applied using the Mantel—Haenszel random effect model. I2[28] was used to determine the heterogeneity of the selected studies. Subgroup analysis and sensitivity analysis were carrying out to assess the potential contributions of different type of tumors and different cutoff values for defining PD-L1 expression in TIICs. Egger’s regression asymmetry test[29] and Begg’s adjusted rank correlation[30] were performed to evaluate the potential publication bias. All tests were two-sided, and P<0.05 was considered statistically significant.
Results
Identification of eligible studies
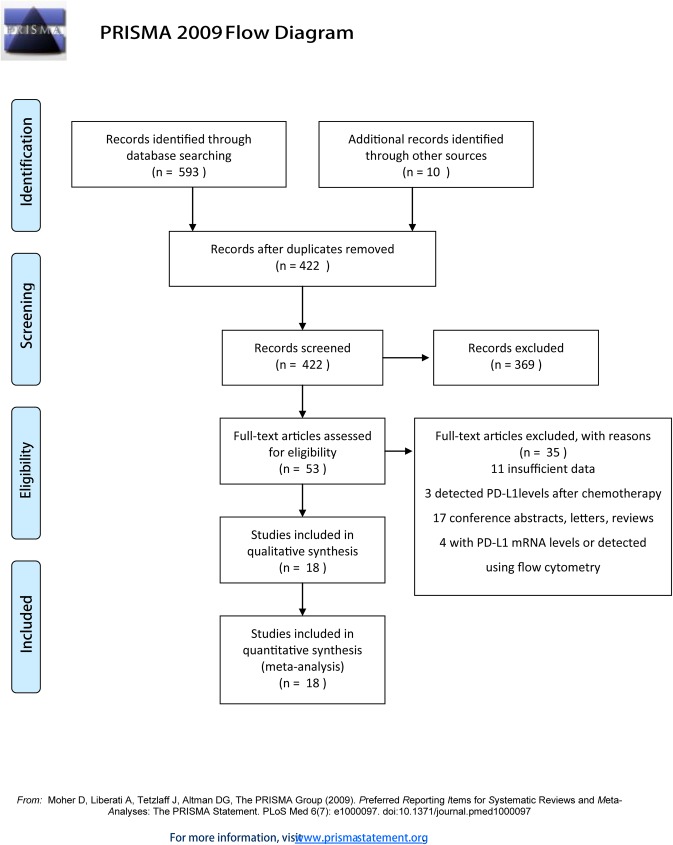
The search results shown in Fig 1 identified 603 studies from the initial database. After careful manual selection and review of these articles, 18 studies with full text and available data according to the inclusion and exclusion criteria were included in the final analysis (Fig 1). And the PRISMA checklist was showed in S1 File.
Fig 1. Flow diagram of the literature search and study selection for the meta-analysis.
The flow diagram shows eligible publications at each stage of the analysis process. The database search was conducted in December 2016.
Description of studies
The characteristics of the 18 studies summarized in Table 1. All of the studies assessed PD-L1 expression in tumor infiltrating immune cells using immunohistochemistry techniques. Seven studies evaluated cancers of the digestive system (5 gastric cancers, 1 esophageal squamous cell carcinoma and 1 colorectal cancer), two evaluated cancers of the urinary system (1 urothelial carcinoma and 1 renal cell carcinoma), two evaluated breast cancer, two evaluated lung cancer, and one each evaluated ovarian high grade serous carcinoma, extranodal NK/T-cell lymphoma, head and neck cancer, diffuse large B-cell lymphoma and spinal chordoma. In total, 3674 subjects were included in our meta-analysis.
Table 1. Characteristics of the studies included.
| No | Study, Year | Cancer types | Tissue slides | No. of patients | Age (range) | Tumor stagesa | Follow up (months) | PD-L1 NO (+/-) |
|---|---|---|---|---|---|---|---|---|
| 1 | Bellmunt J,2015[20] | urothelial carcinoma | TMAs | 89 | NR | IV (metastatic) | 1–24 | 33/56 |
| 2 | Boger C,2016[21] | gastric cancer | Whole | 465 | 68 (median) | I-IV | 0.2–109 | 160/291 |
| 3 | Choueiri TK,2014[22] | nonclear-cell renal cell carcinoma | NR | 101 | 24–81 | I-IV | 60(median) | 57/44 |
| 4 | Darb-Esfahani S,2015[23] | ovarian high grade serous carcinoma | TMAs | 200 | 60 (median) | I-IV (FIGO) | 37.9(median) | 60/140 |
| 5 | Hatogai K,2016[24] | esophageal squamous cell carcinoma | TMAs | 196 | 42–87 | I-IV | 1.2–127.2 | 119/77 |
| 6 | Hou J,2014[25] | gastric cancer | NR | 111 | 18–96 | I-IV | NR | 71/40 |
| 7 | Jo J-C,2016[26] | extranodal NK/T-cell lymphoma | NR | 79 | 19–79 | I-IV | 52.4(median) | 62/17 |
| 8 | Kawazoe A,2016[31] | gastric cancer | TMAs | 383 | 26–92 | III-IV | NR | 241/142 |
| 9 | Kim HR,2016[32] | head and neck cancer | TMAs | 402 | 22–88 | I-IV | 46.3 (median) | 112/290 |
| 10 | Kiyasu J,2015[33] | diffuse large B-cell lymphoma | NR | 239 | 30–92 | I-IV | NR | 53/186 |
| 11 | Li X,2016[34] | triple-negative breast cancer | Whole | 136 | NR | NR | NR | 32/104 |
| 12 | Paulsen E-E,2016[35] | none small cell lung cancer | TMAs | 505 | 28–85 | I-IIIA | 86(34–267) | 182/323 |
| 13 | Saito R,2016[36] | EBV positive gastric cancer | TMAs | 96 | 40–90 | I-IV | 3-262(range) | 43/53 |
| 14 | Sun W-Y,2016[37] | triple negative breast cancer | TMAs | 218 | NR | I-IIIB | 0.2–98 (range) | 80/138 |
| 15 | Thompson ED, 2016[38] | gastric adenocarcinomas | Whole | 33 | 21–92 | I-IV | 40(median) | 15/18 |
| 16 | Wang L,2016[39] | Colorectal cancer | TMAs | 262 | 28–75 | II-III | 43.5(mean) (21–68) | 55/207 |
| 17 | Yang C-Y,2016[40] | pulmonary squamous cell carcinoma | Whole | 105 | 40–84 | IA-IB | 79(mean) | 31/74 |
| 18 | Zou MX,2016[41] | spinal chordoma | Whole | 54 | 23–79 | I-III | 42.39(mean) (5–158) | 12/42 |
NR, not reported; TMAs, tissue microarrays; EBV, Epstein-Barr virus; FIGO: International Federation of Gynecology and Obstetrics
a Unless otherwise noted, Tumor stage was classified according to the AJCC/UICC staging system
Evaluation of PD-L1 expression in TIICs
The antibodies, cutoff values and staining locations used in the evaluation of PD-L1 expression in TIICs of the included studies are shown in Table 2. Clone E1L3N was used in four studies, and Clone SP142 was used in three studies. The cutoff values in assessing the positive expression of PD-L1 in TIICs were divided into 4 types: (1) proportion of stained cells greater than 5%, (2) proportion of stained cells greater than 1%, (3) any expression of PD-L1 in TIICs and (4) others. Most studies considered that the positive staining was located in the cell membrane (12 of 18 studies); whereas others thought both membranous and cytoplasmic staining could be considered as positive expression. The median positive rate of PD-L1 expression in TIICs was 36.37%.
Table 2. Detection of the PD-L1 expression in TIICs in the selected studies.
| No | Study, Year | Antibody | Cutoff value of PD-L1 positive expression in TIICs | Staining location | 3-year OS(+/-)% | 5-year OS (+/-)% |
|---|---|---|---|---|---|---|
| 1 | Bellmunt J,2015 [20] | 405.9A11 | Absent (0), focal (1), mild (2), moderate (3), and severe (4); 2–4 were considered positive | Membrane | NR | NR |
| 2 | Boger C,2016 [21] | E1L3N | The percentage of positive cells: 0 (negative), 1 (1–5% positive), 2 (6–20%) and 3 (>20%); Score >1 were considered positive | Membrane | 39.4/18.8 | 23.8/12.0 |
| 3 | Choueiri TK,2014 [22] | 405.9A11 | According to the percentages of PD-L1 positive TIMC (0% = 0, <5% = 1, ≥5% = 2); Score >0 were considered positive | Membrane | 84.5/94.9 | 73.7/84.1 |
| 4 | Darb-Esfahani S [23] | EPR1161 | >20/mm2 were considered positive | Membrane/ Cytoplasm | NR | NR |
| 5 | Hatogai K,2016 [24] | NR | Any expression of PD-L1 in TIICs in the core were considered positive | Membrane | 58.0/40.9 | 52.9/33.8 |
| 6 | Hou J,2014 [25] | NR (Abcam) | Proportion of stained cells >5% were considered positive | Membrane/ cytoplasm | 42.3/70.0 | NR |
| 7 | Jo J-C,2016 [26] | NR (R&D Systems) | More than 5% cells was stained were considered positive | Membrane/ cytoplasm | 54.3/30.1 | 48.6/30.1 |
| 8 | Kawazoe A,2016 [31] | SP142 | <1% (0), 1% to 9% (2), 10% to 19% (3), ≥20% (4); ≥1% were considered positive | Membrane | 62.9/59.1 | 55.5/48.5 |
| 9 | Kim HR,2016 [32] | SP142 | Proportion of stained cells >5% were considered positive | Membrane/ cytoplasm | 93.7/80.1 | 90.2/75.5 |
| 10 | Kiyasu J,2015 [33] | ab174838 | PD-L1 nonmalignant stromal cells represented 20% or more of the total tissue were considered positive | Membrane/ cytoplasm | 63.3/72.9 | 51.6/61.1 |
| 11 | Li X,2016 [34] | E1L3N | Any stromal PD-L1 expression were considered positive | Membrane | NR | NR |
| 12 | Paulsen E-E,2016 [35] | E1L3N | Absent (0), 1% to 49% (1), 50% to 75% (2), or > 75% (3) >1.5 were considered positive | Membrane/ cytoplasm | NR | 52/44 |
| 13 | Saito R,2016 [36] | E1L3N | Simply classified into negative or positive groups depending on the proportion of stained cells (cutoff value: 1%) | Membrane | 88.0/91.7 | 80.9/91.7 |
| 14 | Sun W-Y,2016 [37] | 28–8 | any immunostaining were considered positive | Membrane | NR | NR |
| 15 | Thompson ED, 2016 [38] | 5H1 | >1% of PD-L1 staining on TIL or TAM was considered positive." | Membrane | 71.2/76.0 | 51.6/61.1 |
| 16 | Wang L,2016[39] | SP142 | <1% (0), 1% to 4% (1), 5% to 9% (2), ≥10% (3); scores of 2 and 3 were considered positive | Membrane | 66.5/80.1 | 57.4/72.5 |
| 17 | Yang C-Y,2016 [40] | 17952–1 -AP | Proportion of stained cells >5% were considered positive | Membrane | NR | NR |
| 18 | Zou MX,2016 [41] | ab174838 | Absent (0), rare/few (1), moderate (2), prominent (3), ≥2 were considered positive | Membrane | 87.6/93.3 | 72.7/32.5 |
NR, not reported; TIICs, tumor infiltrating immune cells; TIL, tumor-infiltrating lymphocyte; TAM, tumor-associated macrophages
PD-L1 expression in TIICs and five-year OS
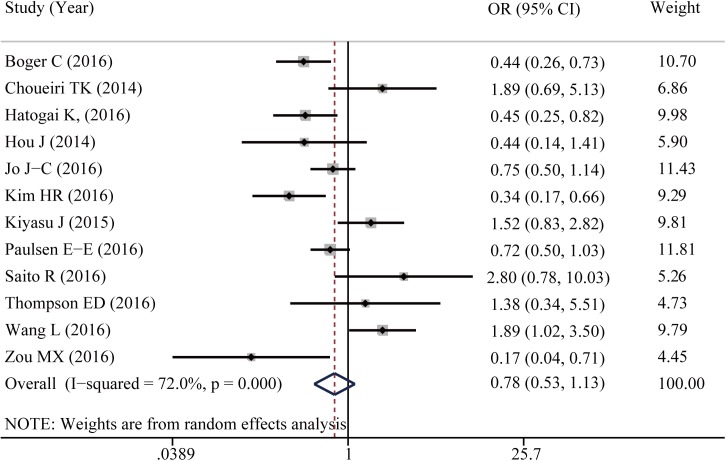
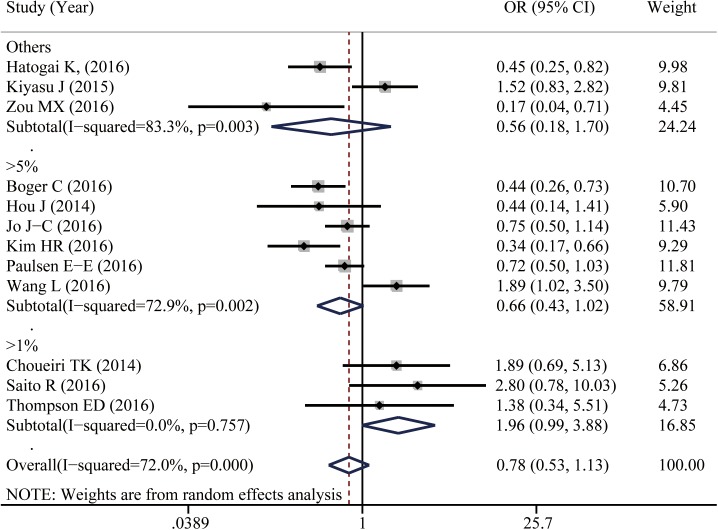
Twelve studies reported data for 5-year OS. As shown in Fig 2, PD-L1 positive expression in TIICs seems to be associated with a better 5-year OS of cancer patients, though it did not reach statistical difference (OR = 0.778, 95%CI: 0.534–1.134, P = 0.192). Because of the significant heterogeneity among studies (I2 = 72%), subgroup analyses were conducted to assess whether the heterogeneity was due to different cancer types and cutoff values. Six studies provided the 5-year OS for digestive system cancers; others reported different types of cancer (S1 Fig). In the stratified analysis by cancer types, PD-L1 positive expression in TIICs of digestive system cancers was not associated with 5-year OS (OR = 0.862, 95%CI: 0.438–1.697, P = 0.667). Further, I2 was calculated to be 76.5%, which indicated that the heterogeneity was not due to different cancer types. We then conducted a subgroup analysis according to different cutoff values. When the cutoff value was defined as ‘proportion of stained cells greater than 5%’, PD-L1 expression in TIICs seems to be associated with a better cancer survival (OR = 0.662, 95% CI: 0.429–1.022, P = 0.062, I2 = 72.9%). When using the cutoff value of ‘proportion of stained cells greater than 1%’ to distinguish the positive and negative expression of PD-L1 in TIICs, the opposite trend has been reported (OR = 1.958, 95%CI: 0.987–1.134, P = 0.055, I2 = 0%; Fig 3).
Fig 2. Forest plot shows the associations between PD-L1 expression in TIICs and five year overall survival of cancer patients.
Fig 3. Subgroup analysis by different cutoff values shows the associations between PD-L1 expression in TIICs and five year overall survival of cancer patients.
PD-L1 expression in TIICs and time-to-event index
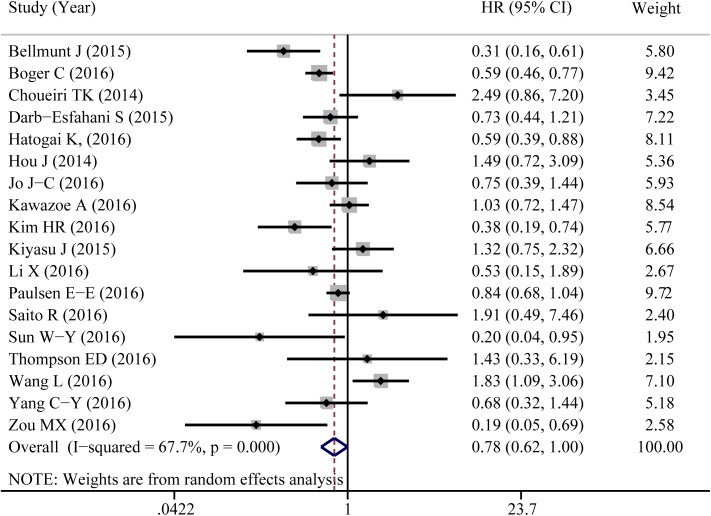
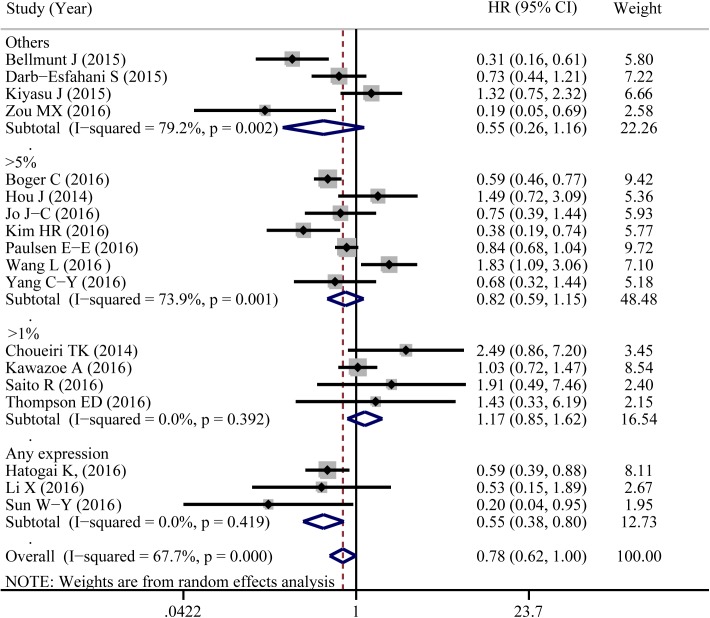
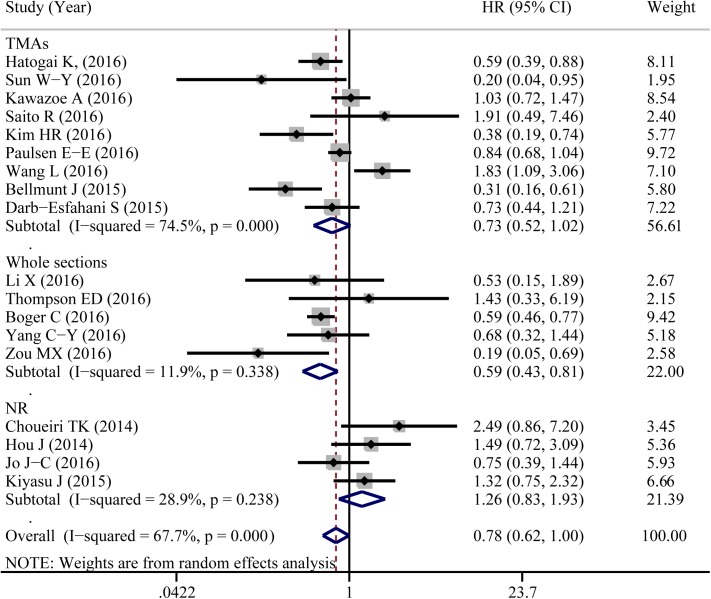
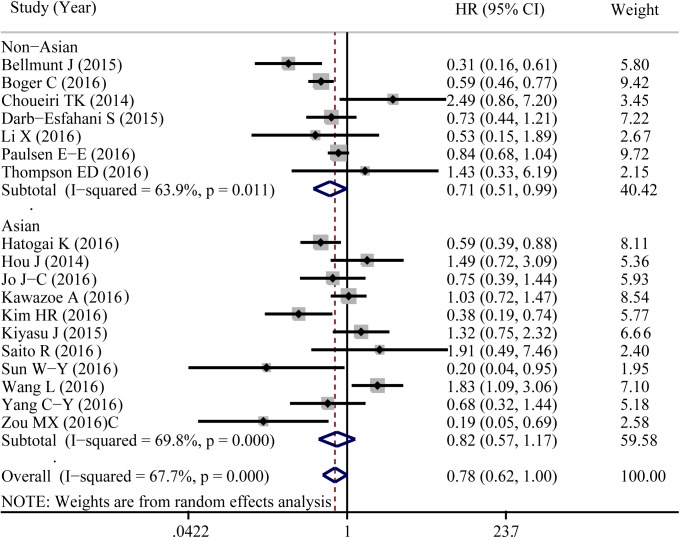
A total of 18 eligible studies were pooled to analyze the predictive value of TIICs expressed PD-L1 in cancer prognosis using HR and 95%CIs. Fig 4 shown that PD-L1 expression in TIICs indicated a decreased risk of death (HR = 0.784, 95%CI: 0.616–0.997, P = 0.047). Similar to 5-year OS, significant heterogeneity was noted (I2 = 67.7%, P<0.001). Exploratory subgroup analysis suggested that PD-L1 expression in TIICs indicted a lower risk of death in patients with breast cancer (HR = 0.359, 95%CI: 0.134–0.961, P = 0.041, I2 = 0%; S2 Fig). As shown in Fig 5, PD-L1 in TIICs was only associated with improved overall survival in those studies using cutoff value of ‘Any positive staining in immune cells’ (HR = 0.549, 95%CI = 0.378–0.798, P = 0.002, I2 = 0%). We also conducted a subgroup analysis according to different types of pathological sections (the whole tissue section slides or tissue microarrays). PD-L1 in TIICs was correlated to a favorable prognosis in those studies using whole tissue section slides in conducting the immunohistochemical stain (HR = 0.587, 95%CI: 0.425–0.810, P = 0.001, I2 = 11.9%, Fig 6). Additionally, genetic differences will contribute to the heterogeneity between individual studies. The ethnicities of included studies were divided into two parts; Asian and non-Asian. Positive expression of PD-L1 in TIICs was an indicator of a favorable prognosis, only in non-Asian cancer patients (HR = 0.709, 95%CI: 0.511–0.985, P = 0.040, I2 = 63.9%, Fig 7).
Fig 4. Forest plot of hazard ratios shows the associations between PD-L1 expression in TIICs and cancer prognosis.
Fig 5. Forest plot of hazard ratios form subgroup analysis by different cutoff values shows the associations between PD-L1 expression in TIICs and cancer prognosis.
Fig 6. Forest plot of hazard ratios form subgroup analysis by different types of pathological sections shows the associations between PD-L1 expression in TIICs and cancer prognosis.
TMAs, tissue microarrays.
Fig 7. Forest plot of hazard ratios form subgroup analysis by different ethnicity shows the associations between PD-L1 expression in TIICs and cancer prognosis.
TMAs, tissue microarrays.
Sensitivity and publication bias analyses
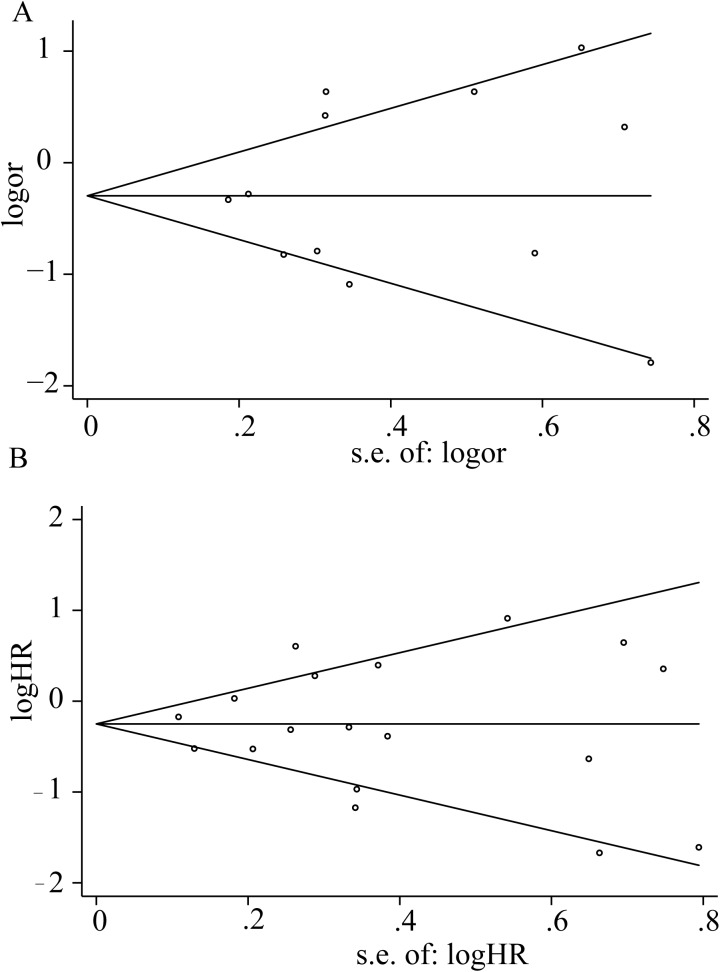
Omitting any individual study did not influence the combined results for 5-year OS or HR (S3 Fig). The funnel plot for the relationship between PD-L1 expression in TIICs and cancer prognosis is presenting in Fig 8. For 5-year OS the P values for Egger’s and Begg’s tests were 0.714 and 0.891. For hazard ratio, the results from Egger’s and Begg’s tests also revealed that there was no publication bias in this meta-analysis (P = 0.986 and P = 0.733, respectively).
Fig 8. Begg’s funnel plots show the publication bias.
(A) Begg’s funnel plot for 5-years OS (B) Begg’s funnel plot for HR.
Discussion
Anti-PD-1/PD-L1 therapy has been discussed as a potential effective strategy for cancer treatment, and numerous studies have reported the positive expression of PD-L1 in tumor cells as a predictive biomarker for the response to PD-1/PD-L1 blocking therapy[42]. Simultaneously, the expression of PD-L1 has always been considered a predicted biomarker in cancer prognosis[14]. Not only tumor cells, but also tumor-infiltrating immune cells could express PD-L1. Previous studies have only focused on PD-L1 expression in tumor cells, but recent studies indicated that the PD-L1 expression in TIICs also played an important role in tumor immune escape and influenced tumor progression[16, 17, 31, 36]. These leading studies highlighted that PD-L1 expression in TIICs could also serve as a prognostic biomarker, and further inform the responses to anti-PD-1/PD-L1 treatment. In this meta-analysis of data from 18 studies, with a cohort of 3674 cancer patients, we firstly provided a quantitative estimate to the prognostic value of PD-L1 expression in TIICs in cancer patients.
Five-year overall survival and hazard ratio were both important indexes in cancer prognostic evaluation. Our results shown that, PD-L1 in TIICs was significantly associated with a decreased risk of death (HR = 0.784, 95%CI: 0.616–0.997, P = 0.047) compared with patients with PD-L1 negative expression in TIICs. A similar trend has observed when using 5-year OS to evaluate the predictive value of PD-L1 in TIICs in cancer prognosis, but the P value was not statistically significant. Our results were quite different from the published meta-analysis which shown that PD-L1 expression in tumor cells was associated with a worse prognosis of cancer. Different mechanisms between PD-L1 expression in TCs and TIICs might explain the inconsistent results. Transcriptome analyses indicated that, PD-L1 expression in TCs was up regulated through the tumor-intrinsic mechanisms, including the activation of endogenous oncogene and related signaling pathway[43]. However, PD-L1 expression in TIICs could be driven by adaptive mechanisms such like exogenous inflammation mediated immune attack and then reflected pre-existing immunity[44, 45]. In other words, comparing with tumor cells, the tumor infiltrating immune cells based PD-L1 expression has stronger relations with cancer immune response, and depends on tumor microenvironments. In fact, PD-L1 positive expression in TIICs was positive correlated to the quantity of multiple tumor-infiltrating immune cells, such as CD4+ T lymphocytes and CD8+T lymphocytes. Since the high expression of CD4+ T lymphocytes and CD8+ T lymphocytes was associated with better outcomes of cancer patients[43, 46], the PD-L1 expression in TIICs was possibly associated with better cancer prognosis. Although PD-L1 expression could mediate the occurrence of cancer immune escape, it also indicated an effective immune response, especially with a favorable profile of immune microenvironments in the early stage of the cancer immune response[40].
According to the results of subgroup analysis, different types of pathological sections and different definition of cutoff values when conducting and assessing the IHC stain could partly explain the large heterogeneity among individual studies. Half of the studies using tissue microarrays (TMAs) to conduct the IHC stain. TMAs usually contain limited tissue (2.0mm) and extract from the central part of tumors. Compare with the whole tissue section slides, TMAs may have less representativeness, especially in assessing the biomarkers expressed in tumor infiltrating immune cells. The subgroup analysis according to different types of pathological sections showed that when using whole section slides to investigate PD-L1 positive expression in TIICs, the heterogeneity among different studies was the lowest. It indicates that using whole section slides to conduct the IHC staining should be recommended in related clinical trials and treatments. Additionally, the appropriate cutoff value in validating the positive expression of PD-L1 remains contentious. Subgroup analysis with different cutoff values has shown that there was a contradictory trend when using the cutoff value of ‘5%’ or ‘1%’ in evaluating the correlations of PD-L1 positive expression in TIICs with survival of cancer patients. Therefore, a multi-classification of cutoff values for assessing PD-L1 expression in TIICs may be feasible and reasonable. Asian and non-Asian cancer patients exhibit distinct tumor immunity signatures. For instance, in gastric cancer, non-Asian patients show significantly higher expression level of T-cell markers, including CD3 and CD8, and lower expression level of immunosuppressive T-regulatory cell markers, such as FOXP3 compared to Asian gastric patients[47]. Immune-related biomarkers differentially expressed between Asian and non-Asian cancer patients who was related to immune function. These differences may affect the associations between PD-L1 expression and survival of cancer patients.
Several limitations should be acknowledged in our study. First, in several studies, when 5-year OS and HRs not provided in the original studies, we derived the indexes from Kaplan–Meier survival curves; six of the 18 studies did not provide 5-year OS or original Kaplan–Meier survival curves. As a result, only 12 studies were available to calculate the association of PD-L1 expression in TIICs with five-year OS, which could affect the level of evidence. Second, not all of the included studies using the multiple Cox regression to estimate the independent prognostic value of PD-L1 expression in TIICs in cancer. Thus, the results from all of the studies could not be further stratified with the same confounding factors, and further studies with more confounding factor adjustments need to be conducted.
Conclusions
Despite these limitations, we have demonstrated that PD-L1 expression in TIICs might serve as a new biomarker for prognosticating the survival of cancer patients. Thus, incorporating the expression of tumor-infiltrating immune cells into the classification of PD-L1 expression is necessary. Our results may be useful supplements when using PD-L1 expression to predict the survival of cancer patients and to select the beneficial patients from anti-PD-L1 treatment.
Supporting information
(TIF)
(TIF)
(A) Sensitivity analysis for 5-years OS (B) Sensitivity analysis for HR.
(TIF)
(DOC)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Finance Department of Jilin Province (No.sczsy201606) [http://czt.jl.gov.cn/jlcz/16/tindex.shtml].
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA: a cancer journal for clinicians. 2017;67(1):7–30. Epub 2017/01/06. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–32. doi: 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 3.Luo D, Carter KA, Miranda D, Lovell JF. Chemophototherapy: An Emerging Treatment Option for Solid Tumors. Advanced science (Weinheim, Baden-Wurttemberg, Germany). 2017;4(1):1600106. Epub 2017/01/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gibney GT, Weiner LM, Atkins MB. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. The Lancet Oncology. 2016;17(12):e542–e51. Epub 2016/12/08. doi: 10.1016/S1470-2045(16)30406-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teng F, Kong L, Meng X, Yang J, Yu J. Radiotherapy combined with immune checkpoint blockade immunotherapy: Achievements and challenges. Cancer letters. 2015;365(1):23–9. Epub 2015/05/20. doi: 10.1016/j.canlet.2015.05.012 [DOI] [PubMed] [Google Scholar]
- 6.Boussiotis VA. Molecular and Biochemical Aspects of the PD-1 Checkpoint Pathway. N Engl J Med. 2016;375(18):1767–78. Epub 2016/11/03. doi: 10.1056/NEJMra1514296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sznol M, Chen L. Antagonist antibodies to PD-1 and B7-H1 (PD-L1) in the treatment of advanced human cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19(5):1021–34. Epub 2013/03/06. PubMed Central PMCID: PMCPMC3702373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costa R, Carneiro BA, Agulnik M, Rademaker AW, Pai SG, Villaflor VM, et al. Toxicity profile of approved anti-PD-1 monoclonal antibodies in solid tumors: a systematic review and meta-analysis of randomized clinical trials. Oncotarget. 2016. Epub 2016/11/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang T, Xie J, Arai S, Wang L, Shi X, Shi N, et al. The efficacy and safety of anti-PD-1/PD-L1 antibodies for treatment of advanced or refractory cancers: a meta-analysis. Oncotarget. 2016;7(45):73068–79. Epub 2016/09/30. doi: 10.18632/oncotarget.12230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gandini S, Massi D, Mandala M. PD-L1 expression in cancer patients receiving anti PD-1/PD-L1 antibodies: A systematic review and meta-analysis. Critical reviews in oncology/hematology. 2016;100:88–98. Epub 2016/02/21. doi: 10.1016/j.critrevonc.2016.02.001 [DOI] [PubMed] [Google Scholar]
- 11.Aguiar PN Jr., Santoro IL, Tadokoro H, de Lima Lopes G, Filardi BA, Oliveira P, et al. The role of PD-L1 expression as a predictive biomarker in advanced non-small-cell lung cancer: a network meta-analysis. Immunotherapy. 2016;8(4):479–88. Epub 2016/03/15. doi: 10.2217/imt-2015-0002 [DOI] [PubMed] [Google Scholar]
- 12.Liu YX, Wang XS, Wang YF, Hu XC, Yan JQ, Zhang YL, et al. Prognostic significance of PD-L1 expression in patients with gastric cancer in East Asia: a meta-analysis. OncoTargets and therapy. 2016;9:2649–54. Epub 2016/05/27. PubMed Central PMCID: PMCPMC4863684. doi: 10.2147/OTT.S102616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, Li M, Lian Z, Zhu H, Kong L, Wang P, et al. Prognostic Role of Programmed Death Ligand-1 Expression in Breast Cancer: A Systematic Review and Meta-Analysis. Targeted oncology. 2016;11(6):753–61. Epub 2016/07/17. doi: 10.1007/s11523-016-0451-8 [DOI] [PubMed] [Google Scholar]
- 14.Wu P, Wu D, Li L, Chai Y, Huang J. PD-L1 and Survival in Solid Tumors: A Meta-Analysis. PloS one. 2015;10(6):e0131403 Epub 2015/06/27. PubMed Central PMCID: PMCPMC4483169. doi: 10.1371/journal.pone.0131403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du Z, Abedalthagafi M, Aizer AA, McHenry AR, Sun HH, Bray MA, et al. Increased expression of the immune modulatory molecule PD-L1 (CD274) in anaplastic meningioma. Oncotarget. 2015;6(7):4704–16. Epub 2015/01/23. PubMed Central PMCID: PMCPMC4467109. doi: 10.18632/oncotarget.3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paiva B, Azpilikueta A, Puig N, Ocio EM, Sharma R, Oyajobi BO, et al. PD-L1/PD-1 presence in the tumor microenvironment and activity of PD-1 blockade in multiple myeloma. Leukemia. 2015;29(10):2110–3. Epub 2015/03/18. doi: 10.1038/leu.2015.79 [DOI] [PubMed] [Google Scholar]
- 17.Koirala P, Roth ME, Gill J, Piperdi S, Chinai JM, Geller DS, et al. Immune infiltration and PD-L1 expression in the tumor microenvironment are prognostic in osteosarcoma. Sci Rep. 2016;6:30093 Epub 2016/07/28. PubMed Central PMCID: PMCPMC4960483. doi: 10.1038/srep30093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Powles T, Eder JP, Fine GD, Braiteh FS, Loriot Y, Cruz C, et al. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature. 2014;515(7528):558–62. Epub 2014/11/28. doi: 10.1038/nature13904 [DOI] [PubMed] [Google Scholar]
- 19.Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515(7528):563–7. Epub 2014/11/28. PubMed Central PMCID: PMCPMC4836193. doi: 10.1038/nature14011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bellmunt J, Mullane SA, Werner L, Fay AP, Callea M, Leow JJ, et al. Association of PD-L1 expression on tumor-infiltrating mononuclear cells and overall survival in patients with urothelial carcinoma. Annals of Oncology. 2015;26(4):812–7. doi: 10.1093/annonc/mdv009 [DOI] [PubMed] [Google Scholar]
- 21.Boger C, Behrens HM, Mathiak M, Kruger S, Kalthoff H, Rocken C. PD-L1 is an independent prognostic predictor in gastric cancer of Western patients. Oncotarget. 2016;7(17):24269–83. Epub 2016/03/25. PubMed Central PMCID: PMCPMC5029700. doi: 10.18632/oncotarget.8169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choueiri TK, Fay AP, Gray KP, Callea M, Ho TH, Albiges L, et al. PD-L1 expression in nonclear-cell renal cell carcinoma. Annals of Oncology. 2014;25(11):2178–84. doi: 10.1093/annonc/mdu445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Darb-Esfahani S, Kunze CA, Kulbe H, Sehouli J, Wienert S, Lindner J, et al. Prognostic impact of programmed cell death-1 (PD-1) and PD-ligand 1 (PD-L1) expression in cancer cells and tumor-infiltrating lymphocytes in ovarian high grade serous carcinoma. Oncotarget. 2016;7(2):1486–99. Epub 2015/12/02. PubMed Central PMCID: PMCPMC4811475. doi: 10.18632/oncotarget.6429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hatogai K, Kitano S, Fujii S, Kojima T, Daiko H, Nomura S, et al. Comprehensive immunohistochemical analysis of tumor microenvironment immune status in esophageal squamous cell carcinoma. Oncotarget. 2016;7(30):47252–64. Epub 2016/06/21. doi: 10.18632/oncotarget.10055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hou J, Yu Z, Xiang R, Li C, Wang L, Chen S, et al. Correlation between infiltration of FOXP3+ regulatory T cells and expression of B7-H1 in the tumor tissues of gastric cancer. Experimental and Molecular Pathology. 2014;96(3):284–91. doi: 10.1016/j.yexmp.2014.03.005 [DOI] [PubMed] [Google Scholar]
- 26.Jo J-C, Kim M, Choi Y, Kim H-J, Kim JE, Chae SW, et al. Expression of programmed cell death 1 and programmed cell death ligand 1 in extranodal NK/T-cell lymphoma, nasal type. Annals of Hematology. 2016;96(1):25–31. doi: 10.1007/s00277-016-2818-4 [DOI] [PubMed] [Google Scholar]
- 27.Newcastle–Ottawa quality assessment scale cohort studies 2013. Available from: http://www.ohri.ca/programs/clinical_epidemiology/nosgen.pdf.
- 28.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ (Clinical research ed). 2003;327(7414):557–60. Epub 2003/09/06. PubMed Central PMCID: PMCPMC192859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ (Clinical research ed). 1997;315(7109):629–34. Epub 1997/10/06. PubMed Central PMCID: PMCPMC2127453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–101. Epub 1994/12/01. [PubMed] [Google Scholar]
- 31.Kawazoe A, Kuwata T, Kuboki Y, Shitara K, Nagatsuma AK, Aizawa M, et al. Clinicopathological features of programmed death ligand 1 expression with tumor-infiltrating lymphocyte, mismatch repair, and Epstein–Barr virus status in a large cohort of gastric cancer patients. Gastric Cancer. 2016. [DOI] [PubMed] [Google Scholar]
- 32.Kim HR, Ha S-J, Hong MH, Heo SJ, Koh YW, Choi EC, et al. PD-L1 expression on immune cells, but not on tumor cells, is a favorable prognostic factor for head and neck cancer patients. Scientific Reports. 2016;6:36956 doi: 10.1038/srep36956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiyasu J, Miyoshi H, Hirata A, Arakawa F, Ichikawa A, Niino D, et al. Expression of programmed cell death ligand 1 is associated with poor overall survival in patients with diffuse large B-cell lymphoma. Blood. 2015;126(19):2193–201. Epub 2015/08/05. PubMed Central PMCID: PMCPMC4635115. doi: 10.1182/blood-2015-02-629600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X, Wetherilt CS, Krishnamurti U, Yang J, Ma Y, Styblo TM, et al. Stromal PD-L1 Expression Is Associated With Better Disease-Free Survival in Triple-Negative Breast Cancer. American Journal of Clinical Pathology. 2016;146(4):496–502. doi: 10.1093/ajcp/aqw134 [DOI] [PubMed] [Google Scholar]
- 35.Paulsen E-E, Kilvaer TK, Khanehkenari MR, Al-Saad S, Hald SM, Andersen S, et al. Assessing PDL-1 and PD-1 in Non–Small Cell Lung Cancer: A Novel Immunoscore Approach. Clinical Lung Cancer. 2016. [DOI] [PubMed] [Google Scholar]
- 36.Saito R, Abe H, Kunita A, Yamashita H, Seto Y, Fukayama M. Overexpression and gene amplification of PD-L1 in cancer cells and PD-L1+ immune cells in Epstein–Barr virus-associated gastric cancer: the prognostic implications. Modern Pathology. 2016. [DOI] [PubMed] [Google Scholar]
- 37.Sun WY, Lee YK, Koo JS. Expression of PD-L1 in triple-negative breast cancer based on different immunohistochemical antibodies. Journal of Translational Medicine. 2016;14(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson ED, Zahurak M, Murphy A, Cornish T, Cuka N, Abdelfatah E, et al. Patterns of PD-L1 expression and CD8 T cell infiltration in gastric adenocarcinomas and associated immune stroma. Gut. 2016:gutjnl-2015-310839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L, Ren F, Wang Q, Baldridge LA, Monn MF, Fisher KW, et al. Significance of Programmed Death Ligand 1 (PD-L1) Immunohistochemical Expression in Colorectal Cancer. Molecular Diagnosis & Therapy. 2016;20(2):175–81. [DOI] [PubMed] [Google Scholar]
- 40.Yang C-Y, Lin M-W, Chang Y-L, Wu C-T, Yang P-C. Programmed cell death-ligand 1 expression is associated with a favourable immune microenvironment and better overall survival in stage I pulmonary squamous cell carcinoma. European Journal of Cancer. 2016;57:91–103. doi: 10.1016/j.ejca.2015.12.033 [DOI] [PubMed] [Google Scholar]
- 41.Zou MX, Peng AB, Lv GH, Wang XB, Li J, She XL, et al. Expression of programmed death-1 ligand (PD-L1) in tumor-infiltrating lymphocytes is associated with favorable spinal chordoma prognosis. American journal of translational research. 2016;8(7):3274–87. Epub 2016/08/11. PubMed Central PMCID: PMCPMC4969465. [PMC free article] [PubMed] [Google Scholar]
- 42.Tie Y, Ma X, Zhu C, Mao Y, Shen K, Wei X, et al. Safety and efficacy of nivolumab in the treatment of cancers: A meta-analysis of 27 prospective clinical trials. International journal of cancer. 2017;140(4):948–58. Epub 2016/11/05. doi: 10.1002/ijc.30501 [DOI] [PubMed] [Google Scholar]
- 43.Zeng DQ, Yu YF, Ou QY, Li XY, Zhong RZ, Xie CM, et al. Prognostic and predictive value of tumor-infiltrating lymphocytes for clinical therapeutic research in patients with non-small cell lung cancer. Oncotarget. 2016;7(12):13765–81. Epub 2016/02/13. PubMed Central PMCID: PMCPMC4924677. doi: 10.18632/oncotarget.7282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12(4):252–64. Epub 2012/03/23. doi: 10.1038/nrc3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim HR, Ha SJ, Hong MH, Heo SJ, Koh YW, Choi EC, et al. PD-L1 expression on immune cells, but not on tumor cells, is a favorable prognostic factor for head and neck cancer patients. Scientific reports. 2016;6:36956 Epub 2016/11/15. PubMed Central PMCID: PMCPMC5107906. doi: 10.1038/srep36956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hwang WT, Adams SF, Tahirovic E, Hagemann IS, Coukos G. Prognostic significance of tumor-infiltrating T cells in ovarian cancer: a meta-analysis. Gynecologic oncology. 2012;124(2):192–8. Epub 2011/11/02. PubMed Central PMCID: PMCPMC3298445. doi: 10.1016/j.ygyno.2011.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin SJ, Gagnon-Bartsch JA, Tan IB, Earle S, Ruff L, Pettinger K, et al. Signatures of tumour immunity distinguish Asian and non-Asian gastric adenocarcinomas. Gut. 2015;64(11):1721–31. Epub 2014/11/12. PubMed Central PMCID: PMCPMC4680172. doi: 10.1136/gutjnl-2014-308252 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(A) Sensitivity analysis for 5-years OS (B) Sensitivity analysis for HR.
(TIF)
(DOC)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.