Abstract
The interactions between proteins or peptides and inorganic materials lead to several interesting processes. For example, combining proteins with minerals leads to the formation of composite materials with unique properties. In addition, the undesirable process of biofouling is initiated by the adsorption of biomolecules, mainly proteins, on surfaces. This organic layer is an adhesion layer for bacteria and allows them to interact with the surface. Understanding the fundamental forces that govern the interactions at the organic-inorganic interface is therefore important for many areas of research and could lead to the design of new materials for optical, mechanical and biomedical applications. This paper demonstrates a single-molecule force spectroscopy technique that utilizes an AFM to measure the adhesion force between either peptides or amino acids and well-defined inorganic surfaces. This technique involves a protocol for attaching the biomolecule to the AFM tip through a covalent flexible linker and single-molecule force spectroscopy measurements by atomic force microscope. In addition, an analysis of these measurements is included.
Keywords: Chemistry, Issue 121, amino acids, peptides, single-molecule force spectroscopy, inorganic surfaces, adhesion force, atomic force microscope
Introduction
The interaction between proteins and inorganic minerals leads to the construction of composite materials with distinctive properties. This includes materials with high mechanical strength or unique optical properties.1,2 For example, the combination of the protein collagen with the mineral hydroxyapatite generates either soft or hard bones for different functionalities.3 Short peptides can also bind inorganic materials with high specificity.4,5,6 The specificity of these peptides has been used for designing new magnetic and electronic materials,7,8,9 fabricating nanostructured materials, growing crystals,10 and synthesizing nanoparticles.11Understanding the mechanism underlying interactions between peptides or proteins and inorganic materials will therefore allow us to design new composite materials with improved adsorptive properties. In addition, since the interphase of implants with an immune response is mediated by proteins, better understanding the interactions of proteins with inorganic materials will improve our ability to design implants. Another important area that involves proteins interacting with inorganic surfaces is the fabrication of antifouling materials.12,13,14,15 Biofouling is an undesirable process in which organisms attach to a surface. It has many detrimental implications on our lives. For example, biofouling of bacteria on medical devices leads to hospital-acquired infections. Biofouling of marine organisms on boats and large ships increases the consumption of fuel.12,16,17,18
Single-molecule force spectroscopy (SMFS), using an AFM, can directly measure the interactions between an amino acid or a peptide with a substrate.19,20,21,22,23,24,25,26 Other methods such as phage display,27,28quartz crystal microbalance (QCM)29or surface plasmon resonance (SPR)29,30,31,32,33 measure the interactions of peptides and proteins to inorganic surfaces in bulk.34,35,36This means that the results obtained by these methods relate to ensembles of molecules or aggregates. In SMFS, one or very few molecules are fixed to the AFM tip and their interactions with the desired substrate is measured. This approach can be expanded to study protein folding by pulling the protein from the surface. In addition, it can be used to measure interactions between cells and proteins and the binding of antibodies to their ligands.37,38,39,40 This paper describes in detail how to attach either peptides or amino acids to the AFM tip using silanol chemistry. In addition, the paper explains how to perform force measurements and how to analyze the results.
Protocol
1. Tip Modification
Purchase silicon nitride (Si3N4) AFM cantilevers with silicon tips (nominal cantilever radius of ~2 nm).
Clean each AFM cantilever by dipping in anhydrous ethanol for 20 min. Dry at room temperature. Then treat the cantilevers by exposing them to O2 plasma for 5 min.
Suspend the clean tips above (3 cm) a solution containing methyltriethoxysilane and 3-(aminopropyl) triethoxysilane in a ratio of 15:1 (v/v) in a desiccator under an inert atmosphere (either nitrogen or argon) and connect the desiccator to a vacuum pump. Vacuum for 2 h to form a monolayer of these two types of mixed silane compounds.
Use a metallic tip holder (fabricated for this process) to place the tips on a hot plate. Then dry the tips for 10 minutes at 70 °C under atmospheric conditions. Before use, clean the hot plate, metallic holder and tweezers using ethanol.
Cool the tips at room temperature, and then immerse the tips into a solution of Fluorenylmethyloxycarbonyl-PEG-N-hydroxysuccinimide (Fmoc-PEG-NHS, MW 5,000 Da) at a concentration of 5 mM in chloroform containing 0.5% (v/v) triethylamine for 1 h at room temperature.
Dip the tips in chloroform for 5 min and then dip it in dimethylformamide (DMF) for additional 5 min. In order to deprotect the Fmoc group of the attached PEG molecules, dip the tips in 20% piperidine (v/v) in DMF for 30 min. Dip the tips in DMF for 4 min and then in N-methyl-2-pyrrolidone (NMP) for additional 4 min. Repeat the sequential dipping three times.
For coupling of amino acids, immerse the tips into a solution containing N-terminal protected amino acid (AA) / diisopropylethylamine (DIPEA) / 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluophosphate (HBTU), in a molar ratio of 1:1:1 at a total concentration of 30 mM in 5 mL NMP for 1.5 h.
For peptide coupling, dip the tips into a 5 mL solution containing 40 mg of the protected peptide (N terminal and side chains, for example Fmoc-Gln(Trt)-Pro-Ala-Ser(tBu)-Ser(tBu)-Arg(pbf)-Tyr(tBu)-COOH.), 15 mg 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU), and 5 mL of DIPEA in NMP for 2 h.
Dip the tips in NMP for 4 min. Then, to protect the reaming free/unreacted NH2 by the acetyl group, dip the tips for 30 min in a mixture of acetic anhydride/DIPEA at a molar ratio 4:1 and a total concentration of 0.65 M in NMP.
- For peptide coupling, perform two additional steps.
- In order to deprotect the side chains of the peptide, immerse the tips into a solution containing 95% TFA, 2.5% triisopropylsilane, and 2.5% water for 1 h, and then wash with chloroform and DMF.
- In order to remove the Fmoc group of the peptide, immerse the tips into 20% piperidine (v/v) in DMF for 30 min.
Sequentially dip the peptide/amino acid-functionalized tips for four min each in DMF (for peptides) or NMP (for amino acids), chloroform, 50% ethanol and water. Finally dry the tip in air.
2. Surface Preparation
Prepare mica. Cleave the substrates (9.9 mm diameter) before each use by using scotch tape. Then, wash the surfaces with triple distilled water (TDW).
- Prepare TiO2-coated silicon.
- Cut the silicon wafer (100) into 2 cm squares using a diamond pen.
- Place the substrate in a 15 mL test tube filled with acetone and sonicate it for five minutes in an ultrasonic bath. Then, place this surface in a 15 mL test tube filled with isopropanol and sonicate it for 5 min. Dry the substrate using nitrogen.
- Dissolve the surfactant (e.g., Byk-348) in ethanol to prepare a 5% (weight/volume) solution. Then, add 0.02 mL of the surfactant solution to a 2 mL of 30% TiO2 dispersion.
- From the resulting solution, drop-cast 0.2 mL on a clean Si substrate.
- Anneal these drop-casted surfaces at 250 °C for 2 h in air.41
3. Single-molecule Force Spectroscopy Measurements
Attach the desired surface to a metallic holder of the AFM with commercially available two-component glue. Place the metallic holder in the glass holder of the AFM, which is shaped as a small Petri dish. Fill this holder with Tris buffer (50 mM pH = 7.4) or any desired medium. Then, place the holder on the AFM stage below the tip holder.
Calibrate the AFM cantilevers with spring constants ranging from 10 to 30 pN/nm using the thermal fluctuation method26 (included in the AFM software) with an absolute uncertainty of about 10%.
Measure the force of the interaction by approaching the amino acid or peptide-functionalized tip to the substrate until it is in contact with the substrate with a compression force of ~200 pN and then immediately retract the tip at various speeds, from 0.1 to 0.8 µm/s, for a distance of ~200 nm.
4. Data Analysis
- Convert the deflection values (V) to force by multiplying the photodiode sensitivity (V/m) and using the experimentally determined spring constant.42 This is done automatically by the AFM software.
- To obtain F-D curves with one single molecule events, record several hundreds of curves (800-1,500). Obtain two peaks in a single-molecule curve: the first peak indicates nonspecific interactions between the tip and the surface and the second peak corresponds to the specific interaction of the molecule with the surface. When, the F-D curve contain more than these two peaks, discard them from the calculation of the most probable force. NOTE: Only single adhesion events are taken into an account (from 10 to 30% of the curves).43
To calculate the apparent loading rate, fit at least 50 force vs. distance curves with the worm-like chain (WLC) model44 just prior to the rupture to obtain a set of loading rates, which are then used for preparing histograms of the apparent loading rates. Derive the unbinding forces between the peptides/amino acids and surface from the jump in force after separating the cantilever from the substrate.
Representative Results
Figure 1 exhibits the tip modification procedure. In the first step, a plasma treatment changes the surface of the silicon nitride tip. The tip presents OH groups. These groups will then react with the silanes. At the end of this step, the surface of the tip will be covered by free -NH2 groups. These free amines will then react with Fmoc -PEG-NHS, a covalent linker. The Fmoc group of the PEG linker is removed by pipyridine, a deprotecting reagent. Finally, the examined amino acid or peptide molecule is coupled through the free amine group of the PEG using the coupling reagent HBTU.
With the modified AFM tip it is possible to examine the interactions of the amino acid or peptide with the surface (Figure 2). The PEG molecule separates the peptide or amino acid from the tip and allows them to freely orient. A typical force measurement results in a Force-distance curve (Figure 3). This curve has a characteristic point of separation of the tip from the surface, and a single molecule adhesion event. The first peak indicates nonspecific interactions between the tip and the surface and the second peak refers to the specific adhesion event. From several hundred F-D curves it is possible to construct a histogram by plotting the number of adhesion events versus force. Applying a Gaussian fit on these histograms will determine the most probable force (MPF).
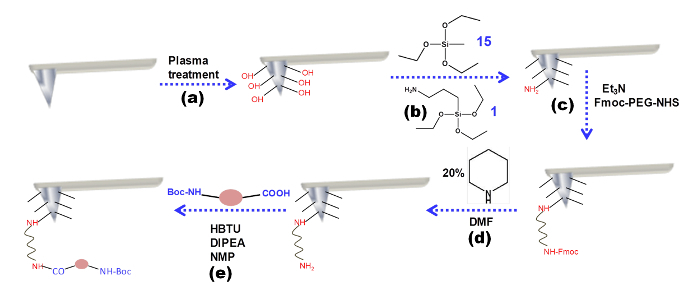 Figure 1: Tip modification procedure. Schematic representation of the chemical modification of the AFM tip. Please click here to view a larger version of this figure.
Figure 1: Tip modification procedure. Schematic representation of the chemical modification of the AFM tip. Please click here to view a larger version of this figure.
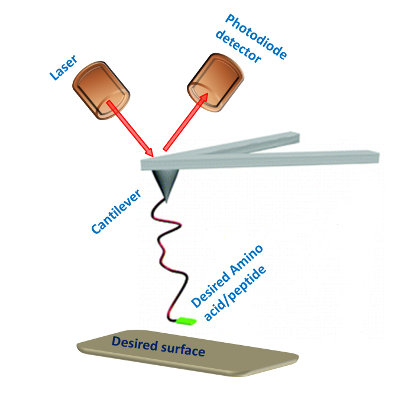 Figure 2: SMFS experimental setup. Schematic illustration of the single-molecule force spectroscopy setup for measuring the interactions between amino acids or peptides and a desired surface. Please click here to view a larger version of this figure.
Figure 2: SMFS experimental setup. Schematic illustration of the single-molecule force spectroscopy setup for measuring the interactions between amino acids or peptides and a desired surface. Please click here to view a larger version of this figure.
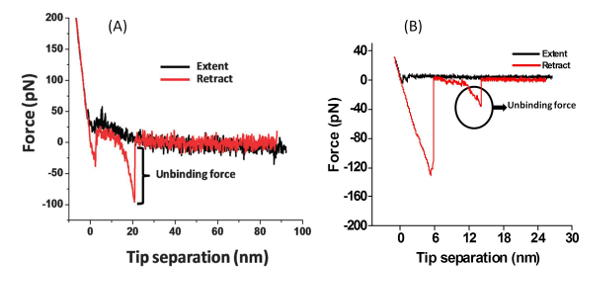 Figure 3: Force-distance Curve. Typical single-molecule F-D curves for the rupture of (A) the peptide Gln-Pro-Ala-Ser-Ser-Arg-Tyr from a mica surface, and (B) the amino acid phenylalanine from a TiO2 surface. Please click here to view a larger version of this figure.
Figure 3: Force-distance Curve. Typical single-molecule F-D curves for the rupture of (A) the peptide Gln-Pro-Ala-Ser-Ser-Arg-Tyr from a mica surface, and (B) the amino acid phenylalanine from a TiO2 surface. Please click here to view a larger version of this figure.
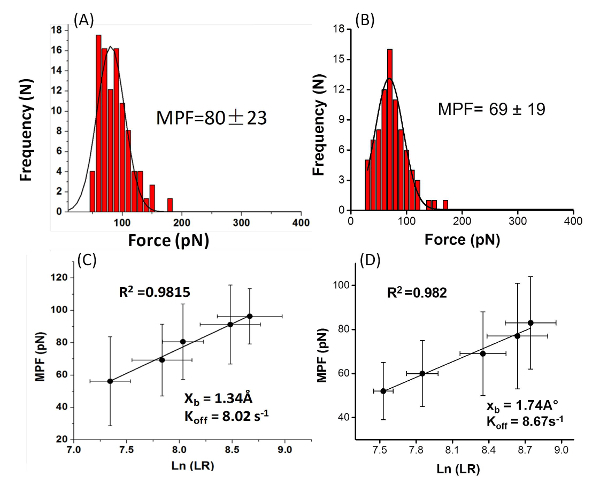 Figure 4: The histograms plot the Most Probable Force (MPF) and the graphs plot the force Vs. loading rate. Typical histograms of the rupture force values of (A) the peptide Gln-Pro-Ala-Ser-Ser-Arg-Tyr from mica (at a loading rate of 3.1 ± 0.6 nN/s (N = 7 8)), (B) the amino acid phenylalanine from TiO2 (at loading rate of 4.2 ± 0.7 nN/s (N = 79)). The most probable force (MPF) value was calculated based on the Gaussian fit (the black lines). Loading-rate dependence for the rupture forces for (C) the peptide Gln-Pro-Ala-Ser-Ser-Arg-Tyr and (D) the amino acid phenylalanine. The kinetic parameters were extrapolated from the linear plot of the force vs. the logarithm of the apparent loading rate. Please click here to view a larger version of this figure.
Figure 4: The histograms plot the Most Probable Force (MPF) and the graphs plot the force Vs. loading rate. Typical histograms of the rupture force values of (A) the peptide Gln-Pro-Ala-Ser-Ser-Arg-Tyr from mica (at a loading rate of 3.1 ± 0.6 nN/s (N = 7 8)), (B) the amino acid phenylalanine from TiO2 (at loading rate of 4.2 ± 0.7 nN/s (N = 79)). The most probable force (MPF) value was calculated based on the Gaussian fit (the black lines). Loading-rate dependence for the rupture forces for (C) the peptide Gln-Pro-Ala-Ser-Ser-Arg-Tyr and (D) the amino acid phenylalanine. The kinetic parameters were extrapolated from the linear plot of the force vs. the logarithm of the apparent loading rate. Please click here to view a larger version of this figure.
Discussion
Steps 1.3, 1.4 and 1.7 in the protocol should be carried out with extensive care and in a very gentle manner. In step 1.3, the tip should not be in contact with the silane mixture and the silanization process should be carried out in an inert atmosphere (moisture free).45 This is done in order to prevent multilayer formation and because silane molecules readily undergo hydrolysis in the presence of moisture.45
In step 1.4, the temperature and time should be kept properly. Before starting step 1.5, the tip should be cooled down to room temperature; otherwise it will be damaged. In the coupling step (1.7), the HBTU and the examined amino acid or peptide should be completely dissolved in the mixture. After coupling, washing the tip with the different solvents should be done in a very gentle manner to avoid any damage to the tip.
The reported technique can be applied to any peptide or amino acid. To modify the silicon tip, we use silanes. This is general chemistry which can be altered. For example, one can use either two or one type of PEGylated silane to modify the tip.23,25,26 If the tip is made of gold, then thiol groups can be used for the modification of the tip. Alternative protocols exploit nitrilotriacetate (NTA)-terminated linkers, able to target histidine, together with recombinant histidine-tagged proteins. Recently, Lyubchenko et al. described the synthesis and examining of a novel polymer linker and showed its application in several SMFS experiments. The synthesis of the linker is based on the well-developed phosphoramidate (PA) chemistry. This chemistry allows a routine synthesis of linkers with predetermined lengths and PA composition. These linkers are homogeneous in length and can be terminated with various functional groups. Furthermore, biomolecules can be anchored on gold substrates or tips through native or engineered thiol groups as gold form covalent bonds with the sulfur atoms.
This method allows the quantitative and detailed measurement of the force needed to unbind a molecule from a surface at the single-molecule level and not in bulk. Future applications of the technique include the attachment of proteins to the tip and the design of new hybrid materials. The design and development of new composite materials and functional surfaces will benefit from obtaining a fundamental understanding of the interactions between proteins or peptides with inorganic materials. The protocol provided here for SMFS with AFM can serve as a powerful tool for studying the interactions between proteins, peptides and amino acids with different surfaces.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
This work was supported by the Marie Curie International Reintegration Grant (EP7). P. D. acknowledges the support of the Israel Council for Higher Education.
References
- Addadi L, Weiner S. Control and design principles in biological mineralization. Angew. Chem., Int. Ed. 1992;31(2):153–169. [Google Scholar]
- Meyers MA, Chen PY, Lin AYM, Seki Y. Biological materials: Structure and mechanical properties. Prog. Mater. Sci. 2008;53(1):1–206. doi: 10.1016/j.jmbbm.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Villee CAJ. Book Review. Engl. J. Med. 1983;309(4):247–248. [Google Scholar]
- Vallee A, Humblot V, Pradier C-M. Peptide interactions with metal and oxide surfaces. Acc. Chem. Res. 2010;43(10):1297–1306. doi: 10.1021/ar100017n. [DOI] [PubMed] [Google Scholar]
- Peelle BR, Krauland EM, Wittrup KD, Belcher AM. Design criteria for engineering inorganic material-specific peptides. Langmuir. 2005;21(15):6929–6933. doi: 10.1021/la050261s. [DOI] [PubMed] [Google Scholar]
- Gabryelczyk B, Szilvay GR, Linder MB. The structural basis for function in diamond-like carbon binding peptides. Langmuir. 2014;30(29):8798–8802. doi: 10.1021/la502396p. [DOI] [PubMed] [Google Scholar]
- Sarikaya M, Tamerler C, Jen AKY, Schulten K, Baneyx F. Molecular biomimetics: Nanotechnology through biology. Nat. Mater. 2003;2(9):577–585. doi: 10.1038/nmat964. [DOI] [PubMed] [Google Scholar]
- Tamerler C, Sarikaya M. Molecular biomimetics: Utilizing nature's molecular ways in practical engineering. Acta Biomater. 2007;3(3):289–299. doi: 10.1016/j.actbio.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Seker UOS, Demir HV. Material binding peptides for nanotechnology. Molecules. 2011;16(2):1426–1451. doi: 10.3390/molecules16021426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JJ, et al. Electrostatic ligand coatings of nanoparticles enable ligand-specific gene delivery to human primary cells. Nano Lett. 2007;7(4):874–879. doi: 10.1021/nl062395b. [DOI] [PubMed] [Google Scholar]
- Grohe B, et al. Control of calcium oxalate crystal growth by face-specific adsorption of an osteopontin phosphopeptide. J. Am. Chem. Soc. 2007;129(48):14946–14951. doi: 10.1021/ja0745613. [DOI] [PubMed] [Google Scholar]
- Maity S, Nir S, Zada T, Reches M. Self-assembly of a tripeptide into a functional coating that resists fouling. Chem. Commun. 2014;50(76):11154–11157. doi: 10.1039/c4cc03578j. [DOI] [PubMed] [Google Scholar]
- Das P, Yuran S, Yan J, Lee PS, Reches M. Sticky tubes and magnetic hydrogels co-assembled by a short peptide and melanin-like nanoparticles. Chem. Commun. 2015;51(25):5432–5435. doi: 10.1039/c4cc07671k. [DOI] [PubMed] [Google Scholar]
- Burg KJL, Porter S, Kellam JF. Biomaterial developments for bone tissue engineering. Biomaterials. 2000;21(23):2347–2359. doi: 10.1016/s0142-9612(00)00102-2. [DOI] [PubMed] [Google Scholar]
- Weiger MC, et al. Quantification of the binding affinity of a specific hydroxyapatite binding peptide. Biomaterials. 2010;31(11):2955–2963. doi: 10.1016/j.biomaterials.2010.01.012. [DOI] [PubMed] [Google Scholar]
- Pettitt ME, Henry SL, Callow ME, Callow JA, Clare AS. Activity of commercial enzymes on settlement and adhesion of cypris larvae of the barnacle Balanus amphitrite, spores of the green alga Ulva linza, and the diatom Navicula perminuta. Biofouling. 2004;20(6):299–311. doi: 10.1080/08927010400027068. [DOI] [PubMed] [Google Scholar]
- Schultz MP, Finlay JA, Callow ME, Callow JA. Three models to relate detachment of low form fouling at laboratory and ship scale. Biofouling. 2003;19:17–26. doi: 10.1080/0892701031000089516. [DOI] [PubMed] [Google Scholar]
- Cao S, Wang J, Chen H, Chen D. Progress of marine biofouling and antifouling technologies. Chinese Science Bulletin. 2010;56(7):598–612. [Google Scholar]
- Wei Y, Latour RA. Correlation between desorption force measured by Atomic Force Microscopy and adsorption free energy measured by surface plasmon resonance spectroscopy for peptide-surface interactions. Langmuir. 2010;26(24):18852–18861. doi: 10.1021/la103685d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, et al. AFM-based force spectroscopy for bioimaging and biosensing. RSC Advances. 2016;6:12893–12912. [Google Scholar]
- Meibner RH, Wei G, Ciacchi LC. Estimation of the free energy of adsorption of a polypeptide on amorphous SiO2 from molecular dynamics simulations and force spectroscopy experiments. Soft Matter. 2015;11(31):6254–6265. doi: 10.1039/c5sm01444a. [DOI] [PubMed] [Google Scholar]
- Xue Y, Li X, Li H, Zhang W. Quantifying thiol-gold interactions towards the efficient strength control. Nat. Commun. 2014;5:4348. doi: 10.1038/ncomms5348. [DOI] [PubMed] [Google Scholar]
- Razvag Y, Gutkin V, Reches M. Probing the interaction of individual amino acids with inorganic surfaces using atomic force spectroscopy. Langmuir. 2013;29:10102–10109. doi: 10.1021/la4015866. [DOI] [PubMed] [Google Scholar]
- Das P, Reches M. Revealing the role of catechol moieties in the interactions between peptides and inorganic surfaces. Nanoscale. 2016;8:15309–15316. doi: 10.1039/c6nr04550b. [DOI] [PubMed] [Google Scholar]
- Das P, Reches M. Review insights into the interactions of amino acids and peptides with inorganic materials using single molecule force spectroscopy. Bioploymers-Pept. Sci. 2015;104:480–494. doi: 10.1002/bip.22655. [DOI] [PubMed] [Google Scholar]
- Maity S, et al. Elucidating the mechanism of interaction between peptides and inorganic surfaces. Phys. Chem. Chem. Phys. 2015;17(23):15305–15315. doi: 10.1039/c5cp00088b. [DOI] [PubMed] [Google Scholar]
- Whaley SR, English DS, Hu EL, Barbara PF, Belcher AM. Selection of peptides with semiconductor binding specificity for directed nanocrystal assembly. Nature. 2000;405(6787):665–668. doi: 10.1038/35015043. [DOI] [PubMed] [Google Scholar]
- Tamerler C, Oren EE, Duman M, Venkatasubramanian E, Sarikaya M. Adsorption Kinetics of an engineered gold binding peptide by surface plasmon resonance spectroscopy and a quartz crystal microbalance. Langmuir. 2006;22(18):7712–7718. doi: 10.1021/la0606897. [DOI] [PubMed] [Google Scholar]
- Santos O, Kosoric J, Hector MP, Anderson P, Lindh L. Adsorption behavior of statherin and a statherin peptide onto hydroxyapatite and silica surfaces by in situ ellipsometry. J. Colloid Interface Sci. 2008;318(2):175–182. doi: 10.1016/j.jcis.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Evans E, Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys. J. 1997;72(4):1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micksch T, Liebelt N, Scharnweber D, Schwenzer B. Investigation of the peptide adsorption on ZrO2, TiZr, and TiO2 surfaces as a method for surface modification. ACS Appl. Mater. Interfaces. 2014;6(10):7408–7416. doi: 10.1021/am500823m. [DOI] [PubMed] [Google Scholar]
- Patwardhan SV, et al. Chemistry of aqueous silica nanoparticle surfaces and the mechanism of selective peptide adsorption. J. Am. Chem. Soc. 2012;134(14):6244–6256. doi: 10.1021/ja211307u. [DOI] [PubMed] [Google Scholar]
- Thyparambil AA, Wei Y, Latour RA. Determination of peptide-surface adsorption free energy for material surfaces not conducive to SPR or QCM using AFM. Langmuir. 2012;28(13):5687–5694. doi: 10.1021/la300315r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnilova M, et al. Effect of molecular conformations on the adsorption behavior of gold-binding peptides. Langmuir. 2008;24(21):12440–12445. doi: 10.1021/la801468c. [DOI] [PubMed] [Google Scholar]
- Sano K, Sasaki H, Shiba K. Utilization of the pleiotropy of a peptidic aptamer to fabricate heterogeneous nanodot-containing multilayer nanostructures. J. Am. Chem. Soc. 2006;128(5):1717–1722. doi: 10.1021/ja057262r. [DOI] [PubMed] [Google Scholar]
- Chen H, Su X, Neoh K-G, Choe W-S. Context-dependent adsorption behavior of cyclic and linear peptides on metal oxide surfaces. Langmuir. 2008;25(3):1588–1593. doi: 10.1021/la8030304. [DOI] [PubMed] [Google Scholar]
- Zlatanova J, Lindsay SM, Leuba SH. Single molecule force spectroscopy in biology using the atomic force microscope. Prog. Biophys. Mol. Biol. 2000;74(1-2):37–61. doi: 10.1016/s0079-6107(00)00014-6. [DOI] [PubMed] [Google Scholar]
- Wang CZ, Yadavalli VK. Investigating biomolecular recognition at the cell surface using atomic force microscopy. Micron. 2014;60:5–17. doi: 10.1016/j.micron.2014.01.002. [DOI] [PubMed] [Google Scholar]
- Galler K, Brautigam K, Grobe C, Popp J, Neugebauer U. Making a big thing of a small cell - recent advances in single cell analysis. Analyst. 2014;139(6):1237–1273. doi: 10.1039/c3an01939j. [DOI] [PubMed] [Google Scholar]
- Carvalho FA, Martins IC, Santos NC. Atomic force microscopy and force spectroscopy on the assessment of protein folding and functionality. Arch. Biochem. Biophys. 2013;531(1-2):116–127. doi: 10.1016/j.abb.2012.11.007. [DOI] [PubMed] [Google Scholar]
- Azoubel S, Magdassi S. Controlling adhesion properties of SWCNT-PET films prepared by wet deposition. ACS Appl. Mater. Interfaces. 2014;6(12):9265–9271. doi: 10.1021/am501488p. [DOI] [PubMed] [Google Scholar]
- Jaschke M, Butt HJ. Height calibration of optical-lever atomic-force microscopes by simple laser interferometry. Rev. Sci. Instrum. 1995;66(2):1258–1259. [Google Scholar]
- Evans E, Kinoshita K, Simon S, Leung A. Long-lived, high-strength states of ICAM-1 bonds to beta(2) integrin, I: Lifetimes of bonds to recombinant alpha(L) beta(2) under force. Biophys. J. 2010;98(8):1458–1466. doi: 10.1016/j.bpj.2009.09.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchiat C, et al. Estimating the persistence length of a Worm-Like Chain molecule from force-extension measurements. Biophys. J. 1999;76(1):409–413. doi: 10.1016/s0006-3495(99)77207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick C, Argento C, Drazer G, Frechette J. Micropatterned Charge Heterogeneities via Vapor Deposition of Aminosilanes. Langmuir. 2015;31(39):10725–10733. doi: 10.1021/acs.langmuir.5b02771. [DOI] [PubMed] [Google Scholar]
- Berquand A, et al. Antigen binding forces of single antilysozyme Fv fragments explored by atomic force microscopy. Langmuir. 2005;21:5517–5523. doi: 10.1021/la050162e. [DOI] [PubMed] [Google Scholar]
- Kienberger F, et al. Recognition Force Spectroscopy Studies of the NTA-His6 Bond. Single Molecules. 2000;1:59–65. [Google Scholar]
- Tong Z, Mikheikin A, Krasnoslobodtsev A, Lv Z, Lyubchenko YL. Novel polymer linkers for single molecule AFM force spectroscopy. Methods. 2013;60:161–168. doi: 10.1016/j.ymeth.2013.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulman A. Formation and Structure of Self-Assembled Monolayers. Chem. Rev. 1996;96:1533–1554. doi: 10.1021/cr9502357. [DOI] [PubMed] [Google Scholar]
- Andolfi L, Bizzarri AR, Cannistraro S. Electron tunneling in a metal-protein-metal junction investigated by scanning tunneling and conductive atomic force spectroscopies. Appl. Phys. Lett. 2006;89:183125. [Google Scholar]


