Abstract
Pluripotent stem cells have the ability to self renew and differentiate to multiple lineages, making them an attractive source for the generation of pancreatic progenitor cells that can be used for the study of and future treatment of diabetes. This article outlines a four-stage differentiation protocol designed to generate pancreatic progenitor cells from human embryonic stem cells (hESCs). This protocol can be applied to a number of human pluripotent stem cell (hPSC) lines. The approach taken to generate pancreatic progenitor cells is to differentiate hESCs to accurately model key stages of pancreatic development. This begins with the induction of the definitive endoderm, which is achieved by culturing the cells in the presence of Activin A, basic Fibroblast Growth Factor (bFGF) and CHIR990210. Further differentiation and patterning with Fibroblast Growth Factor 10 (FGF10) and Dorsomorphin generates cells resembling the posterior foregut. The addition of Retinoic Acid, NOGGIN, SANT-1 and FGF10 differentiates posterior foregut cells into cells characteristic of pancreatic endoderm. Finally, the combination of Epidermal Growth Factor (EGF), Nicotinamide and NOGGIN leads to the efficient generation of PDX1+/NKX6-1+ cells. Flow cytometry is performed to confirm the expression of specific markers at key stages of pancreatic development. The PDX1+/NKX6-1+ pancreatic progenitors at the end of stage 4 are capable of generating mature β cells upon transplantation into immunodeficient mice and can be further differentiated to generate insulin-producing cells in vitro. Thus, the efficient generation of PDX1+/NKX6-1+ pancreatic progenitors, as demonstrated in this protocol, is of great importance as it provides a platform to study human pancreatic development in vitro and provides a source of cells with the potential of differentiating to β cells that could eventually be used for the treatment of diabetes.
Keywords: Developmental Biology, Issue 121, Diabetes, Pancreas, Human Embryonic Stem Cells, Human Pluripotent Stem Cells, Regenerative Medicine, Pancreatic Progenitors, Beta Cell, Differentiation, Flow Cytometry
Introduction
The prevalence of diabetes is increasing and according to the Canadian Diabetes Association, it is estimated that over 11 million individuals in Canada are diabetic or prediabetic, with 5-10% of these individuals having type 1 diabetes (T1D)1. T1D is an autoimmune disease that is caused by the destruction of the insulin producing β cells that are located within the Islets of Langerhans. Currently, individuals living with T1D require exogenous sources of insulin2. Despite advances in insulin therapy, T1D patients continue to have a difficult time regulating their blood glucose levels and continue to suffer both hypo- and hyperglycemia. A promising form of treatment to restore normoglycemia in T1D is the use of human embryonic stem cells (hESCs), which could be used to generate an unlimited supply of insulin producing β cells both in vivo and in vitro3,4,5,6,7. Differentiating hESCs to β-like cells could make it possible to study diabetes in vitro, allowing for the identification of new therapeutic targets for type 2 diabetes and provide cells for transplantation into T1D patients.
The most successful attempt at generating insulin producing cells from hESCs in vitro is to recapitulate the embryonic events that occur during pancreatic development4,5. This involves the manipulation of distinct signaling pathways to accurately model key stages of the developing pancreas. Pancreatic development begins with the induction of the definitive endoderm, which is characterized by the expression of CXCR4 and CD117 (c-KIT)8,9. Precise regulation of definitive endoderm organization is required for the formation of the gut tube, which then undergoes anterior-to-posterior and ventral-dorsal patterning. The dorsal and ventral pancreatic buds emerge from the region of the posterior foregut that expresses the pancreatic and duodenal homeobox gene (Pdx1), which is necessary for pancreatic development10. The dorsal and ventral buds fuse to form the pancreas, which then undergoes extensive epithelial remodeling and expansion11. Commitment to the endocrine and exocrine lineage is accompanied by the generation of multipotent progenitor cells (MPCs) that express, among others, the transcription factors Pdx1, Nkx6.1 and Ptf1a12,13. MPCs that will become endocrine and ductal cells continue to express Nkx6-1 while decreasing Ptf1a expression. Contrary to this, exocrine lineage cells will lose expression of Nkx6-1 and maintain Ptf1a expression12.
The transcription factor Nkx6-1 has a key role in pancreatic development, particularly during the differentiation of endocrine progenitor cells to β cells. As previously described, deletion of Nkx6-1 results in impaired formation of β cells during pancreatic development14. Therefore, generating insulin-producing β cells both in vitro and in vivo requires the efficient induction of Nkx6-1.
We recently developed a protocol to efficiently generate PDX1+/NKX6-1+ pancreatic progenitors from hPSCs. These hPSC-derived pancreatic progenitors generate mature β cells upon transplantation into immunodeficient mice3. The differentiation protocol can be divided into 4 stages characteristic of: 1) definitive endoderm induction, 2) posterior foregut patterning, 3) pancreatic specification and 4) NKX6-1 induction. Here we provide a detailed description of each step of the directed differentiation.
Protocol
1. Preparation of Solutions and Media
NOTE: Prepare all media for cell culture in a sterile environment. Media has to be made and used immediately. Reagent details are provided in the Materials Table.
- Differentiation Media
- Prepare Day 0 Differentiation Media: RPMI Medium with 1% Glutamine, 2 µM CHIR 99021, 100 ng/ml Activin A, 104 M MTG.
- Prepare Day 1-2 Differentiation Media: RPMI Medium with 1% Glutamine, 100 ng/ml Activin A, 104 M MTG, 5 ng/ml bFGF, 50 µg/ml Ascorbic Acid.
- Prepare Day 3-5 Differentiation Media: RPMI Medium with 1% Glutamine, 1% B27, 104 M MTG, 0.75 µM Dorsomorphin, 50 ng/ml FGF10.
- Prepare Day 6-7 Differentiation Media: DMEM with 1% Glutamine, 1% B27, 50 µg/ml Ascorbic Acid, 50 ng/ml FGF10, 0.25 µM SANT-1, 2 µM Retinoic Acid, 50 ng/ml NOGGIN. NOTE: Retinoic Acid should be added to the media last and media should be protected from light to prevent oxidative degradation15.
- Prepare Day 8-12 Differentiation Media: DMEM with 1% Glutamine, 1% B27, 50 µg/ml Ascorbic Acid, 50 ng/ml NOGGIN, 100 ng/ml hEGF, 10 mM Nicotinamide.
Prepare FACS buffer: 10% FBS in PBS, minus calcium and magnesium.
2. HESC Differentiation
NOTE: HESC are thawed, passaged and expanded on irradiated mouse embryonic fibroblasts in the presence of a KOSR-based media supplemented with bFGF16. HESCs are ready for differentiation when the cells reach 80-95% confluency. At this time the colonies should be large with defined borders and a 'domelike' structure (Figure 1A). All cell culture is performed in flat-bottom, tissue culture treated plates coated with 0.1% gelatin. Typically, the cells are grown in 6 or 12-well plates, in media volumes of 2 ml or 1 ml, respectively.
On day 0, begin differentiation by replacing the KOSR-based media with Day 0 Differentiation Media.
On days 1 and 2, gently shake the plate to remove any dead cells from the monolayer before aspirating. Replace with Day 1-2 Differentiation Media.
On day 3, harvest cells for flow cytometry. If the cells express more than 90% CXCR4+/CD117+, proceed to step 2.4.
On days 3 and 5, gently shake the plate to remove any dead cells from the monolayer prior to aspirating. Replace with Day 3-5 Differentiation Media.
On days 6 and 7, replace with Day 6-7 Differentiation Media.
On days 8, 10 and 12, replace with Day 8-12 Differentiation Media.
On day 13, harvest cells for flow cytometry to determine PDX1 and NKX6-1 expression.
3. Harvesting Cells for Flow Cytometry Analysis
Dissociate cells with 1x commercial trypsin solution according to manufacturer's protocol and incubate at 37 °C for 3 min.
Remove commercial trypsin solution and resuspend single cells in 1,000 µl FACS Buffer with 30 µl DNase I.
Filter cells using a 35 µm nylon mesh cell strainer and transfer to a microcentrifuge tube.
Centrifuge cells at 455 x g for 5 min. Prepare cells for either live or fixed staining.
4. Staining for Flow Cytometry
- Flow preparation for live staining on day 3
- Resuspend cells in 1,000 µl FACS Buffer. Transfer 200 µl (approximately 1-3 x 105 cells) into two wells of a 96-well plate (for both unstained and stained samples).
- Centrifuge the plate at 931 x g for 2 min. Remove the supernatant by inverting the plate.
- Stain with primary-conjugated antibodies, CXCR4:APC and CD117:PE (see Materials Table) for 30 min at room temperature protected from light.
- Centrifuge the plate at 931 x g for 2 min. Remove the supernatant by inverting the plate.
- Resuspend samples in 100 µl FACS Buffer.
- Centrifuge the plate at 931 x g for 2 min. Remove the supernatant by inverting the plate.
- Resuspend each sample and transfer in a total volume of 300-500 µl FACS Buffer to 1 ml micro test tubes or 5 ml round-bottom tubes.
- Run samples on the flow cytometer17. If samples cannot be run immediately, store at 4 °C protected from light.
- Flow preparation for fixed staining on day 13
- Resuspend the cell pellet in commercial fixation/permeabilization solution for 24 hr at 4 °C.
- Centrifuge the sample at 455 x g for 5 min. Resuspend in 400 µl Perm/Wash solution.
- Transfer 200 µl of the sample (approximately 0.5-1 x 106 cells) to two wells of a 96-well plate (for IgG control and PDX1/NKX6-1 staining). Centrifuge at 931 x g for 2 min.
- Resuspend the cell pellets in 100 µl Perm/Wash solution containing anti-PDX1 and anti-NKX6-1 primary or isotype control antibodies (see Materials Table). Incubate overnight at 4 °C.
- Centrifuge the plate at 931 x g for 2 min. Remove the supernatant by inverting the plate. Resuspend samples in 100 µl Perm/Wash solution.
- Centrifuge the plate at 931 x g for 2 min. Remove the supernatant by inverting the plate.
- Resuspend the samples in 100 µl Perm/Wash containing secondary antibodies at room temperature for 1 hr protected from light.
- Centrifuge the plate at 931 x g for 2 min. Remove the supernatant by inverting the plate.
- Resuspend samples in 100 µl Perm/Wash Solution.
- Centrifuge the plate at 931 x g for 2 min. Remove the supernatant by inverting the plate.
- Repeat step 4.2.9 and 4.2.10.
- Resuspend each sample and transfer in a total volume of 300-500 µl FACS Buffer to 1 ml micro test tubes or 5 ml round-bottom tubes.
- Run samples on the flow cytometer17. If samples cannot be run immediately, store at 4 °C protected from light.
Representative Results
Efficient generation of pancreatic progenitors relies on the proper growth and maintenance of undifferentiated cells followed by the precise addition of specific signaling molecules during the differentiation protocol, as illustrated in the schematic in Figure 1A. On day 0, undifferentiated cells should be 80-95% confluent and colonies should have defined edges (Figure 1A). During Stage 1, the media will likely appear cloudy since cell death is quite common at this stage. By the end of Stage 1, cells should co-express the definitive endoderm markers, CXCR4 and CD117, at a proportion greater than 90% (Figure 1B).
As cells differentiate throughout the four-stage protocol, their morphology changes from an elongated, oval-like appearance (observed at Stages 1 and 2) to smaller, rounder cells (as seen at Stages 3 and 4) (Figure 1A). By the end of Stage 4, efficient induction of PDX1+/NKX6-1+ is confirmed by flow cytometry. In the representative figure shown in Figure 1C, the expression of NKX6-1+ at day 13 is approximately 88%. As illustrated, all of the Stage 4-derived NKX6-1+ cells co-express PDX-1+, a characteristic of pancreatic progenitor cells. However, NKX6-1 expression varies across different cell lines, and a previous report showed that NKX6-1+ expression ranges from 30.5-91.4% in the 8 different hPSC lines tested3.
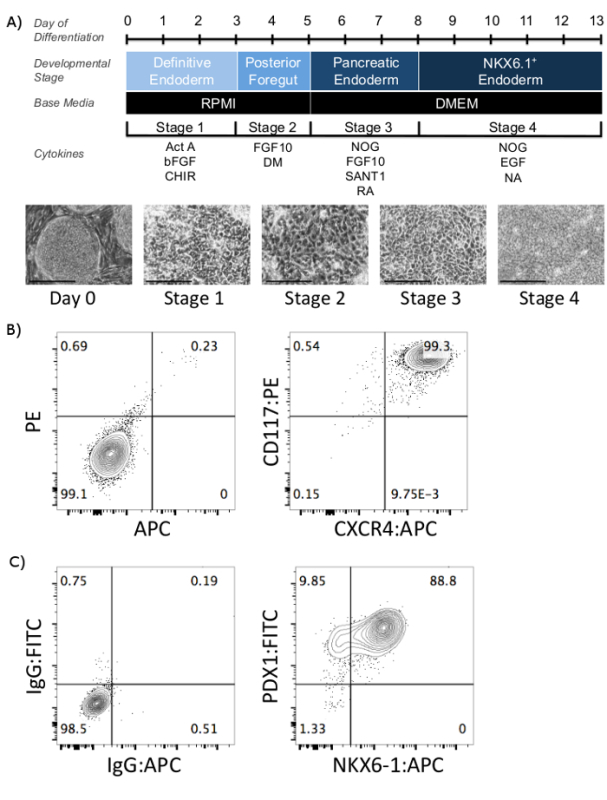 Figure 1: Schematic of pancreatic progenitor differentiation from hESCs and supporting data. (A) Schematic of the induction of pancreatic progenitor from hESCs. Day of differentiation, developmental stage, base media, and the cytokines added at every stage are included. Below, phase contrast images at the key stages of pancreatic differentiation are shown. In all representative figures, H1 cells were differentiated according to the 4-stage differentiation protocol depicted above. Day 0, undifferentiated hESCs; Stage 1, Definitive Endoderm; Stage 2, Posterior Foregut; Stage 3, PDX1+ Endoderm; Stage 4, NKX6-1+ endoderm/pancreatic progenitors. Scale bar = 200 µm. (B) Day 3 representative flow cytometry plots for CXCR4:PE and CD117:APC antibody staining. Unstained sample on left, stained sample on right. (C) Day 13 representative flow cytometry plot for stage-4 derived pancreatic progenitor cells expressing NKX6-1 and PDX1. IgG control on left, stained sample on right. hESC - human embryonic stem cell, Act A - Activin A; bFGF - basic fibroblast growth factor, FGF10 - fibroblast growth factor 10, DM - dorsomorphin, NOG - NOGGIN, RA - retinoic acid, EGF - epidermal growth factor, NA - nicotinamide. Please click here to view a larger version of this figure.
Figure 1: Schematic of pancreatic progenitor differentiation from hESCs and supporting data. (A) Schematic of the induction of pancreatic progenitor from hESCs. Day of differentiation, developmental stage, base media, and the cytokines added at every stage are included. Below, phase contrast images at the key stages of pancreatic differentiation are shown. In all representative figures, H1 cells were differentiated according to the 4-stage differentiation protocol depicted above. Day 0, undifferentiated hESCs; Stage 1, Definitive Endoderm; Stage 2, Posterior Foregut; Stage 3, PDX1+ Endoderm; Stage 4, NKX6-1+ endoderm/pancreatic progenitors. Scale bar = 200 µm. (B) Day 3 representative flow cytometry plots for CXCR4:PE and CD117:APC antibody staining. Unstained sample on left, stained sample on right. (C) Day 13 representative flow cytometry plot for stage-4 derived pancreatic progenitor cells expressing NKX6-1 and PDX1. IgG control on left, stained sample on right. hESC - human embryonic stem cell, Act A - Activin A; bFGF - basic fibroblast growth factor, FGF10 - fibroblast growth factor 10, DM - dorsomorphin, NOG - NOGGIN, RA - retinoic acid, EGF - epidermal growth factor, NA - nicotinamide. Please click here to view a larger version of this figure.
Discussion
Successfully generating NKX6-1+ pancreatic progenitors from hPSCs in vitro relies on the use of high quality cultures of hPSCs and directed differentiation involving the precise regulation of specific signaling pathways that govern key developmental stages during pancreatic development. Although this protocol can be used to induce robust expression of NKX6-1 across a variety of hPSC lines as previously shown3, to ensure efficient NKX6-1 generation the following considerations should be made. It is important that all cell culture and media preparation is performed in a sterile environment in order to prevent contamination. It is important that the hESC colonies have reached appropriate confluency and have not begun to differentiate prior to beginning day 0 of differentiation. Starting differentiation with hESCs that are not optimal can result in inefficient induction of the definitive endoderm and proceeding developmental stages. This is why it is critical to analyze cells at day 3 by flow cytometry to ensure efficient induction of definitive endoderm. The percentage of double positive CXCR4+CD117+ cells at day 3 should be greater than 90% for efficient pancreatic development3. If the percentage of cells expressing both CXCR4 and CD117 is lower than 90%, efficient induction to pancreatic progenitors will likely be compromised (results not shown). Furthermore, contrary to what was previously published, it is not necessary to wash cells with RPMI prior to feeding with stage 1 media16. Gentle shaking of the plate prior to aspirating the media is sufficient to remove the dead cells from the monolayer.
The addition of dorsomorphin at stage 2 has been shown to only be required for specific hPSC lines18. Thus, it is important to determine if dorsomorphin is necessary when working with different hPSC lines. Additionally, the duration of Stage 3 has been shown to be critical for specifying either monohormonal or polyhormonal progenitor cells and is cell line-dependent3. Thus, when working with a new hPSC line it is important to determine the duration that the cells should be cultured in Stage 3 media to determine the optimal conditions for NKX6-1 induction. To determine this, cells should be cultured in Stage 3 media for 1-4 days and analyzed 5 days later by flow cytometry for NKX6-1 expression.
Efficiently generating populations enriched in NKX6-1+ cells is important, as Nkx6-1 is critical for βcell development14. While others have developed protocols that can differentiate hESC to NKX6-1+ cells (approximately 60% H1-derived NKX6-1+ cells and 55% HUES8-derived PDX+/NKX6-1+ cells)4,5, our method consistently generates >80% H1-derived PDX1+/NKX6-1+ cells. However, our differentiation protocol relies on the use of mouse embryonic fibroblasts, which limits its use to research-only, as we have yet to optimize the method in Good Manufacturing Practices. Although we have not attempted to cryopreserve our stage-4 derived PDX1+/NKX6-1+ cells, this approach has been successfully performed by Schulz et al., who demonstrated that cryopreserved pancreatic aggregates were capable of retaining their function in vivo19.
In this paper we describe a 4-stage protocol to efficiently generate PDX1+/NKX6-1+ pancreatic progenitors from a variety of hPSC lines. By differentiating hPSC through four key stages of pancreatic development, this protocol provides a model to study the development of the human pancreas in vitro. Furthermore, since the PDX1+/NKX6-1+ cells have the potential to generate insulin-producing cells both in vivo and in vitro (data not shown)3,4,5, the hESC-derived PDX1+/NKX6-1+ pancreatic progenitors provide a possible source for the treatment of diabetes.
Disclosures
The authors have nothing to disclose.
Acknowledgments
This manuscript was supported by funding from the Toronto General and Western Foundation and the Banting & Best Diabetes Centre-University Health Network Graduate Award.
References
- Canadian Diabetes Association. 2016. Available from: http://www.diabetes.ca/about-diabetes/types-of-diabetes.
- Cogger K, Nostro MC. Recent advances in cell replacement therapies for the treatment of type 1 diabetes. Endocrinology. 2015;156(1):8–15. doi: 10.1210/en.2014-1691. [DOI] [PubMed] [Google Scholar]
- Nostro MC, et al. Efficient generation of NKX6-1+ pancreatic progenitors from multiple human pluripotent stem cell lines. Stem Cell Reports. 2015;4(4):591–604. doi: 10.1016/j.stemcr.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliuca FW, et al. Generation of Functional Human Pancreatic beta Cells In Vitro. Cell. 2014;159(2):428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezania A, et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat Biotechnol. 2014. [DOI] [PubMed]
- Kroon E, et al. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotechnol. 2008;26(4):443–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- Rezania A, et al. Maturation of human embryonic stem cell-derived pancreatic progenitors into functional islets capable of treating pre-existing diabetes in mice. Diabetes. 2012;61(8):2016–2029. doi: 10.2337/db11-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amour KA, et al. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol. 2005;23(12):1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- Gouon-Evans V, et al. BMP-4 is required for hepatic specification of mouse embryonic stem cell-derived definitive endoderm. Nat Biotechnol. 2006;24(11):1402–1411. doi: 10.1038/nbt1258. [DOI] [PubMed] [Google Scholar]
- Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371(6498):606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- Pan FC, Wright C. Pancreas organogenesis: from bud to plexus to gland. Dev Dyn. 2011;240(3):530–565. doi: 10.1002/dvdy.22584. [DOI] [PubMed] [Google Scholar]
- Schaffer AE, Freude KK, Nelson SB, Sander M. Nkx6 transcription factors and Ptf1a function as antagonistic lineage determinants in multipotent pancreatic progenitors. Dev Cell. 2010;18(6):1022–1029. doi: 10.1016/j.devcel.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, et al. A multipotent progenitor domain guides pancreatic organogenesis. Dev Cell. 2007;13(1):103–114. doi: 10.1016/j.devcel.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Sander M, et al. Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of beta-cell formation in the pancreas. Development. 2000;127(24):5533–5540. doi: 10.1242/dev.127.24.5533. [DOI] [PubMed] [Google Scholar]
- Sharow KA, Temkin B, Asson-Batres MA. Retinoic acid stability in stem cell cultures. Int J Dev Biol. 2012;56(4):273–278. doi: 10.1387/ijdb.113378ks. [DOI] [PubMed] [Google Scholar]
- Korytnikov R, Nostro MC. Generation of polyhormonal and multipotent pancreatic progenitor lineages from human pluripotent stem cells. Methods. 2016. pp. 56–64. [DOI] [PubMed]
- Baumgarth N, Roederer M. A practical approach to multicolor flow cytometry for immunophenotyping. J Immunol Methods. 2000;243(1-2):77–97. doi: 10.1016/s0022-1759(00)00229-5. [DOI] [PubMed] [Google Scholar]
- Nostro MC, et al. Stage-specific signaling through TGFbeta family members and WNT regulates patterning and pancreatic specification of human pluripotent stem cells. Development. 2011;138(5):861–871. doi: 10.1242/dev.055236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz TC, et al. A scalable system for production of functional pancreatic progenitors from human embryonic stem cells. PLoS One. 2012;7(5):e37004. doi: 10.1371/journal.pone.0037004. [DOI] [PMC free article] [PubMed] [Google Scholar]


