Abstract
Kirsten-ras-revertant 1 (Krev-1) cDNA encodes a ras-related protein and exhibits an activity of inducing flat revertants at certain frequencies (2-5% of total transfectants) when introduced into a v-K-ras-transformed mouse NIH 3T3 cell line, DT. Toward understanding the mechanism of action of Krev-1 protein, we constructed a series of point mutants of Krev-1 cDNA and tested their biological activities in DT cells and HT1080 human fibrosarcoma cells harboring the activated N-ras gene. Substitutions of the amino acid residues in the putative guanine nucleotide-binding regions (Asp17 and Asn116), in the putative effector-binding domain (residue 38), at the putative acylation site (Cys181), and at the unique Thr61 all decreased the transformation suppressor activity. On the other hand, substitutions such as Gly12 to Val12 and Gln63 to Glu63 were found to significantly increase the transformation suppressor/tumor suppressor activity of Krev-1. These findings are consistent with the idea that Krev-1 protein is regulated like many other G proteins by the guanine triphosphate/guanine diphosphate-exchange mechanism probably in response to certain negative growth-regulatory signals.
Full text
PDF

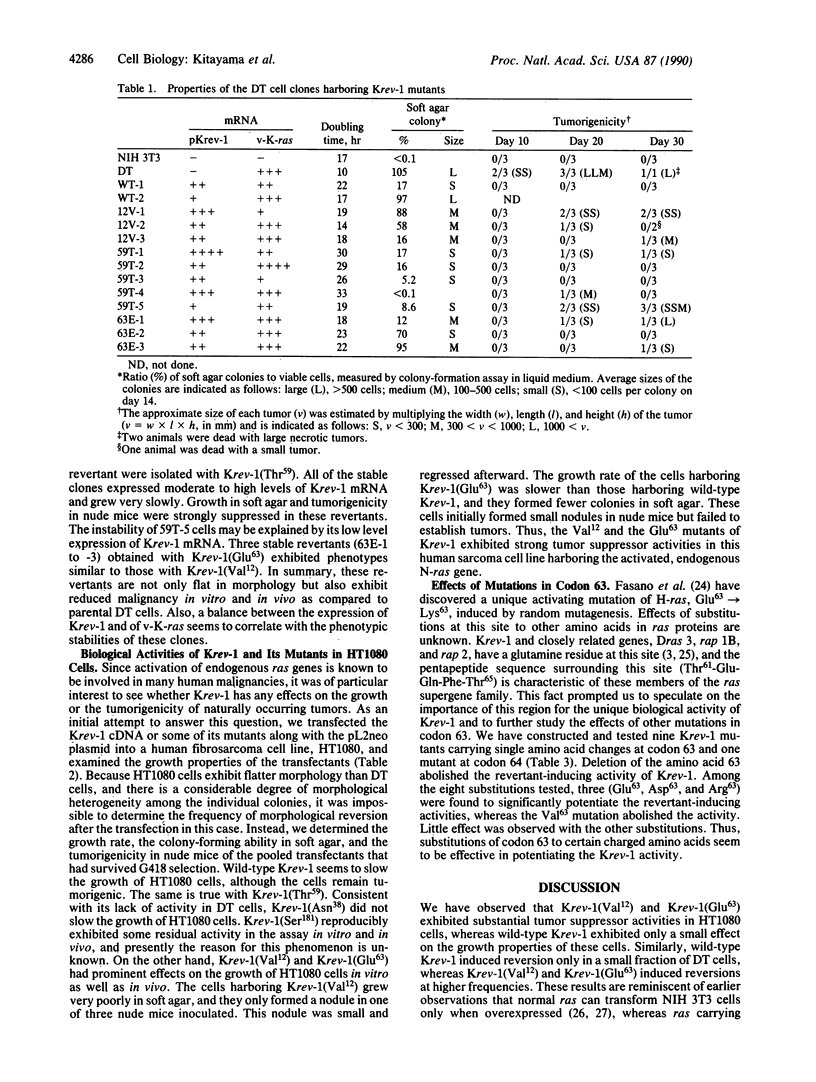
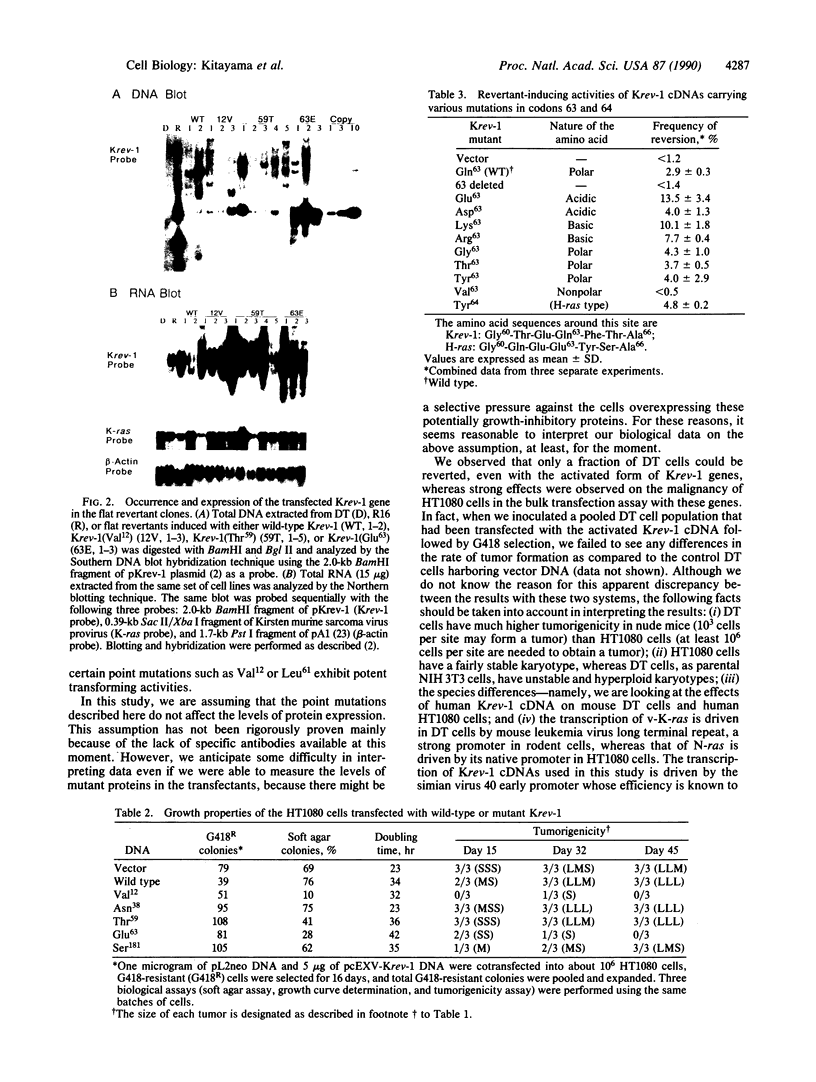

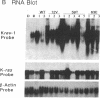
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- Chang E. H., Furth M. E., Scolnick E. M., Lowy D. R. Tumorigenic transformation of mammalian cells induced by a normal human gene homologous to the oncogene of Harvey murine sarcoma virus. Nature. 1982 Jun 10;297(5866):479–483. doi: 10.1038/297479a0. [DOI] [PubMed] [Google Scholar]
- Chen C., Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987 Aug;7(8):2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland D. W., Lopata M. A., MacDonald R. J., Cowan N. J., Rutter W. J., Kirschner M. W. Number and evolutionary conservation of alpha- and beta-tubulin and cytoplasmic beta- and gamma-actin genes using specific cloned cDNA probes. Cell. 1980 May;20(1):95–105. doi: 10.1016/0092-8674(80)90238-x. [DOI] [PubMed] [Google Scholar]
- Der C. J., Finkel T., Cooper G. M. Biological and biochemical properties of human rasH genes mutated at codon 61. Cell. 1986 Jan 17;44(1):167–176. doi: 10.1016/0092-8674(86)90495-2. [DOI] [PubMed] [Google Scholar]
- Fasano O., Aldrich T., Tamanoi F., Taparowsky E., Furth M., Wigler M. Analysis of the transforming potential of the human H-ras gene by random mutagenesis. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4008–4012. doi: 10.1073/pnas.81.13.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig L. A., Cooper G. M. Inhibition of NIH 3T3 cell proliferation by a mutant ras protein with preferential affinity for GDP. Mol Cell Biol. 1988 Aug;8(8):3235–3243. doi: 10.1128/mcb.8.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiyama A., Tamanoi F. Processing and fatty acid acylation of RAS1 and RAS2 proteins in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1266–1270. doi: 10.1073/pnas.83.5.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A., Marshall C. J., Spurr N. K., Weiss R. A. Identification of transforming gene in two human sarcoma cell lines as a new member of the ras gene family located on chromosome 1. Nature. 1983 Jun 2;303(5916):396–400. doi: 10.1038/303396a0. [DOI] [PubMed] [Google Scholar]
- Kawata M., Matsui Y., Kondo J., Hishida T., Teranishi Y., Takai Y. A novel small molecular weight GTP-binding protein with the same putative effector domain as the ras proteins in bovine brain membranes. Purification, determination of primary structure, and characterization. J Biol Chem. 1988 Dec 15;263(35):18965–18971. [PubMed] [Google Scholar]
- Kikuchi A., Sasaki T., Araki S., Hata Y., Takai Y. Purification and characterization from bovine brain cytosol of two GTPase-activating proteins specific for smg p21, a GTP-binding protein having the same effector domain as c-ras p21s. J Biol Chem. 1989 Jun 5;264(16):9133–9136. [PubMed] [Google Scholar]
- Kitayama H., Sugimoto Y., Matsuzaki T., Ikawa Y., Noda M. A ras-related gene with transformation suppressor activity. Cell. 1989 Jan 13;56(1):77–84. doi: 10.1016/0092-8674(89)90985-9. [DOI] [PubMed] [Google Scholar]
- McCormick F., Clark B. F., la Cour T. F., Kjeldgaard M., Norskov-Lauritsen L., Nyborg J. A model for the tertiary structure of p21, the product of the ras oncogene. Science. 1985 Oct 4;230(4721):78–82. doi: 10.1126/science.3898366. [DOI] [PubMed] [Google Scholar]
- McCormick F. ras GTPase activating protein: signal transmitter and signal terminator. Cell. 1989 Jan 13;56(1):5–8. doi: 10.1016/0092-8674(89)90976-8. [DOI] [PubMed] [Google Scholar]
- Molloy C. J., Bottaro D. P., Fleming T. P., Marshall M. S., Gibbs J. B., Aaronson S. A. PDGF induction of tyrosine phosphorylation of GTPase activating protein. Nature. 1989 Dec 7;342(6250):711–714. doi: 10.1038/342711a0. [DOI] [PubMed] [Google Scholar]
- Noda M., Kitayama H., Matsuzaki T., Sugimoto Y., Okayama H., Bassin R. H., Ikawa Y. Detection of genes with a potential for suppressing the transformed phenotype associated with activated ras genes. Proc Natl Acad Sci U S A. 1989 Jan;86(1):162–166. doi: 10.1073/pnas.86.1.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda M., Selinger Z., Scolnick E. M., Bassin R. H. Flat revertants isolated from Kirsten sarcoma virus-transformed cells are resistant to the action of specific oncogenes. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5602–5606. doi: 10.1073/pnas.80.18.5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai E. F., Kabsch W., Krengel U., Holmes K. C., John J., Wittinghofer A. Structure of the guanine-nucleotide-binding domain of the Ha-ras oncogene product p21 in the triphosphate conformation. Nature. 1989 Sep 21;341(6239):209–214. doi: 10.1038/341209a0. [DOI] [PubMed] [Google Scholar]
- Paterson H., Reeves B., Brown R., Hall A., Furth M., Bos J., Jones P., Marshall C. Activated N-ras controls the transformed phenotype of HT1080 human fibrosarcoma cells. Cell. 1987 Dec 4;51(5):803–812. doi: 10.1016/0092-8674(87)90103-6. [DOI] [PubMed] [Google Scholar]
- Pizon V., Chardin P., Lerosey I., Olofsson B., Tavitian A. Human cDNAs rap1 and rap2 homologous to the Drosophila gene Dras3 encode proteins closely related to ras in the 'effector' region. Oncogene. 1988 Aug;3(2):201–204. [PubMed] [Google Scholar]
- Pulciani S., Santos E., Long L. K., Sorrentino V., Barbacid M. ras gene Amplification and malignant transformation. Mol Cell Biol. 1985 Oct;5(10):2836–2841. doi: 10.1128/mcb.5.10.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schejter E. D., Shilo B. Z. Characterization of functional domains of p21 ras by use of chimeric genes. EMBO J. 1985 Feb;4(2):407–412. doi: 10.1002/j.1460-2075.1985.tb03643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal I. S., Gibbs J. B., D'Alonzo J. S., Scolnick E. M. Identification of effector residues and a neutralizing epitope of Ha-ras-encoded p21. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4725–4729. doi: 10.1073/pnas.83.13.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal I. S., Gibbs J. B., D'Alonzo J. S., Temeles G. L., Wolanski B. S., Socher S. H., Scolnick E. M. Mutant ras-encoded proteins with altered nucleotide binding exert dominant biological effects. Proc Natl Acad Sci U S A. 1986 Feb;83(4):952–956. doi: 10.1073/pnas.83.4.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong L., Milburn M. V., de Vos A. M., Kim S. H. Structure of ras proteins. Science. 1989 Jul 21;245(4915):244–244. doi: 10.1126/science.2665078. [DOI] [PubMed] [Google Scholar]
- Willumsen B. M., Christensen A., Hubbert N. L., Papageorge A. G., Lowy D. R. The p21 ras C-terminus is required for transformation and membrane association. Nature. 1984 Aug 16;310(5978):583–586. doi: 10.1038/310583a0. [DOI] [PubMed] [Google Scholar]
- Willumsen B. M., Papageorge A. G., Kung H. F., Bekesi E., Robins T., Johnsen M., Vass W. C., Lowy D. R. Mutational analysis of a ras catalytic domain. Mol Cell Biol. 1986 Jul;6(7):2646–2654. doi: 10.1128/mcb.6.7.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]