Abstract
Mitochondria play a central role for cell metabolism, energy production and control of apoptosis. Inadequate mitochondrial function has been found responsible for very diverse diseases, ranging from neurological pathologies to cancer. Interestingly, mitochondria have recently been shown to display the capacity to be transferred between cell types, notably from human mesenchymal stem cells (MSC) to cancer cells in coculture conditions, with metabolic and functional consequences for the mitochondria recipient cells, further enhancing the current interest for the biological properties of these organelles.
Evaluating the effects of the transferred MSC mitochondria in the target cells is of primary importance to understand the biological outcome of such cell-cell interactions. The MitoCeption protocol described here allows the transfer of the mitochondria isolated beforehand from the donor cells to the target cells, using MSC mitochondria and glioblastoma stem cells (GSC) as a model system. This protocol has previously been used to transfer mitochondria, isolated from MSCs, to adherent MDA-MB-231 cancer cells. This mitochondria transfer protocol is adapted here for GSCs that present the specific particularity of growing as neurospheres in vitro. The transfer of the isolated mitochondria can be followed by fluorescence-activated cell sorting (FACS) and confocal imaging using mitochondria vital dyes. The use of mitochondria donor and target cells with distinct haplotypes (SNPs) also allows detection of the transferred mitochondria based on the concentration of their circular mitochondrial DNA (mtDNA) in the target cells. Once the protocol has been validated with these criteria, the cells harboring the transferred mitochondria can be further analyzed to determine the effects of the exogenous mitochondria on biological properties such as cell metabolism, plasticity, proliferation and response to therapy.
Keywords: Cellular Biology, Issue 120, mitochondria transfer, human mesenchymal stem cells (MSC), glioblastoma stem cells (GSC), metabolism, mitochondrial DNA, neurospheres
Introduction
Mitochondria are organelles found in eukaryotic cells where they play a central role in nutrient uptake as well as in energy and metabolite production. These organelles contain circular mitochondrial DNA (mtDNA), 16.6 kb long, that encodes proteins of the electron transport chain complexes, tRNAs and rRNAs 1. The functionality of these organelles is critical for cell homeostasis and several pathologies have been associated with mitochondria dysfunction 1,2,3. The mitochondria status has for instance been linked to inflammation, infectious diseases and cancer, in this latter case with consequences for metastasis and resistance to therapy 4,5,6,7.
Mitochondria display the remarkable capacity of getting transferred between "donor" and "target" cells. This leads to changes in the energetic metabolism of the target cells as well as in other functional modifications such as tissue repair and resistance to chemotherapeutic agents, as recently shown by different laboratories 8,9,10,11,12,13,14,15,16. The human mesenchymal stem cells (MSCs) display this ability to transfer mitochondria to a wide variety of target cells, including cardiomyocytes, endothelial cells, pulmonary alveolar epithelial cells, renal tubular cells and cancer cells, leading to modifications of the functional properties of these cells 8,9,10,12,17,18.
Mitochondria exchange now appears as a widely used mechanism that allows a number of different cell types to communicate with one another and modify their biological properties. This mitochondria exchange can occur through tunneling nanotubes (TNT) formation, involving connexin 43-containing gap junctions 8 or M-Sec/TNFaip2 and the exocyst complex 19. Alternatively, the mitochondria transfer was also shown to be mediated by arrestin domain-containing protein 1-mediated microvesicles (ARMMs) 20. Interestingly, the efficacy of the mitochondria transfer was linked to the expression rate of the Rho GTPase 1 MIRO1 21, a key factor for explaining the differences in mitochondria transfer efficacies between iPSC-MSCs and adult BM-MSCs 22.
In spite of this wealth of data concerning cell-to-cell mitochondria exchange, relatively little is known about the metabolic and biological outcome of this mitochondria transfer. Therefore, it fully warrants setting up the appropriate tools to fully assess the biological effects of this transfer. Over the years, several technical approaches to transfer mitochondria from donor to acceptor cells have been proposed. This includes direct injection of mitochondria into oocytes 23,24,25, cell fusion to generate transmitochondrial cybrids 26,27and, more recently, transfer of isolated mitochondria using photothermal nanoblades 28.
We and others previously demonstrated the capacity of isolated mitochondria to be internalized by living cells, as observed both in vitro and in vivo29,30,31, through mechanisms proposed to involve macropinocytosis 32. We further developed a method, called MitoCeption, to quantitatively transfer isolated mitochondria (from MSCs) to target cells, as exemplified with the (adherent) MDA-MB-231 breast cancer cell line 31. This protocol was adapted here for the transfer of isolated human MSC mitochondria to glioblastoma stem cells (GSCs).
Glioblastoma are aggressive malignant tumors of the brain that rapidly become resistant to treatment, mainly due to glioblastoma stem cells (GSC) present within the tumor 33. These GSCs grow as neurospheres in vitro and generate tumors in xenograft models. Cancer cells within glioblastoma have the capacity to make cell-to-cell connections, as shown recently for astrocytic brain tumor cells that interconnect via extended microtubes, through which mitochondria (as well as calcium and cell nuclei) can migrate, resulting in radiotherapy-resistant astrocytoma networks 34. Glioblastoma can recruit many different cells within the tumor microenvironment, including MSCs 35,36. We showed that MSCs can make cell-cell connections with GSCs in coculture and transfer their mitochondria (data not shown), which is expected to modify GSC functional properties. The present protocol describes how the MitoCeption technique can be used to transfer mitochondria, isolated beforehand from human MSCs, to human GSCs with the purpose of determining their functional biological outcome. The multipotent and highly tumorigenic Gb4 GSC line 37 was used in this study.
Protocol
Day 1
1. Labeling of the Mesenchymal Stem Cell (MSC) Mitochondria (Optional)
Two days before the mitochondria preparation, seed human MSCs in a 100 mm culture dish, in 10 ml αMEM/FBS 10%, so as to have 4 x 105 MSCs in culture on Day 1.
Rinse MSCs with PBS (4 ml) and add 4 ml αMEM/FBS 1% (prewarmed to 37 °C).
Add the required amount of mitochondria vital dye and incubate cells for 30 min in the 37 °C incubator.
Remove the mitochondria dye solution, rinse cells twice with 4 ml prewarmed (37 °C) αMEM/FBS 1% and add back 4 ml αMEM/FBS 10%. Incubate cells at 37 °C.
Change the culture medium (10 ml αMEM/FBS 10%) after 30 min and, another time, 2 hr later.
Day 1
2. Labeling of the Glioblastoma Stem Cells (GSC) (Optional, See Discussion Section)
| Cell culture medium | Composition |
| GSC basal medium | DMEM/F-12 supplemented with |
| Insulin 20 mg/ml | |
| N2 supplement 1x | |
| Glucose 3 g/L | |
| L-glutamine 2 mM | |
| GSC proliferation medium | Add to the basal medium: |
| B27 supplement 1x | |
| EGF 10 ng/ml | |
| bFGF 10 ng/ml | |
| Fungine 10 mg/ml | |
| Fungizone 0.25 mg/ml | |
| Heparin 2 mg /ml | |
| Ciprofloxacin 2 μg / ml | |
| Gentamicin 2 μg/ml | |
| MSC proliferation medium | αMEM supplemented with |
| L-glutamine 2 mM | |
| 10% FBS | |
| bFGF 2 ng/ml |
Table 1: Culture Media.
Dissociate GSCs (Gb4 cell line 32 (10 x 106 cells) grown as neurospheres on poly-HEMA coated cell culture flasks (see steps 3.1 to 3.12).
Seed GSCs in a 48-well plate at 105 cells/well in GSC proliferation medium (500 µl) (see Table 1).
Centrifuge the plate at 270 x g for 5 min at 20 °C.
Add the required amount of cell vital dye and incubate for 30 min at 37 °C.
Add 500 µl of GSC basal medium (Table 1) per well of the 48-well plate.
Centrifuge the plate at 270 x g at 20 °C for 5 min. Aspirate the supernatant.
Repeat steps 2.5 to 2.6.
Add 500 µl of GSC basal medium and incubate at 37 °C for 30 min.
Centrifuge the plate at 270 x g at 20 °C for 5 min. Aspirate the supernatant.
Add 500 µl of GSC proliferation medium per well of the 48-well plate and incubate in the 37 °C incubator. Note: The amount of GSCs indicated (10 x 106 cells) allows performing the different dose-response experiments and FACS controls. Once the experimental conditions are more precisely defined this amount can be scaled down.
Day 2
3. Seeding of Glioblastoma Stem Cells
Collect the GSC neurospheres (10 x 106 cells) by centrifugation at 270 x g for 5 min, at 20 °C in a 50 ml tube.
Wash cells with 5 ml of HBSS, and centrifuge at 270 x g for 5 min at 20 °C.
Aspirate the supernatant.
Gently resuspend the GSC pellet in 100 µl trypsin-EDTA (0.25%) (per 10 x 106 cells)
Incubate at 37 °C for 3 min.
Add 10 µl CaCl2 (20 mM) and 2 µl DNase I (10 mg/ml).
Dissociate the neurospheres by gently pipetting up and down (30-50x) with a P200 pipette. Avoid bubbles. Check under the microscope that all GSCs are dissociated.
Add 10 µl trypsin inhibitor (5%) and 10 ml HBSS.
Centrifuge GSCs at 270 x g for 7 min at 20 °C.
Discard the supernatant and add 10 ml GSC basal medium (Table 1).
Count GSCs with a Thoma counting chamber and then centrifuge the cells at 270 x g for 7 min at 20 °C.
Add the appropriate volume of GSC proliferation medium (Table 1) to reach the cellular concentration of 106 GSCs/ml.
Seed 105 GSCs (100 µl of the cell suspension) per well of a 96-well plate.
Centrifuge the plates at 270 x g for 7 min at 20 °C to get GSCs at the bottom of wells.
Incubate the 96-well plate at 37 °C until Section 5.
Day 2
4. MSC Mitochondria Isolation
Adjust the microtube centrifuge temperature to 4 °C.
Prepare two 1.5 ml tubes "A" and "C" containing the reagents for the mitochondria extraction (200 µl reagent A and 400 µl reagent C, both containing the EDTA-free protease inhibitors). Prepare 2 other tubes, labeled "MSC" and "Mito". Keep all tubes on ice. Also prepare tubes for the mitochondria serial dilutions.
Wash MSCs with 10 ml prewarmed (37 °C) PBS.
Wash MSCs with 2 ml trypsin (no EDTA) for 10 sec and add 1 ml trypsin (no EDTA). Incubate cells for 5-10 min at 37 °C.
Recover the MSCs by adding 10 ml αMEM/FBS 10%, transfer to a 50 ml tube.
Centrifuge cells at 270 x g for 5 min at 20 °C.
Discard the supernatant and add 10 ml αMEM/FBS 10% to the cell pellet.
Count the MSCs with a Malassez counting chamber.
Centrifuge MSCs (4-5 x 105) at 270 x g for 5 min at 20 °C.
Discard the supernatant, add 1 m ice-cold αMEM/FBS 10% to the cell pellet and transfer the cells to the "MSC"-labeled tube (prepared at step 4.2). Keep the tube on ice.
Centrifuge the tube containing the MSCs at 900 x g for 5 min, at 4 °C.
Remove all residual medium from the tube.
Add 200 µl of Mitochondria Isolation Reagent A (containing the EDTA-free protease inhibitors). Vortex at medium speed for 5 sec and leave tubes on ice for exactly 2 min.
Add 2.5 µl of Mitochondria Isolation Reagent B. Vortex at maximum speed for 10 sec, and then leave tubes on ice. Repeat every 30 sec for 5 min.
Add 200 µl of Mitochondria Isolation Reagent C (containing the EDTA-free protease inhibitors). Mix by tilting the tube (roughly 30 times, do not vortex). Centrifuge the tube at 700 x g for 10 min at 4 °C.
Transfer the supernatant (containing the MSC mitochondria) to the "Mito" tube (prepared at step 4.2).
Centrifuge at 3,000 x g, 15 min at 4 °C, to get the mitochondria pellet. Discard the supernatant. The pellet contains the isolated mitochondria.
Rinse the mitochondria pellet with 200 µl of the Reagent C. Then, centrifuge at 12,000 x g, 5 min at 4 °C, to get the mitochondria pellet.
5. Transfer of Isolated MSC Mitochondria to GSCs (MitoCeption)
Add 200 µl of the pre-cooled (0 °C) GSC proliferation medium to the mitochondria pellet isolated from MSCs (4 x 105).
Dilute the mitochondria preparation (in GSC proliferation medium) to consistently add 20 µl of mitochondrial suspension to the GSCs.
Add the volume of isolated mitochondria into the wells of the 96-well plate containing the GSCs (from step 3.15), at the desired concentration (0.1 to 10 µg). Add the mitochondria slowly, close to the bottom of the well, covering the entire surface at least once.
For controlling MSC mitochondria vital dye leakage, add the same amounts of the mitochondria preparation (0.1 to 10 µg) to wells of a 96-well plate containing GSC proliferation medium only (no GSCs) (see part 6.2 for vital dye leakage detection).
Centrifuge the 96-well plate containing the mitochondria recipient GSCs with the MSC mitochondria (step 5.3) and the control plate (step 5.4) at 1,500 x g for 15 min at 4 °C.
Place the culture plates in the 37 °C cell incubator immediately after centrifugation. Note: The mitochondria transfer protocol relies on the centrifugation of the mitochondria suspension on the cultured cells at the adequate centrifugation force, with a number of centrifugations that can be adjusted depending on the system of mitochondria donor/recipient cells.
Day 3
6. Analysis of the Mitochondria Transfer by FACS and Confocal Imaging
- Preparation of GSC samples for FACS analysis Note: If MSC mitochondria were labeled with a vital dye beforehand (Section 1), the efficiency can be monitored by FACS, 24 hr after the mitochondria transfer.
- Centrifuge the 96-well plate with the MitoCepted GSCs at 270 x g for 5 min at 20 °C.
- Discard the supernatant.
- Add 100 µl trypsin (no EDTA) in each well. Incubate at 37 °C for 3 min. Pipet up and down to dissociate the neurospheres formed in the 24 hr time period.
- Add 100 µl of GSC basal medium.
- Centrifuge the plate at 270 x g for 5 min at 20 °C and discard the supernatant.
- Resuspend GSCs in 300 µl GSC basal medium and transfer to FACS adapted tubes.
- Perform the FACS analysis.
- Control for mitochondria vital dye leakage from the isolated mitochondria (FACS) Note: The purpose of this step is to determine the background FACS signal that would not reflect a genuine MSC mitochondria transfer but, rather, the mere leakage of vital dye from the labeled MSC mitochondria used for MitoCeption (step 5.3).
- Seed GSC cells as described in steps 3.1 to 3.15.
- Incubate the cells at 37 °C for 1 hr.
- Centrifuge the 96-well plate containing mitochondria only (from step 5.4 to 5.6) at 1,500 x g for 5 min at 20 °C.
- Aspirate the culture medium from the GSC 96-well plate (step 6.2.2) and replace it by the medium incubated with mitochondria only (supernatant from step 6.2.3).
- Incubate the 96-well plate containing the GSCs at 37 °C for 2 hr.
- Proceed as in steps 6.1.1 to 6.1.7 for FACS analysis.
- Preparation of GSC samples for confocal imaging Note: If the transfer of MSC mitochondria to GSCs is to be analyzed by fluorescence imaging, both MSCs and GSCs need to be labeled beforehand, respectively with mitochondria (step 1) and cell (step 2) vital dyes.
- Prepare GSCs as in steps 6.1.1 to 6.1.5.
- Resuspend GSCs in 2 ml GSC proliferation medium containing 2% FBS and seed in 35 mm culture dishes (glass bottom).
- Perform confocal imaging on the GSCs with the transferred fluorescent MSC mitochondria 24 hr later.
Representative Results
The procedural steps outlining the isolation of mitochondria from mesenchymal stem cells (MSC) and their transfer to the targeted glioblastoma stem cells (GSC) by MitoCeption are shown in Figure 1. GSCs are cancer stem cells grown as neurospheres to preserve their stem cell properties. For the protocol, GSCs are seeded as single cells a couple of hours before the transfer of mitochondria (step 3) to allow higher mitochondria transfer efficiency (see FACS data Figure 3B). Following Section 5 (day 3), these cells are observed again as forming neurospheres (Figure 1).
Imaging of the Gb4 GSCs, 72 hr after MitoCeption with fluorescently-labeled MSC mitochondria, shows that the MSC mitochondria are localized inside the Gb4 GSCs, as demonstrated by the orthogonal views obtained from confocal images (Figure 2B). The transferred MSC mitochondria appear localized throughout the GSCs, as this is also the case for the endogenous GSC mitochondria (Figure 2A). Confocal imaging of the MitoCepted GSCs was also performed with MSC mitochondria prelabeled with a deep red vital dye so that the same GSC samples were analyzed both by FACS (see Figure 3A) and imaging (Figure 2C). The transfer of MSC mitochondria could be observed for most GSCs (panel a) and 3D reconstruction from GSC confocal images confirmed the intracellular localization of the transferred MSC mitochondria (panels b, c). Prelabeling GSCs (on day 1) with both blue cellular and red mitochondrial vital dyes allowed visualizing the localization of the transferred MSC mitochondria (colored in green) relative to the endogenous GSC mitochondria (in red), following the MSC mitochondria transfer (Figure 2D).
While confocal imaging allows checking the intracellular localization of the transferred MSC mitochondria following MitoCeption, fluorescence-activated cell sorting (FACS) is the tool of choice to quantify the mitochondria transfer, provided MSC mitochondria were prelabeled with a vital dye. As shown in Figure 3A, MitoCeption with increasing amounts of fluorescent MSC mitochondria led to increased FACS signals. Noteworthy, this signal (in red) was several orders of magnitude higher than the signal obtained in the control conditions (in blue, see step 6.2). This further provided evidence that the FACS signal is representative of MSC mitochondria transferred inside GSCs and not merely the result of the fluorescent vital dye leakage from the labeled MSC mitochondria. The mitochondria transfer protocol presented here, performed on single cell GSCs, showed a dose-dependent response (Figure 3B). The efficiency of the MSC mitochondria transfer was also evaluated on neurospheres. Although this also led to MSC mitochondria transfer to the GSCs, the efficacy of the transfer was lower, as shown in a representative experiment (Figure 3B).
The transfer of MSC mitochondria and their maintenance inside the GSCs can be followed by quantifying MSC mitochondrial DNA (mtDNA) in the GSCs. This requires MSCs and GSCs to be obtained from different donors, with distinct haplotypes. Figure 3C shows mtDNA sequences found within the D-loop of two donors with single nucleotide polymorphisms (SNPs) indicated at the 3' end position. For the design of PCR primers, to specifically amplify mtDNA from either donor 1 or donor 2, an additional mutation was introduced at position n-2 relative to the 3' end of the PCR primers. The mtDNA from donor 1 could be amplified with the set of primers: 5'-TAACAGTACATAGCACATACAA-3' and 5'-GAGGATGGTGGTCAAGGGA-3' (universal sequencing primer 38), while the set of primers: 5'-TTAACTCCACCATTAGCACC-3' (universal sequencing primer from 38) and 5'-AGTATTTATGGTACCGTCCG-3' was used to amplify the mtDNA from donor 2.
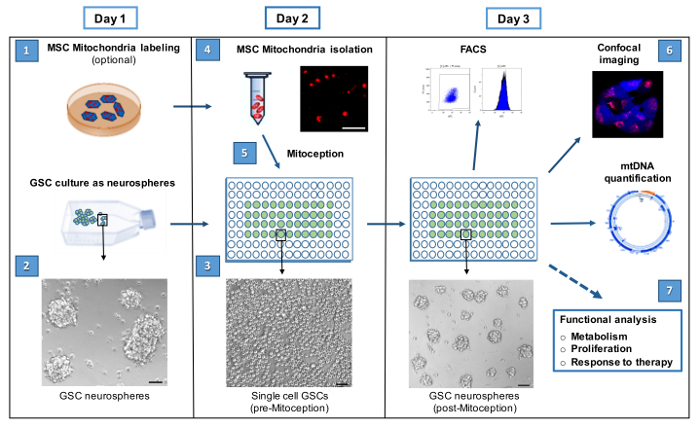 Figure 1: Workflow for the transfer of isolated MSC mitochondria to GSCs by MitoCeption. The 7 steps for the MitoCeption of MSC mitochondria to GSCs and the subsequent GSC analyses are shown. The step numbers are indicated as in the protocol section. If MSC and GSC labeling is not performed, as for functional analyses, steps 1 and 2 can be omitted and the protocol can be followed from day 2 on. Phase contrast images represent Gb4 GSCs as neurospheres (day 1), as unicellular culture ready for the MSC mitochondria transfer (day 2) and 24 hr after the mitochondria transfer (day 3). Scale bar = 100 µm. In step 4, representative photomicrograph of isolated MSC mitochondria, prelabeled with MitoTracker Red CMXRos before isolation from MSCs. Scale bar = 5 µm. Please click here to view a larger version of this figure.
Figure 1: Workflow for the transfer of isolated MSC mitochondria to GSCs by MitoCeption. The 7 steps for the MitoCeption of MSC mitochondria to GSCs and the subsequent GSC analyses are shown. The step numbers are indicated as in the protocol section. If MSC and GSC labeling is not performed, as for functional analyses, steps 1 and 2 can be omitted and the protocol can be followed from day 2 on. Phase contrast images represent Gb4 GSCs as neurospheres (day 1), as unicellular culture ready for the MSC mitochondria transfer (day 2) and 24 hr after the mitochondria transfer (day 3). Scale bar = 100 µm. In step 4, representative photomicrograph of isolated MSC mitochondria, prelabeled with MitoTracker Red CMXRos before isolation from MSCs. Scale bar = 5 µm. Please click here to view a larger version of this figure.
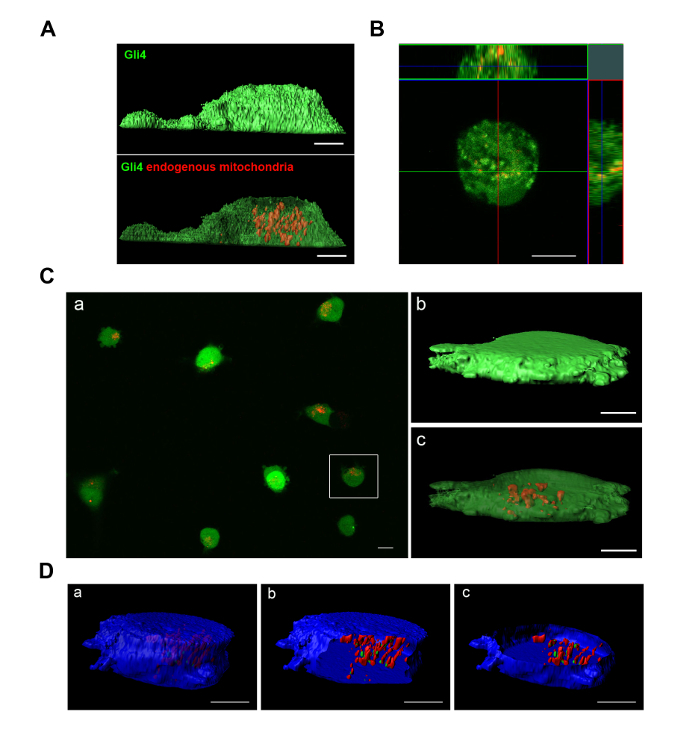 Figure 2: Imaging of Gb4 endogenous mitochondria and of MSC mitochondria after transfer to the Gb4 GSCs by MitoCeption. (A, B, C) Gb4 GSCs were labeled with a green vital dye. (B, C, D) Gb4 GSCs were MitoCepted with 2.5 µg MSC mitochondria preparation (for 105 GSCs). (A) GSCs were labeled with MitoTracker Red CMXRos and cultured for 48 hr (GSC proliferation medium w/o EGF/bFGF). (B) GSC confocal section and orthogonal views, 72 hr after MitoCeption with MitoTracker Red CMXRos-labeled MSC mitochondria (culture medium: GSC proliferation medium/FBS 2%). (C, D) MSCs were prelabeled with a deep red mitochondria vital dye (same conditions as for FACS analysis) before the transfer of MSC mitochondria to GSCs. After the MSC mitochondria transfer, GSCs were seeded in GSC proliferation medium containing FBS 10%. (C) Isosurface views (b, c) with xy plane section (c) of a GSC 3D reconstruction from confocal images (48 hr after MitoCeption). (D) Gb4 GSCs were labeled with a blue cell vital dye and a red mitochondria vital dye before the transfer of MSC mitochondria prelabeled with a deep red mitochondria vital dye (imaged in green). Isosurface views with xz (b) and xy (c) plane sections of a GSC 3D reconstruction from confocal images (72 hr after the MSC mitochondria transfer). All cells were seeded on culture dishes with glass bottom and images were taken on live cells. Scale bars = 5 µm, except for image C panel (a) where Scale bar = 10 µm. Please click here to view a larger version of this figure.
Figure 2: Imaging of Gb4 endogenous mitochondria and of MSC mitochondria after transfer to the Gb4 GSCs by MitoCeption. (A, B, C) Gb4 GSCs were labeled with a green vital dye. (B, C, D) Gb4 GSCs were MitoCepted with 2.5 µg MSC mitochondria preparation (for 105 GSCs). (A) GSCs were labeled with MitoTracker Red CMXRos and cultured for 48 hr (GSC proliferation medium w/o EGF/bFGF). (B) GSC confocal section and orthogonal views, 72 hr after MitoCeption with MitoTracker Red CMXRos-labeled MSC mitochondria (culture medium: GSC proliferation medium/FBS 2%). (C, D) MSCs were prelabeled with a deep red mitochondria vital dye (same conditions as for FACS analysis) before the transfer of MSC mitochondria to GSCs. After the MSC mitochondria transfer, GSCs were seeded in GSC proliferation medium containing FBS 10%. (C) Isosurface views (b, c) with xy plane section (c) of a GSC 3D reconstruction from confocal images (48 hr after MitoCeption). (D) Gb4 GSCs were labeled with a blue cell vital dye and a red mitochondria vital dye before the transfer of MSC mitochondria prelabeled with a deep red mitochondria vital dye (imaged in green). Isosurface views with xz (b) and xy (c) plane sections of a GSC 3D reconstruction from confocal images (72 hr after the MSC mitochondria transfer). All cells were seeded on culture dishes with glass bottom and images were taken on live cells. Scale bars = 5 µm, except for image C panel (a) where Scale bar = 10 µm. Please click here to view a larger version of this figure.
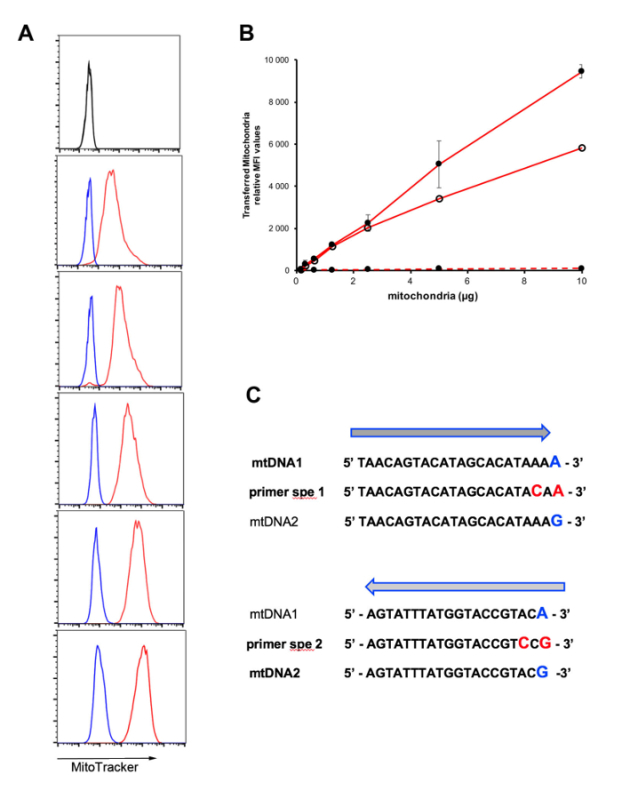 Figure 3: Quantification of the mitochondria transfer by fluorescence-activated cell sorting (FACS) and detection of MSC mitochondrial DNA (mtDNA) in the GSCs. (A, B) GSCs were MitoCepted with MSC mitochondria prelabeled with a deep red mitochondria vital dye and analyzed by FACS. (A) Representative FACS results obtained with MitoCepted GSCs (in red, from top to bottom: 0.6; 1.2; 2.5; 5; 10 µg MSC mitochondria preparation (for 105 GSCs)). In blue, for each panel, control FACS profile of GSCs incubated with the culture medium conditioned with the same amount of MSC mitochondria as used for MitoCeption (see step 5.4). Control conditions with unlabeled GSCs in black (top). (B) Relative mean fluorescence intensity (MFI) obtained after the mitochondria transfer on single cell GSCs (- •) (average of 3 experiments, with SEM), on one-day GSC neurospheres (- o) and with control mitochondria supernatants on GSC single cells (--). (C) DNA sequences within the mtDNA D-loop and SNP differences between donors 1 and 2. SNPs are indicated in blue at the 3' end of the sequences. Design of primers for specific PCR amplification of mtDNA from either donor (primers spe 1 and 2). Please click here to view a larger version of this figure.
Figure 3: Quantification of the mitochondria transfer by fluorescence-activated cell sorting (FACS) and detection of MSC mitochondrial DNA (mtDNA) in the GSCs. (A, B) GSCs were MitoCepted with MSC mitochondria prelabeled with a deep red mitochondria vital dye and analyzed by FACS. (A) Representative FACS results obtained with MitoCepted GSCs (in red, from top to bottom: 0.6; 1.2; 2.5; 5; 10 µg MSC mitochondria preparation (for 105 GSCs)). In blue, for each panel, control FACS profile of GSCs incubated with the culture medium conditioned with the same amount of MSC mitochondria as used for MitoCeption (see step 5.4). Control conditions with unlabeled GSCs in black (top). (B) Relative mean fluorescence intensity (MFI) obtained after the mitochondria transfer on single cell GSCs (- •) (average of 3 experiments, with SEM), on one-day GSC neurospheres (- o) and with control mitochondria supernatants on GSC single cells (--). (C) DNA sequences within the mtDNA D-loop and SNP differences between donors 1 and 2. SNPs are indicated in blue at the 3' end of the sequences. Design of primers for specific PCR amplification of mtDNA from either donor (primers spe 1 and 2). Please click here to view a larger version of this figure.
Discussion
An increasing number of studies show that cells can exchange mitochondria and that these mitochondria have profound effects on the target cell metabolism and functions. Therefore, it is essential to master the tools to quantitatively transfer mitochondria from the donor cells to these target cells to enable an accurate study of their biological effects.
The protocol described here was originally worked out to transfer mitochondria isolated from human mesenchymal stem cells to the adherent cancer cell line MDA-MB-231 33. As shown here, it can be adapted for cancer stem cells that grow as neurospheres in vitro. Although the mitochondria transfer does occur when MSC mitochondria are added to the GSC neurospheres, it was found to be more efficient on single cell GSCs, as described in the present protocol. This protocol could also be extrapolated to other non-adherent cells, like T cells, with the need to adapt the seeded cell concentration for the MitoCeption step.
The mitochondria preparation should be performed on ice to maintain their integrity. To obtain mitochondria preparations with reduced contamination from other cytosol compounds, centrifugation for the recovery of mitochondria is performed at 3,000 x g for 15 min. To monitor the amount of MSC mitochondria used for the transfer to the GSCs, the mitochondria protein content is determined. For this purpose, mitochondria protein extracts are prepared using RIPA buffer supplemented with protease inhibitors. The protein concentration was determined using the bicinchoninic acid assay, according to the manufacturer's instructions. Bovine serum albumin was used as a standard. On average 10 µg of proteins were obtained from 100,000 MSCs.
Labeling MSC mitochondria with a vital dye allows quantifying the efficiency of the mitochondria transfer, by fluorescence-activated cell sorting (FACS), and determining the localization of the transferred mitochondria by confocal imaging. The GSCs can be labelled with the same mitochondria vital dye as the MSCs. These cells will be used as a positive control for the FACS analysis. Alternatively, GSCs can be labeled with a mitochondria vital dye different from that of the MSCs, allowing the localization of the transferred MSC mitochondria relative to the endogenous GSC mitochondria. The deep red mitochondria vital dye is well suited for FACS analysis while the green and red mitochondria dyes are preferred for confocal imaging. Of great importance, MSC exposure to EDTA should be avoided at all steps of the protocol. Therefore, cell trypsinization is recommended with trypsin and no EDTA; the cocktail of protease inhibitors used for the mitochondria preparation should, as well, be devoid of EDTA. In the presence of EDTA, a leakage of the mitochondria vital dye from MSC mitochondria was observed. If the mitochondria transfer to GSCs is to be analyzed by microscopy, the recipient glioblastoma cells should also be labeled with a vital dye. The green and blue cell dyes (see Materials and Reagents Table) were preferred as they give a more homogeneous cell labeling, enabling 3D reconstruction from confocal images.
Once the protocol has been validated by the user with his specific cells, mitochondria labeling will be left out for functional studies, to preclude possible biological biases. Dose-response analyses over a wide range of MSC mitochondria concentrations, with serial dilutions of the mitochondria preparation, is advisable. In fact, it should be noted that biological effects for the transferred mitochondria could be achieved for low mitochondria concentrations, in the lower range for detection by FACS or imaging. These functional studies, including investigations on GSC metabolism, proliferation and response to therapy, are carried out the day following the mitochondria transfer.
If MSCs and GSCs are isolated from different donors (with different mtDNA haplotypes), the transferred MSC mitochondria can be monitored by measuring the concentrations of MSC mtDNA in the targeted GSCs. The discrimination between the MSC and endogenous GSC mtDNAs is based on the presence of SNPs, specific for each cell type, in the mtDNA hypervariable D-loop region. Therefore, among the various applications of the MitoCeption technique, the possibility of transferring mitochondria harboring mitochondrial DNA with different haplotypes not only allows detection of the transferred mitochondria but also provides the tools to study the biology of mtDNA maintenance, including the analysis of haplotype exclusion.
Disclosures
INSERM (Institut National de la Santé et de la Recherche Médicale), with which C.J. and M-L.V. are affiliated, has filed a patent application on the MitoCeption technique (EP14306154.7).
Acknowledgments
We thank Andrea Parmeggiani (L2C and DIMNP, Montpellier), Benoit Charlot (IES, Montpellier) as well as members of the laboratory for helpful discussions, Christophe Duperray for help with the FACS analysis, the Montpellier RIO imaging facility (MRI) for providing the adequate environment for FACS and confocal microscopy. B.N.M was supported by a graduate fellowship from the LabEx Numev (convention ANR-10-LABX-20). A.B. was supported by an undergraduate fellowship from the University of Warsaw and European Union (n° POKL.04.01.02-00-221/12). M.L.V is a staff scientist from the National Center for scientific research (CNRS).
References
- Vafai SB, Mootha VK. Mitochondrial disorders as windows into an ancient organelle. Nature. 2012;491(7424):374–383. doi: 10.1038/nature11707. [DOI] [PubMed] [Google Scholar]
- Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148(6):1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandel NS. Mitochondria as signaling organelles. BMC Biol. 2014;12(1):34. doi: 10.1186/1741-7007-12-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze A, Harris AL. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature. 2012;491(7424):364–373. doi: 10.1038/nature11706. [DOI] [PubMed] [Google Scholar]
- Peiris-Pages M, Martinez-Outschoorn UE, Pestell RG, Sotgia F, Lisanti MP. Cancer stem cell metabolism. Breast Cancer Res. 2016;18(1):55. doi: 10.1186/s13058-016-0712-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBleu VS, et al. PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol. 2014;16(10):992–1003. doi: 10.1038/ncb3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Feng M, Guan W. Mitochondrial DNA sensing by STING signaling participates in inflammation, cancer and beyond. Int J Cancer. 2016;139(4):736–741. doi: 10.1002/ijc.30074. [DOI] [PubMed] [Google Scholar]
- Islam MN, et al. Mitochondrial transfer from bone-marrow-derived stromal cells to pulmonary alveoli protects against acute lung injury. Nat Med. 2012;18(5):759–765. doi: 10.1038/nm.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquier J, et al. Preferential transfer of mitochondria from endothelial to cancer cells through tunneling nanotubes modulates chemoresistance. J Transl Med. 2013;11:94. doi: 10.1186/1479-5876-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikov EY, Khryapenkova TG, Galkina SI, Sukhikh GT, Zorov DB. Cytoplasm and organelle transfer between mesenchymal multipotent stromal cells and renal tubular cells in co-culture. Exp Cell Res. 2010;316(15):2447–2455. doi: 10.1016/j.yexcr.2010.06.009. [DOI] [PubMed] [Google Scholar]
- Spees JL, Olson SD, Whitney MJ, Prockop DJ. Mitochondrial transfer between cells can rescue aerobic respiration. Proc Natl Acad Sci U S A. 2006;103(5):1283–1288. doi: 10.1073/pnas.0510511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, et al. Mesenchymal stem cells rescue injured endothelial cells in an in vitro ischemia-reperfusion model via tunneling nanotube like structure-mediated mitochondrial transfer. Microvasc Res. 2014;92:10–18. doi: 10.1016/j.mvr.2014.01.008. [DOI] [PubMed] [Google Scholar]
- Bukoreshtliev NV, et al. Selective block of tunneling nanotube (TNT) formation inhibits intercellular organelle transfer between PC12 cells. FEBS Lett. 2009;583(9):1481–1488. doi: 10.1016/j.febslet.2009.03.065. [DOI] [PubMed] [Google Scholar]
- Gurke S, et al. Tunneling nanotube (TNT)-like structures facilitate a constitutive, actomyosin-dependent exchange of endocytic organelles between normal rat kidney cells. Exp Cell Res. 2008;314(20):3669–3683. doi: 10.1016/j.yexcr.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Vallabhaneni KC, Haller H, Dumler I. Vascular smooth muscle cells initiate proliferation of mesenchymal stem cells by mitochondrial transfer via tunneling nanotubes. Stem Cells Dev. 2012;21(17):3104–3113. doi: 10.1089/scd.2011.0691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Gerdes HH. Transfer of mitochondria via tunneling nanotubes rescues apoptotic PC12 cells. Cell Death Differ. 2015;22(7):1181–1191. doi: 10.1038/cdd.2014.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikov EY, et al. Cell-to-cell cross-talk between mesenchymal stem cells and cardiomyocytes in co-culture. J Cell Mol Med. 2008;12(5A):1622–1631. doi: 10.1111/j.1582-4934.2007.00205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acquistapace A, et al. Human mesenchymal stem cells reprogram adult cardiomyocytes toward a progenitor-like state through partial cell fusion and mitochondria transfer. Stem Cells. 2011;29(5):812–824. doi: 10.1002/stem.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase K, et al. M-Sec promotes membrane nanotube formation by interacting with Ral and the exocyst complex. Nat Cell Biol. 2009;11(12):1427–1432. doi: 10.1038/ncb1990. [DOI] [PubMed] [Google Scholar]
- Phinney DG, et al. Mesenchymal stem cells use extracellular vesicles to outsource mitophagy and shuttle microRNAs. Nat Commun. 2015;6:8472. doi: 10.1038/ncomms9472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad T, et al. Miro1 regulates intercellular mitochondrial transport & enhances mesenchymal stem cell rescue efficacy. Embo J. 2014;33(9):994–1010. doi: 10.1002/embj.201386030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, et al. iPSC-MSCs with High Intrinsic MIRO1 and Sensitivity to TNF-a Yield Efficacious Mitochondrial Transfer to Rescue Anthracycline-Induced Cardiomyopathy. Stem Cell Reports. 2016;7(4):749–763. doi: 10.1016/j.stemcr.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, et al. Influence of intergeneric/interspecies mitochondrial injection; parthenogenetic development of bovine oocytes after injection of mitochondria derived from somatic cells. J Reprod Dev. 2012;58(3):323–329. doi: 10.1262/jrd.2011-013. [DOI] [PubMed] [Google Scholar]
- Takeda K, et al. Microinjection of cytoplasm or mitochondria derived from somatic cells affects parthenogenetic development of murine oocytes. Biol Reprod. 2005;72(6):1397–1404. doi: 10.1095/biolreprod.104.036129. [DOI] [PubMed] [Google Scholar]
- Van Blerkom J, Sinclair J, Davis P. Mitochondrial transfer between oocytes: potential applications of mitochondrial donation and the issue of heteroplasmy. Hum Reprod. 1998;13(10):2857–2868. doi: 10.1093/humrep/13.10.2857. [DOI] [PubMed] [Google Scholar]
- Ishikawa K, et al. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320(5876):661–664. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- Kaipparettu BA, Ma Y, Wong LJ. Functional effects of cancer mitochondria on energy metabolism and tumorigenesis: utility of transmitochondrial cybrids. Ann N Y Acad Sci. 2010;1201:137–146. doi: 10.1111/j.1749-6632.2010.05621.x. [DOI] [PubMed] [Google Scholar]
- Wu TH, et al. Mitochondrial Transfer by Photothermal Nanoblade Restores Metabolite Profile in Mammalian Cells. Cell Metab. 2016;23(5):921–929. doi: 10.1016/j.cmet.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuzawa A, et al. Transplantation of autologously derived mitochondria protects the heart from ischemia-reperfusion injury. Am J Physiol Heart Circ Physiol. 2013;304(7):H966–H982. doi: 10.1152/ajpheart.00883.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitani T, et al. Direct human mitochondrial transfer: a novel concept based on the endosymbiotic theory. Transplant Proc. 2014;46(4):1233–1236. doi: 10.1016/j.transproceed.2013.11.133. [DOI] [PubMed] [Google Scholar]
- Caicedo A, et al. MitoCeption as a new tool to assess the effects of mesenchymal stem/stromal cell mitochondria on cancer cell metabolism and function. Sci Rep. 2015;5:9073. doi: 10.1038/srep09073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesner EE, Saada-Reich A, Lorberboum-Galski H. Characteristics of Mitochondrial Transformation into Human Cells. Sci Rep. 2016;6:26057. doi: 10.1038/srep26057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29(12):1203–1217. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osswald M, et al. Brain tumour cells interconnect to a functional and resistant network. Nature. 2015;528(7580):93–98. doi: 10.1038/nature16071. [DOI] [PubMed] [Google Scholar]
- Shinojima N, et al. TGF-beta mediates homing of bone marrow-derived human mesenchymal stem cells to glioma stem cells. Cancer Res. 2013;73(7):2333–2344. doi: 10.1158/0008-5472.CAN-12-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velpula KK, Dasari VR, Rao JS. The homing of human cord blood stem cells to sites of inflammation: unfolding mysteries of a novel therapeutic paradigm for glioblastoma multiforme. Cell Cycle. 2012;11(12):2303–2313. doi: 10.4161/cc.20766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichet PO, et al. Cell death and neuronal differentiation of glioblastoma stem-like cells induced by neurogenic transcription factors. Glia. 2013;61(2):225–239. doi: 10.1002/glia.22429. [DOI] [PubMed] [Google Scholar]
- Lyons EA, Scheible MK, Sturk-Andreaggi K, Irwin JA, Just RS. A high-throughput Sanger strategy for human mitochondrial genome sequencing. BMC Genomics. 2013;14:881. doi: 10.1186/1471-2164-14-881. [DOI] [PMC free article] [PubMed] [Google Scholar]


