Abstract
The NLRP3 inflammasome is a central regulator of inflammation in many common diseases, including atherosclerosis and type 2 diabetes, driving the production of pro-inflammatory mediators such as IL-1β and IL-18. Due to its function as an inflammatory gatekeeper, expression and activation of NLRP3 need to be tightly regulated. In this study, we highlight novel post-transcriptional mechanisms that can modulate NLRP3 expression. We have identified the RNA-binding protein Tristetraprolin (TTP) as a negative regulator of NLRP3 in human macrophages. TTP targets AU-rich elements in the NLRP3 3′-untranslated region (UTR) and represses NLRP3 expression. Knocking down TTP in primary macrophages leads to an increased induction of NLRP3 by LPS, which is also accompanied by increased Caspase-1 and IL-1β cleavage upon NLRP3, but not AIM2 or NLRC4 inflammasome activation. Furthermore, we found that human NLRP3 can be alternatively polyadenylated, producing a short 3′-UTR isoform that excludes regulatory elements, including the TTP- and miRNA-223-binding sites. Because TTP also represses IL-1β expression, it is a dual inhibitor of the IL-1β system, regulating expression of the cytokine and the upstream controller NLRP3.
Keywords: inflammasome, inflammation, interleukin 1 (IL-1), macrophage, NLRP3, post-transcriptional regulation, TTP, Tristetraprolin
Introduction
The NLRP3 inflammasome has emerged as a potent driver of many common inflammatory diseases, due to its ability to induce production of the pro-inflammatory cytokines IL-1β and IL-18, as well as pyroptosis, a pro-inflammatory type of cell death (reviewed in Ref. 1). After a “licensing” signal downstream of Toll-like receptor activation, which primes the system transcriptionally (2) and post-translationally (3), NLRP3 can be activated by a vast number of structurally diverse stimuli, from bacterial toxins to extracellular ATP and protein or crystalline aggregates (1). Even though the molecular mechanism of NLRP3 activation is still not fully understood, potassium efflux appears to be a common requirement and ultimately leads to the interaction of NLRP3 with the adaptor protein ASC and Pro-Caspase-1, which aggregate to form the inflammasome (4). Once initiated, the inflammasome provides a platform for autoactivation of Caspase-1, which in turn cleaves downstream effectors. Most notably, Caspase-1 processes the inactive precursors of IL-1β and IL-18 into the active cytokines (5) and cleaves Gasdermin D to initiate pyroptotic cell death (6, 7). In the broadest sense, NLRP3 activators are signs of “disturbed homeostasis,” for example, as the result of infection or tissue damage. Of particular interest is that the deposition of NLRP3 stimuli is a feature of many common inflammatory diseases, including type 2 diabetes, atherosclerosis, gout, and Alzheimer's disease (8–11). Many studies over the last decade have demonstrated that NLRP3 activation is functionally relevant in disease models, because pharmacological or genetic inhibition of NLRP3 activation is protective (12). Due to its central role as an inflammatory gatekeeper, NLRP3 expression and activation are tightly regulated. For example, NFκB can drive NLRP3 transcription downstream of Toll-like receptor or cytokine activation (2, 13). Furthermore, the NLRP3 protein can be post-translationally modified in a number of ways, including ubiquitination, phosphorylation, ADP-ribosylation, and nitrosylation (14). Many modifications inhibit inflammasome formation and need to be removed in the “priming” step to allow for efficient activation (3). Finally, we and others have demonstrated that miRNA-223 can repress NLRP3 expression in myeloid cells (15, 16).
Even though post-transcriptional mechanisms are central to the regulation of innate immune signaling (17), apart from miR-223,3 very little is known about post-transcriptional regulation of NLRP3. In this study, we provide the first detailed characterization of the 3′-untranslated region (UTR) of the NLRP3 gene in mouse and human. We identify two 3′-UTR isoforms in human (a long and a short) and one in mouse, corresponding to the long human isoform. The long human isoform has several AU-rich elements (AREs). We identify the RNA-binding protein Tristetraprolin (TTP, encoded by ZFP36) as an inducible repressor of the long isoform of NLRP3 in human macrophages, inhibition of which likely contributes to inflammasome priming. Because TTP also targets expression of the downstream effector IL-1β, it appears to be particularly important for controlling the IL-1β system, regulating both the cytokine and its key controller, NLRP3.
Results
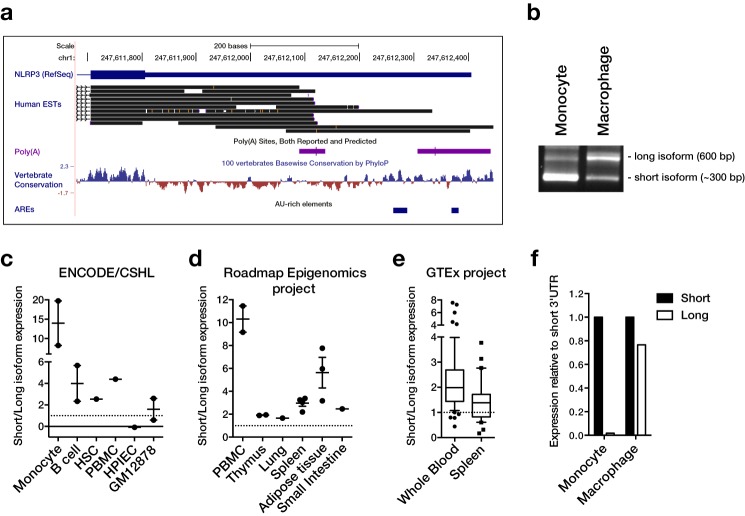
The NLRP3 3′-UTR is alternatively polyadenylated and contains a repressive AU-rich element
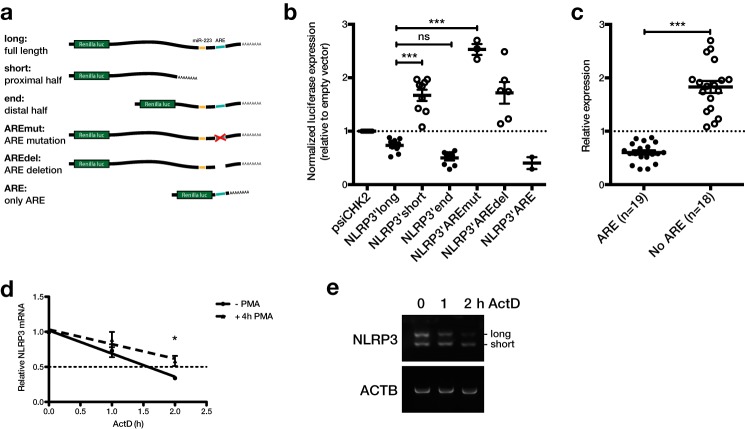
During detailed analysis of the human NLRP3 sequence, we noticed that expressed sequence tags (ESTs) indicated alternative 3′-ends of transcripts, even though only one 3′-UTR is annotated in RefSeq (Fig. 1a). The alternative isoform that is suggested by ESTs comprises almost exactly the first half of the full-length sequence. Polyadenylation sites are located at the 3′-end of both isoforms, suggesting that they could arise from alternative polyadenylation. To verify expression of the isoforms experimentally, we analyzed expression of NLRP3 in monocytes and macrophages by 3′-RACE. This successfully amplified two isoforms of the expected lengths, ∼600 bp for the long and 300 bp for the short isoform (Fig. 1b). We also estimated the relative expression levels of the 3′-UTR isoforms from different RNA sequencing datasets. The average signal intensity of the short and long 3′-UTR isoform in samples generated by the ENCODE consortium, Roadmap Epigenomics project, or GTEx project revealed a big variability of their relative expression among cell types and tissues (Fig. 1, c–e, and supplemental Fig. S1). In most samples, the short isoform was more abundant than the long isoform. However, the ratio of short to long ranged from below 1 to 20. In particular monocytes appear to express predominantly the short 3′-UTR isoform. Higher expression of the short isoform in monocytes could also be confirmed by 3′-RACE and qPCR (Fig. 1, b and f). In contrast, macrophages expressed the isoforms at similar levels (Fig. 1, b and f). In terms of GC content and conservation, the two halves of the human NLRP3 3′-UTR are very different. The more conserved distal half of the 3′-UTR, which is excluded from the short isoform, has a much lower GC content (25 versus 55%) and also contains AREs (Fig. 1a and supplemental Fig. S2, a and d). In particular, a main ARE consisting of three overlapping AUUUA pentamers is present in this region, in close proximity to the target site for miR-223 (15) (supplemental Fig. S2d). Interestingly, these features are only partially conserved in the mouse gene. For a start, even though the proximal polyadenylation site is conserved in many species, it is mutated in the murine sequence (AAUAAA → AAGAAA) (supplemental Fig. S2b). Although the mutated sequence has also been identified as an active polyadenylation site (18), it is a lot weaker than the canonical sequence (19). In agreement with a weak proximal polyadenylation site, we could only detect one isoform of the murine Nlrp3 3′-UTR by 3′-RACE (supplemental Fig. S2c). Similarly, the main human ARE is conserved in many species, but not in the mouse (supplemental Fig. S2d). However, the mouse 3′-UTR also contains AREs and the human and mouse full-length 3′-UTR indeed show a similar enrichment of AREs as quantified by the AREScore algorithm (20) (supplemental Fig. S2a). To test if the isoforms are differentially regulated, we generated a range of luciferase reporters containing the two isoforms or different truncations and mutations (Fig. 2a). When the vectors were expressed in the human monocytic cell line THP-1, we observed that their basal expression broadly fell into two groups (Fig. 2b). On the one hand, the reporters containing the long 3′-UTR isoform, only the distal 3′-UTR half or the ARE only, all of which contain the ARE, showed relatively low expression (Fig. 2b, filled circles). On the other hand, reporters containing the short 3′-UTR isoform or the long 3′-UTR with a mutated or deleted ARE, all of which lack a functional ARE, were expressed at a higher level (Fig. 2b, open circles). This was particularly obvious when the reporters were grouped by presence of the ARE, where ARE-containing reporters showed a significantly lower expression (Fig. 2c). Because AREs are known to destabilize mRNAs (21), we also measured mRNA stability of NLRP3 in THP-1 cells. We found that NLRP3 was basally unstable with a half-life of ∼1.5 h and that it could be stabilized after stimulating the cells with PMA (Fig. 2d). Interestingly, when the samples were amplified by 3′-RACE, the long 3′-UTR isoform appeared to be especially susceptible to degradation. This would be in agreement with the location of the repressive ARE specifically in the long 3′-UTR isoform (Fig. 2e).
Figure 1.
The human NLRP3 3′-UTR is alternatively polyadenylated and the isoforms are differentially expressed. a, the human NLRP3 3′-UTR visualized in the UCSC genome browser (hg19) with ESTs, polyadenylation sites from PolyA_DB, and mammalian conservation tracks. Location of AU-rich elements (AUUUA pentamers) are indicated. b, 3′-RACE of the NLRP3 3′-UTR in CD14+ monocytes or M-CSF-differentiated macrophages. PCR products were analyzed by agarose gel electrophoresis. c–e, expression of the short and long 3′-UTR were quantified in the UCSC genome browser for expression tracks sourced from the ENCODE consortium (CSHL) (c), NIH Roadmap Epigenomics project (d), or GTEx project (e). The ratio for individual samples is plotted. f, qPCR analysis of 3′-UTR isoform expression in primary CD14+ monocytes or M-CSF-differentiated macrophages using primer pairs that amplify specifically within the short or long 3′-UTR isoforms. Isoform expression was normalized to ACTB and expression of representative samples relative to the short isoform is shown.
Figure 2.
The long NLRP3 3′-UTR isoform contains a functional repressive AU-rich element. a, human NLRP3 3′-UTR isoforms, truncations, and mutations cloned into the psiCHECK-2 vector downstream of the Renilla luciferase gene. b, luciferase reporter constructs shown in panel a expressed in THP-1 cells. Open circles represent vectors lacking a functional ARE. Renilla luciferase expression 48 h after transfection was normalized to firefly luciferase activity. Values were calculated relative to empty vector control (psiCHK2). Mean relative luciferase expression ± S.E. of 2–9 independent experiments is shown. c, compiled data points from b. Groups were separated based on whether the 3′-UTR construct contains the functional ARE (filled circles) or not (open circles). d, THP-1 were stimulated with 12.5 ng/ml of PMA for 4 h as indicated. Actinomycin D (ActD) was added and RNA extracted 0, 1, and 2 h after addition. Gene expression was measured by qPCR using SYBR Green primers. NLRP3 was normalized to the geometric mean of ACTB, RPS13, and GAPDH expression. Mean ± S.D. of a representative of three independent experiments is shown. e, 3′-RACE for NLRP3 and ACTB was performed on a representative sample and analyzed by agarose gel electrophoresis. *, p ≤ 0.05; ***, p ≤ 0.001.
TTP targets the ARE in the NLRP3 3′-UTR
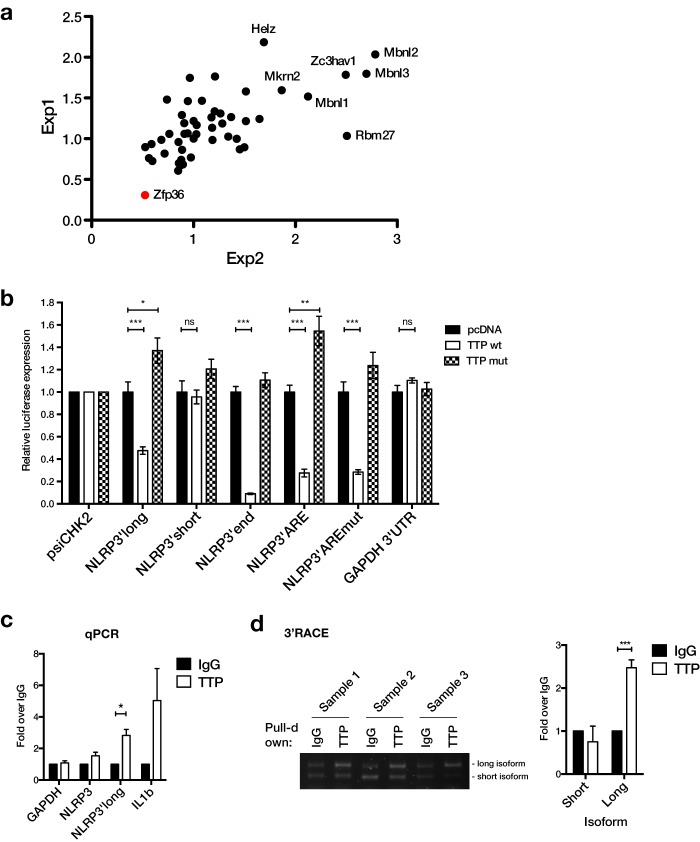
We next further analyzed the 3′-UTR of the NLRP3 gene by first screening for RNA-binding proteins that might impact its expression. For this, we used an expression library of 58 CCCH-type zinc finger proteins (ZFPs) that has been used by Lacey et al. (22) to identify novel regulators of TNFα expression. We co-expressed each ZFP expression vector with a luciferase reporter containing the NLRP3 3′-UTR or an empty vector control. Expression of most ZFPs did not alter reporter expression, indicated by a relative expression around 1 (Fig. 3a and supplemental Fig. S3). Expression of some ZFPs, including three members of the Musclebind-like (MBNL) family, led to an increase of relative reporter expression, indicating they might be positive regulators of NLRP3 expression. Interestingly, a single ZFP, Zfp36 (encoding Tristetraprolin/TTP), potently repressed expression of the 3′-UTR reporter (Fig. 3a and supplemental Fig. S3). Given that TTP is a well known ARE-binding protein that generally represses target mRNAs (23), we decided to test whether TTP could also target the NLRP3 ARE. To validate the result from the screen and to narrow down the region targeted by TTP, we expressed the luciferase constructs containing truncated regions of the NLRP3 3′-UTR with either wild-type (wt) or a mutant of TTP, in which both zinc finger motifs are disrupted (24). Similar to the result of the screen, wild-type TTP was able to repress expression of the reporter containing the full-length 3′-UTR (Fig. 3b). Interestingly, it did not affect expression of the short 3′-UTR isoform, but strongly repressed both the distal 3′-UTR half, which contains the ARE, as well as a reporter containing only the ARE (Fig. 3b). A reporter containing a mutated ARE was also still repressed, probably due to targeting of the smaller second ARE. As a negative control, expression of the GAPDH 3′-UTR was unaffected by TTP overexpression (Fig. 3b). Importantly, overexpression of mutant TTP did not repress expression of any reporter, indicating that the effects of wild-type TTP are mediated by RNA binding (Fig. 3b). For the long 3′-UTR and ARE-only reporters, mutant TTP even increased expression (Fig. 3b). This could be due to dominant-negative effects of the mutant protein, which can still bind proteins of the mRNA degradation machinery but not direct them to the target mRNA. We next wanted to confirm that TTP could also bind to the endogenous NLRP3 mRNA and used RNA immunoprecipitation to test this. In a TTP pulldown, specifically the long NLRP3 3′-UTR isoform was enriched, similar to the IL1B mRNA, which is a known TTP target (25) (Fig. 3c). The specificity of the interaction was also confirmed by 3′-RACE of the RNA-IP samples, which preferentially amplified the long 3′-UTR isoform in TTP pulldown samples compared with IgG controls (Fig. 3d). Taken together, these results suggest that TTP can bind to AREs specifically in the long isoform of the NLRP3 3′-UTR and repress its expression. This is also in agreement with results from two recent reports that measured transcriptome-wide TTP association in mouse macrophages (26, 27). In both studies, TTP was found to interact with ARE sequences in the distal part of the mouse 3′-UTR.
Figure 3.
TTP (encoded by ZFP36), represses NLRP3 3′-UTR luciferase reporters and binds to the long NLRP3 3′-UTR isoform. a, HEK293T cells were transfected with an empty vector or full-length NLRP3 3′-UTR firefly luciferase reporter constructs, an internal TK-Renilla control, and ZFP expression vectors. Firefly luciferase readings were normalized to Renilla activity and the ratio between the expression of the NLRP3 3′-UTR and empty vector in two independent experiments was plotted against each other. Because most data points scatter around 1, outliers represent potential regulators of NLRP3 3′-UTR expression. Zfp36 is highlighted. b, NLRP3 3′-UTR vectors described in the legend to Fig. 1c were co-expressed with an empty vector (pcDNA3.1), wild-type (wt), or zinc finger mutant TTP (mut) in HEK293T cells. Renilla luciferase expression 24 h after transfection was normalized to Firefly luciferase activity. Relative expression normalized to psiCHK-2 for each expression vector and pcDNA ± S.E. of 6 independent experiments is shown. c, TTP RNA-IP from lysate of THP-1 cells stimulated with PMA for 4 h. RNA was extracted from the pulldown and GAPDH, NLRP3, and NLRP3′long (long 3′-UTR isoform) and IL1B were quantified by qPCR using SYBR primers. Enrichment in TTP over IgG control pulldown ± S.E. of three independent experiments is shown. d, RNA-IP samples were analyzed by 3′-RACE for NLRP3 and ACTB. PCR products were visualized by agarose gel electrophoresis and band intensities were quantified. Relative intensity of each isoform relative to IgG is shown. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001.
TTP represses expression of endogenous human NLRP3
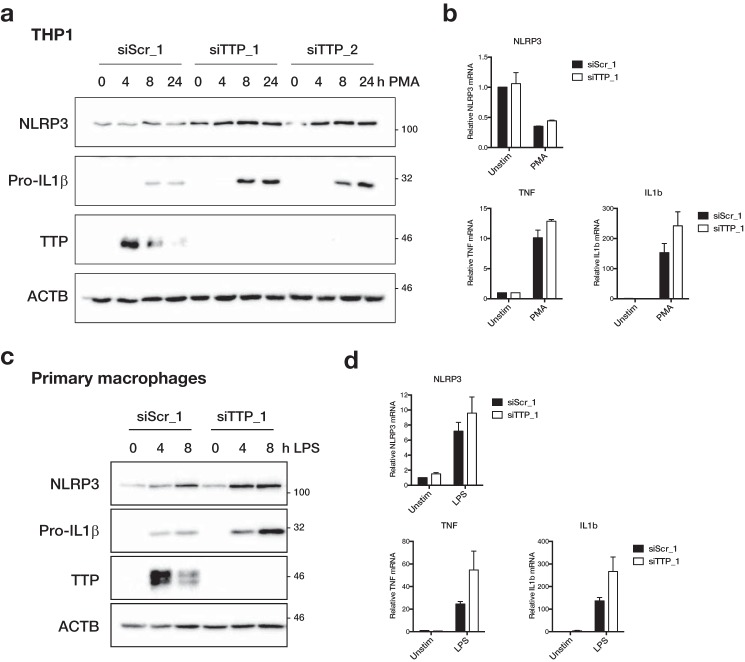
We next aimed to test if TTP can also regulate endogenous NLRP3 expression. To this end we knocked down TTP expression in THP-1 cells and primary macrophages using siRNA and assayed NLRP3 expression in response to PMA or LPS stimulation, respectively. In both cases, TTP expression was efficiently reduced by siRNA transfection (Fig. 4, a and c). Importantly, expression of the known TTP targets TNFα (23) and IL-1β (25) was increased in cells lacking TTP expression. Both in THP-1 and macrophages, PMA- or LPS-induced Pro-IL-1β protein expression was increased (Fig. 4, a and c) and there was also a trend to higher TNF and IL1B mRNA expression (Fig. 4, b and d). In THP-1 cells, knockdown of TTP with two different siRNAs led to an increase in PMA-induced NLRP3 protein expression (Fig. 4a). Similarly, LPS-induced NLRP3 expression was enhanced in primary macrophages that were transfected with a TTP-specific siRNA (Fig. 4c). There was also a trend to increased NLRP3 mRNA expression upon TTP knockdown in macrophages (Fig. 4d). In THP-1 cells, however, mRNA levels were unchanged by TTP knockdown (Fig. 4b), suggesting that TTP primarily affected translation of NLRP3 in these cells.
Figure 4.
TTP regulates endogenous NLRP3 expression. a, THP-1 were transfected with 40 nm control (siScr_1) or two different TTP siRNA (siTTP_1/2). Starting from 8 h after transfection, cells were stimulated with 12.5 ng/ml of PMA for 0, 4, 8, or 24 h. b, cells were transfected as in a, treated with 12.5 ng/ml of PMA for 4 h as indicated. c, primary human macrophages were transfected with 50 nm control (siScr_1) or TTP siRNA (siTTP_1). Starting from 24 h after transfection, cells were stimulated with 10 ng/ml LPS for 0, 4, or 8 h. d, cells were transfected as in c, treated with 10 ng/ml of LPS for 3 h as indicated. a and c, cells were lysed in SDS sample buffer and NLRP3, Pro-IL-1β, TTP, and β-actin (ACTB) were analyzed by Western blot. Representative blots of at least three independent experiments are shown. b and d, RNA was extracted and gene expression was measured by qPCR. NLRP3, TNF, and IL1B expression was normalized to the geometric mean of ACTB, GAPDH, and RPS13. Mean expression of two independent experiments relative to unstimulated siScr_1-transfected cells ± S.E. is shown.
TTP represses NLRP3 inflammasome activation
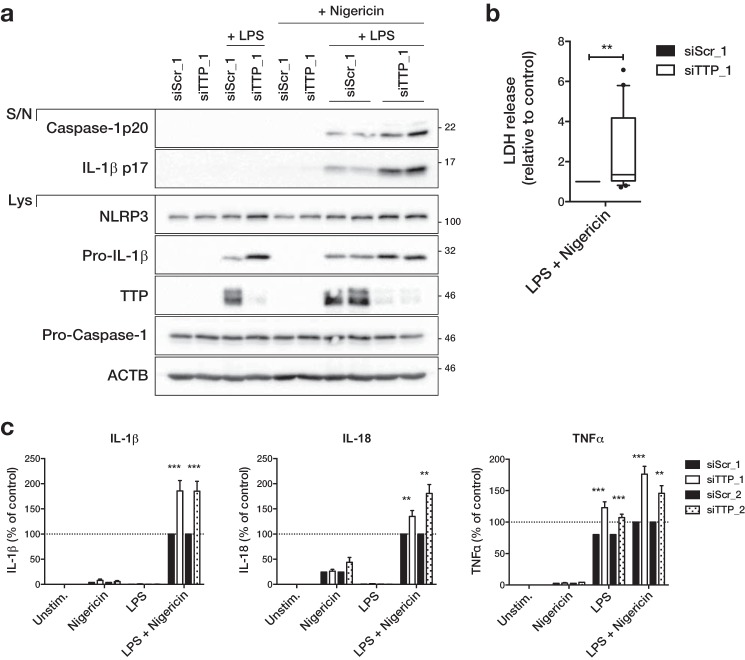
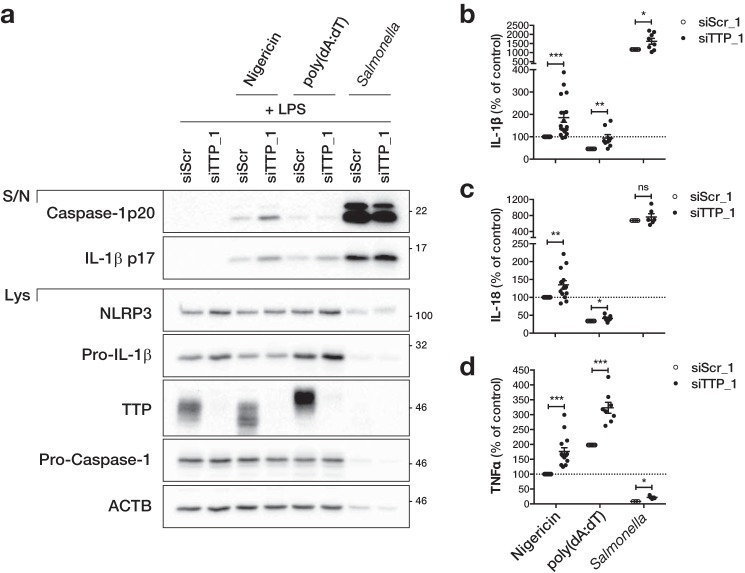
Because TTP knockdown increased NLRP3 expression, we next tested if it could affect NLRP3 inflammasome activation. As we had seen before, TTP knockdown led to an increase in LPS-induced expression of NLRP3 and Pro-IL-1β in primary macrophages (Fig. 5a and supplemental Fig. S4a). Similarly, TTP knockdown increased NLRP3 expression in THP-1 cells (supplemental Fig. S4b). As expected, activation of the NLRP3 inflammasome in primed cells using nigericin led to the cleavage of Caspase-1 and its release into the supernatant (Fig. 5a and supplemental Fig. S4). In human macrophages, cleaved IL-1β was also released upon nigericin stimulation (Fig. 5a and supplemental Fig. S4a). Importantly, all of these readouts were increased in cells transfected with two different TTP-specific siRNA (Fig. 5a and supplemental Fig. S4). Pyroptotic cell death induced by nigericin treatment of LPS-primed macrophages as measured by LDH release was also increased upon TTP knockdown (Fig. 5b). In accordance with increased inflammasome activation, release of IL-1β and IL-18, as measured by ELISA, was higher in cells transfected with two different TTP siRNA (Fig. 5c). Similarly, the LPS-induced release of the known TTP target TNFα was increased in TTP knockdown cells (Fig. 5c). To test the specificity of TTP in regulating other inflammasomes, we also activated the AIM2 and NLRC4 inflammasome by poly(dA:dT) transfection (28, 29) and Salmonella typhimurium infection (30, 31), respectively. Activation of NLRP3, AIM2, or NLRC4 inflammasome in LPS-primed macrophages led to the cleavage and release of IL-1β and Caspase-1 (Fig. 6a). Although both were increased in TTP knockdown cells stimulated with nigericin, only IL-1β release was increased in cells transfected with poly(dA:dT) or infected with Salmonella, probably due to increased expression of Pro-IL-1β (Fig. 6a). This was mirrored by an increased release of the direct TTP targets IL-1β and TNFα in TTP knockdown cells regardless of which inflammasome was activated (Fig. 6, b and c). However, release of IL-18 was only increased in TTP knockdown cells after NLRP3 activation, with little or no increase following AIM2 or NLRC4 activation (Fig. 6c). Of note, inflammasome activation by Salmonella was much stronger than after stimulation with nigericin or poly(dA:dT), resulting in very high release of IL-1β, IL-18, and cleaved Caspase-1 (Fig. 6, a–c). Cell death resulting from strong inflammasome activation is also likely to be responsible for the low expression of proteins in the cell lysate of Salmonella-infected samples (Fig. 6a). Even though Salmonella infection can also activate NLRP3 (32), the lack of difference in Caspase-1 activation and IL-18 release indicates that the contribution of NLRP3 in our protocol is negligible, possibly due to the relatively short incubation time. Overall, our findings suggest that TTP can repress NLRP3 inflammasome activity but does not affect AIM2 or NLRC4. At the same time, TTP directly targets IL-1β and TNFα and represses them irrespective of which inflammasome is activated.
Figure 5.
TTP represses NLRP3 inflammasome activity. Primary human macrophages were transfected with 50 nm control (siScr_1/2) or TTP siRNA (siTTP_1/2). Starting from 24 h after transfection, cells were primed with 10 ng/ml of LPS for 3 h, followed by inflammasome activation with 10 μm nigericin for 1 h as indicated. a, cleaved IL-1β and cleaved Caspase-1 in the supernatant (S/N) and NLRP3, Pro-IL-1β, Pro-Caspase-1, TTP, and β-actin (ACTB) in the cell lysate (Lys) were analyzed by Western blot and representative blots of at least three independent experiments are shown. b, LDH release to the supernatant as a measure of cell death was determined. The 10–90 percentile of LDH release relative to control transfected cells of 10 independent experiments is shown. The average cell death in control transfected cells across all experiments was 10.4%. c, the concentration of IL-1β, IL-18, and TNFα in the supernatant was measured by ELISA. The mean concentration relative to control transfected cells treated with LPS and nigericin ± S.E. of 4–18 independent measurements is shown. The range and average concentrations of LPS and nigericin-treated samples (100%) were: IL-1β (0.27–10.69 ng/ml; average 5.42 ng/ml), IL-18 (0.04–7.32 ng/ml; average 3.21 ng/ml), TNFα (0.44–55.37 ng/ml; average 17.86 ng/ml). **, p ≤ 0.01; ***, p ≤ 0.001.
Figure 6.
TTP does not regulate the AIM2 and NLRC4 inflammasome. Primary human macrophages were transfected with 50 nm control (siScr_1) or TTP siRNA (siTTP_1). Starting from 24 h after transfection, cells were primed with 10 ng/ml of LPS for 2 or 3 h, followed by inflammasome activation with 10 μm nigericin for 1 h (NLRP3), 1 μg/ml of poly(dA:dT) transfection for 6 h (AIM2) or Salmonella infection for 1 h (m.o.i. 20; NLRC4) as indicated. a, cleaved IL-1β and cleaved Caspase-1 in the supernatant (S/N) and NLRP3, Pro-IL-1β, Pro-Caspase-1, TTP, and β-actin (ACTB) in the cell lysate (Lys) were analyzed by Western blot and representative blots of at least three independent experiments are shown. The concentration of IL-1β (b), IL-18 (c), and TNFα (d) in the supernatant was measured by ELISA. The concentration of each sample relative to control transfected cells treated with LPS and nigericin and the mean ± S.E. of 4–18 independent measurements is shown. The range and average concentrations of LPS and nigericin-treated samples (100%) were: IL-1β (0.27–10.69 ng/ml; average 5.42 ng/ml), IL-18 (0.04–7.32 ng/ml; average 3.21 ng/ml), TNFα (0.44–55.37 ng/ml; average 17.86 ng/ml). *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001.
Discussion
In this study, we report a detailed analysis of the NLRP3 3′-UTR and identify TTP as a novel inducible regulator of NLRP3 expression. We found that human NLRP3 can be alternatively polyadenylated, resulting in transcripts containing a long or short 3′-UTR isoform. The isoforms can, furthermore, be differentially expressed in different cell types and tissues. In recent years, alternative polyadenylation has emerged as an important mechanism of gene expression regulation. Alternative 3′-UTR isoform selection has been shown to generate tissue-specific gene expression (33), but also to contribute to activation (34, 35) and malignant transformation of cells (36, 37). More recently, it has even been linked to alternative protein localization depending on which 3′-UTR isoform the encoding transcript contained (38). In the case of NLRP3, the short isoform could contribute to higher gene expression, due to the lack of both TTP and miR-223 regulation. For example, transcripts containing the short isoform could provide basal NLRP3 expression, whereas the long 3′-UTR could contribute more to its inducible expression or differential expression between cell types.
We also demonstrate that the RNA-binding protein TTP can bind to and repress specifically the long 3′-UTR isoform, most likely by binding to a main AU-rich element and possibly more weakly to a shorter ARE. TTP can also repress endogenous NLRP3 expression, since siRNA-mediated TTP knockdown led to increased NLRP3 expression in PMA-stimulated THP-1 cells and LPS-stimulated primary human macrophages. Especially in THP-1 cells, there was little effect on NLRP3 mRNA levels, whereas NLRP3 protein expression was increased. This points to a reduction of translation efficiency in the presence of TTP, which is increasingly recognized as an important feature of TTP-mediated repression (39, 40). Finally, we show that inhibition of NLRP3 expression by TTP is also functionally relevant, since NLRP3 inflammasome activation was exacerbated in TTP knockdown cells, indicated by increased Caspase-1 and IL-1β cleavage, IL-18 release, and cell death. Like Caspase-1 cleavage, increased IL-18 production is likely to be a direct consequence of increased inflammasome activation, since the IL18 3′-UTR does not contain any AU-rich elements and can therefore not be directly targeted by TTP. At the same time, TTP knockdown had little effect on the activation of the AIM2 and NLRC4 inflammasome, suggesting that NLRP3 is targeted specifically. Overall, these findings point to an important role of TTP as a feedback regulator during the priming phase of the NLRP3 inflammasome activation. Importantly, TTP can repress both NLRP3 and Pro-IL-1β, which is a known TTP target (25), and therefore has a combinatorial inhibitory effect.
The role of TTP as a regulator of the inflammatory response has been intensively studied due to the dramatic inflammatory phenotype of TTP-deficient mice, which spontaneously develop cachexia, dermatitis, and erosive polyarthritis (41). This has been linked to overproduction of the TTP target TNFα, since treatment with TNFα neutralizing antibodies (42) or crossing mice to a TNF receptor 1 (Tnfr1)-deficient background (43), rescued most of the TTP knock-out phenotypes. However, the same is true for genetic ablation of the IL-23/-17 axis, suggesting that a wider cytokine network, probably including feedback loops with TNF, is responsible for the phenotype of Zfp36−/− mice (44). Indeed, many cytokines and chemokines other than TNFα have been shown to be direct targets of TTP, including IL-10, IL-1β, IL-3, IL-6, CXCL1, CCL3, and GM-CSF (45). Given our findings, overactivation of the NLRP3 inflammasome could be another contributing factor to the global inflammation in these mice. Indeed, high levels of circulating IL-1β have been reported in Zfp36−/− mice (46, 47).
The regulation of TTP, in particular its close connection to p38 MAPK signaling, has been studied extensively. Upon LPS stimulation of macrophages, p38 signaling leads to the phosphorylation of TTP by the downstream kinase MK2 (48), which impacts activity and expression of TTP in a number of ways (reviewed in Ref. 49). An important effect of TTP phosphorylation is the inhibition of its ability to degrade target mRNAs, probably due to the interaction with 14-3-3 proteins, which interferes with the recruitment of the deadenylases CCR4 and CAF1 (50, 51). This also directly affects TTP expression, because TTP is involved in a negative feedback loop by targeting AREs in its own 3′-UTR (52, 53). Thus, phosphorylation of TTP increases its mRNA stability and expression. Finally, phosphorylation also increases TTP protein stability by protecting it from proteasomal degradation (54). The overall effect of this is rapid accumulation of TTP. Because this pool of TTP is largely phosphorylated and therefore inactive, TTP target mRNAs are relatively stable at this stage. However, prolonged LPS stimulation leads to the induction of yet another negative feedback regulator, the phosphatase DUSP1 (also known as MKP-1). DUSP1 inhibits p38 signaling (55), which allows TTP to be dephosphorylated by PP2A (56). This ultimately reactivates the accumulated pool of TTP and leads to the rapid degradation of target mRNAs. Targeting the phosphorylation state of TTP is an interesting prospect to modulate the severity of inflammatory conditions, such as experimental arthritis (57). The timing of TTP phosphorylation, occurring between 3 and 6 h post-stimulation, is strikingly similar to the priming phase in typical NLRP3 activation protocols. It is therefore tempting to speculate that phosphorylation of TTP could contribute to NLRP3 induction during priming due to temporal NLRP3 mRNA stabilization and/or translational derepression. Interestingly, p38 has been shown to be required for the induction of NLRP3 by LPS (58). At the same time, prolonged LPS stimulation could hinder inflammasome activation because of TTP reactivation. Chronic LPS stimulation has indeed been linked to decreased NLRP3 expression and inflammasome activation (59, 60). Repression of NLRP3 by prolonged LPS stimulation has also been connected to IL-10 induction, which can inhibit NLRP3 expression (59). This provides another possible link to TTP, since IL-10 is a potent inducer of TTP expression and TTP is needed for the full anti-inflammatory effects of IL-10 (61, 62). IL-10 has been found to repress NLRP3 expression in the inflamed joints in a mouse model of arthritis (63) and colonic macrophages (64). Additionally, a regulatory T cell subset, Tr1 cells, can suppress NLRP3 priming in macrophages by producing high levels of IL-10 (65). Furthermore, increased expression and activation of NLRP3 and inflammasome-dependent IL-1β production in IL-10 knock-out mice drive the development of spontaneous colitis (66) and severity of antigen-induced arthritis (63). The same is true for intestinal inflammation in IL-10R knock-out mice or patients harboring a loss-of-function mutation in the IL-10R (67). In colonic macrophages, the suppression of NLRP3 by IL-10 has been shown to happen at the post-transcriptional level, because NLPR3 protein is repressed without an effect on mRNA expression (64). The authors hypothesize that this is due to increased protein degradation, since treatment with the proteasome inhibitor MG132 increases NLRP3 expression. However, it could also be an indirect effect via TTP, because TTP expression is critically regulated by proteasomal degradation (54). IL-10 might chronically stimulate TTP expression in colonic macrophages, which represses NLRP3 and Pro-IL-1β expression. Either IL-10 or proteasome inhibition could then disrupt this regulatory loop and restore inflammasome activation.
TTP and NLRP3 have been shown to have opposing effects in a number of diseases. For example, TTP deficiency has been shown to exacerbate disease progression in the ApoE−/− model of atherosclerosis (47), whereas it is blunted in NLRP3-deficient mice (9). TTP knock-out mice are also more susceptible to DSS-induced colitis (68), which has been shown to involve activation of the NLRP3 inflammasome. Conflicting results have been reported about the role of inflammasomes in DSS colitis, with some reports finding that mice deficient in inflammasome components are protected (69, 70), whereas others report the opposite (71, 72). This is likely due to a complex cross-talk between the different genotypes and the microbiome (73). Given the pathogenic role of NLRP3 also in chronic models of colitis (66, 67), inflammasome activation is likely to be a driver of intestinal inflammation. Furthermore, TTP has been shown to suppress the development of different models of arthritis. For example, prevention of TTP phosphorylation in Zfp36aa/aa mice or by treatment with a PP2A agonist can protect mice from serum transfer-induced arthritis and zymosan-induced inflammatory arthritis (57). Similarly, TTP overexpression in Zfp36ΔARE mice is protective in collagen antibody-induced arthritis (74). Notably, NLRP3 inflammasome activation and IL-1β have been shown to be important drivers of arthritis in A20-deficient mice (75), ANK-deficient mice (76), and antigen-induced arthritis (63). In all examples mentioned above, TTP could suppress chronic inflammation caused by NLRP3 agonists in atherosclerotic plaques, arthritic joints, and the inflamed intestines. Consequently, TTP deficiency could worsen disease at least partially due to increased inflammasome activation. Equally, boosting TTP expression or activity could be beneficial by reducing NLRP3 activity among other anti-inflammatory effects.
In conclusion, we have identified TTP as a negative regulator of NLRP3, which could be particularly important during inflammasome priming and might contribute to IL-10-mediated inflammasome repression. Because TTP also represses IL-1β expression, it is a dual inhibitor of the IL-1β system. TTP-mediated reduction of the NLRP3 inflammasome activation could also reduce the development of chronic inflammatory diseases, such as atherosclerosis, colitis, and arthritis. We also found that human NLRP3 can be alternatively polyadenylated, which enables yet another level of regulation. Switching to predominant expression of the short 3′-UTR isoform could relieve repression not only by TTP but also miR-223. Targeting by TTP and alternative polyadenylation could help to explain how NLRP3 expression is regulated in macrophages and other cell types and during disease. TTP or the modulation of TTP activity via p38 signaling or PP2A might also provide new targets for therapeutic intervention in NLRP3-driven diseases.
Experimental procedures
Cell culture
THP-1, PBMCs, and primary human macrophages were maintained in RPMI 1640 medium and HEK293T were maintained in DMEM (Gibco), both with GlutaMAX-I and supplemented with 10% FCS and 1% (v/v) penicillin/streptomycin solution (both from Sigma). PBMCs were isolated from whole blood or buffy coats (supplied by the Irish Blood transfusion service) of healthy donors using Lymphoprep (Stemcell Technologies) density gradient medium. Primary monocytes were isolated from PBMCs by positive selection using anti-CD14 magnetic beads (Miltenyi Biotec) and the MACS system. Cells were then differentiated into macrophages by cultivation with 50 ng/ml of recombinant human M-CSF (Immunotools) for 7 days. Murine bone marrow-derived macrophages were generated by incubating bone marrow cells of wild-type mice with 10% L929 cell supernatant as a source of M-CSF for 7 days. THP-1 cells were activated with 12.5 ng/ml of PMA for the indicated times. Primary human macrophages were stimulated with 10 ng/ml of LPS as indicated. For inflammasome activation, THP-1 cells were primed with PMA (Sigma) for 8 h and primary human macrophages were primed with ultrapure rough LPS (from Escherichia coli, serotype EH100; Alexis) for 2 (AIM2) or 3 h (NLRP3 or NLRC4). For NLRP3 activation, cells were treated with 10 μm nigericin (Invivogen) for 1 h. AIM2 was activated by transfection of 1 μg/ml of poly(dA:dT) (Invivogen) with Lipofectamine 2000 (Invitrogen) for 6 h. For NLRC4 stimulation, LPS-primed macrophages were grown in antibiotic-free media and infected with S. typhimurium (UK-1) (multiplicity of infection 20). Cells were centrifuged at 300 × g for 5 min and the media changed after 30 min to complete media containing 100 μg/ml of Gentamycin (Sigma) to kill extracellular bacteria. Samples were analyzed after 1 h. For analysis of cleaved Caspase-1 or IL-1β in the supernatant by Western blot, inflammasome activation was performed in serum-free media as not to overload the precipitation step with serum proteins.
Transfection
Plasmid transfection of HEK293T (2 × 105 cells/ml) for luciferase assays was performed with GeneJuice (Novagen) or Lipofectamine 2000 (Invitrogen) in complete media according to the manufacturer's instructions. For psiCHECK-2 vectors, 40 ng were transfected per well of a 96-well plate using GeneJuice. If co-expressed with TTP expression vectors, 30 ng of psiCHK-2 plus 120 ng of expression vector were transfected. For the ZFP screen, cells were transfected with 150 ng of ZFP expression plasmid, 40 ng of pSGG (empty vector or NLRP3 3′-UTR), and 10 ng of TK-Renilla per well using Lipofectamine 2000. THP-1 cells were batch transfected with plasmid DNA using Lipofectamine 2000 (Invitrogen) according to manufacturer's instructions. In short, 3 × 106 cells in 3 ml of antibiotic-free media were transfected with 2.25 μg of DNA and 5.63 μl of Lipofectamine and media changed to complete media 4 h after transfection to minimize toxicity and then distributed to individual wells.
Cells were transfected with siRNA using Viromer Blue (Lipocalyx) according to the manufacturer's instructions. THP-1 cells were transfected with 40 nm and primary macrophages with 50 nm siRNA. Cells were generally used for assays 24 h post-transfection. siTTP_1 was as described before (77). siScr_1 and siTTP_1 were synthezised by Eurofins Genomics, siScr_2 (negative control No. 1) and siTTP_2 (s14979) were Silencer Select from Ambion. siRNA sequences were: siScr_1 (UUCUCCGAACGUGUCACGUdTdT), siTTP_1 (CGACGAUAUAAUUAUUAUAdTdT), and siTTP_2 (CUUUAUUUAUGACGACUUUdTdT).
Luciferase assay
Cells were lysed in 50 μl of 1× Passive lysis buffer (Promega) 48 h after transfection. 20 μl of lysate was combined with 40 μl of Firefly or Renilla luciferase substrate and luminescence was measured using a FLUOStar Optima (BMG Labtech). The substrate solution for measuring Firefly luciferase activity was Firefly luciferase assay mixture (10 mm Tricine, 0.535 mm (MgCO3)4Mg(OH)2·5H2O, 1.335 mm MgSO4·7H2O, 0.05 m EDTA, 16.65 mm DTT, 135 mm acetyl-Coenzyme A, 235 mm d-luciferin, 265 mm ATP) and for Renilla luciferase activity Coelenterazine (1 mg/ml diluted 1:1000 in PBS; Biotium) was added. For each well, luciferase activity was normalized to the internal control (Switchgear plasmids: Firefly reading divided by Renilla reading; psiCHECK-2: Renilla reading divided by Firefly reading). For the ZFP screen, samples with too low internal Renilla readings were excluded from the analysis.
Western blot
Cells were lysed in 2× Laemmli sample buffer and separated by SDS-PAGE and blotted according to standard protocols. For measurement of cleaved IL-1β and Caspase-1 in the supernatant, proteins in the supernatant were precipitated with 1% (v/v) Strataclean Resin (Agilent Technologies) and beads lysed in 2× sample buffer. Proteins were visualized using the HRP substrate WesternBright Spray ECL (Advansta) on a ChemiDoc imaging system (Bio-Rad). Primary antibodies were: NLRP3 (Cell Signaling D2P5E, 1:1000), TTP (Millipore ABE285, 1:1000), β-actin (Sigma AC-74, 1:15000), IL-1β (Pro and cleaved) (R&D Systems AF-401, 1:1000), Caspase-1 p20 (Asp297) (Cell Signaling D57A2, 1:1000), and Pro-Caspase-1 p45 (A-19, Santa Cruz sc-622; 1:2000). HRP-conjugated secondary antibodies were from Jackson ImmunoResearch.
ELISA
Supernatants were collected and cytokine abundance determined by human IL-1β DuoSet ELISA (R&D Systems DY201), human Total IL-18 DuoSet ELISA (R&D Systems DY318) or human TNFα (Immunotools) ELISA kits according to the manufacturer's instructions.
Cell death measurement
Supernatants were collected and cleared of cells by centrifugation. LDH activity as a measure of lytic cell death was determined using the CytoTox 96 Non-Radioactive Cytotoxicity Assay (Promega).
qPCR
RNA was isolated using the PureLink RNA Mini kit (Ambion) or the RNeasy Mini kit (Qiagen) according to the manufacturer's instructions. For measurement of NLRP3 3′-UTR isoforms by qPCR, RNA was DNase-treated using the TURBO DNA-free kit (Ambion). RNA was converted to cDNA using the High Capacity cDNA reverse transcription kit (Applied Biosystems) in duplicate, using a 1:1 mixture of random primers and an anchored oligo(dT) primer (TTTTTTTTTTTTTTTTTTVN, synthesized by Eurofins Genomics). cDNA was then diluted 1:2–1:5 and one qPCR per reverse transcription replicate assembled. Gene expression was determined by SYBR Green qPCR using the KAPA SYBR Fast qPCR Master Mix (KAPA Biosystems) or PowerUp SYBR Green Master Mix (Applied Biosystems) on a 7500 Fast thermocycler (Applied Biosystems). Oligonucleotide primers (Table 1) were designed using PrimerQuest (IDT Technologies) and synthesized by Eurofins Genomics as salt-free oligos. Primers were tested for the presence of a single PCR product and acceptable amplification efficiency. Relative expression was determined by ΔΔCt calculation for one internal control gene. If more than one internal control was used, the geometric mean of all controls was used for normalization.
Table 1.
Primers for SYBR Green qPCR
| qPCR | Forward | Reverse |
|---|---|---|
| hACTB | CACAGAGCCTCGCCTTT | GAGCGCGGCGATATCAT |
| hGAPDH | AGCCGAGCCACATCGCT | GCAACAATATCCACTTTACCAGAGT |
| hIL1B | AGCTGATGGCCCTAAACAGA | TGTCCATGGCCACAACAACTGA |
| hNLRP3 | TGGATCTTCGCTGCGATCAACA | TCAATGCTGTCTTCCTGGCA |
| hNLRP3′long | TCTCTTCTCTGTCTAACTTTCTT | GCGGGAATGATGATATGAGCAAA |
| hNLRP3′short | ATGCCTTCCTGTGCAGAGCTT | AGGCGCTGTGATGACAACAA |
| hRPS13 | CGAAAGCATCTTGAGAGGAACA | TCGAGCCAAACGGTGAATC |
| hTNF | CCAGGGACCTCTCTCTAATCA | TCAGCTTGAGGGTTTGCTAC |
3′-RACE (rapid amplification of cDNA ends)
To amplify 3′-UTR sequences of NLRP3, RNA was first converted to cDNA using polyT_T7 (CAGTGAATTGTAATACGACTCACTATAGGTTTTTTTTTTTTTTTTVN) instead of random primers/oligo(dT). 3′-UTR sequences were amplified by end point PCR (22–35 cycles) using the KAPA2G Fast ReadyMix PCR kit (KAPA Biosystems) in 10-μl reactions with the common reverse primer T7_rev (CAGTGAATTGTAATACGACTCACTATAGG) and gene-specific forward primers (hNLRP3_CDS_f, TGACCGTCGTCTTTGAGCCTTCTT; hACTB_CSF_f, TCCACGAAACTACCTTCAACTC; mNlrp3_CDS_f, CCTTAGAAGCGCTCCAGGAAGAAA). PCR products were separated on an agarose gel and bands were visualized with Nancy-520 (Sigma) on a gel imaging system (UVP) and quantified using ImageJ.
Quantification of 3′-UTR isoforms from sequencing data
Expression tracks from the Encyclopedia of DNA Elements (ENCODE) (78), the NIH Roadmap Epigenomics project (79), or the Gene-Tissue Expression (GTEx) (80) project were loaded into the Genome browser (GRCh37/hg19) using the respective Track Hubs (81). The ENCODE data used in this article (accession numbers GSM758559, GSM758566, GSM981256, GSM984609, GSM981257, GSM984606, and GSM984604) were generated by the Gingeras lab (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY). The Roadmap Epigenomics data used in this article was sourced from the Epigenome Browser or the Roadmap Epigenomics Integrative Analysis Hub. 30 random spleen samples and 56 random whole blood samples were chosen from the GTEx RNA-seq Signal hub. Windows for the short (Window 1: chr1:247,611,808-247,612,121) and long NLRP3 3′-UTR isoform (Window 2: chr1:247,612,120-247,612,407) were defined. For all tracks, the average value of both windows was recorded from the details page. Expression of the short isoform was estimated by subtracting the average value of Window 1 from the average value of Window 2. Expression of the long isoform was estimated as the average value of Window 2. The ratio of short to long isoform expression was plotted for each track.
mRNA stability measurement
THP-1 cells were first treated with PMA as indicated and then 2 μg/ml of Actinomycin D was added to stop transcription. Cells were lysed for RNA analysis at the indicated times after Actinomycin D addition.
Plasmids
NLRP3 3′-UTR and control firefly luciferase reporter vector (Switchgear Genomics) and TK-Renilla (Promega) used for the ZFP screen have been described previously (15). ZFP expression vectors have been described previously (22) and were kindly provided by Dr. Derek Lacey and Dr. Marco Herold (WEHI, Melbourne). Expression vectors for murine wild-type and ZFP mutant (m1,2) TTP have been described previously (82) and were kindly provided by Prof. Georg Stoecklin (University of Heidelberg).
3′-UTR sequences were cloned into the psiCHECK-2 (Promega) empty vector using a nested PCR setup using the KAPA HiFi Hotstart ReadyMix (Kapa Biosystems) PCR kit. In the first PCR step, genomic 3′-UTR regions were amplified for 15 cycles with 60 °C annealing temperature with genomic DNA (human: Promega Human Genomic DNA) as input. The following primer pairs were used: human NLRP3 (forward, TATCTGAAGAGTGCAACCCAGGCT; reverse, ACTCTCAAACCTTTCCCTCCACGA); and human GAPDH (forward, CATGGCCTCCAAGGAGTAAG; reverse, CCAGCAAGAATGTCTCACCT). Next, the 3′-UTRs were further amplified using the primary PCR as input for 3 cycles with 55 °C and 23–27 cycles 60 °C annealing temperature. The primer pairs containing restriction sites were: NLRP3′long (ctgacgctcgagaGAGTGGAAACGGGGCTGCCAGA, cgcatggcggccgcgTTTTTTAAAATTAAGAAAAGGA); NLRP3′short (ctgacgctcgagaGAGTGGAAACGGGGCTGCCAGA, cgcatggcggccgcgAAAATAAAGAAAGTGCTTTATT); NLRP3′end (ctgacgctcgagaGATCTCTTCTCTGTCTAACTTT, cgcatggcggccgcgTTTTTTAAAATTAAGAAAAGGA); NLRP3′ARE (ctgacgctcgagaTTGACTATATATTATGTTGAAAT, cgcatggcggccgcgTTTTCTTATTAGAAAACAAAACT); GAPDH3′ (ctgacgctcgagACCCCTGGACCACCAGCCCC, gcgtgggcggccgcTGGTTGAGCACAGGGTACTT).
PCR products were separated by agarose gel electrophoresis and bands of the correct size were gel purified using the Wizard SV Gel and PCR Clean-Up System (Promega). Gel-purified inserts and 5 μg of vector DNA were digested with XhoI and NotI (New Engalnd Biolabs). Digested vectors were dephosphorylated by adding 1 μl of CIP (New England Biolabs) into the digestion reaction. Digested vectors and inserts were gel-purified and ligated overnight at 4 °C using T4 DNA ligase (Promega), transformed into competent bacteria, and selected on LB-Amp plates. Vectors were sequenced using the sequencing primer GTGCTGAAGAACGAGCAGTA.
NLRP3′AREdel and NLRP3′AREmut were generated from psiCHK2-NLRP3′long by site-directed mutagenesis using the QuikChange Lightning kit (Agilent Technologies) according to manufacturer's instructions. Mutagenesis primers were designed with the QuikChange Primer Design tool and were: AREdel (forward, gactatatattatgttgaaattttatggcagctaattttttgtaacagttttgttttctaataaga; reverse, tcttattagaaaacaaaactgttacaaaaaattagctgccataaaatttcaacataatatatagtc) and AREmut (forward, ttgaaattttatggcagctatCtatCtatCtaaattttttgtaacagttt; reverse, aaactgttacaaaaaatttaGataGataGatagctgccataaaatttcaa).
RNA immunoprecipitation
Per sample, 1–2 × 107 THP-1 cells were treated with PMA for 4 h and then lysed for 10 min on ice in RIP buffer (1% (v/v) Nonidet P-40, 150 mm NaCl, 50 mm Tris-HCl, pH 8.0, 1 mm MgCl2, 10% (v/v) glycerol, 1 mm DTT, supplemented with 50 mm NaF, 1 Na3VO4, 1 mm PMSF, 1 μg/ml of leupeptin, 0.5% (v/v) aprotinin (5–10 TUI/ml)). The lysate was cleared from nuclei by centrifugation for 5 min at 2000 × g at 4 °C. The lysate was then precleared for 30 min at 4 °C with 5% (v/v) equilibrated protein A/G PLUS-agarose beads (Santa Cruz). Beads were removed by centrifugation and an aliquot of the lysate taken as the input sample. The rest of the lysate was then incubated with 2 μg of anti-TTP (ABE285) or rabbit IgG control antibody (both Millipore) for 1.5 h at 4 °C. 5% (v/v) washed protein A/G beads were added for another 1.5 h. The beads were washed 5 times in RIP buffer and pulldown and input samples lysed in Qiazol (Qiagen). RNA was extracted according to the manufacturer's instructions, with the addition of 15 μg of linear polyacrylamide (prepared according to Delaney lab protocol (University of Vermont)) to the aqueous phase prior to RNA precipitation with isopropyl alcohol. Equal volumes of RNA were then converted to cDNA and relative target mRNA enrichment was determined by qPCR.
Statistics
p values were calculated by two-sided Student's t test (not assuming equal variance) in GraphPad Prism. Linear regression analysis of RNA stability measurements was also done in Prism.
Author contributions
M. H. conceived and coordinated the study and wrote the manuscript. L. A. J. O. helped with study design and coordination, interpretation of results, and critically revised the manuscript. M. H. and J. D. O. designed, performed, and analyzed experiments. S. L. M. and A. R. C. helped with coordination of experiments and interpretation of results. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Daniel Johnston (Trinity College Dublin) for assistance with the Salmonella infection experiments and Dr. Derek Lacey and Dr. Marco Herold (WEHI, Melbourne) for providing plasmids for the ZFP screen. We also thank Prof. Georg Stoecklin (University of Heidelberg) for providing TTP expression vectors.
This work was supported by the Science Foundation Ireland and the European Research Council. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S4.
- miR
- microRNA
- ARE
- AU-rich element
- TTP
- Tristetraprolin
- EST
- expressed sequence tag
- 3′-RACE
- 3′-rapid amplification of cDNA ends
- ZFP
- zinc finger protein
- PMA
- phorbol 12-myristate 13-acetate
- qPCR
- quantitative PCR
- DSS
- dextran sulfate sodium
- PBMC
- peripheral blood mononuclear cell
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
References
- 1. He Y., Hara H., and Núñez G. (2016) Mechanism and regulation of NLRP3 inflammasome activation. Trends Biochem. Sci. 41, 1012–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bauernfeind F. G., Horvath G., Stutz A., Alnemri E. S., MacDonald K., Speert D., Fernandes-Alnemri T., Wu J., Monks B. G., Fitzgerald K. A., Hornung V., and Latz E. (2009) Cutting edge: NF-κB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J. Immunol. 183, 787–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Juliana C., Fernandes-Alnemri T., Kang S., Farias A., Qin F., and Alnemri E. S. (2012) Non-transcriptional priming and deubiquitination regulate NLRP3 inflammasome activation. J. Biol. Chem. 287, 36617–36622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Muñoz-Planillo R., Kuffa P., Martínez-Colón G., Smith B. L., Rajendiran T. M., and Núñez G. (2013) K(+) efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity 38, 1142–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Martinon F., Burns K., and Tschopp J. (2002) The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol. Cell 10, 417–426 [DOI] [PubMed] [Google Scholar]
- 6. Kayagaki N., Stowe I. B., Lee B. L., O'Rourke K., Anderson K., Warming S., Cuellar T., Haley B., Roose-Girma M., Phung Q. T., Liu P. S., Lill J. R., Li H., Wu J., Kummerfeld S., et al. (2015) Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526, 666–671 [DOI] [PubMed] [Google Scholar]
- 7. Shi J., Zhao Y., Wang K., Shi X., Wang Y., Huang H., Zhuang Y., Cai T., Wang F., and Shao F. (2015) Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665 [DOI] [PubMed] [Google Scholar]
- 8. Masters S. L., Dunne A., Subramanian S. L., Hull R. L., Tannahill G. M., Sharp F. A., Becker C., Franchi L., Yoshihara E., Chen Z., Mullooly N., Mielke L. A., Harris J., Coll R. C., Mills K. H., et al. (2010) Activation of the NLRP3 inflammasome by islet amyloid polypeptide provides a mechanism for enhanced IL-1β in type 2 diabetes. Nat. Immunol. 11, 897–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Duewell P., Kono H., Rayner K. J., Sirois C. M., Vladimer G., Bauernfeind F. G., Abela G. S., Franchi L., Nuñez G., Schnurr M., Espevik T., Lien E., Fitzgerald K. A., Rock K. L., Moore K. J., Wright S. D., Hornung V., and Latz E. (2010) NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature 464, 1357–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Martinon F., Pétrilli V., Mayor A., Tardivel A., and Tschopp J. (2006) Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 440, 237–241 [DOI] [PubMed] [Google Scholar]
- 11. Heneka M. T., Kummer M. P., Stutz A., Delekate A., Schwartz S., Vieira-Saecker A., Griep A., Axt D., Remus A., Tzeng T. C., Gelpi E., Halle A., Korte M., Latz E., and Golenbock D. T. (2013) NLRP3 is activated in Alzheimer's disease and contributes to pathology in APP/PS1 mice. Nature 493, 674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Broderick L., De Nardo D., Franklin B. S., Hoffman H. M., and Latz E. (2015) The inflammasomes and autoinflammatory syndromes. Annu. Rev. Pathol. 10, 395–424 [DOI] [PubMed] [Google Scholar]
- 13. Franchi L., Eigenbrod T., and Núñez G. (2009) Cutting edge: TNF-α mediates sensitization to ATP and silica via the NLRP3 inflammasome in the absence of microbial stimulation. J. Immunol. 183, 792–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang J., Liu Z., and Xiao T. S. (2017) Post-translational regulation of inflammasomes. Cell Mol. Immunol. 14, 65–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haneklaus M., Gerlic M., Kurowska-Stolarska M., Rainey A. A., Pich D., McInnes I. B., Hammerschmidt W., O'Neill L. A., and Masters S. L. (2012) Cutting edge: miR-223 and EBV miR-BART15 regulate the NLRP3 inflammasome and IL-1β production. J. Immunol. 189, 3795–3799 [DOI] [PubMed] [Google Scholar]
- 16. Bauernfeind F., Rieger A., Schildberg F. A., Knolle P. A., Schmid-Burgk J. L., and Hornung V. (2012) NLRP3 inflammasome activity is negatively controlled by miR-223. J. Immunol. 189, 4175–4181 [DOI] [PubMed] [Google Scholar]
- 17. Carpenter S., Ricci E. P., Mercier B. C., Moore M. J., and Fitzgerald K. A. (2014) Post-transcriptional regulation of gene expression in innate immunity. Nat. Rev. Immunol. 14, 361–376 [DOI] [PubMed] [Google Scholar]
- 18. Tian B., Hu J., Zhang H., and Lutz C. S. (2005) A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 33, 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sheets M. D., Ogg S. C., and Wickens M. P. (1990) Point mutations in AAUAAA and the poly(A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 18, 5799–5805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Spasic M., Friedel C. C., Schott J., Kreth J., Leppek K., Hofmann S., Ozgur S., and Stoecklin G. (2012) Genome-wide assessment of AU-rich elements by the AREScore algorithm. PLoS Genet. 8, e1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Garneau N. L., Wilusz J., and Wilusz C. J. (2007) The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 8, 113–126 [DOI] [PubMed] [Google Scholar]
- 22. Lacey D., Hickey P., Arhatari B. D., O'Reilly L. A., Rohrbeck L., Kiriazis H., Du X. J., and Bouillet P. (2015) Spontaneous retrotransposon insertion into TNF 3′UTR causes heart valve disease and chronic polyarthritis. Proc. Natl. Acad. Sci. U.S.A. 112, 9698–9703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lai W. S., Carballo E., Strum J. R., Kennington E. A., Phillips R. S., and Blackshear P. J. (1999) Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor α mRNA. Mol. Cell. Biol. 19, 4311–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Johnson B. A., Geha M., and Blackwell T. K. (2000) Similar but distinct effects of the tristetraprolin/TIS11 immediate-early proteins on cell survival. Oncogene 19, 1657–1664 [DOI] [PubMed] [Google Scholar]
- 25. Chen Y. L., Huang Y. L., Lin N. Y., Chen H. C., Chiu W. C., and Chang C. J. (2006) Differential regulation of ARE-mediated TNFalpha and IL-1beta mRNA stability by lipopolysaccharide in RAW264.7 cells. Biochem. Biophys. Res. Commun. 346, 160–168 [DOI] [PubMed] [Google Scholar]
- 26. Sedlyarov V., Fallmann J., Ebner F., Huemer J., Sneezum L., Ivin M., Kreiner K., Tanzer A., Vogl C., Hofacker I., and Kovarik P. (2016) Tristetraprolin binding site atlas in the macrophage transcriptome reveals a switch for inflammation resolution. Mol. Syst. Biol. 12, 868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tiedje C., Diaz-Muñoz M. D., Trulley P., Ahlfors H., Laaß K., Blackshear P. J., Turner M., and Gaestel M. (2016) The RNA-binding protein TTP is a global post-transcriptional regulator of feedback control in inflammation. Nucleic Acids Res. 44, 7418–7440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hornung V., Ablasser A., Charrel-Dennis M., Bauernfeind F., Horvath G., Caffrey D. R., Latz E., and Fitzgerald K. A. (2009) AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458, 514–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fernandes-Alnemri T., Yu J. W., Datta P., Wu J., and Alnemri E. S. (2009) AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458, 509–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miao E. A., Alpuche-Aranda C. M., Dors M., Clark A. E., Bader M. W., Miller S. I., and Aderem A. (2006) Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1β via Ipaf. Nat. Immunol. 7, 569–575 [DOI] [PubMed] [Google Scholar]
- 31. Franchi L., Amer A., Body-Malapel M., Kanneganti T. D., Ozören N., Jagirdar R., Inohara N., Vandenabeele P., Bertin J., Coyle A., Grant E. P., and Núñez G. (2006) Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1β in salmonella-infected macrophages. Nat. Immunol. 7, 576–582 [DOI] [PubMed] [Google Scholar]
- 32. Broz P., Newton K., Lamkanfi M., Mariathasan S., Dixit V. M., and Monack D. M. (2010) Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J. Exp. Med. 207, 1745–1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lianoglou S., Garg V., Yang J. L., Leslie C. S., and Mayr C. (2013) Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev. 27, 2380–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sandberg R., Neilson J. R., Sarma A., Sharp P. A., and Burge C. B. (2008) Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320, 1643–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chang J. W., Zhang W., Yeh H. S., de Jong E. P., Jun S., Kim K. H., Bae S. S., Beckman K., Hwang T. H., Kim K. S., Kim D. H., Griffin T. J., Kuang R., and Yong J. (2015) mRNA 3′-UTR shortening is a molecular signature of mTORC1 activation. Nat. Commun. 6, 7218. [DOI] [PubMed] [Google Scholar]
- 36. Mayr C., and Bartel D. P. (2009) Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138, 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Elkon R., Drost J., van Haaften G., Jenal M., Schrier M., Oude Vrielink J. A., and Agami R. (2012) E2F mediates enhanced alternative polyadenylation in proliferation. Genome Biol. 13, R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Berkovits B. D., and Mayr C. (2015) Alternative 3′ UTRs act as scaffolds to regulate membrane protein localization. Nature 522, 363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Qi M. Y., Wang Z. Z., Zhang Z., Shao Q., Zeng A., Li X. Q., Li W. Q., Wang C., Tian F. J., Li Q., Zou J., Qin Y. W., Brewer G., Huang S., and Jing Q. (2012) AU-rich-element-dependent translation repression requires the cooperation of tristetraprolin and RCK/P54. Mol. Cell. Biol. 32, 913–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tao X., and Gao G. (2015) Tristetraprolin recruits eukaryotic initiation factor 4E2 to repress translation of AU-rich element-containing mRNAs. Mol. Cell. Biol. 35, 3921–3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Taylor G. A., Carballo E., Lee D. M., Lai W. S., Thompson M. J., Patel D. D., Schenkman D. I., Gilkeson G. S., Broxmeyer H. E., Haynes B. F., and Blackshear P. J. (1996) A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 4, 445–454 [DOI] [PubMed] [Google Scholar]
- 42. Keffer J., Probert L., Cazlaris H., Georgopoulos S., Kaslaris E., Kioussis D., and Kollias G. (1991) Transgenic mice expressing human tumour necrosis factor: a predictive genetic model of arthritis. EMBO J. 10, 4025–4031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Carballo E., and Blackshear P. J. (2001) Roles of tumor necrosis factor-α receptor subtypes in the pathogenesis of the tristetraprolin-deficiency syndrome. Blood 98, 2389–2395 [DOI] [PubMed] [Google Scholar]
- 44. Molle C., Zhang T., Ysebrant de Lendonck L., Gueydan C., Andrianne M., Sherer F., Van Simaeys G., Blackshear P. J., Leo O., and Goriely S. (2013) Tristetraprolin regulation of interleukin 23 mRNA stability prevents a spontaneous inflammatory disease. J. Exp. Med. 210, 1675–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brooks S. A., and Blackshear P. J. (2013) Tristetraprolin (TTP): interactions with mRNA and proteins, and current thoughts on mechanisms of action. Biochim. Biophys. Acta 1829, 666–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kaplan I. M., Morisot S., Heiser D., Cheng W. C., Kim M. J., and Civin C. I. (2011) Deletion of tristetraprolin caused spontaneous reactive granulopoiesis by a non-cell-autonomous mechanism without disturbing long-term hematopoietic stem cell quiescence. J. Immunol. 186, 2826–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kang J. G., Amar M. J., Remaley A. T., Kwon J., Blackshear P. J., Wang P. Y., and Hwang P. M. (2011) Zinc finger protein tristetraprolin interacts with CCL3 mRNA and regulates tissue inflammation. J. Immunol. 187, 2696–2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chrestensen C. A., Schroeder M. J., Shabanowitz J., Hunt D. F., Pelo J. W., Worthington M. T., and Sturgill T. W. (2004) MAPKAP kinase 2 phosphorylates tristetraprolin on in vivo sites including Ser-178, a site required for 14-3-3 binding. J. Biol. Chem. 279, 10176–10184 [DOI] [PubMed] [Google Scholar]
- 49. Clark A. R., and Dean J. L. (2016) The control of inflammation via the phosphorylation and dephosphorylation of tristetraprolin: a tale of two phosphatases. Biochem. Soc. Trans. 44, 1321–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Clement S. L., Scheckel C., Stoecklin G., and Lykke-Andersen J. (2011) Phosphorylation of tristetraprolin by MK2 impairs AU-rich element mRNA decay by preventing deadenylase recruitment. Mol. Cell. Biol. 31, 256–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Marchese F. P., Aubareda A., Tudor C., Saklatvala J., Clark A. R., and Dean J. L. (2010) MAPKAP kinase 2 blocks tristetraprolin-directed mRNA decay by inhibiting CAF1 deadenylase recruitment. J. Biol. Chem. 285, 27590–27600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Brooks S. A., Connolly J. E., and Rigby W. F. (2004) The role of mRNA turnover in the regulation of tristetraprolin expression: evidence for an extracellular signal-regulated kinase-specific, AU-rich element-dependent, autoregulatory pathway. J. Immunol. 172, 7263–7271 [DOI] [PubMed] [Google Scholar]
- 53. Tchen C. R., Brook M., Saklatvala J., and Clark A. R. (2004) The stability of tristetraprolin mRNA is regulated by mitogen-activated protein kinase p38 and by tristetraprolin itself. J. Biol. Chem. 279, 32393–32400 [DOI] [PubMed] [Google Scholar]
- 54. Brook M., Tchen C. R., Santalucia T., McIlrath J., Arthur J. S., Saklatvala J., and Clark A. R. (2006) Posttranslational regulation of tristetraprolin subcellular localization and protein stability by p38 mitogen-activated protein kinase and extracellular signal-regulated kinase pathways. Mol. Cell. Biol. 26, 2408–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen P., Li J., Barnes J., Kokkonen G. C., Lee J. C., and Liu Y. (2002) Restraint of proinflammatory cytokine biosynthesis by mitogen-activated protein kinase phosphatase-1 in lipopolysaccharide-stimulated macrophages. J. Immunol. 169, 6408–6416 [DOI] [PubMed] [Google Scholar]
- 56. Sun L., Stoecklin G., Van Way S., Hinkovska-Galcheva V., Guo R. F., Anderson P., and Shanley T. P. (2007) Tristetraprolin (TTP)-14-3-3 complex formation protects TTP from dephosphorylation by protein phosphatase 2a and stabilizes tumor necrosis factor-α mRNA. J. Biol. Chem. 282, 3766–3777 [DOI] [PubMed] [Google Scholar]
- 57. Ross E. A., Naylor A. J., O'Neil J. D., Crowley T., Ridley M. L., Crowe J., Smallie T., Tang T. J., Turner J. D., Norling L. V., Dominguez S., Perlman H., Verrills N. M., Kollias G., Vitek M. P., et al. (2017) Treatment of inflammatory arthritis via targeting of tristetraprolin, a master regulator of pro-inflammatory gene expression. Ann. Rheum. Dis. 76, 612–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Moon J. S., Lee S., Park M. A., Siempos I. I., Haslip M., Lee P. J., Yun M., Kim C. K., Howrylak J., Ryter S. W., Nakahira K., and Choi A. M. (2015) UCP2-induced fatty acid synthase promotes NLRP3 inflammasome activation during sepsis. J. Clin. Invest. 125, 665–680 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59. Gurung P., Li B., Subbarao Malireddi R. K., Lamkanfi M., Geiger T. L., and Kanneganti T. D. (2015) Chronic TLR stimulation controls NLRP3 inflammasome activation through IL-10 mediated regulation of NLRP3 expression and caspase-8 activation. Sci. Rep. 5, 14488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hu Y., Mao K., Zeng Y., Chen S., Tao Z., Yang C., Sun S., Wu X., Meng G., and Sun B. (2010) Tripartite-motif protein 30 negatively regulates NLRP3 inflammasome activation by modulating reactive oxygen species production. J. Immunol. 185, 7699–7705 [DOI] [PubMed] [Google Scholar]
- 61. Schaljo B., Kratochvill F., Gratz N., Sadzak I., Sauer I., Hammer M., Vogl C., Strobl B., Müller M., Blackshear P. J., Poli V., Lang R., Murray P. J., and Kovarik P. (2009) Tristetraprolin is required for full anti-inflammatory response of murine macrophages to IL-10. J. Immunol. 183, 1197–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gaba A., Grivennikov S. I., Do M. V., Stumpo D. J., Blackshear P. J., and Karin M. (2012) Cutting edge: IL-10-mediated tristetraprolin induction is part of a feedback loop that controls macrophage STAT3 activation and cytokine production. J. Immunol. 189, 2089–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Greenhill C. J., Jones G. W., Nowell M. A., Newton Z., Harvey A. K., Moideen A. N., Collins F. L., Bloom A. C., Coll R. C., Robertson A. A., Cooper M. A., Rosas M., Taylor P. R., O'Neill L. A., Humphreys I. R., Williams A. S., and Jones S. A. (2014) Interleukin-10 regulates the inflammasome-driven augmentation of inflammatory arthritis and joint destruction. Arthritis Res. Ther. 16, 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Filardy A. A., He J., Bennink J., Yewdell J., and Kelsall B. L. (2016) Posttranscriptional control of NLRP3 inflammasome activation in colonic macrophages. Mucosal. Immunol. 9, 850–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yao Y., Vent-Schmidt J., McGeough M. D., Wong M., Hoffman H. M., Steiner T. S., and Levings M. K. (2015) Tr1 cells, but not Foxp3+ regulatory T cells, suppress NLRP3 inflammasome activation via an IL-10-dependent mechanism. J. Immunol. 195, 488–497 [DOI] [PubMed] [Google Scholar]
- 66. Zhang J., Fu S., Sun S., Li Z., and Guo B. (2014) Inflammasome activation has an important role in the development of spontaneous colitis. Mucosal. Immunol. 7, 1139–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shouval D. S., Biswas A., Kang Y. H., Griffith A. E., Konnikova L., Mascanfroni I. D., Redhu N. S., Frei S. M., Field M., Doty A. L., Goldsmith J. D., Bhan A. K., Loizides A., Weiss B., Yerushalmi B., et al. (2016) Interleukin 1β mediates intestinal inflammation in mice and patients with interleukin 10 receptor deficiency. Gastroenterology 151, 1100–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Joe Y., Uddin M. J., Zheng M., Kim H. J., Chen Y., Yoon N. A., Cho G. J., Park J. W., and Chung H. T. (2014) Tristetraprolin mediates anti-inflammatory effect of carbon monoxide against DSS-induced colitis. PloS One 9, e88776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zaki M. H., Boyd K. L., Vogel P., Kastan M. B., Lamkanfi M., and Kanneganti T. D. (2010) The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity 32, 379–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Allen I. C., TeKippe E. M., Woodford R. M., Uronis J. M., Holl E. K., Rogers A. B., Herfarth H. H., Jobin C., and Ting J. P. (2010) The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J. Exp. Med. 207, 1045–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bauer C., Duewell P., Mayer C., Lehr H. A., Fitzgerald K. A., Dauer M., Tschopp J., Endres S., Latz E., and Schnurr M. (2010) Colitis induced in mice with dextran sulfate sodium (DSS) is mediated by the NLRP3 inflammasome. Gut 59, 1192–1199 [DOI] [PubMed] [Google Scholar]
- 72. Siegmund B., Lehr H. A., Fantuzzi G., and Dinarello C. A. (2001) IL-1 β-converting enzyme (caspase-1) in intestinal inflammation. Proc. Natl. Acad. Sci. U.S.A. 98, 13249–13254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bauer C., Duewell P., Lehr H. A., Endres S., and Schnurr M. (2012) Protective and aggravating effects of Nlrp3 inflammasome activation in IBD models: influence of genetic and environmental factors. Dig. Dis. 30, 82–90 [DOI] [PubMed] [Google Scholar]
- 74. Patial S., Curtis A. D. 2nd, Lai W. S., Stumpo D. J., Hill G. D., Flake G. P., Mannie M. D., and Blackshear P. J. (2016) Enhanced stability of tristetraprolin mRNA protects mice against immune-mediated inflammatory pathologies. Proc. Natl. Acad. Sci. U.S.A. 113, 1865–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Vande Walle L., Van Opdenbosch N., Jacques P., Fossoul A., Verheugen E., Vogel P., Beyaert R., Elewaut D., Kanneganti T. D., van Loo G., and Lamkanfi M. (2014) Negative regulation of the NLRP3 inflammasome by A20 protects against arthritis. Nature 512, 69–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Jin C., Frayssinet P., Pelker R., Cwirka D., Hu B., Vignery A., Eisenbarth S. C., and Flavell R. A. (2011) NLRP3 inflammasome plays a critical role in the pathogenesis of hydroxyapatite-associated arthropathy. Proc. Natl. Acad. Sci. U.S.A. 108, 14867–14872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. King E. M., Kaur M., Gong W., Rider C. F., Holden N. S., and Newton R. (2009) Regulation of tristetraprolin expression by interleukin-1β and dexamethasone in human pulmonary epithelial cells: roles for nuclear factor-κB and p38 mitogen-activated protein kinase. J. Pharmacol. Exp. Ther. 330, 575–585 [DOI] [PubMed] [Google Scholar]
- 78. ENCODE Project Consortium (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Roadmap Epigenomics Consordium, Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M. J., Amin V., Whitaker J. W., Schultz M. D., Ward L. D., et al. (2015) Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. GTEx Consortium (2013) The Genotype-Tissue Expression (GTEx) project. Nature Genet. 45, 580–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., Zahler A. M., and Haussler D. (2002) The human genome browser at UCSC. Genome Res. 12, 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Stoecklin G., Stubbs T., Kedersha N., Wax S., Rigby W. F., Blackwell T. K., and Anderson P. (2004) MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 23, 1313–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.