Abstract
Arginine methylation is a common posttranslational modification that has been shown to regulate both gene expression and extranuclear signaling events. We recently reported defects in protein arginine methyltransferase 1 (PRMT1) activity and arginine methylation in the livers of cirrhosis patients with a history of recurrent infections. To examine the role of PRMT1 in innate immune responses in vivo, we created a cell type-specific knock-out mouse model. We showed that myeloid-specific PRMT1 knock-out mice demonstrate higher proinflammatory cytokine production and a lower survival rate after cecal ligation and puncture. We found that this defect is because of defective peroxisome proliferator-activated receptor γ (PPARγ)-dependent M2 macrophage differentiation. PPARγ is one of the key transcription factors regulating macrophage polarization toward a more anti-inflammatory and pro-resolving phenotype. We found that PRMT1 knock-out macrophages failed to up-regulate PPARγ expression in response to IL4 treatment resulting in 4-fold lower PPARγ expression in knock-out cells than in wild-type cells. Detailed study of the mechanism revealed that PRMT1 regulates PPARγ gene expression through histone H4R3me2a methylation at the PPARγ promoter. Supplementing with PPARγ agonists rosiglitazone and GW1929 was sufficient to restore M2 differentiation in vivo and in vitro and abrogated the difference in survival between wild-type and PRMT1 knock-out mice. Taken together these data suggest that PRMT1-dependent regulation of macrophage PPARγ expression contributes to the infection susceptibility in PRMT1 knock-out mice.
Keywords: differentiation, innate immunity, macrophage, monocyte, sepsis, arginine methylation, cecal ligation and puncture, histone arginine methylation
Introduction
Protein arginine methylation is a common posttranslational modification that plays a role in multiple pathways, including cell cycle control, RNA processing, and DNA replication (1). Protein arginine methyltransferase 1 (PRMT1)2 is responsible for about 85% of total cellular arginine methylation (2). It methylates both histone and non-histone proteins; however, many protein targets are not yet defined (3). Abnormal function of PRMT1 is closely associated with several types of cancer and cardiovascular disease. Arginine methylation affects gene transcription and splicing as well as upstream signal transduction including a number of innate immunity pathways (4).
Peroxisome proliferator-activated receptor γ (PPARγ) has an important role in many biological processes, including fatty acid synthesis, glucose metabolism, adipogenesis, and inflammatory signaling (5–7). PPARγ-mediated macrophage polarization plays a critical role in all stages of the resolution phase of inflammation. PPARγ can shift production from pro- to anti-inflammatory mediators. PPARγ alters macrophage trafficking, increases efferocytosis and phagocytosis, and promotes alternative M2 macrophage activation (8). Mice with a macrophage-specific deletion of PPARγ have shown impairment in the maturation of alternatively activated M2 macrophages (9).
In this study we examined the role of PRMT1 in immune responses using a cell type-specific knock-out mouse model. We found that myeloid-specific PRMT1 knock-out mice have lower survival rates in a cecal ligation and puncture infection model. This defect is largely because of a defect in expression of the PPARγ transcription factor and from the resulting defective M2 differentiation. Supplementing mice with PPARγ agonists was sufficient to abrogate the difference in survival and proinflammatory cytokine production between wild-type and PRMT1 knock-out mice.
Results
Myeloid-specific PRMT1 knock-out mice are more susceptible to infection
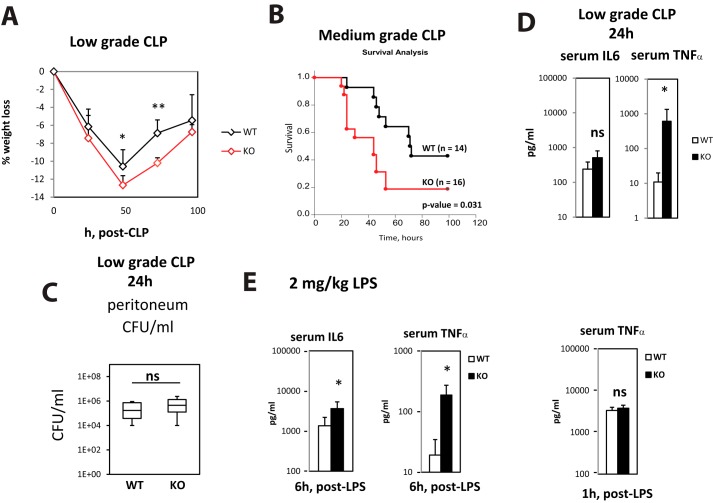
In our previous work we found that PRMT1 can control innate immune responses through regulation of TRAF6 (10). To study the relevance of this mechanism and other PRMT1-dependent mechanisms described by others (11) in vivo, we created myeloid cell-specific PRMT1 knock-out mice, PRMT1fl/fl LysM-Cre (KO), which results in specific deletion in about 40% of monocytes and 90–100% of macrophages (12, 13). We compared their response to wild-type PRMT1fl/fl littermates (WT) in the cecal ligation and puncture (CLP) infection model (14). We used low-grade CLP with ligation of one-third of the cecum to study cytokine production and gene expression (100% of mice survived at 24 h), and medium grade with ligation of one-half of the cecum for survival studies. Fig. 1 shows that knock-out mice suffered more severe weight loss (Fig. 1A) and had a significantly lower survival rate (Fig. 1B). We found no difference in bacterial loads in the peritoneum or blood in those mice (Fig. 1C), but knock-out mice had higher serum TNFα levels 24 h after CLP (Fig. 1D), suggesting that a difference in inflammatory response and not in bacterial clearance was primarily responsible for the survival difference. To further assess differences in inflammatory response, we injected WT and KO mice with an intraperitoneal dose of LPS in the absence of CLP. After a low dose (2 mg/kg) of LPS, knock-out mice had 3- to 4-fold higher levels of serum IL6 and 10-fold higher levels of serum TNFα 6 h after LPS (Fig. 1E). However, at an earlier time point (1 h post LPS), serum TNFα levels were the same, suggesting that PRMT1 knock-out mice do not have defective rapid early responses but may lack an anti-inflammatory mechanism that suppresses cytokine production at later stages of infection.
Figure 1.
Myeloid-specific PRMT1 knock-out mice are more susceptible to infection. A and B, relative weight loss (n = 4–6 per group) (A) and survival (n = 14–16 per group) (B) after cecal ligation and puncture (CLP) procedure in wild-type (WT = PRMT1fl/fl) and myeloid-specific PRMT1 knock-out mice littermates (KO = PRMT1fl/fl LysM-Cre/WT). Data are presented as mean ± S.D. *, p < 0.05, **, p < 0.01. C, bacterial load in peritoneum of wild-type or knock-out mice after 24 h of CLP. D, serum cytokine levels 24 h after CLP measured by ELISA. n = 3–5 per group. Data are presented as mean ± S.D. *, p < 0.05. E, wild-type (WT) and myeloid-specific PRMT1 knock-out mice (KO) were injected intraperitoneally with 2 mg/kg of LPS. Serum cytokine levels in control mice were measured at 6 h or 1 h post LPS. Data are presented as mean ± S.D. n = 4–6 per group. *, p < 0.05.
PRMT1 knock-out macrophages have a defect in IL4-induced differentiation
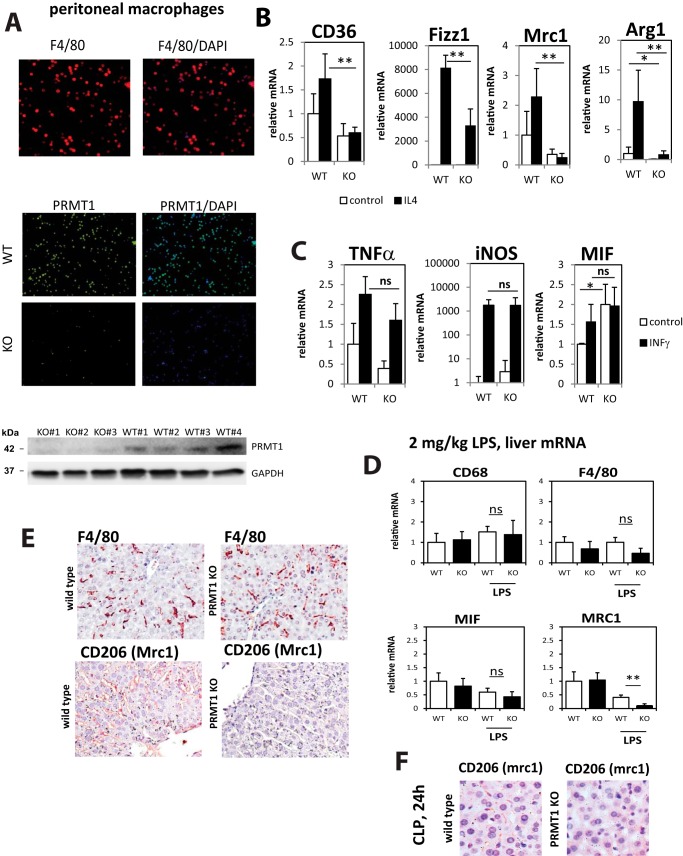
Next we studied the phenotype of wild-type and knock-out macrophages in vitro using peritoneal macrophages isolated from untreated mice. We confirmed that 95 ± 3% of those cells express the F4/80 macrophage marker (Fig. 2A). LysM-Cre expression resulted in PRMT1 deletion in the majority of the cells with only 17 ± 5% of cells expressing PRMT1 (Fig. 2B). We examined the ability of these macrophages to differentiate along the alternative IL4-driven pathway (M2 differentiation) by examining mRNA levels strongly associated with specific macrophage phenotypes. We found that PRMT1 knock-out macrophages are deficient in IL4-induced differentiation as evidenced by failure to up-regulate Arg1, CD36, Mrc1, and Fizz1 in response to IL4 (Fig. 2B). There was no difference between wild type and KO in the ability of INFγ (interferon γ) to induce expression of mRNAs associated with classic (M1) macrophage activation (Fig. 2C).
Figure 2.
PRMT1 knock-out macrophages have a defect in IL4-induced differentiation. A, representative immunofluorescence images of F4/80 staining (red) and PRMT1 staining (green) in wild-type (PRMT1fl/fl) or knock-out (PRMT1fl/fl LysM-Cre) peritoneal macrophages isolated from untreated mice. Western blotting analysis of PRMT1 expression in macrophages is shown at the bottom. B and C, peritoneal macrophages were isolated from wild-type and myeloid-specific PRMT1 knock-out mice. Relative mRNA levels in macrophages differentiated with IL4 (B) for 24 h or relative mRNA levels in macrophages differentiated with INFγ (C) for 24 h are presented as mean ± S.D. n = 4–8 per group, *, p < 0.05, **, p < 0.01 compared with WT. D, wild-type (WT) and myeloid-specific PRMT1 knock-out mice (KO) were injected intraperitoneally with 2 mg/kg of LPS. Relative liver mRNA levels at 24 h post LPS are presented as mean ± S.D. **, p < 0.01 compared with WT, n = 4–6 per group. E, representative images of immunohistochemical staining for total macrophages (F4/80) and anti-inflammatory macrophages (Mrc1) in the livers of wild-type and myeloid-specific PRMT1 knock-out mice at 24 h post LPS. F, representative images of immunohistochemical staining for Mrc1 in the livers of wild-type and myeloid-specific PRMT1 knock-out mice at 24 h post CLP.
We further investigated the phenotype of liver macrophages in vivo after low dose (2 mg/kg) intraperitoneal LPS injection. Fig. 2D shows that a number of macrophage-specific markers were unchanged in the livers; the M2 differentiation marker Mrc1 (CD206) expression was significantly lower in PRMT1 myeloid KO mice at 24 h post LPS. This suggests lack of M2 differentiation. Immunohistochemical staining of liver sections 24 h post LPS confirmed that although the number of macrophages is the same in wild-type and knock-out mice (F4/80 staining), Mrc1 expression is not detectable in sections from PRMT1 KO mice (Fig. 2E). Similar results were seen in macrophage staining 24 h post CLP (Fig. 2F).
The defect in IL4-induced differentiation in PRMT1 knock-out macrophages results in higher cytokine production after LPS challenge
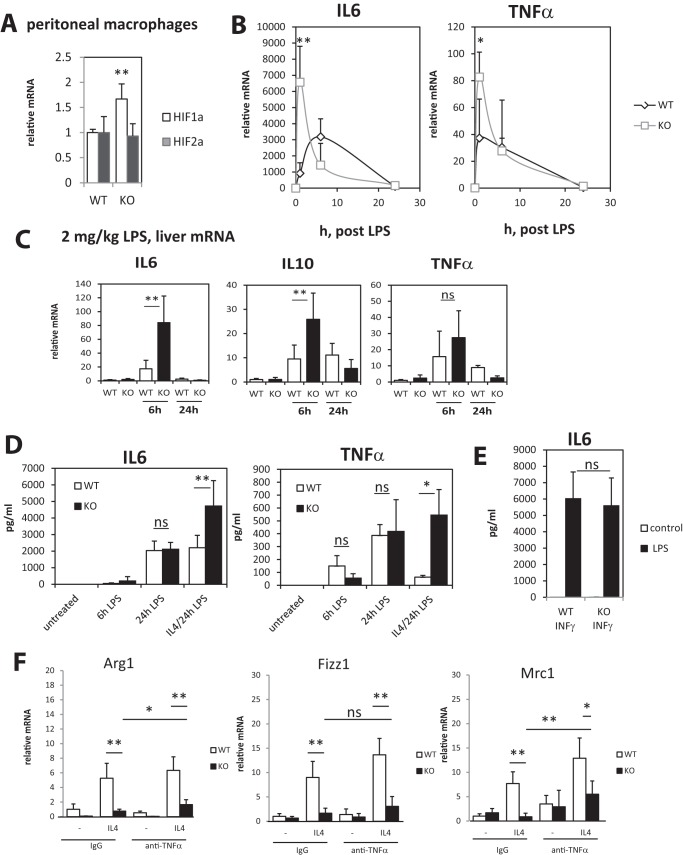
The above data suggest that PRMT1 deficiency results in a defect of M2 differentiation, without altering INFγ-induced macrophage differentiation. Additionally some of the M2 and M1 markers were differentially expressed even in unstimulated cells (Fig. 2, A and B), suggesting that knock-out macrophages are closer to M1 macrophages than are wild-type macrophages. In agreement with that finding we detected elevated levels of Hif1α, a key transcription factor for M1 macrophages, in KO peritoneal macrophages (Fig. 3A). Taken together these data suggest that KO cells might be hyperresponsive to LPS challenge, which could explain higher levels of cytokines after CLP. To test this possibility we treated isolated peritoneal macrophages with LPS for various times (Fig. 3B). We found that KO macrophages produce higher mRNA levels of IL6 and TNFα at 1 h post LPS but at 6 and 24 h the levels were lower or not different compared with wild type (Fig. 3B). A similar time course difference was observed in liver IL6, IL10, and TNFα mRNA levels after low-dose intraperitoneal LPS challenge with higher early cytokine production and lower levels at later time points (Fig. 3C). Next we examined how the differences in the mRNA LPS responses translate to secreted cytokine level, which are related to an integrated value of cytokine production over time. We found that there was no significant difference in secreted cytokine levels between wild-type and knock-out macrophages (Fig. 3D). These data suggested that the more rapid LPS response alone does not explain higher levels of cytokines after CLP.
Figure 3.
Defect in IL4-induced differentiation in PRMT1 knock-out macrophages results in higher cytokine production after LPS challenge. Peritoneal macrophages were isolated from wild-type and myeloid-specific PRMT1 knock-out mice. A, relative mRNA of Hif1α and Hif2α in wild-type and PRMT1 knock-out macrophages are presented as mean ± S.D. n = 4, **, p < 0.01. B, relative cytokine mRNA in macrophages treated with100 ng/ml of LPS for indicated times. Data are presented as mean ± S.D. n = 4–6, *, p < 0.05, **, p < 0.01. C, wild-type (WT) and myeloid-specific PRMT1 knock-out mice (KO) were injected intraperitoneally with 2 mg/kg of LPS. Relative liver mRNA levels at 6 and 24 h post LPS are presented as mean ± S.D. **, p < 0.01 compared with WT, n = 4–6 per group. D and E, peritoneal macrophages were isolated from wild-type and myeloid-specific PRMT1 knock-out mice. Secreted cytokine levels in macrophages not differentiated or differentiated with IL4 or INFγ for 24 h and then treated with 100 ng/ml of LPS for indicated times are presented as mean ± S.D. *, p < 0.05, **, p < 0.01 compared with WT, n = 4–6. F, relative mRNA levels in wild type of PRMT1 knock-out macrophages differentiated with IL4 in the presence of anti-TNFα neutralizing antibody or control IgG (1 μg/ml). Data are presented as mean ± S.D. n = 4, *, p < 0.05, **, p < 0.01.
Next we examined LPS responses under M1 or M2 differentiation conditions. IL4-mediated differentiation of macrophages is known to suppress cytokine production in response to LPS challenge. It is an important step in infection clearance to avoid excessive cytokine production. We found that KO macrophages produced higher levels of IL6 and TNFα when differentiated with IL4 prior to LPS stimulation (Fig. 3D). These data were similar to in vivo data, as in Fig. 1, D and E. In contrast, we did not see any differences in IL6 production when cells were differentiated with INFγ to produce M1 macrophages (Fig. 3E).
Monocyte to macrophage differentiation induces PRMT1 expression and PRMT1 dependent histone arginine methylation
Next we examined the mechanism for the PRMT1 dependence of M2 differentiation. One possible mechanism is that it could be a result of the observed increase in TNFα. TNFα is a potent suppressor of M2 differentiation (15, 16). To address the role of TNFα in the M2 defect seen in PRMT1 knock-out macrophages, we used an in vitro neutralization assay (Fig. 3F). We found that although there was a small increase in M2 differentiation after addition of TNF antibody, the effect of PRMT1 knock out was much greater than that accounted for by TNFα. Thus, PRMT1 can regulate M2 differentiation in a TNFα-independent manner.
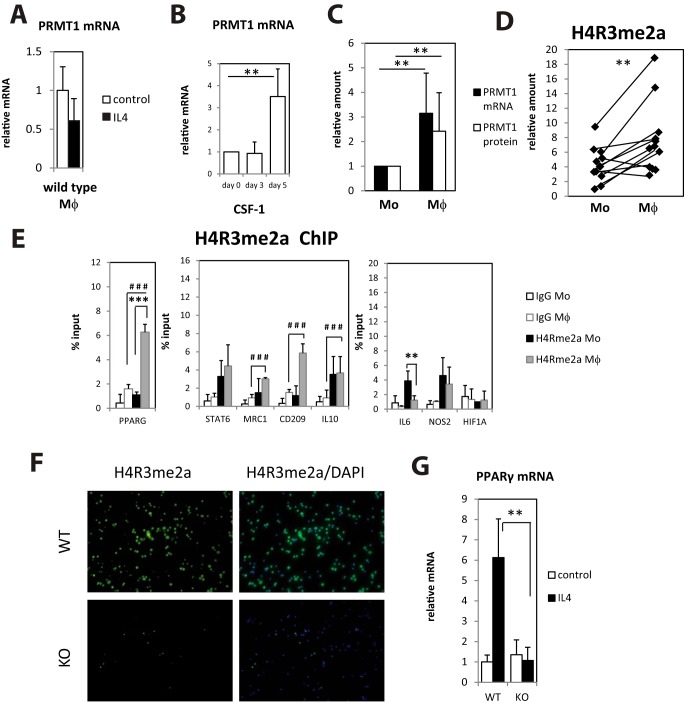
To better define the step at which PRMT1 regulates macrophage phenotype, we first determined if PRMT1 is induced at some stage of macrophage differentiation. IL4 treatment of wild-type peritoneal macrophages did not result in any changes of PRMT1 expression (Fig. 4A). In contrast, we found that PRMT1 expression is induced during monocyte to macrophage differentiation. Fig. 4B shows relative PRMT1 levels in human blood monocytes isolated from control subjects and subsequently differentiated in vitro with CSF-1 (colony-stimulating factor 1) for 5 days. To determine whether PRMT1 also increases after in vivo monocyte differentiation we directly compared blood monocytes and peritoneal macrophages from patients with liver cirrhosis and ascites; a similar increase was observed when comparing PRMT1 expression in monocytes and macrophages from the same patients (Fig. 4C). The difference in PRMT1 expression between macrophages and monocytes derived from the same patient correlated with the level of the PRMT1-specific histone methylation mark H4R3me2a (Fig. 4D). We further examined the presence of these methylated histone marks by chromatin immunoprecipitation at the promoters of genes relevant to M2 macrophage activation in both monocytes and in vitro differentiated monocyte-derived macrophages from healthy individuals (Fig. 4E). PPARγ, CD209, Mrc1, and IL10 promoter methylations in macrophages were significantly above background (IgG), but only PPARγ promoter methylation was significantly increased during differentiation from monocyte to macrophage (Fig. 4E). PPARγ is a key transcription factor necessary for M2 differentiation, and its expression is induced by IL4, likely in a STAT6 dependent manner (17–19). We hypothesized that promoter histone arginine methylation can regulate PPARγ expression in IL4-differentiated cells. We confirmed that PRMT1 knock-out macrophages have dramatically lower levels of the PRMT1-specific histone methylation mark H4R3me2a (Fig. 4F), and we found that PRMT1 knock-out macrophages completely failed to induce PPARγ expression in response to IL4 (Fig. 4G).
Figure 4.
Monocyte to macrophage differentiation induces PRMT1 expression and PRMT1-dependent histone arginine methylation. A, relative mRNA levels of PRMT1 in wild-type macrophages differentiated with IL4 for 24 h. n = 4–6. Data are presented as mean ± S.D. B, relative mRNA levels of PRMT1 in human blood monocytes differentiated with CSF-1 for indicated time. n = 4. Data are presented as mean ± S.D. **, p < 0.01 compared with day 0. C, relative mRNA and protein levels of PRMT1 in blood monocytes and corresponding peritoneal macrophages from the same patients with ascites (n = 13). Data are presented as mean ± S.D. **, p < 0.01. D, relative H4R3me2a levels in blood monocytes and corresponding peritoneal macrophages from patients with ascites (n = 12). **, p < 0.01 by paired t test. E, chromatin immunoprecipitation (ChIP) using H4R3me2a specific antibodies from healthy individuals' blood monocytes and corresponding monocyte-derived macrophages. Each individual's cells were analyzed separately. Data are presented as mean ± S.D. **, p < 0.01, ***, p < 0.001 between paired monocytes and macrophages, ###, p < 0.001 for H4R3me2a pulldown signal in macrophages compared with IgG. F, representative immunofluorescence images of H4R3me2a staining (green) in wild-type (PRMT1fl/fl) of knock-out (PRMT1fl/fl LysM-Cre) peritoneal macrophages isolated from untreated mice. G, relative mRNA levels of PPARγ in wild-type or PRMT1 knock-out macrophages differentiated with IL4 for 24 h, **, p < 0.01. n = 4. Data are presented as mean ± S.D.
The PRMT1 effect on M2 differentiation is PPARγ dependent
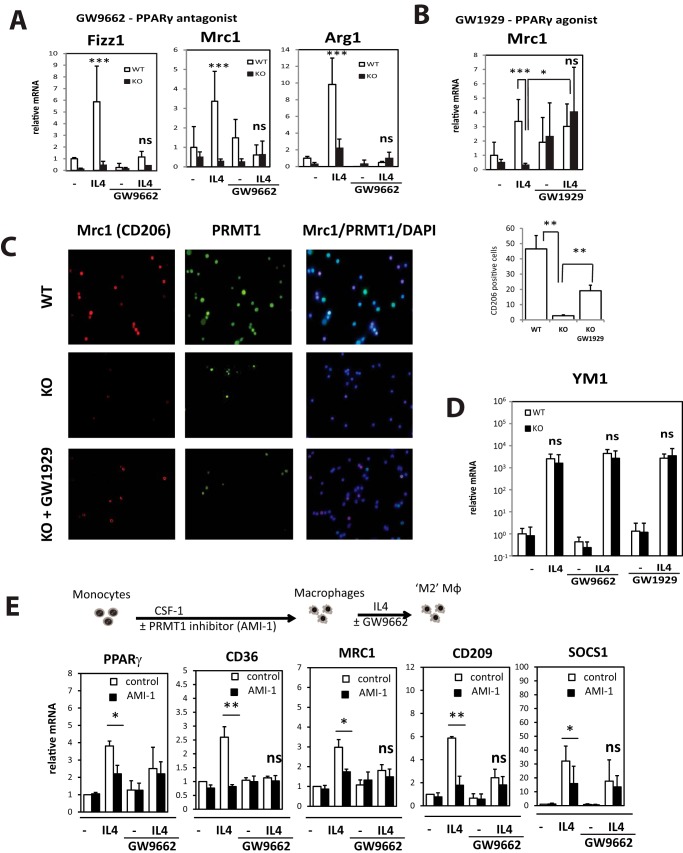
To examine whether a defect in alternative macrophage differentiation can be explained by this loss of PPARγ induction, we isolated peritoneal macrophages from wild-type and myeloid PRMT1 knock-out mice and treated them with IL4 in the presence or absence of the PPARγ agonist GW1929 or the PPARγ antagonist GW9662 (Fig. 5, A and B). Antagonist treatment prevented IL4-induced up-regulation of Mrc1, Fizz1, and Arg1 in wild-type macrophages (Fig. 5A). Thus treatment with PPARγ antagonist resulted in defect in IL4 dependent signaling in wild-type macrophages that made them similar to knock-out macrophages. PPARγ agonist, on the other hand, significantly induced expression of Mrc1 in the presence of IL4 in knock-out macrophages, suggesting that activation of PPARγ can be sufficient to prevent the defect caused by reduced expression of PRMT1 (Fig. 5, B and C). We observed only partial rescue of the number of Mrc1-positive macrophage in the presence of the agonist, possibly because of either lower levels of PPARγ in the knock-out cells or other PPARγ-independent mechanisms. Interestingly, another M2 differentiation marker, Ym1 expression, was not affected by PRMT1 knock-out and was also not changed by PPARγ manipulation (Fig. 5D).
Figure 5.
Defective differentiation of PRMT1 knock-out macrophages is PPARγ dependent. A and B, relative mRNA levels in wild-type or PRMT1 knock-out macrophages differentiated with IL4 for 24 h in the presence or absence of PPARγ antagonist GW9662 (1 μm) (A) or PPARγ agonist GW1929 (1 μm) (B). n ≥ 3. Data are presented as mean ± S.D. ***, p < 0.001 compared with wild type, *, p < 0.05 compared with DMSO control. C, representative immunofluorescence images of Mrc1 (red) and PRMT1 staining (green) in wild type of knock-out peritoneal macrophages differentiated with IL4 for 24 h in the presence or absence of PPARγ agonist GW1929 (1 μm). Diagram shows average percentage of Mrc1-positive cells calculated from 10 random fields from each of n = 3 mice per condition **, p < 0.01. D, relative Ym1 mRNA levels in cells treated as in A and B. E, human blood monocytes were differentiated with CSF-1 for 5 days in the presence or absence of AMI-1. Treatment was removed and after 24 h cells were further differentiated with IL4 for 24 h. Bar graphs compare relative mRNA levels in macrophages differentiated with IL4 for 24 h in the presence or absence of AMI-1 and PPARγ antagonist GW9662 (1 μm). Data are presented as mean ± S.D. n = 3. *, p < 0.05, **, p < 0.01 compared with untreated.
Next we examined whether only histone arginine methylation induced during monocyte to macrophage differentiation might be important for PRMT1-dependent Mrc1 and other gene expression. To investigate if transient inhibition of PRMT1 activity during monocyte to macrophage differentiation can prevent alternative activation as seen in the PRMT1 knock-out phenotype, we isolated blood monocytes from healthy individuals and differentiated them with CSF-1 in the presence or absence of the PRMT1 inhibitor AMI-1. AMI-1 was present only during the differentiation step and was removed prior to further manipulations. Next we assessed the expression of macrophage genes after IL4-induced differentiation. We found that macrophages that were treated with AMI-1 during differentiation are deficient in PPARγ-dependent gene expression induced by IL4 (Fig. 5E). When PPARγ antagonist was present, there was no longer any effect of AMI-1 on expression of the M2-related genes. This confirms that the effects of PRMT1 on M2 differentiation are largely mediated through PPARγ and only important during the monocyte to macrophage differentiation step (Fig. 5D).
Rosiglitazone treatment is sufficient to abrogate differences in survival and inflammatory cytokine production between wild-type and PRMT1 knock-out mice
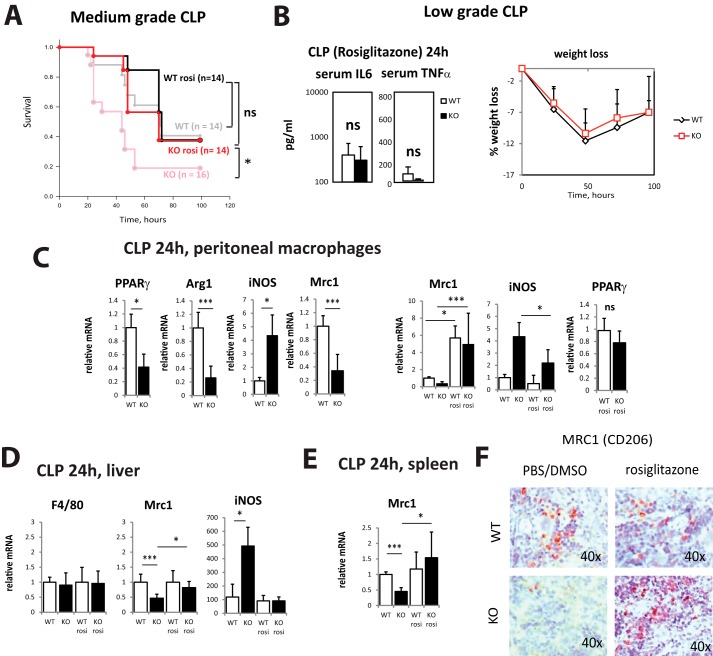
The in vitro data presented in Fig. 5B suggested that PPARγ agonists can efficiently restore M2 differentiation in PRMT1 knock-out macrophages. Thus we examined whether treatment with PPARγ agonist in vivo can reduce the difference in infection susceptibility between wild-type and knock-out mice. We injected 1 mg/kg rosiglitazone i.p. in 10% DMSO/PBS 2 h before cecal ligation and puncture (Fig. 6) and analyzed survival rate in the medium-grade CLP model. In wild-type animals, rosiglitazone improved survival slightly at 72 h, but overall survival was not different. In PRMT1 knock-out mice, rosiglitazone improved the survival to the level of wild-type untreated animals (Fig. 6A) suggesting that PPARγ activation abolishes the difference in infection susceptibility between WT and KO animals (Fig. 1, A–C). Rosiglitazone-treated knock-out mice showed similar weight loss and serum cytokines levels as wild-type untreated mice (Fig. 6B; compare with Fig. 1D).
Figure 6.
Rosiglitazone restores M2 differentiation in PRMT1 knock-out mice. Wild-type or myeloid-specific PRMT1 knock-out mice were injected with 1 mg/kg of rosiglitazone or equal volume of 10% DMSO in PBS i.p. 2 h before performing cecal ligation and puncture. A, survival (n = 14–16 per group) after CLP in mice receiving rosiglitazone or no rosiglitazone group (gray and light red lines represent the same data from Fig. 1). *, p < 0.05. B, serum cytokine levels (left) 24 h after CLP (n = 4) and relative weight loss (right, n = 4) after cecal ligation and puncture in mice receiving 1 mg/kg of rosiglitazone. Data are presented as mean ± S.D. C–E, relative mRNA levels in peritoneal macrophages (C), liver (D), and spleen (E) 24 h post CLP. Data are presented as mean ± S.D. n = 3 per group. *, p < 0.05, ***, p < 0.001 compared with wild type. F, representative images of immunohistochemical staining of anti-inflammatory macrophages (Mrc1) in the spleen of wild-type and myeloid-specific PRMT1 knock-out mice at 24 h post CLP.
Next we confirmed that rosiglitazone treatment resulted in a restored M2 differentiation phenotype in vivo. We injected 1 mg/kg rosiglitazone or 10% DMSO/PBS (vehicle) 2 h before cecal ligation and puncture and analyzed peritoneal macrophages 24 h later (Fig. 6C). In the control group, knock-out macrophages showed reduced expression of PPARγ and its targets Mrc1 and Arg1, as well as an increase in expression of iNOS. These differences were reduced or abolished in the animals that received rosiglitazone (Fig. 6C). PPARγ expression itself was also partially restored in the knock-out animals. These data are in accordance with previously published data on the effect of rosiglitazone in vivo (20, 21). Similar results were observed in liver and spleen (Fig. 6, D and E). Additionally, we examined the presence of M2 macrophages in the spleen by immunohistochemistry in wild-type and knock-out mice that received rosiglitazone or vehicle control (Fig. 6F). We found that in the control group, knock-out macrophages lacked expression of the alternative macrophage marker Mrc1 at 24 h after CLP. However, in the rosiglitazone-treated group we detected a similar number of alternatively activated macrophages (Mrc1 positive) in the spleen of wild-type and PRMT1 knock-out mice (Fig. 6F).
Discussion
Arginine methylation has emerged as an important regulator of the immune response (4, 10, 11). The work presented in this study aimed to define the role of PRMT1 in myeloid cell functions.
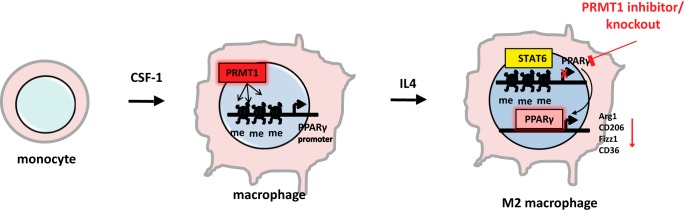
We found that PRMT1 is induced during monocyte to macrophage differentiation. This induction results in the deposition of the PRMT1-dependent histone mark H4R3me2a on the PPARγ promoter. PRMT1 deficiency results in diminished IL4-induced PPARγ expression and lack of PPARγ-dependent M2 differentiation (Fig. 7).
Figure 7.
Model of PRMT1-dependent regulation. PRMT1 is induced during monocyte to macrophage differentiation. This induction results in the deposition of PRMT1-dependent histone marks H4R3me2a on the PPARγ promoter. PRMT1 deficiency results in loss of this modification and a consequent inability of IL4 to induce PPARγ expression. This results in the lack of PPARγ-dependent expression of Arg1, CD206, CD36, Fizz1, and other genes necessary for M2 differentiation.
In addition to its well known role as a regulator of lipid metabolism, PPARγ is also critical for alternative activation of macrophages (5–7). Loss of PPARγ blocks M2 differentiation associated with the resolution phase of inflammation (9). We showed that PRMT1 deficiency in peritoneal macrophages prevented IL4 driven M2 differentiation and suppression of production of proinflammatory cytokines such as TNFα. Using an in vivo model, we found that myeloid PRMT1 deficiency led to lower survival after cecal ligation and puncture (CLP). Polymicrobial sepsis induced by cecal ligation and puncture is a frequently used model as its progression closely resembles the progression and characteristics of human sepsis (14, 22). We found that differences in survival observed in knock-out mice are because of the lack of PPARγ-dependent M2 macrophage differentiation. Enhancing PPARγ activity with agonists was sufficient to abrogate the difference between wild-type and PRMT1 knock-out mice.
We used a LysM-Cre-mediated knock-out strategy which allows achieving a high level of the knock out in the majority of macrophage populations. However, studies have also reported that LysM-Cre affects a subset of about 50% of neutrophils, which were not investigated in this study (12). Neutrophils are important for bacterial clearance but dispensable for survival in the CLP model (23), thus we focused our study on macrophages where we found over 80% knock-out efficiency. Other cell types that might be affected by LysM-Cre-mediated knock out include early bone marrow progenitors and neurons (24, 25) which, in addition to macrophages, might also contribute to the phenotype of the knock-out mice in vivo.
PRMT1 regulates a large number of genes, and its ability to influence immune function may result from other mechanisms as well. We previously identified TRAF6 as a PRMT1 target and NF-κB has been shown to be regulated as well (10, 11). However, in this work we showed that genetic ablation of PRMT1 in myeloid cells did not appear to have the predicted effect of up-regulating TRAF6-dependent NF-κB signaling. This is evidenced by lack of up-regulation of IRAK3 and A20 in knock-out cells (data not shown). This may reflect the existence of other compensatory pathways in these constitutive knock-out animals. Our results do, however, show a profound effect on PPARγ that appears to dominate in vivo over other mechanisms. Possibly in another infection/inflammation model other PRMT1 targets play an important role as well. This issue will require further investigation.
Interestingly, we found that PPARγ expression is different in wild-type and PRMT1 knock-out macrophages only after stimulation with IL4, as well as in later stages of infection clearance when IL4-dependent M2 differentiation is induced. However in the absence of infection, expression levels were similar. We hypothesized that PRMT1-dependent histone modification of the PPARγ promoter might be contributing to induction PPARγ after IL4 stimulation but not at baseline.
Histone arginine methylation is a less well studied part of the “histone code” that directs differentiation and function of many cell types (1, 4). Recent findings that PRMT1-dependent arginine methylation is a dynamic modification have opened a new field of PRMT1-regulated signaling and differentiation events (10, 26, 27). Generally, it is accepted that PRMT1-induced histone modifications promote gene expression (28–30), whereas demethylation by JMJD6 suppresses gene expression (26, 31). Recent studies showed that the H4R3me2a mark is associated with progression of neuronal differentiation (32, 33). Here we show that H4R3me2a is induced during monocyte to macrophage differentiation. Further studies will be necessary to define other histone targets of PRMT1 that can affect macrophage functions.
We observed previously that patients with decompensated cirrhosis have low PRMT1 levels in hepatic macrophages and circulating monocytes (34). Immune dysfunction is a prominent feature of decompensated cirrhosis and contributes to ∼50% of cirrhotic deaths (35). It is known to be associated with susceptibility to bacterial infection, immune paresis, and monocyte dysfunction, but the mechanisms of these widespread immune deficiencies are not well understood (36, 37). Data presented here suggest that PRMT1 deficiency can contribute to immune dysfunction through the failure of M2 differentiation. Although PPARγ agonists have also been shown to have negative impact on sepsis survival in certain conditions, it is believed that PPARγ activation is beneficial during inflammation and sepsis via its anti-inflammatory properties (8, 38–40). We suspect that PPARγ agonists might be promising as a part of a treatment strategy to diminish the inflammatory dysfunction in patients with decompensated cirrhosis.
Materials and methods
Mice
C57BL/6NTac-Prmt1tm1a(EUCOMM)Wtsi/WtsiCnbc mice were obtained from EUCOMM (EUCOMM project, 40181) and bred with Flp recombinase mice to get homozygous Prmt1 floxed breeders. These mice were next crossed with LysM-Cre mice to generate mice lacking Prmt1 in myeloid cells. For experiments, PRMT1fl/fl Cre/WT mice were used together with PRMTfl/fl WT/WT littermates as a control.
All mice were housed in a temperature-controlled, specific pathogen-free environment with 12-h light-dark cycles and fed regular mouse chow and water ad libitum. All animal handling procedures were approved by the Institutional Animal Care and Use Committees at the University of Kansas Medical Center.
Isolation of mouse peritoneal macrophages
Primary peritoneal macrophages were isolated as described previously (41). 8- to 10-week-old mice were killed by CO2 asphyxiation. Briefly, 10 ml of sterile PBS were injected into the caudal half of the peritoneal cavity using a 25-gauge needle (beveled side up), following by gentle shaking of the entire body for 10 s. Saline containing resident peritoneal cells was collected and cells were plated on uncoated tissue culture plates and incubated for 60 min at 37 °C. Nonadherent cells were removed by washing five times with warm PBS. Macrophages were maintained in RPMI medium (Invitrogen) containing 10% FBS.
Cecal ligation and puncture
Cecal ligation and puncture (CLP) was used to produce sepsis and was performed as described previously (42). Male and female 8- to 10-week-old mice were anesthetized with isoflurane (2–3% in O2 with 1.5% maintenance). An abdominal incision was made and the cecum was mobilized. The distal one-third (for low-grade CLP) or one-half of the cecum (for medium-grade CLP) was ligated and punctured through and through (i.e. 2 holes) with a 23-gauge needle, and a small amount of cecal material was expressed before the cecum was replaced, and the incision was closed with 4–0 surgical suture. Following surgery, all animals received a subcutaneous injection of warm sterile 0.9% saline (1 ml) and buprenorphine (sustained release, 0.2 mg/kg, subcutaneously) every 72 h. All mice were individually housed following surgery and given access to water until euthanasia. Mice were monitored for 96–100 h post surgery. All animals were monitored every 12 h for signs of illness or distress including physical appearance, weight loss (over 20%), inactivity, and behavioral responses to external stimuli. Any animals deemed moribund were euthanized. Alternatively, 24 h after the induction of sepsis, mice were euthanized and peritoneal macrophages, liver, spleen, and lymph nodes were collected. Blood was collected from the vena cava in heparinized syringes and centrifuged (10,000 × g for 10 min) for isolation of plasma. Both frozen tissue and plasma were stored at −80 °C until analysis.
Bacterial counts
The bacterial counts were determined in peritoneal lavage fluid and blood samples by colony counting. Serial dilutions were plated on blood agar dishes (Hardy Diagnostics, Santa Maria, CA) and incubated for 24 h at 37 °C.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde for 15 min at room temperature, washed with PBS, and permeabilized with 1% Triton X-100 for 15 min then blocked in immunofluorescence buffer (PBS containing 2.5 mm EDTA, 1% BSA) for 1 h. Cells were then incubated with primary antibody, 1:300 in PBS containing 2.5 mm EDTA, 1% BSA, 0.1% Triton X-100 overnight at 4 °C. After washing with PBS, coverslips were incubated with Alexa Fluor-conjugated secondary antibody (1:500) in 0.1 μg/ml DAPI for 1 h in the dark at room temperature. Coverslips were washed and mounted with FluorSave Reagent (Calbiochem). Slides were observed in a Nikon Eclipse 800 upright epifluorescence microscope (Nikon Instruments, Melville, NY). Images were acquired using a Nikon CoolSNAP camera.
Human specimens
De-identified human blood and peritoneal fluid specimens were obtained from the Liver Center Tissue Bank at the University of Kansas Medical Center. All studies using human samples were approved by the Institutional Review Board (Human Subjects Committee) of the University of Kansas Medical Center. Peripheral blood monocytes were isolated from peripheral blood mononuclear cell (PBMC) fractions using MACS beads (human CD14) (130–050-201, Miltenyi Biotec) according to manufacturer's instructions.
Western blots
Protein extracts (15 μg) were subjected to 10% SDS-PAGE, electrophoretically transferred to nitrocellulose membranes (Amersham Biosciences Hybond ECL, GE Healthcare), and blocked in 3% BSA/PBS at room temperature for 1 h. Primary antibodies were incubated overnight at manufacturer's recommended concentrations. Immunoblots were detected with the ECL Plus Western Blotting Detection System (Amersham Biosciences) or using near-infrared fluorescence with the ODYSSEY Fc, Dual-Mode Imaging system (Li-COR Biosciences). Expression levels were evaluated by quantification of relative density of each band normalized to that of the corresponding β-actin or GAPDH band density.
ELISA
Enzyme-linked immunosorbent assays (ELISAs) were carried out as follows. Plastic 96-well microtiter plates (Nunc-Immuno Microwell MaxiSorp, Sigma) were coated overnight with excess (0.5 μg) of first primary antibody. Unbound protein was washed with PBS. Wells were blocked for 1 h with 0.3 ml 3% BSA (Cohn Fraction V, essentially fatty acid free; Sigma) in PBS. After washing, samples containing the protein of interest (in 50 μl) were added, incubated for 2 h at room temperature, and then washed with PBS. Second primary antibody (0.1 μg in 50 μl) was added, incubated for 1 h at room temperature, washed, and then visualized by incubation with secondary antibody conjugated with horseradish peroxidase (The Jackson Laboratory) in the presence of 3% BSA for 0.5 h, followed by reaction with ABTS (Sigma). The reaction was allowed to proceed for 20 min and then plates were quantitated spectrophotometrically at 410 nm.
ELISA for human cytokines was performed using IL6 ELISA Ready-SET-Go!® and TNFα ELISA Ready-SET-Go!® from Affymetrix/eBioscience according to the manufacturer's instructions.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed as described before (43). Briefly, THP-1 cells (1.5 × 107) were cross-linked by the addition of 1% formaldehyde for 10 min. Cells were lysed with 10 mm Tris-HCl, pH 8.0, 10 mm NaCl, 3 mm MgCl2, 0.5% Nonidet P-40. Nuclei were collected by centrifugation, resuspended in 1% SDS, 5 mmol/liter EDTA, 50 mmol/liter Tris-HCl, pH 8.0, and sonicated to generate chromatin to an average length of ∼100–500 bp. Next, samples in ChIP dilution buffer (1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl, pH 8.1, 150 mm NaCl), were immunoprecipitated overnight at 4 °C with 4 μg ChIP-grade antibody. 20 μl of magnetic beads (Dynabeads M-280, Invitrogen) were used to purify immunocomplexes. Following purification, cross-links were reverted by incubation at 65 °C for 6 h. Samples were purified with Qiagen kit.
Cell culture
AMI-1 was obtained from EMD Biosciences and used at 10 μm and 50 μm for 16–24 h prior to harvest. Cell were treated where indicated with 5 ng/ml of INFγ or 5 ng/ml IL4 in the presence or absence of PPARγ antagonist GW9662 (1 μm) or PPARγ agonist GW1929 (1 μm) for 24 h.
Whole cell lysates were prepared from cells that had been washed and harvested by centrifugation in PBS, pH 7.5. Cell pellets were resuspended in radioimmune precipitation assay buffer that contained 50 mm Tris, pH 7.5, 150 mm sodium chloride, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1 mm EDTA, and 1% protease and phosphatase inhibitors (Sigma-Aldrich). Lysates were centrifuged at 14,000 × g for 15 min; supernatants were collected and protein concentration was measured using the Bio-Rad protein assay kit.
Antibodies used
Primary antibodies
Anti-PRMT1 (F339), anti-Lamin B (C20), anti-Mrc1, and anti-β-actin were from Santa Cruz Biotechnology. Rabbit anti-PRMT1 antibody (against amino acids 300–361) were from Abcam. Anti-asymmetric-dimethyl-arginine H4R3me2a antibodies were from Active Motif. Anti F4/80 antibody was from Novus. Mouse anti-β-actin and mouse monoclonal anti-PRMT1 antibody clone 171 (against amino acids 1–361) were from Sigma-Aldrich. Anti-GAPDH was from Ambion.
Secondary antibodies
IRDye 800CW goat anti-mouse IgG and IRDye 680RD goat anti-rabbit IgG were from Li-COR. General HRP-conjugated secondary antibodies were from Southern Biotechnology Associates.
Immunohistochemistry
Immunostaining on formalin-fixed sections was performed by deparaffinization and rehydration, followed by antigen retrieval by heating in a pressure cooker (121 °C) for 5 min in 10 mm sodium citrate, pH 6.0. Peroxidase activity was blocked by incubation in 3% hydrogen peroxide for 10 min. Sections were rinsed three times in PBS/PBS-T (0.1% Tween 20) and incubated in Dako Protein Block (Dako, Carpinteria, CA) at room temperature for 1 h. After removal of blocking solution, slides were placed into a humidified chamber and incubated overnight with an antibody, diluted 1:300 in Dako Protein Block at 4 °C. Antigen was detected using the SignalStain Boost IHC Detection Reagent (catalog no. 8114; Cell Signaling Technology, Danvers, MA), developed with diaminobenzidene (Dako), counterstained with hematoxylin (Sigma-Aldrich), and mounted.
Real-time PCR
RNA was extracted using the RNeasy Mini Kit (Qiagen). cDNA was generated using the RNA reverse transcription kit (catalog no. 4368814; Applied Biosystems). Quantitative real-time RT-PCR was performed in a CFX96 Real-Time PCR Detection System (Bio-Rad) using specific sense and antisense primers combined with iQ SYBR Green Supermix (Bio-Rad) for 40 amplification cycles: 5 s at 95 °C, 10 s at 57 °C, 30 s at 72 °C.
Statistics
Results are expressed as mean ± S.D. The Student's t test, paired t test, Pearson's correlation, or one-way analysis of variance (ANOVA) with Bonferroni post hoc test was used for statistical analyses. Survival was compared using Kaplan-Meier curves and log rank test. p value < 0.05 was considered significant.
Author contributions
I. T. and S. W. designed experiments, analyzed data, and wrote the paper. I. T., A. A., and J. Z. performed experiments. J. O., R. T., L. M., B. B., and B. R. provided materials. All authors analyzed the results and approved the final version of the manuscript
Acknowledgments
We thank Drs. Jameson Forster, Sean Kumer, Tim Schmitt, and Bashar Abdulkarim for their assistance in obtaining these specimens and Dr. C. Lang for advice on ligation puncture models. The human specimens used in this study were derived from samples provided by the University of Kansas Liver Center Tissue Bank.
This study was supported by National Institute on Alcoholism and Alcohol Abuse Grant AA012863, COBRE Pilot Grant NCRR/NIH P20 RR021940, and the 2016 Pinnacle Research Award from AASLD. The authors declare that they have no conflicts of interest with the contents of this article.The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- PRMT1
- protein arginine methyltransferase 1
- PPARγ
- peroxisome proliferator-activated receptor γ
- CLP
- cecal ligation and puncture
- ABTS
- 2, 2′-Azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt
- INF
- interferon
- CSF
- colony-stimulating factor.
References
- 1. Bedford M. T., and Richard S. (2005) Arginine methylation: an emerging regulator of protein function. Mol. Cell 18, 263–272 [DOI] [PubMed] [Google Scholar]
- 2. Tang J., Frankel A., Cook R. J., Kim S., Paik W. K., Williams K. R., Clarke S., and Herschman H. R. (2000) PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J. Biol. Chem. 275, 7723–7730 [DOI] [PubMed] [Google Scholar]
- 3. Gary J. D., and Clarke S. (1998) RNA and protein interactions modulated by protein arginine methylation. Prog. Nucleic Acid Res. Mol. Biol. 61, 65–131 [DOI] [PubMed] [Google Scholar]
- 4. Bedford M. T., and Clarke S. G. (2009) Protein arginine methylation in mammals: who, what, and why. Mol. Cell 33, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chawla A., Barak Y., Nagy L., Liao D., Tontonoz P., and Evans R. M. (2001) PPAR-γ dependent and independent effects on macrophage-gene expression in lipid metabolism and inflammation. Nat. Med. 7, 48–52 [DOI] [PubMed] [Google Scholar]
- 6. Rigamonti E., Chinetti-Gbaguidi G., and Staels B. (2008) Regulation of macrophage functions by PPAR-α, PPAR-γ, and LXRs in mice and men. Arteriosclerosis. Thromb. Vasc. Biol. 28, 1050–1059 [DOI] [PubMed] [Google Scholar]
- 7. Tontonoz P., Nagy L., Alvarez J. G., Thomazy V. A., and Evans R. M. (1998) PPARγ promotes monocyte/macrophage differentiation and uptake of oxidized LDL. Cell 93, 241–252 [DOI] [PubMed] [Google Scholar]
- 8. Croasdell A., Duffney P. F., Kim N., Lacy S. H., Sime P. J., and Phipps R. P. (2015) PPARγ and the innate immune system mediate the resolution of inflammation. PPAR Res. 2015, 549691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Odegaard J. I., Ricardo-Gonzalez R. R., Goforth M. H., Morel C. R., Subramanian V., Mukundan L., Red Eagle A., Vats D., Brombacher F., Ferrante A. W., and Chawla A. (2007) Macrophage-specific PPARγ controls alternative activation and improves insulin resistance. Nature 447, 1116–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tikhanovich I., Kuravi S., Artigues A., Villar M. T., Dorko K., Nawabi A., Roberts B., and Weinman S. A. (2015) Dynamic arginine methylation of tumor necrosis factor (TNF) receptor-associated factor 6 regulates toll-like receptor signaling. J. Biol. Chem. 290, 22236–22249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hassa P. O., Covic M., Bedford M. T., and Hottiger M. O. (2008) Protein arginine methyltransferase 1 coactivates NF-κB-dependent gene expression synergistically with CARM1 and PARP1. J. Mol. Biol. 377, 668–678 [DOI] [PubMed] [Google Scholar]
- 12. Abram C. L., Roberge G. L., Hu Y., and Lowell C. A. (2014) Comparative analysis of the efficiency and specificity of myeloid-Cre deleting strains using ROSA-EYFP reporter mice. J. Immunol. Methods 408, 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Clausen B. E., Burkhardt C., Reith W., Renkawitz R., and Förster I. (1999) Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 8, 265–277 [DOI] [PubMed] [Google Scholar]
- 14. Baker C. C., Chaudry I. H., Gaines H. O., and Baue A. E. (1983) Evaluation of factors affecting mortality rate after sepsis in a murine cecal ligation and puncture model. Surgery 94, 331–335 [PubMed] [Google Scholar]
- 15. Kratochvill F., Neale G., Haverkamp J. M., Van de Velde L. A., Smith A. M., Kawauchi D., McEvoy J., Roussel M. F., Dyer M. A., Qualls J. E., and Murray P. J. (2015) TNF counterbalances the emergence of M2 tumor macrophages. Cell Rep. 12, 1902–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schleicher U., Paduch K., Debus A., Obermeyer S., König T., Kling J. C., Ribechini E., Dudziak D., Mougiakakos D., Murray P. J., Ostuni R., Körner H., and Bogdan C. (2016) TNF-mediated restriction of arginase 1 expression in myeloid cells triggers type 2 NO synthase activity at the site of infection. Cell Rep. 15, 1062–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pello O. M., De Pizzol M., Mirolo M., Soucek L., Zammataro L., Amabile A., Doni A., Nebuloni M., Swigart L. B., Evan G. I., Mantovani A., and Locati M. (2012) Role of c-MYC in alternative activation of human macrophages and tumor-associated macrophage biology. Blood 119, 411–421 [DOI] [PubMed] [Google Scholar]
- 18. Lefèvre L., Authier H., Stein S., Majorel C., Couderc B., Dardenne C., Eddine M. A., Meunier E., Bernad J., Valentin A., Pipy B., Schoonjans K., and Coste A. (2015) LRH-1 mediates anti-inflammatory and antifungal phenotype of IL-13-activated macrophages through the PPARγ ligand synthesis. Nat. Commun. 6, 6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Szanto A., Balint B. L., Nagy Z. S., Barta E., Dezso B., Pap A., Szeles L., Poliska S., Oros M., Evans R. M., Barak Y., Schwabe J., and Nagy L. (2010) STAT6 transcription factor is a facilitator of the nuclear receptor PPARγ-regulated gene expression in macrophages and dendritic cells. Immunity 33, 699–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jung U. J., Torrejon C., Chang C. L., Hamai H., Worgall T. S., and Deckelbaum R. J. (2012) Fatty acids regulate endothelial lipase and inflammatory markers in macrophages and in mouse aorta: a role for PPARγ. Atherscler. Thromb. Vasc. Biol. 32, 2929–2937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sun X., Tang Y., Tan X., Li Q., Zhong W., Sun X., Jia W., McClain C. J., and Zhou Z. (2012) Activation of peroxisome proliferator-activated receptor-γ by rosiglitazone improves lipid homeostasis at the adipose tissue-liver axis in ethanol-fed mice. Am. J. Physiol. Gastrointest. Liver Physiol. 302, G548–G557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dejager L., Pinheiro I., Dejonckheere E., and Libert C. (2011) Cecal ligation and puncture: the gold standard model for polymicrobial sepsis? Trends Microbiol. 19, 198–208 [DOI] [PubMed] [Google Scholar]
- 23. Ocuin L. M., Bamboat Z. M., Balachandran V. P., Cavnar M. J., Obaid H., Plitas G., and DeMatteo R. P. (2011) Neutrophil IL-10 suppresses peritoneal inflammatory monocytes during polymicrobial sepsis. J. Leukocyte Biol. 89, 423–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Orthgiess J., Gericke M., Immig K., Schulz A., Hirrlinger J., Bechmann I., and Eilers J. (2016) Neurons exhibit Lyz2 promoter activity in vivo: Implications for using LysM-Cre mice in myeloid cell research. Eur. J. Immunol. 46, 1529–1532 [DOI] [PubMed] [Google Scholar]
- 25. Murray P. J. (2017) Macrophage polarization. Annu. Rev. Physiol. 79, 541–566 [DOI] [PubMed] [Google Scholar]
- 26. Chang B., Chen Y., Zhao Y., and Bruick R. K. (2007) JMJD6 is a histone arginine demethylase. Science 318, 444–447 [DOI] [PubMed] [Google Scholar]
- 27. Poulard C., Rambaud J., Hussein N., Corbo L., and Le Romancer M. (2014) JMJD6 regulates ERα methylation on arginine. PloS One 9, e87982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang Y., McBride K. M., Hensley S., Lu Y., Chedin F., and Bedford M. T. (2014) Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Mol. Cell 53, 484–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. An W., Kim J., and Roeder R. G. (2004) Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117, 735–748 [DOI] [PubMed] [Google Scholar]
- 30. Barrero M. J., and Malik S. (2006) Two functional modes of a nuclear receptor-recruited arginine methyltransferase in transcriptional activation. Mol. Cell 24, 233–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Aprelikova O., Chen K., El Touny L. H., Brignatz-Guittard C., Han J., Qiu T., Yang H. H., Lee M. P., Zhu M., and Green J. E. (2016) The epigenetic modifier JMJD6 is amplified in mammary tumors and cooperates with c-Myc to enhance cellular transformation, tumor progression, and metastasis. Clin. Epigenetics 8, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Berthet C., Guehenneux F., Revol V., Samarut C., Lukaszewicz A., Dehay C., Dumontet C., Magaud J.-P., and Rouault J.-P. (2002) Interaction of PRMT1 with BTG/TOB proteins in cell signalling: molecular analysis and functional aspects. Genes Cells 7, 29–39 [DOI] [PubMed] [Google Scholar]
- 33. Chittka A. (2010) Dynamic distribution of histone H4 arginine 3 methylation marks in the developing murine cortex. PloS One 5, e13807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tikhanovich I., Olson J. C., Taylor R., Bridges B., Dorko K., Roberts B. R., and Weinman S. A. (2015) Impaired TRAF6 methylation and TLR responses in liver tissue and circulating monocytes from patients with spontaneous bacterial peritonitis. Hepatology 62, 846a [Google Scholar]
- 35. Borzio M., Salerno F., Piantoni L., Cazzaniga M., Angeli P., Bissoli F., Boccia S., Colloredo-Mels G., Corigliano P., Fornaciari G., Marenco G., Pistarà R., Salvagnini M., and Sangiovanni A. (2001) Bacterial infection in patients with advanced cirrhosis: a multicentre prospective study. Dig. Liver Dis. 33, 41–48 [DOI] [PubMed] [Google Scholar]
- 36. Bernsmeier C., Pop O. T., Singanayagam A., Triantafyllou E., Patel V. C., Weston C. J., Curbishley S., Sadiq F., Vergis N., Khamri W., Bernal W., Auzinger G., Heneghan M., Ma Y., Jassem W., Heaton N. D., Adams D. H., Quaglia A., Thursz M. R., Wendon J., and Antoniades C. G. (2015) Patients with acute-on-chronic liver failure have increased numbers of regulatory immune cells expressing the receptor tyrosine kinase MERTK. Gastroenterology 148, 603–615.e614 [DOI] [PubMed] [Google Scholar]
- 37. Antoniades C. G., Wendon J., and Vergani D. (2005) Paralysed monocytes in acute on chronic liver disease. J. Hepatol. 42, 163–165 [DOI] [PubMed] [Google Scholar]
- 38. Zingarelli B., and Cook J. A. (2005) Peroxisome proliferator-activated receptor-γ is a new therapeutic target in sepsis and inflammation. Shock 23, 393–399 [DOI] [PubMed] [Google Scholar]
- 39. Martin H. (2010) Role of PPAR-γ in inflammation. Prospects for therapeutic intervention by food components. Mutat. Res. 690, 57–63 [DOI] [PubMed] [Google Scholar]
- 40. Reddy A. T., Lakshmi S. P., and Reddy R. C. (2016) PPARγ in bacterial infections: a friend or foe? PPAR Res. 2016, 7963540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Davies J. Q., and Gordon S. (2005) Isolation and culture of murine macrophages. Methods Mol. Biol. 290, 91–103 [DOI] [PubMed] [Google Scholar]
- 42. Steiner J. L., and Lang C. H. (2015) Sepsis attenuates the anabolic response to skeletal muscle contraction. Shock 43, 344–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li Z., Zhao J., Tikhanovich I., Kuravi S., Helzberg J., Dorko K., Roberts B., Kumer S., and Weinman S. A. (2016) Serine 574 phosphorylation alters transcriptional programming of FOXO3 by selectively enhancing apoptotic gene expression. Cell Death Differ. 23, 583–595 [DOI] [PMC free article] [PubMed] [Google Scholar]