Abstract
The biogenesis and maintenance of cell organelles such as mitochondria and chloroplasts require the import of many proteins from the cytosol, a process that is controlled by phosphorylation. In the case of chloroplasts, the import of hundreds of different proteins depends on translocons at the outer and inner chloroplast membrane (TOC and TIC, respectively) complexes. The essential protein TOC159 functions thereby as an import receptor. It has an N-terminal acidic (A-) domain that extends into the cytosol, controls receptor specificity, and is highly phosphorylated in vivo. However, kinases that phosphorylate the TOC159 A-domain to enable protein import have remained elusive. Here, using co-purification with TOC159 from Arabidopsis, we discovered a novel component of the chloroplast import machinery, the regulatory kinase at the outer chloroplast membrane 1 (KOC1). We found that KOC1 is an integral membrane protein facing the cytosol and stably associates with TOC. Moreover, KOC1 phosphorylated the A-domain of TOC159 in vitro, and in mutant koc1 chloroplasts, preprotein import efficiency was diminished. koc1 Arabidopsis seedlings had reduced survival rates after transfer from the dark to the light in which protein import into plastids is required to rapidly complete chloroplast biogenesis. In summary, our data indicate that KOC1 is a functional component of the TOC machinery that phosphorylates import receptors, supports preprotein import, and contributes to efficient chloroplast biogenesis.
Keywords: Arabidopsis, chloroplast, phosphorylation, protein import, receptor, TOC complex, integral membrane kinase
Introduction
Biogenesis and maintenance of the chloroplast require the import of proteins from the cytosol. During evolution, the majority of chloroplast genes were lost or transferred to the nucleus (1). Chloroplast genes that were successfully transferred to the nucleus acquired additional sequences that encode cleavable N-terminal targeting peptides, which are known as transit peptides (2). Cytoplasmic ribosomes synthesize the preproteins (chloroplast protein with a transit peptide). The transit sequence is recognized by the chloroplast protein import machinery, which initiates import. It consists of translocons at the outer (TOC)2 and inner chloroplast membrane (TIC) (3–5).
The TOC core in Arabidopsis contains three components: TOC159, TOC33, and TOC75. TOC159 and TOC33 are exposed to the cytoplasm and function as preprotein co-receptors (6–8). TOC75 embedded in the outer membrane forms a protein-conducting channel (9). The central component of the TIC complex is TIC20 that contributes to the protein-conducting channel across the inner membrane (10). During preprotein translocation, TIC20 associates with other TIC components including TIC110 and TIC40 that recruit chaperones to the exit of the TIC complex (11, 12). The ClpC and cpHsp70 chaperones have both been reported to provide the driving force for preprotein import into the chloroplast (13–15). Recently, a 1-MDa complex has been described: in addition to TIC20, it includes TIC56, TIC100, and the chloroplast-encoded TIC214 (YCF1) (16, 17). The 1-MDa complex associates with the translocating preprotein and forms a preprotein-sensitive channel when reconstituted into planar lipid bilayers. TIC components (both absent and present in the 1-MDa complex) co-purified with TOC159, suggesting that they cooperate in preprotein import (17).
TOC159 and TOC33 belong to a small family of GTP-binding proteins sharing homology in their GTP-binding domains (18, 19). In Arabidopsis, TOC33 has one homolog (TOC34), and TOC159 has three, TOC90, -120, and -132 (20, 21). In addition to the GTP-binding (G-) domain, the TOC159 homologs have a C-terminal membrane-anchoring (M-) domain and an N-terminal acidic (A-) domain (7, 22).
The functions of the A-domain are not completely understood. A domain swapping study (23) analyzing the roles of the A-domains of TOC159 and TOC132 indicated that they mediate preprotein specificity (TOC159 specializing in photosynthesis-associated proteins and TOC132 specializing in housekeeping plastid proteins). Removal of the A-domain reduced preprotein specificity, resulting in receptors with overlapping specificity (23). In contrast to the G- and M-domains, the A-domain is dispensable in vivo (24–26). TOC159 is present with and without the A-domain in isolated chloroplasts and may be cleaved by an unknown protease. The ratio between the cleaved and uncleaved forms is unknown.
Regulation of protein import has been studied in the past. TOC159 and TOC33 are GTP-binding proteins, suggesting regulation of import by a GTPase cycle (19, 27). Specific point mutations in the GTP-binding motifs (reducing binding or hydrolysis of GTP) altered protein import kinetics but did not result in visible phenotypes (26, 28, 29).
In addition to GTP, ubiquitination and phosphorylation affect the TOC159 homologs (22). Ubiquitination and degradation by the ubiquitin-proteasome system (UPS) play an important role during plastid developmental transitions (for example from non-photosynthetic etioplasts in the dark to active chloroplasts in the light) (30). To achieve this, the composition of the protein import machinery is modified to accommodate massive import of photosynthesis-associated proteins. This implicates degradation by the UPS of one kind of TOC159 homolog and replacement by another (31). The chloroplast outer membrane RING-type E3 ligase SP1 mediates the ubiquitination reaction (30). SP1 also mediates the depletion of the TOC components by the UPS under conditions inducing oxidative stress. This diminishes import of photosystem components and thereby limits production of harmful reactive oxygen species (32).
Phosphorylation has been reported to regulate the activity of the pea homologs of both TOC33 (psTOC34) and TOC159 (psTOC159) (33, 34). psTOC34 is the target of a protein kinase at the outer membrane. The phosphorylation of psTOC34 decreases the affinity of psTOC34 for GTP and regulates the GTP-dependent interaction with the preprotein. The interaction between TOC34 and the preprotein is strongly enhanced by phosphorylation of the transit peptide (35). An outer envelope kinase of 98 kDa (OEK98) has been implicated in the phosphorylation of psTOC34 but has not been identified (33). In Arabidopsis, TOC33 can be phosphorylated at Ser-181 (36). However, mutation of this residue either to non-phosphorylable alanine or to phosphomimic aspartate or glutamate had no detectable effect in vivo (36). The G-domain of psTOC159 is the target of an outer envelope kinase of 70 kDa (OEK70). Phosphorylation by OEK70 inhibits association of TOC159 with the TOC complex (33).
Recent large-scale phosphoproteomics projects in Arabidopsis revealed hyperphosphorylation of the A-domain (PhosphAT 4.0) but provided no evidence for the phosphorylation of either the G- or M-domain of TOC159. The A-domain has many predicted and experimentally identified casein kinase II (CKII) sites. CKII efficiently phosphorylates recombinant A-domain in vitro (37). However, some of the phosphorylation sites within the A-domain do not resemble CKII sites, suggesting that additional kinases may be involved. Sucrose nonfermenting 1-related protein kinase 2 (SnRK2) phosphorylates Thr-692 in response to abscisic acid (ABA) activation (38). Interestingly, ABA-dependent phosphorylation of TOC159 homologs TOC132 and -120 was also reported but only in the SnRK2-deficient mutant background. Wang et al. proposed that chloroplast protein import activity may be regulated by ABA via SnRK2-dependent phosphorylation of the A-domain (38). Additional A-domain kinases other than CKII and SnRK2 have been predicted but not identified (37).
Here, a tandem affinity purification (TAP)-tagged version of TOC159 (NTAP-TOC159) was used to co-isolate new interaction partners by IgG-affinity chromatography. This approach has already proven useful in the identification of TIC56 in a TOC159-containing supercomplex (17). Here, we identify KOC1 as a new component in this complex and show that it is required for full import activity and successful de-etiolation.
Results
KOC1 co-purifies with TOC159
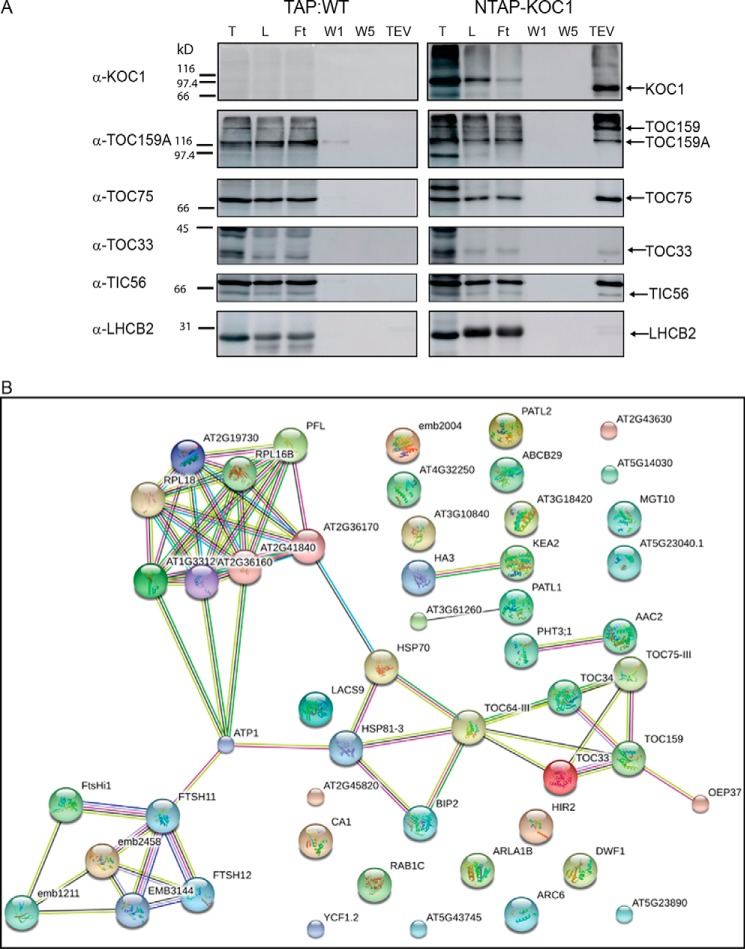
Arabidopsis plants (NTAP-TOC159:ppi2) expressing N-terminally TAP-tagged TOC159 were used to isolate potential TOC159 interactors by IgG-affinity chromatography (17). Plants expressing the TAP tag alone (TAP:WT) were used as a negative control. The TAP tag contains an IgG-binding domain separated from a calmodulin-binding peptide (CBP) sequence by a tobacco etch virus (TEV) protease site. Triton X-100 (TX100) detergent extracts were prepared and subjected to IgG-affinity chromatography. TEV protease elution was applied to release either TOC159 still carrying the CBP (together with any interacting proteins) or the negative control CBP. Proteins in the TEV eluates were identified by mass spectrometry (17). The mass spectrometric data revealed a member of the protein kinase superfamily protein (AT4G32250) that we named kinase at the outer chloroplast membrane 1 (KOC1). KOC1 co-purified with NTAP-TOC159 but not with TAP control (Fig. 1). Specific antibodies against recombinant KOC1 were raised in rabbit and affinity-purified.
Figure 1.
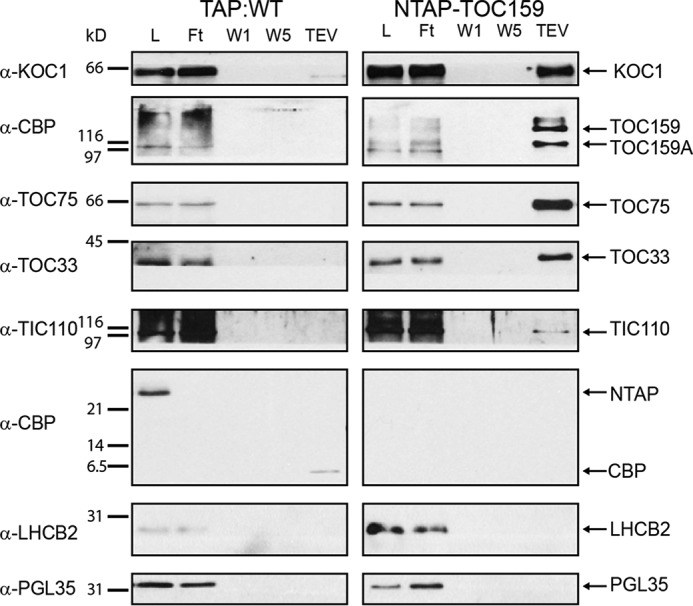
Co-purification of KOC1 with TOC159 and TOC components. Sequential fractions of NTAP-TOC159 IgG-affinity purification were analyzed by Western blotting. The membrane was probed with antibodies against KOC1, CBP, TOC75, TOC33, TIC110, LHCB2, and PGL35 (L, load; Ft, flow-through;W1, first wash; W5, last wash; TEV, eluate). 50 μg of proteins (load, flow-through, and first wash) or 10% of the fractions (last was and eluate) were loaded. Several identical blots were used for immunoblotting. TAP:WT was used as a negative control.
To confirm the association of KOC1 with the TOC159 complex, aliquots of the sequential steps of the IgG-affinity purification experiment were separated by SDS-PAGE and analyzed by Western blotting (Fig. 1). A 66-kDa band corresponding to the expected size of KOC1 was detected in the total detergent extract load and in the flow-through fractions of the NTAP-TOC159 and TAP:WT samples (Fig. 1). The KOC1 band was present only in the TEV eluate of the NTAP-TOC159 purification but not that of the TAP tag negative control (TAP:WT). In addition, TOC75 and -33 as well as TIC110 co-purified with NTAP-TOC159 (Fig. 1). The thylakoid and plastoglobule marker proteins LHCB2 and FBN1a (PGL35) were detected by Western blotting in the load and flow-through fractions but not in the TEV eluates.
To confirm the interaction of KOC1 with the TOC complex proteins, NTAP-KOC1:koc1-1 plants (see below) were used for IgG-affinity purification. KOC1 was present in the TEV eluate (Fig. 2A). Due to TEV cleavage, it had a noticeably smaller size than NTAP-KOC1 in the total extract and load fraction. TOC159, TOC75, TOC33, and TIC56 were also detected in the eluate of NTAP-KOC1 but not in the eluate of the negative control TAP:WT. The thylakoid marker protein LHCB2 was not detectable in the eluate. Altogether these results demonstrate that KOC1 associates with TOC complex in vivo.
Figure 2.
Co-purification of TOC components with KOC1. A, sequential fractions of NTAP-KOC1 IgG-affinity purification were analyzed by Western blotting. The membrane was probed with antibodies against KOC1, TOC159, TOC75, TOC33, TIC56, and LHCB2 (T, total; L, load; Ft, flow-through; W1, first wash; W5, last wash; TEV, eluate). 50 μg of proteins (total, load, flow-through, and first wash) or 10% of the fractions (last wash and eluate) were loaded. Several identical blots were used for immunoblotting. TAP:WT was used as a negative control. B, interaction networks of KOC1 using the list of co-immunopurified proteins and the STRING 10.0 program.
The TEV eluate of the NTAP-KOC1 purification experiment was subjected to quantitative mass spectrometry. The experiment was carried out in two biological replicates. A list of common proteins in the two experiments was created and contained 191 proteins (not shown). Among the 15 most abundant interaction candidates were seven TOC and TIC components. In order of decreasing abundance in femtomoles, these were TOC75, TOC159, YCF1.2, TIC110, TOC33, TOC34, and ClpC. When the protein list established for NTAP-KOC1 was compared with a list for NTAP-TOC159 consisting of 43 proteins weighted for enrichment, a high overlap of 83% was detected. Notably, 11 known components of the TOC and TIC complexes as well as components of the 1-MDa complex were represented in this list for which the overlap between TOC159 and the KOC1 protein list was complete.
We filtered the list of KOC1-interacting proteins by protein abundance and enrichment factor to identify specific interactions at high stringency (supplemental Table S1). We first selected proteins that were identified in both biological replicates but not in the negative controls. We then calculated an enrichment factor from the ratio between protein abundance in isolated chloroplasts and that in the TAP-purified fraction of KOC-interacting proteins and retained those proteins with an enrichment factor >1 in the list. And lastly, we requested interacting proteins to have a minimum abundance of 2% of the bait protein (i.e. KOC1) to select for specific KOC/protein interactions. This filtering resulted in 51 proteins that fulfilled requirements (supplemental Table S1). These proteins were plotted into an interaction network using the STRING database (STRING version 10.0 software). This analysis identified three complexes interacting with KOC1: the TOC complex, an FtsH-FtsHi protease complex, and the cytoplasmic ribosome (Fig. 2B).
KOC1 is a predicted transmembrane kinase
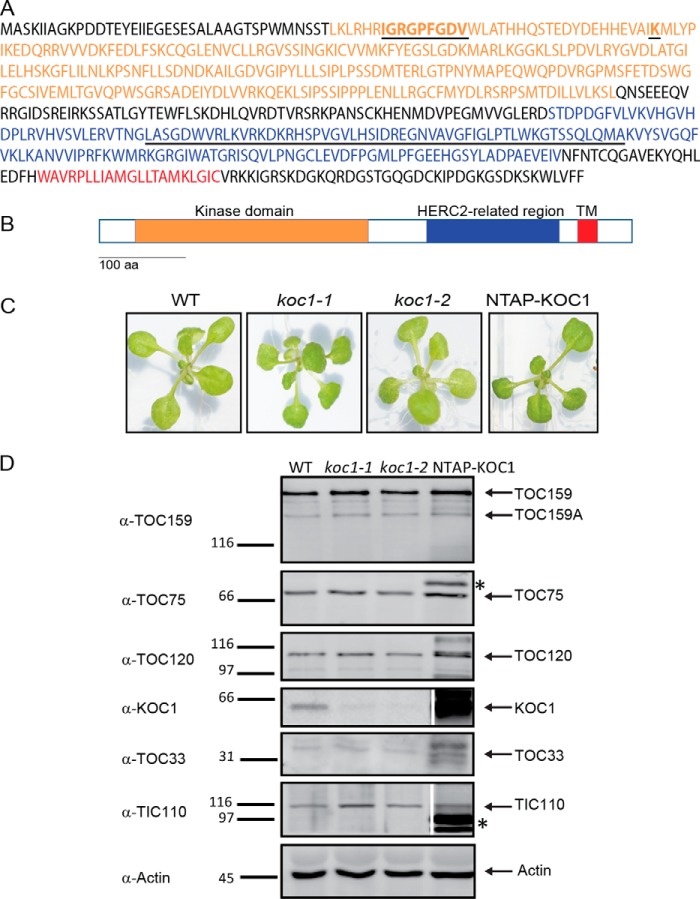
KOC1 has 611 amino acids residues. It contains a kinase domain (amino acids 39–306) predicted by Prosite software. The C-terminal half of KOC1 contains a HERC2-related region (HECT (homologous to the E6-AP C terminus) and RLD (RCC1-like domain)-containing E3 ubiquitin protein ligase 2; amino acids 376–531). The HERC2-related region in KOC1 is followed by a 24-amino-acid stretch (549–572) enriched in hydrophobic amino acids predicted to form a transmembrane helix (Fig. 3A) by TMpred software. ChloroP software does not predict a transit peptide (Fig. 3B). Still, KOC1 was identified in chloroplast proteome studies (39, 40).
Figure 3.
KOC1 sequence analysis and mutant characterization. A and B, the amino acid sequence of KOC1 contains a kinase domain (orange; amino acids (aa) 39–306). Underlined amino acids 45–53 are conserved in the catalytic domain; amino acid 74 is predicted to bind ATP. In the HERC2-related region (blue; amino acids 376–531), underlined amino acids share homology with the KEG protein. A predicted transmembrane stretch (TM; amino acids 549–572) is highlighted in red. C, images of 2-week-old WT, koc1-1, koc1-2, and NTAP-KOC1:koc1-1 plants grown in vitro. D, total protein extracts of 2-week-old plants (WT, koc1-1, koc1-2, and NTAP-KOC1) were separated by SDS-PAGE and transferred to nitrocellulose membrane. Antibodies against TOC159A, TOC75, TOC120, KOC1, TOC33, and TIC110 were used. Actin was used as a loading control. *, bands corresponding to overexpressed NTAP-KOC1.
KOC1 most closely resembles the Keep On Going (KEG) protein (AT5G13530), a RING E3 ubiquitin ligase. In addition to the RING sequence, KEG contains ankyrin and HERC2-related repeats and a kinase domain (41). The RING sequence and the ankyrin repeats are absent from KOC1, but both the kinase domain and the HERC2-related region in KOC1 are homologous to those in KEG. The sequence analysis suggests that KOC1 functions as a kinase, and an implication in ubiquitination also appears remotely possible.
Isolation of koc1 mutants and NTAP-KOC1:koc1-1 plants
To obtain information on the function of KOC1 in vivo, T-DNA mutant collections were searched for insertions in the KOC1 gene (AT4G32250). Two independent mutant lines, SALK_083378 termed koc1-1 and SALK_051823 termed koc1-2, were identified, and homozygous lines were isolated (supplemental Fig. S1 and Fig. 3, C and D). The koc1-1 line contained a single T-DNA insertion at 841 base pairs after the start codon. The koc1-2 line contained a double T-DNA insertion at 2269 base pairs after the start codon (supplemental Fig. S1).
Immunoblotting using KOC1 antibodies demonstrated the absence of KOC1 protein (Fig. 3D) and confirmed the knock-out nature of the mutants. NTAP-KOC1:koc1-1 plants were obtained by introducing a T-DNA construct encoding an N-terminally TAP-tagged KOC1 (NTAP-KOC1) in koc1-1 mutant background. The koc1-1 and koc1-2 mutants as well as the homozygous NTAP-KOC1:koc1-1 plants displayed a wild-type phenotype under standard growth conditions (long day; 16-h light, 8 h-dark) (Fig. 3C).
TOC components accumulate normally in koc1 mutants
Sequence analysis of KOC1 revealed two potential biochemical functions: that of a kinase and potentially that of a factor in ubiquitination and subsequent proteasome-mediated degradation. Both potentially affect assembly and stability of TOC and TIC components. Therefore, the two koc mutants were compared with the wild type- and NTAP-KOC1-overexpressing lines by Western blotting. The levels of the components of the TOC core complex (TOC159, -75, and -33) as well as TOC120 and TIC110 appeared unchanged in the koc1 mutants. This was more difficult to judge in the overexpressing line due to the proximity of NTAP-containing bands that give strong signals with any kind of IgG and that precluded quantification. Visibly, however, no major changes appeared to occur (Fig. 3D). The results suggest that the TOC and TIC components are normally assembled and stable in the koc1 and overexpressing backgrounds.
KOC1 localizes at the outer chloroplast membrane
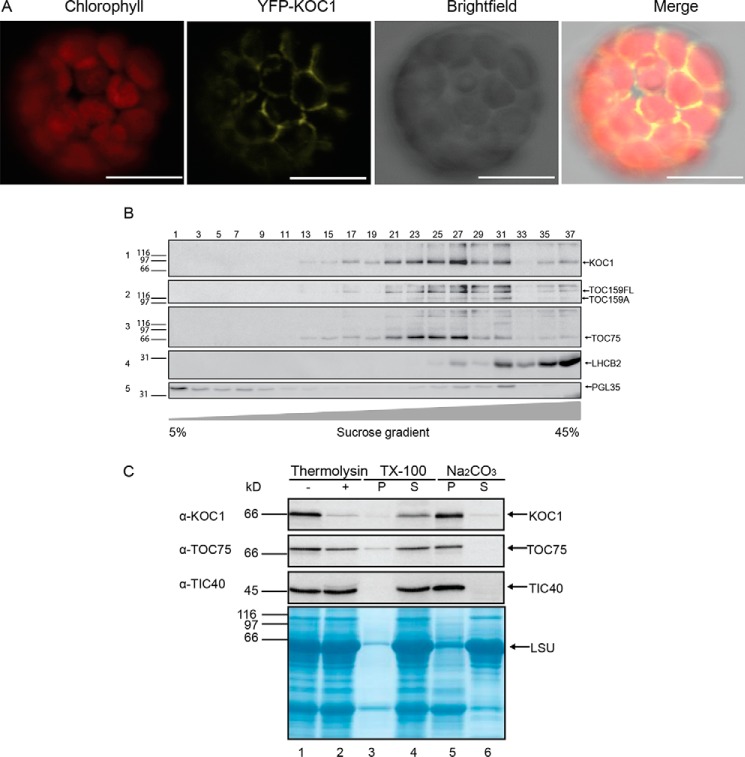
For in vivo localization, isolated Arabidopsis wild-type protoplasts were transformed with a vector (pEG104-N-YFP-KOC1) coding for KOC1 with a N-terminal YFP tag (YFP-KOC1) and with vectors pEG101-C-YFP-EMB2004 and pCL60-GFP for transient expression of EMB2004 (an envelope protein at the inner membrane that also co-isolated with KOC1) fused to YFP (EMB2004-YFP) and GFP alone and analyzed by confocal microscopy. Chloroplasts were identified by red chlorophyll autofluorescence (Fig. 4A, Chlorophyll). YFP-KOC1 gave a ringlike fluorescence pattern (Fig. 4A, YFP-KOC1). The merge between the YFP-KOC1 and chlorophyll signals shows that YFP-KOC1 was localized at the chloroplast periphery (Fig. 4A, Merge). EMB2004-YFP gives a fluorescence pattern strongly resembling that of YFP-KOC1 (supplemental Fig. S3), whereas GFP results in a pattern typical for the cytosol and that is distinct from that of YFP-KOC1 (supplemental Fig. S3).
Figure 4.
Localization of KOC1 at the outer membrane of the chloroplast. A, confocal microscopy images of an isolated representative protoplast expressing YFP-KOC1. Chlorophyll fluorescence in red identifies chloroplasts, the signal of YFP-KOC1 appears yellow, and intact protoplasts were visualized by bright field. The merge shows the overlay of chlorophyll, YFP-KOC1, and bright field images. Scale bars, 10 μm. B, total membranes from NTAP-KOC1 chloroplasts were separated on a continuous sucrose gradient (5–45%), and 37 fractions were collected. Proteins from uneven fractions were separated by SDS-PAGE, transferred to nitrocellulose, and probed with antibodies against CBP (1), TOC159A (2), TOC75 (3), LHCB2 (4), and PGL35 (5). C, Col-0 chloroplasts were subjected (+) to thermolysin treatment or not (−), TX1000 solubilization, and alkaline extraction (Na2CO3) (P, pellet; S, supernatant). Samples were separated by SDS-PAGE, transferred to nitrocellulose, stained with Amido Black (lower part), and probed with antibodies against KOC1, TOC75, and TIC40. LSU, large subunit.
KOC1 is present in the chloroplast envelope fraction
To localize NTAP-KOC1 in chloroplast membrane compartments, chloroplasts were isolated from NTAP-KOC1:koc1-1 plants. A total chloroplast membrane fraction was prepared and separated into thylakoids, envelope membranes, and plastoglobules by floatation on a linear 5–45% sucrose gradient. Fractions were analyzed by Western blotting (Fig. 4B). LHCB2 was present mostly in fractions 27–37, indicating thylakoids. FIB1a/PGL35 was found in fractions 1–13, indicating the presence of plastoglobules. FIB1a/PGL35 was also detected in denser fractions (fractions 23–31) likely due to plastoglobule association with thylakoids. NTAP-KOC1 was detected mostly in fractions 17–31 and was well separated from the fractions enriched in thylakoid and plastoglobule markers. Moreover, TOC159 and TOC75 co-fractionated with NTAP-KOC1 (Fig. 4B), supporting KOC1 localization at the chloroplast envelope.
KOC1 is exposed at the chloroplast surface
To investigate KOC1 localization at the chloroplast envelope, we treated isolated chloroplasts with thermolysin protease (Fig. 4C). Thermolysin degrades surface-exposed proteins but does not access the intermembrane space. Thermolysin-treated chloroplasts were separated by SDS-PAGE and probed by immunoblotting. The KOC1 band was strongly diminished by thermolysin, whereas the known thermolysin-resistant outer membrane protein TOC75 and the inner membrane protein TIC40 were largely unaffected (Fig. 4C), indicating that KOC1 was accessible at the outer surface of the outer envelope membrane.
KOC1 is an integral membrane protein
To analyze the membrane association of KOC1, extraction experiments were carried out. Isolated chloroplasts were extracted with alkaline carbonate buffer (Na2CO3) or solubilized with Triton X-100 (Fig. 4C). Upon centrifugation of the alkaline carbonate extraction, KOC1 remained in the pellet fraction but was found in the supernatant after Triton X-100 solubilization. The known integral membrane proteins TOC75 and TIC40 behaved in the same way. We therefore conclude that KOC1 is an integral membrane protein.
KOC1 phosphorylates the A-domain of TOC159 in vitro
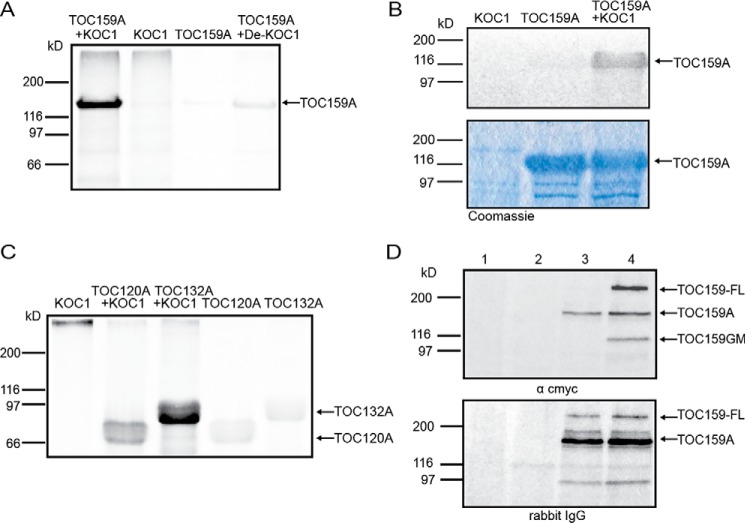
The A-domain of TOC159 is hyperphosphorylated in vivo. To test whether KOC1 kinase phosphorylates TOC159A, we isolated KOC1 from NTAP-KOC1:koc1-1 plants (see above) and performed an in vitro phosphorylation experiment on recombinant TOC159A in the presence of radioactive ATP. The experiment resulted in strong phosphorylation of TOC159A (Fig. 5A, lane 1). The control experiments carried out with KOC1 and TOC159A alone did not result in detectable phosphorylation (Fig. 5A, lanes 2 and 3), whereas denatured KOC1 showed slight residual activity (Fig. 5A, lane 4). Recombinant KOC1 purified from Escherichia coli bacteria also phosphorylated TOC159A in vitro (Fig. 5B), providing additional evidence for the kinase function. The A-domains of the TOC159 homologs TOC120 and TOC132 are also known phosphoproteins, and recombinant TOC120A and -132A were phosphorylated by KOC1 kinase in vitro (Fig. 5C, lanes 2 and 3). Altogether the in vitro phosphorylation experiments demonstrate that TOC159 and its homologs are substrates of KOC1 kinase in vitro.
Figure 5.
Phosphorylation of the A-domain by KOC1 and its presence in TOC159. A, TOC159A was incubated with KOC1 isolated from NTAP-KOC1:koc1-1 plants (lane 1) or the same but heat-denatured KOC1 (De-KOC1) (lane 4) in the presence of [γ-33P]ATP. B, purified TOC159A was incubated with [γ-33P]ATP and with recombinant full-length KOC1 purified from bacteria (lane 3). C, purified TOC120A (lane 2) and TOC132A (lane 3) were incubated with [γ-33P]ATP and KOC1 isolated from NTAP-KOC1:koc1-1 plants. In A–C, the samples were separated by SDS-PAGE and analyzed using a phosphorimaging system (Molecular Imager FX) and Quantity One 4.6 software. KOC1, TOC159A, TOC120A, and TOC132A were incubated alone with [γ-33P]ATP as negative controls. D, total protein extracts of seedlings ppi2 (lane 1), WT (lane 2), NTAP-TOC159:ppi2 (lane 3), and NTAP-TOC159-cmyc:ppi2 (lane 4) were analyzed by Western blotting. The membrane was consecutively probed with antibodies against c-myc and rabbit IgG. The data shown are representative experiments of several replicates.
A large percentage of TOC159 exists as the full-length protein
TOC159 occurs both with and without its A-domain, but the ratio of the two forms is unknown. Regulation of protein import at the A-domain is only plausible if the A-domain is present. To determine to what extent this is the case, we engineered a TOC159 construct encoding an N-terminal NTAP tag and a C-terminal myc tag (Fig. 5D and supplemental Fig. S2). The C-terminal myc tag allows detection of full-length TOC159 (TOC159-FL) as well as TOC159 lacking the A-domain (TOC159GM). By Western blotting, this should result in two bands corresponding to TOC159-FL and TOC159GM. A Western blotting experiment was carried out on equal amounts of total protein of ppi2, WT, NTAP-TOC159:ppi2, and NTAP-TOC159-cmyc:ppi2 plants (Fig. 5D). To detect the myc tag with minimal interference from the TAP tag (which binds IgG), the NTAP tag was first saturated with nonspecific rabbit IgG. In a second incubation, the blot was incubated with mouse anti-myc antibodies and developed with goat anti-mouse IgG coupled to HRP (Fig. 5D, α cmyc). The anti-myc antibodies detected two specific bands in the NTAP-TOC159-cmyc extract that were not present in NTAP-TOC159, one at above 200 kDa and the other at around 100 kDa, corresponding to TOC159-FL and TOC159GM, respectively (Fig. 5D, lane 4).
The ratio between TOC159-FL and TOC159GM was around 2:1, indicating that around two-thirds of TOC159 exists in the full-length form, and around one-third lacks the A-domain. A third band in between the two was observed in the NTAP-TOC159 and NTAP-TOC159-cmyc extracts and most likely corresponded to the free A-domain (TOC159A) that had not been completely saturated by the nonspecific IgG. Finally the blot was incubated with goat anti-rabbit antibodies coupled to HRP to detect the NTAP tag (Fig. 5D, rabbit IgG). The patterns and intensity of bands observed for NTAP-TOC159 and NTAP-TOC159-cmyc were highly similar, suggesting that the two proteins were expressed at similar levels in their respective genetic backgrounds. The anti-rabbit antibody detected levels of free TOC159A that appeared greater than those of TOC159-FL, suggesting that the TOC159A is stable in the cytosol, whereas lower levels of TOC159-FL are maintained at the chloroplast outer membrane.
KOC1 supports import activity
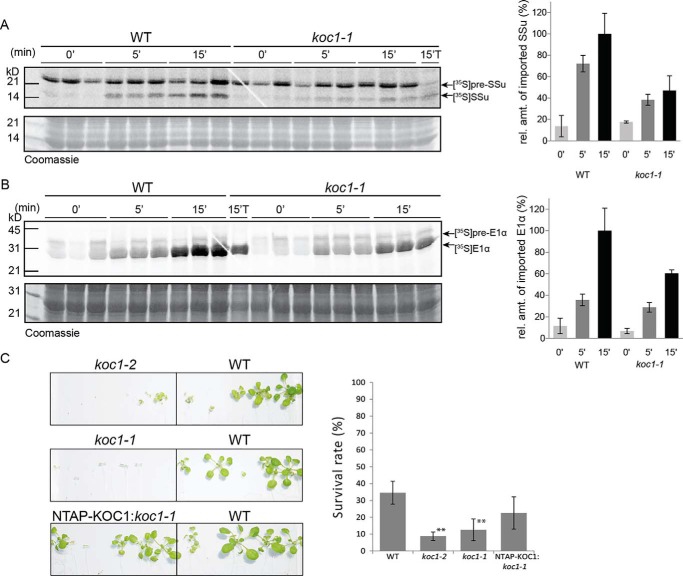
Because KOC1 can interact and phosphorylate TOC components, we tested whether KOC1 plays a role in preprotein import. Chloroplasts were isolated from 2-week-old wild-type and koc1-1 mutant seedlings. The isolated chloroplasts were incubated with either [35S]pSSu (a client preprotein of the TOC159 receptor) (Fig. 6A) or [35S]pE1α (a client preprotein of the TOC120 and -132 receptors) (Fig. 6B) for 0, 5, and 15 min, and one sample was treated with thermolysin after 15 min. The samples were analyzed by SDS-PAGE followed by phosphorimaging analysis. Import was defined as the accumulation of mature [35S]SSu and [35S]E1α. In koc1-1 chloroplasts, the accumulation of both mature SSu (47%) and E1α (60.3%) after 15 min was reduced in comparison with wild-type chloroplasts (100%) (Fig. 6, A and B).
Figure 6.
Requirement of KOC1 for efficient chloroplast protein import and de-etiolation. A and B, isolated chloroplasts from koc1-1 and Col-0 (WT) plants were incubated with [35S]Met-labeled preproteins of pSSu in A and E1-α in B. The preproteins were incubated with chloroplasts, and import was allowed to proceed for 0, 5, and 15 min (0′, 5′, and 15′); one sample was treated with thermolysin after 15 min (15′T). Proteins from chloroplasts were separated by SDS-PAGE followed by phosphorimaging analysis. The graphs show the quantification of the bands corresponding to imported mature SSu and E1α at 0, 5, and 15 min averaged over three technical replicates. The amount of mature protein imported into WT chloroplasts at 15 min was arbitrarily set to 100%. The qualitatively similar results were obtained in five independent experiments. C, images of surviving koc1-2, koc1-1, WT, and NTAP-KOC1:koc1-1 plants upon exposure to long-day conditions for 2 weeks after etiolation for 6 days in dark. The germination and survival rates were calculated. The germination rate was around 100% for all genotypes. The survival rates were as follows: WT, 34.6%; NTAP-KOC1:koc1-1, 22.5%; koc1-2, 8.8%; and koc1-1, 12.5% (Student's t test: **, p value <0.01; *, p value <0.05 (n = 80 for koc1-1 and koc1-2; n = 240 for WT)). Error bars represent S.D. This experiment was repeated three times with comparable results. rel. amt., relative amount.
KOC1 is required for survival during de-etiolation
During de-etiolation, rapid import of proteins occurs to accomplish the transition to photoautotrophic growth. To analyze a potential role for KOC1 in the process, wild-type, koc1-1, koc1-2, and NTAP-KOC1:koc1-1 plants were grown in the dark for 6 days and then exposed to standard light conditions. To guarantee identical seed quality, the parental plants had been grown simultaneously and allowed to set seed in the same growth chamber. For all four genotypes, the germination rate was close to 100%. However, the survival rate differed for the different genotypes. Approximately 34% of the wild-type plants survived. In comparison, only around 9% of the koc1-2 and 13% koc1-1 plants survived, which differed significantly from wild-type survival rates (Fig. 6C). The survival rate of NTAP-KOC1:koc1-1 plants was around 23% and statistically indistinguishable from WT. This provides evidence that NTAP-KOC1 expressed in koc1-1 plants is functional and complements the koc1-1 de-etiolation phenotype.
Discussion
Phosphorylation is emerging as an important mechanism in regulating the assembly and activity of protein import complexes in protein translocation systems. CKII and SnRK2, two cytosolic kinases, are known to phosphorylate the TOC159 A-domain. Here, we identify KOC1, an integral chloroplast envelope kinase that phosphorylates the A-domain. We demonstrate that KOC1 affects import activity but does not interfere with composition and abundance of import components.
N-terminally TAP-tagged TOC159 (NTAP-TOC159) was used to isolate the protein import machinery and to identify new potential interaction partners of TOC159 in vivo (Fig. 1) (17). Among the identified proteins was the predicted kinase AT4G32250 that we named KOC1. In the reverse experiment, NTAP-KOC1 was used as the bait (Fig. 2A). NTAP-KOC1 was most likely fully functional as it 1) complemented the koc1-1 phenotype in the de-etiolation survival assay (Fig. 6C) and 2) phosphorylated recombinant A-domains after affinity purification (Fig. 5, A and B). TOC159 as well as other recognized components of the import machinery were co-isolated together with NTAP-KOC1. Overall the lists of proteins identified by mass spectrometry that associated with NTAP-TOC159 and NTAP-KOC1, respectively, were highly overlapping. The combination of the two co-isolation experiments provided strong evidence for the association of KOC1 with the chloroplast protein import machinery.
By mass spectrometric analysis, not TOC159 but TOC75 gave the highest score in femtomoles of any component of the import machinery associating with KOC1. This indicates that TOC159 may not be the primary interaction partner of KOC1. KOC1 may also associate with multiple TOC complexes containing the different TOC159 homologs as suggested by the ability of KOC1 to phosphorylate the A-domains of TOC132 and -120. The NTAP-KOC1-associated proteins belonged to three distinct networks: the TOC complex, the intrachloroplastic FtsH proteases, and the cytosolic ribosome (Fig. 2B). The interaction with the FtsH network hints at an interaction between the import and protein quality-control systems within the chloroplast. The interaction between KOC1 and FtsH proteins is most likely indirect and may involve interaction with TIC components that also co-isolated with KOC1. Interaction with the cytosolic ribosome suggests coordination of preprotein synthesis with the import process. These hypotheses should be experimentally explored in future studies.
KOC1 lacks a predicted cleavable transit peptide at the N terminus and is therefore not a predicted chloroplast protein (Fig. 3A). The absence of a cleavable transit peptide, however, is typical of chloroplast outer envelope proteins. KOC1 has a single predicted transmembrane region near its C terminus. It is therefore probable that KOC1 belongs to the category of tail-anchored outer membrane proteins (42, 43). Outer envelope localization was supported by three lines of evidence (fluorescence, membrane fractionation, and protease sensitivity) (Fig. 4). Based on resistance to alkaline extraction, KOC1 behaves as an integral membrane protein (Fig. 4C). The predicted transmembrane sequence at the C terminus in combination with protease sensitivity indicated that the bulk of the KOC1 protein faces the cytosol. The only other known organelle-associated kinase facing the cytosol apart from KOC1 is mitochondrial CKI, which phosphorylates TOM20 and stimulates assembly of the TOM complex (44).
The KOC1 protein contains two striking elements in addition to the transmembrane region: a predicted N-terminal kinase domain and a C-terminal HERC2-like region. The sequence of KOC1 is most closely related to that of KEG (AT5G13530). KEG contains a kinase domain and 12 HERC2 repeats with similarity to KOC1. In addition to those domains, KEG also has a RING domain responsible for its function as an E3 ligase (41). The RING domain is absent from the KOC1 sequence, suggesting that it does not function as an E3 ligase. It cannot be excluded, however, that KOC1 functions in conjunction with other E3 ligases under specific conditions. One such candidate is the outer membrane E3 ligase SP1 that is involved in ubiquitination and turnover of TOC159 and -75 (30). However, the components of the TOC complex as well as TIC110 were present in both koc1 lines at similar concentrations as in the wild type (Fig. 3D). This indicated that loss of KOC1 does not affect composition and abundance of the components of the import machinery under standard growth conditions.
A question of central interest was whether KOC1 kinase phosphorylates the A-domains of TOC159 and its homologs. KOC1 purified from transgenic plants phosphorylated the recombinant TOC159, -132, and -120 A-domains (Fig. 5, A and C). The A-domains of TOC159 and TOC132/-120 determine preprotein specificity (TOC159 specializing in photosynthesis-associated proteins and TOC132/-120 specializing in housekeeping proteins) (23). The ability of KOC1 to phosphorylate all three A-domains suggests a function in import of both photosynthesis-associated and housekeeping proteins. It is important to note that the majority of TOC159 is full length (Fig. 5D), and therefore such a scenario is plausible. However, we were not able to reliably identify KOC1-dependent phosphorylation sites in the recombinant TOC159 A-domain using mass spectrometry. Their identification will allow site-specific mutagenesis to test the role of KOC1-dependent phosphorylation in protein import. Although TOC159 family members are likely in vivo targets of KOC1, other targets at the chloroplast outer membrane or in the cytosol could also exist. Such targets could be identified in the future using phosphoproteomics approaches.
OEK70, which phosphorylates the C-terminal G- and M-domains of TOC159 in pea, has been characterized earlier but was never identified at the molecular level (33). Based on the similar mass of Arabidopsis KOC1, it is tempting to speculate that OEK70 may be a KOC1 homolog in pea.
The proposition that KOC1 affects import of TOC159 as well as TOC132/-120-dependent import substrates (pSSu for TOC159 and pE1α for TOC132/-120 (23)) was tested in in vitro protein import assays (Fig. 6, A and B). In both cases, import into koc1-1 mutant chloroplasts was reduced by about 40%. This suggests that KOC1 is required for efficient protein import in both pathways. As the concentrations of the TOC components in koc1 chloroplasts were similar to wild type (Fig. 3D), it appears probable that KOC1 directly regulates the activity of the import receptors. This is likely to implicate phosphorylation, but we cannot exclude other possibilities. Based on the diminished import activity in the mutant chloroplasts, the effect of KOC1 is predicted to be positive. In contrast, PKA-dependent phosphorylation of Tom70 inhibited receptor activity and import of metabolite carriers in mitochondria (45).
Despite the diminished import efficiency, the koc1 mutant had a wild-type phenotype under standard growth conditions. Similarly, TOC159 and TOC33 GTPase mutants had diminished import efficiency in vitro but were phenotypically indistinguishable from the wild type (26, 29). Possibly the mutant plants are able to compensate for the chloroplast protein import deficit over time. A phenotype in such mutants might only occur at specific points in development where high import capacity is required. Such a point in development is the switch from etioplasts (chloroplast precursor organelle in the dark-grown plants) to chloroplasts that develop when plants are moved into the light, a process known as de-etiolation.
In the de-etiolation assay (Fig. 6C) (30), dark-grown seedlings near seed depletion were moved to the light. Under these conditions, chloroplast biogenesis must be completed quickly to prevent starvation and initiate photoautotrophic growth. In the de-etiolation assay, the koc1 seedlings had a lower survival rate than the wild-type and NTAP-KOC1:koc1-1 seedlings, corroborating the proposition that KOC1 activity is required when protein import demand is high.
SnRK2 kinase phosphorylates the A-domain of TOC159 in an ABA-dependent fashion. This suggests hormonal control of TOC159 phosphorylation and import activity. It appears likely that various regulatory pathways converge at TOC159 and its homologs via pathway-associated kinases.
Experimental procedures
DNA constructs
Recombinant KOC1 proteins were expressed from constructs obtained by PCR amplification from AT4G32250 cDNA using the primers KOC1_NheI_F and KOC1_NotI_R for KOC1-FL, KOC1_NcoI_F and KOC1_NcoI_R for KOC1(1–343), and KOC1_NheI_F and KOC1_NcoI_R2 for KOC1(1–547) (see supplemental Table S2) and cloned in fusion with a C-terminal hexahistidinyl tag into the pET21d plasmid. The pCHF8-NTAP-KOC1 vector was obtained by inserting KOC1 (amplified using primers KOC1_FL_NcoI_F and KOC1_FL_XbaI_R from plasmid pET21d-KOC1-FL) (supplemental Table S2), digesting with NcoI, and ligating into the BspHI-XbaI-digested vector pCHF8-NTAPi (37). To obtain the pCHF7-NTAP-TOC159-cmyc construct, a DNA fragment was amplified using primers Toc159-StuI-F and Toc159-cmyc-Gib-R (supplemental Table S2) from plasmid pCHF7-NTAPi-Toc159 (26), and from this fragment a second DNA fragment was amplified using primers Toc159-StuI-F and cmyc-uniGib-R and assembled into the StuI-XbaI-digested vector pCHF7-NTAPi-Toc159 by Gibson technology following the manufacturer's instructions (Gibson Assembly kit, New England Biolabs). For the pEG104-N-YFP-KOC1 construct, the sequence coding for KOC1 was amplified using primers attB-KOC1-FW and attB-KOC1-REV (supplemental Table S2) and recombined in pEG104 using the Gateway® system (Invitrogen Gateway technology with ClonaseTM II). For the pEG101-C-YFP-EMB2004 construct, the sequence coding for EMB2004 (AT1G10510) was amplified using primers attB-EMB2004-FW and attB-EMB2004-REV and recombined in pEG101 vector using the Gateway system.
Plant material
Plants were grown in vitro under long-day conditions (16-h light, 8-h dark) or short-day conditions (8-h light, 16-h dark) at 100–120 μmol·m−2 s−1 and 21 °C. Agar plates contained 0.8% (w/v) Phytoagar (Duchefa); 0.5× Murashige and Skoog medium (Duchefa); and 0, 0.5, 0.8, or 1% (w/v) sucrose (for survival, import, subplastidial fractionation and protoplast isolation, or IgG purification experiments, respectively). The Arabidopsis thaliana mutants NTAP-TOC159:ppi2 (NTAP-TOC159) and TAP:WT were described previously (26). The homozygous T-DNA insertion lines SALK_083378 (koc1-1) and SALK_051823 (koc1-2) (46) were selected on 0.5× Murashige and Skoog medium containing kanamycin (50 mg/liter) and screened by PCR amplification using the primers koc1-2_LP with koc1-2_RP; koc1-1_LP with koc1-1_RP; and koc1-2_LP, koc1-2_RP, or koc1-1_LP with LbB1.3. Wild-type Columbia-0 (Col-0) plants were used. The transgenic plants NTAP-KOC1:koc1-1 (NTAP-KOC1) and NTAP-TOC159-cmyc:ppi2 (NTAP-TOC159-cmyc) were obtained by transformation of homozygous koc1-1 and heterozygous ppi2 plants (25) using the vectors pCHF8-NTAP-KOC1 and pCHF7-NTAP-TOC159-cmyc, respectively. Plants were selected as described (26) and displayed a wild-type phenotype under standard growth conditions.
Protein expression and purification
KOC1-FL, KOC1(1–343) and KOC1(1–547) were overexpressed in E. coli SoluBL21 (Genlantis) transformed with the corresponding expression vector. For antibody (α-KOC1) production and purification, recombinant proteins were purified on Ni2+-nitrilotriacetic acid (Ni2+-NTA)-agarose beads under denaturing conditions according to the manufacturer's instructions (QiaexpressionistTM, Qiagen).
Native recombinant KOC1-FL was purified from bacterial pellets resuspended in buffer L (50 mm NaH2PO4, 300 mm NaCl, 20 mm imidazole, 0.1% (v/v) TX100, 1 mm PMSF, 0.2% (v/v) protease inhibitor mixture (Sigma), pH 8.0). Bacterial cells were disrupted by high pressure using a French press, incubated for 30 min with DNase (Roche Applied Science) (0.2 μl/ml), and centrifuged for 30 min at 40,000 × g. The supernatant was filtered (0.2 μm) and incubated for 1 h with Ni2+-NTA beads. The resin was washed two times with buffer L containing 20 mm imidazole and one time with the same buffer containing 0.5% n-dodecyl β-d-maltoside (DDM) instead of Triton X-100. Recombinant KOC1-FL was eluted with buffer L containing 250 mm imidazole and 0.5% DDM. Eluates were dialyzed against 30 mm Tris-HCl, pH 7.5, 75 mm NaCl, 75 mm KCl, 1 mm PMSF, 0.5% DDM. All procedures were done at 4 °C.
TOC120A (TOC120(1–343)-His6), TOC132A (TOC132(1–431)-His6) (21), and TOC159A (TOC159(1–740)-His6) were overexpressed in transformed E. coli strain BL21(DE3) cells. The cells were lysed in buffer L′ (50 mm Tris-HCl, pH 8, 300 mm NaCl, 5 mm imidazole, 1 mm PMSF) by high pressure using a French press. After centrifugation for 30 min at 40,000 × g the supernatant fraction was passed over an Ni2+-NTA column using an ÄKTA Prime® system (for TOC159A) or incubated for 2 h with Ni2+-NTA beads in microtubes (for TOC132A and TOC120A). The column was washed three times with Buffer L′. The proteins were eluted in a buffer L′ containing 250 mm imidazole. TOC120A and TOC132A were dialyzed against 10 mm Tris-HCl, pH 8, 50 mm NaCl. TOC159A was dialyzed against 20 mm piperazine, pH 5.5, 50 mm NaCl and further purified on a DEAE ion-exchange column using ÄKTA Prime as described previously (47). The proteins were precipitated by CHCl3-methanol extraction methods (48).
Antibodies
Antibodies for TOC and TIC components were described previously (17, 25, 49, 50). Antibodies against PGL35 were described previously (51). Antibodies against the following proteins were obtained as indicated: CBP from Genscript, LHCB2 from Agrisera, and c-myc and IgG from Cell Signaling Technology. For anti-KOC1 antibody production, two forms of recombinant KOC1 (KOC1(1–343) and KOC1(1–547)) purified by Ni2+-NTA-affinity chromatography were pooled and injected into rabbits for polyclonal antibody production (Eurogentec, Seraing, Belgium). To affinity-purify antibodies from the serum, purified recombinant KOC1-FL was cross-linked to Affi-Gel 10 (Bio-Rad) according to the manufacturer's protocol. The serum was incubated on the KOC1-FL Affi-Gel column. The column was washed twice with 10 ml of PBS buffer. The anti-KOC1 antibodies were eluted with 0.2 m glycine pH 2.2 buffer and immediately neutralized with 1 m Tris, pH 8.
IgG-affinity purification
The procedure has already been described (37) and was applied with few modifications. All steps were performed at 4 °C. Plants grown in vitro (10 g fresh weight) were ground in a mortar in a total volume of 18 ml of buffer G (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 1 mm PMSF, 5 mm NaF, 0.2% (v/v) protease inhibitor mixture). The 100,000 × g pellet fraction of the NTAP-KOC1 or NTAP-TOC159 was resuspended in buffer G and centrifuged for an additional 1 h at 100,000 × g. Pellet-associated proteins were solubilized in buffer G containing 1.65% (v/v) TX100 and 10% glycerol (buffer GKOC) for NTAP-KOC1 or 0.375% (v/v) TX100 and 5% glycerol (buffer GTOC) for NTAP-TOC159. Proteins of TAP:WT plants were extracted directly in buffer GKOC or GTOC. Solubilized proteins were incubated overnight with 100 μl of IgG-Sepharose resin. The beads were washed once with 35 ml and six times with 5 ml of buffer GKOC for NTAP-KOC1 or buffer GTOC for NTAP-TOC159. The last wash was done with the same buffer without proteases inhibitors. The proteins were eluted in 50 mm Tris-HCl, pH 8, 0.5 mm EDTA, 100 mm NaCl, 1 mm dithiothreitol (DTT), 1.65 or 0.375% TX100, 5 or 10% glycerol for NTAP-KOC1 or NTAP-TOC159, respectively, for 2 h at 16 °C with 50 units of AcTEVTM protease (Invitrogen). 50 μg of proteins of the “total” fraction (total, load, flow-through, and first wash fractions) or 10% of the fraction (last wash and eluate fractions) were loaded for SDS-PAGE and transferred by Western blotting in Dunn buffer onto nitrocellulose membrane.
Transient expression in Arabidopsis protoplasts
Protoplasts were isolated from Col-0 plants (4 weeks old) grown in short-day conditions and transformed using a polyethylene glycol-based method adapted from Jin et al. (52) and Yoo et al. (53) as described in Köhler et al. (17). Protoplasts were transformed with plasmids pEG104-N-YFP-KOC1 for the localization of YFP-KOC1, pEG101-C-YFP-EMB2004 as an envelope marker, and pCL60 (Stratagene) as a control.
Chloroplast isolation
For the isolation of intact chloroplasts, we used the protocols by Smith et al. (54) and Agne et al. (26) with the following modifications. Chloroplasts were obtained from Arabidopsis plants (2 weeks old) grown in vitro under long-day conditions. The tissue (5–9 g) was enzymatically digested using 1.5% (w/v) cellulase “Onozuka” and 0.375% (w/v) Macerozyme R-10 (Serva). The digestion was extended to 12 h at 19 °C.
Chloroplast protease treatment and extraction
Intact chloroplasts were subjected to thermolysin treatment according to Smith et al. (54). Chloroplast pellet corresponding to 20 μg of chlorophyll was resuspended in 100 μl of HEPES-sorbitol buffer and incubated for 1 h on ice with 20 μl of thermolysin (2 mg/ml). For alkaline extraction, we used chloroplast corresponding to 20 μg of chlorophyll incubated for 10 min on ice with 600 μl of 0.1 m Na2CO3, pH 11. The fractions were separated by centrifugation for 1 h at 100,000 × g.
Chloroplast fractionation
Fractionation of intact chloroplasts was carried out according to Vidi et al. (51) and Hiltbrunner et al. (49) with some modifications. NTAP-KOC1:koc1-1 plants (140 g fresh weight) grown on soil for 8 weeks under short-day conditions were ground in a blender in 400 ml of HB buffer (450 mm sorbitol, 20 mm Tricine-KOH, pH 8.4, 10 mm EDTA, pH 8.4, 10 mm NaHCO3, 1 mm MnCl2, 5 mm sodium ascorbate, 1 mm PMSF). The lysate was filtered through two layers of Miracloth and centrifuged at 4 °C for 5 min at 600 × g. The pellet was resuspended in 10 ml of RB buffer. Intact chloroplasts were purified on a Percoll step gradient (40% (v/v) and 85% (v/v) in RB buffer (300 mm sorbitol, 20 mm Tricine-KOH, pH 8.4, 2.5 mm EDTA, pH 8.4, and 5 mm MgCl2)). Intact chloroplast were washed with 50 ml of RB buffer and centrifuged for 5 min at 700 × g. After centrifugation, the chloroplasts were hypertonically lysed in 0.6 m sucrose TED buffer (500 mm Tricine, pH 7.5, 20 mm EDTA, 20 mm DTT) at −80 °C overnight. The thawed suspension was resuspended using a Potter homogenizer and centrifuged at 100,000 × g for 1 h at 4 °C. The membrane pellet was resuspended in 45% sucrose in TED buffer (3.5 mg/ml) using a Potter homogenizer. The total membrane fraction (corresponding to 12 mg of chlorophyll) was separated on a linear sucrose gradient (5–45%) in TED buffer and centrifugation for 16 h at 100,000 × g.
Chloroplast protein import assay
Chloroplast import experiments were performed according to Agne et al. (26) with some modifications. For each reaction, we used chloroplasts corresponding to 20 μg of chlorophyll and 4 μl of in vitro translated [35S]methionine-labeled preproteins (TnT® T7 Quick-coupled Transcription/Translation System, Promega). The in vitro translocated, 35S-labeled preproteins of the small subunit of Rubisco (pre-SSu) and the α subunit of pyruvate dehydrogenase E1α (preE1α) (pET21d-preE1-α-DHFR-His6) (23) were used as substrates. The proteins of the import experiments were separated by SDS-PAGE, and dried gels were analyzed using a phosphorimaging system (Molecular Imager® FX (Bio-Rad) and Quantity One 4.6 software for quantification.
De-etiolation survival test
koc1-1, koc1-2, WT, and NTAP-KOC1:koc1-1 plants were grown on 0.5× Murashige and Skoog medium, and seeds were cold-treated for 3 days at 4 °C to synchronize germination. The seeds were exposed for 4 h to standard light and then grown in the dark. After 6 days, the plants were exposed to standard light conditions for 2 weeks. The germination and survival rates were calculated.
Phosphorylation assays
KOC1 purified from NTAP-KOC1:koc1-1 plants by IgG-affinity purification was incubated for 30 min at 25 °C with 3–5 μg of either purified TOC159A, TOC120A, or TOC132A in phosphorylation buffer (50 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 5 mm MnCl2, 5 mm CaCl2,1 mm DTT, 50 μm ATP) in the presence of 10 μCi of [γ-33P]ATP. In addition, KOC1 denatured for 10 min at 65 °C was also incubated with TOC159A in the presence of [γ-33P]ATP. Reactions were stopped by diluting in ice-cold phosphorylation buffer followed by CHCl3-methanol precipitation (48). The samples were separated by SDS-PAGE and examined by autoradiography.
Native recombinant KOC1-FL (20 μg) purified on Ni2+-NTA-agarose beads was incubated for 30 min at 25 °C with 10 μg of purified TOC159A in phosphorylation buffer containing 0.2% DDM and [γ-33P]ATP. The reactions were treated as described in the preceding paragraph.
Mass spectrometric identification of KOC-interacting proteins
KOC-interacting proteins were isolated by a TAP-tagged version of KOC compared with a control. Proteins were identified and quantified after nano-LC separation using the data-independent HD-MSE data acquisition method as previously described (55). In brief, LC separation and HD-MSE data acquisition were performed using 1 μl from each of the in solution-digested samples on a nanoACQUITY UPLC system coupled to a Synapt G2-S mass spectrometer (Waters, Eschborn, Germany). MS acquisition range was set to 50–5000 Da. Data analysis was carried out using the ProteinLynx Global Server (PLGS 3.0.1, Apex3D algorithm v.2.128.5.0, 64-bit, Waters) with automated determination of chromatographic peak width as well as MS TOF resolution. The lock mass value for charge state 2 was defined as 785.8426 Da/e, and the lock mass window was set to 0.25 Da. The data bank search query (PLGS workflow) was carried out as follows. Peptide and fragment tolerances were set to automatic (resulting in a maximum mass tolerance of 30 ppm), two fragment ion matches per peptide, five fragment ions for protein identification, and two peptides per protein. The false discovery rate was set to 4% at the protein level. MSE data were searched against the modified A. thaliana database (TAIR10) containing common contaminants such as keratin and glycogen phosphorylase B from rabbit (UniProt ID P00489). Quantification was performed based on the intensity of the three most abundant proteotypic peptides (Hi3 method (56)). Glycogen phosphorylase B was used at 10 fmol/μl as the internal quantification standard.
Author contributions
M. Z., C. M., V. D., and E. D. performed and analyzed experiments. M. Z., B. A., S. B., and F. K. designed the study, analyzed the results, and prepared the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
This work was supported by the University of Neuchâtel and Swiss National Science Foundation Grants 31003A_156998 and IZEBZ0_143169. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S3 and Tables S1 and S2.
- TOC
- translocon at the outer chloroplast membrane
- TIC
- translocon at the inner chloroplast membrane
- A
- acidic
- KOC
- kinase at the outer chloroplast membrane
- G
- GTP-binding
- M
- membrane-anchoring
- UPS
- ubiquitin-proteasome system
- OEK
- outer envelope kinase
- CK
- casein kinase
- SnRK2
- sucrose nonfermenting 1-related protein kinase 2
- ABA
- abscisic acid
- TAP
- tandem affinity purification
- NTAP
- N-terminal TAP tag
- CBP
- calmodulin-binding peptide
- TEV
- tobacco etch virus
- KEG
- Keep On Going
- TOM
- translocase of the mitochondrial outer membrane
- Ni2+-NTA
- Ni2+-nitrilotriacetic acid
- TX100
- Triton X-100
- DDM
- n-dodecyl β-d-maltoside
- SSu
- small subunit
- Rubisco
- ribulose-bisphosphate carboxylase/oxygenase
- HD-MSE
- high-definition mass spectrometry
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
References
- 1. Timmis J. N., Ayliffe M. A., Huang C. Y., and Martin W. (2004) Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5, 123–135 [DOI] [PubMed] [Google Scholar]
- 2. Bruce B. D. (2000) Chloroplast transit peptides: structure, function and evolution. Trends Cell Biol. 10, 440–447 [DOI] [PubMed] [Google Scholar]
- 3. Schnell D. J., Kessler F., and Blobel G. (1994) Isolation of components of the chloroplast protein import machinery. Science 266, 1007–1012 [DOI] [PubMed] [Google Scholar]
- 4. Schnell D. J., Blobel G., Keegstra K., Kessler F., Ko K., and Soll J. (1997) A consensus nomenclature for the protein-import components of the chloroplast envelope. Trends Cell Biol. 7, 303–304 [DOI] [PubMed] [Google Scholar]
- 5. Richardson L. G., Paila Y. D., Siman S. R., Chen Y., Smith M. D., and Schnell D. J. (2014) Targeting and assembly of components of the TOC protein import complex at the chloroplast outer envelope membrane. Front. Plant Sci. 5, 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kessler F., and Schnell D. (2009) Chloroplast biogenesis: diversity and regulation of the protein import apparatus. Curr. Opin. Cell Biol. 21, 494–500 [DOI] [PubMed] [Google Scholar]
- 7. Paila Y. D., Richardson L. G., and Schnell D. J. (2015) New insights into the mechanism of chloroplast protein import and its integration with protein quality control, organelle biogenesis and development. J. Mol. Biol. 427, 1038–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Andrès C., Agne B., and Kessler F. (2010) The TOC complex: preprotein gateway to the chloroplast. Biochim. Biophys. Acta 1803, 715–723 [DOI] [PubMed] [Google Scholar]
- 9. Paila Y. D., Richardson L. G., Inoue H., Parks E. S., McMahon J., Inoue K., and Schnell D. J. (2016) Multi-functional roles for the polypeptide transport associated domains of Toc75 in chloroplast protein import. Elife 5, e12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kovács-Bogdán E., Benz J. P., Soll J., and Bölter B. (2011) Tic20 forms a channel independent of Tic110 in chloroplasts. BMC Plant Biol. 11, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kouranov A., Chen X., Fuks B., and Schnell D. J. (1998) Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J. Cell Biol. 143, 991–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nakai M. (2015) The TIC complex uncovered: the alternative view on the molecular mechanism of protein translocation across the inner envelope membrane of chloroplasts. Biochim. Biophys. Acta 1847, 957–967 [DOI] [PubMed] [Google Scholar]
- 13. Su P.-H., and Li H. (2010) Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell 22, 1516–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shi L.-X., and Theg S. M. (2010) A stromal heat shock protein 70 system functions in protein import into chloroplasts in the moss Physcomitrella patens. Plant Cell 22, 205–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Flores-Pérez Ú., and Jarvis P. (2013) Molecular chaperone involvement in chloroplast protein import. Biochim. Biophys. Acta. 1833, 332–340 [DOI] [PubMed] [Google Scholar]
- 16. Kikuchi S., Bédard J., Hirano M., Hirabayashi Y., Oishi M., Imai M., Takase M., Ide T., and Nakai M. (2013) Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 339, 571–574 [DOI] [PubMed] [Google Scholar]
- 17. Köhler D., Montandon C., Hause G., Majovsky P., Kessler F., Baginsky S., and Agne B. (2015) Characterization of chloroplast protein import without Tic56, a component of the 1-megadalton translocon at the inner envelope membrane of chloroplasts. Plant Physiol. 167, 972–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kessler F., Blobel G., Patel H. A., and Schnell D. J. (1994) Identification of two GTP-binding proteins in the chloroplast protein import machinery. Science 266, 1035–1039 [DOI] [PubMed] [Google Scholar]
- 19. Kessler F., and Schnell D. J. (2004) Chloroplast protein import: solve the GTPase riddle for entry. Trends Cell Biol. 14, 334–338 [DOI] [PubMed] [Google Scholar]
- 20. Hiltbrunner A., Bauer J., Alvarez-Huerta M., and Kessler F. (2001) Protein translocon at the Arabidopsis outer chloroplast membrane. Biochem. Cell Biol. 79, 629–635 [DOI] [PubMed] [Google Scholar]
- 21. Ivanova Y., Smith M. D., Chen K., and Schnell D. J. (2004) Members of the Toc159 import receptor family represent distinct pathways for protein targeting to plastids. Mol. Biol. Cell 15, 3379–3392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Demarsy E., Lakshmanan A. M., and Kessler F. (2014) Border control: selectivity of chloroplast protein import and regulation at the TOC-complex. Front. Plant Sci. 5, 483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Inoue H., Rounds C., and Schnell D. J. (2010) The molecular basis for distinct pathways for protein import into Arabidopsis chloroplasts. Plant Cell 22, 1947–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee K. H., Kim S. J., Lee Y. J., Jin J. B., and Hwang I. (2003) The M domain of atToc159 plays an essential role in the import of proteins into chloroplasts and chloroplast biogenesis. J. Biol. Chem. 278, 36794–36805 [DOI] [PubMed] [Google Scholar]
- 25. Bauer J., Chen K., Hiltbunner A., Wehrli E., Eugster M., Schnell D., and Kessler F. (2000) The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature 403, 203–207 [DOI] [PubMed] [Google Scholar]
- 26. Agne B., Infanger S., Wang F., Hofstetter V., Rahim G., Martin M., Lee D. W., Hwang I., Schnell D., and Kessler F. (2009) A Toc159 import receptor mutant, defective in hydrolysis of GTP, supports preprotein import into chloroplasts. J. Biol. Chem. 284, 8670–8679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schleiff E., Jelic M., and Soll J. (2003) A GTP-driven motor moves proteins across the outer envelope of chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 100, 4604–4609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang F., Agne B., Kessler F., and Schnell D. J. (2008) The role of GTP binding and hydrolysis at the atToc159 preprotein receptor during protein import into chloroplasts. J. Cell Biol. 183, 87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aronsson H., Combe J., Patel R., Agne B., Martin M., Kessler F., and Jarvis P. (2010) Nucleotide binding and dimerization at the chloroplast pre-protein import receptor, atToc33, are not essential in vivo but do increase import efficiency. Plant J. 63, 297–311 [DOI] [PubMed] [Google Scholar]
- 30. Ling Q., Huang W., Baldwin A., and Jarvis P. (2012) Chloroplast biogenesis is regulated by direct action of the ubiquitin-proteasome system. Science 338, 655–659 [DOI] [PubMed] [Google Scholar]
- 31. Kessler F. (2012) Chloroplast delivery by UPS. Science 338, 622–623 [DOI] [PubMed] [Google Scholar]
- 32. Ling Q., and Jarvis P. (2015) Regulation of chloroplast protein import by the ubiquitin E3 ligase SP1 is important for stress tolerance in plants. Curr. Biol. 25, 2527–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fulgosi H., and Soll J. (2002) The chloroplast protein import receptors Toc34 and Toc159 are phosphorylated by distinct protein kinases. J. Biol. Chem. 277, 8934–8940 [DOI] [PubMed] [Google Scholar]
- 34. Jelic M., Sveshnikova N., Motzkus M., Hörth P., Soll J., and Schleiff E. (2002) The chloroplast import receptor Toc34 functions as preprotein-regulated GTPase. Biol. Chem. 383, 1875–1883 [DOI] [PubMed] [Google Scholar]
- 35. Sveshnikova N., Soll J., and Schleiff E. (2000) Toc34 is a preprotein receptor regulated by GTP and phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 97, 4973–4978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Aronsson H., Combe J., Patel R., and Jarvis P. (2006) In vivo assessment of the significance of phosphorylation of the Arabidopsis chloroplast protein import receptor, atToc33. FEBS Lett. 580, 649–655 [DOI] [PubMed] [Google Scholar]
- 37. Agne B., Andrès C., Montandon C., Christ B., Ertan A., Jung F., Infanger S., Bischof S., Baginsky S., and Kessler F. (2010) The acidic A-domain of Arabidopsis Toc159 occurs as a hyperphosphorylated protein. Plant Physiol. 153, 1016–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang P., Xue L., Batelli G., Lee S., Hou Y.-J., Van Oosten M. J., Zhang H., Tao W. A., and Zhu J.-K. (2013) Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. U.S.A. 110, 11205–11210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ferro M., Brugière S., Salvi D., Seigneurin-Berny D., Court M., Moyet L., Ramus C., Miras S., Mellal M., Le Gall S., Kieffer-Jaquinod S., Bruley C., Garin J., Joyard J., Masselon C., and Rolland N. (2010) AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol. Cell. Proteomics 9, 1063–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zybailov B., Rutschow H., Friso G., Rudella A., Emanuelsson O., Sun Q., and van Wijk K. J. (2008) Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One 3, e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stone S. L., Williams L. A., Farmer L. M., Vierstra R. D., and Callis J. (2006) KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18, 3415–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim D. H., and Hwang I. (2013) Direct targeting of proteins from the cytosol to organelles: the ER versus endosymbiotic organelles. Traffic 14, 613–621 [DOI] [PubMed] [Google Scholar]
- 43. Dhanoa P. K., Richardson L. G., Smith M. D., Gidda S. K., Henderson M. P., Andrews D. W., and Mullen R. T. (2010) Distinct pathways mediate the sorting of tail-anchored proteins to the plastid outer envelope. PLoS One 5, e10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gerbeth C., Schmidt O., Rao S., Harbauer A. B., Mikropoulou D., Opalińska M., Guiard B., Pfanner N., and Meisinger C. (2013) Glucose-induced regulation of protein import receptor tom22 by cytosolic and mitochondria-bound kinases. Cell Metab. 18, 578–587 [DOI] [PubMed] [Google Scholar]
- 45. Schmidt O., Harbauer A. B., Rao S., Eyrich B., Zahedi R. P., Stojanovski D., Schönfisch B., Guiard B., Sickmann A., Pfanner N., and Meisinger C. (2011) Regulation of mitochondrial protein import by cytosolic kinases. Cell 144, 227–239 [DOI] [PubMed] [Google Scholar]
- 46. Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., Chen H., Shinn P., Stevenson D. K., Zimmerman J., Barajas P., Cheuk R., Gadrinab C., Heller C., Jeske A., Koesema E., Meyers C. C., et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657 [DOI] [PubMed] [Google Scholar]
- 47. Richardson L. G., Jelokhani-Niaraki M., and Smith M. D. (2009) The acidic domains of the Toc159 chloroplast preprotein receptor family are intrinsically disordered protein domains. BMC Biochem. 10, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wessel D., and Flügge U. I. (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143 [DOI] [PubMed] [Google Scholar]
- 49. Hiltbrunner A., Bauer J., Vidi P. A., Infanger S., Weibel P., Hohwy M., and Kessler F. (2001) Targeting of an abundant cytosolic form of the protein import receptor at Toc159 to the outer chloroplast membrane. J. Cell Biol. 154, 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ma Y., Kouranov A., LaSala S. E., and Schnell D. J. (1996) Two components of the chloroplast protein import apparatus, IAP86 and IAP75, interact with the transit sequence during the recognition and translocation of precursor proteins at the outer envelope. J. Cell Biol. 134, 315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vidi P.-A., Kanwischer M., Baginsky S., Austin J. R., Csucs G., Dörmann P., Kessler F., and Bréhélin C. (2006) Tocopherol cyclase (VTE1) localization and vitamin E accumulation in chloroplast plastoglobule lipoprotein particles. J. Biol. Chem. 281, 11225–11234 [DOI] [PubMed] [Google Scholar]
- 52. Jin J. B., Kim Y. A., Kim S. J., Lee S. H., Kim D. H., Cheong G. W., and Hwang I. (2001) A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13, 1511–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yoo S.-D., Cho Y.-H., and Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572 [DOI] [PubMed] [Google Scholar]
- 54. Smith M. D., Schnell D. J., Fitzpatrick L., and Keegstra K. (2003) In vitro analysis of chloroplast protein import. Curr. Protoc. Cell Biol. Chapter 11, Unit 11.16 [DOI] [PubMed] [Google Scholar]
- 55. Helm S., Dobritzsch D., Rödiger A., Agne B., and Baginsky S. (2014) Protein identification and quantification by data-independent acquisition and multi-parallel collision-induced dissociation mass spectrometry (MSE) in the chloroplast stroma proteome. J. Proteomics. 98, 79–89 [DOI] [PubMed] [Google Scholar]
- 56. Silva J. C., Gorenstein M. V., Li G. Z., Vissers J. P., and Geromanos S. J. (2006) Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol. Cell. Proteomics 5, 144–156 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.