Abstract
Absorption of dietary sphingomyelin (SM) requires its initial degradation into ceramide, a process catalyzed by the intestinal enzyme alkaline sphingomyelinase (alk-SMase, NPP7, ENPP7). alk-SMase belongs to the nucleotide pyrophosphatase/phosphodiesterase (NPP) family, the members of which hydrolyze nucleoside phosphates, phospholipids, and other related molecules. NPP7 is the only paralog that can cleave SM, and its activity requires the presence of bile salts, a class of physiological anionic detergents. To elucidate the mechanism of substrate recognition, we determined the crystal structure of human alk-SMase in complex with phosphocholine, a reaction product. Although the overall fold and catalytic center are conserved relative to other NPPs, alk-SMase recognizes the choline moiety of its substrates via an NPP7-specific aromatic box composed of tyrosine residues. Mutational analysis and enzymatic activity assays identified features on the surface of the protein—a cationic patch and a unique hydrophobic loop—that are essential for accessing SM in bile salt micelles. These results shed new light on substrate specificity determinants within the NPP enzyme family.
Keywords: bile acid, choline, micelle, sphingolipid, sphingomyelinase, ENPP7, NPP7, alk-SMase
Introduction
Sphingomyelin (SM)3 is a major dietary sphingolipid found in dairy products, eggs, meat, and fish (1). Digestion of SM involves its initial degradation to ceramide and phosphocholine (PC) by the enzyme alkaline sphingomyelinase (alk-SMase, NPP7, ENPP7), followed by hydrolysis of ceramide by the neutral ceramidase and subsequent absorption of sphingosine and fatty acids (2). alk-SMase is present in the small intestine on the outer surface of microvilli (3) and in the liver of humans where it is secreted in the bile (4). Intestinal alk-SMase can be dissociated from the brush border by trypsin or bile and act in the lumen (5). In addition to its main substrate SM, the enzyme is also able to cleave lysophosphatidylcholine to monoacylglycerol (6) and inactivate the signaling lipid platelet-activating factor (7). Coupled to the production of ceramide, these functions confer an anti-inflammatory and antiproliferative role to the protein in the intestines and colon (2). Indeed, alk-SMase knock-out mice display increased colonic tumorigenesis (8), and the activity of the enzyme is decreased in human colorectal carcinomas and in familial adenomatous polyposis (9, 10). Providing recombinant alk-SMase has been suggested as potentially beneficial in patients with high risk of colon cancer (8).
alk-SMase is a member of the (ecto)nucleotide pyrophosphatase/phosphodiesterase protein family (NPP), a group of seven extracellular enzymes that act on diverse substrates to carry out various functions (11). NPP1 generates pyrophosphate from ATP to inhibit bone mineralization and ectopic calcification. NPP3 (CD203c) is highly expressed on basophils and hydrolyzes ATP with a negative regulatory role in inflammation and allergy (12). NPP4 participates in the activation of platelet aggregation via cleavage of diadenosine triphosphate (13). In contrast to these enzymes acting on extracellular nucleotides involved in purinergic signaling, other NPP members degrade phospholipid substrates. NPP2 (autotaxin) converts lysophosphatidylcholine to lysophosphatidic acid, a signal mediator that stimulates cell survival, proliferation, differentiation, and migration (14, 15). NPP6 participates in choline metabolism, with glycerophosphocholine as the proposed substrate (16). The specificity and functions of NPP5 are unknown. NPP7 (alk-SMase) is the only NPP that hydrolyzes SM and is thus one of the mammalian SMases, along with the acid SMase and the four neutral SMases, classified by their optimum activity pH. Acid SMase is found in lysosomes, on the outer face of the cell membrane, and in the circulation (17), whereas its neutral counterparts are located on the inner leaflet of the plasma membrane, Golgi, endoplasmic reticulum, nucleus, or mitochondria (18).
Digestion of dietary fats is assisted by their solubilization with bile in the small intestine. Bile contains lipids and bile salts, a class of physiological detergents. Bile salts are cholesterol derivatives with up to three hydroxyl groups on one face of their roughly planar ring system and often bear a conjugated glycine or taurine residue on their tail, conferring to them a negative charge. Their physiological concentration can reach 20 mm (19). Intestinal alk-SMase acts on its substrate solubilized by bile salts, a characteristic distinguishing it from the other mammalian SMases and NPPs but common to other digestive lipases. In vitro, taurocholate (TC) and taurochenodeoxycholate are the most stimulatory bile salts, whereas other detergents commonly used for acid and neutral SMase assays, such as CHAPS and Triton X-100, are not conducive to alk-SMase activity (3). The molecular basis for this bile salt dependence and for the specificity of NPP7 for SM among the NPPs is unknown. First isolated 20 years ago (20), this protein has not yet been structurally characterized, although it has been the subject of molecular modeling studies (21, 22). Here we report the crystal structure of human alk-SMase. The enzyme recognizes PC, the head group of SM, via an NPP7-specific aromatic box composed of tyrosine residues. Structural and functional analysis identified features on the surface of the protein essential for accessing SM in bile salt micelles.
Results
Overall structure
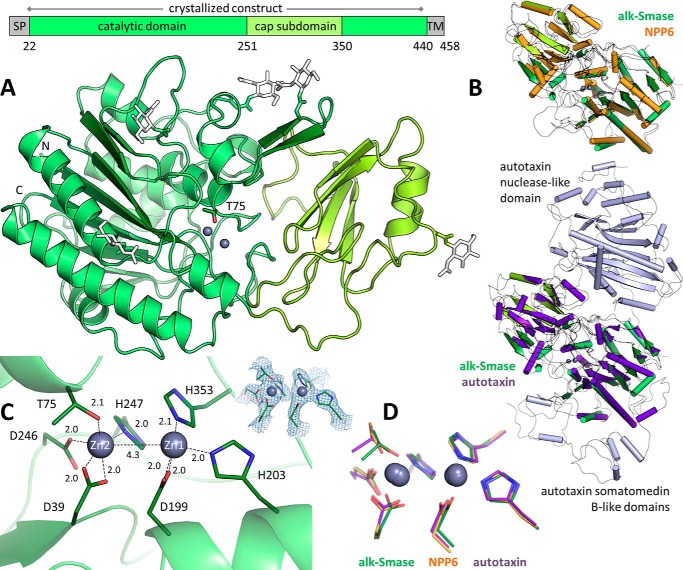
We determined the 2.4 Å-resolution crystal structure of human alk-SMase. The crystallized construct (residues 22–433) excludes the N-terminal signal peptide and the C-terminal transmembrane segment, which normally anchors the protein on the luminal side of the intestinal microvillar membrane but which can be cleaved off by pancreatic trypsin (5). Residues 30–417 of the construct are visible in the electron density, whereas residues 418–433 are disordered; thus the region from residues 414 to 440 likely constitutes a flexible linker of low sequence conservation between the catalytic domain and the C-terminal transmembrane helix.
alk-SMase is composed of a catalytic domain with a “cap” subdomain (Fig. 1A). The core of the catalytic domain is an eight-stranded mixed β-sheet flanked by α-helices on both faces. This α-β-α architecture is characteristic of the alkaline phosphatase superfamily (CATH database code 3.40.720.10). The cap subdomain comprises residues 251–350 and consists of a four-stranded antiparallel β-sheet surrounded mainly by loop segments and a small α-helix. It is packed against one helical side of the catalytic domain, forming a 1,500 Å2 interface. This cap insertion is specific to the NPP family (PFAM code PF00245), which is subdivided into two groups: the single-domain proteins NPP4–7 and the larger members NPP1–3 that contain an additional nuclease-like domain and two somatomedin B-like units (11). The crystal structures of NPP1 (23, 24), NPP2 (autotaxin) (14, 15), NPP4 (13), and NPP6 (16) are known. Despite the rather low sequence identity of ∼30% between alk-SMase and the other NPPs, secondary structure elements are overall conserved (Fig. 1B). alk-SMase contains no disulfide bonds and possesses five N-linked glycosylation sites, all required for full activity (25) and visible in the structure (Fig. 1A), three of which are located on the face of the enzyme harboring the active site.
Figure 1.
Crystal structure of alk-SMase. A, the catalytic domain is in green with the cap subdomain in yellow-green. N-Linked glycans (white sticks) are simplified for clarity. The catalytic threonine (sticks) is labeled. Zinc ions are represented by gray spheres. The N and C termini are indicated. SP, signal peptide; TM, transmembrane helix. B, alk-SMase (green) is superimposed with NPP6 (orange, PDB code 5EGH, 29% identity, root mean square deviation of 1.0 Å over 299 alpha carbons) or autotaxin (purple, PDB code 3NKN, 27% identity, root mean square deviation of 1.1 Å over 270 α carbons). The additional domains of autotaxin are in light blue. C, active site residues of alk-SMase are shown as sticks. Interatomic distances are indicated (Å). The electron density 2Fo − Fc map is contoured at 2 σ. D, the active sites of alk-SMase (green), NPP6 (orange), and autotaxin (purple) are overlaid.
Active site
A depression on the catalytic domain surface encloses two zinc ions held by the side chains of seven residues (Fig. 1C). Zn1 is coordinated by His-353, His-203, and Asp-199 (bidentate), whereas Zn2 is coordinated by His-247, Asp-246, Asp-39 (bidentate), and Thr-75. The catalytic machinery is conserved within the NPP family (Fig. 1D), so the proposed reaction mechanism for these enzymes (26) likely also applies to the cleavage of SM into ceramide and PC by alk-SMase. SM would bind with its phosphate moiety positioned at the zinc ions and undergo a nucleophilic attack on the phosphorus atom by the side chain hydroxyl of Thr-75 activated by Zn2. Departure of the ceramide leaving group results in a covalent phosphocholyl-threonine intermediate. Subsequent nucleophilic attack on this intermediate by a water molecule activated by Zn1 then re-forms Thr-75 and releases PC.
Substrate recognition
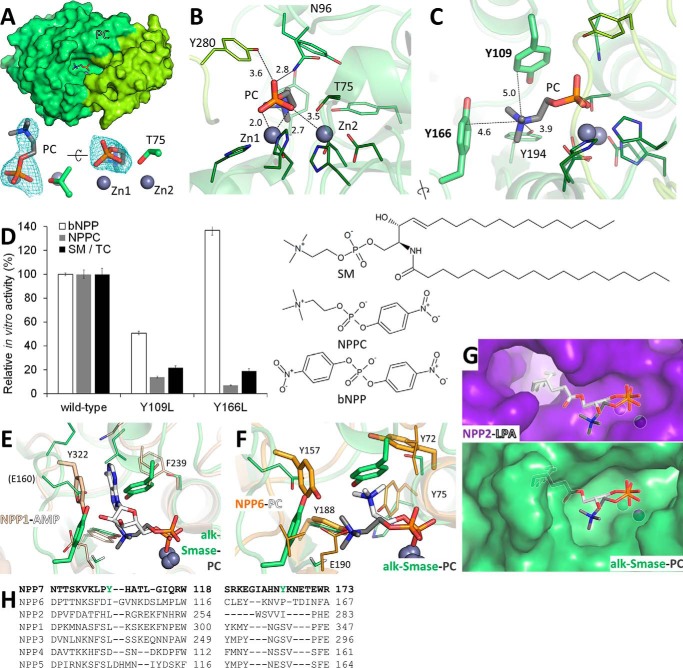
Although we could not obtain a structure of alk-SMase bound to SM, we determined the structure of the enzyme in complex with the lipid head group and reaction product, PC. The molecule is found in the active site with its phosphate moiety positioned at the zinc ions and its choline portion extending sideways into the active site cavity (Fig. 2A). In addition to the strong zinc-phosphate electrostatic interactions, one phosphate oxygen atom is hydrogen-bonded to the side chains of Asn-96, a contact conserved in NPPs, and Tyr-280, specific to alk-SMase (Fig. 2B). The choline quaternary ammonium group forms cation-π interactions with two aromatic side chains oriented perpendicularly to it, Tyr-166 and Tyr-109 (Fig. 2C). Such an arrangement of aromatic residues forming a cation-π “box” is a common choline-binding motif (27). No changes in the protein itself were observed in the PC-bound structure relative to the apo form.
Figure 2.
Recognition of PC by alk-SMase. A, PC (sticks) bound to alk-SMase. Zinc ions (gray spheres) and the catalytic threonine (sticks) are labeled. The electron density Fo − Fc simulated annealing omit map around PC is contoured at 2.5 σ. B, protein residues (sticks) contacting the phosphate group of PC are labeled. C, protein residues (sticks) interacting with the choline moiety of PC are labeled, including the tyrosine side chains forming the aromatic box (bold, thicker sticks). Interatomic distances are indicated (Å). D, enzymatic activity of purified alk-SMase on substrates with chemical structures displayed. 100% activity corresponds to 2.58 and 61.68 μm of p-nitrophenol and 44.72 μm of PC released per nm protein per hour from bNPP, NPPC, and SM in TC micelles, respectively. The values are the means and standard deviations of triplicates representative of one of two experiments. E, superimposition of the PC-alk-SMase complex (gray and green sticks) with the AMP-NPP1 complex (white and beige sticks, PDB code 4GTW). The tyrosine residue of NPP1 stacking with the adenine ring is in thicker sticks. F, superimposition of the PC-alk-SMase complex (gray and green sticks) with the PC-NPP6 complex (white and orange sticks, PDB code 5EGH). For both proteins, the tyrosine side chains forming an aromatic box are in thicker sticks. Key residues of NPP1 and NPP6 are labeled. G, comparison of the substrate binding site of autotaxin (purple, PDB code 3NKN) in complex with lysophosphatidic acid (white sticks) with that of alk-SMase bound to PC (green and gray sticks). Both ligands are superimposed on both proteins. H, structure-assisted sequence alignments of the regions containing the aromatic box of NPP7 (green).
To validate this SM head group recognition mechanism, in vitro enzymatic activity of purified alk-SMase mutants Y109L and Y166L was assayed. These mutants were designed to minimize structural disruption of the protein while removing the aromatic box. The substrates tested were SM, the PC-containing small molecule p-nitrophenylphosphorylcholine (NPPC), and the generic phosphodiesterase substrate bis(4-nitrophenyl) phosphate (bNPP). Both mutants decreased the hydrolysis rate of SM and NPPC 5–10-fold, whereas the activity on bNPP (not PC-based) varied depending on the mutation, although it was 17–24-fold slower than PC-based substrates (Fig. 2D).
NPPs were initially identified as nucleotide phosphodiesterases, but their catalytic repertoire has subsequently been expanded. NTPs, NDPs, dinucleoside polyphosphates, and possibly related molecules such as NAD+ and nucleotide sugars are the major substrates for NPP1, NPP3, and NPP4 (11–13, 23, 24). Crystal structures of NPP1 and NPP4 in complex with the product AMP (13, 24) revealed a binding mode with π-π stacking between the nucleobase and a key tyrosine residue. This amino acid is replaced by a glutamate in alk-SMase (Fig. 2E). Furthermore, other substitutions in surrounding residues sterically prevent positioning of the nucleobase; indeed, alk-SMase is inactive on p-nitrophenyl-TMP (6). On the other hand, NPP2 (autotaxin) and NPP6 can hydrolyze choline-based phospholipids including lysophosphatidylcholine, sphingosylphosphorylcholine, platelet-activating factor, and the small metabolite glycerophosphocholine (11, 14, 16). Nevertheless, their substrate recognition modes are distinct from that of alk-SMase. NPP6 interacts with choline moieties via an aromatic box assisted by an anionic side chain (16); however, an entirely different set of tyrosine residues is involved, not conserved in alk-SMase (Fig. 2F). NPP2 is a phospholipase D, generating lysophosphatidic acid and choline, whereas alk-SMase is a phospholipase C, cleaving the reverse phosphoester bond. NPP2 accommodates the acyl chain of lysophosphatidylcholine inside a hydrophobic tunnel (14), a feature absent from alk-SMase. Therefore, substrates bind to alk-SMase and NPP2 with their head groups in opposite directions (Fig. 2G) to preserve the position of the scissile bond. In summary, the structural and functional data shows that alk-SMase recognizes its SM substrate via an NPP7-specific (Fig. 2H) cation-π box.
Activity with bile salts
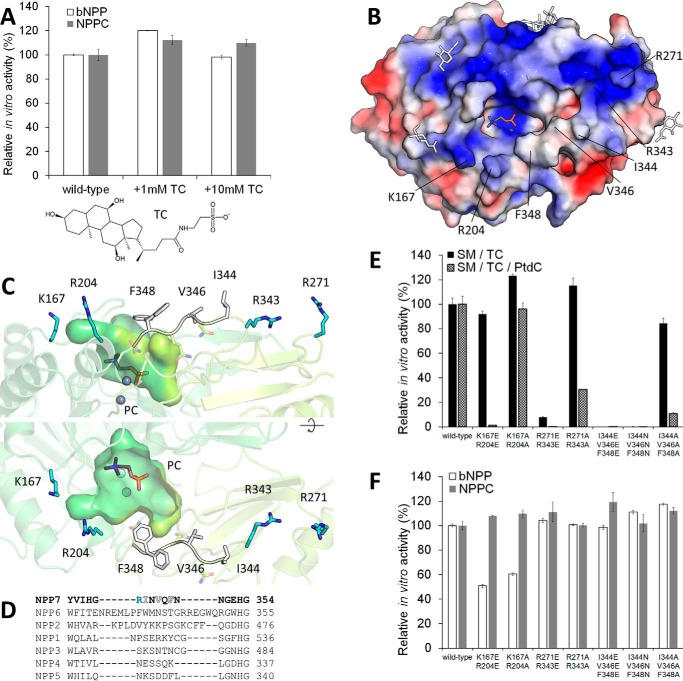
alk-SMase is uniquely dependent on bile salts for activity (2); other detergents such as CHAPS and Triton X-100 are not stimulatory or even inhibit the protein (3). This feature raised the possibility that bile salts may exert a specific stimulatory effect on the enzyme beyond their lipid-solubilizing role, perhaps by directly interacting with the protein (2). To assess this, in vitro enzymatic activity of purified alk-SMase on the non-lipid substrates bNPP and NPPC, which would not rely on bile salts for solubility, was tested in the absence or presence of TC, a common bile salt (Fig. 3A). No stimulation was observed (Fig. 3A) with TC at concentrations below (1 mm) or above (10 mm) its critical micelle concentration of ∼3.2 mm (28) in this experimental system. This result suggests that bile salts do not directly stimulate catalysis by the enzyme.
Figure 3.
Regions of alk-SMase important for bile salt-dependent activity. A, enzymatic activity of purified alk-SMase on substrates in presence of TC (chemical structure displayed). 100% activity corresponds to 2.49 and 50.26 μm of p-nitrophenol released per nm protein per hour from bNPP and NPPC, respectively. B, the electrostatic surface potential of alk-SMase is colored from negative (−3 kT/e, red) to positive (+3 kT/e, blue). PC is shown as gray sticks and N-linked glycans, as simplified white sticks. C, residues near the substrate binding site that form cationic patches (blue sticks) and hydrophobic residues of the loop 342–351 (white sticks) are labeled. Two conformations of Phe-348 are visible. D, loop 344–348 of alk-SMase (NPP7) is aligned with corresponding regions of the other NPPs. E and F, enzymatic activity of purified alk-SMase on substrates. 100% activity corresponds to 2.58 and 61.68 μm of p-nitrophenol and 44.72 and 41.80 μm of PC released per nm protein per hour from bNPP, NPPC, and SM in TC micelles and SM in mixed TC-phosphatidylcholine (PtdC) micelles, respectively. For A, E, and F, the values are the means and standard deviations of triplicates representative of one of two experiments.
Examination of the face of alk-SMase that harbors the active site reveals a mostly positively charged surface (Fig. 3B), a feature that would favor association of the enzyme with anionic bile salt micelles. In addition, a loop (residues 342–351) bearing three solvent-exposed hydrophobic residues is located near the substrate-binding site (Fig. 3C), which could be important for interacting with micellar lipids. This segment is specific to alk-SMase, because its sequence greatly diverges within the NPPs (Fig. 3D). We tested the functional importance of these elements by mutating the hydrophobic loop (Ile-344 + Val-346 + Phe-348), as well as two nearby surface cationic patches (Lys-167 + Arg-204 and Arg-271 + Arg-343; Fig. 3C) and assaying cleavage of SM in anionic TC micelles (Fig. 3E, black bars). Hydrolysis of the non-lipid substrates bNPP and NPPC are largely unaffected by the mutations (Fig. 3F), indicating that the catalytic ability of the protein was not disrupted. Substitution of Lys-167 + Arg-204 had no effect on SM hydrolysis, whereas replacement of Arg-271 + Arg-343 by glutamates decreased activity, suggesting that this patch is located near the micelle adsorption area. The corresponding alanine mutant did not impair SM cleavage, however, indicating that electrostatic interaction of Arg-271 + Arg-343 with the micelle is dispensable for this process. Interestingly, substitution of the three hydrophobic loop residues by glutamates or asparagines abolished SM cleavage, positing this loop as essential for alk-SMase function.
Solubilization of lipids by bile salts results in micelles of markedly different properties than those formed by bile salts alone. In the latter case, micelles are small and composed of 2–10 monomers (28). Presumably, the standard assay conditions used here (0.5 mm SM in 10 mm TC) result in such small micelles because of the low lipid to bile salt ratio. However, at higher ratios of up to 2:1, micelles are over 100 kDa (28) and adopt a disk-like shape with a bilayer of phospholipids whose head groups only are solvent-exposed, surrounded by a ring of bile salt molecules (19). This arrangement is relevant to the digestive system where phospholipids, cholesterol, and fatty acids from bile combine with dietary fats to form mixed micelles. To validate the results described above, the assay was also carried out with 0.4 mm SM in 4 mm TC supplemented with 8 mm phosphatidylcholine (Fig. 3E, checkered bars). Here, mutation of Lys-167 + Arg-204 to glutamates (but not alanines) decreased activity, whereas substitution of Arg-271 + Arg-343 by glutamates or alanines decreased the SM hydrolysis rate by at least two-thirds. This indicates that both cationic patches contact the larger, mixed micelles, but only the electrostatic interaction with Arg-271 + Arg-343 is essential for activity. Furthermore, replacement of the hydrophobic residues Ile-344 + Val-346 + Phe-348 by glutamates, asparagines, or alanines greatly impaired SM cleavage, suggesting that this surface loop is essential for the physiological activity of alk-SMase, perhaps via interactions with the acyl chains of SM or by embedding into micelles. In summary, this mutational analysis reveals features on the surface of alk-SMase important for accessing its substrate in bile salt micelles.
Discussion
The overall structure of alk-SMase is similar to that of the other NPPs, as is the arrangement of its catalytic residues. In the PC-bound alk-SMase structure, the phosphate group position and orientation slightly differ from the other NPP-product complexes (Fig. 2). One explanation could be the relatively poor ligand electron density preventing accurate orientation (Fig. 2A). Alternatively, the hydrogen bond between the phosphate oxygen and the NPP7-specific Tyr-280 could be responsible for this orientation (Fig. 2B). In any case, SM is expected to be positioned closer to the catalytic threonine with respect to its phosphate moiety; the distance between the phosphorus and threonine oxygen atoms is 3.4 Å in the PC complex. Tyr-280 could be involved in interactions with the ceramide portion of the substrate.
Certain digestive lipases, including the gastric lipase (29) and the neutral ceramidase (30), accommodate their substrates inside a deep hydrophobic pocket. In contrast, the PC head group binds in a comparatively shallow cavity on alk-SMase (Fig. 3B); presumably the acyl chains of SM remain mostly buried in the bile salt micelle during activity. To reach their substrate, lipases must first attach to a micelle. In pancreatic triacylglycerol lipase, a hydrophobic loop (β5′) and other parts of the protein, including a “lid” segment, are predicted to contact the lipid-water interface (31). Upon interaction, the conformation of the lid is altered, exposing the active site (31). Loop 342–351 of alk-SMase contains three hydrophobic surface residues necessary for accessing SM in bile salt micelles (Fig. 3C). This loop could either become inserted into the micelle or interact with the acyl chains of an SM molecule.
Bile salts are anionic detergents, and the positively charged surface surrounding the substrate binding site of alk-SMase (Fig. 3B) is likely important for interactions with micelles, including one specific cationic patch (Fig. 3C). Mutation of Arg-271 + Arg-343 disrupted activity on SM in mixed TC-phosphatidylcholine micelles. The importance of electrostatic attraction could explain the in vitro effects of different surfactants on alk-SMase (3). CHAPS is similar in structure to TC, but it is zwitterionic, and Triton X-100 is nonionic; unlike the anionic bile salts, these detergents do not enable SM hydrolysis. Additionally, in the absence of bile salts, the negatively charged lipids phosphatidylserine and phosphatidylinositol, but not the neutral choline or ethanolamine counterparts, stimulate SM hydrolysis to an extent (1).
The bile salt-activated lipase is another intestinal enzyme that requires these detergents for activity. Its unique activation mechanism implicates a specific bile salt-binding site close to a hairpin loop near the active site (32). Upon TC binding, this loop changes conformation and permits access to the active site (32). However, such a mechanism does not seem to be at play in alk-SMase, because its activity on non-lipid substrates is unaffected by the addition of TC (Fig. 3A). Furthermore, we did not find any clear electron density that would indicate a bound bile salt molecule in a 3.2 Å resolution structure of the protein crystallized in presence of 0.33 mm TC (data not shown). Although NPP2 (autotaxin) contains a specific sterol-binding site (33), it serves a different purpose in that paralog.
In conclusion, several features of alk-SMase confer this NPP family member with the ability to hydrolyze SM in the intestinal environment: an aromatic box for PC head group recognition, a hydrophobic loop, and positively charged surfaces for contacting bile salt micelles. Further biophysical characterization of the interactions between the protein and micelles will lead to a better understanding of the activity of this enzyme.
Experimental procedures
Protein expression and purification
Recombinant human alk-SMase was expressed as secreted protein in Sf9 insect cells infected with baculovirus. The endogenous signal peptide comprising the first 21 residues was replaced by the melittin signal peptide MKFLVNVALVFMVVYISYIYA followed by the hexahistidine tag DRHHHHHHKL. The construct encompassed residues 22–433 (UniProt no. Q6UWV6); the last 21 residues forming a transmembrane segment were excluded. All mutants were verified by sequencing. alk-SMase was isolated from expression culture medium using nickel-nitrilotriacetic acid resin (Thermo Fisher Scientific), further purified by size exclusion chromatography on a Superdex 200 column (GE Healthcare) in buffer (15 mm Tris-HCl, pH 7.5, 100 mm NaCl), and concentrated to 10 mg/ml.
Crystallization and structure determination
alk-SMase crystallized in 200 mm NaI with 20% PEG 3350. The crystals were grown by hanging drop vapor diffusion at 22 °C and were flash-frozen. X-ray diffraction data were collected at 100 K on Beamline 08ID-1 with a Rayonix MX300 CCD detector at the Canadian Macromolecular Crystallography Facility of the Canadian Light Source. The data were processed by HKL2000 (34). The apo protein structure was solved by iodine single-wavelength anomalous diffraction using Autosol (35) in Phenix (36) and manually built in Coot (37). Additional crystals were soaked in well solution supplemented with 50 mm PC; the PC-bound alk-SMase structure was obtained by molecular replacement using Phaser (38) in Phenix. Refinement was carried out by phenix.refine (39). Translation-liberation-screw parameters were applied for both structures, and temperature factors for zinc and iodide ions were refined anisotropically for the PC-bound structure. Crystallographic data collection and structure refinement statistics are presented in Table 1. Structural images were prepared with PyMOL (PyMOL Molecular Graphics System, version 1.3; Schrödinger).
Table 1.
Crystallographic data collection and structure refinement statistics
The values in parentheses are for the highest-resolution shell. ASU, asymmetric unit; RMSD, root-mean-square deviation.
| Human alk-SMase with PC | Human alk-SMase | |
|---|---|---|
| PDB code | 5TCD | 5UDY |
| Data collection | ||
| Space group | P3121 | P3121 |
| Unit cell a, b, c (Å) | 104.22, 104.22, 113.89 | 103.25, 103.25, 113.55 |
| Unit cell α, β, γ (°) | 90, 90, 120 | 90, 90, 120 |
| X-ray wavelength (Å) | 0.9795 | 0.9795 |
| Resolution range (Å) | 50–2.40 (2.49–2.40) | 50–2.60 (2.69–2.60) |
| Total reflections | 155,404 (14,219) | 120,607 (11,858) |
| Unique reflections | 28,331 (2,788) | 21,834 (2,156) |
| Multiplicity | 5.5 (5.1) | 5.5 (5.5) |
| Completeness (%) | 100 (100) | 100 (100) |
| Rmeas (%) | 6 (113) | 7 (137) |
| Rpim (%) | 3 (44) | 4 (67) |
| Mean I/σ(I) | 25.8 (1.4) | 19.0 (1.1) |
| CC½ | (0.822) | (0.692) |
| Wilson B factor (Å2) | 31.0 | 38.1 |
| Refinement | ||
| Protein copies per ASU | 1 | 1 |
| Resolution range (Å) | 37.96–2.40 (2.49–2.40) | 35.12–2.60 (2.69–2.60) |
| Reflections used | 23,507 (948) | 18,878 (662) |
| Reflections for Rfree | 1,916 (86) | 1,532 (59) |
| Rwork (%) | 17.3 (23.6) | 17.9 (28.7) |
| Rfree (%) | 20.8 (27.4) | 20.9 (30.7) |
| Non-hydrogen atoms | 3,555 | 3,457 |
| Protein | 3,152 | 3,158 |
| Glycans, ligands, ions | 198 | 136 |
| Water | 205 | 163 |
| Average B factor (Å2) | 39.6 | 43.6 |
| Protein | 37.8 | 43.0 |
| Glycans, ligands, ions | 72.4 | 67.7 |
| Water | 35.8 | 34.6 |
| RMSD bond lengths (Å) | 0.003 | 0.004 |
| RMSD bond angles (°) | 0.60 | 0.72 |
| Ramachandran favored (%) | 96.1 | 97.9 |
| Ramachandran allowed (%) | 3.9 | 2.1 |
| Ramachandran outliers (%) | 0 | 0 |
| Clashscore | 1.1 | 1.4 |
In vitro enzymatic activity assays
The proteins were purified as described above, but the buffer included 10 μm ZnCl2. To measure hydrolysis of SM to PC, the Amplex Red sphingomyelinase assay (Thermo Fisher Scientific, catalog no. A12220) was slightly modified. Proteins at 1 nm were incubated in assay buffer (50 mm Tris-HCl, pH 8, 100 mm NaCl) with substrate: 0.5 mm SM with 10 mm NaTC (Sigma, catalog no. 86339) or 0.4 mm SM with 4 mm NaTC and 8 mm egg phosphatidylcholine (Avanti, catalog no. 840051). After 1 h at 37 °C, the reaction was terminated at 95 °C for 5 min, and an equal volume of the second step solution was added as recommended. To quantify hydrolysis of bNPP and NPPC to 4-nitrophenol, proteins at 100 and 5 nm, respectively, were incubated with 2 mm substrate in assay buffer for 1 h at 37 °C. NaOH was added to 100 mm before measuring absorbance at 405 nm.
Accession codes
Atomic coordinates and structure factors were deposited in the Protein Data Bank under the accession codes 5TCD and 5UDY.
Author contributions
A. G. designed the study and carried out cloning. K. I. conducted baculovirus generation and insect cell culture. F. L. and A. G. purified and crystallized the protein. A. G. performed structural biology and enzymatic work. A. G. and B. N. wrote the manuscript. All authors approved the final version of the manuscript.
Acknowledgment
We thank Dr. Shaun Labiuk for crystallographic data collection performed at Beamline 08ID-1 at the Canadian Light Source.
This work was supported by funds from the Canada Foundation for Innovation, Natural Sciences and Engineering Research Council of Canada, the University of Saskatchewan, the Government of Saskatchewan, Western Economic Diversification Canada, the National Research Council Canada, and the Canadian Institutes of Health Research (to the Canadian Light Source). The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (codes 5TCD and 5UDY) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- SM
- sphingomyelin
- alk-SMase
- alkaline sphingomyelinase
- bNPP
- bis(4-nitrophenyl) phosphate
- (E)NPP
- (ecto)nucleotide pyrophosphatase/phosphodiesterase
- NPPC
- p-nitrophenylphosphorylcholine
- PC
- phosphocholine
- TC
- taurocholate
- PDB
- Protein Data Bank.
References
- 1. Liu J. J., Nilsson A., and Duan R. D. (2000) Effects of phospholipids on sphingomyelin hydrolysis induced by intestinal alkaline sphingomyelinase: an in vitro study. J. Nutr. Biochem. 11, 192–197 [DOI] [PubMed] [Google Scholar]
- 2. Duan R. D. (2006) Alkaline sphingomyelinase: an old enzyme with novel implications. Biochim. Biophys. Acta 1761, 281–291 [DOI] [PubMed] [Google Scholar]
- 3. Duan R. D., Cheng Y., Hansen G., Hertervig E., Liu J. J., Syk I., Sjostrom H., and Nilsson A. (2003) Purification, localization, and expression of human intestinal alkaline sphingomyelinase. J. Lipid Res. 44, 1241–1250 [DOI] [PubMed] [Google Scholar]
- 4. Nyberg L., Duan R. D., Axelson J., and Nilsson A. (1996) Identification of an alkaline sphingomyelinase activity in human bile. Biochim. Biophys. Acta. 1300, 42–48 [DOI] [PubMed] [Google Scholar]
- 5. Wu J., Liu F., Nilsson A., and Duan R. D. (2004) Pancreatic trypsin cleaves intestinal alkaline sphingomyelinase from mucosa and enhances the sphingomyelinase activity. Am. J. Physiol. Gastrointest. Liver Physiol. 287, G967–G973 [DOI] [PubMed] [Google Scholar]
- 6. Duan R. D., Bergman T., Xu N., Wu J., Cheng Y., Duan J., Nelander S., Palmberg C., and Nilsson A. (2003) Identification of human intestinal alkaline sphingomyelinase as a novel ecto-enzyme related to the nucleotide phosphodiesterase family. J. Biol. Chem. 278, 38528–38536 [DOI] [PubMed] [Google Scholar]
- 7. Wu J., Nilsson A, Jönsson B. A., Stenstad H., Agace W., Cheng Y., and Duan R. D. (2006) Intestinal alkaline sphingomyelinase hydrolyses and inactivates platelet-activating factor by a phospholipase C activity. Biochem. J. 394, 299–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen Y., Zhang P., Xu S. C., Yang L., Voss U., Ekblad E., Wu Y., Min Y., Hertervig E., Nilsson Å., Duan R. D. (2015) Enhanced colonic tumorigenesis in alkaline sphingomyelinase (NPP7) knockout mice. Mol. Cancer Ther. 14, 259–267 [DOI] [PubMed] [Google Scholar]
- 9. Hertervig E., Nilsson A., Nyberg L., and Duan R. D. (1997) Alkaline sphingomyelinase activity is decreased in human colorectal carcinoma. Cancer 79, 448–453 [PubMed] [Google Scholar]
- 10. Hertervig E., Nilsson A., Björk J., Hultkrantz R., and Duan R. D. (1999) Familial adenomatous polyposis is associated with a marked decrease in alkaline sphingomyelinase activity: a key factor to the unrestrained cell proliferation? Br. J. Cancer 81, 232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zimmermann H., Zebisch M., and Sträter N. (2012) Cellular function and molecular structure of ecto-nucleotidases. Purinergic Signal. 8, 437–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tsai S. H., Kinoshita M., Kusu T., Kayama H., Okumura R., Ikeda K., Shimada Y., Takeda A., Yoshikawa S., Obata-Ninomiya K., Kurashima Y., Sato S., Umemoto E., Kiyono H., Karasuyama H., et al. (2015) The ectoenzyme E-NPP3 negatively regulates ATP-dependent chronic allergic responses by basophils and mast cells. Immunity 42, 279–293 [DOI] [PubMed] [Google Scholar]
- 13. Albright R. A., Ornstein D. L., Cao W., Chang W. C., Robert D., Tehan M., Hoyer D., Liu L., Stabach P., Yang G., De La Cruz E. M., and Braddock D. T. (2014) Molecular basis of purinergic signal metabolism by ectonucleotide pyrophosphatase/phosphodiesterases 4 and 1 and implications in stroke. J. Biol. Chem. 289, 3294–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nishimasu H., Okudaira S., Hama K., Mihara E., Dohmae N., Inoue A., Ishitani R., Takagi J., Aoki J., and Nureki O. (2011) Crystal structure of autotaxin and insight into GPCR activation by lipid mediators. Nat. Struct. Mol. Biol. 18, 205–212 [DOI] [PubMed] [Google Scholar]
- 15. Hausmann J., Kamtekar S., Christodoulou E., Day J. E., Wu T., Fulkerson Z., Albers H. M., van Meeteren L. A., Houben A. J., van Zeijl L., Jansen S., Andries M., Hall T., Pegg L. E., Benson T. E., et al. (2011) Structural basis of substrate discrimination and integrin binding by autotaxin. Nat. Struct. Mol. Biol. 18, 198–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morita J., Kano K., Kato K., Takita H., Sakagami H., Yamamoto Y., Mihara E., Ueda H., Sato T., Tokuyama H., Arai H., Asou H., Takagi J., Ishitani R., Nishimasu H., et al. (2016) Structure and biological function of ENPP6, a choline-specific glycerophosphodiester-phosphodiesterase. Sci. Rep. 6, 20995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kornhuber J., Rhein C., Müller C. P., and Mühle C. (2015) Secretory sphingomyelinase in health and disease. Biol. Chem. 396, 707–736 [DOI] [PubMed] [Google Scholar]
- 18. Airola M. V., and Hannun Y. A. (2013) Sphingolipid metabolism and neutral sphingomyelinases. Handb. Exp. Pharmacol. 215, 57–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maldonado-Valderrama J., Wilde P., Macierzanka A., and Mackie A. (2011) The role of bile salts in digestion. Adv. Colloid. Interface Sci. 165, 36–46 [DOI] [PubMed] [Google Scholar]
- 20. Duan R. D., and Nilsson A. (1997) Purification of a newly identified alkaline sphingomyelinase in human bile and effects of bile salts and phosphatidylcholine on enzyme activity. Hepatology 26, 823–830 [DOI] [PubMed] [Google Scholar]
- 21. Duan J., Wu J., Cheng Y., and Duan R. D. (2010) Understanding the molecular activity of alkaline sphingomyelinase (NPP7) by computer modeling. Biochemistry 49, 9096–9105 [DOI] [PubMed] [Google Scholar]
- 22. Parrill A. L., Wanjala I. W., Pham T. C., and Baker D. L. (2011) Computational identification and experimental characterization of substrate binding determinants of nucleotide pyrophosphatase/phosphodiesterase 7. BMC Biochem. 12, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jansen S., Perrakis A., Ulens C., Winkler C., Andries M., Joosten R. P., Van Acker M., Luyten F. P., Moolenaar W. H., and Bollen M. (2012) Structure of NPP1, an ectonucleotide pyrophosphatase/phosphodiesterase involved in tissue calcification. Structure 20, 1948–1959 [DOI] [PubMed] [Google Scholar]
- 24. Kato K., Nishimasu H., Okudaira S., Mihara E., Ishitani R., Takagi J., Aoki J., and Nureki O. (2012) Crystal structure of Enpp1, an extracellular glycoprotein involved in bone mineralization and insulin signaling. Proc. Natl. Acad. Sci. U.S.A. 109, 16876–16881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu J., Hansen G. H., Nilsson A., and Duan R. D. (2005) Functional studies of human intestinal alkaline sphingomyelinase by deglycosylation and mutagenesis. Biochem. J. 386, 153–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gijsbers R., Ceulemans H., Stalmans W., and Bollen M. (2001) Structural and catalytic similarities between nucleotide pyrophosphatases/phosphodiesterases and alkaline phosphatases. J. Biol. Chem. 276, 1361–1368 [DOI] [PubMed] [Google Scholar]
- 27. Cheng J., Goldstein R., Gershenson A., Stec B., and Roberts M. F. (2013) The cation-π box is a specific phosphatidylcholine membrane targeting motif. J. Biol. Chem. 288, 14863–14873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Carey M. C., and Small D. M. (1972) Micelle formation by bile salts: physical-chemical and thermodynamic considerations. Arch. Intern. Med. 130, 506–527 [PubMed] [Google Scholar]
- 29. Roussel A., Miled N., Berti-Dupuis L., Rivière M., Spinelli S., Berna P., Gruber V., Verger R., and Cambillau C. (2002) Crystal structure of the open form of dog gastric lipase in complex with a phosphonate inhibitor. J. Biol. Chem. 277, 2266–2274 [DOI] [PubMed] [Google Scholar]
- 30. Airola M. V., Allen W. J., Pulkoski-Gross M. J., Obeid L. M., Rizzo R. C., and Hannun Y. A. (2015) Structural basis for ceramide recognition and hydrolysis by human neutral ceramidase. Structure 23, 1482–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Freie A. B., Ferrato F., Carrière F., and Lowe M. E. (2006) Val-407 and Ile-408 in the β5′-loop of pancreatic lipase mediate lipase-colipase interactions in the presence of bile salt micelles. J. Biol. Chem. 281, 7793–7800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang X., Wang C. S., Tang J., Dyda F., and Zhang X. C. (1997) The crystal structure of bovine bile salt activated lipase: insights into the bile salt activation mechanism. Structure 5, 1209–1218 [DOI] [PubMed] [Google Scholar]
- 33. Keune W. J., Hausmann J., Bolier R., Tolenaars D., Kremer A., Heidebrecht T., Joosten R. P., Sunkara M., Morris A. J., Matas-Rico E., Moolenaar W. H., Oude Elferink R. P., and Perrakis A. (2016) Steroid binding to Autotaxin links bile salts and lysophosphatidic acid signalling. Nat. Commun. 7, 11248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 35. Terwilliger T. C., Adams P. D., Read R. J., McCoy A. J., Moriarty N. W., Grosse-Kunstleve R. W., Afonine P. V., Zwart P. H., and Hung L. W. (2009) Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 65, 582–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., and Adams P. D. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 [DOI] [PMC free article] [PubMed] [Google Scholar]