Abstract
The transforming growth factor β isoforms, TGF-β1, -β2, and -β3, are small secreted homodimeric signaling proteins with essential roles in regulating the adaptive immune system and maintaining the extracellular matrix. However, dysregulation of the TGF-β pathway is responsible for promoting the progression of several human diseases, including cancer and fibrosis. Despite the known importance of TGF-βs in promoting disease progression, no inhibitors have been approved for use in humans. Herein, we describe an engineered TGF-β monomer, lacking the heel helix, a structural motif essential for binding the TGF-β type I receptor (TβRI) but dispensable for binding the other receptor required for TGF-β signaling, the TGF-β type II receptor (TβRII), as an alternative therapeutic modality for blocking TGF-β signaling in humans. As shown through binding studies and crystallography, the engineered monomer retained the same overall structure of native TGF-β monomers and bound TβRII in an identical manner. Cell-based luciferase assays showed that the engineered monomer functioned as a dominant negative to inhibit TGF-β signaling with a Ki of 20–70 nm. Investigation of the mechanism showed that the high affinity of the engineered monomer for TβRII, coupled with its reduced ability to non-covalently dimerize and its inability to bind and recruit TβRI, enabled it to bind endogenous TβRII but prevented it from binding and recruiting TβRI to form a signaling complex. Such engineered monomers provide a new avenue to probe and manipulate TGF-β signaling and may inform similar modifications of other TGF-β family members.
Keywords: cancer, cell signaling, fibrosis, inhibitor, protein engineering, transforming growth factor beta (TGF-B), dominant negative
Introduction
The transforming growth factor β isoforms, TGF-β1, -β2, and -β3, are small secreted signaling proteins. Their overall structures are similar and consist of two cystine-knotted monomers tethered together by a single inter-chain disulfide bond (1). They coordinate wound healing, modulate immune cell function, maintain the extracellular matrix, and regulate epithelial and endothelial cell growth and differentiation (2). The TGF-βs are synthesized as pre-pro-proteins, and after maturation, secretion, and release from their pro-domains (3), the mature homodimeric growth factors (GFs)3 bind and bring together two single-pass transmembrane receptors, known as TβRI and TβRII, to form the signaling-competent TβRI2-TβRII2 heterotetramer (4, 5). TGF-β GFs assemble TβRI2-TβRII2 heterotetramer in a sequential manner, first by binding TβRII followed by recruitment of TβRI (6, 7). The stepwise assembly of TβRII and TβRI into a heterotetramer is driven by binding of TβRI to a composite TGF-β/TβRII interface (Fig. 1A) (8, 9).
Figure 1.

Structure of the TGF-β signaling complex and sequences of the engineered TGF-β variants lacking the heel helix, α3. A, schematic representation of the TGF-β signaling complex formed between human TGF-β3 homodimer (magenta and blue ribbons) and the extracellular ligand binding domains of the human TGF-β type I and type II receptors, TβRI (red ribbon) and TβRII (tan ribbon) (PDB 2PJY) (8). The disulfide bonds, including the single inter-chain disulfide connecting the TGF-β monomers, are depicted in yellow. The TGF-β monomers are described as curled left hands, with the heel formed by a 3½ turn α-helix (α3) and the four fingers formed by the β-strands that extend from the cystine knot that stabilizes each monomer. B, expanded view illustrating packing interactions formed by hydrophobic residues that emanate from the heel α-helix (blue ribbon) of one TGF-β3 monomer with hydrophobic residues from the palm region of the opposing TGF-β3 monomer (magenta ribbon with transparent magenta surface). C, expanded view illustrating ionic, hydrogen bonding, and hydrophobic interactions that stabilize TβRI (red ribbon) at the composite interface formed by both monomers of TGF-β3 (magenta and blue ribbons) and TβRII (tan ribbon). D, sequence alignment of TGF-β1, -β2, and -β3 with monomeric variants in which Cys-77, which normally forms the inter-chain disulfide bond, is substituted with serine (mTGF-β2 and mTGF-β3) or mini-monomeric variants in which Cys-77 is substituted with serine, residues 52–71 have been deleted, and two or three additional residues (highlighted in red) have been substituted (mmTGF-β1, mmTGF-β2, and mmTGF-β3). Calculated net charge of the corresponding monomers at pH 7.0 is shown on the right. E, sequence alignment of TGF-β1, -β3, -β2, mmTGF-β2, and mmTGF-β2-7M in the TβRII-binding region. Residues in the TβRII binding interface are indicated by yellow shading. Residues substituted in mmTGF-β2-7M relative to mmTGF-β2 are highlighted in red, and include K25R, I92V, and N94R, which were shown previously to be necessary and sufficient for high affinity TβRII binding (39, 40). F, interface between TGF-β3 and TβRII, with Arg-25, Val-92, and Arg-94 highlighted by red labels.
The disruption or dysregulation of the TGF-β pathway is responsible for several human diseases. These include connective tissue disorders, such as Marfan's disease and Loeys-Dietz syndrome, which are caused by increased or decreased signaling due to mutations in the matrix protein fibrillin-1 or TβRII, respectively (10, 11). The dysregulation of the pathway is also responsible for fibrotic disorders (12) and soft tissue cancers (13). The fibrotic disorders are a result of hyperactive TGF-β signaling following tissue injury or disease progression that leads to the accumulation of extracellular matrix proteins. TGF-β's role in cancer is complex, with loss of its potent growth inhibitory activity being responsible for cancer initiation (14), and excessive TGF-β signaling, in the context of growth refractory advanced cancers, potently stimulating cancer progression and metastasis (13).
TGF-β's disease promoting activities, together with animal studies that have demonstrated beneficial effects of inhibiting TGF-β in models of cancer and fibrosis (15–22), have made them important targets for the development of inhibitors. However, despite clinical trials ongoing for nearly 2 decades using receptor kinase inhibitors, neutralizing antibodies, and other approaches, no TGF-β inhibitors have been approved for clinical use in humans (23, 24). One of the main challenges involves finding the correct dosing and pharmacodynamics for the particular disease to enable an effective therapeutic response, but sparing or minimally impacting TGF-β signaling, or other signaling pathways, in normal cells and tissues. TGF-β kinase inhibitors have posed some challenges in this respect as they have significant inhibitory activity against other type I receptors of the TGF-β superfamily, as well as other related kinases (25–27), and may further lead to rapid development of resistance (28). Pan-isoform TGF-β neutralizing antibodies, such as Sanofi's humanized mouse monoclonal antibody, GC1008, are specific, although tissue residence times are long and some concerning side effects, such as keratoacanthoma and squamous cell carcinoma, have been reported in clinical trials (29).
Thus, alternative approaches are needed to target the TGF-β pathway. The objective of this study was to investigate whether it might be possible to design an engineered TGF-β GF that functioned as a dominant negative to potently and specifically inhibit TGF-β signaling. This approach offers several potential advantages over existing therapies. Relative to kinase inhibitors, engineered GFs would be expected to have much higher specificity, especially if they function by binding and blocking TβRII, which is known to only bind and transduce signals for TGF-β1, -β2, and -β3, but not other TGF-β family GFs (1, 30). Another potential advantage over kinase inhibitors is increased bioavailability because, unlike the kinase inhibitors, engineered GFs would not have to cross the plasma membrane to reach their target. Relative to monoclonal antibodies, the engineered GFs, because of their smaller size, would be expected to have shorter tissue lifetimes, which would limit sustained inhibition in normal cells and tissues and may alleviate undesirable side effects. The smaller size of engineered GFs may also lead to improved penetration of diseased tissues, particularly solid tumors, relative to 150-kDa monoclonal antibody molecules (31, 32). Engineered ligands have been successfully used to target other signaling pathways, such as the VEGF pathway (33), and thus represent a largely undeveloped but potentially very effective therapeutic modality for treating disease.
Through previous studies, monomeric forms of TGF-β1 and TGF-β3, formed by substituting the cysteine residue that forms the inter-chain disulfide to serine (C77S), were shown to have diminished signaling activity compared with their disulfide-linked counterparts, but nonetheless were still quite potent, with EC50 values for stimulation of TGF-β reporter gene activity in the range of 100 pm (7, 34). Amatayakul-Chantler et al. (34), and later Zúñiga and co-workers (7), suggested this residual activity might arise from assembly of a dimeric complex of a GF homodimer and two bound TβRIs and two bound TβRIIs, but without the disulfide linkage between the GF monomers. This model was attractive for two reasons: first, structures of the TGF-βs show there are in fact extensive hydrophobic contacts between the TGF-β monomers that could promote non-covalent self-association of the monomers (Fig. 1B) (35, 36); second, once formed, these non-covalent dimers would be stabilized as the receptors bind, because crystal structures show that at least one them, TβRI, binds by straddling the TGF-β homodimer interface (Fig. 1, A and C) (8, 9).
The objective of this study was to design an engineered TGF-β monomer that still retained its full capacity to bind the high affinity TGF-β receptor, TβRII, but was fully impaired in its ability to bind and recruit TβRI. This type of engineered monomer would be expected to function as a dominant negative and thus inhibit TGF-β signaling, because it would bind and thus occupy cell surface TβRII, but in turn be unable to recruit TβRI to form a signaling complex. The results presented here document the generation of such an engineered monomer and demonstrate that such monomers function as potent inhibitors of TGF-β signaling in cultured cells. The results further show that unlike dimeric TGF-βs, as well as their C77S monomeric counterparts, engineered monomers are highly soluble. These properties, together with the high intrinsic specificity of TGF-βs for TβRII, should engender this novel inhibitor with favorable properties for treating human diseases, such as Marfan's disease, fibrotic disorders, and soft tissue cancers that are driven by excessive TGF-β signaling.
Results
Design of engineered mini-monomeric TGF-β (mmTGF-β)
The structures of the TGF-β receptor complexes (8, 9), as well as accompanying binding and cross-linking studies with TGF-β3 C77S (7, 8, 37), suggested that the signaling capacity of monomeric TGF-βs (TGF-β1 C77S or mTGF-β1 and TGF-β3 C77S or mTGF-β3) arise from their ability to non-covalently dimerize and in turn bind their receptors (Fig. 1, A and C). This led to our hypothesis that it should be possible to diminish or completely eliminate receptor complex assembly with monomeric TGF-βs by removing or altering residues responsible for dimer formation and binding of TβRI. The structural motif that likely contributes the greatest to self-association of the monomers is the “heel” α-helix, α-helix 3 (Fig. 1A). This helix is highly amphipathic and has numerous hydrophobic interactions with residues that line the “palm” of the opposing monomer (Fig. 1B). This helix also forms a large portion of the binding surface for TβRI (Fig. 1C). Thus, it was hypothesized that elimination of α-helix 3 should interfere with both self-association of the monomers and binding of TβRI, but it should not impair TβRII binding as this occurs through the ligand fingertips far away from α-helix 3 (Fig. 1A).
To evaluate this hypothesis, bacterial expression constructs were generated for TGF-β1, TGF-β2, and TGF-β3 in which residues 52–71 were eliminated and Cys-77 was substituted with serine. This corresponds to deletion of all of α-helix 3, as well as five flanking residues on the N-terminal end and three flanking residues on the C-terminal end (Fig. 1D). The length of the deletion was chosen so as to leave a sufficient number of residues between the last residue of β-strand 4 (Gly-48) and the first residue of β-strand 5 (Cys-77/Ser-77) to form an unconstrained loop that bridges β-strands 4 and 5. Although a secondary consideration, either two (TGF-β2) or three (TGF-β1 and -β3) of the loop-forming residues were also substituted so as to increase the net overall charge at pH 7.0 for the full-length TGF-β1, -β2, and -β3 monomers from −0.9, +1.1, and +4.4 to −3.1, +3.9, and +6.1 for the constructs in which α-helix 3 was deleted (Fig. 1D). The rationale for this was that the solubility of the monomers, which like the homodimers are poor from pH 4.5 to 9.5 (see Fig. 4, A and B, below), might be improved by both removing hydrophobic α-helix 3 and by artificially increasing the net charge at pH 7.0.
Figure 4.
Solubility of TGF-β2 and monomeric variants. A and C, TGF-β2 and mTGF-β2 (A) and mmTGF-β2 and mmTGF-β2-7M (C) were diluted from a concentrated stock in 100 mm acetic acid into either PBS at 7.4 (Neutral pH) or 100 mm acetic acid (Acidic pH), and the light scattering at 340 nm was measured. B and D. TGF-β2 and mTGF-β2 (B) and mmTGF-β2 and mmTGF-β2-7M (D) samples diluted into either PBS or 100 mm acetic acid were centrifuged for 5 min at 20,000 × g, and the protein absorbance at 280 nm was measured.
Isolation and physical characterization of mmTGF-β2
The TGF-β1, -β2, and -β3 “mini-monomers” described above, designated mmTGF-β1, mmTGF-β2, and mmTGF-β3, were expressed in Escherichia coli and accumulated in the form of insoluble inclusion bodies. The inclusion bodies were isolated, and after reconstitution and purification in denaturant, the mini-monomers were renatured by dilution into CHAPS-containing buffer at pH 9.0 as described previously (38). The folding of the mini-monomers differed greatly; a large portion of the mmTGF-β2 remained soluble during the folding and yielded large amounts of monomeric protein after purification by cation exchange chromatography, whereas only a small amount of mmTGF-β1 and mmTGF-β3 remained soluble during the folding, and either no monomeric protein (TGF-β1) or a very small amount of monomeric protein (TGF-β3) was obtained after purification by cation exchange chromatography. This pattern mirrors the pattern previously observed for the folding of TGF-β homodimers from full-length wild type monomers (38) and likely reflects differences in the intrinsic propensity of the monomers to properly form the four intramolecular disulfides characteristic of each monomer. mmTGF-β2 was the least desired variant, due to the expected low affinity for binding TβRII. However, this was considered an addressable concern based on our prior studies, which demonstrated that substitution of Lys-25, Ile-92, and Lys-94 in TGF-β2 with the corresponding residues in TGF-β1 and TGF-β3 engendered TGF-β2 with the ability to bind TβRII with high affinity (39, 40).
To determine whether mmTGF-β2 was suitable for further development in the manner described above, it was characterized in terms of its folding, solubility, and receptor binding properties. To assess folding, a 15N-labeled sample of mmTGF-β2 was prepared and examined by recording a two-dimensional 1H-15N shift correlation spectrum (Fig. 2A). This revealed a highly dispersed spectrum characteristic of natively folded protein. The spectrum could be fully assigned, and analysis of the assigned chemical shifts to identify secondary structure propensities showed that the protein had the expected secondary structure, particularly in the palm region formed by the cystine knot and the finger region where TβRII binds (supplemental Fig. S2A). This analysis further showed that the newly created loop between residues 47 and 56 had near zero probability of forming either an α-helix or β-strand, suggesting that it is likely flexible as would be expected for a loop of this length connecting two antiparallel β-strands. This was directly confirmed by an analysis of backbone 15N T2 values. These values provide information about motions on fast (nanosecond-picosecond) and intermediate (microsecond-millisecond) time scales and were significantly elevated in the region corresponding to the newly created loop relative to the other parts of the protein (supplemental Fig. S2B), which, except for the N terminus and the short loop connecting α-helix 1 and β-strand 1, are expected to be structurally well-ordered.
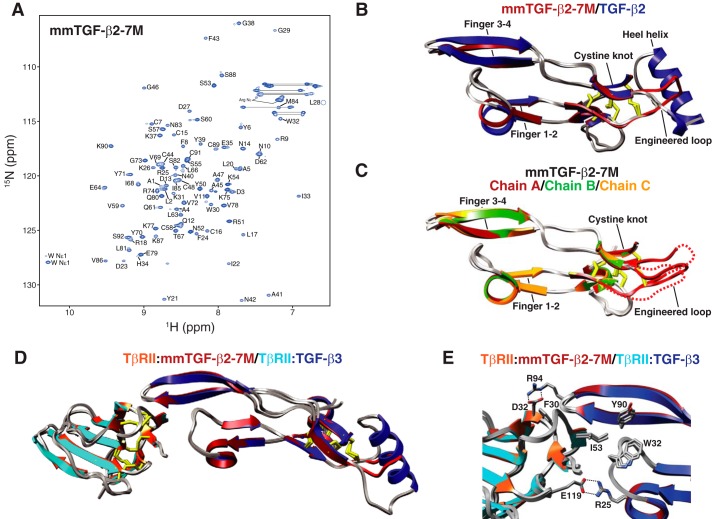
Figure 2.
Structure of mmTGF-β2. A, assigned 1H-15N HSQC spectrum of mmTGF-β2 recorded in 10 mm sodium phosphate, 10 mm CHAPS, 5% 2H2O, pH 4.7, 37 °C, 800 MHz. Assigned backbone amide signals are indicated by their residue number and one-letter amino acid code. B, overlay of 1.8 Å crystal structure of mmTGF-β2 (orange ribbon) with one of the monomers from the 1.8 Å crystal structure of TGF-β2 (PDB 2TGI, blue ribbon). Major structural features are indicated, along with the newly created loop in mmTGF-β2 (red), which takes the place of the heel (α3) helix in TGF-β2. C, overlay of the two mmTGF-β2 chains (chain A and B shown in orange and green ribbon, respectively) from the crystallographic asymmetric unit. Other details as in B. D, overlay of mmTGF-β2 and TGF-β2 as in B, but with the aligned positions restricted to the residues 18–45 and 61–87 in fingers 1/2 and 3/4, respectively.
To directly examine the three-dimensional structure, mmTGF-β2 was crystallized, and its structure was determined to a resolution of 1.8 Å using molecular replacement (Table 1). The overall fold of mmTGF-β2 was shown to be highly similar to that previously determined for TGF-β2, with the exception of the newly created loop, which was shown to take the place of α-helix 3 as anticipated (Fig. 2B). Superposition of the mmTGF-β2 with the monomer from the structure of TGF-β2 shows that there is a systematic displacement of up to about 1.5 Å of the finger region of mmTGF-β2 relative to TGF-β2. Such differences appear to be the result of bending of the monomer near the center of the finger region and not a change in the structure of the finger region, as superimposition of the fingers alone show that they correspond closely, with a backbone r.m.s.d. of under 0.2 Å and similar orientations of the side chains of several residues that pack and stabilize the fingers (Fig. 2D). Such bending is also supported by an overlay of the two molecules of mmTGF-β2 present in the crystallographic asymmetric unit, which also exhibit a smaller but still noticeable displacement of the finger regions relative to one another (Fig. 2C). Consistent with the NMR analysis, not only was the electron density noticeably weaker in the region corresponding to the newly created loop, but also it was shown to adopt different orientations for the two molecules from the asymmetric unit (Fig. 2C).
Table 1.
X-ray data collection and refinement statistics
| Data collection | |||
| Molecule | mmTGF-β2 | mmTGF-β2–7 m | mmTGF-β2-7M·TβRII |
| X-ray source | Advanced Photon Source 24-ID-C | Rigaku 007 generator and Saturn 944 CCD detector | Advanced Photon Source SER-CAT 22-ID-D |
| Space group | C2 | P3121 | P212121 |
| Cell dimensions | |||
| a, b, c (Å) | 99.5, 33.4, 54.1 | 81.74, 81.74, 80.93 | 39.0, 70.8, 77.1 |
| α, β, γ (°) | 90, 109.6, 90 | 90, 90, 120 | 90, 90, 90 |
| Wavelength (Å) | 0.9795 | 1.542 | 0.97949 |
| Resolution (Å) | 51.01–1.82 (1.92–1.82)a | 36.48–2.75 (2.89–2.75)a | 35.39–1.88 (1.97–1.88)a |
| Rsym | 0.050 (0.443) | 0.132 (0.463) | 0.143 (0.97) |
| Rpim | 0.038 (0.307) | 0.055 (0.232) | 0.058 (0.522) |
| I/σI | 12.7 (2.2) | 16.4 (4.0) | 15.17 (2.02) |
| Completeness (%) | 98.4 (98.4) | 99.9 (99.8) | 99.6 (99.4) |
| Redundancy | 3.6 (3.5) | 12.3 (8.9) | 6.8 (6.6) |
| Wilson value (Å2) | 28.9 | 30.23 | 30.08 |
| Refinement | |||
| Resolution (Å) | 51.01–1.82 | 36.48–2.75 | 35.39–1.88 |
| No. of reflections | 15,027 | 8493 | 17,715 |
| Rwork/Rfree | 0.209/0.252 | 0.2127/0.2716 | 0.1955/0.2216 |
| No. of atoms | |||
| Protein | 1462 | 2086 | 1570 |
| Water | 107 | 63 | 82 |
| B-factors (Å2) | |||
| Protein | 33.3 | 40.2 | 43.6 |
| Water | 36.4 | 22.2 | 41.22 |
| Root mean square deviations | |||
| Bond lengths (Å) | 0.012 | 0.003 | 0.011 |
| Bond angles (°) | 1.030 | 0.763 | 1.143 |
| Ramachandran statistics: favored, allowed, outliers (%) | 94.4, 5.0, 0.6 | 93.2, 6.8, 0.0 | 96.39, 3.09, 0.52 |
a Highest resolution shell is shown in parentheses.
The similar folding of mmTGF-β2 relative to TGF-β2, especially in the TβRII-binding finger region, suggested that it would also bind TβRII in a similar manner. To evaluate this, surface plasmon resonance (SPR) experiments were performed in which the same concentration series of TβRII was injected over TGF-β2 and mmTGF-β2 immobilized on separate flow cells (Fig. 3, A and B). Although it was not possible to quantitate affinity due to weak binding, the sensorgrams nonetheless showed similar shapes and concentration dependence. These sensorgrams show that mmTGF-β2 binds TβRII weakly, consistent with earlier reports (39), and that it does so in a manner qualitatively similar to TGF-β2.
Figure 3.
Binding properties of mmTGF-β2 and mmTGF-β2-7M. A and B, SPR sensorgrams for injection of a 2-fold dilution series from 3 to 0.047 μm TβRII over immobilized TGF-β2 (A) or mmTGF-β2 (B). Responses shown were normalized for the surface density of the immobilized TGF-βs. C–H, SPR sensorgrams for injection of a 2-fold dilution series from 3 to 0.012 μm TβRII (C and D), 1.024 to 0.008 μm TβRI (E and F), or 1.024 to 0.008 μm TβRI in the presence of 2 μm TβRII in both the running buffer and injected samples (G and H) over immobilized avi-TGF-β3 (C, E, and G) or avi-mmTGF-β2-7M (D, F, and H). Sensorgrams shown in C, D, and G were fitted to a 1:1 binding model; raw data are shown in black, and the fitted curve is shown in red. TGF-β2 and mmTGF-β2 were immobilized by direct carbodiimide-based amine coupling to the sensor surface, whereas avi-TGF-β3 or avi-mmTGF-β2-7M were immobilized by capturing the enzymatically biotinylated proteins onto the surface of sensor chip coated with streptavidin at high (∼8000 resonance units) density.
The solubility of mmTGF-β2 appeared to be significantly better than that of TGF-β2 and the full-length TGF-β2 monomer, mTGF-β2, as samples of the former could be readily prepared at concentrations of 2–3 mg ml−1 without noticeable precipitation at pH 7.0, whereas samples of the latter two proteins were completely precipitated under these same conditions. To quantitate solubility, TGF-β2, mTGF-β2, and mmTGF-β2 were prepared as concentrated stocks in 100 mm acetic acid, pH 2.9, where they were readily soluble and then diluted into PBS, pH 7.4. The light scattering at 340 nm was measured to assess precipitation, and then the samples were centrifuged, and the absorbance at 280 nm was measured to assess the protein concentration. This demonstrated that TGF-β2 and mTGF-β2 were both effectively insoluble at neutral pH over the entire concentration range evaluated (7–100 μm) (Fig. 4, A and B). This is consistent with the known poor solubility of the TGF-β homodimers (41), but it shows that this property also extends to full-length monomeric TGF-βs. The mini-monomeric TGF-β2, mmTGF-β2, in contrast, exhibited modest light scattering and a corresponding modest reduction in the amount of soluble protein relative to that expected when the protein concentration was 40 μm or higher, indicating that indeed mmTGF-β2 was reasonably soluble at neutral pH, although not perfectly so. This was reflected in NMR spectra, which showed that although 100–200 μm 15N mmTGF-β2 samples could be readily prepared, the spectrum was nonetheless poor, with the only detectable signals arising from residues in the flexible parts of the protein, namely the N terminus, the exposed loop between α-helix 1 and β-strand 1, and the newly created loop between β-strands 4 and 5. The fact that signals could only be detected from the flexible parts of the protein suggested that mmTGF-β2 forms large soluble aggregates under these conditions. Through trial and error, it was found that these soluble aggregates could be eliminated by addition of the zwitterionic detergent CHAPS, with the majority of the NMR signals appearing at the concentration of 5 mm CHAPS and all of the NMR signals appearing at 10 mm CHAPS. Thus, all NMR spectra, including that shown in Fig. 2A, were recorded in the presence of 10 mm CHAPS.
Isolation and physical characterization of mmTGF-β2-7M
The results presented above show that whereas mmTGF-β2 is natively folded, it nonetheless possesses low intrinsic affinity for binding TβRII. To confer mmTGF-β2 with the ability to bind TβRII with high affinity comparable with that of TGF-β1 and TGF-β3, the three residues in mouse TGF-β2 shown previously to differ in the interface with TβRII, Lys-25, Ile-92, and Asn-94 (40, 42), were substituted with the corresponding residues from TGF-β1 and -β3, Arg-25, Val-92, and Arg-94 (Fig. 1, E and F). In previous studies, substitution of these three residues was shown to be sufficient to confer TGF-β2 with a TβRII binding affinity comparable with TGF-β1 and TGF-β3 (39, 40). Despite this, four additional residues peripheral to the TβRII-binding site that differed in TGF-β2 relative to TGF-β1 were also substituted with the corresponding residues from TGF-β1 (R26K, L89V, T95K, and I98V) (Fig. 1, E and F). Although previous results suggested this was not strictly necessary, it was nonetheless done to ensure that the precise orientation of residues in the mmTGF-β2-binding site for TβRII matched as closely as possible with that in the high affinity TGF-β isoforms, TGF-β1 and TGF-β3. The resulting construct bearing these seven amino acid substitutions, designated mmTGF-β2-7M (Fig. 1E, supplemental Fig. S1, and supplemental Table S1), was expressed in E. coli in the form of insoluble inclusion bodies. As with mmTGF-β2, most of the protein remained in solution after reconstitution and dilution into native folding buffer, and large amounts of homogenous monomer could be isolated (4–5 mg/liter of E. coli culture medium).
The folding and homogeneity of the isolated mmTGF-β2-7M was evaluated by NMR, and as with mmTGF-β2, the protein was found to have the expected number of signals in a 2D 1H-15N shift correlation spectrum (Fig. 5A) as well as secondary structure, as determined by an analysis of the NMR secondary shifts (supplemental Fig. S3A). The solubility of mmTGF-β2-7M was evaluated as before, and as shown, its behavior was comparable or perhaps slightly better than that of mmTGF-β2 (Fig. 4, C and D). This slight improvement in the macroscopic solubility did not however change the microscopic solubility as NMR analysis showed that it was still necessary to include 10 mm CHAPS in the sample buffer to detect signals from all of the backbone amide resonances in the protein.
Figure 5.
Structure of mmTGF-β2-7M and mmTGF-β2-7M·TβRII complex. A, assigned 1H-15N HSQC spectrum of mmTGF-β2-7M recorded in 10 mm sodium phosphate, 10 mm CHAPS, 5% 2H2O, pH 4.70, 37 °C, 800 MHz. Assigned backbone amide signals are indicated by their residue number and one-letter amino acid code. B, overlay of 1.8 Å crystal structure of mmTGF-β2-7M (dark red ribbon) with one of the monomers from the 1.8 Å crystal structure of TGF-β2 (PDB 2TGI, blue ribbon). Major structural features are indicated, along with the newly created loop in mmTGF-β2 (red), which takes the place of the heel (α3) helix in TGF-β2. C, overlay of the three mmTGF-β2-7M chains (chain A, B, and C shown in dark red, green, and orange ribbon, respectively) from the crystallographic asymmetric unit. Dashed line corresponds to missing segments in the newly created loop in chains A and C due to weak electron density. Other details as in B. D, overlay of the 1.8 Å crystal structure of mmTGF-β2-7M·TβRII complex (dark red and orange ribbons, respectively) with one of the TGF-β3 monomers and its bound TβRII from the 3.0 Å crystal structure of the TGF-β3·TβRII·TβRI complex (PDB 2PJY, TGF-β3 monomer and TβRII shown in dark blue and cyan ribbon, respectively; TβRI not shown for clarity). Newly created loop in mmTGF-β2 (red), which takes the place of the heel (α3) helix in TGF-β2, is depicted in red. E, overlay as in B, but expanded to show the near identity of critical hydrophobic and hydrogen-bonding/electrostatic interactions shown previously to be essential for high affinity TGF-β3·TβRII binding (39, 40).
The three-dimensional structure of mmTGF-β2-7M was determined by crystallography to a resolution of 2.75 Å (Table 1), and as before the overall fold was preserved relative to TGF-β2, with the only difference being a slight hinge bending of the monomer as described for mmTGF-β2 (Fig. 5, B and C). The increase in the 15N T2 relaxation times in the region corresponding to the newly formed loop in mmTGF-β2-7M was comparable with that in mmTGF-β2 (supplemental Fig. S3B). This suggested that the missing density in the region corresponding to the newly formed loop in mmTGF-β2-7M, which among the three molecules in the asymmetric unit was observed for part of chain A and most of chain C, was not due to increased dynamics, but other factors, most likely the lower resolution of the mmTGF-β2-7M structure compared with the mmTGF-β2 structure (Table 1).
To determine whether mmTGF-β2-7M bound TβRII with high affinity, variants of mmTGF-β2-7M and TGF-β3 were produced bearing an N-terminal avitag, and after biotinylation and immobilization onto a streptavidin-coated SPR sensor, their binding affinity for TβRII was measured by performing kinetic SPR experiments (Fig. 3, C and D). The sensorgrams obtained differed greatly from that previously obtained for mmTGF-β2 and TGF-β2, in that they exhibited a clear pattern of saturation. The sensorgrams were furthermore shown to have similar shapes as well as fitted parameters, including KD values (Table 2), which were within experimental error of one another and consistent, although on the high end, with KD values reported earlier for TβRII binding to TGF-β1 and TGF-β3 (37, 39, 40).
Table 2.
SPR binding parameters for TβRII and TβRI binding to TGF-β3 and mmTGF-β2-7M
| Immobilized ligand | Injected receptor | Buffer | kaa | kda | KDa | Rmaxa |
|---|---|---|---|---|---|---|
| m−1 s−1 | s−1 | μm | RU | |||
| avi-mmTGF-β2-7M | TβRII | HBS-EP | 1.16 × 105 (1.48 × 103) | 5.46 × 10−2 (3.78 × 10−4) | 0.47 (0.07) | 256 (2) |
| avi-TGF-β3 | TβRII | HBS-EP | 2.64 × 105 (3.97 × 103) | 1.132 × 10−1 (6.94 × 10−4) | 0.43 (0.05) | 128 (1) |
| avi-TGF-β3 | TβRI | HBS-EP + 2 μm TβRII | 4.64 × 104 (1.27 × 103) | 2.05 × 10−2 (3.42 × 10−4) | 0.44 (0.11) | 44 (2) |
| avi-mmTGF-β2–7M | TβRI | HBS-EP + 2 μm TβRII | NDb | NDb | NDb | NDb |
a Error estimates are shown in parentheses.
b ND indicates No detectable response.
To determine whether the interactions that enabled high affinity TβRII binding were preserved in mmTGF-β2-7M compared with TGF-β1 and TGF-β3, the mmTGF-β2-7M·TβRII complex was crystallized, and its structure was determined to a resolution of 1.88 Å (Table 1). The overall structure of the mmTGF-β2-7M·TβRII complex is shown to be very similar to that of one of the TβRII-bound monomers from the structure of the TGF-β3·TβRII·TβRI complex, with TβRII bound to the mmTGF-β2-7M fingertips in a manner that is essentially indistinguishable from that of TGF-β3 (Fig. 5D). The interactions known to contribute most significantly to high affinity binding are furthermore shown to be fully preserved in the mmTGF-β2-7M·TβRII complex relative to TGF-β1·TβRII and TGF-β3·TβRII complexes that have been previously determined (the TGF-β3·TβRII complex determined to 1.8 Å (42) is shown as this is the highest resolution structure determined to date) (Fig. 5E). This includes the packing of Ile-53 from TβRII in the hydrophobic pocket between the TGF-β fingers, and the hydrogen-bonded ion pairs formed between TGF-β Arg-25 and Arg-94 on the tips of the loops connecting fingers 1/2 and 3/4, respectively, and the carboxylate groups of Glu-119 and Asp-32 on TβRII (Fig. 5E).
Inhibitory activity of mmTGF-β2-7M and the underlying mechanism
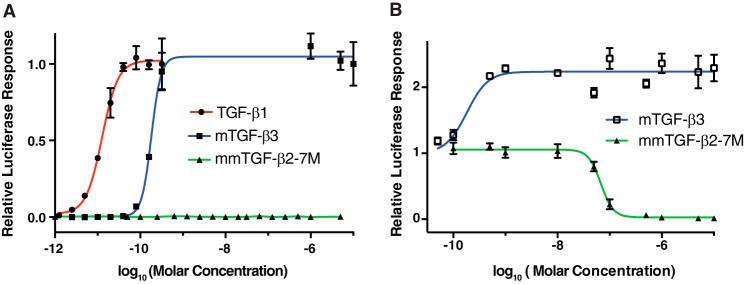
The results presented above show that mmTGF-β2-7M possesses one of the essential attributes required to function as a dominant negative inhibitor of TGF-β signaling, which is the ability to bind TβRII with high affinity comparable with that of TGF-β1 and TGF-β3. To directly assess whether mmTGF-β2-7M might signal and, if not, whether it might function as an inhibitor, TGF-β signaling was assessed by treating HEK293 cells stably transfected with a TGF-β luciferase reporter under the control of a CAGA12 promoter (43) with increasing concentrations of TGF-βs. The results showed that dimeric TGF-β1 (TGF-β1) and full-length monomeric TGF-β3 (mTGF-β3) resulted in a sigmoidal increase in the luciferase response, with concentrations of roughly 25 pm TGF-β1 and 250 pm mTGF-β3 leading to no further increase in the measured luciferase response. This is consistent with earlier reports that showed that (full-length) monomeric TGF-β1 and -β3 were 5–15-fold less potent than their dimeric counterparts (7, 34). The normalized luciferase responses could be readily fitted to a standard model for ligand-dependent activation and yielded EC50 values of 12.4 ± 1.5 pm for TGF-β1 and 182 ± 16 pm for mTGF-β3. The values for TGF-β1 and mTGF-β3 are in close accord with the values previously reported by Amatayakul-Chantler et al. (34) for TGF-β1 and by Zúñiga et al. (7)for mTGF-β3. The potent sub-nanomolar signaling activity observed for TGF-β1 and mTGF-β3 stands in contrast to that of mmTGF-β2-7M, which had no detectable signaling activity at the concentration that led to a saturating response for mTGF-β3 (ca. 200 pm) or at concentrations that were up to four orders of magnitude higher (Fig. 6A). Thus, mmTGF-β2-7M was either completely devoid of signaling activity or it possessed signaling activity, but with a potency more than a 10,000-fold less than that of mTGF-β3.
Figure 6.
Signaling activity of TGF-β dimers and monomers. A, TGF-β luciferase reporter activity for TGF-β1, mTGF-β3, and mmTGF-β2-7M shown in solid circles, squares, and triangles, respectively. The solid lines, colored red and blue, correspond to the fitted curves to derive the EC50 (green line for mmTGF-β2-7M was not fit due to the lack of signaling activity for this variant). B, TGF-β luciferase reporter activity for cells treated with a sub-saturating concentration of TGF-β1 (8 pm) with increasing concentration of the indicated monomeric TGF-β variant added (mTGF-β3 and mmTGF-β2-7M shown in open squares and closed triangles, respectively). The solid blue line corresponds to the fitted curve for mTGF-β3 to derive the EC50. The solid green line corresponds to the fitted curve for mTGF-β2-7M to derive the IC50.
To further investigate the properties of mmTGF-β2-7M, a competition experiment was performed in which the same HEK293 luciferase reporter cell line was stimulated with a constant sub-EC50 concentration of dimeric TGF-β1 (8.0 pm) and increasing concentrations of mTGF-β3 or mmTGF-β2-7M. The results showed that mTGF-β3 further stimulated signaling with a midpoint concentration similar to that of mTGF-β3 alone (Fig. 6B). The fitted EC50 values confirm this, with an EC50 of 182 ± 16 pm for the data shown in Fig. 6A and EC50 of 194 ± 36 pm for the data shown in Fig. 6B. The behavior of mmTGF-β2-7M was very different, with no detectable change in the signaling activity when added up to concentrations of 10 nm, but with a sharp decrease to no detectable signaling activity when the concentration was increased to 100 nm (Fig. 6B). This shows that mmTGF-β2-7M indeed possesses no signaling activity and that it can function to completely block and inhibit TGF-β signaling. The normalized luciferase responses could be readily fitted to a standard model for ligand-dependent inhibition and yielded an IC50 value of 68 ± 7 nm. Similar experiments showed that mmTGF-β2-7M also functioned as a potent competitive inhibitor against the other TGF-β isoforms, TGF-β2 and TGF-β3, with measured IC50 values (TGF-β2 IC50 19 ± 3 nm and TGF-β3 IC50 21 ± 8 nm) within a factor of 2–3 of that measured for TGF-β1 (supplemental Fig. S4, A and B). These IC50 values are on the lower end of the range of affinities that have been reported for binding of the high affinity TGF-β isoforms to TβRII, including mmTGF-β2-7M reported here (Table 2). This suggests that mmTGF-β2-7M functions to inhibit TGF-β signaling in the manner anticipated, which is by binding to and blocking endogenous TβRII. The fact that the measured potency is greater than the greatest affinity previously reported for TGF-β1 and TGF-β3 binding to TβRII (140 nm) (9), suggests that other factors, such as nonspecific association of mmTGF-β2-7M with the plasma membrane, may serve to potentiate its inhibitory activity.
The finding that mmTGF-β2-7M possesses no apparent signaling activity, and in fact functions as a low nanomolar inhibitor of TGF-β signaling, suggests that the elimination of α-helix 3 in fact diminished non-covalent association of the monomers and greatly attenuated or abrogated TβRI binding. To assess this directly, SPR experiments were performed to determine whether mmTGF-β2-7M could recruit TβRI in the presence of TβRII. To accomplish this, increasing concentrations of TβRI and the same concentration series of TβRI in the presence of near-saturating amounts of TβRII (2 μm) were injected over the same TGF-β3 and mmTGF-β2-7M SPR chip surfaces used for the TβRII binding measurements described above. This showed that TβRI alone binding is negligible to both TGF-β3 and mmTGF-β2-7M (Fig. 3, E and F), but unlike TGF-β3, TβRII-bound mmTGF-β2-7M is unable to recruit TβRI (Fig. 3, G and H). This is consistent with the earlier result reported by Huang et al. (37) that TβRII-bound mTGF-β3 was significantly or completely impaired in terms of its ability to bind and recruit TβRI. This also provides further evidence that TβRII-bound TGF-β monomers are incapable of binding and recruiting TβRI, but because the mmTGF-β2-7M was immobilized on the surface of the sensor, it alone does not provide any insight as to whether mmTGF-β2-7M might be capable of non-covalently dimerizing and binding and recruiting TβRI.
To address these questions directly, two solution-based techniques were used, analytical ultracentrifugation (AUC) and time-resolved fluorescence resonance energy transfer (TR-FRET). The AUC experiments were performed by measuring the total UV absorbance at 280 nm as a function of the radial position and time as mTGF-β3, mmTGF-β2, and mmTGF-β2-7M were sedimented under acidic conditions, pH 3.8, where the monomers are fully soluble. The AUC data revealed parabolically shaped van Holde-Weischet sedimentation coefficient distribution plots for all three monomers (data not shown), consistent with each undergoing reversible self-association to form a dimer or other higher order oligomer. To determine more precisely which species might be present in solution, the data were fitted to the simplest model possible, a discrete monomer-dimer equilibrium, using finite element analysis as described under “Experimental procedures.” The fitting procedure resulted in near-perfect fits for all three monomers to the simple monomer-dimer model, as shown by (a) the close overlays between the fitted curves (red) with the raw data, after the time and radially-invariant noise was removed (black) and (b) the absence of any systemic deviations in the residuals (supplemental Figs. S5–S7). The fitted parameters further showed that KD for self-association was 1 order of magnitude greater for mTGF-β3 compared with mmTGF-β2 and mmTGF-β2-7M. Thus, the removal of the heel helix, α3, does diminish self-association of the monomers to form dimers, but it does not completely abrogate dimer formation.
TR-FRET was used to assess the ability of dimeric and monomeric TGF-βs to bind and bring TβRI and TβRII together. This was accomplished by generating differentially tagged forms of TβRII and TβRI and in turn binding to these tags with proteins labeled with fluorescent donors and acceptors. TβRII was tagged with a C-terminal His tag and was bound by a terbium cryptate-labeled anti-His monoclonal antibody fluorescent donor, and TβRI was tagged with an N-terminal avitag, which after enzymatic biotinylation was bound to a dye-labeled (XL-665) streptavidin fluorescent acceptor (Fig. 7A). The addition of TGF-β to the tagged receptors brings them together and leads to a large increase in the ΔF value, which is defined as the ratio of the acceptor and donor emission fluorescent intensities. The TR-FRET assay is demonstrated by the data presented in supplemental Fig. S8 and was used here to compare the ability of the TGF-β3 full-length monomer, mTGF-β3, and the TGF-β2 mini-monomer that binds TβRII with high affinity, mmTGF-β2-7M, to bind and bring TβRI and TβRII together. The TR-FRET signal for mTGF-β3 was shown to be comparable with that of TGF-β3, and this did not depend on whether the TGF-β concentration was 100 or 250 nm (Fig. 7B). The TR-FRET signal of mmTGF-β2-7M was, in contrast, within the error limits of the buffer control, and this did not depend on the TGF-β concentration (Fig. 7B). These results demonstrate that under these conditions, mTGF-β3 retains full capacity to assemble a non-covalent dimeric complex with TβRI and TβRII, but under these same conditions, mmTGF-β2-7M has no capacity to do so. These results, together with the AUC results, suggest that the removal of the heel helix had the effects anticipated; its removal appears to have reduced, although did not eliminate, dimer formation, and even though dimers are still formed, they are evidently unable to bind and recruit TβRI.
Figure 7.
TR-FRET assay for ligand-mediated assembly of TβRI·TβRII complexes. A, structure of the TGF-β3·TβRII·TβRI complex with tags appended to the C terminus of TβRI and TβRII and fluorescently labeled donor and acceptor proteins that associate with the tags. TβRII has a C-terminal hexahistidine tag (His6) and is bound by an Tb3+-cryptate-labeled antihexahistidine tag antibody (CisBio, Bedford, MA). TβRI has a C-terminal biotinylated avitag and is bound by XL665-labeled streptavidin (CisBio, Bedford, MA). The single lysine residue in the TβRI C-terminal avitag that is biotinylated is labeled as K-B. B, preassembled TGF-β3·TβRII-His (1:2), mTGF-β3·TβRII-His (1:1), and mmTGF-β2-7M·TβRII-His (1:1) complexes at a concentration of 100 nm (blue bars) or 250 nm (gray bars) were incubated with 50 nm biotinylated TβRI-ΔC-Avi and 2 nm terbium-anti-His and 30 nm SA-665 for 2 h at room temperature. Buffer control (orange bars) contained only 2 nm terbium-anti-His and 30 nm SA-665. Measurements were performed using a BMG Labtech Pherastar FS. ΔF for each sample was determined by assigning two buffer control assays as the negative control as described under “Experimental procedures.”
Discussion
The TGF-βs are responsible for promoting the progression of numerous human diseases (11–13, 44), yet despite nearly 2 decades of preclinical studies and clinical trials, no inhibitors have been approved for use in humans. The results presented here demonstrate that an engineered TGF-β monomer, lacking Cys-77 and the heel α-helix (α3), functions to potently block and inhibit signaling of the TGF-β1, -β2, and -β3 with IC50 values in the range of 20–70 nm (Fig. 6B and supplemental Fig. S4). This novel inhibitor has several attributes that may overcome limitations that have been encountered with other classes of inhibitors, for example the natural high specificity of TGF-β and thus the inhibitor for TβRII may engender it with much greater specificity, and thus fewer undesirable side effects, compared with the much more promiscuous TGF-β kinase inhibitors. The small size of the inhibitor (∼10 kDa) may further engender it with a much greater ability to penetrate tumors and other dense tissues where the TGF-βs drive disease progression, a distinct advantage compared with IgG antibodies, which are much larger (∼150 kDa) and tend to occupy only the vascular and interstitial space of well perfused organs (31, 32). The other advantages of this novel inhibitor include its high intrinsic stability, because of the four intramolecular disulfide bonds that tie the four fingers together, and the fact that it is highly soluble in water at neutral pH, unlike native TGF-β dimers or full-length TGF-β monomers.
The structures of TGF-β receptor complexes, together with the previously published chemical cross-linking data, suggested that the potent signaling activity of TGF-β1 C77S and TGF-β3 C77S was due to the ability of the monomers to non-covalently dimerize and in turn assemble a (TβRI·TβRII)2 heterotetramer. The results presented here, namely the AUC experiments that were used to assess non-covalent dimer formation and the TR-FRET experiments that were used to assess assembly of complexes with TβRI and TβRII, provided further evidence for this. The AUC data showed that full-length monomeric TGF-β3, mTGF-β3, self-associates to form dimers with a dimerization constant of 4.1 μm (Table 3). The TR-FRET data showed that at a concentration of 0.1 or 0.25 μm and in the presence of comparable concentrations of the TβRI and TβRII ectodomains, mTGF-β3 assembles TβRI·TβRII complexes to the same extent as dimeric TGF-β3 (Fig. 7B). That this occurs, even under conditions where the mTGF-β3 concentrations (0.1–0.25 μm, Fig. 7B) were more than an order of magnitude below the KD value for self-association (4.1 μm, Table 3), indicates that receptor binding also contributes significantly to assembly of TβRI·TβRII complexes. The assembly of TβRI·TβRII complexes with mTGF-β3, and presumably mTGF-β1 as well, therefore appears to be a cooperative process, much like protein folding, in which multiple weaker interactions, including monomer-monomer, non-covalent dimer-receptor, and receptor-receptor interactions, cooperate to enable formation of a thermodynamically stable TGF-β·TβRI·TβRII complex. This manner of cooperative assembly is likely responsible for the ability of mTGF-β1 and mTGF-β3 to induce signaling at concentrations that are more than 4 orders of magnitude below the KD value for self-association of the monomers (EC50 values of about 0.1 nm versus KD values for self-association of 4.1 μm).
Table 3.
Fitting results for the finite element monomer-dimer model for TGF-β monomers
r.m.s.d. means root mean square deviation.
| Parameter | mTGF-β3 | mmTGF-β2 | mmTGF-β2–7 m |
|---|---|---|---|
| r.m.s.d. of the fit (OD280 nm) | 0.00253 | 0.00276 | 0.00361 |
| KD, 1–2 (m) | 4.1 × 10−6 (1.9 × 10−6, 6.2 × 10−6) | 4.4 × 10−5 (3.9 × 10−5, 4.8 × 10−5) | 4.9 × 10−5 (4.5 × 10−5, 5.3 × 10−5) |
| Loading concentration (m) | 1.25 × 10−5 | 1.58 × 10−5 | 1.57 × 10−5 |
| Frictional ratio, monomer | 1.04 (0.99, 1.09) | 1.18 (1.16, 1.19) | 1.30 (1.29, 1.31) |
| Frictional ratio, dimer | 1.37 (1.29, 1.44) | 1.30 (1.29, 1.31) | 1.44 (1.43, 1.45) |
| Partial specific volume, monomer, dimer (v̄, ml g−1) | 8.10 × 10−1 (7.99 × 10−1, 8.21 × 10−1) | 7.70 × 10−1 (7.67 × 10−1, 7.72 × 10−1) | 7.07 × 10−1 (7.05 × 10−1, 7.10 × 10−1) |
| Sedimentation coefficient, monomer (s, ×10−13) | 1.29 (1.26, 1.32) | 1.24 (1.23, 1.25) | 1.46 (1.45, 1.46) |
| Sedimentation coefficient, dimer (s, ×10−13) | 1.56 (1.54, 1.58) | 1.78 (1.75, 1.81) | 2.08 (2.07, 2.10) |
a Parameters in parentheses denote the 95% confidence interval obtained from Monte Carlo analysis.
The elimination of the heel helix from the TGF-β monomer was shown to be very effective in terms of blocking the cooperative assembly of TβRI·TβRII complexes as shown by the TR-FRET data (Fig. 7B) and the cell based signaling data (Fig. 6, A and B). The AUC data showed that elimination of the heel helix led to the weakening of the monomer-monomer interaction by 1 order of magnitude (Table 3). The SPR data shown in Fig. 3, G and H, further showed that the TβRII-bound form of mmTGF-β2-7M was incapable of binding and recruiting TβRI, which is completely expected based on published structures of TGF-β receptor complexes that show that TβRI binds to a composite interface formed by both chains of TGF-β, as well as TβRII (8, 9). Thus, the data show that the reduced propensity of the engineered monomer to self-associate, together with what would be expected to be very weak binding of TβRI to any dimers that do form, is responsible for the inability of mmTGF-β2-7M to assemble a TβRI·TβRII complex. This accounts for the lack of signaling activity, and this together with the retention of high affinity TβRII binding accounts for the inhibitory activity.
The other type II receptors of the family, activin type II receptor II, activin type IIB receptor, BMP type II receptor, and anti-Müllerian hormone type II receptor, have either been shown or are predicted to bind the GF knuckle and not the GF fingertips, as does TβRII (1). Nonetheless, they share the same property as TβRII in that they bind only by contacting residues from a single GF monomer and not both monomers as has been shown or is predicted for all type I receptors of the family (1). This, together with the structures reported here that show that it is possible to remove α3 without affecting the overall structure of the monomer (Figs. 2, B–D, and 5, B–E), suggests that it might be possible to generate monomers of other GFs of the family lacking the heel helix that function as inhibitors. These types of inhibitors have numerous potential applications, ranging from research tools for probing roles of specific ligands in vivo to clinically useful inhibitors for treating disease, which are driven by hyperactive signaling by other ligands of the family, such as cancer cachexia by activin (45).
Experimental procedures
Protein expression and purification
TGF-β1 was expressed as a secreted protein bound to its prodomain in stably transfected CHO cells. The cell line used to produce TGF-β1, and the accompanying procedure to isolate the mature disulfide-linked TGF-β1 homodimer from the conditioned medium, has been described previously (46) and was kindly provided from Dr. Peter Sun (NIAID, National Institutes of Health, Rockville, MD). Mouse homodimeric TGF-β2 (TGF-β2), human homodimeric TGF-β3 (TGF-β3), and variants, including homodimeric N-terminal avi-tagged (47) TGF-β3 (avi-TGF-β3), monomeric TGF-β2 (mTGF-β2), monomeric TGF-β3 (mTGF-β3), mini-monomeric TGF-β1 (mmTGF-β1), mini-monomeric TGF-β2 (mmTGF-β2), mini-monomeric TGF-β3 (mmTGF-β3), mini-monomeric TGF-β2 with seven substitutions to enable high affinity TβRII binding (mmTGF-β2-7M), and mini-monomeric N-terminal avi-tagged (47) TGF-β2 with seven substitutions to enable high affinity TβRII binding (avi-mmTGF-β2-7M), were expressed in E. coli, refolded from inclusion bodies into native folded disulfide-linked homodimers (TGF-β2, TGF-β3, avi-TGF-β3) or monomers (mTGF-β1, mTGF-β2, mTGF-β3, mmTGF-β1, mmTGF-β2, mmTGF-β3, mmTGF-β2-7M, avi-mmTGF-β2-7M), and purified to homogeneity using high resolution cation exchange chromatography (Source Q, GE Healthcare) as described previously (38). The nomenclature and features of the dimeric and monomeric TGF-βs used in this study are summarized in the supplemental Table S1, and the complete sequences are shown in supplemental Fig. S1.
The human TβRI ectodomain (TβRI), spanning residues 1–101 of the mature receptor, or a variant spanning residues 1–88 of the mature receptor with a 15-amino acid avitag (47) appended to the C terminus (TβRI-ΔC-Avi) was expressed in E. coli, refolded from inclusion bodies, and purified to homogeneity as described previously (7). The human TβRII ectodomain (TβRII), spanning residues 15–136 of the mature receptor, or the same but with a C-terminal hexahistidine tag (TβRII-His) was expressed in E. coli, refolded from inclusion bodies, and purified to homogeneity as described previously (48).
Solubility assays
TGF-β dimers and monomers were prepared in 100 mm acetic acid to concentrations of 300 μm or higher and diluted to the desired concentration in either 100 mm acetic acid or phosphate-buffered saline (PBS, 10 mm Na2HPO4, 1.8 mm KH2PO4, 137 mm NaCl, 2.7 mm KCl, pH 7.4). The pH of the samples diluted into PBS were adjusted with small aliquots of NaOH to ensure a final pH of 7.4. The light scattering at 340 nm of the samples was measured in a 1-cm quartz cuvette using a HP 8452 diode array spectrophotometer (Hewlett-Packard, Palo Alto, CA). The samples were transferred to a microcentrifuge tube and centrifuged at 20,000 × g for 5 min, and the absorbance at 280 nm of the supernatant was measured using a Nanodrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA).
NMR spectroscopy
mmTGF-β2 and mmTGF-β2-7M samples isotopically labeled with 15N or 15N and 13C for NMR were prepared by growing bacterial cells in M9 media containing 0.1% (w/v) 15NH4Cl or 0.1% (w/v) 15NH4Cl and 0.03% (w/v) 13C-labeled glucose. All NMR samples were prepared in 10 mm sodium phosphate, 10 mm CHAPS, and 5% 2H2O at a protein concentration of 0.2 mm, pH 4.7. All NMR data were acquired at a sample temperature of 37 °C at either 700 or 800 MHz using Bruker AV-I or AV-II spectrometers equipped with a 5-mm 1H-{13C,15N} TCI cryogenically cooled probe (Bruker, Billerica, MA). Backbone resonance assignments of mmTGF-β2 and mmTGF-β2-7M were obtained by collecting and analyzing sensitivity-enhanced HNCACB (49), CBCA(CO)NH (50), C(CO)NH (51), and HNCO (52) data sets with 25% non-uniform sampling of the points in the 13C,15N acquisition grid. Backbone amide 15N T2 relaxation parameters were measured in an interleaved manner at 300 K at a 15N frequency of 70.95 MHz using 1H-detected pulse schemes previously described (53). The T2 data sets were each collected using 8–10 delay times, varying between 16 and 192 ms. The T2 relaxation times were obtained by fitting relative peak intensities as a function of the T2 delay time to a two-parameter decaying exponential. Data were processed using NMRPipe (54), with the SMILE algorithm used for prediction of the missing points in the 13C and 15N dimensions of the non-uniform sampling data sets (55). Data analysis was performed using NMRFAM-SPARKY (56).
SPR binding measurements
SPR measurements with TGF-β2 and mmTGF-β2 shown in Fig. 3, A and B, were performed using a Biacore 3000 SPR (GE Healthcare) instrument with direct immobilization of TGF-β2 or mmTGF-β2 on the surface of a CM5 sensor chip (GE Healthcare) using an amine (carbodiimide-based) coupling kit (GE Healthcare). SPR experiments shown in Fig. 3, C, E, and G and in D, F, and H with TGF-β3 and mmTGF-β2-7M, respectively, were performed using a Biacore X100 SPR instrument (GE Healthcare) with biotinylated ligands captured at a moderate density (50–200 resonance units) onto a streptavidin-coated CM5 sensor chip (GE Healthcare). Biotinylated TGF-β3 or mmTGF-β2-7M was generated by expressing TGF-β3 or mmTGF-β2-7M with an N-terminal 15-amino acid avitag (47). avi-TGF-β3 or avi-mmTGF-β2-7M was bound to TβRII in 10 mm Bicine, pH 8.0, and biotinylated by incubating with a catalytic amount of bacterially expressed BirA recombinase, biotin, and ATP at 37 °C for 2 h as described (38). Biotinylated avi-tagged TGF-β3 or avi-tagged TGF-β2-7M was bound to a C4 reverse phase column equilibrated with 94.9% water, 5% acetonitrile, 0.1% trifluoroacetic acid and eluted with a linear acetonitrile gradient.
SPR measurements shown in Fig. 3, A–F, were performed in HBS-EP buffer (10 mm Hepes, pH 7.4, 150 mm NaCl, 3 mm EDTA, 0.005% surfactant P-20 (GE Healthcare)) with the receptor indicated injected over a series of 2-fold dilutions over the concentration range shown. Injections were carried out in duplicate and included 10 buffer blank injections at the start of the experiment. Injections were performed for 2–3 min at a flow rate of 100 μl min−1, followed by a dissociation for 1 min or longer in which buffer alone was injected. Each cycle of injection was followed by a 30-s injection of 4 m guanidine·HCl, 2 m NaCl. Data were processed by subtracting both the response from a blank flow cell as well as buffer blanks using the program Scrubber2 (Biologic software, Campbell, Australia). Kinetic fitting of the data was performed with Scrubber2 assuming a simple 1:1 binding model. SPR measurements shown in Fig. 3, G and H, were performed similarly, except 2 μm TβRII was included in both the running buffer and the injected samples.
Crystallization, structure determination, and refinement
Crystals of mmTGF-β2 were formed in sitting drops at 25 °C by combining 0.2 μl of a 7.9 mg ml−1 protein stock solution in 10 mm MES, pH 5.5, with 0.2 μl of the precipitant from the well, 20% polyethylene glycol 3350, 0.2 m sodium thiocyanate. Harvested crystals were mounted in undersized nylon loops with excess mother liquor wicked off, followed by flash-cooling in liquid nitrogen prior to data collection. Data were acquired at the Advanced Photon Source NE-CAT beamline 24-ID-C and integrated and scaled using XDS (57). The structure was determined by the molecular replacement method implemented in PHASER (58) using a truncated version of PDB entry 2TGI (59) as the search model. Coordinates were refined using PHENIX (60), including simulated annealing with torsion angle dynamics, and alternated with manual rebuilding using COOT (61). Data collection and refinement statistics are shown in Table 1.
Crystals of the mmTGF-β2-7M·TβRII complex were formed in hanging drops at 25 °C by combining 1.0 μl of a 7.4 mg ml−1 stock solution of the complex in 10 mm Tris, pH 7.4, with 1.0 μl of 0.1 m Hepes, pH 7.5, 60% v/v (±)-2-methyl-2,4-pentanediol. Harvested crystals were mounted in nylon loops, followed by flash-cooling in liquid nitrogen prior to data collection. Data were acquired at the Advanced Photon Source 22-ID-D and integrated and scaled using HKL2000 (62). The structure was determined by the molecular replacement method implemented in PHASER (58) using TβRII (PDB 1M9Z (63)) and mmTGF-β2 as search models. Coordinates were refined using PHENIX (60), alternated with manual rebuilding using COOT (61). Data collection and refinement statistics are shown in Table 1.
Crystals of mmTGF-β2-7M were formed in hanging drops at 25 °C by combining 1.0 μl of a 10 mg ml−1 protein stock solution in 20 mm acetic acid with 0.8 μl of the precipitant from the well, 100 mm sodium acetate dibasic trihydrate, pH 4.6, 25% 2-propanol, and 400 mm calcium chloride dehydrate, and 0.2 μl of 5% n-octyl-β-d-glucoside. Harvested crystals were mounted in nylon loops and cryoprotected in well buffer containing 20% glycerol and flash-cooled in a nitrogen stream. Data were collected at 100 K using a Rigaku FR-E Superbright generator equipped with a Saturn 944 CCD detector and processed using MOSFLM (64) in CCP4 (65). The structure of mmTGF-β2-7M was solved via molecular replacement using the structure of mmTGF-β2-7M from its co-crystal structure with TβRII. Iterative model building and refinement were performed using COOT (61) and PHENIX4, respectively. Data collection and refinement statistics are shown in Table 1.
Luciferase assays
HEK293 cells stably transfected with the CAGA12 TGF-β reporter were used for the luciferase reporter assays (43) and were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Cells were treated for 16 h with a TGF-β (TGF-β1, mTGF-β3, or mmTGF-β2-7M) concentration series or a mmTGF-β2-7M concentration series in the presence of a constant sub-saturating concentration of TGF-β (TGF-β1, 8 pm; TGF-β2, 20 pm; or TGF-β3, 10 pm). Proteins were diluted in DMEM containing 0.1% w/v BSA. After 16 h, cells were lysed with Tropix lysis buffer (Thermo Fisher Scientific), and luciferase activity was read with a Promega GloMax luminometer (Promega, Madison, WI). Luciferase activity was normalized to total protein levels determined by bicinchoninic acid (BCA) protein assay. GraphPad Prism 6 was used to fit the data to standard models for ligand activity (EC50) and ligand inhibitory activity (IC50) (GraphPad, La Jolla, CA).
Time-resolved FRET assays
The following purified proteins were used to address the ligand requirements for the formation of complexes containing TβRI and TβRII: TGF-β3, mTGF-β3, mmTGF-β2-7M, biotinylated TβRI-ΔC-Avi, and TβRII-His. Initially, 20 μm binary complexes of TGF-β3·TβRII-His, mTGF-β3·TβRII-His, and mmTGF-β2-7M·TβRII-His were formed in a 50 mm Tris, pH 7.5, buffer and stored at 4 °C. A TR-FRET assay based on the proximity-dependent transfer of fluorescence from the donor terbium cryptate-labeled anti-His mAb (terbium-anti-His, CisBio, Bedford, MA) to the acceptor XL665-labeled streptavidin (SA-665, CisBio, Bedford, MA) was used to monitor the assembly of ternary ligand·TβRII-His·biotinylated TβRI-ΔC-Avi complexes. 50-μl assays containing 100 or 250 nm TGF-β3·TβRII-His (1:2), mTGF-β3·TβRII-His (1:1), and mmTGF-β2-7M·TβRII-His (1:1) complexes were incubated with 50 nm biotinylated TβRI-ΔC-Avi. Each 50-μl ternary complex formation assay also contained 2 nm terbium-anti-His and 30 nm SA-665 and was incubated at room temperature for 2 h. Each condition was tested in replicates of six. Buffer control (n = 6) contained only 2 nm terbium-anti-His and 30 nm SA-XL665. The buffer conditions for each assay were 50 mm Tris, 50 mm NaCl, pH 7.5. The assays were performed in Corning black 384 well low flange microplates (Thermo Fisher Scientific). After a 2-h incubation, the assay plate was measured for terbium/XL-665 TR-FRET on a BMG Labtech Pherastar FS multimode plate reader (BMG Labtech Inc., Cary, NC). An optic module containing 337-, 490-, and 665-nm filters was used to monitor TR-FRET producing raw data for 337/490 (terbium emission) and 337/665 (XL-665) emission. The ratio of 665 emission/490 emission was determined for each condition and was subsequently used to calculate ΔF, which is a measure that reflects the signal of the sample versus the background. ΔF was calculated using the following equation: (ratiosignal − rationegative/rationegative) × 100. The ratiosignal refers to the assays containing the trimeric complexes or buffer control. The rationegative refers to two buffer control assays (2 nm terbium-anti-His and 30 nm SA-665). For the buffer control, 2 of the 6 replicates were assigned as negative controls for the purpose of calculating ΔF. ΔF was calculated for the remaining four buffer control replicates.
Analytical ultracentrifugation
mTGF-β3, mmTGF-β2, and mmTGF-β2-7M were analyzed by sedimentation velocity to establish equilibrium constants for self-association of monomeric TGF-βs to form homodimers. mTGF-β3, mmTGF-β2, and mmTGF-β2-7M were each measured at 280 nm in an Epon two-channel centerpiece fitted with quartz windows, and centrifuged at 20 °C and 42,000 rpm for 27 h in a 15 mm sodium phosphate buffer adjusted to pH 3.8, containing 100 mm NaCl. 300 scans were collected in intensity mode on a Beckman Optima XL-I analytical ultracentrifuge at the CAUMA facility at the University of Texas Health Science Center at San Antonio. Data analysis was performed with UltraScan release 2142 (66, 67); calculations were performed at the San Diego Supercomputing Center on Comet and Gordon. The sedimentation velocity data were initially fitted with the two-dimensional spectrum analysis, as described previously (66), to remove time- and radially-invariant noise from the raw data and to fit the meniscus position. Subsequently, the data were fitted to a discrete monomer-dimer model using the adaptive space-time finite element method (67) and genetic algorithms for the parameter optimization (68). The monomer-dimer model accounts for mass action and the reversible association behavior, fitting the thermodynamic and hydrodynamic parameters, as well as the partial specific volume while assuming the predicted molar mass for either wild type or mutant. A Monte Carlo analysis (69) with 100 iterations was performed for each dataset to obtain fitting statistics. Buffer density and viscosity were estimated with UltraScan based on buffer composition, and all hydrodynamic values were corrected for standard conditions (20 °C and water). The fitting results provided an excellent fit with random residuals and very low r.m.s.d. values (see supplemental material and Figs. 4–6). All results are summarized in Table 3.
Author contributions
S. K. K. crystallized mmTGF-β2 and the mmTGF-β2-7M·TβRII complex, performed the solubility measurements, performed a portion of the SPR experiments, and wrote the initial draft of the paper. L. B. and C. S. H. performed the luciferase assays. C. S. H. together with M. B., M. V., B. L., B. R., and K. E. C. established the expression and purification of mmTGF-β2-7M. A. T., B. I., and K. E. C. performed the NMR assignment of mmTGF-β2. E. M. P. crystallized and determined the structure of mmTGF-β2-7M. A. B. T. and P .J. H. determined the structure of mmTGF-β2. R. K. performed some of the SPR experiments. S. D., C. O. B., and G. C. determined the structure of mmTGF-β2-7M·TβRII. M. J. H. performed the TR-FRET experiments, and B. D. performed the AUC experiments. A. P. H. conceived the design of the dominant negative TGF-β inhibitors, in consultation with T. S. A. P. H. also performed the NMR assignment of mmTGF-β2-7M and wrote the final draft of the paper. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Peter Sun, NIAID, National Institutes of Health, for providing the stably transected CHO cell line overexpressing TGF-β1; Dr. William Furey for guidance on the refinement of the mmTGF-β2-7M and mmTGF-β2-7M·TβRII structures; Doowon Lee for assistance with the X-ray instrumentation; Mike Delk for assistance with the NMR instrumentation; and Liping Wang for technical assistance with the analytical ultracentrifugation measurements. UltraScan software development was supported by National Institutes of Health Grant GM120600 (to B. D.) and National Science Foundation Grant ACI-1339649 (to B. D.). The calculations at San Diego Supercomputer Center (SDSC) were supported by Extreme Science and Engineering Discovery Environment (XSEDE) Community Allocation Grant MCB070039 (to B. D.).
This work was supported in part by National Institutes of Health Grants GM58670 and CA172886 (to A. H.) and Robert A. Welch Foundation Grant AQ1842 (to A. H.). Additional support was provided by University of Texas Health Science Center Cancer Therapy and Research Center Macromolecular Structure and Interactions Core supported by National Institutes of Health Grant P30 CA54174 from NCI, University of Texas Health Science Center San Antonio (UTHSCSA) Center for Macromolecular Interactions Core Facility supported by the UTHSCSA and UTHSCSA/University of Texas San Antonio Center for Innovative Drug Discovery and High Throughput Screening Facility supported by National Institutes of Health Grant NCATS UL1 TR001120 (to M. J. H). A. P. H. and T. S. are co-inventors of a provisional patent (United States Patent Application 62/423,920) that covers the dominant negative TGF-β, mmTGF-β2-7M. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as one of our Editors' Picks.
This article contains Figs. S1–S8 and Table S1.
The atomic coordinates and structure factors (codes 5TX2, 5TX6, and 5TX4) have been deposited in the Protein Data Bank (http://wwpdb.org/).
The assigned chemical shifts for mmTGF-β2 and mmTGF-β2-7M were deposited to BioMagResBank (BMRB) under accession codes 26943 and 26944, respectively.
- GF
- growth factor
- TβRII
- TGF-β type II receptor
- TβRI
- TGF-β type I receptor
- SPR
- surface plasmon resonance
- HSQC
- heteronuclear single quantum shift correlation
- r.m.s.d.
- root mean square deviation
- TR-FRET
- time-resolved fluorescence resonance energy transfer
- AUC
- analytical ultracentrifugation
- BMP
- bone morphogenetic protein
- Bicine
- N,N-bis(2-hydroxyethyl)glycine
- PDB
- Protein Data Bank.
References
- 1. Hinck A. P., Mueller T. D., and Springer T. A. (2016) Structural biology and evolution of the TGF-β family. Cold Spring Harb. Perspect. Biol. 8, a022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Massagué J. (1998) TGF-β signal transduction. Annu. Rev. Biochem. 67, 753–791 [DOI] [PubMed] [Google Scholar]
- 3. Robertson I. B., and Rifkin D. B. (2013) Unchaining the beast; insights from structural and evolutionary studies on TGFβ secretion, sequestration, and activation. Cytokine Growth Factor Rev. 24, 355–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wrana J. L., Attisano L., Cárcamo J., Zentella A., Doody J., Laiho M., Wang X. F., and Massagué J. (1992) TGF β signals through a heteromeric protein kinase receptor complex. Cell 71, 1003–1014 [DOI] [PubMed] [Google Scholar]
- 5. Wrana J. L., Attisano L., Wieser R., Ventura F., and Massagué J. (1994) Mechanism of activation of the TGF-β receptor. Nature 370, 341–347 [DOI] [PubMed] [Google Scholar]
- 6. Laiho M., Weis F. M., Boyd F. T., Ignotz R. A., and Massagué J. (1991) Responsiveness to transforming growth factor-β (TGF-β) restored by genetic complementation between cells defective in TGF-β receptors I and II. J. Biol. Chem. 266, 9108–9112 [PubMed] [Google Scholar]
- 7. Zúñiga J. E., Groppe J. C., Cui Y., Hinck C. S., Contreras-Shannon V., Pakhomova O. N., Yang J., Tang Y., Mendoza V., López-Casillas F., Sun L., and Hinck A. P. (2005) Assembly of TβRI:TβRII:TGFβ ternary complex in vitro with receptor extracellular domains is cooperative and isoform-dependent. J. Mol. Biol. 354, 1052–1068 [DOI] [PubMed] [Google Scholar]
- 8. Groppe J., Hinck C. S., Samavarchi-Tehrani P., Zubieta C., Schuermann J. P., Taylor A. B., Schwarz P. M., Wrana J. L., and Hinck A. P. (2008) Cooperative assembly of TGF-β superfamily signaling complexes is mediated by two disparate mechanisms and distinct modes of receptor binding. Mol. Cell 29, 157–168 [DOI] [PubMed] [Google Scholar]
- 9. Radaev S., Zou Z., Huang T., Lafer E. M., Hinck A. P., and Sun P. D. (2010) Ternary complex of transforming growth factor-β1 reveals isoform-specific ligand recognition and receptor recruitment in the superfamily. J. Biol. Chem. 285, 14806–14814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Loeys B. L., Schwarze U., Holm T., Callewaert B. L., Thomas G. H., Pannu H., De Backer J. F., Oswald G. L., Symoens S., Manouvrier S., Roberts A. E., Faravelli F., Greco M. A., Pyeritz R. E., Milewicz D. M., et al. (2006) Aneurysm syndromes caused by mutations in the TGF-β receptor. N. Engl. J. Med. 355, 788–798 [DOI] [PubMed] [Google Scholar]
- 11. Dietz H. C., Cutting G. R., Pyeritz R. E., Maslen C. L., Sakai L. Y., Corson G. M., Puffenberger E. G., Hamosh A., Nanthakumar E. J., and Curristin S. M. (1991) Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature 352, 337–339 [DOI] [PubMed] [Google Scholar]
- 12. Biernacka A., Dobaczewski M., and Frangogiannis N. G. (2011) TGF-β signaling in fibrosis. Growth Factors 29, 196–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Massagué J. (2008) TGFβ in cancer. Cell 134, 215–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Markowitz S. D., and Roberts A. B. (1996) Tumor suppressor activity of the TGF-β pathway in human cancers. Cytokine Growth Factor Rev. 7, 93–102 [DOI] [PubMed] [Google Scholar]
- 15. Bandyopadhyay A., Agyin J. K., Wang L., Tang Y., Lei X., Story B. M., Cornell J. E., Pollock B. H., Mundy G. R., and Sun L. Z. (2006) Inhibition of pulmonary and skeletal metastasis by a transforming growth factor-β type I receptor kinase inhibitor. Cancer Res. 66, 6714–6721 [DOI] [PubMed] [Google Scholar]
- 16. Ganapathy V., Ge R., Grazioli A., Xie W., Banach-Petrosky W., Kang Y., Lonning S., McPherson J., Yingling J. M., Biswas S., Mundy G. R., and Reiss M. (2010) Targeting the transforming growth factor-β pathway inhibits human basal-like breast cancer metastasis. Mol. Cancer 9, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Muraoka R. S., Dumont N., Ritter C. A., Dugger T. C., Brantley D. M., Chen J., Easterly E., Roebuck L. R., Ryan S., Gotwals P. J., Koteliansky V., and Arteaga C. L. (2002) Blockade of TGF-β inhibits mammary tumor cell viability, migration, and metastases. J. Clin. Invest. 109, 1551–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nam J. S., Terabe M., Mamura M., Kang M. J., Chae H., Stuelten C., Kohn E., Tang B., Sabzevari H., Anver M. R., Lawrence S., Danielpour D., Lonning S., Berzofsky J. A., and Wakefield L. M. (2008) An anti-transforming growth factor β antibody suppresses metastasis via cooperative effects on multiple cell compartments. Cancer Res. 68, 3835–3843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang Y. A., Dukhanina O., Tang B., Mamura M., Letterio J. J., MacGregor J., Patel S. C., Khozin S., Liu Z. Y., Green J., Anver M. R., Merlino G., and Wakefield L. M. (2002) Lifetime exposure to a soluble TGF-β antagonist protects mice against metastasis without adverse side effects. J. Clin. Invest. 109, 1607–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miao Z. F., Zhao T. T., Wang Z. N., Miao F., Xu Y. Y., Mao X. Y., Gao J., Wu J. H., Liu X. Y., You Y., Xu H., and Xu H. M. (2014) Transforming growth factor-β1 signaling blockade attenuates gastric cancer cell-induced peritoneal mesothelial cell fibrosis and alleviates peritoneal dissemination both in vitro and in vivo. Tumour Biol. 35, 3575–3583 [DOI] [PubMed] [Google Scholar]
- 21. Di Sabatino A., Jackson C. L., Pickard K. M., Buckley M., Rovedatti L., Leakey N. A., Picariello L., Cazzola P., Monteleone G., Tonelli F., Corazza G. R., MacDonald T. T., and Pender S. L. (2009) Transforming growth factor β signalling and matrix metalloproteinases in the mucosa overlying Crohn's disease strictures. Gut 58, 777–789 [DOI] [PubMed] [Google Scholar]
- 22. Yamada M., Kuwano K., Maeyama T., Yoshimi M., Hamada N., Fukumoto J., Egashira K., Hiasa K., Takayama K., and Nakanishi Y. (2007) Gene transfer of soluble transforming growth factor type II receptor by in vivo electroporation attenuates lung injury and fibrosis. J. Clin. Pathol. 60, 916–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Connolly E. C., Freimuth J., and Akhurst R. J. (2012) Complexities of TGF-β targeted cancer therapy. Int. J. Biol. Sci. 8, 964–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hawinkels L. J., and Ten Dijke P. (2011) Exploring anti-TGF-β therapies in cancer and fibrosis. Growth Factors 29, 140–152 [DOI] [PubMed] [Google Scholar]
- 25. Ge R., Rajeev V., Subramanian G., Reiss K. A., Liu D., Higgins L., Joly A., Dugar S., Chakravarty J., Henson M., McEnroe G., Schreiner G., and Reiss M. (2004) Selective inhibitors of type I receptor kinase block cellular transforming growth factor-β signaling. Biochem. Pharmacol. 68, 41–50 [DOI] [PubMed] [Google Scholar]
- 26. Singh J., Ling L. E., Sawyer J. S., Lee W. C., Zhang F., and Yingling J. M. (2004) Transforming the TGFβ pathway: convergence of distinct lead generation strategies on a novel kinase pharmacophore for TβRI (ALK5). Curr. Opin. Drug Discov. Dev. 7, 437–445 [PubMed] [Google Scholar]
- 27. Yingling J. M., Blanchard K. L., and Sawyer J. S. (2004) Development of TGF-β signalling inhibitors for cancer therapy. Nat. Rev. Drug. Discov. 3, 1011–1022 [DOI] [PubMed] [Google Scholar]
- 28. Connolly E. C., Saunier E. F., Quigley D., Luu M. T., De Sapio A., Hann B., Yingling J. M., and Akhurst R. J. (2011) Outgrowth of drug-resistant carcinomas expressing markers of tumor aggression after long-term TβRI/II kinase inhibition with LY2109761. Cancer Res. 71, 2339–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lonning S., Mannick J., and McPherson J. M. (2011) Antibody targeting of TGF-β in cancer patients. Curr. Pharm. Biotechnol. 12, 2176–2189 [DOI] [PubMed] [Google Scholar]
- 30. Hinck A. P. (2012) Structural studies of the TGF-βs and their receptors–insights into evolution of the TGF-β superfamily. FEBS Lett. 586, 1860–1870 [DOI] [PubMed] [Google Scholar]
- 31. Meibohm B. (2012) in Therapeutic Proteins: Strategies to Modulate Their Plasma Half-Lives (Kontermann R., ed) pp. 23–38, Wiley-Blackwell, Weinheim, Germany [Google Scholar]
- 32. Meibohm B., and Braeckman R. A. (2008) Pharmacokinetics and Pharmacodynamics of Peptides and Protein Drugs. in Pharmaceutical Biotechnology: Fundamentals and Applications (Crommelin D. J. A., Sindelar R. D., and Meibohm B., eds) pp. 95–123, Informa Healthcare, New York [Google Scholar]
- 33. Papo N., Silverman A. P., Lahti J. L., and Cochran J. R. (2011) Antagonistic VEGF variants engineered to simultaneously bind to and inhibit VEGFR2 and αvβ3 integrin. Proc. Natl. Acad. Sci. U.S.A. 108, 14067–14072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Amatayakul-Chantler S., Qian S. W., Gakenheimer K., Böttinger E. P., Roberts A. B., and Sporn M. B. (1994) [Ser77]transforming growth factor-β 1. Selective biological activity and receptor binding in mink lung epithelial cells. J. Biol. Chem. 269, 27687–27691 [PubMed] [Google Scholar]
- 35. Hinck A. P., Archer S. J., Qian S. W., Roberts A. B., Sporn M. B., Weatherbee J. A., Tsang M. L., Lucas R., Zhang B. L., Wenker J., and Torchia D. A. (1996) Transforming growth factor β 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor β 2. Biochemistry 35, 8517–8534 [DOI] [PubMed] [Google Scholar]
- 36. Mittl P. R., Priestle J. P., Cox D. A., McMaster G., Cerletti N., and Grütter M. G. (1996) The crystal structure of TGF-β3 and comparison to TGF-β2: implications for receptor binding. Protein Sci. 5, 1261–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huang T., David L., Mendoza V., Yang Y., Villarreal M., De K., Sun L., Fang X., López-Casillas F., Wrana J. L., and Hinck A. P. (2011) TGF-β signalling is mediated by two autonomously functioning TβRI:TβRII pairs. EMBO J. 30, 1263–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huang T., and Hinck A. P. (2016) Production, isolation, and structural analysis of ligands and receptors of the TGF-β superfamily. Methods Mol. Biol. 1344, 63–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baardsnes J., Hinck C. S., Hinck A. P., and O'Connor-McCourt M. D. (2009) TβR-II discriminates the high- and low-affinity TGF-β isoforms via two hydrogen-bonded ion pairs. Biochemistry 48, 2146–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. De Crescenzo G., Hinck C. S., Shu Z., Zúñiga J., Yang J., Tang Y., Baardsnes J., Mendoza V., Sun L., López-Casillas F., O'Connor-McCourt M., and Hinck A. P. (2006) Three key residues underlie the differential affinity of the TGFβ isoforms for the TGFβ type II receptor. J. Mol. Biol. 355, 47–62 [DOI] [PubMed] [Google Scholar]
- 41. Pellaud J., Schote U., Arvinte T., and Seelig J. (1999) Conformation and self-association of human recombinant transforming growth factor-β3 in aqueous solutions. J. Biol. Chem. 274, 7699–7704 [DOI] [PubMed] [Google Scholar]
- 42. Hart P. J., Deep S., Taylor A. B., Shu Z., Hinck C. S., and Hinck A. P. (2002) Crystal structure of the human TβR2 ectodomain–TGF-β3 complex. Nat. Struct. Biol. 9, 203–208 [DOI] [PubMed] [Google Scholar]
- 43. Thies R. S., Chen T., Davies M. V., Tomkinson K. N., Pearson A. A., Shakey Q. A., and Wolfman N. M. (2001) GDF-8 propeptide binds to GDF-8 and antagonizes biological activity by inhibiting GDF-8 receptor binding. Growth Factors 18, 251–259 [DOI] [PubMed] [Google Scholar]
- 44. Loeys B. L., Mortier G., and Dietz H. C. (2013) Bone lessons from Marfan syndrome and related disorders: fibrillin, TGF-B and BMP at the balance of too long and too short. Pediatr. Endocrinol. Rev. 10, 417–423 [PubMed] [Google Scholar]
- 45. Coerver K. A., Woodruff T. K., Finegold M. J., Mather J., Bradley A., and Matzuk M. M. (1996) Activin signaling through activin receptor type II causes the cachexia-like symptoms in inhibin-deficient mice. Mol. Endocrinol. 10, 534–543 [DOI] [PubMed] [Google Scholar]
- 46. Zou Z., and Sun P. D. (2004) Overexpression of human transforming growth factor-β1 using a recombinant CHO cell expression system. Protein Expr. Purif. 37, 265–272 [DOI] [PubMed] [Google Scholar]
- 47. Cull M. G., and Schatz P. J. (2000) Biotinylation of proteins in vivo and in vitro using small peptide tags. Methods Enzymol. 326, 430–440 [DOI] [PubMed] [Google Scholar]
- 48. Hinck A. P., Walker K. P. 3rd., Martin N. R., Deep S., Hinck C. S., and Freedberg D. I. (2000) Sequential resonance assignments of the extracellular ligand binding domain of the human TGF-β type II receptor. J. Biomol. NMR 18, 369–370 [DOI] [PubMed] [Google Scholar]
- 49. Wittekind M., and Mueller L. (1993) HNCACB, a high-sensitivity 3-D NMR experiment to correlate amide-proton and nitrogen resonances with the α-carbon and β-carbon resonances in proteins. J. Magn. Reson. Ser. B. 101, 201–205 [Google Scholar]
- 50. Grzesiek S., and Bax A. (1993) Amino acid type determination in the sequential assignment procedure of uniformly 13C/15N-enriched proteins. J. Biomol. NMR 3, 185–204 [DOI] [PubMed] [Google Scholar]
- 51. Grzesiek S., Anglister J., and Bax A. (1993) Correlation of backbone amide and aliphatic side-chain resonances in C-13/N-15-enriched proteins by isotropic mixing of C-13 magnetization. J. Magn. Reson. Ser. B. 101, 114–119 [Google Scholar]
- 52. Kay L. E., Ikura M., Tschudin R., and Bax A. (1990) 3-Dimensional triple-resonance Nmr-spectroscopy of isotopically enriched proteins. J. Magn. Reson. 89, 496–514 [DOI] [PubMed] [Google Scholar]
- 53. Kay L. E., Torchia D. A., and Bax A. (1989) Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry 28, 8972–8979 [DOI] [PubMed] [Google Scholar]
- 54. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., and Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 55. Ying J., Delaglio F., Torchia D. A., and Bax A. (2016) Sparse multidimensional iterative lineshape-enhanced (SMILE) reconstruction of both non-uniformly sampled and conventional NMR data. J. Biomol. NMR 10.1007/s10858-016-0072-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee W., Tonelli M., and Markley J. L. (2015) NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Daopin S., Piez K. A., Ogawa Y., and Davies D. R. (1992) Crystal structure of transforming growth factor-β2: an unusual fold for the superfamily. Science 257, 369–373 [DOI] [PubMed] [Google Scholar]
- 60. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 63. Boesen C. C., Radaev S., Motyka S. A., Patamawenu A., and Sun P. D. (2002) The 1.1 angstrom crystal structure of human TGF-β type II receptor ligand binding domain. Structure 10, 913–919 [DOI] [PubMed] [Google Scholar]
- 64. Battye T. G., Kontogiannis L., Johnson O., Powell H. R., and Leslie A. G. (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., and Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Demeler B. (2010) Methods for the design and analysis of sedimentation velocity and sedimentation equilibrium experiments with proteins. Curr. Protoc. Protein Sci. Chapter 7, Unit 7.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cao W., and Demeler B. (2008) Modeling analytical ultracentrifugation experiments with an adaptive space-time finite element solution for multicomponent reacting systems. Biophys. J. 95, 54–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Demeler B., Brookes E., Wang R., Schirf V., and Kim C. A. (2010) Characterization of reversible associations by sedimentation velocity with UltraScan. Macromol. Biosci. 10, 775–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Demeler B., and Brookes E. (2008) Monte Carlo analysis of sedimentation experiments. Colloid Polym. Sci. 286, 129–137 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.