Abstract
Most Gram-negative bacteria respond to excessive levels of H2O2 using the peroxide-sensing transcriptional regulator OxyR, which can induce the expression of antioxidant genes to restore normality. Vibrio vulnificus has two distinct OxyRs (OxyR1 and OxyR2), which are sensitive to different levels of H2O2 and induce expression of two different peroxidases, Prx1 and Prx2. Although OxyR1 has both high sequence similarity and H2O2 sensitivity comparable with that of other OxyR proteins, OxyR2 exhibits limited sequence similarity and is more sensitive to H2O2. To investigate the basis for this difference, we determined crystal structures and carried out biochemical analyses of OxyR2. The determined structure of OxyR2 revealed a flipped conformation of the peptide bond before Glu-204, a position occupied by glycine in other OxyR proteins. Activity assays showed that the sensitivity to H2O2 was reduced to the level of other OxyR proteins by the E204G mutation. We solved the structure of the OxyR2-E204G mutant with the same packing environment. The structure of the mutant revealed a dual conformation of the peptide bond before Gly-204, indicating the structural flexibility of the region. This structural duality extended to the backbone atoms of Gly-204 and the imidazole ring of His-205, which interact with H2O2 and invariant water molecules near the peroxidatic cysteine, respectively. Structural comparison suggests that Glu-204 in OxyR2 provides rigidity to the region that is important in H2O2 sensing, compared with the E204G structure or other OxyR proteins. Our findings provide a structural basis for the higher sensitivity of OxyR2 to H2O2 and also suggest a molecular mechanism for bacterial regulation of expression of antioxidant genes at divergent concentrations of cellular H2O2.
Keywords: antioxidant, bacterial transcription, crystal structure, crystallography, hydrogen peroxide, reactive oxygen species (ROS), OxyR, Vibrio vulnificus, sensitivity
Introduction
Most aerobic organisms encounter reactive oxygen species, including hydrogen peroxide (H2O2), during aerobic respiration processes (1, 2). At higher than critical concentrations of H2O2, highly toxic hydroxyl radicals, which can damage essential cellular components, can be generated (3). To cope with H2O2 stress, many bacteria have developed elaborate sensing and scavenging systems. In particular, pathogenic bacteria have sophisticated systems to overcome host immune responses that produce a high level of H2O2 (4).
Many LysR-type transcriptional regulators are found in various bacteria, and they are typically homotetramers that contain an N-terminal DNA-binding domain (DBD)5 and C-terminal regulatory domain (RD) (5). OxyR, a member of the LysR-type transcriptional regulator family, recognizes H2O2 above a certain threshold concentration and induces a diverse array of antioxidant genes, including catalase-peroxidase katG and peroxiredoxins (prx genes) (6, 7). OxyR has a DBD for recognizing target gene promoters and an RD containing two conserved cysteine residues (peroxidatic and resolving cysteines) for sensing H2O2 (8). Many crystal structures have been reported since the first descriptions of Escherichia coli OxyR structures revealed the redox-state-dependent conformational change of RDs (8). Recently elucidated full-length OxyR crystal structures from Pseudomonas aeruginosa have revealed the homotetrameric arrangement of the protein, where the two dimeric RDs face each other, and the two dimeric DBDs are oriented in the same direction. The crystal structures have also provided insight into how this redox-dependent structural alteration of RDs is propagated to the DBDs, leading to inward or outward motion between dimeric DBDs (9).
Reduced OxyRs that have two free thiols in the two conserved cysteine residues are converted into oxidized forms via two steps in the presence of H2O2 (10, 11). The peroxidatic cysteine residue of OxyR is first oxidized by H2O2 to cysteine-sulfenic acid (Cys-SOH), in which a bound H2O2 molecule and two water molecules near the peroxidatic cysteine are thought to play crucial roles via a so-called “H2O2-driven oxidation mechanism” (9). The Cys-SOH ultimately forms a disulfide bond via the resolving cysteine in a certain concentration range of H2O2, resulting in formation of the oxidized (or disulfidized) form of OxyR that activates the induction of target genes (11, 12).
Vibrio vulnificus is a facultative aerobic pathogen that can cause fatal septicemia (13, 14) and has two different OxyR proteins (VvOxyR1 and VvOxyR2) and two different Prx proteins (VvPrx1 and VvPrx2) (15, 16). VvOxyR1 has high sequence and functional similarities to typical types of OxyR proteins and induces VvPrx1 expression at a certain H2O2 concentration (∼5 μm) (16). VvOxyR2 exhibits limited sequence similarity to other typical OxyR types and is activated by a lower H2O2 concentration (∼0.5 μm) than VvOxyR1. VvOxyR2 induces Vvprx2 encoding VvPrx2, which has higher activity than VvPrx1 at low H2O2 levels, although VvPrx2 is more susceptible to irreversible inactivation caused by overoxidation of the peroxidatic cysteine above the working concentration of H2O2 (>5 μm) (16). Recently, a three-state activation mechanism for VvOxyR2 was proposed (11). In this mechanism, above the working concentration of H2O2 for VvPrx2, VvOxyR2 undergoes a new conformational change to prevent non-functional gene expression. In this third state, the peroxidatic cysteine residue of VvOxyR2 is further oxidized to cysteine-sulfinic acid (Cys-SO2H) or cysteine-sulfonic acid (Cys-SO3H), yielding the overoxidized form of OxyR that lacks transcriptional activity (11). To understand how OxyR proteins with similar structures have differing sensitivities to different levels of H2O2, we determined the crystal structures of OxyR2 RD from V. vulnificus and performed biochemical analyses.
Results
Structural analysis of VvOxyR2-RD
To investigate the relationships between the structural features and the higher H2O2 sensitivity of OxyR2 from V. vulnificus (VvOxyR2), the crystal structures of wild-type VvOxyR2-RD (residues 86–301) grown under reduced conditions were determined using the zinc-anomalous signals from a ZnCl2-soaked crystal (17). The resulting electron-density maps were of sufficient quality to allow us to build an almost complete model. The structure was refined against the 1.6 Å-resolution data set. The asymmetric unit contained two protomers of VvOxyR2-RD, and the final model produced 19.2% Rfree and 15.9% Rfactor with reasonable stereochemistry (Table 1). Like other OxyR-RDs, the protomer consists of two subdomains (RD-I and RD-II; Fig. 1A). Two notable dimers generated by the crystallographic 2-fold axis were found in the crystal, where each protomer individually participated in dimer formation. These observations indicate that VvOxyR2 is also a dimer in solution, as are other OxyR proteins. The overall structure of VvOxyR2-RD was similar to other OxyR-RD structures previously determined in the reduced state (Fig. 1, A and B). The two conserved cysteine residues (Cys-206 and Cys-215) remained as free thiol forms separated by the inter-cysteine helix (residues 206–215). The relative orientation between the dimers is similar to those of OxyR proteins from E. coli, Neisseria meningitidis, P. aeruginosa, and Porphyromonas gingivalis in the reduced state (8, 9, 18, 19). Thus, it was concluded that VvOxyR2-RD exhibited the typical reduced conformation of OxyR.
Table 1.
X-ray diffraction and refinement statistics
| VvOxyR2-RD | WT (SO4-free) | WT (ZnCl2-soaked) | E204G (SO4-free) (high resolution) | E204G (SO4-free) (P3121) | E204G (SO4-bound) |
|---|---|---|---|---|---|
| Data collection | |||||
| Beam line | PAL 5C | PAL 5C | PAL 5C | PAL 7A | PAL 7A |
| Wavelength (Å) | 0.97940 | 1.28230 | 1.00820 | 0.97933 | 0.97933 |
| Space group | C2221 | C2221 | C2221 | P3121 | C2221 |
| Cell dimensions | |||||
| a, b, c (Å) | 70.2, 138.3, 97.5 | 70.3, 138.4, 97.4 | 71.2, 138.2, 97.1 | 141.4, 141.4, 109.7 | 72.7, 137.4, 96.7 |
| α, β, γ (degrees) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 120 | 90, 90, 90 |
| Resolution (Å) | 50.0–1.60 (1.63–1.60)a | 50.0–1.96 (1.99–1.96) | 50.0–1.55 (1.58–1.55) | 50.0–2.3 (2.32–2.30) | 50.0–1.52 (1.55–1.52) |
| Rpim | 0.027 (0.164) | 0.034 (0.113) | 0.028 (0.163) | 0.089 (0.368) | 0.020 (0.131) |
| I/σI | 18.6 (2.45) | 51.2 (12.6) | 19.3 (3.2) | 9.7 (2.46) | 31.3 (3.8) |
| Completeness (%) | 98.7 (95.5) | 99.6 (99.4) | 99.4 (96.0) | 96.6 (88.0) | 99.7 (99.1) |
| Redundancy | 8.0 (4.2) | 5.4 (4.7) | 9.5 (6.2) | 4.8 (3.1) | 10.8 (6.7) |
| Refinement | |||||
| Resolution (Å) | 48.7–1.60 | 19.9–1.55 | 33.9–2.30 | 19.8–1.52 | |
| No. of reflections | 55,578 | 64,561 | 53,827 | 72,860 | |
| Rwork/Rfree | 0.159/0.192 | 0.155/0.188 | 0.228/0.267 | 0.157/0.183 | |
| No. of total atoms | 3836 | 4010 | 6752 | 4132 | |
| Wilson B-factor (Å) | 11.22 | 11.21 | 29.7 | 11.9 | |
| Root mean square deviations | |||||
| Bond lengths (Å) | 0.015 | 0.012 | 0.003 | 0.008 | |
| Bond angles (degrees) | 1.310 | 1.209 | 0.586 | 0.964 | |
| Ramachandran plot | |||||
| Favored (%) | 98.1 | 98.3 | 97.6 | 98.8 | |
| Allowed (%) | 1.9 | 1.7 | 2.4 | 1.2 | |
| Outliers (%) | 0 | 0 | 0 | 0 | |
| PDB code | 5X0V | 5X0Q | 5B70 | 5B7D | |
a Values in parentheses are for the highest-resolution shell.
Figure 1.
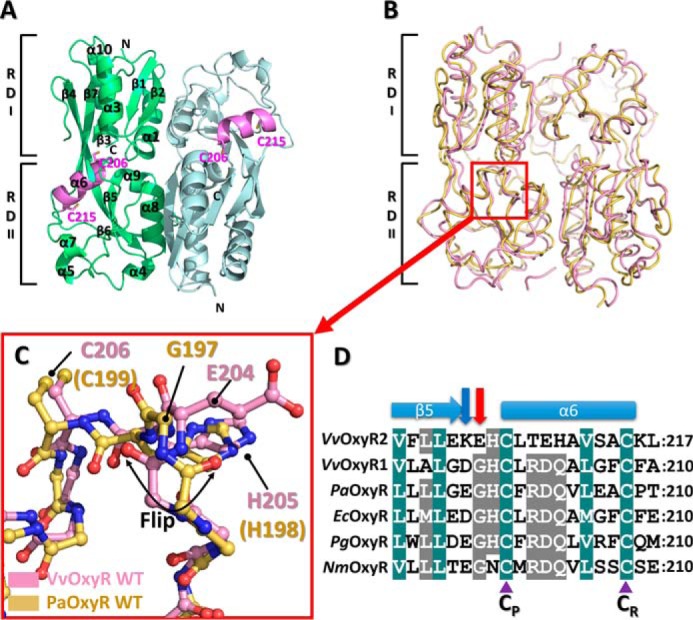
Structure of the wild-type VvOxyR2-RD. A, dimeric assembly of wild-type VvOxyR2-RD under reduced conditions. Protomers are highlighted in green or light blue, except in the inter-cysteine α-helical regions (pink). The conserved cysteines (Cys-206 and Cys-215) are represented as sticks. Secondary structural elements and the subdomains RD-I and RD-II are labeled. B, comparison of VvOxyR2-RD dimers (pink) with P. aeruginosa OxyR-RD in the reduced state (yellow; PDB code 4Y0M): The red box indicates the region near the peroxidatic cysteine (Cys-206) and the Glu-204-containing loop. C, close-up view of the region indicated by the red box in B. The regions near the peroxidatic cysteine of VvOxyR2-RD (pink) and PaOxyR-RD (yellow) are shown in a ball-and-stick representation. The flipped peptide bond is indicated with a double-headed arrow. D, sequence alignment of VvOxyR2 with other OxyRs, focusing on the region containing the conserved cysteine residues (V. vulnificus (Vv), P. aeruginosa (Pa), and N. meningitidis (Nm)). The secondary structures are displayed above the sequence. Glu-204 or its equivalent residue and Lys-203 or its equivalent residue are indicated by a red or a blue arrow, respectively. Two conserved cysteine residues (peroxidatic cysteine (CP) and resolving cysteine (CR)) are indicated by purple triangles. The strictly conserved amino acid residues are indicated by cobalt blue boxes, and the moderately conserved amino acid residues are indicated by gray boxes.
To analyze the structure in more detail, we superposed the VvOxyR2-RD structure on P. aeruginosa OxyR (PaOxyR) in the reduced state (Fig. 1B). Whereas the RD-I subdomain showed minimal deviation among the structures, the RD-II subdomain had a particular variation in the location of the resolving cysteine (Cys-215 in VvOxyR2, Cys-208 in PaOxyR) (Fig. 1C). Compared with cysteine positions in other OxyR proteins, the resolving cysteine Cys-215 is buried in the interior of the RD-II subdomain along with the inter-cysteine α-helix. However, the peroxidatic cysteine is located at the same position as in other OxyR proteins (Fig. 1).
VvOxyR2 has a noncanonical conformation of the peptide bond between Lys-203 and Glu-204
Notably, the structural superposition of VvOxyR2 with other OxyR proteins revealed large conformational differences at the peptide bond (or peptide flipping) between Lys-203 and Glu-204 (Fig. 1C). However, neither the Lys-203 nor Glu-204 side chains was involved in an interaction with other residues or the bound H2O2 and water molecules near the peroxidatic cysteine residue. Sequence alignment revealed that Glu-204 of VvOxyR2 was replaced with a glycine residue observed in most OxyR proteins and that the glycine residue usually provided conformational flexibility to the adjacent peptide bonds (Fig. 1D). Thus, Glu-204 of VvOxyR2 is more likely than the Lys-203 residue to be responsible for the noncanonical conformation of the peptide bond.
Glu-204 is involved in the hypersensitivity of VvOxyR2 to H2O2
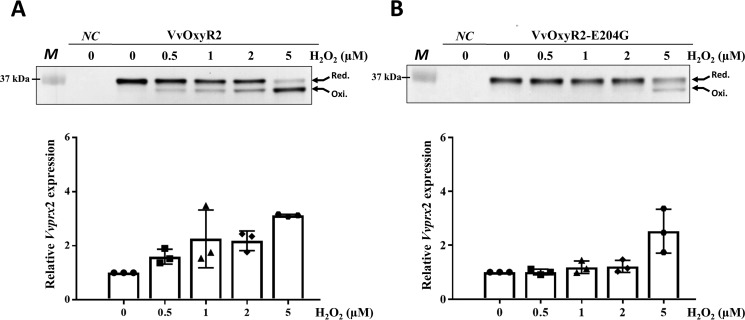
To examine the role of Glu-204, we generated a mutant V. vulnificus that harbors the VvoxyR2 (E204G) variant gene instead of wild-type VvoxyR2 and measured the minimum concentrations of H2O2 required for oxidation of VvOxyR2 in a bacterial cell. The bacterial strains expressing either the wild-type VvOxyR2 or the VvOxyR2 (E204G) variant were grown anaerobically and then were briefly exposed to various concentrations of H2O2. To facilitate analyses to determine the number of free thiols in the cellular VvOxyR2 proteins, the cells were treated with a 0.5-kDa alkylating agent, 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS), that specifically reacts with a free thiol group. AMS linking would cause upshifts in the protein bands when visualized on SDS-polyacrylamide gel; the upshifts were proportional to the number of attached AMSs on the protein. To estimate the number of free thiols in VvOxyR2, the cells were treated with AMS, and all cellular proteins were then resolved on non-reducing SDS-PAGE and immunoblotted using anti-OxyR2 antibody to visualize the VvOxyR2 protein bands. The number of free thiols in VvOxyR2 cellular proteins was estimated by the upshift in protein bands on SDS-polyacrylamide gel. Upon oxidation, the molecular size of the resulting AMS-alkylated VvOxyR2 decreased by ∼1 kDa because a disulfide bond that forms between the two redox-sensitive cysteines, Cys-206 and Cys-215, prevents alkylation of the two cysteine thiols (16). Oxidation of wild-type VvOxyR2 occurred within 30 s of exposure to 0.5 μm H2O2 (Fig. 2A, top), as reported previously (16). In contrast, the VvOxyR2 (E204G) variant was not oxidized until it was exposed to 5 μm H2O2 (Fig. 2B, top), suggesting that a Glu-204 to glycine mutation resulted in a significant increase in H2O2 level that can be sensed by VvOxyR2.
Figure 2.
Effects of the Glu-204 mutation and transition to glycine on H2O2 sensing at low levels in VvOxyR2. The VvoxyR2 mutants containing plasmids expressing VvOxyR2 (A) or VvOxyR2 (E204G), in which Glu204 was mutated to glycine (B), were grown anaerobically to A600 = 0.3 and exposed to various concentrations of H2O2, as indicated, for 30 s (top panels) or 3 min (bottom panels) before harvesting. The cells (top panels) were then precipitated with TCA and alkylated with fresh AMS buffer for 1 h at 37 °C. Alkylated VvOxyR2 proteins were resolved by non-reducing SDS-PAGE and immunoblotted using anti-VvOxyR2 antibody. The two redox states for each VvOxyR2 (reduced and oxidized) are shown with arrows. The negative control (NC) is the VvoxyR2 mutant expressing an empty vector. M, protein size marker. Total RNAs (bottom panels) were isolated, and the relative Vvprx2 transcript levels were estimated by quantitative real-time PCR analyses. The Vvprx2 mRNA level under anaerobic conditions (H2O2 of 0) was set to 1. Error bars from three independent experiments represent S.D.
To assess the transcriptional activities of wild-type VvOxyR2 and the E204G variant, we next measured the transcript level of Vvprx2, a target gene of VvOxyR2 in V. vulnificus, using quantitative real-time PCR. The results demonstrated that Vvprx2 expression was activated by VvOxyR2 when exposed to exogenous H2O2 exceeding concentrations of 0.5 μm (Fig. 2A, bottom). In contrast, the VvOxyR2 (E204G) variant poorly activated Vvprx2 expression until exposed to 5 μm H2O2 (Fig. 2B, bottom). Collectively, these results indicate that Glu-204 of VvOxyR2 is a key residue for sensitivity to H2O2.
Crystal structures of the E204G variant
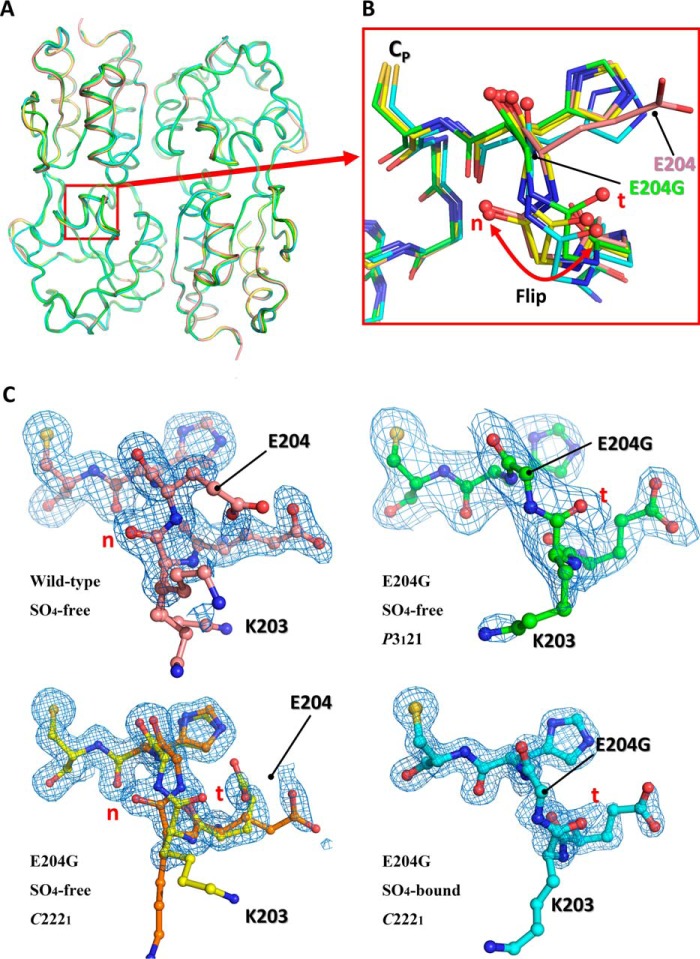
To ascertain the structural role of Glu-204, we determined three crystal structures of the VvOxyR2-RD (E204G) variant. Of these, the structure of the crystals grown under crystallization conditions similar to those of the wild-type protein exhibited the same space group (C2221) and packing constraints at a resolution of 1.55 Å, resulting in a SO4-free high-resolution structure. Another structure was determined in a different space group (P3121) at a moderate resolution (2.3 Å). We additionally determined another crystal structure at 1.5 Å resolution, designated the SO4-bound form, because it contains a sulfate ion near the peroxidatic cysteine residue. The overall structures of the variant were very similar to those of the wild-type protein except for the region around Gly-204 (Fig. 3, A and B).
Figure 3.
Structural comparison of the wild-type VvOxyR-RD and the VvOxyR2-RD (E204G) variant. A, structural superposition of the wild-type VvOxyR2-RD (pink) and the three VvOxyR2-RD (E204G) variants that are named SO4-free P3121 (green), SO4-free high-resolution (yellow and orange), and SO4-bound (cyan). Regions containing the peroxidatic cysteine and the Glu-204-containing loop are indicated by a red box. B, a close-up view of the red boxed region in A; all residues are drawn as ball-and-stick representations. A flipped peptide bond is indicated with a double-headed arrow. t and n letters in red indicate the typical conformation observed in the most OxyR proteins and the noncanonical conformation only observed in the wild-type VvOxyR2, respectively. C, 2Fo − Fc electron density maps (blue mesh) around the Glu-204-containing of the wild-type (top left, pink), SO4-free high-resolution E204G (bottom left, yellow and orange), SO4-free P3121 E204G (top right, green), and SO4-bound E204G variant (bottom right, cyan) are contoured at 1.0 σ. t and n letters in red indicate the typical conformation observed in the most OxyR proteins and the noncanonical conformation only observed in the wild-type VvOxyR2, respectively.
In the SO4-free high-resolution structure of the E204G variant, the peptide bond at the mutated region exhibited a dual conformation containing both the noncanonical conformation observed in wild-type VvOxyR2 and the typical conformation observed in most OxyR proteins (Fig. 3, B and C). The other structures (SO4-free P3121 and SO4-bound forms) of the variant presented only the typical conformation or the flipped conformation compared with the wild-type VvOxyR2 structure (Fig. 3, B and C). Taken together, our finding suggests that the substitution to a glycine residue provides structural flexibility favoring the typical conformation at the peptide bond between residues 203 and 204.
The peptide bonds around Glu-204 are structurally linked to the environment of the active site
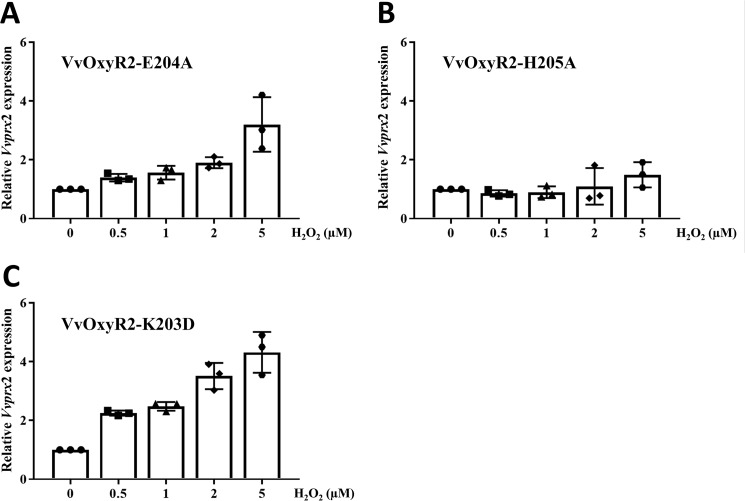
To analyze the structural role of Glu-204 of VvOxyR2, we generated a V. vulnificus variant strain harboring a VvoxyR2 (E204A) that would allow the role of the side chain to be excluded. This E204A variant VvOxyR2 was expected to have a peptide conformation similar to that of the wild-type VvOxyR2 having Glu-204, because only a glycine residue endows flexibility to the peptide bonds in proteins. As shown in Fig. 4A, this substitution exhibited an expression profile of Vvprx2 similar to the wild-type strain. Vvprx2 expression was induced by the mutant VvOxyR2 when treated with H2O2 exceeding concentrations of 0.5 μm, as observed with wild-type VvOxyR2 (Fig. 2A). This observation indicates that the peptide bond conformation of Glu-204 rather than the side chain of the Glu-204 is responsible for the hypersensitivity of VvOxyR2. We next excluded the role of the positive charge at Lys-203, which, like Glu-204, seems to be unique in VvOxyR2 (Fig. 1D, blue arrow). When the lysine residue was changed to a negatively charged Asp, a similar Vvprx2 expression profile was found (Fig. 4C).
Figure 4.
Effects of the Glu-204 and His-205 mutations on H2O2 sensing at low levels in VvOxyR2. The VvoxyR2 mutants containing plasmids expressing VvOxyR2 (E204A) (A), VvOxyR2 (H205A) (B), and VvOxyR2 (K203D) (C) were grown anaerobically to an A600 of 0.3 and exposed to various concentrations of H2O2, as indicated, for 3 min. Total RNA isolation and quantitative real-time PCR analyses were performed as described previously. The Vvprx2 mRNA level under anaerobic conditions (H2O2 = 0) was set as 1. All data are presented as mean ± S.D. (error bars); n = 3.
We examined the structures of VvOxyR2 by focusing on the interactions involved in the peptides around Glu-204 or Gly-204. We noted that the carbonyl group of the peptide bond after Glu-204 or Gly-204 interacts directly with a chloride ion, which corresponds to the H2O2 molecule in the H2O2-bound PaOxyR C199D structure (Fig. 5, A and B) (9). More importantly, the peptide bond after Gly-204 exhibited a minor conformational duality, caused by the dual conformation of the peptide bond before Gly-204 in the SO4-free high-resolution E204G structure that has the same packing constraints as the wild-type protein (Fig. 3C). The backbone carbonyl group of Gly-204 (or the carbonyl group of the peptide bond between Gly-204 and His-205) exhibited a 0.7-Å difference between the two conformations. This observation indicates motion or positional uncertainty of 0.7 Å in the carbonyl group involved in the binding of H2O2 near the peroxidatic cysteine residue (Figs. 3B and 5C). The conformational duality or flexibility in this carbonyl group contributes to the lower sensitivity of E204G variant VvOxyR2. In other words, the wild-type VvOxyR2 has a higher sensitivity to H2O2 due to the more rigid conformation resulting from employing Glu-204 instead of glycine with its intrinsic conformational flexibility.
Figure 5.
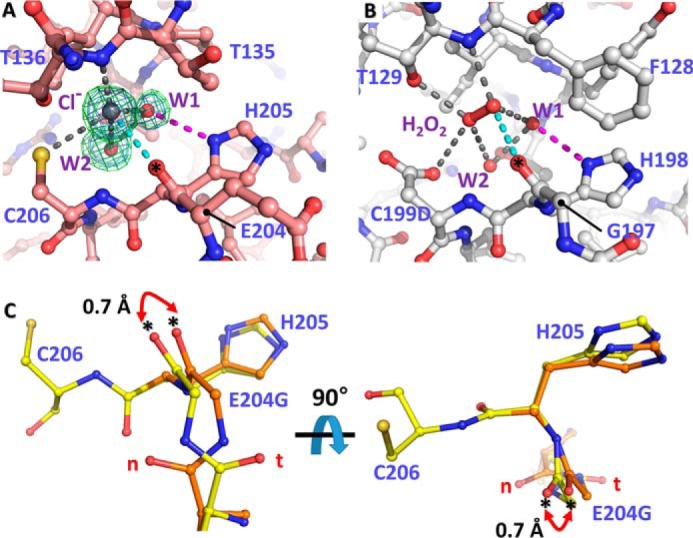
Structural variation of VvOxyR2 His-205 at the H2O2 and water binding sites near the peroxidatic cysteine residue. The putative chloride ion in VvOxyR2 (A) and the H2O2 molecule in P. aeruginosa OxyR (B; PDB code 4X6G) are bound near the peroxidatic cysteine (C206) or the mutated aspartic acid (C199D) residue. The polar interactions involved in binding the chloride ion (gray ball) or H2O2 (red ball and stick) are indicated by dotted lines. The bound water molecules (W1 and W2) are indicated by red balls. The 2Fo − Fc (blue mesh) and Fo − Fc (green mesh) omit maps of chloride ion and water molecules are contoured at 1.0 σ and 3.0 σ, respectively. C, two orthogonal views of the SO4-free high-resolution structure of VvOxyR2 E204G variant. The alternative residues are presented by yellow or orange ball-and-stick representations. The difference points in dual conformation are indicated by red dihedral arrows. t and n letters in red indicate the typical conformation observed in the most OxyR proteins and the noncanonical conformation only observed in the wild-type VvOxyR2, respectively. *, carbonyl atom between Glu-204 (or Gly-204) and His-205.
His-205 is important in sensing H2O2
Due to the structural variation in the peptide bond after Glu-204 or between Glu-204 and His-205, the position of the side chain of His-205 was also affected by the conformation of the peptide bond (Fig. 5C). We thus investigated the role of the side chain of His-205. A V. vulnificus variant strain harboring the oxyR2 (H205A) mutant gene failed to express the target gene Vvprx2 at the indicated concentrations of H2O2, demonstrating the crucial role of His-205 in sensing H2O2 (Fig. 4B). Because His-205 interacts with an invariant water molecule (W1 in Fig. 5, A and B), this conformational variation at the side chain of His-205 also contributes to the sensitivity of VvOxyR2.
VvOxyR2 has a second-order reaction rate constant 2 times higher than that of VvOxyR1
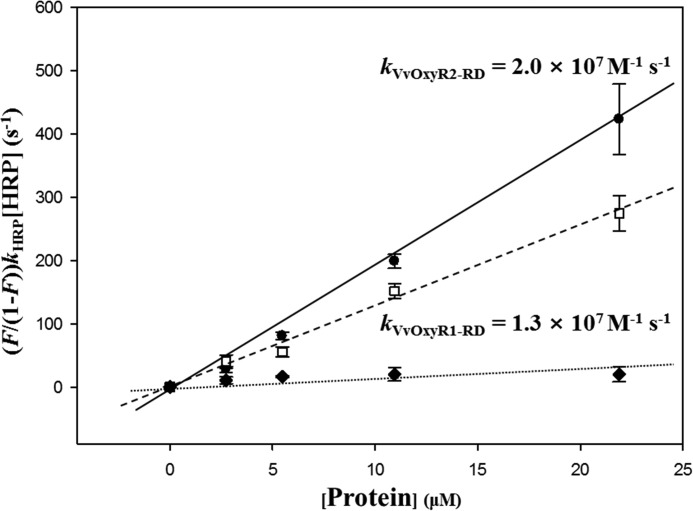
To gain mechanistic insight into the relationship between the structural variations of VvOxyR2 and the sensitivity to H2O2, we measured the second-order reaction rate constants of VvOxyR2-RD and VvOxyR1-RD by analyzing competition kinetics with horseradish peroxidase-like peroxiredoxins (20). In the control, BSA and its 35 cysteine residues (one free cysteine residue) were employed. As shown in Fig. 6, both OxyR proteins exhibited constants of 107 m−1 s−1, and BSA displayed a negligible value in this assay. The rate constants for the OxyR RDs were similar to those for peroxiredoxins, indicating that the peroxidatic cysteines of OxyRs are oxidized to Cys-SOH by H2O2 as fast as occurs with peroxiredoxins. Noticeably, VvOxyR2-RD was about 2 times faster than VvOxyR1-RD, which may account for the higher sensitivity of VvOxyR2 than VvOxyR1. The faster reaction rate implies a lower activation energy of the reactions, and it would be more possible to perform the reaction even at a very low concentration of H2O2.
Figure 6.
Determination of the second-order rate constants of VvOxyR2-RD and VvOxyR1-RD. Reaction mixtures containing HRP and various concentrations of VvOxyR2-RD (●) or VvOxyR1-RD (□) (0–21.9 μm) were exposed to H2O2. BSA (♦) was used as a negative control. The ratio of HRP oxidation was determined by measuring the absorbance at 403 nm. kVvOxyR1-RD and kVvOxyR2-RD were determined from the slope of a plot of the equation, (F/(1-F))kHRP[HRP] versus [VvOxyR1-RD or VvOxyR2-RD]. The fitting lines were calculated by the least square methods, and the error bars reflect the S.D. of three independent experiments.
Discussion
OxyR2 from V. vulnificus has a distinctive amino acid sequence relative to other OxyRs and has a higher sensitivity to H2O2. However, the molecular underpinnings of the higher sensitivity of VvOxyR2 were unknown. To investigate the structural basis of the higher sensitivity of the protein, we determined the crystal structures of wild-type and an E204G variant of VvOxyR2-RD. In particular, we identified a VvOxyR2-specific residue, Glu-204, near the peroxidatic cysteine residue. The plane of the peptide bond that occurs before Glu-204 was rotated compared with the other OxyR proteins that have a glycine residue at the Glu-204 site. Further analyses revealed that a VvOxyR2 variant harboring an E204G mutation exhibited decreased sensitivity to H2O2 in V. vulnificus that was comparable with that of other OxyR proteins. Structural comparisons of the wild-type and the E204G variant VvOxyR2 provided a reasonable structural explanation for the higher sensitivity of VvOxyR2. The E204G mutation resulted in significant conformational flexibility in the region of Gly-204 and His-205. The carbonyl group of the peptide bond between His-205 and Gly-204 is involved in binding of a H2O2 molecule, and the imidazole ring of His-205 interacts with the invariant water molecule near the peroxidatic cysteine. Thus, our study suggests that the OxyR proteins regulate their sensitivity to H2O2 by altering the structural constraints of the peptide bond after Glu-204, which are involved in binding of H2O2 and a water molecule via the peptide bond with and the side chain of the His-205 residue.
What then is the mechanistic relationship between the structural variation of the OxyR proteins and the sensitivity to H2O2? The higher sensitivity could be accounted for by the higher affinity for H2O2 and the water molecules at the site near the peroxidatic cysteine residue. Unfortunately, we were not able to measure the binding constant of H2O2 due to the rapid reaction of H2O2 with the peroxidatic cysteine residue of the OxyR proteins. Instead, we measured the reaction rate constant for the formation of Cys-SOH at the peroxidatic cysteine residue, where H2O2 reacts. The results indicated that the structural alteration of VvOxyR2 may be related to the augmented reaction rate for the decomposition of the H2O2 molecule, where the structural alteration caused by Glu-204 provides a more favorable local environment for the reduction of the activation energy for the oxidation of the peroxidatic cysteine residue.
In many ways, OxyRs share the binding and reaction mechanisms with the peroxide-decomposing enzymes peroxiredoxins. OxyRs and peroxiredoxins have two invariant cysteine residues, which rapidly form a disulfide bond when exposed to H2O2. A H2O2 molecule is specifically bound near the peroxidatic cysteine (9, 21). Two-step oxidation mechanisms were proposed for both proteins. During the first step, disulfide formation in the proteins begins with the nucleophilic attack on the peroxide by the thiolate moiety in the peroxidatic cysteine residue, resulting in the Cys-SOH form (9, 12, 21). During the second step, the Cys-SOH form at the peroxidatic cysteine establishes a disulfide bond with the resolving cysteine in both proteins.
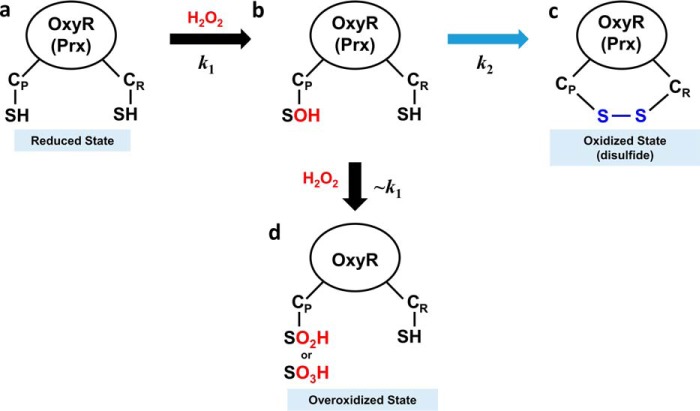
The second-order reaction rate constant for the disulfide bond formation of E. coli OxyR, measured by differential cysteine modification followed by SDS-PAGE, was previously reported to be ∼105 m−1 s−1 (22). However, in this study using competitive kinetics, the rate constants for VvOxyR2 and VvOxyR1 were on the order of ∼107 m−1 s−1. This large discrepancy could be explained by differences under “Experimental procedures.” This study measured the second-order rate constant for the first step (k1) in oxidation mechanism (Fig. 7) because the competition experiment with horseradish peroxidase measured the consumption of H2O2. In the previous experiment, because they focused on the transition between the reduced form and the oxidized (or disulfide) form on SDS-polyacrylamide gels, the reaction rate constant for the overall reaction was measured. Thus, the previous values may reflect the rate constant (k2) for the second step of disulfide bond formation, which is the rate-determining step in the two-step oxidation mechanism of OxyR (Fig. 7) (9, 12).
Figure 7.
Schematic representation of an oxidation mechanism for OxyR and Prx protein. a, the OxyR or Prx in the reduced state has two free thiols at the CP and CR. b, H2O2 rapidly reacts with CP, resulting in CP-SOH at the second-order reaction rate constant of k1. c, CP-SOH then forms a disulfide bond with CR, resulting in the oxidized state with a disulfide bond at the reaction rate constant of k2. d, in an alternative to c, CP-SOH is further oxidized by additional H2O2, resulting in the overoxidized state with CP-SO2H or CP-SO3H. Due to the structural similarity with OxyR in the reduced state, the second-order reaction rate constant would be close to k1 (∼k1).
The higher second-order reaction rate constant also has implications for the high propensity for overoxidation of VvOxyR2. The Cys-SOH form at the peroxidatic cysteine has an alternative reaction pathway for overoxidation of the peroxidatic cysteine (Fig. 7). As proposed previously (9, 11), H2O2 could enter the H2O2 binding site even when the peroxidatic cysteine residue is oxidized to Cys-SOH, suggesting a similar reaction rate constant to k1. Thus, our finding suggests that the more sensitive OxyR with the higher k1 value is more likely to enter the pathway leading to the overoxidation state instead of the oxidized state with the disulfide bond (Fig. 7). A similar explanation can also be applied to peroxiredoxins. In terms of gene regulation, the mechanistic common properties are important to maintain targeted sensitivity for optimal gene expression of peroxiredoxins by OxyR, depending on the cellular H2O2 level. Furthermore, the pathways leading to overoxidation of VvOxyR2 and VvPrx2 are also important in the fine regulation of gene expression.
This study shows that the thiol-based sensor OxyR regulates sensitivity to H2O2 by modulating the conformation of the residues required for binding of an H2O2 molecule. The VvOxyR2 strategy has implications for engineering other thiol-based sensor proteins with different sensitivities.
Experimental procedures
Expression and purification of proteins
VvOxyR2 (accession code: ADV87476) RD containing the amino acids 86–301 (VvOxyR2-RD) was amplified by PCR using the pair of primers described in Table 2. The PCR products were ligated into a His6-tagged protein expression vector, pProEx-HTa (Invitrogen) (Table 3). The expression vector for VvOxyR2-RD (E204G) was constructed using overlapping PCR techniques (Table 2). His6-tagged VvOxyR2-RD and VvOxyR2-RD (E204G) were expressed in E. coli BL21 (DE3) by the addition of 0.5 mm isopropyl-β-d-thiogalactopyranoside for 6 h at 30 °C. The expressed proteins, including the wild-type and mutant proteins, were purified using the same procedure described previously for P. aeruginosa OxyR RD (9). Briefly, the overexpressed VvOxyR2-RDs were purified by nickel-nitrilotriacetic acid chromatography, anion-exchange chromatography (HiTrapQ, GE Healthcare), and size-exclusion chromatography (HiLoad 26/600 Superdex 200 pg, GE Healthcare). All purified proteins were concentrated to 30 mg/ml and stored at −80 °C until used for crystallization. Expression and purification of VvOxyR1-RD (accession code AIL71797, residues 80–299) used the same procedures as used for VvOxyR2-RD (Table 3).
Table 2.
Primers used in this study
| Name | Oligonucleotide sequence (5′ → 3′)a | Use |
|---|---|---|
| For expression of VvOxyR RDs | ||
| OXYR2-RD-F | GGGCAAGACTCCATGGAACTGGGTAG | Overexpression of VvOxyR2 RD |
| OXYR2-RD-R | GTTAGCAGCCGTCGACCTAAAGCAGTTC | |
| OXYR1-RD-F | GGTCCATGGCAGAGATGGCAAGTGGTCAAAGTG | Overexpression of VvOxyR1 RD |
| OXYR1-RD-R | GGTCTCGAGTTACTGCTCTGAACCGCTCAC | |
| For mutagenesis | ||
| OXYR2E204G-F | GTCAGTATTTTTATTGGAAAAAGGGCATTGTCTGACTGAACACGC | VvOxyR2 (E204G) variant |
| OXYR2E204G-R | GCGTGTTCAGTCAGACAATGCCCTTTTTCCAATAAAAATACTGAC | |
| OXYR2_E204A_F | GAGTCAGTATTTTTATTGGAAAAAGCACATTGTCTGACTGAACACGCGG | VvOxyR2 (E204A) variant |
| OXYR2_E204A_R | CCGCGTGTTCAGTCAGACAATGTGCTTTTTCCAATAAAAATACTGACTC | |
| OXYR2_H205A_F | GTCAGTATTTTTATTGGAAAAAGAGGCATGTCTGACTGAACACGCGGTGTC | VvOxyR2 (H205A) variant |
| OXYR2_H205A_R | GACACCGCGTGTTCAGTCAGACATGCCTCTTTTTCCAATAAAAATACTGAC | |
| OXYR2K203D-F | GTTCAGTCAGACAATGCTCGTCTTCCAATAAAAATACTGACTCGTCAGG | VvOxyR2 (K203D) variant |
| OXYR2K203D-R | CCTGACGAGTCAGTATTTTTATTGGAAGACGAGCATTGTCTGACTGAAC | |
| For quantitative RT-PCR | ||
| PRX2QRT-F | GTTGCTTTCCGTGGCTCTTTCC | Quantification of the prx2 expression |
| PRX2QRT-R | TACTTCGCCGTGCTTCTGGTG | |
a The oligonucleotides were designed using the V. vulnificus MO6–24/O genomic sequence (GenBankTM accession number CP002469). Regions of oligonucleotides that are not complementary to the corresponding genes are underlined.
Table 3.
Bacterial strains and plasmids used in this study
| Strains or plasmids | Relevant characteristicsa | Reference or source |
|---|---|---|
| Bacterial strains | ||
| V. vulnificus | ||
| MO6-24/O | Clinical isolate; virulent | Laboratory collection |
| OH0703 | MO6–24/O with VvoxyR2::nptI; Kmr | Kim et al. (16) |
| E. coli | ||
| SM10 λ pir | thi thr leu tonA lacY supE recA::RP4–2-Tc::Mu λpir; Kmr; host for π-requiring plasmids; conjugal donor | Miller and Mekalanos (27) |
| BL21 (DE3) | F− ompT hsdS (rB−mB−) gal (DE3) | Laboratory collection |
| Plasmids | ||
| pProEx-HTa | His6-tag fusion protein expression vector; Apr | Invitrogen |
| pBANG1408 | pProEx-HTa with wild-type VvoxyR2 regulatory domain; Apr | This study |
| pPRO-OxyR2RD-E204G | pProEx-HTa with the mutant VvoxyR2 regulatory domain encoding VvOxyR2-RD-E204G; Apr | This study |
| pPRO-OxyR1RD | pProEx-HTa with the wild-type VvoxyR1 regulatory domain; Apr | This study |
| pJH0311 | 0.3-kb MCS of pUC19 cloned into pCOS5; Apr, Cmr | Goo et al. (30) |
| pDY1025 | pJH0311 with wild-type VvoxyR2; Apr, Cmr | Kim et al. (16) |
| pBANG1414 | pJH0311 with the mutant VvoxyR2 encoding VvOxyR2-E204G; Apr, Cmr | This study |
| pDY1607 | pJH0311 with the mutant VvoxyR2 encoding VvOxyR2-E204A; Apr, Cmr | This study |
| pDY1608 | pJH0311 with the mutant VvoxyR2 encoding VvOxyR2-H205A; Apr, Cmr | This study |
| pDY1618 | pJH0311 with the mutant VvoxyR2 encoding VvOxyR2-K203D; Apr, Cmr | This study |
a Apr, ampicillin-resistant; Cmr, chloramphenicol-resistant; Kmr, kanamycin-resistant.
Structural determination
Purified VvOxyR2-RD proteins (wild-type and E204G variants) were crystallized using the hanging-drop vapor diffusion method at 14 °C after mixing 1 μl of protein solution and 1 μl of precipitation solution. The wild-type VvOxyR2 RD protein was crystallized at 14 °C in a precipitation solution containing 0.2 m ammonium citrate (pH 6.5), 14% (w/v) PEG 3350, 2 mm tris(2-carboxyethyl)phosphine (TCEP), and 5 mm DTT. To obtain phase information, we soaked the crystals in 10 mm ZnCl2-supplemented precipitation solution. The native and zinc-soaked crystals were flash-frozen in a nitrogen steam at −173 °C for X-ray diffraction using each crystallization solution supplemented with 20% (v/v) glycerol as a cryoprotectant. Native and single-wavelength anomalous diffraction (Zn-SAD) data were collected in Pohang Accelerator Laboratory beam lines. Phasing information and the initial model were obtained using a combination of molecular replacement and single-wavelength anomalous diffraction methods (MR-SAD) with the PHENIX Phaser-EP program and the Zn-SAD data set (17, 23). The final structure was refined with the PHENIX program against the native data set to a 1.8 Å resolution (24).
The E204G variant proteins of VvOxyR2-RD were also crystallized at 14 °C. The VvOxyR2-RD (E204G) protein was crystallized in three different conditions. One crystal for the SO4-free, high-resolution structure of E204G variant was crystallized under the same conditions as wild-type crystals. Another crystal for the SO4-free, P3121 structure was crystallized in a solution containing 0.2 m sodium acetate, 0.1 m Tris-HCl (pH 9.0), 14% PEG 4000, and 2 mm TCEP. The other crystal for the SO4-bound structure was grown in a solution containing 0.2 m ammonium sulfate, 0.1 m sodium citrate (pH 5.6), 11% PEG 4000, and 2 mm TCEP. The E204G variant crystals were flash-frozen in a liquid nitrogen stream at −173 °C for X-ray diffraction using each crystallization solution supplemented with 20% (v/v) glycerol as a cryoprotectant. All structures of VvOxyR2 variants were determined by the molecular replacement method using the wild-type VvOxyR2-RD structure. The MOLREP program in the CCP4 package (25) was used for molecular replacement, and COOT (26) and PHENIX (24) were used to rebuild and refine the models. Crystallographic data and refinement statistics are shown in Table 1.
Site-directed mutagenesis of VvoxyR2
To determine VvOxyR2s mutant (E204G, E204A, H205A, and K203D) expression in V. vulnificus, a QuikChange® site-directed mutagenesis kit (Agilent Technologies) was used, according to previously described protocols (16). The complementary mutagenic primers, listed in Table 2, were used in conjunction with the pDY1025 plasmid (VvoxyR2 cloned into a broad host-range vector, pJH0311, as template DNA) to create pBANG1414, pDY1607, pDY1608, and pDY1618 (for VvOxyR2 E204G, E204A, H205A, and K203D, respectively; Table 2). E. coli SM10 λ pir, tra (27) harboring pJH0311, pDY1025, pBANG1414, pDY1607, pDY1608, and pDY1618 was used as a conjugal donor to the VvoxyR2 mutant (OH0703). The conjugation was conducted as described previously (16).
In vivo alkylation of VvOxyR2 and Western blotting analysis
The VvoxyR2 deletion mutant OH0703 with either pDY1025-expressing VvoxyR2 or pBANG1414-expressing VvoxyR2-E204G was used for Western blotting analysis of VvOxyR2 (Table 3), as described previously (16). Bacterial cells were grown anaerobically to an A600 of 0.3, aliquoted to an equal volume, and exposed to various concentrations of H2O2. To alkylate free thiols in the proteins with 0.5-kDa AMS (Invitrogen), the cells were immediately precipitated with 10% (w/v) ice-cold trichloroacetic acid, and the resulting pellets were dissolved in 50 μl of fresh AMS buffer (15 mm AMS, 1 m Tris (pH 8.0), 0.1% (w/v) SDS) (16). After incubation at 37 °C for 1 h, 3.5 μg of pelleted total protein was resolved on SDS-PAGE under non-reducing conditions and immunoblotted with anti-VvOxyR2 polyclonal antibody, as described previously (15).
RNA purification and transcript analysis
Total cellular RNAs from the cultures grown anaerobically to an A600 of 0.3 were isolated using RNAprotect® bacteria reagent and the RNeasy® minikit (all from Qiagen). The cells were exposed to given concentrations of H2O2 for 30 s or for 3 min in culture media and then harvested. cDNA was synthesized using the iScriptTM cDNA synthesis kit (Bio-Rad), and the Vvprx2 expression level was measured by quantitative RT-PCR amplification of the cDNA using the Chromo 4 real-time PCR detection system (Bio-Rad). The primer pair sequences are listed in Table 2. Relative expression levels of the specific transcripts were calculated using the 16S rRNA expression level as the internal reference for normalization (28). Quantitative RT-PCR data are presented as mean ± S.D. of three independent experiments.
Competitive kinetics with horseradish peroxidase (HRP)
The purified proteins VvOxyR2-RD and VvOxyR1-RD were reduced by incubation with a buffer (20 mm Tris-Cl (pH 8.0), 150 mm NaCl, 1 mm EDTA, 1 mm DTT) for 60 min at room temperature. Residual DTT was removed by ultrafiltration using 10-kDa Amicon Ultra centrifugal filter devices (Merck Millipore, Darmstadt, Germany). The reaction mixtures containing 5.84 μm HRP (Sigma) and various concentrations (0–21.9 μm) of the reduced VvOxyR2-RD or VvOxyR1-RD were treated with 4 μm H2O2 at room temperature in reaction buffer containing 20 mm Tris-HCl (pH 8.0), 150 mm NaCl, and 1 mm EDTA. The concentration of HRP was calculated by absorbance measurement at 403 nm (ϵ403 = 1.02 × 105 m−1 cm−1), and the concentrations of other proteins were determined spectrophotometrically based on the molar extinction coefficients at 280 nm. The competitive kinetics of VvOxyR1 and VvOxyR2-RD were determined using a similar procedure, as described previously (20, 29). In brief, the ratio of inhibition of horseradish peroxidase oxidation (F/(1 − F)) was measured at 403 nm using a Tecan Infinite M200 reader (Tecan, Mannedorf, Switzerland). The second-order rate constants (k) of VvOxyR1-RD and VvOxyR2-RD were determined from the slope of the plot of the equation, (F/(1 − F))kHRP[HRP]/[VvOxyR1-RD or VvOxyR2-RD] (kHRP = 1.7 × 107 m−1 s−1).
Author contributions
I. J., D. K., Y.-J. B., S. H. C., and N.-C. H. designed research; I. J., D. K., and Y.-J. B. performed research; I. J., D. K., Y.-J. B., J. A., S. H. C., and N.-C. H. analyzed data; and I. J., D. K., S. H. C., and N.-C. H. wrote the paper.
Acknowledgment
This study used beam lines 5C and 7A from Pohang Light Sources (Pohang, Korea).
This research was supported by the R&D Convergence Center Support Program (to S. H. C. and N.-C. H.), funded by Ministry of Agriculture, Food, and Rural Affairs Republic of Korea, and by Mid-career Researcher Program Grant 2015R1A2A1A13001654 (to S. H. C.) through the National Research Foundation, funded by the Ministry of Science, ICT, and Future Planning, Republic of Korea. The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (codes 5X0V, 5X0Q, 5B70, and 5B7D) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- DBD
- DNA-binding domain
- RD
- regulatory domain
- AMS
- 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid
- TCEP
- tris(2-carboxyethyl)phosphine
- SAD
- single-wavelength anomalous diffraction
- Prx
- peroxidase
- CP
- peroxidatic cysteine
- CR
- resolving cysteine
- PDB
- Protein Data Bank.
References
- 1. Storz G., and Imlay J. A. (1999) Oxidative stress. Curr. Opin. Microbiol. 2, 188–194 [DOI] [PubMed] [Google Scholar]
- 2. Stadtman E. R. (2001) Protein oxidation in aging and age-related diseases. Ann. N.Y. Acad. Sci. 928, 22–38 [DOI] [PubMed] [Google Scholar]
- 3. Imlay J. A., Chin S. M., and Linn S. (1988) Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science 240, 640–642 [DOI] [PubMed] [Google Scholar]
- 4. Storz G., Tartaglia L. A., Farr S. B., and Ames B. N. (1990) Bacterial defenses against oxidative stress. Trends Genet. 6, 363–368 [DOI] [PubMed] [Google Scholar]
- 5. Schell M. A. (1993) Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47, 597–626 [DOI] [PubMed] [Google Scholar]
- 6. Storz G., and Tartaglia L. A. (1992) OxyR: a regulator of antioxidant genes. J. Nutr. 122, 627–630 [DOI] [PubMed] [Google Scholar]
- 7. Hishinuma S., Yuki M., Fujimura M., and Fukumori F. (2006) OxyR regulated the expression of two major catalases, KatA and KatB, along with peroxiredoxin, AhpC in Pseudomonas putida. Environ. Microbiol. 8, 2115–2124 [DOI] [PubMed] [Google Scholar]
- 8. Choi H., Kim S., Mukhopadhyay P., Cho S., Woo J., Storz G., and Ryu S. E. (2001) Structural basis of the redox switch in the OxyR transcription factor. Cell 105, 103–113 [DOI] [PubMed] [Google Scholar]
- 9. Jo I., Chung I. Y., Bae H. W., Kim J. S., Song S., Cho Y. H., and Ha N. C. (2015) Structural details of the OxyR peroxide-sensing mechanism. Proc. Natl. Acad. Sci. U.S.A. 112, 6443–6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zheng M., Aslund F., and Storz G. (1998) Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279, 1718–1721 [DOI] [PubMed] [Google Scholar]
- 11. Bang Y. J., Lee Z. W., Kim D., Jo I., Ha N. C., and Choi S. H. (2016) OxyR2 functions as a three-state redox switch to tightly regulate production of Prx2, a peroxiredoxin of Vibrio vulnificus. J. Biol. Chem. 291, 16038–16047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee C., Lee S. M., Mukhopadhyay P., Kim S. J., Lee S. C., Ahn W. S., Yu M. H., Storz G., and Ryu S. E. (2004) Redox regulation of OxyR requires specific disulfide bond formation involving a rapid kinetic reaction path. Nat. Struct. Mol. Biol. 11, 1179–1185 [DOI] [PubMed] [Google Scholar]
- 13. Horseman M. A., and Surani S. (2011) A comprehensive review of Vibrio vulnificus: an important cause of severe sepsis and skin and soft-tissue infection. Int. J. Infect. Dis. 15, e157–e166 [DOI] [PubMed] [Google Scholar]
- 14. Lee M. A., Kim J. A., Yang Y. J., Shin M. Y., Park S. J., and Lee K. H. (2014) VvpM, an extracellular metalloprotease of Vibrio vulnificus, induces apoptotic death of human cells. J. Microbiol. 52, 1036–1043 [DOI] [PubMed] [Google Scholar]
- 15. Bang Y. J., Oh M. H., and Choi S. H. (2012) Distinct characteristics of two 2-Cys peroxiredoxins of Vibrio vulnificus suggesting differential roles in detoxifying oxidative stress. J. Biol. Chem. 287, 42516–42524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim S., Bang Y. J., Kim D., Lim J. G., Oh M. H., and Choi S. H. (2014) Distinct characteristics of OxyR2, a new OxyR-type regulator, ensuring expression of Peroxiredoxin 2 detoxifying low levels of hydrogen peroxide in Vibrio vulnificus. Mol. Microbiol. 93, 992–1009 [DOI] [PubMed] [Google Scholar]
- 17. Cha S. S., An Y. J., Jeong C. S., Kim M. K., Lee S. G., Lee K. H., and Oh B. H. (2012) Experimental phasing using zinc anomalous scattering. Acta Crystallogr. D Biol. Crystallogr. 68, 1253–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sainsbury S., Ren J., Nettleship J. E., Saunders N. J., Stuart D. I., and Owens R. J. (2010) The structure of a reduced form of OxyR from Neisseria meningitidis. BMC Struct. Biol. 10, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Svintradze D. V., Peterson D. L., Collazo-Santiago E. A., Lewis J. P., and Wright H. T. (2013) Structures of the Porphyromonas gingivalis OxyR regulatory domain explain differences in expression of the OxyR regulon in Escherichia coli and P. gingivalis. Acta Crystallogr. D Biol. Crystallogr. 69, 2091–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ogusucu R., Rettori D., Munhoz D. C., Netto L. E., and Augusto O. (2007) Reactions of yeast thioredoxin peroxidases I and II with hydrogen peroxide and peroxynitrite: rate constants by competitive kinetics. Free Radic. Biol. Med. 42, 326–334 [DOI] [PubMed] [Google Scholar]
- 21. Hall A., Karplus P. A., and Poole L. B. (2009) Typical 2-Cys peroxiredoxins: structures, mechanisms and functions. FEBS J. 276, 2469–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aslund F., Zheng M., Beckwith J., and Storz G. (1999) Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol-disulfide status. Proc. Natl. Acad. Sci. U.S.A. 96, 6161–6165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zwart P. H., Afonine P. V., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., McKee E., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Storoni L. C., Terwilliger T. C., and Adams P. D. (2008) Automated structure solution with the PHENIX suite. Methods Mol. Biol. 426, 419–435 [DOI] [PubMed] [Google Scholar]
- 24. Echols N., Grosse-Kunstleve R. W., Afonine P. V., Bunkóczi G., Chen V. B., Headd J. J., McCoy A. J., Moriarty N. W., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., and Adams P. D. (2012) Graphical tools for macromolecular crystallography in PHENIX. J. Appl. Crystallogr. 45, 581–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., and Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 27. Miller V. L., and Mekalanos J. J. (1988) A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170, 2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lim J. G., Bang Y. J., and Choi S. H. (2014) Characterization of the Vibrio vulnificus 1-Cys peroxiredoxin Prx3 and regulation of its expression by the Fe-S cluster regulator IscR in response to oxidative stress and iron starvation. J. Biol. Chem. 289, 36263–36274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Winterbourn C. C., and Peskin A. V. (2016) Kinetic approaches to measuring peroxiredoxin reactivity. Mol. Cells 39, 26–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goo S. Y., Lee H. J., Kim W. H., Han K. L., Park D. K., Lee H. J., Kim S. M., Kim K. S., Lee K. H., and Park S. J. (2006) Identification of OmpU of Vibrio vulnificus as a fibronectin-binding protein and its role in bacterial pathogenesis. Infect. Immun. 74, 5586–5594 [DOI] [PMC free article] [PubMed] [Google Scholar]