Abstract
The Rab family of small GTPases functions in multiple aspects of cellular membrane trafficking. Proteins bearing a differentially expressed in normal and neoplastic cells (DENN) domain have emerged as the largest family of Rab-activating guanine nucleotide exchange factors (GEFs). Rab12 functions in the initiation of starvation-induced autophagy, and our previous work revealed that its activator, DENN domain-containing protein 3 (DENND3), is phosphorylated and activated upon starvation. However, how the GEF activity of DENND3 toward Rab12 is regulated at the molecular level is still not understood. Here, we combine size-exclusion chromatography, Förster resonance energy transfer, pulldown, and in vitro GEF assays to demonstrate that regulation of GEF activity is achieved through an intramolecular interaction that is controlled by a key residue in DENND3, tyrosine 940. Our study sheds light on the regulation of Rab12 activation and lays the groundwork for characterizing the regulation of other DENN domain-containing proteins.
Keywords: guanine nucleotide exchange factor (GEF), membrane trafficking, oligomerization, phosphorylation, phosphotyrosine, DENN domain, DENND3, Rab12, autophagy, intramolecular interaction
Introduction
The Rab family of small GTPases functions in all aspects of cellular membrane trafficking ranging from vesicle budding to transport along the cytoskeleton and fusion with acceptor membranes. Rabs switch between an inactive GDP-bound form and an active GTP-bound form that interacts with effectors to mediate trafficking functions. Guanine nucleotide exchange factors (GEFs)3 activate Rabs by facilitating the exchange of GDP for GTP (1). There are at least four families of GEFs for Rabs: TRAPP, Rabin8/Sec2, Vps9 domain-containing proteins, and DENN domain-containing proteins (2). With 26 distinct members, DENN domain-containing proteins are the largest family of Rab GEFs (3–8). They are involved in diverse biological functions (4), and mutations in several DENN domain-bearing proteins are linked to human diseases, including the tumor suppressor folliculin associated with Birt-Hogg-Dubé syndrome (9), C9orf72 linked to familial frontotemporal dementia and amyotrophic lateral sclerosis (10, 11), and DENND1B associated with childhood asthma (12).
We recently demonstrated that DENN domain-containing protein 3 (DENND3) functions in starvation-induced macroautophagy (13, 14). Macroautophagy, which we will hereafter refer to as autophagy, is a conserved cellular process in which various physiological signals, including nutrient starvation, lead to the recruitment of organelles or cytosolic proteins into double membrane vesicles called autophagosomes. The autophagosomes eventually fuse with lysosomes for degradation of the internalized cargo. The resulting building blocks, such as amino acids, are then released back to the cytosol for reuse, helping cells survive starvation. Unc-51-like kinase (ULK) is the most upstream kinase for autophagy initiation (15). Upon starvation, ULK phosphorylates DENND3 at Ser-554 and Ser-572, recruiting the adapter protein 14-3-3, and this process leads to up-regulation of DENND3 GEF activity toward its substrate Rab12. Rab12 on recycling endosomes (16) then incorporates into forming autophagosomes and facilitates autophagosome trafficking (13, 14). Although starvation-mediated phosphorylation of DENND3 regulates its GEF activity toward Rab12, the mechanism underlying this regulation is unknown.
Here, we report an intramolecular interaction accounting for the regulation of the GEF activity of DENND3. We demonstrate that phosphorylation of a specific tyrosine residue in DENND3 regulates this intramolecular interaction, suggesting that a signaling pathway involving a tyrosine kinase impinges upon the intramolecular interaction to regulate DENND3 GEF activity toward Rab12.
Results and discussion
Characterization of an intramolecular interaction within DENND3
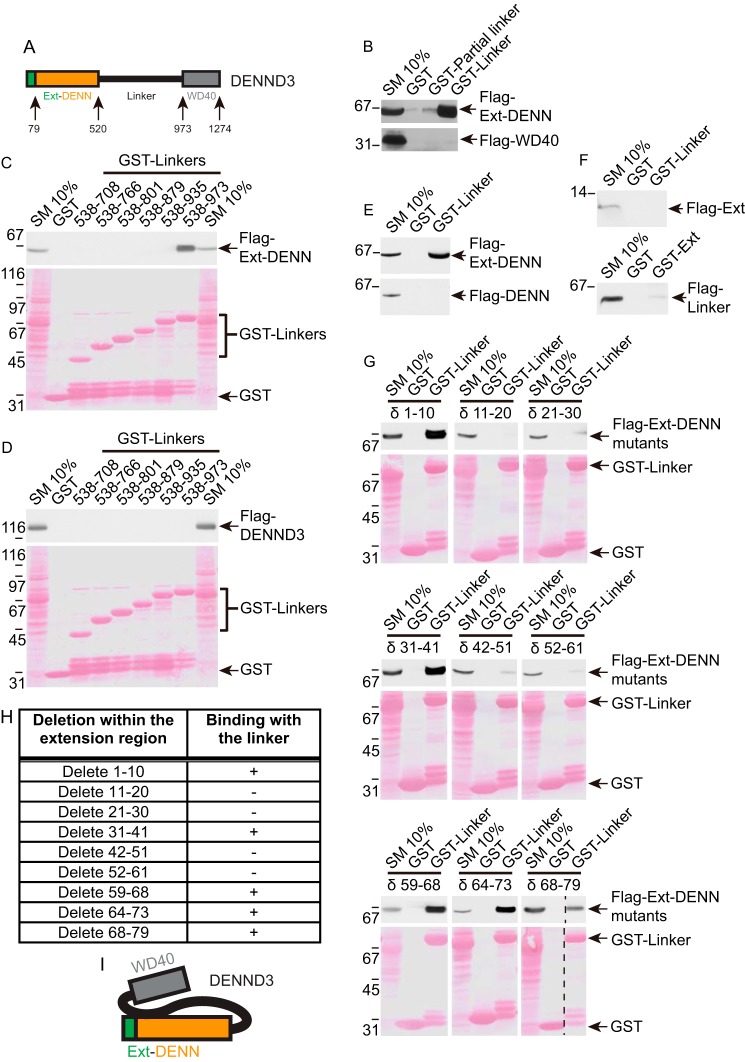
DENND3 is composed of an uncharacterized N-terminal region of 79 amino acids followed by a DENN domain, an ∼50-kDa linker region, and a C-terminal WD40 repeat domain (Fig. 1A). We demonstrated that signals stimulating autophagy up-regulate DENND3-mediated activation of Rab12 (13, 14). In many cases, activation of GEFs is mediated by release of autoinhibition (17–21). We thus hypothesized that DENND3 forms an intramolecular interaction that regulates the GEF activity and that this interaction involves the linker region, which contains the ULK phosphorylation sites.
Figure 1.
Characterization of an intramolecular interaction within DENND3. A, domain diagram of DENND3. Note the DENN domain between amino acids 80 and 520 and the N-terminal extension between amino acids 1 and 79. B, HEK-293T cells were transfected with FLAG-DENN domain and the N-terminal extension of DENND3 or FLAG-WD40 repeats, and lysates were incubated with GST, GST-partial linker (amino acids 538–611), or GST-linker (538–973) coupled to glutathione-Sepharose beads. Proteins specifically bound to the beads were processed for Western blotting with anti-FLAG antibody. An aliquot of the lysate (starting material; SM) equal to 10% of that added to the beads was analyzed in parallel. C and D, HEK-293T cells were transfected with FLAG-DENN domain with its N-terminal extension (C) or FLAG full-length DENND3 (D). Lysates were incubated with GST or GST-linker constructs coupled to glutathione-Sepharose beads and processed as described in B. E, HEK-293T cells were transfected with FLAG-DENN domain with its N-terminal extension (top panel) or FLAG-DENN domain alone (bottom panel). Lysates were incubated with GST or GST-linker coupled to glutathione-Sepharose beads and processed as described in B. F, HEK-293T cells were transfected with FLAG N-terminal extension, and lysates were incubated with GST or GST-linker coupled to glutathione-Sepharose beads (top panel), or HEK-293T cells were transfected with FLAG-linker, and lysates were incubated with GST or GST N-terminal extension coupled to glutathione-Sepharose beads (bottom panel) and processed as described in B. G, HEK-293T cells were transfected with FLAG-Ext-DENN with a series of deletions within the N-terminal extension. Lysates were incubated with GST or GST-linker coupled to glutathione-Sepharose beads and processed as described in B. H, summary table for the deletion constructs used in G. I, intramolecular interaction occurs between the C-terminal region of the linker and the Ext-DENN.
To explore this hypothesis, we first tested for an intramolecular interaction. We generated GST fusion proteins from amino acids 538–611 (partial linker), covering the N terminus of the linker around the ULK phosphorylation sites, and amino acids 538–973 (linker), covering the full linker region, and we performed pulldown experiments with cell lysates expressing the FLAG-tagged DENN domain with the N-terminal extension (hereafter called the extension and DENN or Ext-DENN) (Fig. 1A). These experiments were based on the rationale that because the DENN domain encodes the catalytic GEF activity, if there is autoinhibition mediated by the intramolecular interaction, it would most likely involve an interaction with the DENN domain. Interestingly, the GST-linker fusion protein bound robustly to Ext-DENN, although the partial linker had little or no binding (Fig. 1B, top panel). There was no binding of either construct to the WD40 domain (Fig. 1B, bottom panel). Thus, DENND3 has an intramolecular interaction involving Ext-DENN and a site on the linker between residues 612 and 973.
We next sought to map the intramolecular interaction site in the linker. We generated a series of GST-linker fusion proteins with increasing C-terminal deletions, which we used in pulldown assays with cell lysates expressing FLAG-tagged Ext-DENN. Binding between the linker and Ext-DENN was lost after truncating 38 amino acids from the C terminus of the GST-linker (Fig. 1C). When we used the same set of GST fusions for pulldown with full-length DENND3, there was no detectable binding, even for the full linker (Fig. 1D). Thus, the intramolecular interaction within full-length DENND3 is sufficiently robust as to prevent binding to the fusion protein.
We next sought to map the interaction site on Ext-DENN. Surprisingly, whereas the linker fusion protein binds strongly to FLAG-tagged Ext-DENN, it does not bind the DENN domain alone (Fig. 1E), indicating that the N-terminal extension is necessary for the intramolecular interaction. However, there was no binding of the GST-linker to the isolated N-terminal extension (Ext) tagged with FLAG nor was their binding of the GST-Ext to the linker tagged with FLAG (Fig. 1F). Thus, both the Ext and the DENN domain are required for the intramolecular interaction with the linker. We next sought to map the site(s) within Ext required for the intramolecular interaction. We generated a series of FLAG-tagged Ext-DENN constructs with different deletions within the N-terminal extension, which we screened for interaction with the GST-linker. Interestingly, we found two patches within Ext, residues 11–30 and 42–59, necessary for interacting with the linker (Fig. 1, G and H). Thus, there is an intramolecular interaction within DENND3 mediated by the C-terminal region of the linker and the Ext-DENN, which requires two patches within the N-terminal extension (Fig. 1I).
Identification of Tyr-940 as a key residue involved in the intramolecular interaction
We mapped the intramolecular interaction site in the linker to a region between amino acids 935 and 973 (Fig. 1C). Phosphorylation regulates protein interactions in a fast and reversible manner, and we hypothesized that a phosphorylation event within this region could be involved in the intramolecular interaction. We aligned the sequence between 935 and 973 across species to identify conserved Ser, Thr, and Tyr residues, and for each we created Asp or Glu phosphomimetics as well as Ala or Phe phosphorylation-defective mutants. Binding between the linker and Ext-DENN was not altered with any of the Ser/Thr phosphomimetic or phosphorylation-defective mutations (Fig. 2A). Intriguingly, a Y940D mutation almost completely abolished the interaction, although a phosphorylation-defective Y940F mutation continued to show strong binding with Ext-DENN (Fig. 2B). These data indicate that Tyr-940 is a key residue involved in the intramolecular interaction, and consistently it was reported that Tyr-940 of DENND3 is phosphorylated in rat brain (22). Because of the aromatic ring of tyrosine, there is no natural amino acid substitute that creates an ideal phosphomimetic mutant, but many examples do exist in which mutation of Tyr to Asp or Glu has been used successfully to mimic phosphorylated tyrosine (23–27). Theoretically, if the effect of the phosphorylation is due primarily to the negative charge of the phospho group, Asp or Glu should mimic phosphorylated tyrosine. Secondary structure predictions indicate that the residues flanking Tyr-940 have no secondary structure and are located between α-helices (supplemental Fig. S1). As such, this region would be flexible, perhaps forming a loop. Thus, loss of binding of the linker to Ext-DENN after introducing the Y940D mutation is unlikely to be caused by disruption of a secondary or tertiary structure in the linker.
Figure 2.
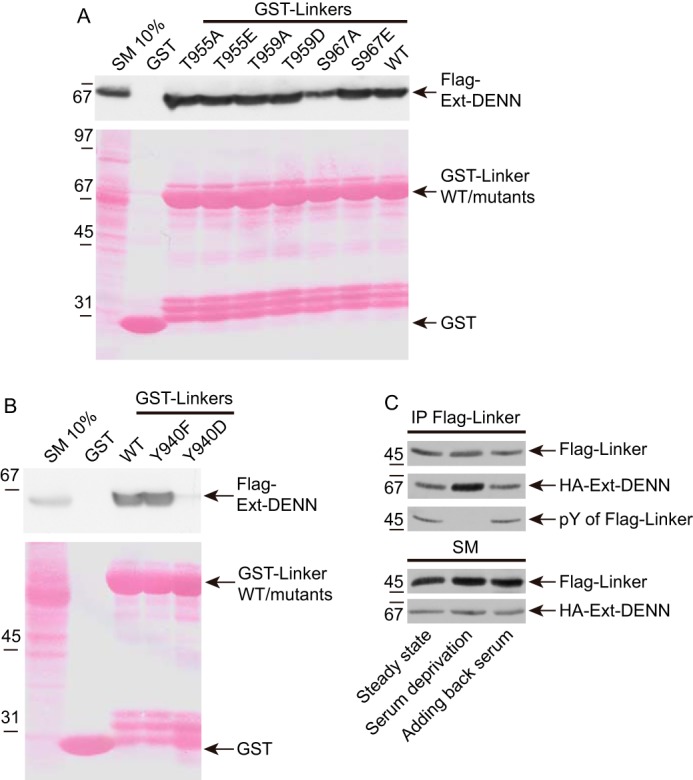
Tyr-940 is a key residue involved in the intramolecular interaction of DENND3. A, cells were transfected with FLAG-Ext-DENN. Lysates were incubated with GST, GST-linker, or GST-linker with phosphomimetic or phosphorylation-defective mutants at conserved serine or threonine residues within amino acids 936–973 of DENND3, coupled to glutathione-Sepharose beads. Proteins specifically bound to the beads were processed for Western blotting with anti-FLAG antibody. An aliquot of the lysate (starting material; SM) equal to 10% of that added to the beads was analyzed in parallel. B, cells were transfected with FLAG-Ext-DENN. Lysates were incubated with GST, GST-linker, or GST-linker with phosphomimetic Y940D or phosphorylation-defective Y940F mutants, coupled to glutathione-Sepharose beads, and processed as described in A. C, cells co-transfected with FLAG-linker and HA-Ext-DENN domain were left unstarved, or were deprived of cell culture serum for 1.5 h, or were re-fed with serum for 0.5 h following the deprivation. Lysates were incubated with protein G beads coupled to anti-FLAG antibody (IP-FLAG). Proteins bound specifically to the beads were processed for Western blotting with anti-FLAG, anti-HA, or anti-phosphotyrosine antibodies. An aliquot of the cell lysate (starting material, SM) was analyzed in parallel.
Because the GST-linker binds to the isolated Ext-DENN, but not the full-length DENND3 (Fig. 1, C and D), we wondered whether full-length DENND3 with the Y940D mutation would disrupt the cis interaction allowing for an interaction in trans. However, as shown in supplemental Fig. S2, Y940D does not have better binding to the linker as compared with wild-type or Y940F. We think it likely that when the Ext-DENN and the linker are within full-length DENND3, although the Y940D mutation abolishes the binding between the Ext-DENN and the linker, somehow steric hindrance of the linker or the WD40 repeats in cis blocks access of linker in trans to the Ext-DENN.
Tyrosine phosphorylation events are often mediated by tyrosine kinase receptors activated by growth factors or hormones in serum. To gain insight into the regulation of Tyr-940 phosphorylation, we expressed FLAG-tagged linker and HA-tagged Ext-DENN in cells, and after depriving cells of serum, or adding back serum following the deprivation, we performed immunoprecipitation experiments to measure the tyrosine phosphorylation on the linker and the corresponding intramolecular interaction. Interestingly, the interaction is stronger when the cells are deprived of serum compared with cells at steady state or those re-fed with serum (Fig. 2C). Moreover, as determined by a phosphotyrosine antibody, the level of tyrosine phosphorylation of the immunoprecipitated linker negatively correlated with the binding intensity (Fig. 2C), indicating that the tyrosine phosphorylation may be the cause of the suppression of the intramolecular interaction. These data reveal that there is an activity in serum, even at steady state, that evokes tyrosine phosphorylation on the linker of DENND3.
Because ULK-mediated DENND3 phosphorylation at Ser-554 and Ser-572 increases DENND3 GEF activity (14), we wondered whether there is cross-talk between this event and Tyr-940 phosphorylation. One potential model is that Tyr-940 phosphorylation opens the intramolecular interaction, allowing easier access for either ULK-mediated phosphorylation or 14-3-3 binding to the phosphorylated Ser-554/Ser-572 residues. One way to test this model was to determine whether the Y940D mutant that disrupts the intramolecular interaction alters 14-3-3 binding. As shown in supplemental Fig. S3A, when compared with wild type, the non-phosphorylatable DENND3 S554A mutant loses 14-3-3 binding, and GST-14-3-3 K50E, a mutant of 14-3-3 that suppresses the interaction between 14-3-3 and its binding partners (28), does not bind to the phosphorylated substrate, which serves as a control for the 14-3-3 pulldown. 14-3-3 binding is unaltered when comparing the Y940D mutant to wild type or the Y940F mutant. Thus, the phosphorylation status of Tyr-940 does not lead to better access of 14-3-3 to DENND3. Moreover, ULK phosphorylation on DENND3 is equivalent when comparing wild-type protein to the Y940D and Y940F mutants (supplemental Fig. S3B). In summary, Tyr-940 is a key residue for regulating an intramolecular interaction between the linker region and the Ext-DENN region of DENND3. Phosphorylation of Tyr-940 is triggered by an activity in serum, and it appears that Tyr-940 phosphorylation is a separate process from ULK-mediated phosphorylation.
Phosphorylation of Tyr-940 in the linker alters DENND3 conformation
The Y940F and Y940D mutants of DENND3 provide useful tools to test the hypothesis that Tyr-940 phosphorylation abolishes the intramolecular interaction leading to a conformation that is open to Rab12 (Fig. 3A). To test this concept, we first performed size-exclusion chromatography, which separates proteins and protein complexes by their native mass and hydrodynamic volume. As shown in Fig. 3B, wild-type DENND3 elutes with a peak at fraction 27 on a Superose 6 10/300 GL column, the same as the globular marker ferritin, an ∼440-kDa oligomer consisting of 24 subunits. This is surprising because the molecular mass of DENND3 is ∼140 kDa, which is marked by the elution peak of GAPDH. This suggests DENND3 exists as an oligomer. Interestingly, the Y940D mutant peaked at an earlier fraction (fraction 23) (Fig. 3C). This is consistent with an open conformation providing a larger hydrodynamic radius (Stokes radius), leading to an apparently larger size and earlier elution than wild type (Fig. 3A). In contrast, the Y940F mutant had a similar elution profile as wild-type DENND3, likely reflecting the closed conformation (Fig. 3D).
Figure 3.
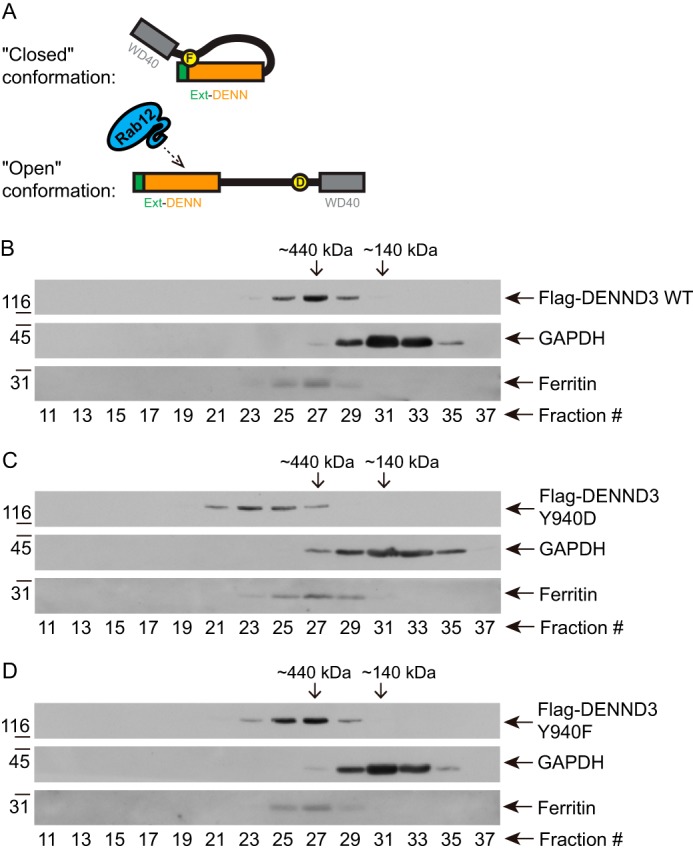
Phosphorylation of Tyr-940 in the linker alters the conformation of DENND3. A, model depicting the hypothesis of the closed and open conformations. B–D, cells were transfected with FLAG-DENND3 wild-type (B), Y940D (C), or Y940F (D) mutants. Lysates were subjected to size-exclusion chromatography. The collected fractions were analyzed by Western blotting with the indicated antibodies.
To further examine whether Y940D leads to an open conformation, we performed gradient centrifugation, which separates proteins based on protein mass and shape. As demonstrated in supplemental Fig. S4, in which the fractions are collected from the top of the gradient, wild-type DENND3 and Y940F mutant emerged and peaked in fraction 10 (supplemental Fig. S4, A and C), whereas Y940D emerged in fraction 8 (supplemental Fig. S4B). This would indicate an open conformation with a larger frictional coefficient. This further supports that the Tyr-940 phosphorylation alters DENND3 conformation.
DENND3 forms an oligomer
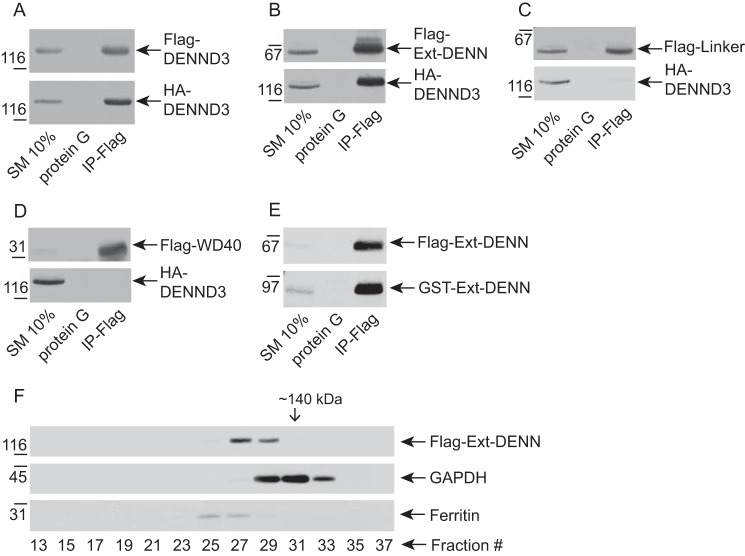
The data from size-exclusion chromatography and gradient centrifugation suggest that DENND3 exists as an oligomer, and consistently FLAG-tagged DENND3 robustly co-immunoprecipitates HA-tagged DENND3 (Fig. 4A). Because the linker interacts with Ext-DENN of DENND3, we wondered whether the intramolecular interaction is in fact an intermolecular interaction, as demonstrated in the hypothetical model of supplemental Fig. S5A. However, HA-tagged DENND3 Y940F and Y940D co-immunoprecipitated with FLAG-tagged DENND3 Y940F and Y940D to an equal extent as the wild-type proteins (supplemental Fig. S5B). These data demonstrate that the interaction between Tyr-940 in the linker and Ext-DENN, which is the hallmark of the intramolecular interaction, is not required for the oligomerization. We thus sought to map the region responsible for the oligomerization of DENND3 using co-immunoprecipitation of various domains of DENND3 tagged with FLAG and full-length DENND3 tagged with HA. Ext-DENN co-immunoprecipitated efficiently with full-length DENND3 (Fig. 4B), whereas neither the linker nor WD40 repeats co-immunoprecipitated (Fig. 4, C and D). Moreover, GST-tagged Ext-DENN co-immunoprecipitated with FLAG-tagged Ext-DENN (Fig. 4E), supporting that Ext-DENN is sufficient to mediate the oligomerization, which is distinct from the intramolecular interaction.
Figure 4.
Characterization of DENND3 oligomerization. A, lysates from HEK-293T cells co-transfected with FLAG-DENND3 and HA-DENND3 were incubated with protein G beads alone or protein G beads coupled to anti-FLAG antibody (IP-FLAG). Proteins specifically bound to the beads were processed for Western blotting with anti-FLAG and anti-HA antibodies. An aliquot of the cell lysate (starting material, SM) equal to 10% of that added to the beads was analyzed in parallel. B, lysates from HEK-293T cells co-transfected with FLAG-Ext-DENN and HA-DENND3 were incubated with protein G beads alone or protein G beads coupled to anti-FLAG antibody (IP-FLAG). Proteins bound specifically to the beads were processed as described in A. C, lysates from HEK-293T cells co-transfected with FLAG-linker and HA-DENND3 were incubated with protein G beads alone or protein G beads coupled to anti-FLAG antibody (IP-FLAG). Proteins bound specifically to the beads were processed as described in A. D, lysates from HEK-293T cells co-transfected with FLAG-WD40 and HA-DENND3 were incubated with protein G beads alone or protein G beads coupled to anti-FLAG antibody (IP-FLAG). Proteins bound specifically to the beads were processed as described in A. E, lysates from HEK-293T cells co-transfected with FLAG-Ext-DENN and GST-Ext-DENN were incubated with protein G beads alone or protein G beads coupled to anti-FLAG antibody (IP-FLAG). Proteins bound specifically to the beads were processed as described in A. F, HEK-293T cells were transfected with FLAG-Ext-DENN. Lysates were subjected to size-exclusion chromatography. The collected fractions were analyzed by Western blotting with the indicated antibodies.
Size-exclusion chromatography further supports that Ext-DENN is sufficient for the oligomerization. As shown in Fig. 4F, Ext-DENN eluted at fractions corresponding to a molecular mass greater than the size of monomer of Ext-DENN (∼70 kDa).
Tyr-940 phosphorylation leads to an open conformation with enhanced GEF activity
To further examine the role of Tyr-940 in the intramolecular interaction of DENND3, we turned to Förster resonance energy transfer (FRET) microscopy. We generated constructs of DENND3 (minus the WD40 domain) with or without Tyr-940 mutations, flanked by fluorescent reporters Cerulean and Venus at the N and C termini, respectively (Fig. 5A). We transiently expressed these constructs and performed FRET imaging experiments. As shown in Fig. 5, B and C, the FRET signal with the Y940D mutation was suppressed compared with wild-type and the Y940F mutant, with the Y940F mutant displaying the greatest FRET signal. These data further support that the Tyr-940 phosphorylation suppresses the intramolecular interaction rendering DENND3 in an open conformation.
Figure 5.
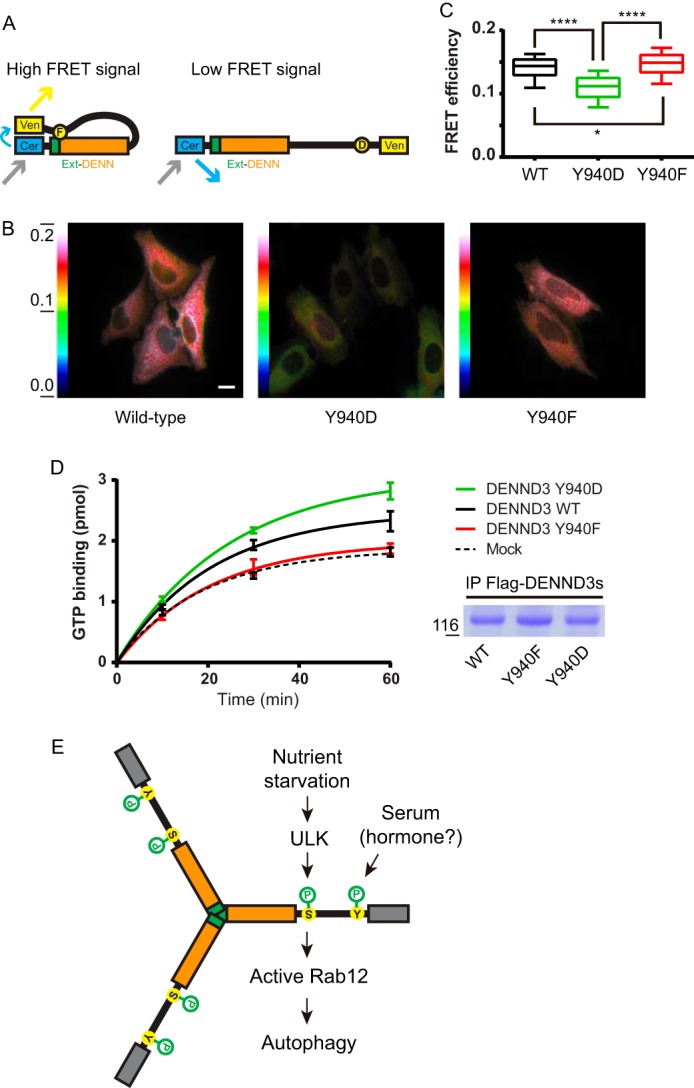
With Tyr-940 phosphorylation DENND3 assumes an open conformation with enhanced GEF activity. A, diagram showing the principle underlying the test of the hypothesis of open/closed conformations through FRET experiments. B, HeLa cells were transfected with a FRET construct of DENND3 wild-type, Y940D, or Y940F mutant before FRET imaging. The color-coded images indicate the measured FRET signal. Warm colors represent high FRET signal; cool colors represent low FRET signal. The color scale bar represents the efficiency of the FRET signal. The mean values of FRET efficiency for wild type, Y940D, and Y940F are 0.1397, 0.1109, and 0.1460, respectively. Scale bar, 5 μm. C, FRET efficiency measurements from images as in B were quantified from n > 180 cells for each group over three repeats. The data are presented with box and whisker graphs. The box extends from the 25th to 75th percentiles. The line inside the box is plotted at the median. The whiskers are from the 10th to the 90th percentile. Statistical analysis employed one-way analysis of variance followed by Tukey's post-test. ****, p < 0.0001, *, p < 0.05. D, FLAG-DENND3 wild-type, Y940D, or Y940F mutants were immunoprecipitated from HEK-293T cells. Immunoprecipitations from mock-transfected cells were used as mock for the GDP/GTP exchange assay. To measure GEF activity, GDP-loaded Rab12 was incubated with immunoprecipitated DENND3 wild type, mutant or mock in the presence of GTPγS and [35S]GTPγS. At the time points indicated, an aliquot of the reaction was analyzed for nucleotide exchange. The enzymatic activity based on the amount of Rab12 loaded with GTPγS over time is shown. The curves were fit with a nonlinear regression one-phase association. The points represent mean ± S.E., n = 3. Duplicates of the immunoprecipitated DENND3 proteins were processed by SDS-PAGE and stained with Coomassie Blue dye, showing that equal amounts of DENND3 wild type and mutants were used for the GDP/GTP exchange assay (right bottom panel). E, model summarizing under nutrient starvation ULK-mediated phosphorylation and Tyr-940 phosphorylation on DENND3 and their functional consequence.
To test whether the open conformation has greater GEF activity toward Rab12, we performed in vitro GEF assays. Overexpressed DENND3 proteins (wild type, Y940D, and Y940F) were immunoprecipitated from HEK-293T cells and added to purified Rab12 (preloaded with GDP) in the presence of [35S]GTPγS. As shown in Fig. 5D, the amount of GTP loaded on Rab12 was greater when Rab12 is incubated with the Y940D mutant compared with wild type. Moreover, the Y940F mutant was not able to promote GEF activity (Fig. 5D). These data indicate that the open conformation of DENND3 has greater GEF activity.
We hypothesized that the increased GEF activity of the Tyr-940 mutant results from enhanced accessibility of Rab12 for the DENN domain when the intramolecular interaction is abolished. To test this hypothesis, we took advantage of the fact that binding with nucleotide-free Rab is a hallmark of GEF domains, and we conducted a pulldown experiment using nucleotide-free GST-Rab12. As shown in supplemental Fig. S6, Rab12 binds to both FLAG-tagged full-length Ext-DENN and FLAG-Ext-DENN with deletion from residues 21 to 30, which does not interact with the linker region due to the deletion. Interestingly, addition of purified linker inhibited the binding of Rab12 to Ext-DENN but not the Ext-DENN with deletion. This indicates that the intramolecular interaction excludes Rab12 from accessing the DENN domain.
We next explored whether there is a difference in subcellular localization between wild-type DENND3 and the Y940D mutation. As shown in the immunofluorescence experiments (supplemental Fig. S7A), expressed wild-type DENND3 has a cytosolic distribution, regardless of whether the FLAG tag is at the N or C terminus of the protein. The Y940D and Y940F mutants also display cytosolic localization, as does the S554A mutant, which no longer binds to 14-3-3 (14). Considering that overexpression of the protein may contribute to the cytosolic pattern, we limited the expression level by reducing the duration of expression following transfection from 16 to 10 h. Under this condition, the wild-type DENND3 and its mutants still kept the cytosolic distribution, but a few fine puncta were observed within the cytosolic pool (supplemental Fig. S7A), which suggests some membrane-associated DENND3 was captured. Further subcellular fractionation experiments were employed to examine the membrane pool of DENND3. The post-nuclear supernatant of cell lysates was centrifuged at 200,000 × g to separate membrane fractions from cytosolic fractions. As shown in supplemental Fig. S7B, whereas RME-8, a peripheral membrane protein, and Na+K+-ATPase, an integral membrane protein, were found in the pellet and GAPDH, a cytosolic protein, was found in the supernatant, wild-type DENND3 is found in both fractions, as are the Y940D and Y940F mutants. This indicates that DENND3 has both membrane-associated and cytosolic pools, but there is seemingly no difference in the distributions of the wild type and mutants. Considering that the cytosolic pool may mask the membrane-associated pool in the immunocytochemical experiments, we pre-permeabilized the cells with saponin to release cytosolic DENND3 before fixing the cells for immunofluorescence experiments. As shown in supplemental Fig. S7C, after the pre-permeabilization the fluorescent signal from wild-type DENND3 and its mutants became faint. As a result, a few puncta emerged. However, the wild type and the Y940D/Y940F mutants still display the same pattern (supplemental Fig. S7C). Thus, based on the data of supplemental Fig. S7, A–C, the Y940D mutation does not facilitate the translocation of DENND3 to the membrane where the GEF activity is conducted. We consider that the recruitment of DENND3 to the membrane for activating Rab12 is a transient process.
We previously demonstrated that upon starvation ULK phosphorylates DENND3 at Ser-554 and Ser-572, increasing its GEF activity toward Rab12, which promotes autophagy by facilitating autophagosome trafficking (13, 14). Here we investigated the molecular mechanisms underlying the regulation of DENND3. We found there is an intramolecular interaction within DENND3 that impinges upon the GEF activity. Unexpectedly, instead of residues Ser-554 and Ser-572, Tyr-940 in the linker of DENND3 is a key residue regulating the intramolecular interaction.
Based on our mapping data, it seems that the ULK-mediated phosphorylation is not involved in regulating the intramolecular interaction between the linker and Ext-DENN. Because the phosphorylation by ULK recruits 14-3-3 proteins (14), perhaps upon nutrient starvation the ULK-mediated phosphorylation changes the GEF activity of DENND3 through translocating DENND3 to a subcellular localization, such as recycling endosome, where it can meet and activate its substrate Rab12 (13, 14). Alternatively, the 14-3-3 binding to the DENND3 oligomer may lead to a favorable conformation, which facilitates GEF activity toward Rab12.
Our data suggest that an activity in serum phosphorylates Tyr-940, suppressing the intramolecular interaction (Fig. 5E). Phosphorylation of Tyr-940 is crucial for regulating the GEF activity of DENND3 through impinging on the intermolecular interaction. Considering the role of DENND3 in autophagy, Tyr-940 phosphorylation at steady state, enabling DENND3 basal GEF activity, may be required for basal levels of autophagy (14). One potential model is that under nutrient starvation a stress signal triggers the release of a specific hormone or serum factor that results in the up-regulated phosphorylation of Tyr-940 and thus the open conformation of DENND3. Together with ULK-mediated phosphorylation and the resulting 14-3-3 binding, there is an additive increase of DENND3 GEF activity toward Rab12 (Fig. 5E).
It is interesting to note that some DENN domain-containing proteins form hetero-oligomers, such as C9ORF72 and SMCR8 (29, 30) and folliculin and folliculin-interacting proteins (31). Here we report for the first time that a DENN domain-containing protein also forms a homo-oligomer.
Our findings provide an example in which the GEF activity of a DENN domain-containing protein is regulated by an intramolecular interaction. Because the interaction involves the conserved DENN domain, the mechanism of the regulation we report here may be shared among DENN domain proteins, the vast majority of which remain to be characterized. Considering the importance of Rab-activating GEFs in membrane trafficking and the pathogenic role of mutations in DENN domain proteins (9–12, 32), further characterization of DENN domain proteins will not only reveal fundamental membrane trafficking processes but will also shed light on related pathological pathways.
Experimental procedures
Antibodies and reagents
Mouse monoclonal and rabbit polyclonal FLAG antibodies were from Sigma. Rabbit polyclonal antibody against GFP was from Life Technologies, Inc. Mouse DENND3 cDNA was from Imagenes. The coding sequence was amplified by PCR and cloned into pCMV vector creating FLAG-DENND3. The DENN domain with the N-terminal extension (amino acids 1–520) and the DENN domain (amino acids 79–520) were cloned into pCMV vector creating FLAG-Ext-DENN and FLAG-DENN domain, respectively. Linker region constructs were cloned into pGEX vector generating GST-linkers coding different regions of the linker. GST-linker Y940F or Y940D, FLAG-DENND3 Y940F or Y940D, and FLAG-Ext-DENN with a series of deletions within the N-terminal extension were created by the QuikChange site-directed mutagenesis kit from Stratagene. The His-tagged Rab12 construct used for GDP/GTP exchange assay was from Addgene (catalog no. 25512). DENND3 constructs used for FRET microscopy were generated by inserting DENND3 coding sequence (11–953, with or without Y940D/Y940F mutation) into MCS of C17V construct (Addgene catalog no. 26395), which is between Cerulean and Venus coding sequences. GST-14-3-3ϵ wild-type and K50E mutant constructs (rat coding sequence in pGEX-4T1) were gifts from Dr. Philippe Roux, Université de Montréal.
Immunoprecipitation
Cells were collected in HEPES lysate buffer (20 mm HEPES, pH 7.4, 10 mm sodium fluoride, 0.5 mm sodium orthovanadate, 60 nm okadaic acid, 100 mm sodium chloride, 1% Triton X-100, 0.5 μg/ml aprotinin, 0.5 μg/ml leupeptin, 0.83 mm benzamidine, and 0.23 mm phenylmethylsulfonyl fluoride). Following 10 min at 4 °C, lysates were spun at 200,000 × g for 15 min. The supernatant was incubated for ∼3 h at 4 °C with antibodies coupled to protein A- or G-Sepharose. Beads were subsequently washed three times with HEPES lysates buffer and analyzed by SDS-PAGE. Samples were then processed for Western blotting.
Pulldown assay
Cell lysates prepared in the same way as described in immunoprecipitation were incubated for ∼3 h at 4 °C with GST or GST fusion proteins coupled to glutathione-Sepharose. The samples were subsequently washed and prepared for Western blotting as described under “Immunoprecipitation.”
Size-exclusion chromatography
HEK-293T cells were transfected with FLAG-tagged DENND3 wild-type or mutant construct. After 2 days the cells were collected in HEPES lysate buffer (20 mm HEPES, pH 7.4, 10 mm sodium fluoride, 0.5 mm sodium orthovanadate, 60 nm okadaic acid, 100 mm sodium chloride, 0.5 μg/ml aprotinin, 0.5 μg/ml leupeptin, 0.83 mm benzamidine, and 0.23 mm phenylmethylsulfonyl fluoride). Following 10 min at 4 °C, lysates were spun at 200,000 × g for 15 min. The supernatant was loaded on Superose 6 10/300 GL column (GE Healthcare). The collected fractions were analyzed by SDS-PAGE. Samples were then processed for Western blotting.
Glycerol gradient centrifugation
Cells were transfected with FLAG-tagged DENND3, wild type, or mutants. After 2 days the cells were washed in PBS and collected in HEPES lysate buffer (20 mm HEPES, pH 7.4, 10 mm sodium fluoride, 0.5 mm sodium orthovanadate, 60 nm okadaic acid, 100 mm sodium chloride, 0.5 μg/ml aprotinin, 0.5 μg/ml leupeptin, 0.83 mm benzamidine, and 0.23 mm phenylmethylsulfonyl fluoride). Following snap freezing and thawing to further lyse the cells, lysates were spun at 200,000 × g for 15 min. The supernatant was loaded on glycerol gradients (10–30%) in the HEPES lysate buffer. Samples were then centrifuged in a swinging-bucket rotor at 103,000 × g for 16 h at 4 °C. The fractions were collected and processed by SDS-PAGE. Samples were then analyzed by Western blotting.
Subcellular fractionation
HEK-293T cells were transfected with FLAG-tagged wild-type DENND3, wild type, or mutants. After 1 day the cells were washed in PBS and collected in HEPES buffer (20 mm HEPES, pH 7.4, 0.5 μg/ml aprotinin, 0.5 μg/ml leupeptin, 0.83 mm benzamidine, and 0.23 mm phenylmethylsulfonyl fluoride). After a 10-min incubation on ice, lysates were spun at 800 × g for 10 min. The supernatant was adjusted to 2 mg/ml and then subjected to centrifugation at 200,000 × g for 30 min at 4 °C. The resulting supernatant and pellet were processed for SDS-PAGE and Western blotting.
GDP/GTP exchange assay
His-tagged Rab12 was purified by nickel-nitrilotriacetic acid-agarose (Qiagen). The His tag was then removed by thrombin-mediated cleavage between the tag and the Rab12. Purified 3.6 μm Rab12 was loaded with 20 μm GDP by incubation for 10 min in loading buffer (20 mm Tris, pH 7.5, 100 mm NaCl, 5 mm EDTA). MgCl2 (10 mm) was added and incubated for another 10 min to stabilize the loaded GDP. Exchange reactions were carried out in 65-μl total volume containing 0.36 μm loaded Rab12, 0.55 μm immunoprecipitated FLAG-tagged DENND3 wild type or mutant, 5 μm GTPγS, 0.2 mCi/mmol [35S]GTPγS, and 0.5 mg/ml BSA in reaction buffer (20 mm Tris, pH 7.5, 100 mm NaCl, 30 mm MgCl2, 0.5 mm DTT). Following the indicated incubation time, 13 μl of the reaction was removed, added to 1 ml of 4 °C wash buffer (20 mm Tris, pH 7.5, 100 mm NaCl, 30 mm MgCl2), and passed through nitrocellulose filters. The filters were washed with 5 ml of wash buffer and counted with a scintillation counter.
Förster resonance energy transfer (FRET) microscopy
HeLa cells were plated on MatTek plates (MatTek Corp.). When the cells reached 50% confluency, DENND3 FRET constructs and constructs expressing Cerulean/Venus alone were transfected into HeLa cells by jetPRIME (Polyplus transfection). After ∼15 h, live cells were used for FRET microscopy. The images were taken by Zeiss Observer.Z1 microscope, with 25% light intensity. Images from cells expressing Cerulean/Venus alone were used as control measurement for subtracting bleed-through. All cells were imaged in the configurations of cyan fluorescent protein, YFP, and FRET in which 50 ms of exposure time was kept fixed for all groups. The FRET signal was analyzed with AxioVision software. To subtract bleed-through from cyan and yellow channels, the FRET Xia formula was used (33).
Immunofluorescence
HeLa cells were plated on coverslips coated with poly-l-lysine. Cells with/without pre-permeabilization for 1 min by 0.033% saponin were fixed with 4% paraformaldehyde in 37 °C PBS for 10 min, followed by standard protocols of immunofluorescence. Images were taken by Zeiss LSM 710 confocal microscope.
Statistical evaluation
Experiments were repeated at least three times, from which values, expressed as mean ± S.E., were obtained if needed. Statistical analysis of the results was carried out by one-way analysis of variance, followed by Tukey's multiple comparison test when appropriate. p < 0.05 was considered significant. For GDP/GTP exchange assays, data were plotted in GraphPad Prism, and curves were fitted by a nonlinear regression one-phase association.
Author contributions
J. X. conceived and performed the experiments and wrote the manuscript. P. S. M. conceived the experiments and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Kalle Gehring and Tin Pan for discussion and access to their fast protein liquid chromatography (FPLC); Dr. Khanh Huy Bui and Shunkai Yang for Superose 6 10/300 GL column; and Drs. Wayne Sossin, Matthew Tang, Carole Abi Farah, and Margaret Hastings for Zeiss Observer.Z1 microscope and AxioVision software.
This work was supported by Canadian Institutes of Health Research Grant MOP-62684 (to P. S. M.). The authors declare that they have no conflicts of interest with the contents of this article.
This article was selected as one of our Editors' Picks.
This article contains supplemental Figs. S1–S7.
- GEF
- guanine nucleotide exchange factor
- DENN domain
- differentially expressed in normal and neoplastic cells domain
- DENND3
- DENN domain-containing protein 3
- ULK
- Unc-51-like kinase
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate.
References
- 1. Stenmark H. (2009) Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10, 513–525 [DOI] [PubMed] [Google Scholar]
- 2. Cherfils J., and Zeghouf M. (2013) Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 93, 269–309 [DOI] [PubMed] [Google Scholar]
- 3. Levine T. P., Daniels R. D., Gatta A. T., Wong L. H., and Hayes M. J. (2013) The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab-GEFs. Bioinformatics 29, 499–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marat A. L., Dokainish H., and McPherson P. S. (2011) DENN domain proteins: regulators of Rab GTPases. J. Biol. Chem. 286, 13791–13800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang D., Iyer L. M., He F., and Aravind L. (2012) Discovery of novel DENN proteins: implications for the evolution of eukaryotic intracellular membrane structures and human disease. Front. Genet. 3, 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sato M., Sato K., Liou W., Pant S., Harada A., and Grant B. D. (2008) Regulation of endocytic recycling by C. elegans Rab35 and its regulator RME-4, a coated-pit protein. EMBO J. 27, 1183–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Allaire P. D., Marat A. L., Dall'Armi C., Di Paolo G., McPherson P. S., and Ritter B. (2010) The Connecdenn DENN domain: a GEF for Rab35 mediating cargo-specific exit from early endosomes. Mol. Cell 37, 370–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yoshimura S., Gerondopoulos A., Linford A., Rigden D. J., and Barr F. A. (2010) Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J. Cell Biol. 191, 367–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nickerson M. L., Warren M. B., Toro J. R., Matrosova V., Glenn G., Turner M. L., Duray P., Merino M., Choyke P., Pavlovich C. P., Sharma N., Walther M., Munroe D., Hill R., Maher E., et al. (2002) Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dube syndrome. Cancer Cell 2, 157–164 [DOI] [PubMed] [Google Scholar]
- 10. DeJesus-Hernandez M., Mackenzie I. R., Boeve B. F., Boxer A. L., Baker M., Rutherford N. J., Nicholson A. M., Finch N. A., Flynn H., Adamson J., Kouri N., Wojtas A., Sengdy P., Hsiung G. Y., Karydas A., et al. (2011) Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Renton A. E., Majounie E., Waite A., Simón-Sánchez J., Rollinson S., Gibbs J. R., Schymick J. C., Laaksovirta H., van Swieten J. C., Myllykangas L., Kalimo H., Paetau A., Abramzon Y., Remes A. M., Kaganovich A., et al. (2011) A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang C. W., Hojer C. D., Zhou M., Wu X., Wuster A., Lee W. P., Yaspan B. L., and Chan A. C. (2016) Regulation of T cell receptor signaling by DENND1B in TH2 cells and allergic disease. Cell 164, 141–155 [DOI] [PubMed] [Google Scholar]
- 13. Xu J., and McPherson P. S. (2015) DENND3: a signaling/trafficking interface in autophagy. Cell Cycle 14, 2717–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xu J., Fotouhi M., and McPherson P. S. (2015) Phosphorylation of the exchange factor DENND3 by ULK in response to starvation activates Rab12 and induces autophagy. EMBO Rep. 16, 709–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mizushima N., Yoshimori T., and Ohsumi Y. (2011) The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132 [DOI] [PubMed] [Google Scholar]
- 16. Matsui T., Itoh T., and Fukuda M. (2011) Small GTPase Rab12 regulates constitutive degradation of transferrin receptor. Traffic 12, 1432–1443 [DOI] [PubMed] [Google Scholar]
- 17. Delprato A., and Lambright D. G. (2007) Structural basis for Rab GTPase activation by VPS9 domain exchange factors. Nat. Struct. Mol. Biol. 14, 406–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hussain N. K., Jenna S., Glogauer M., Quinn C. C., Wasiak S., Guipponi M., Antonarakis S. E., Kay B. K., Stossel T. P., Lamarche-Vane N., and McPherson P. S. (2001) Endocytic protein intersectin-l regulates actin assembly via Cdc42 and N-WASP. Nat. Cell Biol. 3, 927–932 [DOI] [PubMed] [Google Scholar]
- 19. He X., Kuo Y. C., Rosche T. J., and Zhang X. (2013) Structural basis for autoinhibition of the guanine nucleotide exchange factor FARP2. Structure 21, 355–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mitin N., Betts L., Yohe M. E., Der C. J., Sondek J., and Rossman K. L. (2007) Release of autoinhibition of ASEF by APC leads to CDC42 activation and tumor suppression. Nat. Struct. Mol. Biol. 14, 814–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Z., Zhang T., Wang S., Gong Z., Tang C., Chen J., and Ding J. (2014) Molecular mechanism for Rabex-5 GEF activation by Rabaptin-5. eLife 2014 3, e02687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hornbeck P. V., Kornhauser J. M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., and Sullivan M. (2012) PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40, D261–D270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yu B., Martins I. R., Li P., Amarasinghe G. K., Umetani J., Fernandez-Zapico M. E., Billadeau D. D., Machius M., Tomchick D. R., and Rosen M. K. (2010) Structural and energetic mechanisms of cooperative autoinhibition and activation of Vav1. Cell 140, 246–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cuevas B. D., Lu Y., Mao M., Zhang J., LaPushin R., Siminovitch K., and Mills G. B. (2001) Tyrosine phosphorylation of p85 relieves its inhibitory activity on phosphatidylinositol 3-kinase. J. Biol. Chem. 276, 27455–27461 [DOI] [PubMed] [Google Scholar]
- 25. Fisslthaler B., Loot A. E., Mohamed A., Busse R., and Fleming I. (2008) Inhibition of endothelial nitric oxide synthase activity by proline-rich tyrosine kinase 2 in response to fluid shear stress and insulin. Circ. Res. 102, 1520–1528 [DOI] [PubMed] [Google Scholar]
- 26. Zimnicka A. M., Husain Y. S., Shajahan A. N., Sverdlov M., Chaga O., Chen Z., Toth P. T., Klomp J., Karginov A. V., Tiruppathi C., Malik A. B., and Minshall R. D. (2016) Src-dependent phosphorylation of caveolin-1 Tyr-14 promotes swelling and release of caveolae. Mol. Biol. Cell 27, 2090–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Martinez-Quiles N., Ho H. Y., Kirschner M. W., Ramesh N., and Geha R. S. (2004) Erk/Src phosphorylation of cortactin acts as a switch on-switch off mechanism that controls its ability to activate N-WASP. Mol. Cell. Biol. 24, 5269–5280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang L., Wang H., Liu D., Liddington R., and Fu H. (1997) Raf-1 kinase and exoenzyme S interact with 14-3-3ζ through a common site involving lysine 49. J. Biol. Chem. 272, 13717–13724 [DOI] [PubMed] [Google Scholar]
- 29. Sellier C., Campanari M. L., Julie Corbier C., Gaucherot A., Kolb-Cheynel I., Oulad-Abdelghani M., Ruffenach F., Page A., Ciura S., Kabashi E., and Charlet-Berguerand N. (2016) Loss of C9ORF72 impairs autophagy and synergizes with polyQ Ataxin-2 to induce motor neuron dysfunction and cell death. EMBO J. 35, 1276–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Amick J., Roczniak-Ferguson A., and Ferguson S. M. (2016) C9orf72 binds SMCR8, localizes to lysosomes, and regulates mTORC1 signaling. Mol. Biol. Cell 27, 3040–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Baba M., Hong S. B., Sharma N., Warren M. B., Nickerson M. L., Iwamatsu A., Esposito D., Gillette W. K., Hopkins R. F. 3rd., Hartley J. L., Furihata M., Oishi S., Zhen W., Burke T. R. Jr., Linehan W. M., et al. (2006) Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proc. Natl. Acad. Sci. U.S.A. 103, 15552–15557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Han C., Alkhater R., Froukh T., Minassian A. G., Galati M., Liu R. H., Fotouhi M., Sommerfeld J., Alfrook A. J., Marshall C., Walker S., Bauer P., Scherer S. W., Riess O., Buchert R., et al. (2016) Epileptic encephalopathy caused by mutations in the guanine nucleotide exchange factor DENND5A. Am. J. Hum. Genet. 99, 1359–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xia Z., and Liu Y. (2001) Reliable and global measurement of fluorescence resonance energy transfer using fluorescence microscopes. Biophys. J. 81, 2395–2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.