Abstract
Biophysical signals act as potent regulators of stem cell function, lineage commitment, and epigenetic status. In recent years, synthetic biomaterials have been used to study a wide range of outside-in signaling events, and it is now well appreciated that material cues modulate the epigenetic status of cells. Here, we review the role of extracellular signals in guiding stem cell behavior via epigenetic regulation, and stress the role of physicochemical material properties as an often-overlooked modulator of intracellular signaling. We also highlight promising new research tools for ongoing interrogation of the stem cell-material interface.
Introduction
The stem cell niche is much more than a set of soluble factors and cell-cell interactions; it is also a physical space with definable mechanical and chemical properties that influence a variety of intracellular events. Both in vivo developmental studies and in vitro manipulations using substrates with defined mechanical properties have made it increasingly clear that the mechanosensitivity of cells strongly influences their decision-making, and that the substrate upon which a stem cell is grown is therefore itself a potent stimulus. While tissue culture-treated plastic is invaluable in research as a reproducible, standardized culture substrate, it possesses physical properties - high stiffness and surface homogeneity – that are non-physiological and known to affect cell fate decisions (Dalby et al., 2007; Engler et al., 2006).
Studies using materials specifically designed to recapitulate individual aspects of a cell’s complex physical and mechanical environment have repeatedly shown that a number of stimuli strongly affect cell behavior (Stevens and George, 2005). These include factors such as material stiffness (Engler et al., 2006), microstructure (Dalby et al., 2007; McMurray et al., 2011), and three-dimensionality (Levenberg et al., 2003; Mabry et al., 2016). There has already been a highly productive focus on developing and defining stem cell culture conditions in terms of biomolecular cues; a decade-long refinement has allowed the field to move away from the usage of xenogeneic feeders and undefined serum towards fully defined culture media such as 2i + LIF. These innovations have led to greatly improved experimental reproducibility, which is critical for basic biological understanding and eventual clinical translation. In a similar fashion, defined material systems with tunable parameters have provided a framework for studying how (stem) cell fate can be influenced through changes in the extracellular space. These factors are more influential than might be generally appreciated, and the physicochemical properties of culture substrates used for stem cells and their progeny therefore merit additional attention. The application of materials in the biological realm will continue to supplement the role of conventional cues in specifying desirable stem cell behavior.
In this review, we discuss the biophysical relationship between a cell and its surroundings, particularly focusing on how epigenetic status is influenced by extracellular stimuli. We first describe some of the key mechanisms by which cells sense physical signals from their microenvironment, and examine the current model for physical linkage of the nuclear envelope to the extracellular space. We then categorize the external inputs that experimentalists have introduced to cells, review the application of materials systems to studying (stem) cell biology and epigenetics, and discuss the intracellular machinery implicated in signal transduction in each case. Finally, we highlight key research tools that we believe hold great promise for ongoing investigations at the interface of stem cell biology and materials science.
Extracellular Mechanosensing
From a cell’s perspective, biophysical cues ultimately result in a change in protein conformation in response to tension or compression. Conversion of mechanical inputs to biological responses occurs at several levels, each with varying layers of complexity and often happening simultaneously. At the level of the plasma membrane, cell-matrix and cell-cell adhesions are formed mostly by integrins and cadherins, respectively; these transmembrane adhesive structures are tethered between the cytoskeleton and an external anchor, physically linking the extra- and intra-cellular compartments. In response to tension, integrins and cadherins undergo a conformational change, which initiates a variety of cytosolic signaling cascades such as via the kinases Src and PI3K (Tzima et al., 2005). For a comprehensive review of cell-ECM homeostasis and integrin signaling, the reader is referred to (Humphrey et al., 2014). Mechanosensitive ion channels may be similarly activated by tension between the extracellular matrix and cytoskeleton (Ko et al., 2001). Heterotrimeric G-proteins (Gudi et al., 1998) and ion channels (Maroto et al., 2005) can also respond directly to changes in membrane tension or fluidity caused by fluid shear stress or changes in cell shape. Alternatively, although its components constantly turn over, the cytoskeleton forms a rigid network that transmits physical forces to the cell as a whole. From the cell-extracellular interface, forces can be transduced through these stiff linkages directly to other sites such as the mitochondria (Wang et al., 2001), or the nucleus (Maniotis et al., 1997). Although there are various mechanisms through which extracellular signals generate gene-, protein-, and whole cell-level changes, we focus primarily on mechanotransduction and the downstream behaviors generated in response to external cues for the scope of this review.
The Nucleus is Physically Linked to the Extracellular Space
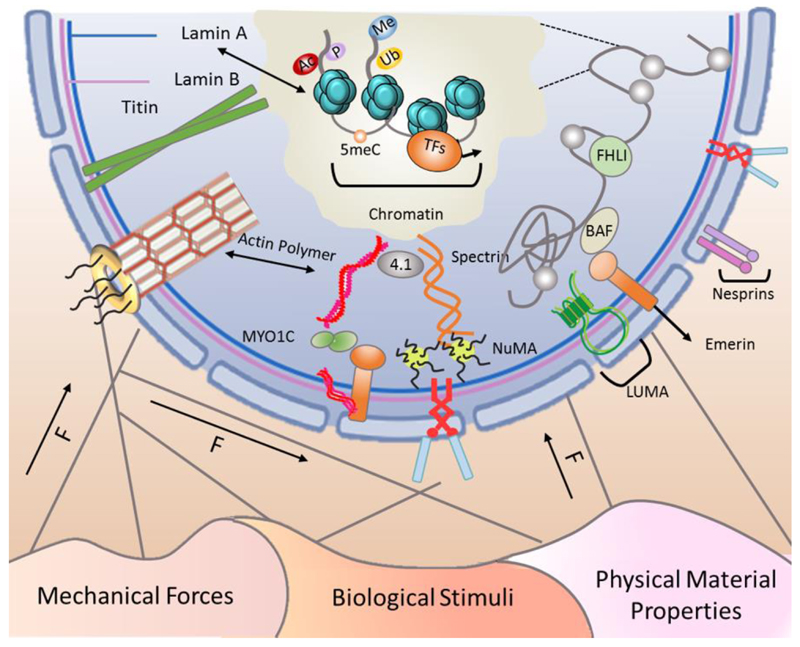
The nucleus is physically linked to the ECM and other cells, acting as a part of a continuous, transcellular tensile network composed of extra- and intra-cellular fibers. External biophysical cues, such as the classifications listed in Figure 1, can mechanically alter the nuclear matrix via cytoskeletal filaments, impacting gene expression through recruitment of epigenetic modifiers or reorganisation of chromatin and the nuclear lamina (reviewed in (Wang et al., 2009a)). The nucleoskeleton is a multicomponent, intra-nuclear scaffold comprised of several classes of proteins - including A- and B-type lamins, spectrin, titin, and nuclear actin – that couple the nuclear envelope to chromatin, and whose combined effects impart viscoelastic properties to the nucleus (Swift et al., 2013). The nucleoskeleton is physically connected to the cytoskeleton by LINC (Linker of Nucleoskeleton and Cytoskeleton) complex proteins (Lu et al., 2012), which include emerin and various isoforms of SUN and nesprin. The LINC complex helps to transduce biophysical forces into functional, intra-nuclear changes, including the expression of mechanosensitive genes (Lammerding et al., 2005). Nesprin proteins connect the nucleus to cytoskeletal components, with the four isoforms exhibiting different binding partners. Nesprin-1 and -2 are the largest (giant) isoforms, connecting to nuclear envelope proteins via C-terminal spectrin repeats and to polymerized actin via N-terminal calponin homology (CH) domains (Padmakumar et al., 2004; Zhen et al., 2002). In contrast, the smaller nesprin-3 and -4 isoforms lack the N-terminal CH domain and link the nuclear envelope to either intermediate filaments via plectin (Wilhelmsen et al., 2005) or the motor protein kinesin-1 (Roux et al., 2009), respectively.
Figure 1. Extracellular forces influence the cell’s epigenetic status through interactions with the nuclear membrane.
The cell is an interconnected entity with cytoskeletal components linking the membrane to the nucleus. Cells sense mechanical forces in their environment and propagate the forces along the cytoskeleton to the nucleus in order to alter epigenetic status and gene expression profile in response to different biophysical stimuli. Here, we classify these cues as mechanical forces, biological stimuli, and physical material properties. The nuclear envelope contains components of the LINC (Linker of Nucleoskeleton and Cytoskeleton) complex including nesprin and SUN proteins, emerin, and LUMA. These LINC complex components act as receivers and transmitters of mechanical forces to the chromatin and the nucleoskeleton, which includes polymerized actin, nuclear mitotic apparatus protein (NuMA), intermediate filaments, spectrins, protein 4.1, titin, A- and B-type lamins, and nuclear pore complex (NPC)-linked filaments. External forces induce mechanosensitive changes in the nucleoskeletal complexes that, in turn, alter the epigenome and chromatin accessibility. Therefore, these extracellular signals are perceived at the nuclear level as the cell adapts its transcriptome in response to the signals it perceives.
Material-derived Cues Regulate Epigenetic Status
Direct linkage of the nuclear envelope to chromatin implies that physical forces exerted on the nucleus should have direct consequences on chromatin organization and accessibility of DNA for transcription. Epigenetics define the various changes in gene expression that may occur without alteration in the DNA sequence; these include chromatin modifications, DNA methylation, and non-coding RNAs (reviewed in (Flynn and Chang, 2014; Laugesen and Helin, 2014; Lee et al., 2014), respectively). To date, few studies have investigated how synthetic material-derived cues can regulate epigenetic status, although available data suggest exciting possibilities. For example, when mesenchymal stem cells (MSCs) were cultured on parallel microgrooves, the level of histone H3 acetylation was significantly enhanced relative to the flat material control. In addition, when either compressive or tensile force was exerted perpendicular to the alignment of cells within the microgrooves, MSCs demonstrated a decrease of histone deacetylase (HDAC) activity (Li et al., 2011b). In a follow-up study, Downing et al. demonstrated that forced elongation improves the reprogramming efficiency of embryonic fibroblasts by decreasing HDAC activity, which modulates the acetylation pattern of histone H3 and the subsequent susceptibility to reprogramming factors (Downing et al., 2013). This discovery directly demonstrated the ability of a seemingly-subtle material cue to influence the epigenetic signature, as well as the functional consequence in priming cells for reprogramming. Recently, our group has shown that parallel microgrooves also enhance the trans-differentiation of cardiac progenitors into cardiomyocyte-like cells. We showed that the grooved substrates increase histone H3 acetylation and promote sumoylation of myocardin, a modification that strongly activates cardiogenic gene activity (Morez et al., 2015). Therefore, the ability to regulate the epigenetic landscape and subsequent stem cell function with relatively simple material cues suggests that a wealth of unexplored intracellular processes might be controlled by individual or combined extracellular cues.
Categorization of Cues
Each cell is presented with enormous spatiotemporal complexity that results from the summation of soluble and insoluble inputs from the extracellular matrix (ECM) and neighboring cells. Biophysical signals are a particularly interesting category of cue that encompasses both extracellular-derived inputs (fluid shear stress, cyclic strain, cell elongation) and cell-generated responses (intracellular tension, focal adhesion formation). At the cellular level, however, discrimination between types or sources of biophysical cues might not be as clear as the experimentalist intended. The cell may not independently sense complex physical cues such as cell shape, microtopography, or force application, but rather these cues may differentially impact overlapping tension-sensitive signaling pathways. Although tremendous work has been done in these fields, the mechanisms by which a cell distinguishes these signals as distinct information are yet to be fully understood.
Here, we categorize the “cues” presented to a cell from a materials-based standpoint, and classify them simply, but acknowledge through our discussion that the resultant cellular response(s) can overlap. Broadly, we identify three main categories (Figure 2): (1) Direct application of mechanical force, such as fluid shear stress or cyclic stretching; (2) Conventional biological stimuli, covering the incorporation of soluble cues and tethered adhesion molecules; and (3) Physical material properties at varying length scales (macro-, micro-, and nano-scales), covering stiffness, topography, and control of cell shape. By organizing our discussions into these three categories, we can explore the intended efforts of researchers to study specific biological responses while emphasizing how seemingly-different cues can generate similar intracellular responses.
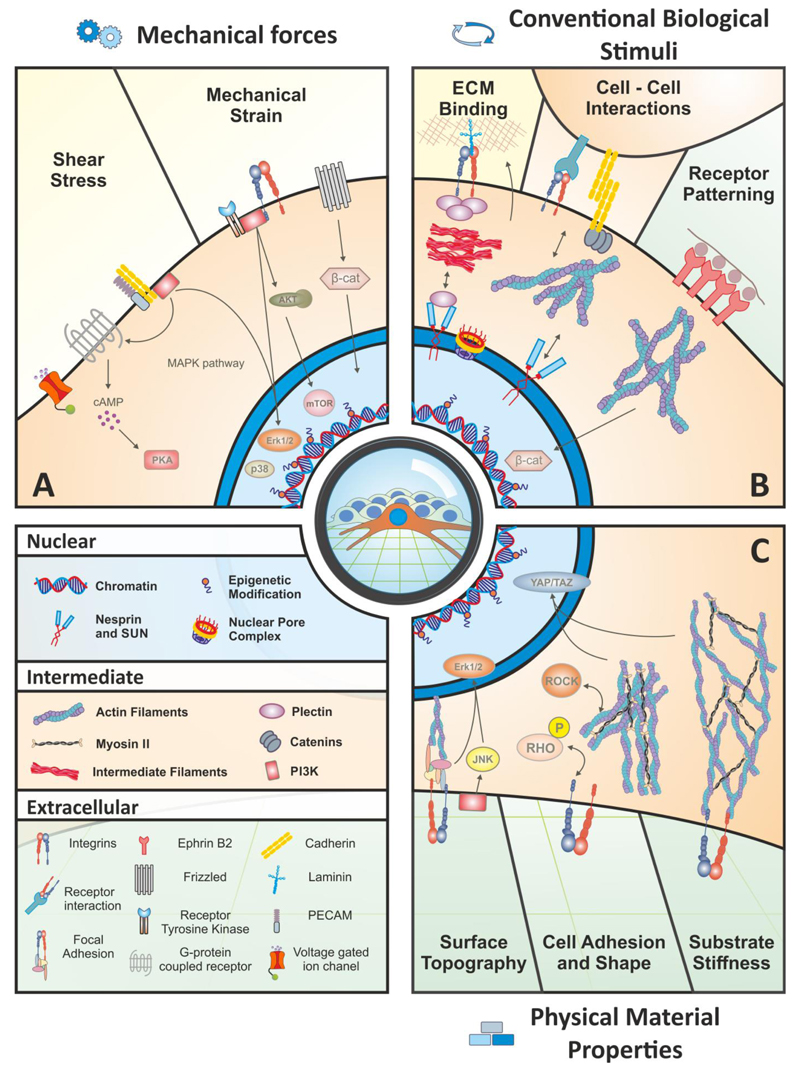
Figure 2. External biophysical regulation of cell fate and function.
Extrinsic cues can be divided into three general categories: (A) Direct application of mechanical forces, such as shear stress and mechanical strain are relatively well-characterized examples of how biophysical signals from the microenvironment activate specific signaling pathways and alter the epigenetic status. Shear stress is transduced through multiple mechanisms, and a few key examples listed here include voltage-gated ion channels, G protein-coupled receptors (GPCRs), and cell adhesion molecules such as cadherins and platelet-endothelial cell adhesion molecule (PECAM). These sensors activate classical elements such as the MAPK pathway, which leads to the nuclear translocation of ERK1/2 and p38 to act on downstream target genes, and activation of PI3K and PKA. Mechanical strain sensing involves a myriad of receptor tyrosine kinases as well as the Wnt receptor, Frizzled, and an array of integrins. These extracellular mediators can activate the PI3K/AKT pathway, which stimulates mTOR activity. (B) Conventional biological stimuli include cell-ECM and cell-cell interactions, and receptor patterning/clustering. These events exhibit a feedback response: the cell receives specific input from one of these effectors, and in turn changes the signal by altering the composition or organization of the ECM or responds with an additional signal to a neighboring cell and propagating the message into something dynamic. Cell-ECM interactions are dominated by integrins at the cell surface that exert their effect directly on the nucleus via cytoskeletal filaments. Cell-cell interactions act in a similar manner but also include cell adhesion molecules such as cadherins. Receptor patterning occurs naturally in the sense that the spatial distribution of signaling or adhesion molecules affects cell behavior. In a synthetic system, receptors such as Ephrin-B2 can be encouraged to cluster by presenting the ligand at varying density along a physically-linked backbone, which stimulates a stronger response than a single ligand alone; in this case, Ephrin-B2 stimulates the stabilization and nuclear translocation of β-catenin. (C) Physical material properties at the macro, micro and nano scales include surface topography, cell adhesion and shape, and substrate stiffness. Topography of the material surface, such as the distribution and regularity of nanopit spacing, activates signaling pathways such as JNK and Erk1/2 and affects cytoskeletal tension. Cell adhesion and shape can be controlled by altering the adhesive sites on a synthetic substrate. This effect regulates cell fate decisions in hMSCs through the Rho/ROCK pathway, which is necessary for the activity of the YAP and TAZ transcription factors. Finally, the physical stiffness of the extracellular substrate heavily influences cell behaviors, such as fate decisions, and this signal is also transduced through YAP/TAZ and the actin cytoskeleton.
Category 1 – Direct Application of Mechanical Forces
The effects of mechanical strain and fluid shear stress on cell behavior are well-characterized examples of “outside-in” signaling that affect epigenetic status. Although these forces are not material-derived, they have provided useful information for monitoring how extracellular cues can potently regulate intracellular signaling and the epigenome. Mechanical strain and fluid shear stress are sensed by cells cooperatively – the organization of the cytoskeleton physically links adjacent cells and allows for a concerted response to mechanical forces, often through mechanically-gated ion channels or transmembrane proteins (Figure 2A). These interactions converge by regulating the activity of the small GTPase Rho at the RhoGAP and RhoGEF domains, and are then further transduced via the downstream effector Rho-associated kinase (ROCK) to increase myosin II contractility and intracellular tension. Yes-associated protein (YAP) and transcriptional coactivator with PDZ-binding domain (TAZ), both members of the Hippo signaling pathway, are subsequently translocated to the nucleus where they co-activate multiple transcription factors and regulate a host of target genes and cell fate decisions (Dupont et al., 2011). Cells respond to various extracellular stimuli via YAP/TAZ, as will be discussed in more detail in later sections.
The endothelium is a particularly pertinent model for exploring how fluid shear stress affects epigenetic status. Because alterations to the vascular flow profile are associated with the onset and progression of atherosclerosis, changing the type of fluid shear stress experienced by endothelial cells leads to alterations of their epigenetic profile. Laminar fluid shear stress, or unidirectional flow that occurs in healthy vasculature, induces phosphorylation of H3S10, H3K14 acetylation, and nuclear export of HDAC5, which together result in a more accessible genome because of increased acetylation (Illi et al., 2003). Conversely, oscillatory fluid shear stress, which occurs in diseased vasculature, induces the expression and nuclear accumulation of class I and class II HDACs, and increases DNA methyltransferase 1 (Dnmt1) expression, which subsequently leads to DNA hypermethylation (Lee et al., 2012; Zhou et al., 2014). Laminar fluid shear stress has been shown to guide mouse embryonic stem cells (ESCs) to differentiate toward the cardiovascular lineage, as evidenced by enhanced transcriptional activity of lineage-specific markers, via altered histone methylation profile, including enhanced acetylation of H3K14 (Illi et al., 2005). Shear stress sensing is an intricate process that requires various mechanisms but some notable examples are protein kinase A (PKA)-dependent phosphorylation of cAMP-responsive elements (Boo, 2006), extracellular signal-related kinase (ERK)1/2 activation (Huddleson et al., 2005), and PI3K-dependent chromatin remodeling (Takahashi and Berk, 1996).
A wide variety of cells experience mechanical strain and compression in their native physiological location. Cardiac rhythm produces cyclic strain not only on cardiac tissue but also on the constituents of the arteries that experience pulsatile force following systole. Vascular smooth muscle cells respond to cyclic, isotropic strain by up-regulating HDAC7 and down-regulating the level of HDAC3/4, which lead to H3 hyperacetylation and a block in cell migration (Yan et al., 2009). As shown in Figure 2, mechanical strain can be modulated by the Wnt/β-catenin pathway (Case et al., 2008), PI3K-protein kinase B (Akt) (Danciu et al., 2003), and mitogen-activated protein kinase (MAPK)-mediated p38 signaling (Peng et al., 2010).
Furthermore, LINC complex components have been shown to be critical for transmitting mechanical forces. Knockdown of nesprin-1 reveals its importance in the propagation of mechanical strain from the actomyosin components to the nucleus, as nesprin-1-depleted endothelial cells are unable to reorient in the presence of cyclic strain (Chancellor et al., 2010). Disruption of SUN or nesprin proteins also prevents the physical propagation of ultra-low magnitude mechanical perturbation in MSCs, indicating that LINC-mediated mechanocoupling between perinuclear actin and the nucleus is required (Uzer et al., 2015). These data illustrate the sensitive epigenetic changes that occur in response to external forces, as well as the intricate signaling pathways involved, and are a driving force for conceptualizing and designing materials for regulating epigenetic status.
Category 2 - Conventional Biological Stimuli
The balance between cell-cell and cell-matrix interactions is a critical theme of developmental biology, tissue homeostasis, and disease progression, and materials can be leveraged to bias cells toward one interaction over the other. Attempts to simplify these relationships, as well as to study their fine balance, have used an array of (pseudo)synthetic techniques mimicking the in vivo relationship between cells and their environment (Figure 2B). In a physiological setting, integrin-mediated interactions with the ECM are indispensable for normal development and cell function (De Arcangelis and Georges-Labouesse, 2000). In vitro, arrays of self-assembled peptides have been used as cell culture substrates, demonstrating that heparin-binding peptides that interact with glycosaminoglycans (GAGs) on the cell surface most effectively promote human pluripotent stem cell (hPSC) self-renewal (Klim et al., 2010). Further investigation revealed that combinatorial surfaces that engage both GAGs and integrins promote differentiation of hPSCs toward ectoderm, whereas surfaces that interact exclusively with GAGs enhanced differentiation to mesoderm and endoderm (Wrighton et al., 2014). Materials can also be designed to respond specifically to a remodeling enzyme. A common strategy is to crosslink a hydrogel with an enzymatically-degradable peptide so the cellular production of proteolytic remodeling enzymes, in particular MMPs, specifically induces localized degradation (Lutolf et al., 2003). This design allows for sequestered biomolecules to be released only when a certain enzyme is present to cleave the crosslink and free the molecule.
The stem cell niche involves interactions with supporting cell types and this has been well-defined in experimentally-tractable model organisms like Drosophila and C. elegans (Li and Xie, 2005), but is also known to be true in mammalian systems. For example, maintenance of the hematopoietic stem cell (HSCs) niche in the bone marrow requires interactions with multiple supporting cell types, and HSC attachment to osteoblasts is required, in part, to maintain the hematopoietic pool (Calvi et al., 2003). Engineering materials that promote cell-cell interaction through aggregation can be useful for enhancing the soluble and insoluble factors that drive the therapeutic phenotype. A straightforward example is the use of microwells for MSC aggregation; MSCs cultured in hanging drops form spheroids but exhibit practical issues with scalability. By contrast, 100 μm-deep, microwell-patterned surfaces can be seeded with a single MSC in each well which then proliferates to produce a large quantity of uniformly-sized aggregates (Wang et al., 2009b). Encapsulation of hMSC spheroids is another approach for producing homogeneous populations, a strategy which is particularly beneficial for the delivery of therapeutically-active aggregates (Chan et al., 2013). The synthetic recapitulation of cell-cell interactions using bioactive peptides can also induce beneficial regenerative effects. Poly(ethylene glycol) (PEG) hydrogels are a useful tool for these experiments due to their biocompatibility, ease of chemical modification, aqueous solubility, and active sites for tethering peptides and growth factors. In particular, the experimentalist can incorporate an adhesive RGDS sequence into a 3D PEG gel to encourage cell spreading, along with varying other mimetic peptides. For example, incorporating a peptide that mimics N-cadherin into three-dimensional hydrogels enhances the chondrogenic potential of hMSCs (Bian et al., 2013) and exemplifies how physicochemical and biological cues can be productively combined.
Clustering multiple versions of the same cell surface receptor, including integrins or growth factor receptors, can strengthen or activate the effective signal from a physical signal or soluble ligand, materials can be engineered that present ligands to enhance or permit this clustering (Figure 2B). For example, by controlling the density of the ligand ephrin-B2 along a soluble biopolymer backbone, the differentiation of neural progenitors was enhanced both in vitro and in vivo, and similar conjugates were able to promote the differentiation of hPSCs toward a dopaminergic phenotype (Conway et al., 2013). Materials that are engineered to present bioactive molecules in a defined spatial manner are also able to enhance signaling potency (Figure 2B). The spatial control of stem cell fate was achieved by co-presentation of two artificial peptides that bound subunits of the TGF-β receptor II without blocking their ligand-binding domain, causing pre-assembly of the mature complex and sensitising the cell (Li et al., 2011a).
Category 3 - Physical Material Properties with Varying Length Scales
Surface Topography
Altered topographical cues within the natural extracellular environment are linked to changes in cell function. For example, the architecture of the in vivo microenvironment of cardiomyocytes consists of highly-aligned, uniform fibers upon which the healthy cells can contract and transduce signals; in contrast, disease states, such as post-myocardial infarction that disrupt this architecture via scar tissue deposition, interrupt normal function. These microstructural cues influence function at the multicellular level by defining organ architecture but also affect mechanotransduction pathways in individual cells. Similarly, manipulation of nano- and micro-scale topography of a two-dimensional (2D) culture substrate surface has a profound effect on the cellular response (Figure 2C). Cues at the cell-material interface can be as simple as the organization of nanosized pits; surfaces with a disordered pattern of 120 nm-diameter pits bias hMSCs toward osteogenic differentiation (Dalby et al., 2007) while highly-ordered 120 nm pits maintain long-term, undifferentiated cultures (McMurray et al., 2011). Exactly how artificial stimuli such as these relate to in vivo stimuli is not clear, but the authors postulated that differences in physical interactions at the interface might reflect the types of signals that hMSCs receive in their native niche. The effects of the surface topography order extended to changes in metabolism, with undifferentiated cells exhibiting a reduced metabolic load relative to their differentiating counterparts that correlated with regulation by ERK1/2, c- Jun N-terminal kinase (JNK), and low-density lipoprotein (LDL) effectors (Tsimbouri et al., 2012). On the sub-nanometre scale, the cell’s recognition of the surface chemistry and shape of ECM proteins may be partially imitated by the structure of a substrate polymer itself (Celiz et al., 2015), which enables it to act as a growth or adhesion factor substitute.
Cell Shape
Similarly, materials can be used to force a cell to adopt a particular shape. Culturing a cell in 3D by encapsulation in a gel allows retention of a rounded morphology rather than the spreading that occurs on a 2D surface. This is critical for retention of a chondrocyte phenotype (Takahashi et al., 2007) and also encourages MSCs to adopt a chondrogenic fate (Mauck et al., 2006). Finer control of cell shape on a 2D surface can be accomplished by microcontact printing of cell adhesion molecules in particular patterns so that a single cell adhering to the material surface spreads into the desired shape. In an early study, McBeath and coworkers demonstrated that MSCs could be directed towards and adipogenic or osteogenic fate by providing a small or large surface area for spreading, respectively; this behavior was shown to be regulated by Rho/ROCK signaling (McBeath et al., 2004). Further evidence suggests that cell shape-dependent contractility of hMSCs stimulates the expression of JNK, ERK1/2, and Wnt effectors (Kilian et al., 2010). The control of cell shape by the ECM also has profound effects on chromatin structure and gene transcription. Three-dimensional culture of human mammary epithelial cells leads to deacetylation of histones H3 and H4, chromatin condensation, and reduced gene expression, and this effect can be recapitulated by forced cell rounding or by inhibiting actin polymerization (Le Beyec et al., 2007). In parallel, fibroblast spreading and actin fiber formation were shown to inversely correlate with the nuclear localization of HDAC3, serum response element (SRE), and myocardin-related transcription factor A (MRTF-A), as well as increased H3K9 acetylation and transcription of cell homeostasis-related genes (Jain et al., 2013). These data indicate that spreading leads to less acetylated histones in two different cell types, demonstrating that a complex combination of intracellular contraction/tension related signaling activity can regulate acetylation profile depending on a cell’s shape/adhesion status.
Matrix Stiffness
Tissues throughout the body display a wide range of stiffness values – from relatively soft neuronal tissue to far stiffer cortical bone. The composition and organization of the tissue dictate the resulting mechanical properties and influence the behavior of resident cells. Monitoring tissue stiffness changes in vivo during development have proven difficult but evidence from Drosophila indicates that developmental ECM dynamics result in changing elasticity which drives neuronal migration and fate decisions (Kim et al., 2014). Similarly, the developing chick heart exhibits increasing stiffness, which correlates with the deposition of type I collagen as well as the contractile capability of maturing cardiomyocytes (Majkut et al., 2013). However, the stiffness of fully-formed, healthy tissues (i.e. those not undergoing wound healing or fibrosis) remains relatively constant until aging or disease onset. Recapitulation of matrix elasticity has been a focus in the biomaterials community owing mainly to the versatility of substrates with tunable mechanical properties. In a seminal 2006 paper, Engler et al. demonstrated the ability of matrix stiffness alone to drive fundamental changes in stem cell behavior. hMSCs cultured on polyacrylamide hydrogels of varying stiffness differentiated into specific cell types that corresponded to the compliance of their underlying matrices (Engler et al., 2006). Soft gels ranging from 0.1-1 kPa have been reported to enable adipogenic differentiation while very stiff gels resembling bone tissue drove them toward the osteogenic lineage (Dupont et al., 2011). When the mechanosensitive signal transduction mechanism was examined, YAP/TAZ were found to be responsible for translating extracellular mechanical cues into functional responses (Dupont et al., 2011), similar to their role in mediating shear stress and extrinsic mechanical forces. Chromatin organisation is also regulated by extracellular elasticity: stiff matrix substrates ( >50kPa) that allow for generation of cellular tension lead to increased nuclear area, decondensed chromatin, and H3 acetylation relative to the soft matrix counterparts (Kocgozlu et al., 2010). How these specific extracellular signals stimulate changes in the nucleus has been an open topic of debate, with one promising explanation that physical forces on the nucleus (dis)allow the transport of key transcription factors across the nuclear envelope (Wang et al., 2009a).
In a landmark finding, nuclear stiffness, as determined by the ratio of the intermediate filaments Lamin A and B, was shown to correlate with bulk stiffness of tissues throughout the body, and altering this ratio by knocking down or overexpressing lamin A enhanced the elasticity-induced differentiation of hMSCs (Swift et al., 2013). Matrix-permitted cytoskeletal tension stabilizes lamin A at the inner nuclear envelope and stimulates expression of YAP, TAZ, and retinoic acid receptor gamma (RARG) target genes, promoting osteogenic differentiation in hMSCs. Recently, the accessibility of specific epitopes of lamin A/C was shown to be mechanoresponsive; the Ig-domain of lamin A/C is less accessible in the basal nuclear envelope, relative to apical, when undergoing osteogenic differentiation, during cell spreading, or in the presence of compressive force (Ihalainen et al., 2015). Of note, this epitope was equally available in both apical and basal locations during adipogenic differentiation, or for cells cultured on soft hydrogels, and overlaps with important binding sites for maintaining efficient mechanotransduction.
In hematopoietic stem and progenitor cells (HSPCs) maintenance and cell fate decisions, matrix elasticity and composition, as well as the actomyosin contractile machinery, play a significant role. Soft culture substrates fabricated from full-length tropoelastin (a naturally-occurring elastic material) promote the expansion of murine HSPCs (Lin-Sca-1+c-kit+, LSK cells) and human progenitors (Holst et al., 2010). However, this effect was reduced when HSPCs were cultured on stiffer gels of truncated or crosslinked tropoelastin, or in the presence of inhibitors of myosin heavy or light chains, indicating a role of the contractile machinery. Furthermore, the specific myosin II heavy chain isoforms A and B exhibit divergent roles is HPSC regulation in vitro and in vivo. Myosin IIB regulates asymmetric cell division and is downregulated during differentiation of human HSPCSs; partial knockdown of myosin IIB promotes symmetric division and increases the number human CD34+ progenitors expanded in culture (Shin et al., 2014). In contrast, dephosphorylation of myosin IIA (pS1943), which renders it more active, increases during cytokine-induced differentiation or following culture on stiff substrates (34 kPa). In vivo, myosin IIA was shown to be required for sustained hematopoiesis in an allogenic transplantation model, as LSK cells were ten-times less abundant in the marrow after eight weeks when myosin IIA was deleted from donor cells; myosin IIB, however, contributes to differentiation, but not survival, of xenogenic transplanted cells (Shin et al., 2014). These data demonstrate the fundamental role of mechanosensing and mechanoregulation in HSPC fate decisions.
Promising Research Tools for the Studying the Cell-Material Interface
Increasingly sophisticated cell-influencing materials are under constant development, and concurrent advances in techniques that aim to analyze the interactions between a cell and its environment have provided methods to monitor these behaviors. Below, we have selected examples of some of the most promising aspects of ongoing research that will contribute to the evolution of our field.
Atomic Force Microscopy
The ability to reproducibly image nanoscale features with high spatial resolution in living cells, as well as measure the associated physical properties, has been extensively explored through atomic force microscopy (AFM). These microscopes are now accessible to many biologists and interdisciplinary researchers, and the development of combinatorial AFMs with other types of microscopes (i.e. confocal fluorescent) has provided a range of tools that can explore cellular processes in real-time. For example, the stiffness of patient-derived metastatic cancer cells was measured by AFM and shown to be 70% softer than local benign cells, ranging from ~0.50 kPa to ~2.0 kPa for cancer and healthy cells, respectively (Cross et al., 2007). The soft characteristic was common for cancer cells derived from different tumour types, suggesting that nanomechanical analysis could be used as part of a multifaceted approach to cancer diagnosis. AFM has also been used extensively to measure the surface modulus (stiffness) of synthetic culture scaffolds as well as the force required to detach an antibody from its ligand (Wen et al., 2014), allowing for investigation of how the cell interacts with microenvironment at relevant length and force scales.
Scanning Ion Conductance (SICM) Microscopy
An emerging technique that offers similar advantages to AFM in terms of high spatial resolution and versatility is scanning ion conductance microscopy (SICM). Although SICM is not nearly as available to biology researchers as AFM, the customisable features are promising for a range of applications. Non-invasive imaging by SICM is achieved using a nanopipette in aqueous, conducting media through which changes in the ion current are measured and then used as feedback to prevent the tip from physically interacting with the surface. This information is then converted into measurements of the proximity and physical properties of the cell surface, providing a high-resolution image of the sample-of-interest without direct interaction (Korchev et al., 1997). This approach has been leveraged to image the precise location of individual ATP- regulated K+ channels in living cardiac myocytes with ~50nm resolution, volumes of whole cells and their subcellular protrusions with 10-20 liter resolution, and the organization of single proteins or complexes at the cell surface with a smaller nanopipette that allowed for 3-6 nm lateral resolution (Shevchuk et al., 2006).
Super-resolution Microscopy
Advances in light microscopy have resulted in unprecedented resolution and image quality that has enhanced our understanding of how cellular process and how cells interact with their microenvironment. The diffraction limit of light has long been considered an inherent roadblock to enhancing spatial resolution. Various forms of super-resolution microscopy exist, including lattice light-sheet microscopy, structured illumination microscopy (SIM), photoactivation localization microscopy (PALM), and stochastic optical reconstruction microscopy (STORM) (Chen et al., 2014) (reviewed in (Sydor et al., 2015)). Here, we discuss stimulated emission depletion (STED) microscopy, which has overcome the diffraction limit by quenching the excited molecules at the periphery of a focal spot, providing a smaller excitation point which results in higher resolution. This approach confers a four-fold improvement in x-y resolution and a 16-fold reduction in focal-spot cross-sectional area compared to confocal microscopy, which allows for unprecedented observation of cellular events at the nanoscale. Illustrating the particular significance of enhanced spatial resolution, the arrangement of organizational proteins in the mitochondria was measured for the first time using STED, and defining features with no known function such as a highly-periodic spatial distribution of the proteins were observed (Jans et al., 2013).
Raman Spectroscopy
Raman spectroscopy (RS) provides information about the types and relative abundance of chemical bonds present in a sample by calculating shifts in photon frequency due to inelastic scattering. This provides a global snapshot of the distribution of biomolecules in a cell or tissue such as proteins, lipids, and DNA, and can be used to characterize spatiotemporal changes in cell composition, deposited extracellular matrix, and heterogeneous cell populations. Some of the earliest applications of RS to biology focused on monitoring stem cell phenotype during differentiation as a means to identify discernible differences such as mRNA levels in mouse ESCs (Notingher et al., 2004). In 2009, our group used RS to capture ‘fingerprints’ of mineralized bone nodules derived from osteoblasts, MSCs, and ESCs (Gentleman et al., 2009). Following thorough characterization, we concluded that hMSCs and osteoblasts could generate nodules similar to native bone in terms of their composition and nanoscale architecture, but ESCs deposited bone that was reminiscent of mechanically-compromised, aging tissue. RS has also shown promise for non- destructive, label-free identification of cell phenotypes such as cell lines that most closely resemble the in vivo counterpart (Swain et al., 2010). An emerging application of RS will be the non-destructive observation and characterization of stem cell dynamics during culture on advanced material platforms such as those listed above; however, only very recent studies have demonstrated the feasibility of these processes (El-Said et al., 2015; von Erlach et al., 2015) and interdisciplinary collaboration to achieve these goals will undoubtedly be necessary.
Next-generation Chromatin Imaging
It is well accepted that genome organization regulates the transcriptional profile by enabling direct physical interactions between gene promoters, distal regulatory elements, and transcriptional proteins. Electron microscopy and fluorescence in situ hybridization (FISH) assays have been employed to study the spatial organization of chromatin; however these methods are still currently limited in throughput, analyzing only a few loci at a time. Molecular assays based upon chromosome conformation capture (3C) have allowed for advanced understanding of how physical interactions between distant genomic loci affect the biophysical properties of chromatin (Dekker et al., 2002) as well as the functional relevance of chromatin interactions with the proteome (Alabert et al., 2014). For example, a modified version of 3C (Hi-C) applied to monitor higher-order chromatin dynamics in hESCs during differentiation revealed extensive chromatin reorganization occurs during lineage specification and provided new insights for allelic expression biases that have been previously observed (Dixon et al., 2015). The application of high-resolution, time-resolved chromatin rearrangement, as well as interactome information, for (stem) cells in contact with materials would shed more light on how external biophysical forces result in whole cell-level changes.
Developing New Materials Systems - Discover or Design?
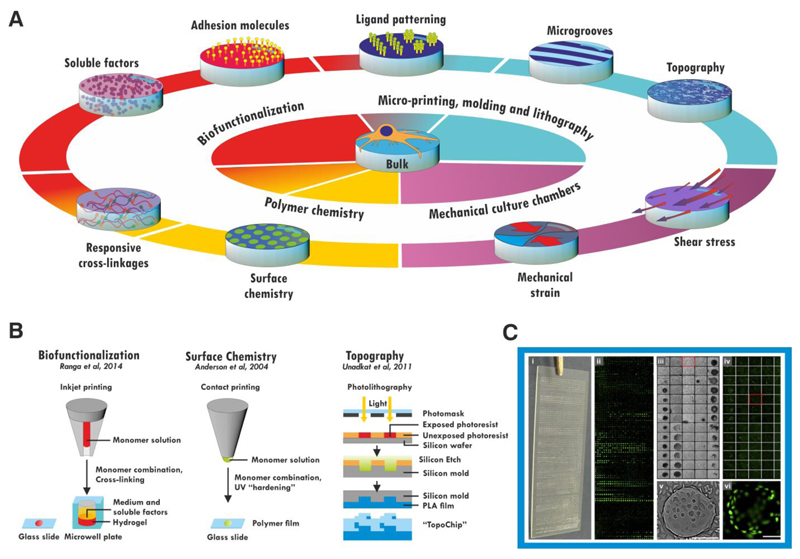
The development of new materials can either follow design- or discovery-driven approaches. Discovery-driven approaches are most appropriate when the underlying mechanisms of an effect are unclear, as is the case for the how polymer chemistry influences cell behavior (Celiz et al., 2014) (Simon and Lin-Gibson, 2011). The approaches used and discoveries made have been extensively reviewed elsewhere; briefly, combinations of monomers are typically printed on a glass side and polymerized in situ to form microarrays of films (Anderson et al., 2004; Celiz et al., 2015) (Figure 3b). Combinations of biological functionalisations of hydrogels have also been screened in microwell plates (Ranga et al., 2014), and topologies by algorithmic design of a poly-lactic acid chip (Unadkat et al., 2011).
Figure 3. Accessing complexity.
A bulk material can be further functionalized by a number of routes (A). Soluble biomolecules can be incorporated into the bulk of the material or presented on the surface, and hydrogels may be made biologically responsive by use of protease-degradable crosslinkages. Micropatterning techniques allow these cues to be patterned within the material, and also allow the material itself to be patterned into grooves or more complex topographical cues on the micro- or nano-scales. The chemistry of the material itself can be altered (e.g. by blending with other polymers) to confer responsiveness or alter the cell-material surface interface. Materials may also be used to subject the cells on them to mechanical strain or shear stress in combination with appropriate culture chambers. These modifications can be made in combination to generate combinatorial materials. Some of these cues have been screened in high-throughput studies (B). Surface chemistry can be investigated by microcontact printing and UV crosslinking of monomer combinations on a glass slide. Biofunctionalizations have been screened using micro-inkjet systems to generate hydrogels within a 1536-well plate or on a glass slide. Topographical cues have been investigated using photolithography to etch microscale geometric cues onto a poly-lactic acid (PLA) chip. The response of cells is screened by imaging (C) the entire surface, generating values for cell attachment or proliferation (iii, v) and fluorescent reporter expression (ii, iv, vi) for each individual material spot. Figure C adapted with permission from (Zhang et al., 2009), scale bar: 200 μm.
As a problem involving the synthesis of complex polymers whose properties and biological effects cannot be fully predicted a priori from the properties of their monomers, materials HTS is comparable to the directed evolution of proteins. Well-established key criteria for successful directed evolution experiments which may be relevant to materials HTS studies include i) appropriate selection criteria, ii) a sufficiently large and functionally diverse library, and iii) appropriate intergenerational candidate mutation and recombination (Romero and Arnold, 2009). As cell response in materials HTS studies is typically evaluated by one or two parameters – cell number (attachment and survival) and expression of a single fluorescent marker, other effects and responses may change if unchecked. Following their initial screenning, Ranga and colleagues found that differently biofunctionalized hydrogels that were apparently similar in their ability to support ESC proliferation and Oct4 expression were varied in their induction of other pluripotency markers such as SSEA-1 and Rex1. The physical properties of materials are also susceptible to this effect – the final polymer identified by Celiz and coworkers cracked after coating on tissue culture flasks – fortuitously, they had also identified blends with similar properties that did not. So far, the largest materials libraries have been many orders of magnitude smaller than most protein libraries (Celiz et al., 2015; Romero and Arnold, 2009). This may limit movement away from local fitness optima; that is, moving away from well-performing homopolymers in favor of co-polymers that might perform better is difficult when poorly-performing candidates are in between (Celiz et al., 2015; Hook et al., 2013; Mei et al., 2010). Unlike proteins, a material’s properties are not necessarily acutely dependent on an exact sequence of monomers. Altering higher-order structure and organisation may be a productive, if challenging, route to library enlargement and diversification, in addition to screening multiple copolymers. Multigenerational biomaterials screens have so far manually curated candidates for each stage, while directed evolution (DE) experiments diversify and recombine the fittest candidates in a stochastic fashion. Adoption of this strategy, coupled with less stringent selection criteria over more generations, may permit tolerance of intermediate, low-fitness stages as the screen moves away from local optima, and a more effective exploration of the potential polymer space.
By contrast, design-based approaches are most useful when an underlying structure-function relationship is already known or hypothesized. New classes of materials that exhibit complex physical behaviours are emerging and can be used to further probe how cells respond their biophysical microenvironment. For example, structural components of cells and ECM are viscoelastic in nature, yet most synthetic materials used in cell culture display only the elastic component. Materials that exhibit stress relaxation (reduced stress under constant strain) promote cell spreading on soft substrates comparable to that of cells on stiff, elastic substrates (Chaudhuri et al., 2015a), and hMSCs cultured in 3D gels of increasing stress relaxation rates exhibit enhanced osteogenic differentiation (Chaudhuri et al., 2015b). Synthetic materials that exhibit stress stiffening (increased stiffness when a particular stress is applied) mimic the temporal response of gels formed from cytoskeletal and ECM components, and the onset of stiffening can be tuned by changing the polymer length (Kouwer et al., 2013). When used as 3D culture substrates, stress-stiffening gels of the same bulk stiffness can determine commitment of hMSCs toward either adipogenic or osteogenic lineages, depending upon the point at which stress-stiffening is experienced (Das et al., 2015). Furthermore, scaffolds fabricated from methacrylated dextran (DexMA) present fibers at the cell-material interface, instead of a flat surface like a classical hydrogel, but still offer user-defined control over cell adhesiveness and mechanical properties (Baker et al., 2015). When cultured on DexMA gels of low fiber stiffness, hMSCs exhibited enhanced spreading and focal adhesion formation due to the ability to physically reorganize the matrix by bringing fibers and the associated adhesive sites into closer proximity to the cell; these effects were strikingly similar to behaviors observed for cells on fibrous collagen substrates, but not homogenous, non-fibrillar hydrogels (Baker et al., 2015). It is difficult to pinpoint a specific relationship between the mechanical properties of these gels and the cellular response, but elucidation of exactly how this outside-in signaling occurs will continue to progress the status of the field.
Dynamic systems that allow for real-time manipulation of substrate properties have demonstrated that cells respond to immediate alterations in the extracellular microenvironment. Hydrogel chemistry can be exploited to increase stiffness by exposure to specific wavelengths of light; when gels underwent an increase in stiffness from 3 to 30 kPa, the spreading of hMSCs was increased along with bias toward osteogenic differentiation (Guvendiren and Burdick, 2012). Taken one step further, the ability to soften hydrogels by exploiting photodegradable crosslinkers has revealed that hMSCs possess a ‘mechanical memory’ by which the duration of culture on a stiff substrate influences the subsequent ability of the cells to adapt and respond to culturing on a soft substrate (Yang et al., 2014). YAP/TAZ responsiveness in this hydrogel system could be attenuated after seven days pre-culture on a stiff substrate and was lost completely if the cells were cultured on a stiff substrate for 10 days. When translated into 3D scaffolds, additional responsive cues can be added to hydrogel systems to allow for the study and exploitation of cell behavior. The ability to degrade the surrounding matrix directly affects hMSC fate decisions by (dis)allowing force generation; non-degradable gels inhibit cell spreading and traction force generation, biasing the cells toward adipogenic differentiation, while matrix metalloproteinase (MMP)-degradable gels allow for cell spreading and force generation in a microenvironment that favors osteogenesis (Khetan et al., 2013). Patterning the 3D environment using elegant chemistry and two-photon microscopy has provided a means for specific tailoring of material properties and protein presentation with high spatial resolution. This technique has proven useful for controlling the dynamic addition and removal of the Notch ligand, Delta, and vitronectin to guide stem cell fate (DeForest and Tirrell, 2015). Next-generation material systems that are tailorable in real- time are promising platforms for elucidating the response of cells cultured in/on them and will allow for more complex biological questions to be addressed.
Advances in biomaterial fabrication techniques have made it possible to create nanostructures with precise geometry, textures and rigidity. An emerging class of very interesting nanomaterials for cell-interfacing are nanoneedles and nanopillars. These can be produced in different formats from nanostraws with a hollow core (VanDersarl et al., 2012) to high-aspect ratio porous needles (Chiappini et al., 2015b). These materials have shown tremendous potential for biological applications such as delivery of biomolecules, electrophysiological measurements of neuronal cells, and pH sensing (Chiappini et al., 2015a; VanDersarl et al., 2012). We found that our newly developed class of porous silicon nanoneedles was not only effective at cellular interfacing for delivery to the cells and biosensing applications, but its application in vivo did not elicit any notable inflammatory response. (Chiappini et al., 2015b; Chiappini et al., 2015c). Since nanoneedles can modify the shape of both the cell and the nuclear membrane, they will be of future interest in studying associated effects on epigenetics.
Conclusion
Within solid tissues, there is no such thing as an isolated cell; all cells exist interconnected in a tensile network that they both secrete and comprise. Unified movements of cell sheets and deposition of extracellular matrix are critical events in development. For tissue stem cells such as MSCs, and even for cells such as ESCs and iPSCs that lack true stable physiological counterparts, the finding that mechanosensing and other physical cues exert a control on fate is, in retrospect, hardly surprising. As the field progresses, details of the signal transduction pathways and epigenetic changes underlying these events are starting to emerge. Cross-disciplinary collaboration and increased adoption of tractable materials systems by biologically focused research groups are both likely to accelerate development of an increasingly complete understanding of these events. In parallel, new emerging materials systems will provide increasingly sophisticated investigative and therapeutic tools. We suggest that together, these strands of research have great potential to both illuminate basic biological questions on stem cell decision-making, and inform progress towards increasingly relevant models of disease and effective regenerative therapies.
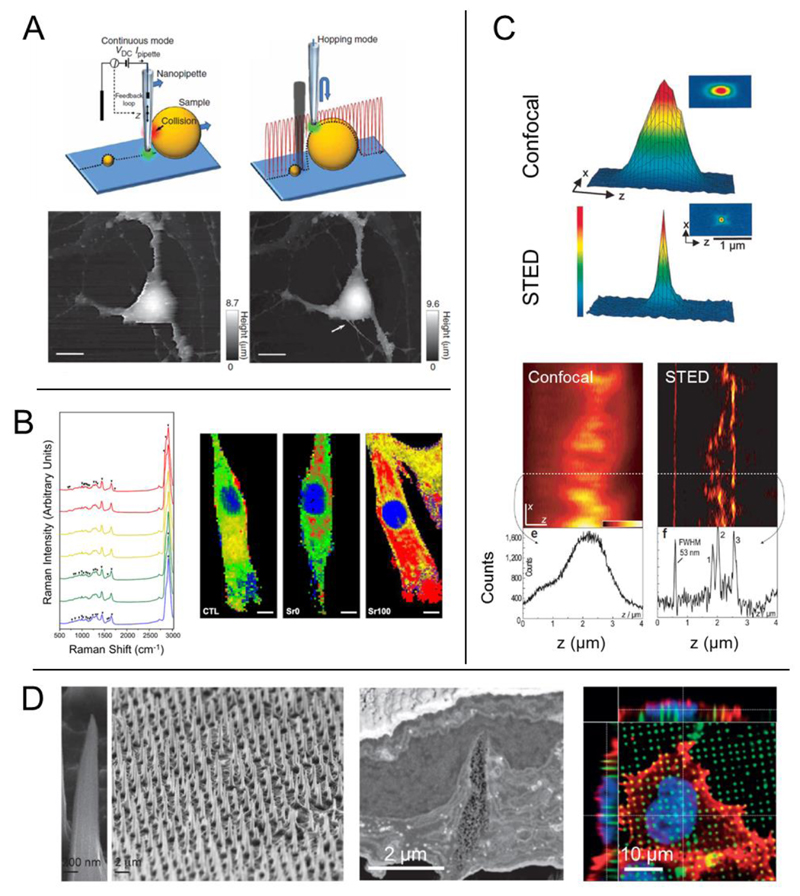
Figure 4. Promising research tools – characterization techniques, imaging modalities, and novel materials.
(A) Scanning ion conductance microscopy (SICM) uses a nanopipette to produce an image of a living cell without physical interaction. Two different modes of SICM are shown here: continuous mode in which the probe does not significantly change its z-position, and hopping mode in which the probe is constantly approaching the sample and then retreating. In hopping mode, small features that are not observed with continuous mode can become spatially resolved (Scale bars, 10 μm). (B) Raman spectroscopy produces a chemical ‘fingerprint’ of the cell(s) or tissue that is studied, providing information about the specific chemical bonds present in the sample. This information can then be used to produce a 2D or 3D map that shows the localization of certain classes of molecules. For example, bioactive glass materials with a significant amount of strontium ion (Sr100) lead to a shift in the lipid distribution in MSCs (lipid, red) (Scale bars, 10 μm). (C) Stimulated emission depletion (STED) microscopy overcomes the diffraction limit typically associated with light-based imaging techniques. The focal point of a STED microscope is much smaller (bottom graph) than that of a standard confocal (top graph). This allows for enhanced spatial resolution, such as imaging the microtubule network in monolayer cell culture. Typical confocal (left image) produces an image in which the network cannot be observed, but sub-diffraction STED imaging allows for crisp visualization of the subcellular processes. (D) High-aspect ratio nanoneedles can be produced into high density, 2D arrays to act as cell culture substrates. HeLa cells cultured on nanoneedles interact directly with the underlying material, and the needles physically deform both the cell membrane as well as the nucleus. Figures adapted with permission from: (A) Nature Publishing Group (NPG) Ref: (Novak et al., 2009); (B) Ref: (Autefage et al., 2015); (C) NPG Ref: (Dyba et al., 2003); (D) Reprinted with permission from Ref: (Chiappini et al., 2015c). Copyright 2015 American Chemical Society; NPG Ref: (Chiappini et al., 2015b), and Ref: (Chiappini et al., 2015a).
Figure 5.
Acknowledgments
M.M.S. acknowledges support from the United Kingdom Medical Research Council Hub “Acellular Approaches for Therapeutic Delivery” (MR/K026682/1), which is funded by the Medical Research Council, the Engineering and Physical Sciences Research Council, and the Biotechnology and Biological Sciences Research Council. M.M.S. is also funded by a Wellcome Trust Senior Investigator Award (098411/Z/12/Z) and the ERC Seventh Framework Programme Consolidator grant “Naturale CG” under grant agreement no. 616417. S.W.C. was also supported by the Whitaker International Program, Institute of International Education, United States of America. T.W. is supported by an EPSRC DTA Ph.D. award.
References
- Alabert C, Bukowski-Wills JC, Lee SB, Kustatscher G, Nakamura K, de Lima Alves F, Menard P, Mejlvang J, Rappsilber J, Groth A. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat Cell Biol. 2014;16:281–293. doi: 10.1038/ncb2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DG, Levenberg S, Langer R. Nanoliter-scale synthesis of arrayed biomaterials and application to human embryonic stem cells. Nat Biotechnol. 2004;22:863–866. doi: 10.1038/nbt981. [DOI] [PubMed] [Google Scholar]
- Autefage H, Gentleman E, Littmann E, Hedegaard MA, Von Erlach T, O'Donnell M, Burden FR, Winkler DA, Stevens MM. Sparse feature selection methods identify unexpected global cellular response to strontium-containing materials. Proc Natl Acad Sci U S A. 2015;112:4280–4285. doi: 10.1073/pnas.1419799112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BM, Trappmann B, Wang WY, Sakar MS, Kim IL, Shenoy VB, Burdick JA, Chen CS. Cell-mediated fibre recruitment drives extracellular matrix mechanosensing in engineered fibrillar microenvironments. Nat Mater. 2015;14:1262–1268. doi: 10.1038/nmat4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian L, Guvendiren M, Mauck RL, Burdick JA. Hydrogels that mimic developmentally relevant matrix and N-cadherin interactions enhance MSC chondrogenesis. Proc Natl Acad Sci U S A. 2013;110:10117–10122. doi: 10.1073/pnas.1214100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boo YC. Shear stress stimulates phosphorylation of protein kinase A substrate proteins including endothelial nitric oxide synthase in endothelial cells. Exp Mol Med. 2006;38:453. doi: 10.1038/emm.2006.53. [DOI] [PubMed] [Google Scholar]
- Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC, Martin RP, Schipani E, Divieti P, Bringhurst FR, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- Case N, Ma M, Sen B, Xie Z, Gross TS, Rubin J. Beta-catenin levels influence rapid mechanical responses in osteoblasts. The Journal of biological chemistry. 2008;283:29196–29205. doi: 10.1074/jbc.M801907200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celiz AD, Smith JG, Langer R, Anderson DG, Winkler DA, Barrett DA, Davies MC, Young LE, Denning C, Alexander MR. Materials for stem cell factories of the future. Nature materials. 2014;13:570–579. doi: 10.1038/nmat3972. [DOI] [PubMed] [Google Scholar]
- Celiz AD, Smith JG, Patel AK, Hook AL, Rajamohan D, George VT, Flatt L, Patel MJ, Epa VC, Singh T, et al. Discovery of a Novel Polymer for Human Pluripotent Stem Cell Expansion and Multilineage Differentiation. Advanced materials. 2015 doi: 10.1002/adma.201501351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan HF, Zhang Y, Ho YP, Chiu YL, Jung Y, Leong KW. Rapid formation of multicellular spheroids in double-emulsion droplets with controllable microenvironment. Sci Rep. 2013;3:3462. doi: 10.1038/srep03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chancellor TJ, Lee J, Thodeti CK, Lele T. Actomyosin tension exerted on the nucleus through nesprin-1 connections influences endothelial cell adhesion, migration, and cyclic strain-induced reorientation. Biophys J. 2010;99:115–123. doi: 10.1016/j.bpj.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri O, Gu L, Darnell M, Klumpers D, Bencherif SA, Weaver JC, Huebsch N, Mooney DJ. Substrate stress relaxation regulates cell spreading. Nat Commun. 2015a;6:6364. doi: 10.1038/ncomms7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri O, Gu L, Klumpers D, Darnell M, Bencherif SA, Weaver JC, Huebsch N, Lee HP, Lippens E, Duda GN, et al. Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nat Mater. 2015b doi: 10.1038/nmat4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BC, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, Janetopoulos C, Wu XS, Hammer JA, 3rd, Liu Z, et al. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science. 2014;346:1257998. doi: 10.1126/science.1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappini C, Campagnolo P, Almeida CS, Abbassi-Ghadi N, Chow LW, Hanna GB, Stevens MM. Mapping Local Cytosolic Enzymatic Activity in Human Esophageal Mucosa with Porous Silicon Nanoneedles. Advanced materials. 2015a doi: 10.1002/adma.201501304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappini C, De Rosa E, Martinez JO, Liu X, Steele J, Stevens MM, Tasciotti E. Biodegradable silicon nanoneedles delivering nucleic acids intracellularly induce localized in vivo neovascularization. Nat Mater. 2015b;14:532–539. doi: 10.1038/nmat4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappini C, Martinez JO, De Rosa E, Almeida CS, Tasciotti E, Stevens MM. Biodegradable nanoneedles for localized delivery of nanoparticles in vivo: exploring the biointerface. ACS Nano. 2015c;9:5500–5509. doi: 10.1021/acsnano.5b01490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway A, Vazin T, Spelke DP, Rode NA, Healy KE, Kane RS, Schaffer DV. Multivalent ligands control stem cell behaviour in vitro and in vivo. Nat Nanotechnol. 2013;8:831–838. doi: 10.1038/nnano.2013.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross SE, Jin YS, Rao J, Gimzewski JK. Nanomechanical analysis of cells from cancer patients. Nat Nanotechnol. 2007;2:780–783. doi: 10.1038/nnano.2007.388. [DOI] [PubMed] [Google Scholar]
- Dalby MJ, Gadegaard N, Tare R, Andar A, Riehle MO, Herzyk P, Wilkinson CD, Oreffo RO. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nat Mater. 2007;6:997–1003. doi: 10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]
- Danciu TE, Adam RM, Naruse K, Freeman MR, Hauschka PV. Calcium regulates the PI3K-Akt pathway in stretched osteoblasts. FEBS Lett. 2003;536:193–197. doi: 10.1016/s0014-5793(03)00055-3. [DOI] [PubMed] [Google Scholar]
- Das RK, Gocheva V, Hammink R, Zouani OF, Rowan AE. Stress-stiffening-mediated stem-cell commitment switch in soft responsive hydrogels. Nat Mater. 2015 doi: 10.1038/nmat4483. [DOI] [PubMed] [Google Scholar]
- De Arcangelis A, Georges-Labouesse E. Integrin and ECM functions: roles in vertebrate development. Trends Genet. 2000;16:389–395. doi: 10.1016/s0168-9525(00)02074-6. [DOI] [PubMed] [Google Scholar]
- DeForest CA, Tirrell DA. A photoreversible protein-patterning approach for guiding stem cell fate in three-dimensional gels. Nat Mater. 2015;14:523–531. doi: 10.1038/nmat4219. [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W, et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518 doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing TL, Soto J, Morez C, Houssin T, Fritz A, Yuan F, Chu J, Patel S, Schaffer DV, Li S. Biophysical regulation of epigenetic state and cell reprogramming. Nat Mater. 2013;12:1154–1162. doi: 10.1038/nmat3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- Dyba M, Jakobs S, Hell SW. Immunofluorescence stimulated emission depletion microscopy. Nat Biotechnol. 2003;21:1303–1304. doi: 10.1038/nbt897. [DOI] [PubMed] [Google Scholar]
- El-Said WA, Kim SU, Choi JW. Monitoring in vitro neural stem cell differentiation based on surface-enhanced Raman spectroscopy using a gold nanostar array. J Mater Chem C. 2015;3:3848–3859. [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Flynn RA, Chang HY. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 2014;14:752–761. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman E, Swain RJ, Evans ND, Boonrungsiman S, Jell G, Ball MD, Shean TA, Oyen ML, Porter A, Stevens MM. Comparative materials differences revealed in engineered bone as a function of cell-specific differentiation. Nat Mater. 2009;8:763–770. doi: 10.1038/nmat2505. [DOI] [PubMed] [Google Scholar]
- Gudi S, Nolan JP, Frangos JA. Modulation of GTPase activity of G proteins by fluid shear stress and phospholipid composition. Proc Natl Acad Sci U S A. 1998;95:2515–2519. doi: 10.1073/pnas.95.5.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guvendiren M, Burdick JA. Stiffening hydrogels to probe short- and long-term cellular responses to dynamic mechanics. Nat Commun. 2012;3:792. doi: 10.1038/ncomms1792. [DOI] [PubMed] [Google Scholar]
- Holst J, Watson S, Lord MS, Eamegdool SS, Bax DV, Nivison-Smith LB, Kondyurin A, Ma L, Oberhauser AF, Weiss AS, et al. Substrate elasticity provides mechanical signals for the expansion of hemopoietic stem and progenitor cells. Nat Biotechnol. 2010;28:1123–1128. doi: 10.1038/nbt.1687. [DOI] [PubMed] [Google Scholar]
- Hook AL, Chang CY, Yang J, Atkinson S, Langer R, Anderson DG, Davies MC, Williams P, Alexander MR. Discovery of novel materials with broad resistance to bacterial attachment using combinatorial polymer microarrays. Advanced materials. 2013;25:2542–2547. doi: 10.1002/adma.201204936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huddleson JP, Ahmad N, Srinivasan S, Lingrel JB. Induction of KLF2 by fluid shear stress requires a novel promoter element activated by a phosphatidylinositol 3-kinase-dependent chromatin-remodeling pathway. The Journal of biological chemistry. 2005;280:23371–23379. doi: 10.1074/jbc.M413839200. [DOI] [PubMed] [Google Scholar]
- Humphrey JD, Dufresne ER, Schwartz MA. Mechanotransduction and extracellular matrix homeostasis. Nature reviews Molecular cell biology. 2014;15:802–812. doi: 10.1038/nrm3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihalainen TO, Aires L, Herzog FA, Schwartlander R, Moeller J, Vogel V. Differential basal-to-apical accessibility of lamin A/C epitopes in the nuclear lamina regulated by changes in cytoskeletal tension. Nat Mater. 2015;14:1252–1261. doi: 10.1038/nmat4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illi B, Nanni S, Scopece A, Farsetti A, Biglioli P, Capogrossi MC, Gaetano C. Shear stress-mediated chromatin remodeling provides molecular basis for flow-dependent regulation of gene expression. Circulation research. 2003;93:155–161. doi: 10.1161/01.RES.0000080933.82105.29. [DOI] [PubMed] [Google Scholar]
- Illi B, Scopece A, Nanni S, Farsetti A, Morgante L, Biglioli P, Capogrossi MC, Gaetano C. Epigenetic histone modification and cardiovascular lineage programming in mouse embryonic stem cells exposed to laminar shear stress. Circulation research. 2005;96:501–508. doi: 10.1161/01.RES.0000159181.06379.63. [DOI] [PubMed] [Google Scholar]
- Jain N, Iyer KV, Kumar A, Shivashankar GV. Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility. Proc Natl Acad Sci U S A. 2013;110:11349–11354. doi: 10.1073/pnas.1300801110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans DC, Wurm CA, Riedel D, Wenzel D, Stagge F, Deckers M, Rehling P, Jakobs S. STED super-resolution microscopy reveals an array of MINOS clusters along human mitochondria. Proc Natl Acad Sci U S A. 2013;110:8936–8941. doi: 10.1073/pnas.1301820110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khetan S, Guvendiren M, Legant WR, Cohen DM, Chen CS, Burdick JA. Degradation-mediated cellular traction directs stem cell fate in covalently crosslinked three-dimensional hydrogels. Nat Mater. 2013;12:458–465. doi: 10.1038/nmat3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian KA, Bugarija B, Lahn BT, Mrksich M. Geometric cues for directing the differentiation of mesenchymal stem cells. Proc Natl Acad Sci U S A. 2010;107:4872–4877. doi: 10.1073/pnas.0903269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SN, Jeibmann A, Halama K, Witte HT, Walte M, Matzat T, Schillers H, Faber C, Senner V, Paulus W, et al. ECM stiffness regulates glial migration in Drosophila and mammalian glioma models. Development. 2014;141:3233–3242. doi: 10.1242/dev.106039. [DOI] [PubMed] [Google Scholar]
- Klim JR, Li L, Wrighton PJ, Piekarczyk MS, Kiessling LL. A defined glycosaminoglycan-binding substratum for human pluripotent stem cells. Nat Methods. 2010;7:989–994. doi: 10.1038/nmeth.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko KS, Arora PD, McCulloch CA. Cadherins mediate intercellular mechanical signaling in fibroblasts by activation of stretch-sensitive calcium-permeable channels. The Journal of biological chemistry. 2001;276:35967–35977. doi: 10.1074/jbc.M104106200. [DOI] [PubMed] [Google Scholar]
- Kocgozlu L, Lavalle P, Koenig G, Senger B, Haikel Y, Schaaf P, Voegel JC, Tenenbaum H, Vautier D. Selective and uncoupled role of substrate elasticity in the regulation of replication and transcription in epithelial cells. Journal of cell science. 2010;123:29–39. doi: 10.1242/jcs.053520. [DOI] [PubMed] [Google Scholar]
- Korchev YE, Bashford CL, Milovanovic M, Vodyanoy I, Lab MJ. Scanning ion conductance microscopy of living cells. Biophys J. 1997;73:653–658. doi: 10.1016/S0006-3495(97)78100-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouwer PH, Koepf M, Le Sage VA, Jaspers M, van Buul AM, Eksteen-Akeroyd ZH, Woltinge T, Schwartz E, Kitto HJ, Hoogenboom R, et al. Responsive biomimetic networks from polyisocyanopeptide hydrogels. Nature. 2013;493:651–655. doi: 10.1038/nature11839. [DOI] [PubMed] [Google Scholar]
- Lammerding J, Hsiao J, Schulze PC, Kozlov S, Stewart CL, Lee RT. Abnormal nuclear shape and impaired mechanotransduction in emerin-deficient cells. The Journal of cell biology. 2005;170:781–791. doi: 10.1083/jcb.200502148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugesen A, Helin K. Chromatin repressive complexes in stem cells, development, and cancer. Cell Stem Cell. 2014;14:735–751. doi: 10.1016/j.stem.2014.05.006. [DOI] [PubMed] [Google Scholar]
- Le Beyec J, Xu R, Lee SY, Nelson CM, Rizki A, Alcaraz J, Bissell MJ. Cell shape regulates global histone acetylation in human mammary epithelial cells. Experimental cell research. 2007;313:3066–3075. doi: 10.1016/j.yexcr.2007.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DY, Lee CI, Lin TE, Lim SH, Zhou J, Tseng YC, Chien S, Chiu JJ. Role of histone deacetylases in transcription factor regulation and cell cycle modulation in endothelial cells in response to disturbed flow. Proc Natl Acad Sci U S A. 2012;109:1967–1972. doi: 10.1073/pnas.1121214109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Hore TA, Reik W. Reprogramming the methylome: erasing memory and creating diversity. Cell Stem Cell. 2014;14:710–719. doi: 10.1016/j.stem.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenberg S, Huang NF, Lavik E, Rogers AB, Itskovitz-Eldor J, Langer R. Differentiation of human embryonic stem cells on three-dimensional polymer scaffolds. Proc Natl Acad Sci U S A. 2003;100:12741–12746. doi: 10.1073/pnas.1735463100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Klim JR, Derda R, Courtney AH, Kiessling LL. Spatial control of cell fate using synthetic surfaces to potentiate TGF-beta signaling. Proc Natl Acad Sci U S A. 2011a;108:11745–11750. doi: 10.1073/pnas.1101454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Xie T. Stem cell niche: structure and function. Annual review of cell and developmental biology. 2005;21:605–631. doi: 10.1146/annurev.cellbio.21.012704.131525. [DOI] [PubMed] [Google Scholar]
- Li Y, Chu JS, Kurpinski K, Li X, Bautista DM, Yang L, Sung KL, Li S. Biophysical regulation of histone acetylation in mesenchymal stem cells. Biophys J. 2011b;100:1902–1909. doi: 10.1016/j.bpj.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Schneider M, Neumann S, Jaeger VM, Taranum S, Munck M, Cartwright S, Richardson C, Carthew J, Noh K, et al. Nesprin interchain associations control nuclear size. Cell Mol Life Sci. 2012;69:3493–3509. doi: 10.1007/s00018-012-1034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutolf MP, Lauer-Fields JL, Schmoekel HG, Metters AT, Weber FE, Fields GB, Hubbell JA. Synthetic matrix metalloproteinase-sensitive hydrogels for the conduction of tissue regeneration: engineering cell-invasion characteristics. Proc Natl Acad Sci U S A. 2003;100:5413–5418. doi: 10.1073/pnas.0737381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabry KM, Payne SZ, Anseth KS. Microarray analyses to quantify advantages of 2D and 3D hydrogel culture systems in maintaining the native valvular interstitial cell phenotype. Biomaterials. 2016;74:31–41. doi: 10.1016/j.biomaterials.2015.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majkut S, Idema T, Swift J, Krieger C, Liu A, Discher DE. Heart-specific stiffening in early embryos parallels matrix and myosin expression to optimize beating. Curr Biol. 2013;23:2434–2439. doi: 10.1016/j.cub.2013.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci U S A. 1997;94:849–854. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroto R, Raso A, Wood TG, Kurosky A, Martinac B, Hamill OP. TRPC1 forms the stretch-activated cation channel in vertebrate cells. Nat Cell Biol. 2005;7:179–185. doi: 10.1038/ncb1218. [DOI] [PubMed] [Google Scholar]
- Mauck RL, Yuan X, Tuan RS. Chondrogenic differentiation and functional maturation of bovine mesenchymal stem cells in long-term agarose culture. Osteoarthritis Cartilage. 2006;14:179–189. doi: 10.1016/j.joca.2005.09.002. [DOI] [PubMed] [Google Scholar]
- McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- McMurray RJ, Gadegaard N, Tsimbouri PM, Burgess KV, McNamara LE, Tare R, Murawski K, Kingham E, Oreffo RO, Dalby MJ. Nanoscale surfaces for the long-term maintenance of mesenchymal stem cell phenotype and multipotency. Nat Mater. 2011;10:637–644. doi: 10.1038/nmat3058. [DOI] [PubMed] [Google Scholar]
- Mei Y, Saha K, Bogatyrev SR, Yang J, Hook AL, Kalcioglu ZI, Cho SW, Mitalipova M, Pyzocha N, Rojas F, et al. Combinatorial development of biomaterials for clonal growth of human pluripotent stem cells. Nat Mater. 2010;9:768–778. doi: 10.1038/nmat2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morez C, Noseda M, Paiva MA, Belian E, Schneider MD, Stevens MM. Enhanced efficiency of genetic programming toward cardiomyocyte creation through topographical cues. Biomaterials. 2015;70:94–104. doi: 10.1016/j.biomaterials.2015.07.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notingher I, Bisson I, Bishop AE, Randle WL, Polak JM, Hench LL. In situ spectral monitoring of mRNA translation in embryonic stem cells during differentiation in vitro. Anal Chem. 2004;76:3185–3193. doi: 10.1021/ac0498720. [DOI] [PubMed] [Google Scholar]
- Novak P, Li C, Shevchuk AI, Stepanyan R, Caldwell M, Hughes S, Smart TG, Gorelik J, Ostanin VP, Lab MJ, et al. Nanoscale live-cell imaging using hopping probe ion conductance microscopy. Nat Methods. 2009;6:279–281. doi: 10.1038/nmeth.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmakumar VC, Abraham S, Braune S, Noegel AA, Tunggal B, Karakesisoglou I, Korenbaum E. Enaptin, a giant actin-binding protein, is an element of the nuclear membrane and the actin cytoskeleton. Experimental cell research. 2004;295:330–339. doi: 10.1016/j.yexcr.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Peng XQ, Damarla M, Skirball J, Nonas S, Wang XY, Han EJ, Hasan EJ, Cao X, Boueiz A, Damico R, et al. Protective role of PI3-kinase/Akt/eNOS signaling in mechanical stress through inhibition of p38 mitogen-activated protein kinase in mouse lung. Acta Pharmacol Sin. 2010;31:175–183. doi: 10.1038/aps.2009.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranga A, Gobaa S, Okawa Y, Mosiewicz K, Negro A, Lutolf MP. 3D niche microarrays for systems-level analyses of cell fate. Nat Commun. 2014;5:4324. doi: 10.1038/ncomms5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero PA, Arnold FH. Exploring protein fitness landscapes by directed evolution. Nature reviews Molecular cell biology. 2009;10:866–876. doi: 10.1038/nrm2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux KJ, Crisp ML, Liu Q, Kim D, Kozlov S, Stewart CL, Burke B. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc Natl Acad Sci U S A. 2009;106:2194–2199. doi: 10.1073/pnas.0808602106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchuk AI, Frolenkov GI, Sanchez D, James PS, Freedman N, Lab MJ, Jones R, Klenerman D, Korchev YE. Imaging proteins in membranes of living cells by high-resolution scanning ion conductance microscopy. Angew Chem Int Ed Engl. 2006;45:2212–2216. doi: 10.1002/anie.200503915. [DOI] [PubMed] [Google Scholar]
- Shin JW, Buxboim A, Spinler KR, Swift J, Christian DA, Hunter CA, Leon C, Gachet C, Dingal PC, Ivanovska IL, et al. Contractile forces sustain and polarize hematopoiesis from stem and progenitor cells. Cell Stem Cell. 2014;14:81–93. doi: 10.1016/j.stem.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon CG, Jr, Lin-Gibson S. Combinatorial and high-throughput screening of biomaterials. Advanced materials. 2011;23:369–387. doi: 10.1002/adma.201001763. [DOI] [PubMed] [Google Scholar]
- Stevens MM, George JH. Exploring and engineering the cell surface interface. Science. 2005;310:1135–1138. doi: 10.1126/science.1106587. [DOI] [PubMed] [Google Scholar]
- Swain RJ, Kemp SJ, Goldstraw P, Tetley TD, Stevens MM. Assessment of cell line models of primary human cells by Raman spectral phenotyping. Biophys J. 2010;98:1703–1711. doi: 10.1016/j.bpj.2009.12.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PC, Pinter J, Pajerowski JD, Spinler KR, Shin JW, Tewari M, et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 2013;341:1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sydor AM, Czymmek KJ, Puchner EM, Mennella V. Super-Resolution Microscopy: From Single Molecules to Supramolecular Assemblies. Trends in cell biology. 2015;25:730–748. doi: 10.1016/j.tcb.2015.10.004. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Berk BC. Mitogen-activated protein kinase (ERK1/2) activation by shear stress and adhesion in endothelial cells. Essential role for a herbimycin-sensitive kinase. J Clin Invest. 1996;98:2623–2631. doi: 10.1172/JCI119083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Ogasawara T, Asawa Y, Mori Y, Uchinuma E, Takato T, Hoshi K. Three-dimensional microenvironments retain chondrocyte phenotypes during proliferation culture. Tissue Eng. 2007;13:1583–1592. doi: 10.1089/ten.2006.0322. [DOI] [PubMed] [Google Scholar]
- Tsimbouri PM, McMurray RJ, Burgess KV, Alakpa EV, Reynolds PM, Murawski K, Kingham E, Oreffo RO, Gadegaard N, Dalby MJ. Using nanotopography and metabolomics to identify biochemical effectors of multipotency. ACS Nano. 2012;6:10239–10249. doi: 10.1021/nn304046m. [DOI] [PubMed] [Google Scholar]
- Tzima E, Irani-Tehrani M, Kiosses WB, Dejana E, Schultz DA, Engelhardt B, Cao G, DeLisser H, Schwartz MA. A mechanosensory complex that mediates the endothelial cell response to fluid shear stress. Nature. 2005;437:426–431. doi: 10.1038/nature03952. [DOI] [PubMed] [Google Scholar]
- Unadkat HV, Hulsman M, Cornelissen K, Papenburg BJ, Truckenmuller RK, Carpenter AE, Wessling M, Post GF, Uetz M, Reinders MJ, et al. An algorithm-based topographical biomaterials library to instruct cell fate. Proc Natl Acad Sci U S A. 2011;108:16565–16570. doi: 10.1073/pnas.1109861108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzer G, Thompson WR, Sen B, Xie Z, Yen SS, Miller S, Bas G, Styner M, Rubin CT, Judex S, et al. Cell Mechanosensitivity to Extremely Low-Magnitude Signals Is Enabled by a LINCed Nucleus. Stem Cells. 2015;33:2063–2076. doi: 10.1002/stem.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDersarl JJ, Xu AM, Melosh NA. Nanostraws for direct fluidic intracellular access. Nano Lett. 2012;12:3881–3886. doi: 10.1021/nl204051v. [DOI] [PubMed] [Google Scholar]
- von Erlach TC, Hedegaard MA, Stevens MM. High resolution Raman spectroscopy mapping of stem cell micropatterns. Analyst. 2015;140:1798–1803. doi: 10.1039/c4an02346c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Naruse K, Stamenovic D, Fredberg JJ, Mijailovich SM, Tolic-Norrelykke IM, Polte T, Mannix R, Ingber DE. Mechanical behavior in living cells consistent with the tensegrity model. Proc Natl Acad Sci U S A. 2001;98:7765–7770. doi: 10.1073/pnas.141199598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Tytell JD, Ingber DE. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nature reviews Molecular cell biology. 2009a;10:75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- Wang W, Itaka K, Ohba S, Nishiyama N, Chung UI, Yamasaki Y, Kataoka K. 3D spheroid culture system on micropatterned substrates for improved differentiation efficiency of multipotent mesenchymal stem cells. Biomaterials. 2009b;30:2705–2715. doi: 10.1016/j.biomaterials.2009.01.030. [DOI] [PubMed] [Google Scholar]
- Wen JH, Vincent LG, Fuhrmann A, Choi YS, Hribar KC, Taylor-Weiner H, Chen S, Engler AJ. Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nat Mater. 2014;13:979–987. doi: 10.1038/nmat4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmsen K, Litjens SH, Kuikman I, Tshimbalanga N, Janssen H, van den Bout I, Raymond K, Sonnenberg A. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. The Journal of cell biology. 2005;171:799–810. doi: 10.1083/jcb.200506083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrighton PJ, Klim JR, Hernandez BA, Koonce CH, Kamp TJ, Kiessling LL. Signals from the surface modulate differentiation of human pluripotent stem cells through glycosaminoglycans and integrins. Proc Natl Acad Sci U S A. 2014;111:18126–18131. doi: 10.1073/pnas.1409525111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan ZQ, Yao QP, Zhang ML, Qi YX, Guo ZY, Shen BR, Jiang ZL. Histone deacetylases modulate vascular smooth muscle cell migration induced by cyclic mechanical strain. Journal of biomechanics. 2009;42:945–948. doi: 10.1016/j.jbiomech.2009.01.012. [DOI] [PubMed] [Google Scholar]
- Yang C, Tibbitt MW, Basta L, Anseth KS. Mechanical memory and dosing influence stem cell fate. Nat Mater. 2014;13:645–652. doi: 10.1038/nmat3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Liberski A, Sanchez-Martin R, Bradley M. Microarrays of over 2000 hydrogels--identification of substrates for cellular trapping and thermally triggered release. Biomaterials. 2009;30:6193–6201. doi: 10.1016/j.biomaterials.2009.07.055. [DOI] [PubMed] [Google Scholar]
- Zhen YY, Libotte T, Munck M, Noegel AA, Korenbaum E. NUANCE, a giant protein connecting the nucleus and actin cytoskeleton. Journal of cell science. 2002;115:3207–3222. doi: 10.1242/jcs.115.15.3207. [DOI] [PubMed] [Google Scholar]
- Zhou J, Li YS, Wang KC, Chien S. Epigenetic Mechanism in Regulation of Endothelial Function by Disturbed Flow: Induction of DNA Hypermethylation by DNMT1. Cellular and molecular bioengineering. 2014;7:218–224. doi: 10.1007/s12195-014-0325-z. [DOI] [PMC free article] [PubMed] [Google Scholar]