Abstract
Cells migrate in vivo within three-dimensional (3D) extracellular matrices. Cells also migrate through 3D longitudinal channels formed between the connective tissue and the basement membrane of muscle, nerve, and epithelium. Although traction forces have been measured during 2D cell migration, no assay has been developed to probe forces during migration through confined microenvironments. We thus fabricated a novel microfluidic device consisting of deflectable PDMS microposts incorporated within microchannels of varying cross-sectional areas. Using NIH-3T3 fibroblasts and human osteosarcoma (HOS) cells as models, we found that the average traction forces per post decreased upon increasing confinement. Inhibition of myosin-II function by blebbistatin in HOS cells decreased traction forces in unconfined (wide) channels but failed to alter them in confined spaces. Myosin activation by calyculin A also failed to affect traction forces in confining channels but increased them in wide channels. These observations underlie the importance of the physical microenvironment in the regulation of cell migration and cellular traction forces.
Introduction
Cell migration plays a key role in numerous physiological and pathological processes, including inflammation, angiogenesis, wound healing, and cancer metastasis.1–4 On 2-dimensional (2D) surfaces, mesenchymal cell migration is characterized by dorsoventral polarization, adhesion-stabilized protrusion of actin-based structures, traction-mediated forward movement, and detachment and retraction of the trailing edge.5 The forces exerted by cells on planar substrates are generated in the cell by the action of myosin-II on the actin cytoskeleton through integrin–extracellular matrix (ECM) linkages (focal adhesions; FAs).6 Proper regulation of cellular traction forces is indispensable for mechanosensing phenomena, including responses of cell migration.7,8
Due to the ubiquity of mechanical interactions between cells and their underlying substrates and the role these forces play in cell migration, it is important to characterize and quantify cell traction forces. Adherent cells exert nanonewton (nN)-scale traction forces that vary locally over the cell.9,10 Quantification of traction forces provides valuable insights about cellular response to mechanical perturbations and available spreading area.9,11–13 Two current methods to measure 2D cell traction forces rely on quantifying (a) the deflection of microposts on which cells spread and migrate,9–11,13 or (b) the displacement of fluorescent marker beads within a deformable gel.12,14–16 Using these assays, it is possible to track the time-dependent forces exerted by migrating cells.9,17,18
Extension of traction force assays to 2.5D and 3D settings is important to understand migration in the complex micro-environments found in the body. While recent progress has been made in measuring 2.5D traction forces exerted by cells sandwiched between two hydrogels19,20 or within 3D collagen gels,21,22 no assay has yet been developed to probe forces during physiologically-relevant migration through 3D longitudinal channels. These 3D channels are formed between the connective tissue and the basement membrane of muscle, nerve, and epithelium.23 3D longitudinal channels are also formed between adjacent bundled collagen fibers in fibrillar interstitial tissues.23 Visualization and quantification of collagen organization in mouse and human samples by confocal reflection microscopy and second harmonic generation imaging reveal pore diameters ranging from less than 1 μm up to 20 μm, a wider range than that found in ECM gels.24 Importantly, cells have been reported to migrate through 3D channels in vivo.25 For example, clefts in the ECM bordered by largely parallel collagen fibers24 create lanes to facilitate cell migration.26,27 Muscle fibers and lymph vessels also create potential paths of least resistance for cancer cell movement.25 Co-culture studies have demonstrated that leader fibroblasts remodel the matrix to create tracks through which follower cancer cells move independently of Rho-mediated contractility.27,28 Mesenchymal cancer cells producing active matrix metalloproteinases (MMPs) can function similarly to create pro-migratory tunnels.26,29 Collectively, these studies establish that cells in vivo may also migrate through channels of varying degrees of physical confinement.
We herein developed a microfluidic device which consists of deflectable PDMS microposts incorporated within microchannels of varying cross-sectional areas (Fig. 1). As such, our device is capable of measuring the traction forces exerted by cells migrating through wide (unconfined) and narrow (confined) channels towards a chemotactic stimulus. Importantly, it enables us to investigate how different physical microenvironments akin to those encountered in vivo regulate cell migration and cellular traction forces.
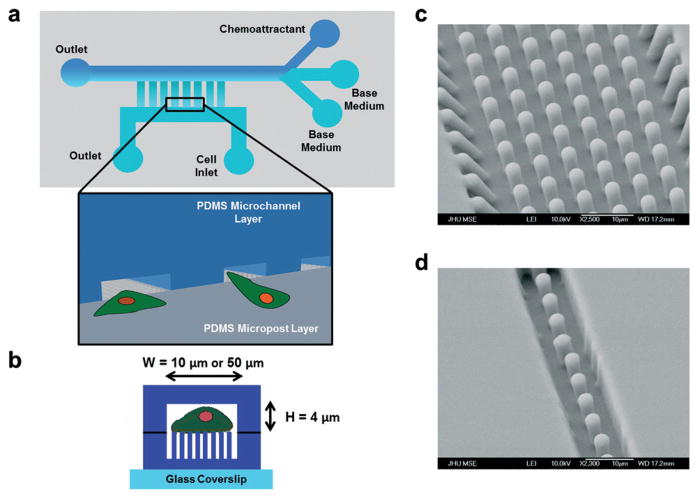
Fig. 1.
Development of a microfluidic device for probing traction forces during cell migration through unconfined and confined microenvironments. The microfluidic device consists of microposts arrayed within microchannels. (a) Schematic of the microfluidic device. Microchannels were aligned in a ladder-like configuration perpendicularly to and connected to two larger main channels serving as the cell seeding inlet and chemokine reservoir. Inset shows a 3D schematic of cells entering the microchannels from the cell seeding area. (b) Side view of a microchannel containing an array of microposts. The microposts were completely enclosed by the channel side walls. Cells migrated within the microchannel (of width W and height H) and on top of the microposts. (c, d) Representative scanning electron micrographs of 3 μm-diameter PDMS microposts within (c) 50 μm-wide and (d) 10 μm-wide microchannels.
Experimental
Cell culture and reagents
Human osteosarcoma cells (HOS.pBABE-puro) were obtained from the NIH AIDS Research and Reference Reagent Program (Division of AIDS, NIAID, NIH, Betheshda, MD). NIH-3T3 cells were obtained from American Type Culture Collection (ATCC; Manassas, VA, USA). Both cell lines were cultured in Dulbecco’s modified Eagle medium (DMEM; Life Technologies, Carlsbad, CA, USA) supplemented with 10% FBS and 1% penicillin–streptomycin (100 U ml−1–100 μg ml−1; Life Technologies) and maintained in a humidified incubator with 5% CO2/95% air at 37 °C. Basal medium was supplemented with 50 μM blebbistatin (Sigma, St. Louis, MO, USA) or 0.1 nM calyculin A (Cell Signaling Technology, Beverly, MA, USA) to inhibit myosin-II or myosin phosphatase, respectively.
Fabrication of the microfluidic device for probing traction forces in unconfined and confined microenvironments
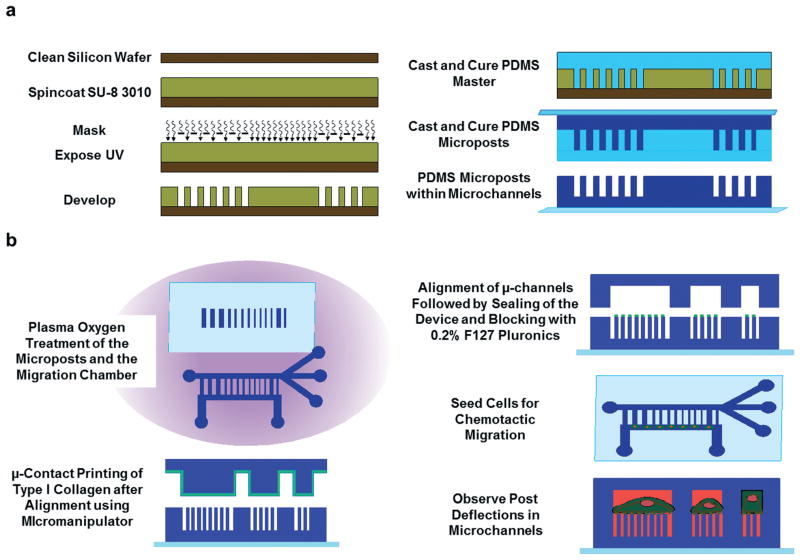
The microfluidic device was fabricated using multilayer photolithography and replica molding; the fabrication scheme is shown schematically in Fig. 2a. Photolithography masks were designed using AutoCAD (Autodesk, McLean, VA) and produced by HTA Photomask (San Jose, CA) or the Photoplot Store (Colorado Springs, CO). The master for the bottom portion of the device was fabricated as a positive replica of the micropost arrays within microchannels30 using SU-8 3010 negative photoresist (Microchem, Newton, MA). SU-8 3010 was spin coated (Single Wafer Spin Processor, Model WS-400A-6NPP-LITE, Laurell Technologies, North Wales, PA) on a clean silicon wafer (University Wafer, South Boston, MA) to create a 9 μm-thick film. The film was soft baked on a hot plate and exposed to 155 mJ cm−2 of UV light energy through the chrome-on-glass light field mask using an EVG620 mask aligner (EVG, Austria). The wafer was post exposure baked to crosslink the pattern before development with SU-8 developer (Fig. 2a). The master was hard baked at 150 °C for several hours to increase the mechanical strength of the photoresist, followed by treatment with a fluorinated silane [(tridecafluoro-1,1,2,2-tetrahydrooctyl)-1-trichlorosilane] (Pfaltz & Bauer, Waterbury, CT) overnight in a vacuum desiccator to facilitate the detachment of PDMS molds from the master.
Fig. 2.
Fabrication of the microfluidic device for probing traction forces. Schematic of the procedure used to (a) fabricate the PDMS microposts and (b) microcontact print collagen type I on the top of microposts, seal the device, and track post deflections.
The master for the top portion of the device contained a negative mold of the microchannels and microfluidic feed lines and was designed to generate parallel microchannels of height HC = 4 μm and length LC = 200 μm spaced 50 μm apart, with channel widths of WC = 10, 20, and 50 μm. To fabricate the master, a 4 μm-thick layer of SU-8 3005 was spin coated on a silicon wafer prior to exposure with 110 mJ cm−2 of UV light energy through a mask defining the microchannels. This step determined the height and width of the micro-channels, as described previously.31 Following post exposure baking and development, a 50 μm-thick SU-8 3025 film was spun onto the wafer. A mask defining the medium and chemo-attractant feed lines was aligned by microscopy with the channels, and the photoresist was exposed to 250 mJ cm−2 of energy. The final master was developed, hard baked, and passivated as described above. A third master had the same design as the microchannel master, but all features were 10 μm high and formed using SU-8 3010. This master was used to create a stamp for microcontact printing, as described below.
Devices were formed from the SU-8 masters using replica molding with PDMS (Sylgard® 184 Silicone Elastomer Kit, Dow Corning, Midland, MI). The top portion of the device (microchannel layer) was formed in a single molding step by curing PDMS at a 10 : 1 prepolymer : curing agent ratio on the master. A 5 mm hole punch was used to create flow ports in the final PDMS device. The bottom portion of the device (PDMS micropost structures) was fabricated using a double casting procedure. First, a negative mold was cast from the master to create an inverse of the microposts (PDMS master, right side of Fig. 2A). PDMS molds were treated with oxygen plasma using a Harrick PDC-32G plasma cleaner (Harrick Plasma, Ithaca, NY) for 2 min at 18 W and then passivated with the fluorinated silane overnight. Passivation prevents cured PDMS from permanently bonding to the PDMS negative mold during a second casting. Next, a clean glass slide or cover glass was pretreated with oxygen plasma and then coated with a thin layer of degassed PDMS. A drop of degassed PDMS was also placed on top of the negative mold, and the PDMS-coated glass was placed on the mold and cured on a hot plate at 110 °C for 20 h to ensure maximum crosslinking of the PDMS polymer. The microposts bound to the top cover glass or slide were then peeled gently away from the negative mold (Fig. 1c,d; Fig. 2a). The same double casting procedure was used to generate the PDMS stamp used for selective microcontact printing of proteins on the micropost tops, described below.
To restrict the migration of the cells to the tops of the microposts without compromising adhesion between the top and bottom PDMS portions of the device, PDMS stamps were used to selectively transfer ECM proteins to the tops of the posts, as well as the medium and chemoattractant feed lines (Fig. 2b). The top and bottom portions of the migration device were briefly treated with oxygen plasma (30 W, 0.3 Torr, 30 s). The PDMS stamp was incubated with the ECM protein of interest, collagen type I, for 1 h, washed, and precisely aligned with the micropost layer using a custom-built micromanipulator setup. The stamp and the substrate were brought in conformal contact and held there for 1 min to transfer the ECM protein. Subsequently, the top portion of the device was precisely aligned using the micromanipulator setup such that the channels were placed over the micro-channel outlines containing microposts, and the layers were brought together and sealed (Fig. 2b). The chamber was washed with DI water and stained with lipophilic tracer DiD (5 μg ml−1) to visualize the microposts. The microchannels were then blocked with 0.2% F-127 Pluronics for 1 h to prevent the adhesion of the cells to the shafts of the microposts.
Cell seeding and live cell migration experiments
HOS or NIH-3T3 cells were grown to confluency, trypsinized, and resuspended in serum-free medium at 2 × 106 cells ml−1. Microchannels were washed with DPBS prior to the addition of 50 μl of cell suspension to the cell inlet well. Cells were incubated in the device for 5–10 min at 37 °C to allow initial cell seeding (Fig. 2b). The cell suspension was then removed from the cell inlet port and replaced with serum-free medium (50 μl). The topmost inlet port of the chemokine gradient generator was filled with 50 μl of medium containing 10% FBS, while the other two inlet ports were filled with 50 μl of serum-free medium, as described previously.31,32 The migration chamber was moved to a temperature- and CO2-controlled stage-top live cell incubator (Okolab, Italy) mounted on the motorized stage of an inverted Nikon Eclipse Ti microscope (Nikon, Tokyo, Japan) with automated controls (NIS-Elements, Nikon). Migrating cells and post deflections were visualized every 10 s with a DS-Fil camera head and a 40× objective for the duration of each live cell experiment (15–30 minutes per microchannel. Frequent imaging (every 10 s) was required to capture time-dependent deflections of individual microposts, which fluctuated by as much as 10 nN per post as the cell migrated through the microchannel (fluctuating micropost deflections as different segments of a cell move over a micropost can be seen in Supplementary Video, ESI†). However, the average force per post did not vary significantly over the duration of a typical experiment (15–30 minutes) or even for longer observation periods (data not shown). For experiments quantifying cell migration speed, cells were seeded at the entrances of 4 μm-tall, 200 μm-long, and 6, 10, 20, or 50 μm-wide microchannels coated with 20 μg ml−1 collagen type I, as described previously.31 A multipoint image capture was set up, and images were taken at 10× every 20 minutes.
Image and data analysis
To quantify the traction forces exerted during cell movement on top of microposts, the difference in position between the ideal undeflected post centroid and the observed centroid of the microposts was measured. Post deflections were converted to traction forces by multiplying the beam top centroid displacement, x, by the spring constant of the microposts, k.10 According to the beam bending theory, the force F is given by:
| (1) |
where E is the modulus of elasticity of PDMS (~2.5 MPa), D is the post diameter, and L is the height of the micropost. Calculation of this relationship provides the conversion from tip displacement (μm) to traction force (nN). Time-lapse videos were exported to ImageJ (NIH, Bethesda, MD), and ideal post grids were generated. The centroid of the microposts was tracked using the MtrackJ Plugin33 to obtain time-dependent post deflections due to the traction forces exerted by migrating cells. Background forces were measured by tracking the centroid of free posts (posts without cells attached) and calculating the force that would be required to cause the observed post deflections. Overall cell speed was calculated by tracking the midpoint of the cell using MTrackJ and averaging the frame-to-frame cell speed. Only cells that were fully within the channels were tracked.
Statistics
Time-dependent force curves for various cell types and channel widths were generated, and the average forces obtained at each time point were calculated. Average forces were compared using an unpaired Student’s t-test at each time point. Forces were considered statistically significantly different for p < 0.05. Average speeds for a given channel width were compared by ANOVA followed by Tukey’s HSD test. At least three independent repeats were performed for each cell speed experiment.
Results and discussion
Fabrication of the microfluidic device for probing cell traction forces
We engineered a microfluidic device to measure cell traction forces during chemotaxis-driven migration on 2D-like substrates versus confined microenvironments. This device consists of microchannels of prescribed dimensions that encompass an array of deflectable PDMS microposts as their bases (Fig. 1). The microchannels were aligned in a ladder-like configuration orthogonally to and connected to two larger main channels serving as the cell seeding inlet and chemokine reservoir (Fig. 1; Fig. S1, ESI†). The deflection of a post due to cell-generated traction forces was used to calculate the force exerted tangentially on the top of that post, as described previously.9–11,13,17 We employed a variety of sophisticated microfabrication techniques to create the device, including photolithography, multistep replica molding, sandwich molding,34 microcontact printing, and precise microalignment.
We fabricated the bottom portion of the microfluidic chamber, which contains an array of 3 μm-diameter, 9 μm-tall microposts with 6 μm center-to-center spacing incorporated within microchannels, using standard lithographic techniques and dual replica molding with PDMS (Fig. 1c,d; Fig. 2a). The two-step replica molding process was required to prevent clogging of PDMS in SU-8 holes, which led to degradation of the negative master. The microposts were located within microchannels of 50, 20, or 10 μm width. On a single device, 30 microchannels were present for each channel width. This design enabled us to impose varying degrees of confinement on a population of migrating cells in a single experiment. Scanning electron microscopy revealed intact microposts of the desired feature dimensions (Fig. 1c,d). The small number of posts per microchannel, limited by the geometry of the channel, necessitated pristine post fabrication. The top portion of the device, an array of microchannels that mirrored those created on the bottom layer, albeit without microposts (Fig. 2b), was fabricated similarly. The spacing between the top of the microposts and the ceiling of the microfluidic device was set to 4 μm.
To ensure that the cells spread prior to their entry to microchannels and migrated only on top of the microposts, we selectively transferred an ECM protein to the seeding area and micropost tops using microcontact printing (Fig. 2b). This is in clear contrast to previous work that measured traction forces of cells moving in between microposts.35 Selective stamping with precise alignment was required to ensure PDMS–PDMS sealing, which was needed to create four-walled microchannels. A flat PDMS stamp could not be used since the presence of proteins on the flat faces that connect the top and bottom portions of the microfluidic channels compromised their seal. Following microcontact printing, the top portion of the microfluidic device was aligned precisely over the bottom portion using the micromanipulator and sealed to the base, thereby defining the channels. Post shafts were then blocked with F-127 Pluronics to limit cell adhesion to the tops of the posts. Cells were seeded at the channel entrances and imaged as they migrated (Fig. 3a,b; Fig. S2, ESI;† Supplementary Video, ESI†).
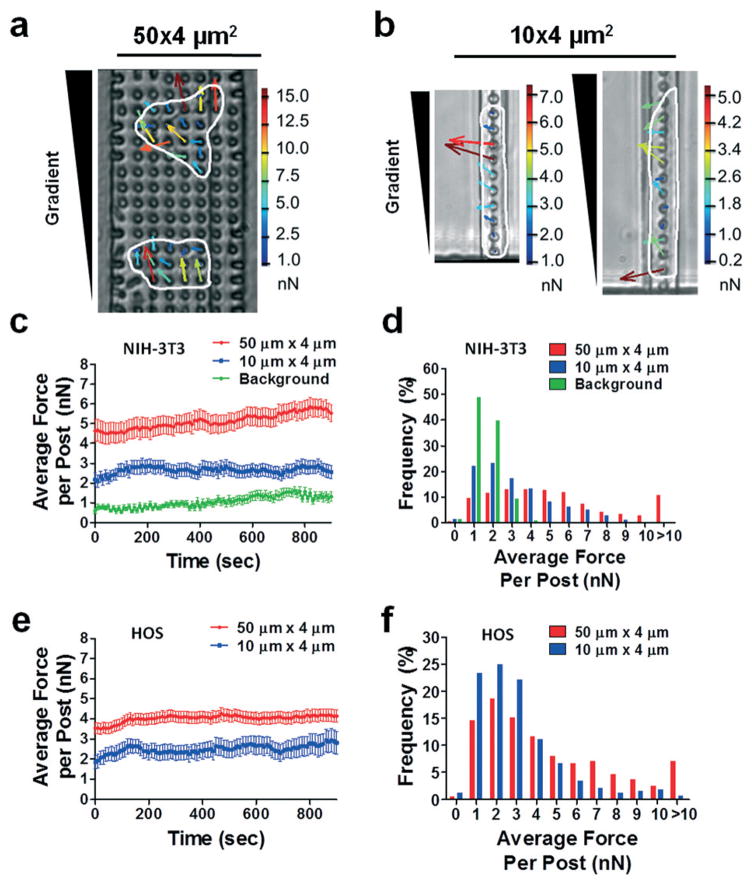
Fig. 3.
Measurement of traction forces exerted by NIH-3T3 and HOS cells in wide and narrow microchannels. Representative phase contrast images of HOS cells migrating through (a) 50 × 4 μm2 and (b) 10 × 4 μm2 microchannels. Cells are outlined for clarity. An FBS gradient was generated by diffusion from an FBS source at the top of the images. Force vectors generated for each micropost are overlaid on the cells. Vector length and color correspond to the magnitude of the traction force, which is indicated on the scale bar to the right of each image. (c, e) Plot of average traction force per post and (d, f) distribution of the traction forces per post for (c, d) NIH-3T3 fibroblasts and (e, f) HOS cells in 50 × 4 μm2 and 10 × 4 μm2 microchannels. For all experiments, micropost tops were coated with 50 μg ml−1 collagen type I. Data represent mean ± SEM from at least n = 20 posts. Background forces remained at ~1 nN per post through remaining experiments and are not indicated on the following plots.
Taken together, sophisticated microfabrication techniques were employed to fabricate a device capable of both traction force measurement and topographic control. Alignment of the top array of microchannels over the micropost arrays is vital to obtaining sealed microchannels for studying chemotactic migration through unconfined and confined microenvironments. We have achieved a high degree of alignment, with alignment precisions of up to ±1 μm between the top and bottom portions of the device. To our knowledge, this is among the highest degrees of alignment yet reported for multiple PDMS microfluidic layers.34,36 Alignment accuracies of 15 μm over a 6.5 cm2 area for a three layer device have been reported using an alignment setup similar to ours.34 Use of a more complicated alignment system has generated an offset of as little as 2 μm between PDMS layers.36 Microfabrication and microfluidic techniques have been utilized to measure 2D cellular traction forces9–12,17 or fabricate devices to study cell migration through confined spaces in vitro.31,32,37–39 However, to our knowledge, ours is the first device to couple these technologies in order to measure traction forces exerted by cells migrating through unconfined and confined microenvironments, as described below.
Migration through 50 μm-wide channels displays the earmarks of 2D migration
To demonstrate the general utility of our migration device for traction force measurement, we first performed experiments with murine fibroblasts (NIH-3T3), which have been commonly used to study 2D and 3D cell migration. We have recently shown that wide channel migration (W × H = 50 × 10 μm2 or 20 × 10 μm2) displays the earmarks of 2D migration.32,39 As a first step, we quantified cell traction forces in 50 μm-wide by 4 μm-tall microchannels stamped with type I collagen. The average force per post was ~4–5 nN per post (Fig. 3c), which is similar to that found in a previous study using NIH-3T3 cells on a 2D micropost array.11 Background forces measured by tracking free posts, that is, posts within the microchannels but not attached to cells, remained at ~1 nN per post through all experiments, significantly less than forces exerted by migrating cells (Fig. 3c,d). Similarly, HOS cells migrating on type I collagen-stamped microposts within 50 μm-wide microchannels in response to an FBS gradient exerted forces of 3–4 nN per post (Fig. 3e). Maximal forces exerted by HOS cells were 2–3 times that of the average forces and were directed towards the chemotactic stimulus (Fig. 3a). The average magnitude of traction forces exerted by migrating HOS cells was similar to the reported maximal forces exerted by individual migrating Madin–Darby canine kidney (MDCK) epithelial cells following application of HGF/scatter factor.9 Furthermore, the distribution of the forces under the cells, with regions of higher stresses punctuating the periphery of the cell (Fig. 3a), match force distributions reported in earlier studies for chemokinetic9 or chemotactic17 2D migration. Therefore, microposts arrayed within 50 μm-wide channels reliably measure average traction forces relevant to 2D chemotactic migration, complementing an existing assay.17
Confinement reduces cell traction forces
We next quantified the average traction forces per post exerted by cells migrating within confined microenvironments. Specifically, forces exerted by cells migrating through 10 μm-wide by 4 μm-tall microchannels stamped with type I collagen were measured (Fig. 3b). We have recently reported that cells contact all 4 walls of microchannels with cross-sectional areas between 30 and 60 μm2.32,39 Because we cannot account for forces exerted on the ceiling or side walls of the microchannels, forces exerted within 50 μm- and 10 μm-wide microchannels were compared on a force per post basis, allowing evaluation of the average force exerted by a cell on its substrate for a given unit of area. This method of reporting is similar to that used to report traction stresses for cells within gel matrices19–22 or confined to 1D lines.40 The forces exerted by NIH-3T3 fibroblasts migrating through 10 × 4 μm2 channels averaged 2–3 nN per post, which were significantly reduced relative to those in 50 × 4 μm2 channels (Fig. 3c; Fig. S3a, ESI;† Supplementary Video, ESI†). Furthermore, the frequency of forces per post greater than 5 nN was lower in comparison to the wider channels (Fig. 3d). Forces per post in both wide and narrow channels were significantly higher than the background signal (Fig. 3c,d), indicating that both unconfined and confined modes of cell migration involve generation of cell traction forces. Our data are in concert with prior observations showing that NIH-3T3 fibroblasts migrating on narrow 1D lines exert lower traction stresses than those on wide lines, possibly due to the inhibition of large focal adhesion complexes and stress fibers by geometric constraints.40
In tissues, mesenchymal fibroblasts are often responsible for invasion and tissue remodeling, two tasks which are dependent on Rho/ROCK function27,28 and likely involve active matrix remodeling. However, microchannel assays better simulate the movement of cells through remodeled ECM. Motility through 3D longitudinal tracks appears to be particularly important for tumor cell migration.25–27 In light of recent work suggesting a reduced role for actomyosin contractility in the migration of metastatic MDA-MB-231 breast cancer cells through confined microenvironments,32 we used our device to probe the traction forces of metastatic tumor cells during confined migration. The magnitude of traction forces per post exerted by HOS cells migrating through 10 × 4 μm2 channels were ~40% lower than those in 50 × 4 μm2 channels (Fig. 3e; Fig. S3b, ESI†). A lower frequency of forces per post greater than 4 nN was noted in narrow versus wide microchannels (Fig. 3f). Interestingly, while the net direction of forces exerted by cells migrating through confined microenvironments was towards the chemoattractant, appreciable forces were also directed towards the side wall of the micro-channel (Fig. 3b).
Collectively, these data suggest that physical confinement modulates the adhesion-mediated traction forces that drive cells forward towards a chemotactic source. The observation that 3T3 fibroblasts and metastatic HOS cells moved through confined microenvironments efficiently and exerted markedly lower traction forces indicates the presence of a distinct migration mechanism in confinement. Therefore, the development of a device to quantitatively measure traction forces during confined migration represents an important step in the mechanistic understanding of cell migration in physiologically-relevant microenvironments.
Inhibition of myosin-II function does not affect cell traction forces or migration speed in confined microenvironments
Blebbistatin, a specific inhibitor of muscle and non-muscle myosin-II, inhibits actin-detached myosin and prevents the rigid cross-linking of actomyosin.41 Inhibition of myosin-II activity by blebbistatin does not suppress the migration speed of MDA-MB-231 breast cancer cells through narrow microchannels with a cross-sectional area of ~30 μm2,32 suggesting that blebbistatin does not affect cell traction forces in confined microenvironments. We thus probed the effects of blebbistatin treatment on cell traction forces in narrow versus wide microchannels.
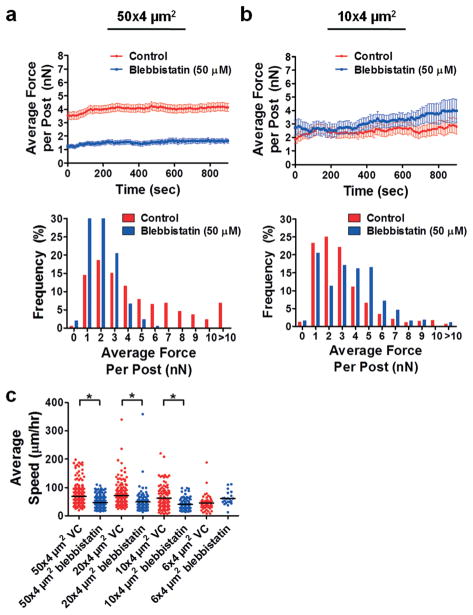
HOS cell treatment with 50 μM blebbistatin drastically suppressed the average force per post in 50 × 4 μm2 microchannels compared to control cells (Fig. 4a; Fig. S4a, ESI†), illustrating the importance of myosin-II in 2D cell migration. Similar effects have been observed in unconfined (2D) microenvironments, where inhibition of myosin-II function by blebbistatin decreases the average traction forces exerted by NIH-3T342 and human pulmonary artery endothelial cells (HPAECs).43 In marked contrast, there was no significant difference in the average traction force per post in 10 × 4 μm2 microchannels between blebbistatin versus control treatment (Fig. 4b; Fig. S4a, ESI†). In line with our previous work,32 blebbistatin treatment resulted in a reduction of HOS cell speed in wide but not narrow microchannels, which were devoid of microposts (Fig. 4c). Interestingly, a previous study reported that inhibition of actomyosin contractility by blebbistatin or Y-27632 does not decrease the migration speed of NIH-3T3 cells on 1D lines.44 Collectively, these findings suggest that myosin-II may play a reduced role in migration through confined microenvironments, as neither speed nor cell traction forces were affected by blebbistatin treatment.
Fig. 4.
Effects of myosin-II inhibition by blebbistatin on traction forces of HOS cells. Plot of average traction force per post and distribution of traction forces per post for HOS cells migrating through (a) 50 × 4 μm2 or (b) 10 × 4 μm2 microchannels in the absence and presence of 50 μM blebbistatin. For all experiments, micropost tops were coated with 50 μg ml−1 collagen type I. Data represent mean ± SEM from at least n = 20 posts. (c) Plot of average cell speed for control and blebbistatin-treated (50 μM) HOS cells migrating through microchannels coated with 20 μg ml−1 collagen type I. *, P < 0.01 by one-way ANOVA followed by Tukey’s HSD post-test.
Activation of myosin-II does not alter cell traction forces in confined microenvironments but increases them in wide microchannels
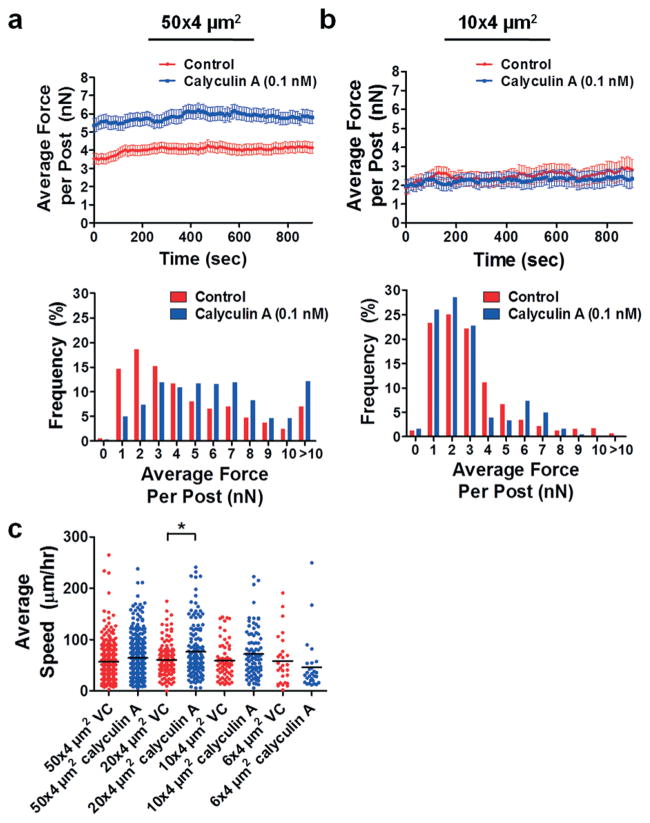
To further elucidate the effect of actomyosin contractility on migration in confined spaces, we treated cells with calyculin A, a serine/threonine phosphatase inhibitor that causes an increase in myosin light chain phosphorylation and, therefore, in cellular contraction.45,46 In wide channels, the average traction force per post increased significantly upon calyculin A treatment (Fig. 5a; Fig. S4b, ESI†). Activation of myosin-II by calyculin A also increases the traction forces of NIH-3T342 and HPAECs43 in 2D microenvironments.
Fig. 5.
Effects of myosin-II activation by calyculin A on traction forces of HOS cells. Plot of average traction force per post and distribution of traction forces per post for HOS cells migrating through (a) 50 × 4 μm2 or (b) 10 × 4 μm2 microchannels in the absence and presence of 0.1 nM calyculin A. For all experiments, micropost tops were coated with 50 μg ml−1 collagen type I. Data represent mean ± SEM from at least n = 20 posts. (c) Plot of average cell speed for control and calyculin A-treated (0.1 nM) HOS cells migrating through microchannels coated with 20 μg ml−1 collagen type I. *, P < 0.01 by one-way ANOVA followed by Tukey’s HSD post-test.
In narrow channels, the magnitudes of cell traction forces exerted by control and calyculin A-treated cells were indistinguishable (Fig. 5b; Fig. S3b, ESI†). Calyculin A treatment tended to increase cell speed in wider channels, while migration speed was unaffected in narrow channels (Fig. 5c). These experiments provide further evidence that actomyosin contractility may play a reduced role in cell migration through confined microenvironments, as activation of myosin-II did not significantly change cell speed or force exertion in narrow channels.
Conclusions
To fully comprehend the combinatorial effects involved in the interplay between physical and biochemical properties in cell migration (e.g., between physical confinement and chemotaxis-induced biochemical signaling), tools are required to facilitate the control and manipulation of the microenvironment with concurrent monitoring of cell response. Here, we describe the fabrication of a novel device to measure the traction forces of cells migrating through a confined microenvironment towards a chemoattractant source. We have demonstrated the utility of this device through experiments with NIH-3T3 fibroblasts, in which we measured lower traction forces in confinement. Furthermore, we have found that distinct mechanisms govern metastatic cancer cell migration through unconfined versus confined spaces. While migration and force exertion of HOS cells on 2D surfaces were highly dependent on actomyosin contractility, migration through confining microchannels was not modulated upon myosin-II inhibition by blebbistatin or myosin-II activation by calyculin A. Specifically, in 10 × 4 μm2 microchannels, it appears that myosin II-mediated traction forces may not play a key role in force generation, as the magnitude of traction forces remains essentially the same in these narrow microchannels upon inhibition or activation of myosin-II. However, non-myosin-II-mediated forces responsible for moving the cell must still be present in confined spaces, as measured forces were significantly greater than the background force. Whether the cell uses a migration mechanism to push off the side walls, as suggested by Fig. 3b, lessening adhesion-based traction forces generated through the actomyosin pathway but still exerting sufficient force to move efficiently, is an important question going forward. For example, bleb- or pressure-based forces resulting from intra-cellular contractility have been suggested as important adhesion-independent modes for cell migration in confinement.47,48 These forces could be responsible for micropost deflections in our device and may explain why the distribution of forces in confined migration (Fig. 3b) is punctuated by forces directed toward the microchannel walls. This force distribution is in contrast to 2D migration, where forces are directed to the center of the cell or along the direction of movement.
Our data underlie the importance of appreciating the full range of cell migration mechanisms in the development of therapeutic strategies aiming to inhibit the migration of cancerous cells away from a primary tumor. This could be particularly important in the development of drugs targeting cell contractility, which have shown contradictory effects on tumor progression. For example, a recent report has shown that treating primary colon cancer cells with blebbistatin or ROCK inhibitors promotes tumor spheroid formation and expression of cancer stem cell markers.49 Conversely, others have advocated inhibiting ROCK to lessen cancer cell invasion and remodeling of the tumor microenvironment.50 Our novel device is an important tool for the determination of which drug targets can effectively interfere with migration in different physical microenvironments encountered in vivo.23
Supplementary Material
Acknowledgments
We thank Huy Vo for his help with microfabrication and the development of the in situ micromanipulator setup. We would also like to thank Mark Koontz for his assistance with SEM imaging, and Dr. Chris Chen of the University of Pennsylvania for sharing the micropost analysis code on which our force analysis was based. PSR was supported through an American Heart Association Predoctoral Fellowship. CDP was supported by National Science Foundation Graduate Research Fellowship. KMS was supported by National Cancer Institute T32-CA130840 and F32-CA177756 awards. This work was supported by grants from the National Cancer Institute (U54-CA143868, CA101135), National Science Foundation (NSF-1159823), and Kleberg Foundation.
Footnotes
Electronic supplementary information (ESI) available: Four supplementary figures and one supplementary video are available. See DOI: 10.1039/c3lc50802a
Notes and references
- 1.Christofori G. Nature. 2006;441:444–450. doi: 10.1038/nature04872. [DOI] [PubMed] [Google Scholar]
- 2.Hagedorn EJ, Sherwood DR. Curr Opin Cell Biol. 2011;23:589–596. doi: 10.1016/j.ceb.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakaya Y, Sukowati EW, Wu Y, Sheng G. Nat Cell Biol. 2008;10:765–775. doi: 10.1038/ncb1739. [DOI] [PubMed] [Google Scholar]
- 4.Konstantopoulos K, Wu PH, Wirtz D. Biophys J. 2013;104:279–280. doi: 10.1016/j.bpj.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gardel ML, Schneider IC, Aratyn-Schaus Y, Waterman CM. Annu Rev Cell Dev Biol. 2010;26:315–333. doi: 10.1146/annurev.cellbio.011209.122036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roca-Cusachs P, Iskratsch T, Sheetz MP. J Cell Sci. 2012;125:3025–3038. doi: 10.1242/jcs.095794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mierke CT, Rosel D, Fabry B, Brabek J. Eur J Cell Biol. 2008;87:669–676. doi: 10.1016/j.ejcb.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stroka KM, Aranda-Espinoza H. FEBS J. 2010;277:1145–1158. doi: 10.1111/j.1742-4658.2009.07545.x. [DOI] [PubMed] [Google Scholar]
- 9.du Roure O, Saez A, Buguin A, Austin RH, Chavrier P, Silberzan P, Ladoux B. Proc Natl Acad Sci U S A. 2005;102:2390–2395. doi: 10.1073/pnas.0408482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. Proc Natl Acad Sci U S A. 2003;100:1484–1489. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han SJ, Bielawski KS, Ting LH, Rodriguez ML, Sniadecki NJ. Biophys J. 2012;103:640–648. doi: 10.1016/j.bpj.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rape AD, Guo WH, Wang YL. Biomaterials. 2011;32:2043–2051. doi: 10.1016/j.biomaterials.2010.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tee SY, Fu J, Chen CS, Janmey PA. Biophys J. 2011;100:L25–L27. doi: 10.1016/j.bpj.2010.12.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butler JP, Tolic-Norrelykke IM, Fabry B, Fredberg JJ. Am J Physiol: Cell Physiol. 2002;282:C595–605. doi: 10.1152/ajpcell.00270.2001. [DOI] [PubMed] [Google Scholar]
- 15.Dembo M, Wang YL. Biophys J. 1999;76:2307–2316. doi: 10.1016/S0006-3495(99)77386-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rape A, Guo WH, Wang YL. J Cell Sci. 2011;124:4233–4240. doi: 10.1242/jcs.090563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ricart BG, Yang MT, Hunter CA, Chen CS, Hammer DA. Biophys J. 2011;101:2620–2628. doi: 10.1016/j.bpj.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stroka K, Vaitkus J, Aranda-Espinoza H. Eur Biophys J. 2012;41:939–947. doi: 10.1007/s00249-012-0851-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Legant WR, Miller JS, Blakely BL, Cohen DM, Genin GM, Chen CS. Nat Methods. 2010;7:969–971. doi: 10.1038/nmeth.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Legant WR, Choi CK, Miller JS, Shao L, Gao L, Betzig E, Chen CS. Proc Natl Acad Sci U S A. 2013;110:881–886. doi: 10.1073/pnas.1207997110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koch TM, Münster S, Bonakdar N, Butler JP, Fabry B. PLoS One. 2012;7:e33476. doi: 10.1371/journal.pone.0033476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franck C, Maskarinec SA, Tirrell DA, Ravichandran G. PLoS One. 2011;6:e17833. doi: 10.1371/journal.pone.0017833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedl P, Alexander S. Cell. 2011;147:992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 24.Wolf K, Alexander S, Schacht V, Coussens LM, von Andrian UH, van Rheenen J, Deryugina E, Friedl P. Semin Cell Dev Biol. 2009;20:931–941. doi: 10.1016/j.semcdb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexander S, Koehl GE, Hirschberg M, Geissler EK, Friedl P. Histochem Cell Biol. 2008;130:1147–1154. doi: 10.1007/s00418-008-0529-1. [DOI] [PubMed] [Google Scholar]
- 26.Fisher KE, Sacharidou A, Stratman AN, Mayo AM, Fisher SB, Mahan RD, Davis MJ, Davis GE. J Cell Sci. 2009;122:4558–4569. doi: 10.1242/jcs.050724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaggioli C, Hooper S, Hidalgo-Carcedo C, Grosse R, Marshall JF, Harrington K, Sahai E. Nat Cell Biol. 2007;9:1392–1400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- 28.Shieh AC, Rozansky HA, Hinz B, Swartz MA. Cancer Res. 2011;71:790–800. doi: 10.1158/0008-5472.CAN-10-1513. [DOI] [PubMed] [Google Scholar]
- 29.Wolf K, Wu YI, Liu Y, Geiger J, Tam E, Overall C, Stack MS, Friedl P. Nat Cell Biol. 2007;9:893–904. doi: 10.1038/ncb1616. [DOI] [PubMed] [Google Scholar]
- 30.Yang MT, Fu J, Wang YK, Desai RA, Chen CS. Nat Protoc. 2011;6:187–213. doi: 10.1038/nprot.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tong ZQ, Balzer EM, Dallas MR, Hung WC, Stebe KJ, Konstantopoulos K. PLoS One. 2012;7:e29211. doi: 10.1371/journal.pone.0029211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balzer EM, Tong Z, Paul CD, Hung WC, Stroka KM, Boggs AE, Martin SS, Konstantopoulos K. FASEB J. 2012;26:4045–4056. doi: 10.1096/fj.12-211441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meijering ODE, Smal I. Methods Enzymol. 2012:183–200. doi: 10.1016/B978-0-12-391857-4.00009-4. [DOI] [PubMed] [Google Scholar]
- 34.Moraes C, Sun Y, Simmons CA. J Micromech Microeng. 2009;19:065015. [Google Scholar]
- 35.Ghibaudo M, Di Meglio JM, Hersen P, Ladoux B. Lab Chip. 2011;11:805–812. doi: 10.1039/c0lc00221f. [DOI] [PubMed] [Google Scholar]
- 36.Kim JY, Baek JY, Lee KA, Lee SH. Sens Actuators, A. 2005;119:593–598. [Google Scholar]
- 37.Chen SH, Hung WC, Wang P, Paul C, Konstantopoulos K. Sci Rep. 2013;3:1870. doi: 10.1038/srep01870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Irimia D, Charras G, Agrawal N, Mitchison T, Toner M. Lab Chip. 2007;7:1783–1790. doi: 10.1039/b710524j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hung WC, Chen SH, Paul CD, Stroka KM, Lo YC, Yang JT, Konstantopoulos K. J Cell Biol. 2013;202:807–824. doi: 10.1083/jcb.201302132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang SS, Guo WH, Kim Y, Wang YL. Biophys J. 2013;104:313–321. doi: 10.1016/j.bpj.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kovacs M, Toth J, Hetenyi C, Malnasi-Csizmadia A, Sellers JR. J Biol Chem. 2004;279:35557–35563. doi: 10.1074/jbc.M405319200. [DOI] [PubMed] [Google Scholar]
- 42.Lemmon CA, Chen CS, Romer LH. Biophys J. 2009;96:729–738. doi: 10.1016/j.bpj.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Z, Tan JL, Cohen DM, Yang MT, Sniadecki NJ, Ruiz SA, Nelson CM, Chen CS. Proc Natl Acad Sci U S A. 2010;107:9944–9949. doi: 10.1073/pnas.0914547107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo WH, Wang YL. Mol Biol Cell. 2012;23:1657–1663. doi: 10.1091/mbc.E11-06-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chartier L, Rankin LL, Allen RE, Kato Y, Fusetani N, Karaki H, Watabe S, Hartshorne DJ. Cell Motil Cytoskeleton. 1991;18:26–40. doi: 10.1002/cm.970180104. [DOI] [PubMed] [Google Scholar]
- 46.Peterson LJ, Rajfur Z, Maddox AS, Freel CD, Chen Y, Edlund M, Otey C, Burridge K. Mol Biol Cell. 2004;15:3497–3508. doi: 10.1091/mbc.E03-09-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tozluoğlu M, Tournier AL, Jenkins RP, Hooper S, Bates PA, Sahai E. Nat Cell Biol. 2013;15:751–762. doi: 10.1038/ncb2775. [DOI] [PubMed] [Google Scholar]
- 48.Hawkins RJ, Piel M, Faure-Andre G, Lennon-Dumenil AM, Joanny JF, Prost J, Voituriez R. Phys Rev Lett. 2009;102:058103. doi: 10.1103/PhysRevLett.102.058103. [DOI] [PubMed] [Google Scholar]
- 49.Ohata H, Ishiguro T, Aihara Y, Sato A, Sakai H, Sekine S, Taniguchi H, Akasu T, Fujita S, Nakagama H, Okamoto K. Cancer Res. 2012;72:5101–5110. doi: 10.1158/0008-5472.CAN-11-3812. [DOI] [PubMed] [Google Scholar]
- 50.Rath N, Olson MF. EMBO Rep. 2012;13:900–908. doi: 10.1038/embor.2012.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.