Abstract
In the last decades, a group of viruses has received great attention due to its relationship with cancer development and its wide distribution throughout the vertebrates: the papillomaviruses. In this article, we aim to review some of the most relevant reports concerning the use of bovines as an experimental model for studies related to papillomaviruses. Moreover, the obtained data contributes to the development of strategies against the clinical consequences of bovine papillomaviruses (BPV) that have led to drastic hazards to the herds. To overcome the problem, the vaccines that we have been developing involve recombinant DNA technology, aiming at prophylactic and therapeutic procedures. It is important to point out that these strategies can be used as models for innovative procedures against HPV, as this virus is the main causal agent of cervical cancer, the second most fatal cancer in women.
Keywords: bovine papillomavirus (BPV), bovine papillomatosis, carcinogenesis, natural history
A brief history of the papillomavirus (PVs) on carcinogenesis
In the last decades, novel diagnostic methods and therapies have been implemented in an attempt to combat cancer. However, the number of patients that succumb to the disease has increased globally (Varga et al., 2014). This negative result emphasizes the complexity of the oncogenic process, which has a multifactorial cause. Among the etiological factors associated to cancer are the infectious agents, such as bacteria and viruses.
The association between cancer and infectious agents has been discussed for centuries (Graner, 2000). In 1858, George B. Wood stated in his book Practice of Medicine that cancer could be disseminated as an infectious disease (Graner, 2000). However, the association between cancer and infectious agents was only implied in the second half of 19th century by Rudolf Maier (Graner, 2000; zur Hausen, 2009). The major difficulty in demonstrating this association can be attributed to the time-lapse of 15-40 years between the infection and the development of the first clinical signs that would allow cancer diagnosis (zur Hausen, 2009). Yet, in the last decades, the involvement of infectious agents with cancer has aroused great attention, as one in ten human malignancies is caused by these pathogens (Ribeiro-Müller and Müller, 2014).
Current studies estimate that 23% of all human malignancies are associated with infectious agents (zur Hausen, 2009; WHO 2013; Brücher and Jamall, 2014; Bravo and Felez-Sanchez, 2015). Among them, human papillomavirus (HPV) is responsible for 27.9% (zur Hausen, 2009) to 30.0% (Bravo et al., 2010) of all incident cancer cases in the world. This data is very concerning, once 75% of the global population lives in developing countries (Marrazzo and Holmes, 2013), where the lack of information about the HPV and others sexually transmitted diseases (STD) contributes to the increase of HPV-associated cancers. According to the World Health Organization (WHO) (http://www.who.int/mediacentre/factsheets/fs380/en/), 85% of cervical cancer cases occurs in less developed countries.
Papillomaviruses (PVs) are not only associated to human cancers, but also with animal malignancies. Although there is no epidemiological data about the number of PV-associated incident animal cancers, this association is recognized since 1932 (Shope and Hurst, 1933; Graner, 2000). Moreover, veterinary research demonstrates an increase in both benign and malignant tumors (Misdorp, 1996), particularly in domestic animals (cats and dogs). Furthermore, animal neoplasms are important models for the study of human oncogenic process (Misdorp, 1996), by allowing the identification of molecular mechanisms associated to carcinogenesis (Cotchin, 1962, 1976) and novel therapeutics (Misdorp, 1996), and emphasizing the importance of comparative oncology.
In this review, we summarize relevant data and advances in papillomaviruses biology, including viral evolution, pathogenic mechanism of viral proteins and oncoproteins, ways of transmission, pathogenesis and oncogenesis. We also discuss the importance of BPV as a study model for HPV-associated oncogenic process.
Genome organization of PVs
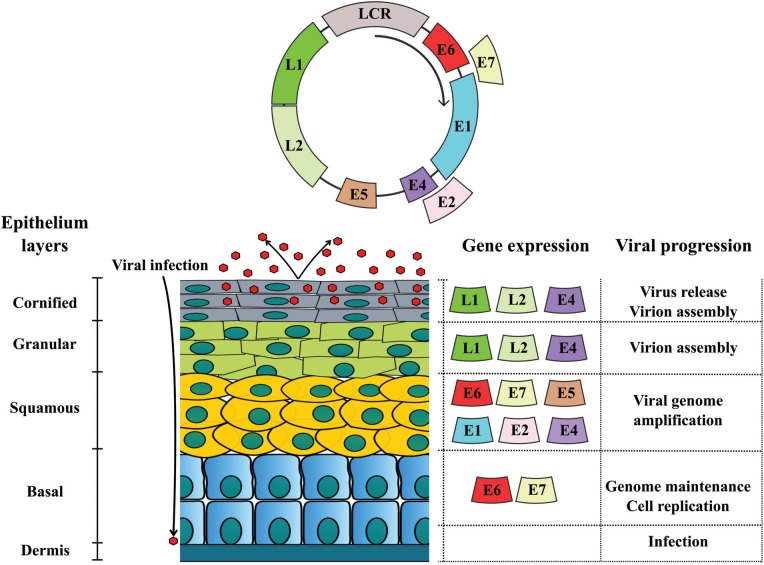
PVs are small, circular, double-stranded DNA viruses, able to infect all vertebrates (zur Hausen, 2009), as shown in Table 1. PVs belong to Papillomaviridae family (Bravo and Felez-Sanchez, 2015), which presents tropism for epithelial and mucous tissue (Cotchin, 1962). These viruses have genomes between 6,953 bp (CmPV1 - Chelonia mydas papillomavirus type 1) to 8,607 bp (CRPV1 - Cotton rabbit papillomavirus type 1), divided in three regions: early (E), late (L) and long codon region (LCR), as showed in Figure 1 (van Doorslaer, 2013). The E region codifies replication proteins (E1, E2, E4), and the oncoproteins E5, E6 and E7 (Bocaneti et al., 2014). The L region codifies capsid proteins (L1 and L2) (van Doorslaer, 2013). The LCR does not codify any protein, but has the origin of replication (ori) (van Doorslaer, 2013). Currently, more than 280 different types of PVs are described. More than 200 types infect humans (Munday, 2014), which are classified in 49 genera according to the International Committee on Taxonomy of Viruses (ICTV) (Araldi et al., 2015b). Phylogenetic classification of PVs are based on the L1 open reading frame (ORF) sequence homology, since this is the most conserved ORF among the different PV types (de Villiers et al., 2004, de Villiers, 2013; Melo et al., 2014; Munday et al., 2015). According to this system, differences over 10% on L1 ORF sequence determine a novel virus type, while differences between 2-10%, a novel virus subtype (de Villiers, 2013).
Table 1. Papillomaviruses identified in different vertebrates.
| Species | Virus | References |
|---|---|---|
| Bovines | Bos taurus papillomavirus or Bovine papillomavirus (BPV) | Araldi et al. (2015b) |
| Araldi et al. (2014a) | ||
| Carvalho et al. (2013) | ||
| Stocco dos Santos et al. (1998) | ||
| Canines | Canine oral papillomavirus (COPV) or Cannis familiaris papillomavirus (CfPV) | Nicholls et al. (2001) |
| Zaugg et al. (2005) | ||
| Deer | Capreolus capreolus papillomavirus (CcPV) or Cervus elaphus papillomavirus (CePV) | Kràl et al. (2015) |
| Felines | Felis catus papillomavirus (FcaPV) | Munday (2014) |
| Rabbit | Cotton rabbit papillomavirus (CRPV) | Shope and Hurst (1933) |
| Danos et al. (1985) | ||
| Rashad and Evans (1967) | ||
| Raccoon | Procyon loto papillomavirus (PlPV) | Ng et al. (2015) |
| Seaotter | Enhydralutris papillomavirus (ElPV) | Ng et al. (2015) |
| Sheep | Ovis aries papillomavirus (OaPV) | Alberti et al. (2010) |
| Mice | Mus musculus papillomavirus (MmPV) | Handisurya et al. (2013) |
| Birds | Fulmarus glaciallis papillomavirus (FgPV) | Gaynor et al. (2015a) |
| Francolinusleucoscepuspapillomavirus (FlPV) | Moreno-Lopez et al. (1984) | |
| Fringillacoelebspapillomavirus (FcPV) | Van Bressem et al. (2007) | |
| Psittacuserithacuspapillomavirus (PePV) | ||
| Rat | Rattus norvegicus papillomavirus (RnPV) | Hansen et al. (2015) |
| Monkey | Rhesus macaca papillomavirus(RmPV) | Roberts (2015) |
| Human | Human papillomavirus (HPV) | zur Hausen (2009) |
| Herbster et al. (2012) |
Figure 1. BPV genome organization and differential protein expression: double strand DNA circular genome divided in early (E), late (L) and long codon region (LCR). Early region codifies replication proteins (E1, E2, E4, E5, E6 and E7). These proteins are expressed from basal to cornified layer, being involved with virus replication and virion release (E4). Late region codifies L1 and L2 capsid proteins. Once these structural proteins are involved with virus assembly, they are expressed in the most differenciated epithelium layers (granular and cornified).
Evolutionary history of papillomaviruses
Although the virus origin is still uncertain (Bernard, 1994; Holland and Domingo, 1998), studies about PVs suggest that they arose concomitantly with tetrapods in the Carboniferous period of the Paleozoic era (330 million years ago) (Rector and Van Ranst, 2013). This makes the PVs one of the oldest and largest known virus family (Cubie, 2013; Rector and Van Ranst, 2013).
Studies based on molecular phylogeny suggest that these viruses originated in Africa, from where they disseminated to all continents (Bernard, 1994). It was not a pandemic dissemination, occurring over one million years (Bernard, 1994).
PVs genomic diversification occurred together with mammals diversification, being influenced by multiple evolutionary forces (Bravo et al., 2010), such as the addition of sequence boxes, previously present in their host (García-Vallvé et al., 2005). Thus, PVs co-evolved with their respective host (Gottschling et al., 2007). An evidence of this co-evolution is the similarity in guanine and cytosine (G + C) content; HPVs have 41-49% of G + C, and humans 40-42% (Black, 1968). Another evidence of this co-evolution is seen with the use of in situ hybridization probe for Shope papilloma virus (CRPV), which presents homology with the rabbit genome sequences (Black, 1968). Moreover, replicative mechanisms of PVs and their host are similar, reinforcing co-evolution (Leatherwood, 1998). These data suggest that PVs could have originated from genomic fragments of amniotes' common ancestor (van Doorlaer and Burk, 2010; Rector and Van Ranst, 2013; van Doorslaer, 2013).
Animal domestication favored the enzootic transmission of PVs (Gottschling et al., 2007). In this process, novel strategies of adaptation were required to guarantee the infection of novel hosts (Gottschling et al., 2011). These adaptations favored PVs evolution, conferring specificity in terms of host to all members of Papillomaviridae family, except for BPV, which is able to promote cross-infection (Bernard, 1994).
Based on this, García-Vallvé et al. (2005) described a hypothetical model to recreate PVs evolution. This model supports the existence of a proto-papillomavirus, comprised by URR-E1-E2-L2-L1 genomic regions, able to infect primitive amniotes. Along mammals divergence (150 million years ago), this proto-papillomavirus was added with E6 and E7 ORFs. From this moment, PVs' interaction with their host became more specific, resulting in a co-evolution. This process also resulted in the addition of E5 ORF in the hot spot between E2 and L2 ORFs (Bravo and Felez-Sanchez, 2015). Phylogenetic analyses point out that PVs only acquired oncogenic potential after they infected humans (García-Vallvé et al., 2005). This suggests that BPV could have originated from HPV transmission to bovines, as a consequence of animal domestication (García-Vallvé et al., 2005)., which would justify the similarities between these viruses, making the BPV a useful model to HPV studies (Campo, 2006; Borzacchiello and Roperto, 2008; Munday, 2014).
Non-oncogenic early (E) proteins expressed by PVs: E1, E2 and E4
E1 and E2 ORFs are expressed after PV infection, once their genetic products are essential to virus replication (Ferraro et al., 2011). The E1 ORF is the second most conserved sequence among the PVs (Forslund et al., 1999; Enemark et al., 2000). and codifies the E1 protein, which has three functional domains: (1) N-terminal, that induces Cdk2 phosphorylation, (2) C-terminal, that acts as ATP-dependent helicase and (3) central, that binds to E2 protein, resulting in E1-E2 complex (García-Vallvé et al., 2005; Wallace and Galloway, 2014). The E1-E2 complex binds to viral replication origin (ori) present in LCR (Enemark et al., 2000; Schuck and Stenlund, 2015). Next, the E1 protein forms the di-hexameric complex in ori (Gauson et al., 2016), and recruits replication proteins such as topoisomerase I, DNA polymerase α and replication protein A (RPA), that need to replicate (Schuck and Stenlund, 2015). Due to its helicase activity, the E1 protein can induce simple strand breaks (SSBs) and double strand breaks (DSBs) in host DNA (Schuck and Stenlund, 2015).
E2 is a modular protein, composed by two domains: (1) C-terminal and (2) transactivation N-terminal (Wallace and Galloway, 2014). The E2 C-terminal binds to Brd4 protein (Gauson et al., 2016). The E2-Brd4 complex interacts with lysine residues in acetylated histones (Schweiger et al., 2006, 2007), resulting in an equitable distribution of virus copies after cytokinesis (Campo, 2006; Wallace and Galloway, 2014).
The E2 protein also acts as an E6 and E7 transcriptional regulator (García-Vallvé et al., 2005; Bogaert et al., 2008; Cai et al., 2013). In high levels, E2 binds to 5′-ACCG(N)4CGGT-3′ palindromic sequence present in four E2 binding sites (E2BSs) in E6 and E7 promoter (P97) (Cai et al., 2013). This inhibits RNA polymerase II binding, repressing E6 and E7 transcription (García-Vallvé et al., 2005). However, under low expression levels, E2 recruits transcription factors necessary to form the open transcription complex (Helfer et al., 2014; Jang et al., 2014). In addition, E2 is an important epigenetic regulator, since the protein interacts with the p300/CBP-p/CAF global transcription activator (Wallace and Galloway, 2014). This interaction leads to TP53 hypo-acetylation, reducing p53 expression levels (Wallace and Galloway, 2014).
The E4 ORF codifies a family of proteins produced by splicing followed by post-translational modifications (Campo, 1997a). The E4 protein is the most expressed protein of PVs (Doorbar, 2013). For this reason, E4 is easily detected in suprabasal and granulosum layers of epidermis (Rampias et al., 2013), being recognized as important hallmark of PVs pathogenic activity (Doorbar, 2013). The E4 protein interacts with cytokeratin filamentous, contributing to viral replication (Campo, 1997a). Moreover, E4 is associated to virus maturation and extracellular matrix (MEC) remodeling (Ferraro et al., 2011).
Late proteins: L1 and L2
The L1 ORF is the most conserved among PVs (Bernard et al., 2006; Haga et al., 2013), and for this reason it is employed in virus classification (Haga et al., 2013; de Villiers, 2013). The L1 protein has 55 kDa (Buck et al., 2,013). and is able to self-organize in pentameric structures that compose the viral capsid (Ribeiro-Müller and Müller, 2014). It has a central role in viral infection mechanism, allowing the capsid anchorage to heparin sulfate receptors present in cell membrane (Florin et al., 2012). Considering that L1 is a late protein, it is expressed in the most differentiated epithelium layers (Buck et al., 2004). Therefore, L1 immunodetection has been considered the main evidence of productive infection (Nasir and Reid, 1999; Costa and Medeiros, 2014; Melo et al., 2015; Araldi et al., 2015b), which is characterized by the virus assembly (Green, 1972).
The L2 protein has 64-78 kDa (Wang and Roden, 2013). The molecular weight variation is a consequence of post-translational modifications (Wang and Roden, 2013). During the assembly of PV particles, 2 binds to viral DNA, contributing to encapsidation and then to viral release (García-Vallvé et al., 2005; Campo, 2006).
A third structural protein (L3) has been described as present exclusively in BPV-4 (Catroxo et al., 2013). However, its function remains unclear.
Oncogenic proteins expressed by PVs: E5, E6 and E7
E5 oncoprotein
The E5 is the most studied BPV oncoprotein and its transforming potential is known since 1960 (Roberts, 2015). The E5 oncoprotein can induce both in vivo and in vitro transformation (Campo, 2006; Silvestre et al., 2009; Rampias et al., 2013; DiMaio, 2014). Furthermore, it is responsible for the fibrotropism verified in fibropapillomas and equine sarcoids (Chambers et al., 2003a; DiMaio, 2014).
PV-1 E5 is a transmembrane protein (Costa and Medeiros, 2014), with 43-44 amino acids, characterized by the presence of a central hydrophobic region, which acts as a transmembrane domain (Burkhardt et al., 1989; Tomita et al., 2007; DiMaio, 2014). E5 also presents two cysteine residues in the C-terminal region that confers stability for the homodimer composed by two E5 monomers (DiMaio, 2014).
The E5 oncoprotein induces cell membrane composition and dynamic changes, affecting the Golgi complex (Burkhardt et al., 1989; García-Vallvé et al., 2005; Balcos et al., 2008). This occurs because E5 is able to bind to the H+-ATPase vacuolar subunit (Burnett et al., 1992), promoting Golgi inner membrane alkalinization (Krawczyk et al., 2010). This leads to major histocompatibility complex II (MHC-II) heavy chain sequestration, conferring an evolutionary mechanism of immune evasion (Venuti et al., 2011). In addition, the E5 oncoprotein inhibits the expression of MHC-I and cyclooxygenase (COX) (Borzacchiello, 2007; Tomita et al., 2007; Venuti et al., 2011). These mechanisms contribute to viral infection persistence.
The E5 oncoprotein can bind to ductin, a conexon component, inhibiting the gap junction formation and therefore cell communication (Campo, 1997a; Borzacchiello and Roperto, 2008). E5 also contributes to focal adhesion loss, affecting cell differentiation (Rampias et al., 2013).
The E5 oncoprotein is able to bind to platelet-derived growth factor receptor beta (PDGFβR) (Chambers et al., 2003a; DiMaio, 2014; Costa and Medeiros, 2014). PDGFβR is a tyrosine-kinase receptor present in the cell surface (Nicolas et al., 2013). Under normal conditions, PDGFβR is activated by PDGF, which promotes receptor dimerization (DiMaio, 2014) and activation of different kinases, such as: A-cdk2, MAPK, JNK, PI3K and C-Src (Chambers et al., 2003a; Borzacchiello, 2007). However, E5 binding to PDGFβR can lead to phosphatidylinositol-3-kinase (PI3K)-mediated Akt pathway activation (Costa and Medeiros, 2014). This activates the D3 cyclin, promoting cell cycle deregulation (Venuti et al., 2011; Costa and Medeiros, 2014). The PI3K/Akt pathway activation also induces metabolic deregulation in the host cell (Vivanco and Sawyers, 2002; Hennessy et al., 2005). PDGFβR activation also recruits pericytes, stimulating angiogenesis (Pietras and Ostman, 2010).
E6 oncoprotein
E6 is a small oncoprotein with 151-158 (HPV) (Ristriani et al., 2000; Boulet et al., 2007) or 137 amino acids (BPV-1) (Tong et al., 1998; Mazzuchelli-de-Souza et al., 2013), without enzymatic activity (Tan et al., 2012a). The E6 oncoprotein is characterized by the presence of a class I PDZ domain (PSD-95/Dlg/ZO-1), which is located in the C-terminal, and four PVs conserved motifs (Cys-X-X-Cys) (Chen et al., 1997; Tong et al., 1998; Nominé et al., 2006; Boon et al., 2015). The PDZ domain is found in BPV (Mischo et al., 2013) and high-risk HPVs (Boon et al., 2015) and can interact with different proteins, promoting loss of intercellular contact and cell polarity (Boon et al., 2015).
The HPV E6 oncoprotein can form a trimeric complex with E6AP ubiquitin ligase and p53 (Zanier et al., 2005, 2007; Tan et al., 2012a). This complex is responsible for the proteolytic degradation of p53 (Zanier et al., 2005; Liu and Baleja, 2008), which directs the p53 to 26S proteasome (Cai et al., 2013). The HPV-5 and 8 as well as BPV E6 oncoprotein interact with the CBP/p300 complex (Werness et al., 1990; Zimmermann et al., 2000; Liu et al., 2005), promoting p53 downregulation (Zimmermann et al., 1999). The loss of p53 contributes to genomic instability verified in cells infected by PVs (Araldi et al., 2013, 2015b), as well as in cells treated with BPV-1 E6 recombinant oncoprotein (Araldi et al., 2015a).
The overexpression of p53 represents a threat to viral replication (Shamanna et al., 2013). For this reason, different oncogenic viruses express proteins able to promote p53 deregulation. Among these proteins are antigen T of SV40, adenovirus E1B, hepatitis B virus (HBV) HBx and PVs E6 (Shamanna et al., 2013; Cuninghame et al., 2014). Thus, p53 deregulation is a characteristic shared by oncogenic viruses.
The E6 protein can interact with paxillin (Turner, 2000; Zanier et al., 2005) and AP-1 gamma subunit clathrin adapter (Tong et al., 1998). These interactions can lead to cytoskeleton alterations, affecting the vesicular traffic. E6 can also interact with fibulin 1, contributing to invasiveness phenotype (Moody and Laimins, 2010).
Studies have also demonstrated that E6 oncoprotein can induce cell transformation (Liu et al., 2002) and immortalization (Boon et al., 2015) due to the up-regulation of telomerases (Cuninghame et al., 2014). This occurs because E6 promotes FOXM1 overexpression, resulting in cyclin B1, D1 and cdc25 expression and, therefore, cell proliferation (Chen and Lee, 2015). In addition, E6 is able to bind to LXXL motif of MAML1 transcriptional regulator, inhibiting the Notch signaling (Tan et al., 2012a; White and Howley, 2013). The E6 oncoprotein of both HPV and BPV also promotes energy metabolic deregulation, contributing to cell transformation (Araldi et al., 2016).
E6 inhibits the SSB repair system, leading to genomic instability. This occurs because the oncoprotein interacts with XRCC1 and O6-methylguanosine-DNA-methyltransferase protein, which is recruited during the SSB repair (Wallace and Galloway, 2014). E6 also leads to cytogenetic damages (Wallace and Galloway, 2014) and stimulates neosis (Araldi et al., 2015a).
E7 oncoprotein
The E7 oncoprotein has 127 amino acids (Borzacchiello, 2007), and is able to bind to LXCXE conserved motif of pRb tumor suppressor protein (Moody and Laimins, 2010; White and Howley, 2013). This binding results in pRb phosphorylation, leading to E2F factor translocation to the nucleus (Longo, 2013). The E2F factor recruits different chromatin modifiers, including histone deacetylases (HDAC) (Moody and Laimins, 2010). Thus, the E7 oncoprotein induces the constitutive expression of E2F-responsive genes, such as cyclin A and E (Moody and Laimins, 2010), leading to S and G2/M cell cycle phase increase (Sacco et al., 2003; Ferraro et al., 2011). The interaction between viral oncoproteins and pRb is not exclusive of PVs, being also observed with adenovirus E1A and antigen T of SV40 (White et al., 2015).
Studies have demonstrated that E7 induces DNA breaks, contributing to cell cycle deregulation (Park et al., 2014; Araldi 2015; Araldi et al., 2015b). In addition, the E7 oncoprotein increases the Sirt1 deacetylase expression levels, modulating γ-H2AX activity (Park et al., 2014), a protein that participates in DSB repair (Sedelnikova and Pilch, 2003; Chowdhury et al., 2005). Thus, the E7 oncoprotein inhibits DSB repair, resulting in genomic instability.
Bovine papillomavirus (BPV): first reports and questions
In 1986 in an intriguing paper by Campo and Jarret (1986) reported the description of six different BPV types (BPV-1 to BPV-6) classified into two subgroups: subgroup A, that promotes fibropapillomas, and subgroup B, which leads to true epithelial papillomas. The report also showed that the BPV-4, a member of subgroup B, was the etiological agent of papillomas of the upper digestive tract, which could become carcinomas in animals feeding on a specific bracken fern pasture (Pteridium aquilinum). Later, Walter-Moura et al. (1988) verified the increase of chromosome aberrations in cells obtained from short term peripheral lymphocytes collected from bovines afflicted with bovine enzootic hematuria (BHE) that were exposed to pastures with bracken fern. These studies were reinforced by following reports, demonstrating that carcinogenesis is associated to the interaction between BPV and carcinogens present in the fern, such as quercetin and ptaquilosides (Pennie and Campo, 1992; Shahin et al., 1999; Potter and Baird, 2000; Beniston et al., 2001; Leal et al., 2003; Lioi et al., 2004). These studies brought new questions to be answered: how can the co-factor act in synergism with the virus? Why can these effects be detected in peripheral blood, considering that the virus is epithelial? Or, could the effect on chromosomes be only related to bracken fern compounds?
BPV and bovine papillomatosis (BP)
BPV is a cosmopolitan virus, worldwide distributed, independently of the level of expertise on livestock exploration (He et al., 2014). It is estimated that 60% of Brazilian cattle herd is infected by BPV (Stocco dos Santos, et al., 1998). However, that rate can be higher, once virus infection can be asymptomatic (Araldi et al., 2013; Silva et al., 2013a). Furthermore, the absence of epidemiological studies about BPV distribution could underestimate the real percentage of infected animals, representing a notorious difficulty in attempting to develop vaccines, once we do not know the prevalent virus types in each country.
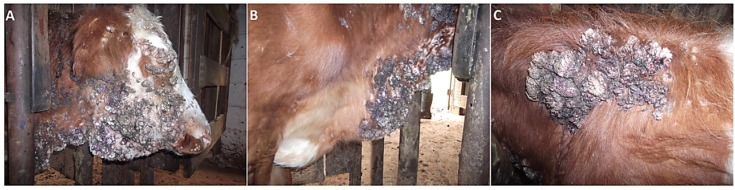
BPV infection is endemic in both dairy and beef cattle breeding (Claus et al., 2009; Araldi, 2015,; Araldi et al., 2015b). However, BPV presents a predilection for dairy cattle (Araldi, 2015). The virus is the causal agent of bovine papillomatosis (BP) (Muro et al., 2008), an infectious, contagious and neoplastic disease, characterized by the presence of multiple benign tumors (papillomas) (Figure 2) that can regress spontaneously or progress to malignant neoplasms (Campo, 2002; Turk et al., 2005; Yaguiu et al., 2006; Borzacchiello and Roperto, 2008; Araldi et al., 2014b; Bocaneti et al., 2014). Although BP affects preferably young cattle, the disease can occur at all ages (Börkü et al., 2007). The persistence of papillomas can lead to feeding and breathing difficulties, requiring the animal's euthanasia (Campo, 2002). Moreover, BP decreases growth rate and induces weight loss (Munday, 2014). The disease also predisposes bovines to opportunist bacterial infections (Munday, 2014), mainly in breast and mammary glands, which can result in mastitis, causing pain, reducing milk production (Campo, 2002; Muro et al., 2008; Lindsey et al., 2009; Santos et al., 2014) and causing depreciation of leather value (Monteiro et al., 2008).
Figure 2. Adult bovine (Simmental breed) with bovine papillomatosis (BP): skin papillomas on face (A), dewlap (B) and back (C).
Cutaneous papillomas are proliferative benign tumors, with complex pathogenesis (Börkü et al., 2007). These neoplasms commonly arise in vascularized areas exposed to physical attrition (Özsoy et al., 2011). Histologically, papillomas are characterized by epithelium hyperplasia, showing an enlargement of interpapillary ridge that extend above dermis (Monteiro et al., 2008). Morphologically, they are classified in: (1) typical - exophytic masses, with “cauliflower” aspect, presenting a wide or narrow insertion base, (2) pedunculated - exophytic masses connected by a narrow base with a peduncle shape, (3) atypical or flat - dense and flat exophytic masses entirely connected with the tissue, (4) filamentous - exophytic masses with highly keratinized surface and a thin base, present in mammary glands and (5) rice-form - small papillomas with rice-like shape (Monteiro et al., 2008).
Currently, 15 types of BPV are described (Munday et al., 2015), and classified in four genera: Deltapapillomavirus (BPV-1, 2, 13 and 14), Epsilonpapillomavirus (BPV-5 and 8), Xipapillomavirus (BPV-3, 4, 6, 9, 10, 11, 12 and 15) and Dyoxipapillomavirus (BPV-7) (Melo et al., 2014; Grindatto et al., 2015; Munday et al., 2015; Silva et al., 2016). Delta and Epsilonpapillomavirus are associated with both papillomas and fibropapillomas, while Xipapillomavirus, only to squamous papillomas (Tomita et al., 2007; Yuan et al., 2007; Tan et al., 2012b; Araldi, 2015; Araldi et al., 2015b).
Although there is no epidemiological study that allows the definition of BPV distribution, BPV-1 and 2 seem to be the most frequently identified virus types (Pathania et al., 2012; Melo et al., 2014; Santos et al., 2014; Alcântara et al., 2015; Araldi, 2015; Cota et al., 2015; Araldi et al., 2015b). Moreover, these viruses are associated with both benign and malignant neoplasms (Gopalkrishna et al., 1995; Nasir and Campo, 2008; Cota et al., 2015). However, the global distribution of BPV is not homogenous (Santos et al., 2014). BPV-1 and 2 have closely related serotypes (Shafti-Keramat et al., 2009), associated with urinary bladder malignant neoplasms (Wosiacki et al., 2005; Balcos et al., 2008; Roperto et al., 2008; Maiolino et al., 2013; Cota et al., 2015). BPV-4 infection is an important cause of upper digestive tract cancer development (Tsirimonaki et al., 2003; Nasir and Campo, 2008; Lucena et al., 2011). Studies also show that BPV-13 is associated to urothelial carcinomas (Roperto et al., 2015).
Diagnostic methods
Studies about BPV diversity and prevalence are mandatory to develop novel therapeutic methods (Silva et al., 2013b), since immunity is species-specific (Claus et al., 2007). Therefore, diagnosis is crucial.
Different methods have been implemented to identify PVs, such as: Southern blot (Leto et al., 2011), immunohistochemistry (IHC) (Elzein et al., 1991; Araldi et al., 2015b), chromogenic in situ hybridization (CISH) (Melo et al. 2015), electron microscopy (Araldi et al., 2014b) and PCR using specific and/or degenerate primers (Stocco dos Santos et al., 1998; Araldi et al., 2013, 2014a, 2015b; Melo et al., 2014). Among these techniques, PCR is the most used due to its high sensitivity (Leto et al., 2011). Moreover, using the restriction fragment length polymorphism of PCR products (PCR-RFLP) allows to identify BPV type (Carvalho et al., 2013), since this method shows a correlation of 95% with the results obtained using DNA sequencing (Martens et al., 2001a; Carvalho et al., 2013; Kawauchi et al., 2015).
Although real-time PCR (qRT-PCR) allows to determine the number of viral copies, this method has the lowest reproducibility (Guo et al., 2012). For this reason, PCR followed by DNA sequencing represents the most common method to identify and typify BPVs (Melo et al., 2014; Araldi et al., 2014b). However, there is no “gold-standard” primer employed in PVs identification (Antonsson et al., 2010).
On the one hand, although the specific primers have higher sensitivity than degenerate primers, they cannot identify the 14 BPV types simultaneously (Araldi et al., 2014b). In addition, specific primers do not allow to identify novel virus types and subtypes (Araldi et al., 2014b). Moreover, evidence indicates that a specific primer for BPV-1 can anneal to BPV-2 (Haga et al., 2013), once these virus types are considered serotypes-like (Shafti-Keramat et al., 2009). However, in a comparative study using complete genomes of BPV-1-6 in 2014 we demonstrated the BPV-1 primer specificity (Araldi et al., 2014b).
Among the degenerate primers described in the literature, FAP59/64 (Forslund et al., 1999) is the most employed in both BPV and HPV identification. This primer was designed based on the L1 ORF homology of HPV (Forslund et al., 1999) and later optimized to identify BPV DNA sequences (Ogawa et al., 2004). Furthermore, the use of FAP59/64 primer already allowed to identity novel BPV types, including BPV-9 and 10 (Hatama et al., 2008). However, despite of these advantages of degenerate primers, there are several reports showing their low sensitivity. For instance, MY09/11, another degenerate primer frequently used in HPV diagnosis, was unable to identify BPV and HPV sequences in clinical samples (Martelli-Marzagão et al., 2010; Zhu et al., 2014) or copies of complete cloned genome (Araldi et al., 2014b).
Although PCR is commonly used to identify PV DNA sequences, the method does not allow to identify their localization and physical state (episomal or integrated). Therefore, CISH represents and additional method used to demonstrate the physical state of these viruses (Black, 1968; Munday, 2014; Melo et al., 2015). Another additional technique to identify PVs is L1 immunodetection (Nasir and Reid, 1999; Longworth and Laimins. 2004; Roperto et al., 2011; Munday, 2014; Melo et al., 2015; Araldi et al., 2015b)., which not only allows identification of the viral presence, but also provides an important evidence of productive infection (Nasir and Reid, 1999; Roperto et al., 2011; Melo et al., 2015).
Histopathological analysis of BPV-infected lesions is a differential diagnosis method, which allows to identify intra-epithelial neoplasms with oncogenic potential (Monteiro et al., 2008; Araldi et al., 2015b).
BPV infection pathway and histopathological alterations
BPV transmission can occur by direct (animal-animal) or indirect contact with contaminated surfaces (Muro et al., 2008; Cubie, 2013). Studies also show that the virus can be transmitted by flies (Finlay et al., 2009) and ticks (Muro et al., 2008).
Studies of PV infection are based on the prototype BPV-1 (Florin et al., 2012). According to the literature, the viral infection requires a tissue micro-injury, exposing heparin sulfate proteoglycan receptors (Ljubojevic and Skerlev, 2014; Greber, 2016), which are needed for BPV L1 anchorage (Hartl et al., 2011; Buck et al., 2013)., This is confirmed by treatment with heparinase preventing viral infection (Kines et al., 2016). The micro-injury is necessary for the virus to access basal keratinocytes (Liu and Baleja, 2008), where the virus cycle begins (Campo, 1997a). Current studies also show that integrin α6 (CD49f) and integrin 332 (laminin 5) are targets for L1 binding (Sibbet et al., 2000; Florin et al., 2012).
The L1 binding to heparin sulfate leads to conformational changes in capsid icosaedric structure (Buck et al., 2013). This exposes the L2 N-terminal to be cleaved by furin protein, present in the cell membrane (Buck et al., 2013). This cleavage induces a second capsid conformational change, allowing L2 to bind to different receptors, such as integrin α2β4 (Buck et al., 2013). Next, the virions are internalized by an clathrin-dependent endocytose mechanism, resulting in cytoplasmic vesicles that associate to lysosomes (Day et al., 2003). The lysosomal acid content release promotes pH alterations in capsid proteins, resulting in viral DNA release (Day et al., 2003). The BPV genome is found in epissomal form (Campo, 2002; Munday, 2014; Cota et al., 2015), while HPV can integrate in fragile sites of the host genome (Monte and Peixoto, 2010; Moody and Laimins, 2010; Munday, 2014). A current study based on qRT-PCR, showed that cutaneous papillomas have about 2.2104 viral copies (Cota et al., 2015).
PVs do not codify polymerase (Moody and Laimins, 2010). For this reason, these viruses induce the S-phase entry, which was verified in BPV-infected cells (Potocki et al., 2014), in a process known as amplification (Moody and Laimins, 2010). Due to the stimulation of cell proliferation, BPV induces mitotic stress (Potocki et al., 2014), resulting in cytogenetic aberrations (Stocco dos Santos et al., 1998; Melo et al., 2011; Araldi et al., 2013; Campos et al., 2013). Moreover, the viral hyperproliferative action leads to the exophytic mass development as a consequence of acanthosis (Cubie, 2013; Munday, 2014; Araldi et al., 2015b). As occurs with epithelium differentiation and virus assembly, a keratinization process is verified (Ferraro et al., 2011). This process is histologically characterized by the increase of keratin granules in the granular layer (Ferraro et al., 2011; Araldi et al., 2015b).
Virus assembly is observed in the most differentiated epithelium layers (Munday, 2014), where the virion release occurs by cell degeneration (Brobst and Hinsman, 1966; Buck et al., 2013). This process results in koilocyte formation. The term koilocyte comes from the Greek word koillos, which means “cavity” (Ferraro et al., 2011). Koilocyte formation results from the cytopathic effect of E5 and E6 oncoproteins, although the molecular mechanism that results in cell vacuolization remains unclear (Krawczyk et al., 2008). However, the cytoplasmic vacuolization contributes to cell fragility and virion release (Krawczyk et al., 2008; Wang and Kieff, 2013). In this sense, koilocytes are cells destined to apoptosis, which emerges as a consequence of DNA replication and macromolecule synthesis inhibition.
After viral assembly, virions are released in the corneum layer, allowing infiltration in the keratin matrix (Brobst and Hinsman, 1966; Buck et al., 2013). This mechanism confers an immune evasion, since the icosaedric morphology of PVs is immunoreactive (Buck et al., 2013). In addition, keratin confers physical protection for virions, as they are non-enveloped.
BPV as a model for HPV
HPV is species-specific, infecting exclusively humans (Koller and Olson, 1972; Campo, 2002). This specificity was demonstrated in the 1970s, when calves, hamsters, ponies and Rhesus monkeys were inoculated with BPV and HPV virions isolated by ultracentrifugation (Koller and Olson, 1972). However, only BPV was able to infect the species (Koller and Olson, 1972). Due to its capability to infect different species and the pathogenic and morphological characteristics shared with HPV (Munday, 2014), BPV has been used as a prototype to study PV biology and oncology (Koller and Olson, 1972; Campo, 2002; Liu et al., 2005; Campo, 2006; Costa and Medeiros, 2014). Therefore, research involving BPV has contributed with the understanding of viral oncogenesis (Costa and Medeiros, 2014; Munday, 2014). Based on these data, the next sections will focus on BPV and its host interaction.
Equine sarcoid: an example of cross-infection
Although considered epithelial- and mucous-tropic, some BPV types, especially Deltapapillomavirus, can infect fibroblasts causing sarcoma-like lesions, known as equine sarcoid (Chambers et al., 2003b; Geisshüsler et al., 2016). Equine sarcoid was first described by Jackson (1936), being considered a biphasic neoplasia, since it affects both epidermis and dermis. Equine sarcoid is the most frequent benign skin neoplasia observed in horses that are 1-6 years old (Jackson, 1936; Otten et al., 1993; Nasir and Reid, 1999; Martens et al., 2000; Bergvall, 2013; Mosseri et al., 2014), affecting 11.5% of all horses (Knottenbelt, 2005). Differently from papillomas, sarcoids rarely present a spontaneous regression (Angelos et al., 1991).
Equine sarcoid is an invasive but not metastatic neoplasia (Nasir and Reid, 1999), causing substantial morbidity and economic loss due to aesthetic and functional impairment (Angelos et al., 1991; Otten et al., 1993). Six different sarcoid morphotypes are known: occult, verrucous, nodular, fibroblastic, mixed and malignant (Knottenbelt, 2005). However, 84% of affected equines have more than one sarcoid morphotype (Goodrich et al., 1998).
Equine sarcoids are lesions with intense fibroblastic proliferation, in which fibroblasts are disposed in fusiform bundles or spirally organized, presenting a morphology of fibropapilloma-like (Martens et al., 2000). Another characteristic of this neoplasia is the presence of anaplastic and pleomorphic fibroblasts, perpendiculary orientated in relation to basal membrane, and being observed in the dermo-epidermal junction (Bogaert et al., 2010). The epidermal component is only present in verrucous and mixed sarcoids (Bogaert et al., 2010). The sarcoid invasiveness capability can be attributed to the high levels of expression of metalloproteinase. MMP1 promotes the laminin and collagen IV degradation, resulting in extracellular matrix remodeling (Mosseri et al., 2014)
Equine sarcoid has a multifactorial cause (Bergvall, 2013). However, the Deltapapillomavirus is recognized as an etiological factor (Nasir and Reid, 1999; Chambers et al., 2003b; Bogaert et al., 2008b; Bergvall, 2013). The association between BPV and equine sarcoid was first described by Olson and Cook in 1951 (Brandt et al., 2008). BPV-1 and 2 DNA sequences are identified in 100% of equine sarcoids (Martens et al., 2001a; Gaynor et al., 2015b). Furthermore, DNA sequences of these virus types are identified in 2/3 of asymptomatic horses (Bravo et al., 2010). These data demonstrate that the virus can be asymptomatic, remaining latent in epidermis and dermis (Bogaert et al., 2008b; Brandt et al., 2008; Bergvall, 2013).
Viral latency is a characteristic shared by BPV (Bogaert et al., 2008b; Araldi et al., 2013; Silva et al., 2013a) and HPV (Maran et al., 1995; Astori et al., 1998; Forslund et al., 2004). However, the virus infection induces DNA damages and genomic instability (Stocco dos Santos, et al., 1998; Melo et al., 2011; Araldi et al., 2013; Campos et al., 2013; Melo et al., 2015). Moreover, studies show that 70% of BPV-infected asymptomatic horses live in contact with cattle (Bergvall, 2013). However, although the presence of BPV-1 L1 transcripts was already verified in equine sarcoids by RT-PCR (Nasir and Reid, 1999), these neoplasias are considered abortive infection sites, since virion presence was not described. (Bogaert et al., 2008b; Brandt et al., 2008). The absence of productive infection in equine sarcoids suggests that the BPV cross-infection is a consequence of an erratic virus cycle (Otten et al., 1993).
As verified in PB, sarcoids are frequently seen in sites most susceptible to traumatism (Angelos et al., 1991; Otten et al., 1993; Martens et al., 2000), confirming the need of tissue micro-injury for virus infection. In addition, as in bovines, it is believed that insects can contribute to BPV transmission, because BPV-1 DNA sequences were already identified in Musca automnalis, Fannia carnicularis and Stomoxys calcinatrans flies (Bergvall, 2013).
Studies also show that the Arabic breed is the most susceptible to sarcoid development (Knottenbelt, 2005; Bogaert et al., 2008b). The reason for this is the presence of W3 and B1 MHC-II haplotypes that facilitate BPV infection persistence (Chambers et al., 2003b; Bogaert et al., 2008b).
Currently, treatments for sarcoid, as well as for PB, are almost inefficient (Bergvall, 2013). Treatment methods are: (1) surgical excision of the neoplasia, with recurrence in 50-64% of the cases within six months (Lancaster et al., 1977; Martens et al., 2001b; Bergvall, 2013; Mosseri et al., 2014), (2) laser therapy, where recurrence is 38% (Martens et al., 2001b); cryotherapy, which is inefficient for lesions larger than 2 cm2 (Carr, 2009), and chemotherapy using cisplatin or 5'-fluouracyl (5-FU), that can cause nephro and hepatotoxicity (Stewart et al., 2006). The recurrence of disease is argued to be a consequence of BPV presence in the surgical margin (Martens et al., 2001a). However, a current study showed a lack of correlation between BPV DNA in surgical margins and recurrence of equine sarcoids (Taylor et al., 2014). Meanwhile, Brandt et al. (2008) identified BPV DNA sequences in peripheral blood of sarcoid-affected equines. These data suggest that, as verified in bovines (Stocco dos Santos et al., 1998; Roperto et al., 2011; Araldi et al., 2013; Melo et al., 2015), the peripheral blood can be argued as being a vehicle of viral dissemination.
Breaking paradigms
After the first reports (Campo and Jarret, 1986; Walter-Moura et al., 1988), our purpose of investigation was to examine the role of peripheral blood as a potential BPV-transmitting agent. The first data came from Stocco dos Santos et al. (1998), which showed high levels of chromosomal aberrations in lymphocytes infected by BPV-2. That study also described the presence of BPV-2 DNA sequences in peripheral blood of donor and recipient animals and in the progeny of recipient animals. These results were the first evidence of vertical transmission (Freitas et al., 2003). In this study, we verified the presence of BPV DNA sequences and DNA damages in peripheral blood samples of animals from different areas of Brazil (Diniz et al., 2009; Melo et al., 2011; Araldi et al., 2013). In addition, we also demonstrated the presence of different BPV types in peripheral blood and cutaneous papilloma (Araldi et al., 2014a). All these data suggest a viral activity in blood cells (Stocco dos Santos et al., 1998; Araldi et al., 2013). The presence of BPV DNA sequences in peripheral blood mononuclear cells (PBMCs) was also described in the literature in both bovines (Roperto et al., 2008; Silva et al., 2013a) and equines (Brandt et al., 2008, 2011), reinforcing our results. Following studies also showed the presence of BPV transcripts and the L1 capsid protein in PBMCs, demonstrating the productive infection in blood cells (Roperto et al., 2011; Melo et al., 2015). We also described the presence of BPV DNA sequences in different non-epithelial tissues such as spermatozoa, urine and milk (Lindsey et al., 2009). These data demonstrate the need to review the natural history of papillomavirus, as currently proposed by Munday (2014).
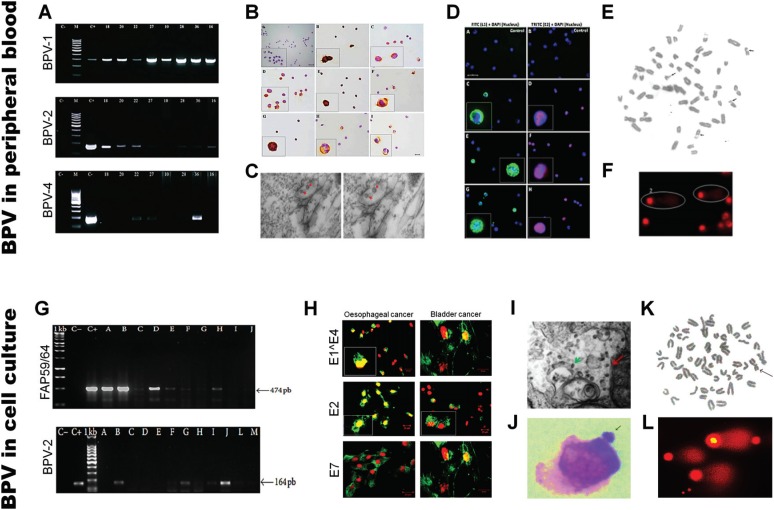
We verified that primary culture cell lines from BPV-infected cutaneous and esophageal papilloma have chromosomal aberrations similar to those verified in peripheral blood (Campos et al., 2013). In addition, using BPV-4 E7 oncoprotein transformed PALF cell lines, we demonstrated the mutagenic potential of quercetin, a flavonoid found in bracken fern P. aquilinum, which is recognized as a co-factor to BPV-associated upper gastric cancer (Leal et al., 2003). In a current study, we also showed that the BPV induced metabolic alteration in host cells, increasing reactive oxygen species and DNA damages (Araldi, 2016; Araldi et al., 2016). A summary of our main results is shown in Figure 3. These data suggest that BPV-infected cell lines are a useful model to study the pathogenic mechanisms that lead to cancer. Our contribution has allowed to know the BPV prevalence and distribution in Brazil (Diniz et al., 2009; Carvalho et al., 2013; Lunardi et al., 2013; Melo et al., 2014; Araldi et al., 2014b; Cota et al., 2015; Gaynor et al., 2015a; Grindatto et al., 2015; Dong et al., 2016; Martano et al., 2016). These studies showed that the co-infection of at least two BPV types is frequent (Araldi et al., 2013, 2014a; Carvalho et al., 2013), demonstrating the importance to develop multivalent vaccines.
Figure 3. Evidences of BPV presence in peripheral blood and cell culture showing: A) amplicon of BPV identified using specific primers for BPV-1, 2 and 4 (Araldi et al., 2013); B) BPV genome identification by chromogenic in situ hybridization (Melo et al., 2015); C) BPV virions identified in cytoplasmic vesicles of PBMCs (Melo et al., 2015); D) Immunodetection of BPV L1 and E2 in lymphocytes (Melo et al., 2015); E) cytogenetic aberrations (breaks) observed in BPV-infected lymphocyte (Stocco dos Santos et al., 1998), F) class 2 nucleoids observed in BPV-infected lymphocytes (Araldi et al., 2013); G) BPV DNA sequences identified in cell culture, using FAP59/64 degenerate primers; H) Immunodetection of BPV early proteins (E1^E4, E2, E7) in both oesophageal and urinary bladder carcinoma cell line (Melo et al., data not published); I) BPV virion, with 55 nm, in cytoplasmic vesicle of cutaneous papilloma cell line, total magnification of 60000X (Melo et al., data not published); J) bovine epithelial cell line (CRIB) treated with E6 recombinant oncoprotein showing micronucleus presence (Araldi et al., 2015a); K) cytogenetic aberration observed in BPV-infected cell line (Campos et al., 2013); L) class 2 nucleoids observed in cutaneous papilloma cell line (Araldi et al., data not published).
BPV-associated malignant neoplasms
Urinary bladder carcinoma
Urinary bladder carcinoma represents 0.01% of all bovine cancers (Roperto et al., 2015). It is estimated that urinary bladder cancer has caused economic losses of € 4 million between 2000-2006 in the Azores (Costa and Medeiros, 2014).
The etiopathogenic role of BPV in urinary bladder carcinoma was first described in 1955 in Brazil and South Africa (Plowrigh, 1955). Currently, the BPV promoter action in urothelial carcinoma is well established. This is because sequences of BPV-1, 2, 13 and 14, as well as the expression of E5 oncoprotein are detected in this neoplasms (Wosiacki et al., 2006; Balcos et al., 2008; Roperto et al., 2016; Russo et al., 2016).
Urinary bladder carcinoma is clinically characterized by bovine enzootic hematuria (BHE) (Wosiacki et al., 2002), which is verified in more than 90% of cattle with urinary bladder cancer (Resendes et al., 2011). BHE is characterized by intermittent hematuria, which can lead to anemia and weight loss (Wosiacki et al., 2002). The disease affects preferably cattle between 3-5 years old, without breed preference (Wosiacki et al., 2002). BHE is associated to dietary intake of carcinogenic compounds present in P. aquilinum, P. esculetum, P. revolution, Chelanthes seiberi, Encephalartos hildebrandti, Ranunaelus montana, R. acris, Climatis vitalbi (Wosiacki et al., 2002). Among these species, P. aquilinum stands out for its wide biogeographical distribution (Oliveira, 2012). Bracken fern is commonly observed in the South of Brazil, where there is a high incidence of BEH (Dias et al., 2012).
P. aquilinum presents high levels of immunosuppressor and carcinogenic compounds, including quercetin, ptaquilosides and shikimic acid (Shahin et al., 1999; Beniston et al., 2001; Bonadies et al., 2004). The immunosuppressor activity of these metabolites contributes to BPV infection persistence and represents an additional source of DNA mutation (Leal et al., 2003). According to Tokarnia et al. (2000), the diary intake of 10 kg of Pteridum ssp. in a year can lead to BEH. Moreover, the consumption of this bracken fern can result in urinary bladder and esophageal carcinoma (Masuda et al., 2011).
Therefore, BPV-1 and 2 are considered important cofactors for BEH development (Campo, 1997a; Roperto et al., 2005; Pavelski et al., 2014), once BPV leads to epidermal and dermal hyperproliferation at the same time that it induces DNA damages, contributing to cancer initiation (Stocco dos Santos et al., 1998; Araldi et al., 2013; Campos et al., 2013).
Esophageal carcinoma (EC)
While the association between HPV and EC remains under discussion despite all evidences (Antonsson et al., 2016), the etiological role of BPV in EC is well established (Borzacchiello et al., 2003). EC in cattle is a self-limiting disease, being directly associated to BPV-4 infection (Borzacchiello et al., 2003; Masuda et al., 2011).
Although there is no report about BPV transmission in humans, a study performed in Germany showed a high incidence of warts in veterinarians that had contact with bovines (Bosse and Christophers, 1964). Another study in Central Asia discussed the association between milk consumption and EC (Nasrollahzadeh et al., 2015). Considering that BPV DNA sequences were already detected in milk (Lindsey et al., 2009) and the thermal resistance of viral capsid (Módolo et al., data not published), it seems that the virus can survive to pasteurization process (zur Hausen, 2012). Therefore, more efforts are necessary to verify this possible cross-infection.
EC is the eighth most prevalent human malignancy, being considered the sixth causa mortis for cancer globally (Antonsson et al., 2010; Herbster et al., 2012). Considered the third most common gastrointestinal cancer (Felin et al., 2008), EC has a high incidence in men (Bjørge et al., 1997). In 2002, 462,000 novel cases of EC in the world were reported (Antonsson et al., 2010). Brazil had 10,780 novel cases of EC in 2014 and 7,636 deaths due to the disease in 2011 (INCA, 2016).; its incidence has increased in the last years (INCA, 2016). This data leads to concern, since EC has a mortality rate that is 25% higher than cervical cancer (Han et al., 1996; Kahrilas and Hirano, 2013).
Among the clinical signs of EC in humans are progressive dysphagia, weight loss, odynophagia, anorexia, fever and retrosternal pain (Felin et al., 2008; Haster and Owyang, 2013), which are similar to those verified in bovines (Borzacchiello et al., 2003). Scarce epidemiological data about EC in cattle are available. In humans, EC has a variable incidence according to geographic area (Bjørge et al., 1997; Syrjänen, 2002; Guo et al., 2012; Nasrollahzadeh et al., 2015; Antonsson et al., 2016). Among countries with high EC incidence are China, Singapore, Iran, South Africa and Brazil (Bey et al., 1976; Han et al., 1996; Syrjänen, 2002; Guo et al., 2012). Due to the high number of EC cases in Central Asia (100/100,000) in relation to North America and Western Europe (5-10/100,000), the region is known as the Asian Esophageal Cancer Belt (Nasrollahzadeh et al., 2015). Although the reason for EC incidence variation is unknown (Syrjänen, 2002; Nasrollahzadeh et al., 2015), studies indicate the contribution of environmental factors, in addition to infectious agents, in the oncogenic process (Chang et al., 1992; Antonsson et al., 2010).
Smoking and alcohol consumption are pointed out as etiological factors for human EC (Han et al., 1996; Lagergren et al., 1999; Syrjänen, 2002; Haster and Owyang, 2013). However, due to sociocultural reasons, smoking does not justify the high incidence of EC in Central Asia (Nasrollahzadeh et al., 2015), reinforcing the contribution of infectious agent in esophageal carcinogenesis, and evidence shows the etiopathogenic contribution of different infectious agents in EC, such as cytomegalovirus (CMV), Epstein-Barr virus (EBV), herpes simplex and HPV (Syrjänen, 2002).
Although the association between HPV and pre-malignant cervical lesions is known since the 1970s (Syrjänen, 2002), the participation of this virus in EC had its history marked by controversial reports in the literature (Lavergne and de Villiers, 1999). The association between HPV and EC was first proposed by Syrjänen (1982). On the one hand, HPV protein expression was observed in samples by IHC (Winkler et al., 1985; Hille et al., 1986; Kulski et al., 1986) and viral DNA sequences were identified by CISH in EC (Chang et al., 1993; Togawa et al., 1994; Cooper et al., 1995),. On the other hand, serological studies contested these results (Dillner et al., 1995; Han et al., 1996; Lagergren et al., 1999). However, despite these controversial results, the association between HPV and EC is currently accepted, being discussed in the 18th edition of Harrison's Principles of Internal Medicine (Kahrilas and Hirano, 2013). Thus, the absence of HPV detection by serological methods reflects a problem still observed nowadays: the use of antibodies against late (L1) proteins (Dillner et al., 1995; Lagergren et al., 1999). However, considering that EC does not present an HPV productive infection, the late protein expression is not expected. This occurs because HPV presents a “hit and run” mechanism (Han et al., 1996; Bjørge et al., 1997).
Not only serological studies are controversial. Studies based on PCR also show a low correlation between HPV and EC, mainly in clinical samples from Australia (Antonsson et al., 2010; Antonsson et al., 2016). These results can be attributed to the origin of clinical samples, since Australia is not considered a high-risk area for HPV-associated EC. In addition, problems involving primer sensitivity are extensively discussed in the literature of both BPV and HPV (Lavergne and de Villiers, 1999; Silva et al., 2013b; de Villiers, 2013; Araldi et al., 2014b). However, evidence of PVs etiopathogenic action in EC has accumulated along the last 20 years. Among these are: (1) the presence of koilocytes in EC biopsies (Bjørge et al., 1997; Syrjänen, 2002; Vieira et al., 2013), (2) loss or mutation in p53 as a consequence of the PV E6 oncoprotein (Chang et al., 1994, 1995; Herbster et al., 2012), (3) increase in telomerase activity (Syrjänen, 2002), and detection of DNA sequences of HPV-6, 11, 18, 31 and 33 in EC (Vieira et al., 2013).
Prophylactic and therapeutic vaccines against BPV and HPV
Currently, there are few forms of treatment against BP available (Muro et al., 2008). Among the possibilities is the papilloma surgical excision (Muro et al., 2008). Although frequently employed, this method is inefficient in cattle with high incidence of BPV, because it is impracticable to perform the excision of papillomas of all cattle. Another strategy frequently employed is self-hemotherapy (Leto et al., 2011). This method consists in the removal and intramuscular reinjection of a volume of 10 mL of venous blood, inducing a nonspecific immune stimulus that can promote the “shedding of the warts” (Leto et al., 2011). However, this technique does not avoid BP re-occurrence, thus being a palliative method. Another possibility to reduce the incidence of BP is to control ectoparasite populations, since it was demonstrated that the biological control of ticks reduces the incidence of BPV (William et al., 1992).
Few medical interventions proposed in the last century can match the effects that immunization exerts on longevity (Schuchat and Jackson, 2013). For this reason, vaccination is considered the best form of prevention, control and eradication of viral etiology diseases (Ribeiro-Müller and Müller, 2014). Moreover, immunization reduces both transmission and dissemination of the infectious agent (Schuchat and Jackson, 2013).
Two prophylactic vaccines against HPV are available in the market since 2006: (1) Cervarix, produced by Glaxo-Smith Klein (GSK) and (2) Gardasil, produced by Merck (Ribeiro-Müller and Müller, 2014). These vaccines are based on virus-like particles (VLPs) of the L1 structural protein (Marigliani et al., 2012). Cervarix is a bivalent vaccine able to confer protection against HPV-16 and 18, employing L1 VLPs produced in Baculovirus in Trichoplusia ni insect cells, using aluminum hydrophosphate as adjuvant (Ribeiro-Müller and Müller, 2014). Gardasil is able to confer protection against two high-risk HPV types (HPV-16 and 18) and two low-risk ones (HPV-6 and 11), associated to genital warts. This vaccine is composed by of L1 VLPs produced in Saccharomyces cerevisae, employing lipid A 3-O-diacilete-44-monophosphoryl (ASO4) as adjuvant‥ Both vaccines are considered safe and well tolerated (Ribeiro-Müller and Müller, 2014). For these reasons, more than 30 countries, including Brazil, adopted immunization programs against HPV based on these vaccines.
Australia, the first country to adopt the vaccination against HPV, observed a reduction of 70% in HPV-6, 11, 16 and 18 infection incidence. Similar results were also verified in Denmark, Finland and Sweden (Ribeiro-Müller and Müller, 2014). However, available vaccines are not able to protect against all HPV types, since there are more than 200 described. Moreover, these vaccines have a high cost of production. Thus, it is necessary to invest in novel multivalent vaccines, with lower production cost. Vaccines based on recombinant protein expressed in Escherichia coli have demonstrated to be a useful alternative, because they have a lower cost, and not requiring the L1 VLPs, they are more stable (Ribeiro-Müller and Müller, 2014).
Although there are two vaccines against HPV, there is no vaccine yet against BPV available to date. The idea of developing a vaccine to combat BPV began with the infection of Shope papillomas extract in the 1940 decade (Shope, 1937). Since then, different vaccine models were proposed (Jarret et al., 1991; Gaukroger et al., 1996; Góes et al., 2008; Love et al., 2012; Mazzuchelli-de-Souza et al., 2013). However, none of them became a commercial product. This denotes the notorious difficulty to obtain a safe and efficient vaccine, and reflects the reduced number of research groups dedicated to develop a vaccine against BPV.
Studies have demonstrated that BPV early proteins (E6 and E7) show a therapeutic action, while later proteins (L1 and L2) have a prophylactic action (Campo, 1997b). Vaccines based on E6 and E7 have also been discussed against HPV (Borysiewicz et al., 1996; He et al., 2000; Yao et al., 2013), reinforcing the usefulness of the BPV model not only for comprehending the pathogenic mechanisms of HPV, but also for vaccine biotechnology. However, in a current study based on BPV-1 E6 recombinant oncoprotein, we demonstrated that this oncoprotein is able to induce clastogenesis and neosis per se (Araldi et al., 2015a). This data emphasizes the necessity of an in silico analysis of E6 and E7 oncoproteins, aiming to obtain novel variants that are more antigenic and less mutagenic. In order to guarantee the immunization and safety of products, our group is now focusing on the development of a prophylactic vaccine based on L1 recombinant protein (Módolo et al., data not published).
Conclusions
Papillomaviridae comprises the most extensive known family of viruses, able to infect all vertebrates including humans, in which it is responsible for 27-30% of all infectious agent-associated incident cancer cases. Moreover, PVs represent an important problem in the veterinary field, inducing papillomas in dogs, felines and cattle. Also, BPV infects equines, resulting in sarcoids. Although novel discoveries contributed to the understanding of the PVs oncogenic role, some carcinogenic mechanisms remain unknown, especially those following cancer initiation (Araldi et al., 2016). In addition, current studies have collected evidence of BPV productive infection in sites earlier considered as not permissive, such as PBMCs (Roperto et al., 2011; Melo et al., 2015), placenta (Roperto et al,. 2012) and primary cell cultures (Campos et al., 2008; Campos et al., 2013). Similar results have also been described for HPV (Chiou et al., 2003; Foresta et al., 2013; Pessoa, 2014). However, despite of these advances, the natural history of PVs remains dependent on cell differentiation, emphasizing the need to review the replication cycle of these viruses (White and Howley, 2013; Munday, 2014). In addition, the available vaccines against HPV do not confer protection against all virus types, providing only a limited protection. The veterinary field lives a most dramatic scene, since there is no vaccine available against BPV. Over the last 30 years, our group has dedicated efforts to elucidate the pathogenic mechanisms of PVs, focusing on BPV, once the virus is considered a prototype for HPV studies. Although our contributions brought important advances, more studies are necessary to propose efficacious and safe prophylactic and therapeutic measures.
Dedication
This paper is dedicated to Dr. Maria Luiza Beçak on occasion of her 80th birthday. She and her husband were the founders of the Laboratory of Genetics of the Instituto Butantan, in 1961. They were Brazil's pioneers in introducing human and vertebrate cytogenetics and the first to use cytogenetics as an important tool in human genetic counseling. Dr. Beçak's contribution in classic and recent papers in the genetic literature is very important and remarkable.
Acknowledgments
The authors thank the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, process 2014/20617-5) and Fundação Butantan for the financial support.
Footnotes
Associate Editor: Carlos F. M. Menck
References
- Alberti A, Pirino S, Pintore F, Addis MF, Chessa B, Cacciotto C, Cubeddu T, Anfossi A, Benenati G, Coradduzza E, et al. Ovis aries Papillomavirus 3: A prototype of a novel genus in the family Papillomaviridae associated with ovine squamous cell carcinoma. Virology. 2010;407:352–359. doi: 10.1016/j.virol.2010.08.034. [DOI] [PubMed] [Google Scholar]
- Alcântara B, Alfieri A, Headley S, Rodrigues W, Otonel R, Lunardi M, Alfieri A. Molecular characterization of bovine Deltapapillomavirus (BPV-1, 2, and 13) DNA in equine sarcoids. Pesqui Veterinária Bras. 2015;35:431–436. [Google Scholar]
- Angelos J, Marti E, Lazary S, Carmichael L. Characterization of BPV-like DNA in equine sarcoids. Arch Virol. 1991;119:95–109. doi: 10.1007/BF01314326. [DOI] [PubMed] [Google Scholar]
- Antonsson A, Knight L, Whiteman D. Human papillomavirus not detected in esophageal adenocarcinoma tumor specimens. Cancer Epidemiol. 2016;41:96–98. doi: 10.1016/j.canep.2016.01.014. [DOI] [PubMed] [Google Scholar]
- Antonsson A, Nancarrow D, Brown I, Green A, Drew P, Watson D, Hayward N, Whiteman D. High-risk human papillomavirus in esophageal squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2010;19:2080–2087. doi: 10.1158/1055-9965.EPI-10-0033. [DOI] [PubMed] [Google Scholar]
- Araldi R. Bovine papillomavirus: What We Know and What We Should Know. Lambert Academic Publishing; Germany: 2015. 124 p [Google Scholar]
- Araldi R. Papillomaviruses: From mutation to metastasis. 6th Euro Virology Congress and Expo. 2016:4172–4172. [Google Scholar]
- Araldi R, Melo T, Diniz N, Carvalho R, Beçak W, Stocco R. Bovine papillomavirus clastogenic effect analyzed in comet assay. Biomed Res Int. 2013;2013:1–7. doi: 10.1155/2013/630683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araldi R, Carvalho R, Melo T, Diniz N, Sant’Ana T, Mazzuchelli-de-Souza J, Spadacci-Morena D, Beçak W, Stocco R. Bovine papillomavirus in beef cattle: First description of BPV-12 and putative type BAPV8 in Brazil. Genet Mol Res. 2014a;13:5644–5653. doi: 10.4238/2014.July.25.20. [DOI] [PubMed] [Google Scholar]
- Araldi R, Giovanni D, Melo T, Diniz N, Mazzuchelli-de-Souza J, Sant’Ana T, Carvalho R, Beçak W, Stocco R. Bovine papillomavirus isolation by ultracentrifugation. J Virol Methods. 2014b;208:119–124. doi: 10.1016/j.jviromet.2014.07.029. [DOI] [PubMed] [Google Scholar]
- Araldi R, Mazzuchelli-de-Souza J, Modolo D, Souza E, Melo T, Spadacci-Morena D, Magnelli R, Rocha M, De-Sá-Júnior P, Carvalho R, et al. Mutagenic potential of Bos taurus papillomavirus type 1 E6 recombinant protein: First description. Biomed Res Int. 2015a;2015:1–15. doi: 10.1155/2015/806361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araldi R, Melo T, Neves A, Spadacci-Morena D, Magnelli R, Módulo D, De-Sá-Júnio P, Mazzuchelli-de-Souza J, Carvalho R, Beçak W, et al. Hyperproliferative action of bovine papillomavirus (BPV): Genetics and histopathological aspects. Genet Mol Res. 2015b;14:12942–12954. doi: 10.4238/2015.October.21.15. [DOI] [PubMed] [Google Scholar]
- Araldi R, Módolo D, De-Sá-Júnior P, Consonni S, Carvalho R, Roperto F, Beçak W, Stocco R. Genetics and metabolic deregulation following cancer initiation: A world to explore. Biomed Pharmacother. 2016;82:449–458. doi: 10.1016/j.biopha.2016.05.031. [DOI] [PubMed] [Google Scholar]
- Astori G, Lavergne D, Benton C, Hockmayr B, Egawa K, Garbe C, de Villiers E. Human papillomavirus are commonly found in normal skin of immunocompetent hosts. J Invest Dermatol. 1998;110:752–755. doi: 10.1046/j.1523-1747.1998.00191.x. [DOI] [PubMed] [Google Scholar]
- Balcos L, Borzacchiello G, Russo V, Popescu O, Roperto S, Roperto F. Association of bovine papillomavirus type-2 and urinary bladder tumours in cattle from Romania. Res Vet Sci. 2008;85:145–148. doi: 10.1016/j.rvsc.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Beniston RG, Morgan IM, O’Brien V, Campo MS. Quercetin, E7 and p53 in papillomavirus oncogenic cell transformation. Carcinogenesis. 2001;22:1069–1076. doi: 10.1093/carcin/22.7.1069. [DOI] [PubMed] [Google Scholar]
- Bergvall K. Sarcoids. Vet Clin Equine. 2013;29:657–671. doi: 10.1016/j.cveq.2013.09.002. [DOI] [PubMed] [Google Scholar]
- Bernard H. Coevolution of papillomaviruses with human populations. Trends Microbiol. 1994;2:18–21. doi: 10.1016/0966-842x(94)90602-5. [DOI] [PubMed] [Google Scholar]
- Bernard H, Calleja-Macias I, Dunn S. Genome variation of human papillomavirus types: Phylogenetic and medical implications. Int J Cancer. 2006;118:1071–1076. doi: 10.1002/ijc.21655. [DOI] [PubMed] [Google Scholar]
- Bey E, Alexander J, Whitcutt J, Hunt J, Gear J. Carcinoma of the esophagus in Africans: Establishment of a continuously growing cell line from a tumor specimen. In Vitro. 1976;12:107–114. doi: 10.1007/BF02796356. [DOI] [PubMed] [Google Scholar]
- Bjørge T, Hakulinen T, Engeland A, Jellum E, Koskel P, Lehtinen M, Luostarinen T, Paavonen J, Sapp M, Schiller J, et al. A prospective, seroepidemiological study of the role of human papillomavirus in esophageal cancer in Norway. Cancer Res. 1997;57:3989–3992. [PubMed] [Google Scholar]
- Black P. The oncogenic DNA viruses: A review of in vitro transformation studies. Annu Rev Microbiol. 1968;22:391–426. doi: 10.1146/annurev.mi.22.100168.002135. [DOI] [PubMed] [Google Scholar]
- Bocaneti F, Altamura G, Corteggio A, Velescu E, Roperto F, Borzacchiello G. Bovine papillomavirus: New insights into an old disease. Transbound Emerg Dis. 2014;63:1–10. doi: 10.1111/tbed.12222. [DOI] [PubMed] [Google Scholar]
- Bogaert L, Martens A, Van Poucke M, Ducatelle R, De Cock H, Dewulf J, De Baere C, Peelman L, Gasthuys F. High prevalence of bovine papillomaviral DNA in the normal skin of equine sarcoid-affected and healthy horses. Vet Microbiol. 2008a;129:58–68. doi: 10.1016/j.vetmic.2007.11.008. [DOI] [PubMed] [Google Scholar]
- Bogaert L, Martens A, Kast W, Van Marck E, De Cock H. Bovine papillomavirus DNA can be detected in keratinocytes of equine sarcoid tumors. Vet Microbiol. 2010;146:269–275. doi: 10.1016/j.vetmic.2010.05.032. [DOI] [PubMed] [Google Scholar]
- Bogaert L, Martens A, Van Poucke M, Ducatelle R, De Cock H, Dewulf J, De Baere C, Peelman L, Gasthuys F. High prevalence of bovine papillomaviral DNA in the normal skin of equine sarcoid-affected and healthy horses. Vet Microbiol. 2008b;129:58–68. doi: 10.1016/j.vetmic.2007.11.008. [DOI] [PubMed] [Google Scholar]
- Bonadies F, Borzacchiello G, Dezzi S, Nicoletti R, Roperto S. Mass spectrometric analysis of ptaquiloside, the toxic sesquiterpene from bracken fern. Rapid Commun Mass Spectometry. 2004;18:825–828. doi: 10.1002/rcm.1410. [DOI] [PubMed] [Google Scholar]
- Boon S, Tomaic V, Thomas M, Roberts S, Banks L. Cancer-causing human papillomavirus E6 proteins display major differences in the phospho-regulation of their PDZ interactions. J Virol. 2015;89:1579–1586. doi: 10.1128/JVI.01961-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börkü M, Atalay O, Kibar M, Cam Y, Atasever A. Ivermectin is an effective treatment for bovine cutaneous papillomatosis. Res Vet Sci Sci. 2007;83:360–363. doi: 10.1016/j.rvsc.2007.01.016. [DOI] [PubMed] [Google Scholar]
- Borysiewicz L, Fiander A, Nimako M, Man S, Wilkinson G, Westmoreland D, Evans A, Adams M, Stacey S, Boursnell M, et al. A recombinant vaccinia virus encoding human papillomavirus types 16 and 18, E6 and E7 proteins as immunotherapy for cervical cancer. Lancet. 1996;347:1523–1527. doi: 10.1016/s0140-6736(96)90674-1. [DOI] [PubMed] [Google Scholar]
- Borzacchiello G. Bovine papillomavirus infections in animals. Commun Curr Res Educ Top Trends Appl Microbiol. 2007:673–679. [Google Scholar]
- Borzacchiello G, Ambrosio V, Roperto S, Poggiali F, Tsirimonakis E, Venuti A, Campo M, Roperto F. Bovine papillomavirus type 4 in oesophageal papillomas of cattle from the South of Italy. J Comp Pathol. 2003;128:203–206. doi: 10.1053/jcpa.2002.0626. [DOI] [PubMed] [Google Scholar]
- Borzacchiello G, Roperto F. Bovine papillomaviruses, papillomas and cancer in cattle. Vet Res. 2008;39:45–45. doi: 10.1051/vetres:2008022. [DOI] [PubMed] [Google Scholar]
- Bosse K, Christophers E. Beitrag zur Epidemiologie der Warzen. Hautarzt. 1964;15:80–80. [PubMed] [Google Scholar]
- Boulet G, Horvath C, Vanden B, Sahebali S, Bogers J. Human papillomavirus: E6 and E7 oncogenes. Int J Biochem Cell Biol. 2007;39:2006–2011. doi: 10.1016/j.biocel.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Brandt S, Haralambus R, Schoster A, Kirnbauer R, Stanek C. Peripheral blood mononuclear cells represent a reservoir of bovine papillomavirus DNA in sarcoid-affected equines. J Gen Virol. 2008;89:1390–1395. doi: 10.1099/vir.0.83568-0. [DOI] [PubMed] [Google Scholar]
- Brandt S, Schoster A, Tober R, Kainzbauer C, Burgstaller JP, Haralambus R, Steinborn R, Hinterhofer C, Stanek C. Consistent detection of bovine papillomavirus in lesions, intact skin and peripheral blood mononuclear cells of horses affected by hoof canker. Equine Vet J. 2011;43:202–209. doi: 10.1111/j.2042-3306.2010.00147.x. [DOI] [PubMed] [Google Scholar]
- Bravo I, Felez-Sanchez M. Papillomaviruses: Viral evolution, cancer and evolutionary medicine. Evol Med Public Health. 2015;2015:32–51. doi: 10.1093/emph/eov003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo I, Sanjosé S, Gottschling M. The clinical importance of understanding the evolution of papillomaviruses. Trends Microbiol. 2010;18:432–438. doi: 10.1016/j.tim.2010.07.008. [DOI] [PubMed] [Google Scholar]
- Brobst D, Hinsman E. Electron microscopy of the bovine cutaneous papilloma. Vet Pathol. 1966;3:196–207. doi: 10.1177/030098586600300304. [DOI] [PubMed] [Google Scholar]
- Brücher B, Jamall I. Epistemology of the origin of cancer: A new paradigm. BMC Cancer. 2014;15:1–15. doi: 10.1186/1471-2407-14-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck C, Day P, Trus B. The papillomavirus major capsid protein L1. Virology. 2013;445:169–174. doi: 10.1016/j.virol.2013.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck C, Pastrana D, Lowy D, Schiller J. Efficient intracellular assembly of papillomaviral vectors. J Virol. 2004;78:751–751. doi: 10.1128/JVI.78.2.751-757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt A, Willingham M, Gay C, Jeang K, Schlegel R. The E5 oncoprotein of bovine papillomavirus is oriented asymmetrically in Golgi and plasma membranes. Virology. 1989;339:334–339. doi: 10.1016/0042-6822(89)90391-7. [DOI] [PubMed] [Google Scholar]
- Burnett S, Jareborg N, Dimaiot D. Localization of bovine papillomavirus type 1 E5 protein to transformed basal keratinocytes and permissive differentiated cells in fibropapilloma tissue. Proc Natl Acad Sci U S A. 1992;89:5665–5669. doi: 10.1073/pnas.89.12.5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Lv L, Shao Q, Li X, Dian A. Human papillomavirus early proteins and apoptosis. Arch Gynecol Obstet. 2013;287:541–548. doi: 10.1007/s00404-012-2665-z. [DOI] [PubMed] [Google Scholar]
- Campo M. Bovine papillomavirus and cancer. Vet J. 1997a;154:175–188. doi: 10.1016/s1090-0233(97)80019-6. [DOI] [PubMed] [Google Scholar]
- Campo M. Vaccination against papillomavirus in cattle. Clin Dermatol. 1997b;15:275–283. doi: 10.1016/s0738-081x(96)00165-4. [DOI] [PubMed] [Google Scholar]
- Campo M. Animal models of papilloma virus pathogenesis. Virus Res. 2002;89:249–261. doi: 10.1016/s0168-1702(02)00193-4. [DOI] [PubMed] [Google Scholar]
- Campo M. Papillomavirus research: From natural history to vaccine and beyond. Caister Academic Press; Norfolk: 2006. Bovine papillomavirus: Old system, new lessons? pp. 373–387. [Google Scholar]
- Campo M, Jarret WF. Papillomavirus infection in cattle: Viral and chemical cofactors in naturally occurring and experimentally induced tumours. Ciba Found Symp. 1986;120:117–135. doi: 10.1002/9780470513309.ch9. [DOI] [PubMed] [Google Scholar]
- Campos S, Trindade C, Ferraz O, Giovanni D, Lima A, Caetano HA, Carvalho R, Birgel E, Dagli M, Mori E, et al. Can established cultured papilloma cells harbor bovine papillomavirus? Genet Mol Res. 2008;7:1119–1126. doi: 10.4238/vol7-4gmr498. [DOI] [PubMed] [Google Scholar]
- Campos S, Melo T, Assaf S, Araldi R, Mazzuchelli-de-Souza J, Sircili M, Carvalho R, Roperto F, Beçak W, Stocco R. Chromosome aberrations in cells infected with bovine papillomavirus: Comparing cutaneous papilloma, esophagus papilloma, and urinary bladder lesion cells. ISRN Oncol. 2013;2013:910849–910849. doi: 10.1155/2013/910849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr E. New developments in diagnosis and treatment of equine sarcoids. In: Robinson N, Sprayberry K, editors. Current Therapy in Equine Medicine. 6th ed. Sauders Elsevier; St. Louis: 2009. pp. 698–705. [Google Scholar]
- Carvalho R, Sakata S, Giovanni D, Mori E, Brandão P, Richtzenhain L, Pozzi C, Arcaro J, Miranda M, Mazzuchelli-de-Souza J, et al. Bovine papillomavirus in Brazil: Detection of coinfection of unusual types by a PCR-RFLP method. Biomed Res Int. 2013;2013:270898–270898. doi: 10.1155/2013/270898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catroxo M, Martins A, Petrella S, Souza F, Nastari B. Ultrastructural study of bovine papillomavirus during outbreaks in Brazil. Int J Morphol. 2013;31:777–784. [Google Scholar]
- Chambers G, Ellsmore V, O’Brien P, Reid S, Love S, Campo M, Nasir L. Sequence variants of bovine papillomavirus E5 detected in equine sarcoids. Virus Res. 2003a;96:141–145. doi: 10.1016/s0168-1702(03)00175-8. [DOI] [PubMed] [Google Scholar]
- Chambers G, Ellsmore V, O’Brien P, Reid S, Love S, Campo M, Nasir L. Association of bovine papillomavirus with the equine sarcoid. J Gen Virol. 2003b;84:1055–1062. doi: 10.1099/vir.0.18947-0. [DOI] [PubMed] [Google Scholar]
- Chang F, Syrjanen S, Wang L, Syrjanen K. Infectious agents in the etiology of esophageal cancer. Gastroenterology. 1992;103:1336–1348. doi: 10.1016/0016-5085(92)91526-a. [DOI] [PubMed] [Google Scholar]
- Chang F, Syrjanen S, Shen Q, Wang L, Syrjanen K. Screening for human papillomavirus infections in esophageal squamous cell carcinomas by in situ hybridization. Cancer. 1993;72:2525–2530. doi: 10.1002/1097-0142(19931101)72:9<2525::aid-cncr2820720902>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Chang F, Syrjanenl S, Tervahauta A, Kurvinen K, Wang L, Syrjänen K. Frequent mutations of p53 gene in oesophageal squamous cell carcinomas with and without human papiliomavirus (HPV) involvement suggest the dominant role of environmental carcinogens in oesophageal carcinogenesis. Br J Cancer. 1994;70:346–351. doi: 10.1038/bjc.1994.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, Syrjänen S, Syrjänen K. Implications of the p53 tumor-suppressor gene in clinical oncology. J Clin Oncol. 1995;13:1009–1022. doi: 10.1200/JCO.1995.13.4.1009. [DOI] [PubMed] [Google Scholar]
- Chen J, Hong Y, Androphy E. Mutational analysis of transcriptional activation by the bovine papillomavirus type 1 E6. Virology. 1997;236:30–36. doi: 10.1006/viro.1997.8725. [DOI] [PubMed] [Google Scholar]
- Chen P, Lee H. FOXM1 induced by E6 oncoprotein promotes tumor invasion and chemoresistance in HPV-infected lung cancer. Cancer Cell Microenviron. 2015;2:6–9. [Google Scholar]
- Chiou H, Wu M, Liaw Y, Cheng Y, Wong R, Chen C, Lee H. The presence of human papillomavirus type 16/18 DNA in blood circulation may act as a risk marker of lung cancer in Taiwan. Cancer. 2003;97:1558–1563. doi: 10.1002/cncr.11191. [DOI] [PubMed] [Google Scholar]
- Chowdhury D, Keogh M, Ishii H, Peterson C, Buratowski S, Lieberman J. gamma-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol Cell. 2005;20:801–809. doi: 10.1016/j.molcel.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Claus M, Vivian D, Lunardi M, Alfieri A, Alfieri A. Análise filogenética de papilomavírus bovino associado com lesões cutâneas em rebanhos do Estado do Paraná. Pesqui Veterinária Bras. 2007;27:314–318. [Google Scholar]
- Claus M, Lunardi M, Alfieri A, Sartori D, Fungaro H, Alfieri A. Identification of the recently described new type of bovine papillomavirus (BPV-8) in a Brazilian beef cattle herd. Pesqui Veterinária Bras. 2009;29:25–28. [Google Scholar]
- Cooper K, Taylor L, Govind S. Human papillomavirus DNA in oesophageal carcinomas in South Africa. J Pathol. 1995;175:273–277. doi: 10.1002/path.1711750304. [DOI] [PubMed] [Google Scholar]
- Costa R, Medeiros R. Bovine papillomavirus: Opening new trends for comparative pathology. Arch Virol. 2014;159:191–198. doi: 10.1007/s00705-013-1801-9. [DOI] [PubMed] [Google Scholar]
- Cota J, Peleteiro M, Petti L, Tavares L, Duarte A. Detection and quantification of bovine papillomavirus type 2 in urinary bladders and lymph nodes in cases of bovine enzootic hematuria from the endemic region of Azores. Vet Microbiol. 2015a;178:138–143. doi: 10.1016/j.vetmic.2015.03.026. [DOI] [PubMed] [Google Scholar]
- Cotchin E. Problems of comparative oncology. Vet Pathol. 1962;26:633–648. [PMC free article] [PubMed] [Google Scholar]
- Cotchin E. Comparative oncology: The veterinary contribution. Proc R Soc Med. 1976;69:649–656. doi: 10.1177/003591577606900906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubie H. Diseases associated with human papillomavirus infection. Virology. 2013;445:21–34. doi: 10.1016/j.virol.2013.06.007. [DOI] [PubMed] [Google Scholar]
- Cuninghame S, Jackson R, Zehbe I. Hypoxia-inducible factor 1 and its role in viral carcinogenesis. Virology. 2014:456–457. doi: 10.1016/j.virol.2014.02.027. 370-383. [DOI] [PubMed] [Google Scholar]
- Danos O, Georges E, Orth G, Yaniv M. Fine structure of the cottontail rabbit papillomavirus mRNAs expressed in the transplantable VX2 carcinoma. J Virol. 1985;53:735–741. doi: 10.1128/jvi.53.3.735-741.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day P, Lowy D, Schiller J. Papillomaviruses infect cells via a clathrin-dependent pathway. Virology. 2003;307:1–11. doi: 10.1016/s0042-6822(02)00143-5. [DOI] [PubMed] [Google Scholar]
- de Villiers E. Cross-roads in the classification of papillomaviruses. Virology. 2013;445:2–10. doi: 10.1016/j.virol.2013.04.023. [DOI] [PubMed] [Google Scholar]
- de Villiers E, Fauquet C, Broker T, Bernard H, zur Hausen H. Classification of papillomaviruses. Virology. 2004;324:17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Dias J, Ignacchiti M, Giuriato P, Nunes L, Pereira-Júnior O. Detecção do papilomavírus bovino tipo 2 em bexigas de bovinos com hematúria enzoótica pela técnica de reaçao em cadeia de polimerase no Sul do Espirito Santo, Brasil. Rev Bras Med Veterinária. 2012;34:146–151. [Google Scholar]
- Dillner J, Knekt P, Schiller J, Hakulinen T. Prospective seroepidemiological evidence that human papillomavirus type 16 infection is a risk factor for oesophageal squamous cell carcinoma. BMJ. 1995;311:1346–1346. doi: 10.1136/bmj.311.7016.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMaio D. Viral miniproteins. Annu Rev Microbiol. 2014;68:21–43. doi: 10.1146/annurev-micro-091313-103727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diniz N, Melo T, Santos J, Mori E, Brandão P, Richtzenhain L, Freitas A, Beçak W, Carvalho R, Stocco R. Simultaneous presence of bovine papillomavirus in blood and in short-term lymphocyte cultures from dairy cattle in Pernambuco, Brazil. Genet Mol Res. 2009;8:1474–1480. doi: 10.4238/vol8-4gmr668. [DOI] [PubMed] [Google Scholar]
- Dong J, Zhu W, Haga T. Papillomavirus in yaks: The isolates of bovine papillomavirus type 1 have a high possibility of belonging to novel type. J Vet Med Sci. 2016;78:1059–1061. doi: 10.1292/jvms.15-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doorbar J. The E4 protein; structure, function and patterns of expression. Virology. 2013;445:80–98. doi: 10.1016/j.virol.2013.07.008. [DOI] [PubMed] [Google Scholar]
- Elzein E, Sundberg J, Housawi F, Gameel A, Ramadan R, Hassanein M. Genital bovine papillomavirus infection in Saudi Arabia. J Vet Diagnostic Investig. 1991;3:36–38. doi: 10.1177/104063879100300108. [DOI] [PubMed] [Google Scholar]
- Enemark E, Chen G, Vaughn D, Stenlund A, Joshua-Tor L, Brook S, York N. Crystal structure of the DNA binding domain of the replication initiation protein E1 from papillomavirus. Mol Cell. 2000;6:149–158. [PubMed] [Google Scholar]
- Felin I, Grivicich I, Felin C, Regner A, Rocha A. Expressão de p53, p16 e COX-2 em carcinoma escamoso de esôfago e associaçao histopatológica. Arq Gastroenterol. 2008;45:308–312. doi: 10.1590/s0004-28032008000400009. [DOI] [PubMed] [Google Scholar]
- Ferraro C, Canedo M, Oliveira S, Carvalho M, Dias E. Infecção oral pelo HPV e lesões epiteliais proliferativas associadas. J Bras Patol Médica e Lab. 2011;47:451–459. [Google Scholar]
- Finlay M, Yuan Z, Burden F, Trawford A, Morgan I, Campo M, Nasir L. The detection of bovine papillomavirus type 1 DNA flies. Virus Res. 2009;144:315–317. doi: 10.1016/j.virusres.2009.04.015. [DOI] [PubMed] [Google Scholar]
- Florin L, Sapp M, Spoden G. Host-cell factors involved in papillomavirus entry. Med Microbiol Immunol. 2012;201:437–448. doi: 10.1007/s00430-012-0270-1. [DOI] [PubMed] [Google Scholar]
- Foresta C, Bertoldo A, Garolla A, Pizzol D, Mason S, Lenzi A, De Toni L. Human papillomavirus proteins are found in peripheral blood and semen Cd20+ and Cd56+ cells during Hpv-16 semen infection. BMC Infectious Dis. 2013;13:e593. doi: 10.1186/1471-2334-13-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forslund O, Antonsson A, Nordin P, Stenquist B, Hansson BG. A broad range of human papillomavirus types detected with a general PCR method suitable for analysis of cutaneous tumours and normal skin. J Gen Virol. 1999;80:2437–2443. doi: 10.1099/0022-1317-80-9-2437. [DOI] [PubMed] [Google Scholar]
- Forslund O, Lidelof B, Hrail E, Norin P, Stenquist B, Kimbauer P, Sluptzky K, Dillner J. High prevalence of cutaneous human papillomavirus DNA on the top of skin tumors but not in “stripped” biopsies from the same tumors. J Invest Dermatol. 2004;123:388–394. doi: 10.1111/j.0022-202X.2004.23205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas AC De, Carvalho C De, Brunner O, Birgel-Junior EH, Maria A, Paiva M, Benesi FJ, Gregory L, Beçak W, De Cassia R. Viral DNA sequences in peripheral blood and vertical transmission of the virus: A discussion about BPV-1. Braz J Microbiol. 2003;34:76–78. [Google Scholar]
- García-Vallvé S, Alonso A, Bravo I. Papillomaviruses: Different genes have different histories. Trends Microbiol. 2005;13:514–521. doi: 10.1016/j.tim.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Gaukroger JM, Chandrachud LM, O’Neil BW, Grindlay GJ, Knowles G, Campo MS. Vaccination of cattle with bovine papillomavirus type 4 L2 elicits the production of virus-neutralizing antibodies. J Gen Virol. 1996;77:1577–1583. doi: 10.1099/0022-1317-77-7-1577. [DOI] [PubMed] [Google Scholar]
- Gauson E, Wang X, Dornan E, Herzyk P, Bristol M, Morgan I. Failure to interact with Brd4 alters the ability of HPV16 E2 to regulate host genome expression and cellular movement. Virus Res. 2016;211:1–8. doi: 10.1016/j.virusres.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor A, Fish S, Duerr R, Cruz F, Pesavento P. Identification of a novel papillomavirus in a Northern Fulmar (Fulmarus glacialis) with viral production in cartilage. Vet Pathol. 2015a;52:553–561. doi: 10.1177/0300985814542812. [DOI] [PubMed] [Google Scholar]
- Gaynor A, Zhu K, Cruz F, Affolter V, Pesavento P. Localization of bovine papillomavirus nucleic acid in equine sarcoids. Vet Pathol. 2015b;53:567–573. doi: 10.1177/0300985815594852. [DOI] [PubMed] [Google Scholar]
- Geisshüsler H, Marti E, Stoffel M, Kühni K, Stojiljkovic A, von Tscharner C, Vidondo B, Gerber V, Koch C. Quantitative analysis of infiltrating immune cells and bovine papillomavirus type 1 E2-positive cells in equine sarcoids. Vet J. 2016;216:45–52. doi: 10.1016/j.tvjl.2016.06.016. [DOI] [PubMed] [Google Scholar]
- Góes L, Freitas A, Ferraz O, Rieger T, Santosa J, Pereira A, Beçak W, Lindsey C, Stocco R. Bovine papillomavirus type 4 L1 gene transfection in a Drosophila S2 cell expression system: Absence of L1 protein expression. Braz J Microbiol. 2008;39:1–4. doi: 10.1590/S1517-83822008000100001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich L, Gerber H, Marti E, Antczak D. Equine sarcoids. Vet Clin North Am Equine Pract. 1998;14:607–623. doi: 10.1016/s0749-0739(17)30189-x. [DOI] [PubMed] [Google Scholar]
- Gopalkrishna V, Srivastava A, Hedau S, Sharma J, Das B. Detection of human papillomavirus DNA sequences in cancer of the urinary bladder by in situ hybridisation and polymerase chain reaction. Sex Transm Infect. 1995;71:231–233. doi: 10.1136/sti.71.4.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling M, Stamatakis A, Nindl I, Stockfleth E, Alonso A, Bravo I. Multiple evolutionary mechanisms drive papillomavirus diversification. Mol Biol Evol. 2007;24:1242–1258. doi: 10.1093/molbev/msm039. [DOI] [PubMed] [Google Scholar]
- Gottschling M, Goker M, Stamatakis A, Bininda-Emonds O, Nindl I, Bravo I. Quantifying the phylodynamic forces driving papillomavirus evolution. Mol Biol Evol. 2011;28:2101–2113. doi: 10.1093/molbev/msr030. [DOI] [PubMed] [Google Scholar]
- Graner J. History of infectious diseases oncology, from Galen to Rous. In: Goedert JJ, editor. Infectious Causes of Cancer. Humana Press; Nova Jersey: 2000. pp. 3–30. [Google Scholar]
- Greber U. Virus and host mechanics support membrane penetration and cell entry. J Virol. 2016;90:3802–3805. doi: 10.1128/JVI.02568-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. Molecular basis for the attack on cancer. Proc Natl Acad Sci U S A. 1972;69:1036–1041. doi: 10.1073/pnas.69.4.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindatto A, Ferraro G, Varello K, Crescio M, Miceli I, Bozzetta E, Goria M, Nappi R. Molecular and histological characterization of bovine papillomavirus in North West Italy. Vet Microbiol. 2015;180:113–117. doi: 10.1016/j.vetmic.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Guo F, Liu Y, Wang X, He Z, Weiss N, Madeleine M, Liu F, Tian X, Song Y, Pan Y, et al. Human papillomavirus infection and esophageal squamous cell carcinoma: A case-control study. Cancer Epidemiol Biomarkers Prev. 2012;21:780–785. doi: 10.1158/1055-9965.EPI-11-1206. [DOI] [PubMed] [Google Scholar]
- Haga T, Dong J, Zhu W, Burk R. The many unknown aspects of bovine papillomavirus diversity, infection and pathogenesis. Vet J. 2013;197:122–123. doi: 10.1016/j.tvjl.2013.05.006. [DOI] [PubMed] [Google Scholar]
- Han C, Qiao G, Hubbert N, Li L, Sun C, Wang Y, Yan M, Xu D, Li Y, Lowy D, et al. Serologic association between human papillomavirus type 16 infection and esophageal cancer in Shaanxi Province, China. J Natl Cancer Inst. 1996;88:1467–1471. doi: 10.1093/jnci/88.20.1467. [DOI] [PubMed] [Google Scholar]
- Handisurya A, Day P, Thompson C, Buck C, Pang Y, Lowy D, Schiller J. Characterization of Mus musculus papillomavirus 1 infection in situ reveals an unusual pattern of late gene expression and capsid protein localization. J Virol. 2013;87:13214–13225. doi: 10.1128/JVI.02162-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T, Fridholm H, Frøslev T, Kjartansdóttir K, Willerslev E, Nielsen L, Hansen A. New type of papillomavirus and novel circular single stranded DNA virus discovered in urban Rattus norvegicus using circular DNA enrichment and metagenomics. PLoS One. 2015;10:e0141952. doi: 10.1371/journal.pone.0141952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl B, Hainisch E, Shafti-Keramat S, Kirnbauer R, Corteggio A, Borzacchiello G, Tober R, Kainzbauer C, Pratscher B, Brandt S. Inoculation of young horses with bovine papillomavirus type 1 virions leads to early infection of PBMCs prior to pseudo-sarcoid formation. J Gen Virol. 2011;92:2437–2445. doi: 10.1099/vir.0.033670-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haster W, Owyang C. Abordagem do paciente com doença gastrointestinal. In: Longo D, Kasper D, Jamenson J, Fauci A, Hauser D, Loscalzo J, editors. Medicina Interna de Harrison. 18ª ed. Artmed; Porto Alegre: 2013. pp. 2402–2426. [Google Scholar]
- Hatama S, Nobumoto K, Kanno T. Genomic and phylogenetic analysis of two novel bovine papillomaviruses, BPV-9 and BPV-10. J Gen Virol. 2008;89:158–163. doi: 10.1099/vir.0.83334-0. [DOI] [PubMed] [Google Scholar]
- He Z, Meng Q, Qiao J, Peng Y, Xie K, Liu Y, Cai X, Zhang J, Chen C. Mixed nipple infections caused by variant of BPV3 and a putative new subtype of BPV in cattle. Transbound Emerg Dis. 2014;63:e140–e143. doi: 10.1111/tbed.12238. [DOI] [PubMed] [Google Scholar]
- He Z, Wlazlo A, Kowalczyk D, Cheng J, Xiang Z, Giles-Davis W, Ertl H. Viral recombinant vaccines to the E6 and E7 antigens of HPV-16. Virology. 2000;270:146–161. doi: 10.1006/viro.2000.0271. [DOI] [PubMed] [Google Scholar]
- Helfer C, Yan J, You J. The cellular bromodomain protein Brd4 has multiple functions in E2-mediated papillomavirus transcription activation. Viruses. 2014;6:3228–3249. doi: 10.3390/v6083228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy B, Smith D, Ram P, Lu Y, Mills G. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- Herbster S, Ferraro C, Koff N, Rossini A, Kruel C, Andreollo N, Rapozo D, Blanco T, Faria P, Santos P, et al. HPV infection in Brazilian patients with esophageal squamous cell carcinoma: Interpopulational differences, lack of correlation with surrogate markers and clinicopathological parameters. Cancer Lett. 2012;326:52–58. doi: 10.1016/j.canlet.2012.07.018. [DOI] [PubMed] [Google Scholar]
- Hille J, Margolius K, Markowitz S, Isaacson C. Human papillomavirus infection related to oesophageal carcinoma in black South Africans. South Africa Med J. 1986;69:417–420. [PubMed] [Google Scholar]
- Holland J, Domingo E. Origin and evolution of viruses. Virus Genes. 1998;16:13–21. doi: 10.1023/a:1007989407305. [DOI] [PubMed] [Google Scholar]
- INCA (Instituto Nacional de Câncer) Estimativa 2016. Ministério da Saúde/Inst Nac Cancer José Alencar Gomes da Silva; Rio de Janeiro: 2016. 122 p [Google Scholar]
- Jackson C. The incidence and pathology of tumours of domestic animals in South Africa. Onderstepoort J Vet Sci Anim Ind. 1936;6:378–385. [Google Scholar]
- Jang M, Shen K, McBride A. Papillomavirus genomes associate with BRD4 to replicate at fragile sites in the host genome. PLoS Pathog. 2014;10:e1004117. doi: 10.1371/journal.ppat.1004117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarret W, Smith K, O’Neil B, Gaukroger J, Chandrachud L, Grindlay G, McGarvie G, Campo M. Studies on vaccination against Papillomavirus: Prophylactic and therapeutic vaccination with recombinant structural proteins. Virology. 1991;184:2–42. doi: 10.1016/0042-6822(91)90819-w. [DOI] [PubMed] [Google Scholar]
- Kahrilas P, Hirano E. Doenças do esôfago. In: Longo D, Kasper D, Jamenson J, Fauci A, Hauser S, Loscalzo J, editors. Medicina Interna de Harrison. 18ª ed. Artmed; Porto Alegre: 2013. pp. 2437–2437. [Google Scholar]
- Kawauchi K, Takahashi C, Ishihara R, Hatama S. Development of a novel PCR-RFLP assay for improved detection and typing of bovine papillomaviruses. J Virol Methods. 2015;218:23–26. doi: 10.1016/j.jviromet.2015.03.005. [DOI] [PubMed] [Google Scholar]
- Kines R, Cerio R, Roberts J, Thompson C, Pinos E, Lowy D, Schiller J. Human papillomavirus capsides prefentially bind and infect tumor cells. Int J Cancer. 2016;138:901–911. doi: 10.1002/ijc.29823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knottenbelt D. A suggested clinical classification for the equine sarcoid. Clin Tech Equine Pract. 2005;4:278–295. [Google Scholar]
- Koller L, Olson C. Attempted transmission of warts from man, cattle, and horses and of deer fibroma, to selected hosts. J Invest Dermatol. 1972;58:366–368. doi: 10.1111/1523-1747.ep12540579. [DOI] [PubMed] [Google Scholar]
- Kràl J, Band'ouchová H, Brichta J, Kovácová V, Ondracék K, Osicková J, Hrubá H, Hutarová Z, Komínkova M, Cernei N, et al. Papillomavirus infection of roe deer in the Czech Republic and fibropapilloma-associated levels of metallothionein, zinc and oxidative stress. Acta Vet Brno. 2015;84:105–111. [Google Scholar]
- Krawczyk E, Suprynowicz F, Liu X, Dai Y, Hartmann D, Hanover J, Schlegel R. Koilocytosis: A cooperative interaction between the human papillomavirus. Am J Pathol. 2008;173:682–688. doi: 10.2353/ajpath.2008.080280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczyk E, Suprynowicz F, Sudarshan S, Schlegel R. Membrane orientation of the human papillomavirus type 16 E5 oncoprotein. J Virol. 2010;84:1696–1696. doi: 10.1128/JVI.01968-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulski J, Demeter T, Sterrett G, KB S. Human papilloma virus DNA in oesophageal carcinoma. Lancet. 1986;2:683–684. doi: 10.1016/s0140-6736(86)90187-x. [DOI] [PubMed] [Google Scholar]
- Lagergren J, Wang Z, Bergstro R, Dillner J, Nyre O. Human papillomavirus infection and esophageal cancer: A nationwide seroepidemiologic case-control study in Sweden. J Natl Cancer Inst. 1999;91:156–162. doi: 10.1093/jnci/91.2.156. [DOI] [PubMed] [Google Scholar]
- Lancaster WD, Olson C, Meinke W. Bovine papilloma virus: Presence of virus-specific DNA sequences in naturally occurring equine tumors. Proc Natl Acad Sci USA. 1977;74:524–528. doi: 10.1073/pnas.74.2.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavergne D, de Villiers E. Papillomavirus in esophageal papillomas and carcinomas. Int J Cancer. 1999;80:681–684. doi: 10.1002/(sici)1097-0215(19990301)80:5<681::aid-ijc8>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Leal A, Ferraz O, Carvalho C, Freitas A, Beniston R, Beçak W, Campo M, Stocco dos Santos R. Quercetin induces structural chromosomal aberrations and uncommon rearrangements in bovine cells transformed by the E7 protein of bovine papillomavirus type 4. Vet Comp Oncol. 2003;1:15–21. doi: 10.1046/j.1476-5829.2003.00008.x. [DOI] [PubMed] [Google Scholar]
- Leatherwood J. Emerging mechanisms of eukaryotic DNA replication initiation. Curr Opin Cell Biol. 1998;10:742–748. doi: 10.1016/s0955-0674(98)80117-8. [DOI] [PubMed] [Google Scholar]
- Leto M, Santos-Júnior G, Porro A, Tomimori J. Human papillomavirus infection?: Etiopathogenesis, molecular biology and clinical manifestations. An Bras Dermatol. 2011;86:306–317. doi: 10.1590/s0365-05962011000200014. [DOI] [PubMed] [Google Scholar]
- Lindsey C, Almeida M, Vicari C, Carvalho C, Yaguiu A, Freitas A, Beçak W, Stocco R. Bovine papillomavirus DNA in milk, blood, urine, semen, and spermatozoa of bovine papillomavirus-infected animals. Genet Mol Res. 2009;8:310–318. doi: 10.4238/vol8-1gmr573. [DOI] [PubMed] [Google Scholar]
- Lioi MB, Barbieri R, Borzacchiello G, Dezzi S, Roperto S, Santoro A, Russo V, Roperto F. Chromosome aberrations in cattle with chronic enzootic haematuria. J Comp Pathol. 2004;131:233–236. doi: 10.1016/j.jcpa.2004.01.011. [DOI] [PubMed] [Google Scholar]
- Liu Y, Baleja J. Structure and function of the papillomavirus E6 protein and its interacting proteins. Front Biosci. 2008;1:121–134. doi: 10.2741/2664. [DOI] [PubMed] [Google Scholar]
- Liu Y, Liu Z, Gao H, Zhou Y, Androphy E, Chen J. Opposing effects of bovine papillomavirus type 1 E6 and E7 genes on Fas-mediated apoptosis. Oncogene. 2005;24:3942–3953. doi: 10.1038/sj.onc.1208542. [DOI] [PubMed] [Google Scholar]
- Liu Z, Liu Y, Hong Y, Rapp L, Androphy E, Chen J. Bovine papillomavirus type 1 E6-induced sensitization to apoptosis is distinct from its transforming activity. Virology. 2002;295:230–237. doi: 10.1006/viro.2001.1351. [DOI] [PubMed] [Google Scholar]
- Ljubojevic S, Skerlev M. HPV-associated diseases. Clin Dermatol. 2014;32:227–234. doi: 10.1016/j.clindermatol.2013.08.007. [DOI] [PubMed] [Google Scholar]
- Longo D. Biologia celular e angiogênese do câncer. In: Longo D, Fauci A, Kasper D, Hauser S, Jameson J, Loscalzo J, editors. Medicina Interna de Harrison. 18ª ed. Artmed; Porto Alegre: 2013. pp. 672–688. [Google Scholar]
- Longworth M, Laimins L. Pathogenesis of human papillomaviruses in differentiating epithelia. Microbiol Mol Biol Rev. 2004;68:362–372. doi: 10.1128/MMBR.68.2.362-372.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love A, Chapman S, Matic S, Noris E, Lomonossoff G, Taliansky M. In planta production of a candidate vaccine against bovine papillomavirus type 1. Planta. 2012;236:1305–1313. doi: 10.1007/s00425-012-1692-0. [DOI] [PubMed] [Google Scholar]
- Lucena R, Rissi D, Kommers G, Pierezan F, Oliveira-Filho J, Macêdo J, Flores M, Barros C. A retrospective study of 586 tumours in Brazilian cattle. J Comp Pathol. 2011;145:20–24. doi: 10.1016/j.jcpa.2010.11.002. [DOI] [PubMed] [Google Scholar]
- Lunardi M, Alcântara B, Otonel R, Rodrigues W, Alfieri A, Alfieri A. Bovine papillomavirus type 13 DNA in equine sarcoids. J Clin Microbiol. 2013;51:2167–2171. doi: 10.1128/JCM.00371-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiolino P, Ozkul A, Sepici-Dincel A, Roperto F, Yücel G, Russo V, Urraro C, Lucà R, Riccardi M, Martano M, et al. Bovine papillomavirus type 2 infection and microscopic patterns of urothelial tumors of the urinary bladder in water buffaloes. Biomed Res Int. 2013;2013:937918–937918. doi: 10.1155/2013/937918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maran A, Amella C, Dilorenzo T, Auborn K, Taichman L, Steinberg B. Human papillomavirus type-11 transcripts are present at low abundance in latently infected respiratory tissue. Virology. 1995;212:285–294. doi: 10.1006/viro.1995.1486. [DOI] [PubMed] [Google Scholar]
- Marigliani B, Kavati EA, Sakauchi D, Oliveira H, Canali R, Sasaki A. Intracellular distribution of recombinant human papillomavirus capsid proteins. Curr Micros Contrib Adv Sci Technol. 2012:678–684. [Google Scholar]
- Marrazzo J, Holmes K. Infecções sexualmente transmissívies: Consideraçoes gerais e abordagem clinica. In: Longo D, Fauci A, Kasper D, Hauser S, Jameson J, Loscalzo J, editors. Medicina Interna de Harrison. 18ª ed. Artmed; Porto Alegre: 2013. pp. 1095–11111. [Google Scholar]
- Martano M, Corteggio A, Restucci B, Biase M, Borzacchiello G, Maiolino P. Extracellular matrix remodeling in equine sarcoid: An immunohistochemical and molecular study. BMC Vet Res. 2016;12:e24. doi: 10.1186/s12917-016-0648-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelli-Marzagão F, Yamashiro A, Ogawa M, Santos-Júnior G, Tomimori J, Porro A. Caracterização clínica e histopatológica e tipagem do papilomavírus humano das verrugas vulgares nos receptores de transplante renal. An Bras Dermatol. 2010;85:743–746. doi: 10.1590/s0365-05962010000500027. [DOI] [PubMed] [Google Scholar]
- Martens A, De Moor A, Demeulemeester J, Ducatelle R. Histopathological characteristics of five clinical types of equine sarcoid. Res Vet Sci. 2000;69:295–300. doi: 10.1053/rvsc.2000.0432. [DOI] [PubMed] [Google Scholar]
- Martens A, De Moor A, Ducatelle R. PCR detection of bovine papilloma virus DNA in superficial swabs and scrapings from equine sarcoids. Vet J. 2001a;161:280–286. doi: 10.1053/tvjl.2000.0524. [DOI] [PubMed] [Google Scholar]
- Martens A, De Moor A, Vlaminck L, Pille F, Steenhaut M. Evaluation of excision, cryosurgery and local BCG vaccination for the treatment of equine sarcoids. Vet Rec. 2001b;49:665–669. doi: 10.1136/vr.149.22.665. [DOI] [PubMed] [Google Scholar]
- Masuda E, Kommers G, Rosa F, Barros C, Fighera R, Piazer J. Relação entre a linfopenia e a persistência da papilomatose alimentar em bovinos intoxicados crônica e espontaneamente por samambaia (Pteridium aquilinum) Pesqui Vet Bras. 2011;31:383–388. [Google Scholar]
- Mazzuchelli-de-Souza J, Carvalho R, Ruiz R, Melo T, Araldi R, Carvalho E, Thompson C, Sircili M, Beçak W, Stocco R. Expression and in silico analysis of the recombinant bovine papillomavirus E6 protein as a model for viral oncoproteins studies. Biomed Res Int. 2013 doi: 10.1155/2013/421398. 2013. 421398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo TC, Diniz N, Campos SRC, Ferraz OP, Lindsey CJ, Rieger TT, Beçak W, Stocco RC. Cytogenetic studies in peripheral blood of bovines afflicted by papillomatosis. Vet Comp Oncol. 2011;9:269–274. doi: 10.1111/j.1476-5829.2011.00263.x. [DOI] [PubMed] [Google Scholar]
- Melo T, Carvalho R, Mazzucchelli-de-Souza J, Diniz N, Vasconcelos S, Assaf S, Araldi R, Ruiz R, Kerkis I, Beçak W, et al. Phylogenetic classification and clinical aspects of a new putative Deltapapillomavirus associated with skin lesions in cattle. Genet Mol Res. 2014;13:2458–2469. doi: 10.4238/2014.April.3.18. [DOI] [PubMed] [Google Scholar]
- Melo T, Araldi R, Pessoa N, De-Sá-Júnior P, Carvalho R, Beçak W, Stocco Bos taurus papillomavirus activity in peripheral blood mononuclear cells?: Demonstrating a productive infection. Genet Mol Res. 2015;14 doi: 10.4238/2015.December.11.19. 16712-1627. [DOI] [PubMed] [Google Scholar]
- Mischo A, Ohlenschläger O, Hortschansky P, Ramachandran R, Görlach M. Structural insights into a wildtype domain of the oncoprotein E6 and its interaction with a PDZ domain. PLoS One. 2013;8:e6284. doi: 10.1371/journal.pone.0062584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misdorp W. Veterinary cancer epidemiology. Vet Q. 1996;18:32–36. doi: 10.1080/01652176.1996.9694610. [DOI] [PubMed] [Google Scholar]
- Monte T, Peixoto G. A incidência de papilomavírus humano em mulheres no Hospital Universitário Sul Fluminense. Rev Bras Análises Clínicas. 2010;42:131–139. [Google Scholar]
- Monteiro V, Coelho M, Carneiro A, Silva R, Teixeira M, Wanderley A, Wanderley E, Franco E. Descrição clínica e histopatológica da papilomatose cutânea bovina (BPV) Ciênc Anim Bras. 2008;10:550–560. [Google Scholar]
- Moody C, Laimins L. Human papillomavirus oncoproteins: Pathways to transformation. Nat Rev Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- Mosseri S, Hetzel U, Hahn S, Michaloupoulou E, Sallabank H, Knottenbelt D, Kipar A. Equine sarcoid: In situ demonstration of matrix metalloproteinase expression. Vet J. 2014;202:279–285. doi: 10.1016/j.tvjl.2014.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munday J. Bovine and human papillomaviruses: A comparative review. Vet Pathol. 2014;51:1063–1075. doi: 10.1177/0300985814537837. [DOI] [PubMed] [Google Scholar]
- Munday J, Thomson N, Dunowska M, Knight C, Laurie R, Hills S. Genomic characterisation of the feline sarcoid-associated papillomavirus and proposed classification as Bos taurus papillomavirus type 14. Vet Microbiol. 2015;177:289–295. doi: 10.1016/j.vetmic.2015.03.019. [DOI] [PubMed] [Google Scholar]
- Muro L, Botteira C, Piccinin A. Papilomatose bovina. Rev Cient Eletr Med Vet 10 2008 [Google Scholar]
- Nasir L, Campo M. Bovine papillomaviruses: Their role in the aetiology of cutaneous tumours of bovids and equids. Vet Dermatol. 2008;19:243–254. doi: 10.1111/j.1365-3164.2008.00683.x. [DOI] [PubMed] [Google Scholar]
- Nasir L, Reid S. Bovine papillomaviral gene expression in equine sarcoid tumours. Virus Res. 1999;61:171–175. doi: 10.1016/s0168-1702(99)00022-2. [DOI] [PubMed] [Google Scholar]
- Ng T, Miller M, Kondov N, Dodd E, Batac F, Manzer M, Ives S, Saliki J, Deng X, Delwart E. Oral papillomatosis caused by Enhydra lutris papillomavirus 1 (ElPV-1) in southern sea otters (Enhydra lutris nereis) in California, USA. J Wildl Dis. 2015;51:446–453. doi: 10.7589/2014-06-152. [DOI] [PubMed] [Google Scholar]
- Nasrollahzadeh D, Ye W, Shakeri R, Sotoudeh M, Merat S, Kamangar F, Abnet C, Islami F, Boffetta P, Dawsey S, et al. Contact with ruminants is associated with esophageal squamous cell carcinoma risk. Int J Cancer. 2015;136:1468–1474. doi: 10.1002/ijc.29109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls PK, Moore PF, Anderson DM, Moore RA, Parry NR, Gough GW, Stanley MA. Regression of canine papillomas is associated with infiltration of CD4+ and CD8+ lymphocytes. Virology. 2001;283:31–39. doi: 10.1006/viro.2000.0789. [DOI] [PubMed] [Google Scholar]
- Nicolas G, Pottier C, Maltête D, Coutant S, Rovelet-Lecrux A, Legallic S, Rousseau S, Vaschalde Y, Guyant-Maréchal L, Augustin J, et al. Mutation of the PDGFRB gene as a cause of idiopathic basal ganglia calcification. Neurology. 2013;80:181–187. doi: 10.1212/WNL.0b013e31827ccf34. [DOI] [PubMed] [Google Scholar]
- Nominé Y, Masson M, Charbonnier S, Zanier K, Ristriani T, Deryckère F, Sibler A, Desplancq D, Atkinson R, Weiss E, et al. Structural and functional analysis of E6 oncoprotein: Insights in the molecular pathways of human papillomavirus-mediated pathogenesis. Mol Cell. 2006;21:665–678. doi: 10.1016/j.molcel.2006.01.024. [DOI] [PubMed] [Google Scholar]
- Moreno-Lopez J, Ahola H, Stenlund A, Osterhaus A, Pettersson U. Genome of an avian papillomavirus. J Virol. 1984;51:872–875. doi: 10.1128/jvi.51.3.872-875.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Tomita Y, Okada M, Shinozaki K, Kubonoya H, Kaiho I, Shirasawa H. Broad-spectrum detection of papillomaviruses in bovine teat papillomas and healthy teat skin. J Gen Virol. 2004;85:2191–2197. doi: 10.1099/vir.0.80086-0. [DOI] [PubMed] [Google Scholar]
- Oliveira R. Studies on the Biopathological Actions of Pteridium aquilinum. Universidade do Porto; Porto: 2012. 150 p [Google Scholar]
- Otten N, von Tscharner C, Lazary S, Antczak D, Gerber H. DNA of bovine papillomavirus type 1 and 2 in equine sarcoids: PCR detection and direct sequencing. Arch Virol. 1993;132:121–131. doi: 10.1007/BF01309847. [DOI] [PubMed] [Google Scholar]
- Özsoy S, Özyildiz Z, Güzel M. Clinical, pathological and immunohistochemical findings of bovine cutaneous papillomatosis. Ankara Üniv Vet Fak Derg. 2011;58:161–165. [Google Scholar]
- Park J, Nickel K, Torres A, Lee D, Lambert P, Kimple R. Human papillomavirus type 16 E7 oncoprotein causes a delay in repair of DNA damage. Radiother Oncol. 2014;113:337–344. doi: 10.1016/j.radonc.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathania S, Dhama K, Saikumar G, Shahi S, Somvanshi R. Detection and quantification of bovine papilloma virus type 2 (BPV-2) by real-time PCR in urine and urinary bladder lesions in enzootic bovine haematuria (EBH)-affected cows. Transboud Emerg Dis. 2012;59(1):79–84. doi: 10.1111/j.1865-1682.2011.01248.x. [DOI] [PubMed] [Google Scholar]
- Pavelski M, Ollhoff R, Barros-Filho I, Deconto I, Biondo A, Dornbusch P. Evaluation of urine dipstick and cystoscopy in bovine enzootic haematuria. Semin Ciênc Agr. 2014;35:1369–1369. [Google Scholar]
- Pennie W, Campo M. Synergism between bovine papillomavirus type 4 and the flavanoid quercetin in cell transformation in vitro. Virology. 1992;190:19–24. doi: 10.1016/0042-6822(92)90926-g. [DOI] [PubMed] [Google Scholar]
- Pessoa N. Estudos Sobre a Expressão do Papilomavirus Humano (HPV): Avaliação Comparativa Sobre Lesões Cervicais, Sangue Periférico e Retinoblastomas. Universidade de São Paulo; 2014. 140 p. Dissertation. Avaliable in http://www.teses.usp.br/teses/disponiveis/87/87131/tde-07052014-090937/pt-br.php. [Google Scholar]
- Pietras K, Ostman A. Hallmarks of cancer: Interactions with the tumor stroma. Exp Cell Res. 2010;316:1324–1331. doi: 10.1016/j.yexcr.2010.02.045. [DOI] [PubMed] [Google Scholar]
- Plowrigh W. Malignant neoplasia of the oesophagus and rumen of cattle in Kenya. J Comp Pathol. 1955;65:108–114. doi: 10.1016/s0368-1742(55)80010-0. [DOI] [PubMed] [Google Scholar]
- Potocki L, Lewinska A, Klukowska-Rötzler J, Bielak-Zmijewska A, Grabowska W, Rzeszutek I, Kaminska P, Roga E, Bugno-Poniewierska M, Slota E, et al. Sarcoid-derived fibroblasts: Links between genomic instability, energy metabolism and senescence. Biochimie. 2014;92:163–172. doi: 10.1016/j.biochi.2013.10.010. [DOI] [PubMed] [Google Scholar]
- Potter D, Baird M. Carcinogenic effects of ptaquiloside in bracken fern and related compounds. Br J Cancer. 2000;83:914–920. doi: 10.1054/bjoc.2000.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampias T, Sasaki C, Psyrri A. Molecular mechanisms of HPV induced carcinogenesis in head and neck. Oral Oncol. 2013;50:356–363. doi: 10.1016/j.oraloncology.2013.07.011. [DOI] [PubMed] [Google Scholar]
- Rashad AL, Evans CA. Histologic features of virus-rich and virus-poor Shope papillomas of cottontail rabbits. Cancer Res. 1967;27:1855–1860. [PubMed] [Google Scholar]
- Rector A, Van Ranst M. Animal papillomaviruses. Virology. 2013;445:213–223. doi: 10.1016/j.virol.2013.05.007. [DOI] [PubMed] [Google Scholar]
- Resendes A, Roperto S, Trapani F, Urraro C, Rodrigues A, Roperto F, Borzacchiello G. Association of bovine papillomavirus type 2 (BPV-2) and urinary bladder tumours in cattle from the Azores archipelago. Res Vet Sci. 2011;90:526–529. doi: 10.1016/j.rvsc.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Ribeiro-Müller L, Müller M. Prophylactic papillomavirus vaccines. Clin Dermatol. 2014;32:235–247. doi: 10.1016/j.clindermatol.2013.08.008. [DOI] [PubMed] [Google Scholar]
- Ristriani T, Masson M, Nominé Y, Laurent C, Lefevre JF, Weiss E, Travé G. HPV oncoprotein E6 is a structure-dependent DNA-binding protein that recognizes four-way junctions. J Mol Biol. 2000;296:1189–1203. doi: 10.1006/jmbi.2000.3527. [DOI] [PubMed] [Google Scholar]
- Roberts S. Papillomaviruses. eLS. 2015;2015:1–11. [Google Scholar]
- Roperto S, Ambrosio V, Borzacchiello G, Galati P, Paciello O, Russo V, Roperto F. Bovine papillomavirus type-2 (BPV-2) infection and expression of uroplakin IIIb, a novel urothelial cell marker, in urinary bladder tumors of cows. Vet Pathol. 2005;42:812–818. doi: 10.1354/vp.42-6-812. [DOI] [PubMed] [Google Scholar]
- Roperto S, Brun R, Paolini F, Urraro C, Russo V, Borzacchiello G, Pagnini U, Raso C, Rizzo C, Roperto F, et al. Detection of bovine papillomavirus type 2 in the peripheral blood of cattle with urinary bladder tumours: Possible biological role. J Gen Virol. 2008;89:3027–3033. doi: 10.1099/vir.0.2008/004457-0. [DOI] [PubMed] [Google Scholar]
- Roperto S, Comazzi S, Ciusani E, Paolini F, Borzacchiello G, Esposito I, Luca R, Russo V, Urraro C, Venuti A, et al. PBMCs are additional sites of productive infection of bovine papillomavirus type 2. J Gen Virol. 2011;92:1787–1794. doi: 10.1099/vir.0.031740-0. [DOI] [PubMed] [Google Scholar]
- Roperto S, Borzacchiello G, Esposito I, Riccardi M, Urraro C, Cermola M, Paciello O, Corteggio A, Tate R. Productive infection of bovine papillomavirus type 2 in the placenta of pregnant cows affected with urinary bladder tumors. PLoS One. 2012;7:e33569. doi: 10.1371/journal.pone.0033569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roperto S, Russo V, Leonardi L, Martano M, Corrado F, Riccardi M, Roperto F. Bovine papillomavirus type 13 expression in the urothelial bladder tumours of cattle. Transbound Emerg Dis. 2015;63(6):628–634. doi: 10.1111/tbed.12322. [DOI] [PubMed] [Google Scholar]
- Roperto S, Munday J, Corrado F, Goria M, Roperto F. Detection of bovine papillomavirus type 14 DNA sequences in urinary bladder tumors in cattle. Vet Microbiol. 2016;190:1–4. doi: 10.1016/j.vetmic.2016.04.007. [DOI] [PubMed] [Google Scholar]
- Russo V, Roperto F, Esposito I, Ceccarelli D, Zizzo N, Leonardi L, Capparelli R, Borzacchiello G, Roperto S. ERas protein is overexpressed and binds to the activated platelet-derived growth factor β receptor in bovine urothelial tumour cells associated with papillomavirus infection. Vet J. 2016;212:44–47. doi: 10.1016/j.tvjl.2016.01.004. [DOI] [PubMed] [Google Scholar]
- Sacco A, Siepi F, Crescenzi M. HPV E7 expression in skeletal muscle cells distinguishes initiation of the postmitotic state from its maintenance. Oncogene. 2003;22:4027–4034. doi: 10.1038/sj.onc.1206353. [DOI] [PubMed] [Google Scholar]
- Santos E, Silva M, Pontes N, Coutinho L, Paiva S, Castro R, Freitas A. Detection of different bovine papillomavirus types and co-infection in bloodstream of cattle. Transbound Emerg Dis. 2014;63:e103–e108. doi: 10.1111/tbed.12237. [DOI] [PubMed] [Google Scholar]
- Schuchat A, Jackson L. Princípios de imunização e uso de vacinas. In: Longo D, Fauci A, Kasper D, Hauser S, Jameson J, Loscalzo J, editors. Medicina Interna de Harrison. 18ª ed. Artmed; Porto Alegre: 2013. pp. 1031–1041. [Google Scholar]
- Schuck S, Stenlund A. A conserved regulatory module at the C-terminus of the papillomavirus E1 helicase domain controls E1 helicase assembly. J Virol. 2015;89:1129–1142. doi: 10.1128/JVI.01903-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiger M, You J, Howley P. Bromodomain protein 4 mediates the papillomavirus E2 transcriptional activation function. J Virol. 2006;80:4276–4285. doi: 10.1128/JVI.80.9.4276-4285.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiger M, Ottinger M, You J, Howley P. Brd4-independent transcriptional repression function of the papillomavirus e2 proteins. J Virol. 2007;81:9612–9622. doi: 10.1128/JVI.00447-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedelnikova O, Pilch D. Histone H2AX in DNA damage and repair. Cancer Biol Ther. 2003;2:233–235. doi: 10.4161/cbt.2.3.373. [DOI] [PubMed] [Google Scholar]
- Shafti-Keramat S, Schellenbacher C, Handisurya A, Christensen N, Reininger B, Brandt S, Kirnbauer R. Bovine papillomavirus type 1 (BPV1) and BPV2 are closely related serotypes. Virology. 2009;393:1–6. doi: 10.1016/j.virol.2009.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahin M, Smith B, Prakash A. Bracken carcinogens in the human diet. Mutat Res - Genet Toxicol Environ Mutagen. 1999;443:69–79. doi: 10.1016/s1383-5742(99)00011-3. [DOI] [PubMed] [Google Scholar]
- Shamanna R, Hoque M, Pe'ery T, Mathews M. Induction of p53, p21 and apoptosis by silencing the NF90/NF45 complex in human papilloma virus-transformed cervical carcinoma cells. Oncogene. 2013;32:5176–5185. doi: 10.1038/onc.2012.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shope R. Immunization of rabbits to infectious papillomatosis. J Exp Med. 1937;65:219–231. doi: 10.1084/jem.65.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shope R, Hurst E. Infectious papillomatosis of rabbits. J Exp Med. 1933;58:607–624. doi: 10.1084/jem.58.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibbet G, Romero-Graillet C, Meneguzzi G, Campo MS. Alpha6 integrin is not the obligatory cell receptor for bovine papillomavirus type 4. J Gen Virol. 2000;81:327–334. doi: 10.1099/0022-1317-81-2-327. [DOI] [PubMed] [Google Scholar]
- Silva F, Cibulski S, Daudt C, Weber M, Guimarães L, Streck A, Mayer F, Riehe P, Canal C. Novel bovine papillomavirus type discovered by rolling-circle amplification coupled with next-generation sequencing. PLoS One. 2016;11:e0162345. doi: 10.1371/journal.pone.0162345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MAR, De Albuquerque BMF, Pontes NE, Coutinho LCA, Leitão MCG, Reis MC, Castro RS, Freitas AC. Detection and expression of bovine papillomavirus in blood of healthy and papillomatosis-affected cattle. Genet Mol Res. 2013a;12:3150–3156. doi: 10.4238/2013.February.28.31. [DOI] [PubMed] [Google Scholar]
- Silva M, Batista M, Pontes N, Santos E, Coutinho L, Castro R, Blabino V, Freitas A. Comparison of two PCR strategies for the detection of bovine papillomavirus. J Virol Methods. 2013b;192:55–58. doi: 10.1016/j.jviromet.2013.04.017. [DOI] [PubMed] [Google Scholar]
- Silvestre O, Borzacchiello G, Nava D, Iovane G, Russo V, Vecchio D, D’Ausilio F, Gault EA, Campo MS, Paciello O. Bovine papillomavirus type 1 DNA and E5 oncoprotein expression in water buffalo fibropapillomas. Vet Pathol. 2009;46:636–641. doi: 10.1354/vp.08-VP-0222-P-FL. [DOI] [PubMed] [Google Scholar]
- Stewart A, Rush B, Davis E. The efficacy of intratumoral 5-fluorouracil for the treatment of equine sarcoids. Aust Vet J. 2006;84:101–106. doi: 10.1111/j.1751-0813.2006.tb12239.x. [DOI] [PubMed] [Google Scholar]
- Stocco dos Santos RC, Lindsey CJ, Ferraz OP, Pinto JR, Mirandola RS, Benesi FJ, Birgel EH, Pereira CA, Beçak W. Bovine papillomavirus transmission and chromosomal aberrations: An experimental model. J Gen Virol. 1998;79:2127–2135. doi: 10.1099/0022-1317-79-9-2127. [DOI] [PubMed] [Google Scholar]
- Syrjänen K. Histological changes identical to those condylomatous lesions found in esophageal squamous cell carcinomas. Arch Geschwulstforsch. 1982;52:283–294. [PubMed] [Google Scholar]
- Syrjänen K. HPV infections and oesophageal cancer. J Clin Pathol. 2002;55:721–728. doi: 10.1136/jcp.55.10.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, White E, Sowa M, Harper J, Aster J, Howley P. Cutaneous β-human papillomavirus E6 proteins bind Mastermind-like coactivators and repress Notch signaling. Proc Natl Acad Sci U S A. 2012a;109:E1473–E1480. doi: 10.1073/pnas.1205991109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Yildirim Y, Mahmut S, Dagalp S, Yilmaz V, Kirmizigul A, Gokce E. A histopathological, immunohistochemical and molecular study of cutaneous bovine papillomatosis. Kafkas Univ Vet Derg. 2012b;18:739–744. [Google Scholar]
- Taylor S, Toth B, Baseler L, Charney V, Miller M. Lack of correlation between papillomaviral DNA in surgical margins and recurrence of equine sarcoids. J Equine Vet Sci. 2014;34:722–725. [Google Scholar]
- Togawa K, Jaskiewicz K, Takahashi H, Meltzer S, Rustgi A. Human papillomavirus DNA in esophageal squamous cell carcinoma. Gastroenterology. 1994;107:128–136. doi: 10.1016/0016-5085(94)90070-1. [DOI] [PubMed] [Google Scholar]
- Tokarnia C, Doberreiner J, Peixoto P. Plantas de ação radiomimética. In: Tokarnia C, Doberreiner J, Peixoto P, editors. Plantas Tóxicas do Brasil. Helianthus; Rio de Janeiro: 2000. pp. 178–187. [Google Scholar]
- Tomita Y, Ogawa T, Jin Z, Shirasawa H. Genus specific features of bovine papillomavirus E6, E7, E5 and E8 proteins. Virus Res. 2007;124:231–236. doi: 10.1016/j.virusres.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Tong X, Boll W, Kirchhausen T, Peter M. Interaction of the bovine papillomavirus E6 protein with the clathrin adaptor complex AP-1. J Virol. 1998;72:476–476. doi: 10.1128/jvi.72.1.476-482.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsirimonaki E, Neil B, Williams R, Campo M. Extensive papillomatosis of the bovine upper gastrointestinal tract. J Comp Pathol. 2003;129:93–99. doi: 10.1016/s0021-9975(03)00007-0. [DOI] [PubMed] [Google Scholar]
- Turk N, Zupancic Z, Staresina V, Kovac S, Babic T, Kreszinger M, Curic S, Barbic L, Milas Z. Severe bovine papillomatosis?: Detection of bovine papillomavirus in tumour tissue and efficacy of treatment using autogenous vaccine and parammunity inducer. Vet Arh. 2005;75:391–397. [Google Scholar]
- Turner C. Paxillin interactions. J Cell Sci. 2000;113:4139–4140. doi: 10.1242/jcs.113.23.4139. [DOI] [PubMed] [Google Scholar]
- Van Bressem M, Cassonnet P, Rector A, Desaintes C, Van Waerebeek K, Alfaro-Shigueto J, Van Ranst M, Orth G. Genital warts in Burmeister's porpoises: Characterization of Phocoena spinipinnis papillomavirus type 1 (PsPV-1) and evidence for a second, distantly related PsPV. J Gen Virol. 2007;88:1928–1233. doi: 10.1099/vir.0.82694-0. [DOI] [PubMed] [Google Scholar]
- van Doorlaer K, Burk R. Evolution of humana papillomavirus carcinogenicity. Adv Virus Res. 2010;77:41–62. doi: 10.1016/B978-0-12-385034-8.00002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorslaer K. Evolution of the papillomaviridae. Virology. 2013;445:11–20. doi: 10.1016/j.virol.2013.05.012. [DOI] [PubMed] [Google Scholar]
- Varga J, Oliveira T, Greten F. The architect who never sleeps: Tumor-induced plasticity. FEBS Lett. 2014;588:2422–2427. doi: 10.1016/j.febslet.2014.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venuti A, Paolini F, Nasir L, Corteggio A, Roperto S, Campo M, Borzacchiello G. Papillomavirus E5?: The smallest oncoprotein with many functions. Mol Cancer. 2011;10:140–140. doi: 10.1186/1476-4598-10-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira C, Lopes J, Velosa J. A case of esophageal squamous cell carcinoma with positive HPV 11. Gastroenterol Hepatol. 2013;36:311–315. doi: 10.1016/j.gastrohep.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Vivanco I, Sawyers C. The phosphatidylinositol 3-Kinase-AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- Wallace N, Galloway D. Manipulation of cellular DNA damage repair machinery facilitates propagation of human papillomaviruses. Semin Cancer Biol. 2014;26:30–42. doi: 10.1016/j.semcancer.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter-Moura J, Stocco-dos-Santos R, Dagli M, D’Angelino J, Birgel E, Beçak W. Chromosome aberrations in cattle raised on bracken fern pasture. Experientia. 1988;44:785–788. doi: 10.1007/BF01959166. [DOI] [PubMed] [Google Scholar]
- Wang F, Kieff E. Virologia médica. In: Longo D, Fauci A, Kasper D, Hauser S, Jameson J, Loscalzo J, editors. Medicina Interna de Harrison. 18th ed. Artmed; Porto Alegre: 2013. pp. 1432–1441. [Google Scholar]
- Wang J, Roden R. L2, the minor capsid protein of papillomavirus. Virology. 2013;445:175–186. doi: 10.1016/j.virol.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werness B, Levine A, Howley P. Association of human papillomavirus types 16 and 18 E6 proteins with p5. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- White E, Howley P. Proteomic approaches to the study of papillomavirus - host interactions. Virology. 2013;435:57–69. doi: 10.1016/j.virol.2012.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E, Kramer R, Hwang J, Pores F, Naetar N, Hahn W, Roberts T, Schaffhausen B, Livingston D, Howley P. Papillomavirus E7 oncoproteins share functions with polyomavirus small T antigens. J Virol. 2015;89:2857–2865. doi: 10.1128/JVI.03282-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Global cancer rates could increase by 50% to 15 million by 2020. 2013. www.who.int/mediacente/news/releases/2003/pr27/en.
- William J, Kirubaharan J, Uthumann K. Survey on incidence and complications of bovine cutaneous papillomatosis. Indian J Vet. 1992;69:842–844. [Google Scholar]
- Winkler B, Capo V, Reumann W, Ma A, La Porta R, Reilly S, Green P, Richart R, Crum C. Human papillomavirus infection of the esophagus. A clinicopathologic study with demonstration of papillomavirus antigen by the lmmunoperoxidase technique. Cancer. 1985;55:149–155. doi: 10.1002/1097-0142(19850101)55:1<149::aid-cncr2820550123>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Wosiacki S, Barreiro M, Alfieri A, Alfieri A. Semi-nested PCR for detection and typing of bovine Papillomavirus type 2 in urinary bladder and whole blood from cattle with enzootic haematuria. J Virol Methods. 2005;126:215–219. doi: 10.1016/j.jviromet.2005.01.021. [DOI] [PubMed] [Google Scholar]
- Wosiacki S, Carlos A, Alfieri A, Alfieri A. Bovine papillomavirus type 2 in enzootic haematuria aetiology. Semin Ciênc Agr. 2002;1:121–130. [Google Scholar]
- Wosiacki S, Claus M, Alfieri A, Alfieri A. Bovine papillomavirus type 2 detection in the urinary bladder of cattle with chronic enzootic haematuria. Mem Inst Oswaldo Cruz. 2006;101:635–638. doi: 10.1590/s0074-02762006000600009. [DOI] [PubMed] [Google Scholar]
- Yaguiu A, Carvalho C, Freitas A, Gustavo L, Góes B, Dagli L, Birgel-Júnior E, Beçak W, Stocco dos Santos R. Papillomatosis in cattle: In situ detection of bovine papillomavirus DNA sequences in reproductive tissues. Brazi J Morphol Sci. 2006;23:525–529. [Google Scholar]
- Yao Y, Huang W, Yang X, Sun W, Liu X, Cun W, Ma Y. HPV-16 E6 and E7 protein T cell epitopes prediction analysis based on distributions of HLA-A loci across populations: An in silico approach. Vaccine. 2013;31:2289–2294. doi: 10.1016/j.vaccine.2013.02.065. [DOI] [PubMed] [Google Scholar]
- Yuan Z, Gallagher A, Gault EA, Campo MS, Nasir L. Bovine papillomavirus infection in equine sarcoids and in bovine bladder cancers. Vet J. 2007;174:599–604. doi: 10.1016/j.tvjl.2006.10.012. [DOI] [PubMed] [Google Scholar]
- Zanier K, Charbonnier S, Baltzinger M, Nominé Y, Altschuh D, Travé G. Kinetic analysis of the interactions of human papillomavirus E6 oncoproteins with the ubiquitin ligase E6AP using surface plasmon resonance. J Mol Biol. 2005;349:401–412. doi: 10.1016/j.jmb.2005.03.071. [DOI] [PubMed] [Google Scholar]
- Zanier K, Nominé Y, Charbonnier S, Ruhlmann C, Schultz P, Schweizer J, Travé G. Formation of well-defined soluble aggregates upon fusion to MBP is a generic property of E6 proteins from various human papillomavirus species. Protein Expr Purif. 2007;51:59–70. doi: 10.1016/j.pep.2006.07.029. [DOI] [PubMed] [Google Scholar]
- Zaugg N, Nespeca G, Hauser B, Ackermann M, Favrot C. Detection of novel papillomaviruses in canine mucosal, cutaneous and in situ squamous cell carcinomas. Vet Dermatol. 2005;16:290–298. doi: 10.1111/j.1365-3164.2005.00467.x. [DOI] [PubMed] [Google Scholar]
- Zhu X, Dong J, Uchida K, Watanabe K, Nakayama H, Goto Y, Haga T. Bovine papillomavirus type 10 with a deletion associated with a lingual papilloma in a cow. Vet J. 2014;199:303–305. doi: 10.1016/j.tvjl.2013.11.016. [DOI] [PubMed] [Google Scholar]
- Zimmermann H, Degenkolbe R, Bernard H, O’Connor M. The human papillomavirus type 16 E6 oncoprotein can down-regulate p53 activity by targeting the transcriptional coactivator CBP/p300. J Virol. 1999;73:6209–6209. doi: 10.1128/jvi.73.8.6209-6219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann H, Koh C, Degenkolbe R, O’Connor M, Muller A, Steger G, Chen J, Lui Y, Androphy E, Bernard H. Interaction with CBP/p300 enables the bovine papillomavirus type 1 E6 oncoprotein to downregulate CBP/p300-mediated transactivation by p53. J Gen Virol. 2000;81:2617–2623. doi: 10.1099/0022-1317-81-11-2617. [DOI] [PubMed] [Google Scholar]
- zur Hausen H. The search for infectious causes of human cancers: Where and why. Virology. 2009;392:1–10. doi: 10.1016/j.virol.2009.06.001. [DOI] [PubMed] [Google Scholar]
- zur Hausen H. Red meat consumption and cancer?: reasons to suspect involvement of bovine infectious factors in colorectal cancer. Int J Cancer. 2012;2483:2475–2483. doi: 10.1002/ijc.27413. [DOI] [PubMed] [Google Scholar]