Abstract
Red swamp crayfish is an important model organism for research of the invertebrate innate immunity mechanism. Its excellent disease resistance against bacteria, fungi, and viruses is well-known. However, the antiviral mechanisms of crayfish remain unclear. In this study, we obtained high-quality sequence reads from normal and white spot syndrome virus (WSSV)-challenged crayfish gills. For group normal (GN), 39,390,280 high-quality clean reads were randomly assembled to produce 172,591 contigs; whereas, 34,011,488 high-quality clean reads were randomly assembled to produce 182,176 contigs for group WSSV-challenged (GW). After GO annotations analysis, a total of 35,539 (90.01%), 14,931 (37.82%), 28,221 (71.48%), 25,290 (64.05%), 15,595 (39.50%), and 13,848 (35.07%) unigenes had significant matches with sequences in the Nr, Nt, Swiss-Prot, KEGG, COG and GO databases, respectively. Through the comparative analysis between GN and GW, 12,868 genes were identified as differentially up-regulated DEGs, and 9,194 genes were identified as differentially down-regulated DEGs. Ultimately, these DEGs were mapped into different signaling pathways, including three important signaling pathways related to innate immunity responses. These results could provide new insights into crayfish antiviral immunity mechanism.
Keywords: Procambarus clarkii, WSSV, gills, Illumina sequencing, de novo assembly, comparative transcriptomics
Introduction
Unlike vertebrates, invertebrates lack an acquired immune system, but they develop the innate immune system, including cellular and humoral immune responses (Du et al., 2010). When hosts suffer insults or infections from pathogens, these genes can be synergistically mobilized to play their respective roles in cellular defense, especially in the humoral immune response (Taffoni and Pujol, 2015). Further study of the coordination mechanisms of the invertebrate innate immune system by immune-related genes is crucial.
As a typical invertebrate, the red swamp crayfish is used as a model organism to research the response principles of the invertebrate innate immune system. This species is native to Northeastern Mexico and South America and was introduced into China from Japan in the 1930s (Shen et al., 2014). Because of its good fitness characteristics, strong adaptability to a changing environment, and high fecundity, the red swamp crayfish has been widely aquacultured in China (Manfrin et al., 2015). Currently, this crayfish has become one of the economically most important aquacultured species. Additionally, the excellent resistances of red swamp crayfish against bacteria, fungi, and viruses are well-known. Recent studies have shown that antibacterial and antifungal mechanisms, such as the Toll and Imd pathways, are strongly conserved (Ermolaeva and Schumacher, 2014). However, the antiviral mechanism in this species remains unclear (Ramos and Fernandez-Sesma, 2015). Systematically identifying the antiviral genes and antivirus-related signaling pathways in this species through transcriptome sequencing is crucial.
Recently, several reports have described transcriptome sequencing results of crayfish tissues, including the hepatopancreas, muscle, ovary, testis, eyestalk, spermary, epidermis, branchia, intestines, and stomach (Shen et al., 2014; Manfrin et al., 2015). However, the transcriptome of crayfish gills or white spot syndrome virus (WSSV)-challenged tissues have not been reported. More importantly, the crayfish gills are in direct contact with the external environment, and gill cells are crucial in the response to external biotic and abiotic factors, especially pathogenic microorganisms (Clavero-Salas et al., 2007). Due to their large number of hemocytes, gills play an important role in cellular host defense against pathogens (Egas et al., 2012). The gills are a vital organ that can remove invasive pathogens through an efficient and specific immune response. The study of the gill transcriptome will define an important part of the research field of the innate immune response mechanism.
Next-generation sequencing (NGS) technology has been widely used to explore and uncover vast genetic information in model organisms (Jiang et al., 2014). NGS technology is superior to the traditional Sanger sequencing technology in many aspects. For example, NGS technology can provide enormous amounts of sequence data in much shorter times and at a much lower cost (Martin and Wang, 2011). The expressed sequences produced using NGS technologies are usually 10-to100-fold greater than the number identified by traditional Sanger sequencing technologies (Christie et al., 2015). In this study, the HiSeq sequencing technology was used to sequence the transcriptomes of normal and WSSV-challenged crayfish gills. This approach was used to generate expression profiles and to discover differentially expressed genes (DEGs) between normal and WSSV-challenged crayfish gills. The functions of DEGs were annotated and classified by the Gene Ontology (GO), Cluster of Orthologous Groups (COG), and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. We believe the data obtained from this study could provide an important resource for research about genes functions, molecular events, and signaling pathways related to the invertebrate antiviral immune response.
Materials and Methods
Preparation of crayfish tissues and immune challenge
P. clarkii (weighing approximately 15–20 g) were purchased from an aquaculture commercial market in Hangzhou, Zhejiang Province, China. The collected crayfish were originally cultured in water tanks at 26–28°C for 10 days and fed twice a day with artificial food throughout the whole experiment (Li Y et al., 2012). To mimic WSSV infection, WSSV (3.2 107 copies per crayfish) was injected into the abdominal segment of each crayfish (Du et al., 2009). Thirty-six hours after the viral challenge, gills were collected from at least ten WSSV-challenged crayfish (Group WSSV-challenged (GW)). Gills were also collected from ten controls, uninfected crayfish designated as the Group normal (GN). All gills were immediately frozen in liquid nitrogen after collection, and samples were temporarily stored at −80°C until total RNA extraction (Du and Jin, 2015).
RNA isolation and Illumina sequencing
The two types of gill tissue samples (GN and GW) previously frozen in liquid nitrogen were delivered to the Beijing Genomics Institute-Shenzhen (BGI, Shenzhen, China) for total RNA extraction. In brief, total RNA from crayfish gills was extracted with TRIzol reagent in accordance with manufacturer's protocol (Invitrogen, Life Technologies, Grand Island, NY, USA). The quality of RNA samples treated with DNase I (Invitrogen) was examined for subsequent procedures, including mRNA purification, cDNA library construction and transcriptome sequencing. Approximately 5 μg of DNase-treated total RNA was used to construct a cDNA library following the protocols of the Illumina TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA, USA). After necessary quantification and qualification, the library was sequenced using an Illumina HiSeq 2000 instrument with 100 bp paired-end (PE) reads for both groups.
Transcriptome de novo assembly and analysis
Transcriptome de novo assembly for all samples was carried out by the RNA-Seq de novo assembly program Trinity (Dedeine et al., 2015). In brief, raw reads generated by the Illumina HiSeq 2000 sequencer were originally trimmed by removing the adapter sequences. After low-quality reads with quality scores ≤ 20 and short reads with a length ≤ 10 bp were removed, high-quality clean reads were obtained to execute transcriptome de novo assembly using the Trinity software with the default parameters. Generally, three steps were performed, including Inchworm, Chrysalis and Butterfly (He et al., 2015). Initially, high-quality clean reads were processed by Inchworm to form longer fragments, called contigs. Then, contigs were connected by Chrysalis to obtain unigenes, which could not be extended on either end. Unigenes resulted in de Bruijn graphs. Finally, the de Bruijn graphs were processed by Butterfly to obtain transcripts (Li et al., 2013).
Transcriptome annotation and Gene Ontology analysis
After transcriptome de novo assembly was finished, transcripts were used to carry out annotation tasks, including protein functional annotation, COG functional annotation, GO functional annotation, and pathways annotation. These tasks were based on sequence similarity with known genes. In detail, assembled contigs were annotated with sequences available in the NCBI database, using the BLASTx and BLASTn algorithms (Leng et al., 2015). The unigenes were aligned by a BLASTx search against NCBI protein databases, including the non-redundant (Nr) sequence, Swiss-Prot, KEGG, and COG databases (Li et al., 2015). However, none of the BLASTx hits were aligned by a BLASTn search against the NCBI non-redundant nucleotide database (Nt). The above alignments were executed to establish the homology of sequences with known genes (with a cutoff E-value ≤ 10-5) (Li ST et al., 2012). Then, the best alignment results were used to decide the sequence orientation and the protein coding region prediction (CDS) of the unigenes. Functional annotation was executed with Gene Ontology (GO) terms (www.geneontology.org) that were analyzed using the Blast2GO software (http://www.blast2go.com/b2ghome) (Conesa et al., 2005). Based on the KEGG database, the complex biological behavior of the genes was analyzed through pathway annotation.
Identification of differentially expressed genes
To acquire the expression profiles of transcripts in crayfish gills, cleaned reads were first mapped to all transcripts using Bowtie software (Langmead and Salzberg, 2012). Then, DEGs were obtained on the basis of fragments per kilobase of exon per million fragments mapped (FPKM) of the genes, followed by a False Discovery Rate (FDR) control, to correct for the P-value (Mortazavi et al., 2008). DEGs were identified using EDGER software (empirical analysis of digital gene expression data in R) (Robinson et al., 2010). In the analysis process, the filtering threshold was set as a 0.5 FDR control. Ultimately, an FDR ≤ 0.001 and the absolute value of the log2Ratio ≥ 1 were used as the filtering threshold to determine the significance of DEGs (Banani et al., 2016). DEGs (GW vs GN) were identified through a comparative analysis of the above data.
Quantitative real-time PCR
Quantitative real-time PCR (qRT-PCR) methods were used to determine the RNA levels for 15 selected genes related to innate immune response (Wang et al., 2015). For the qRT-PCR analysis, cDNA templates from the samples were diluted 20-fold in nuclease-free water and used as templates for the PCR reaction. Gene-specific primers sequences were designed using Primer Premier 6 software on the basis of each identified gene sequence from the transcriptome library (Lu et al., 2014). The specific primersPc-18 S RNA-qRT-F (5'-tct tct tag agg gat tag cgg-3') and Pc-18 S RNA-qRT-R (5'-aag ggg att gaa cgg gtt a-3') were used to amplify the 18 S RNA gene as an internal control. qRT-PCR was performed following the manufacturer's instructions of SYBR Premix Ex Taq (Takara, Dalian, China) using a real-time thermal cycler (Bio-Rad, USA) in a total volume of 10 μl containing 5 μl of 2 Premix Ex Taq, 1 μl of the 1:20 diluted cDNA, and 2 μl (1 μM) each of the forward and reverse primers. The amplification procedure was comprised of an initial denaturation step at 95°C for 3min, followed by 40 cycles of 95°C for 15s, 58°C for 40 s, and melting from 65°C to 95°C. Three parallel experiments were performed (Du et al., 2015). Furthermore, the differentially expressed levels of the target genes between the groups were calculated by the 2-ΔΔCT analysis method (Livak and Schmittgen, 2001). Data obtained were subjected to a statistical analysis, followed by an unpaired sample t-test. A significant difference was assigned when P < 0.05 or P < 0.01.
Results
Illumina sequencing of the crayfish gills transcriptome
After the GN sample was cleaned and quality-tested, 39,390,280 clean reads were identified from 40,384,330 raw reads, which corresponded to 3,939,028,000 total nucleotides (nt). The Q20 percentage (percentage of bases whose quality was ≥ 20 in the clean reads), N percentage (percentage of uncertain bases after filtering) and GC percentage were 98.02%, 0.00% and 41.52%, respectively (Table 1). For the GW sample, 34,011,488 clean reads were identified from 34,840,334 raw reads, which corresponded to 3,401,148,800 total nucleotides (nt). The Q20 percentage, N percentage and GC percentage were 98.04%, 0.00% and 41.98%, respectively (Table 1). All these sequences were used for further analysis.
Table 1. Summary of Illumina sequencing output for the GN and the GW.
| Sample | Total raw reads | Total clean reads | Total clean nucleotides (nt) | Q20 percentage | N percentage | GC percentage |
|---|---|---|---|---|---|---|
| GN-gills | 40,384,330 | 39,390,280 | 3,939,028,000 | 98.02% | 0.00% | 41.52% |
| GW-gills | 34,840,334 | 34,011,488 | 3,401,148,800 | 98.04% | 0.00% | 41.98% |
De novo assembly of the transcriptome
After the low-quality and short length reads were removed, high-quality clean reads were obtained, and transcriptome de novo assembly was performed using Trinity software with its default parameters (Ali et al., 2015). For GN, 39,390,280 high-quality clean reads were randomly assembled to produce 172,591 contigs with an N50 of 690 bp. The contigs were further assembled and clustered into 94,479 unigenes with a mean length of 644 nt, which were composed of 10,931 distinct clusters and 83,548 distinct singletons. The N50 of the above unigenes was 1,345 bp (Table 2). For GW, 34,011,488 high-quality clean reads were randomly assembled to produce 182,176 contigs with an N50 of 528 bp. The contigs were further assembled and clustered into 95,959 unigenes with a mean length of 558 nt, which were composed of 8,299 distinct clusters and 87,660 distinct singletons. The N50 of the above unigenes was 1,007 bp (Table 2).
Table 2. Summary of the assembly analysis for GN and GW.
| Dataset name | Group normal (GN) | Group WSSW-challenged (GW) | ||
|---|---|---|---|---|
| Contigs | Unigenes | Contigs | Unigenes | |
| Total number | 172,591 | 94,479 | 182,176 | 95,959 |
| Total length (nt) | 60,746,050 | 60,823,435 | 57,529,732 | 53,499,561 |
| Mean length (nt) | 352 | 644 | 316 | 558 |
| N50 | 690 | 1,345 | 528 | 1,007 |
| Total consensus sequences | - | 94,479 | - | 95,959 |
| Distinct clusters | - | 10,931 | - | 8,229 |
| Distinct singletons | - | 83,548 | - | 87,660 |
For GN, the lengths of unigenes were primarily distributed between 200 nt and 2,000 nt. The percentage of unigenes with lengths from 201–500 bp was 70.30% (66,423), from 501–1,000 bp was 14.07% (13,294), from 1,001–1,500 bp was 5.40% (5,099), from 1,501–2,000 bp was 3.23% (3,048), and > 2,000 bp was 7.00% (6,615). For GW, the percentage of unigenes with lengths from 201–500 bp was 74.12% (71,124), from 501–1,000 bp was 13.44% (12,897), from 1,001–1,500 bp was 4.91% (4,711), from 1,501–2,000 bp was 2.58% (2,479), and > 2,000 bp was 4.95% (4,748).
Functional annotation of predicted proteins
Transcripts were combined with data from two other transcriptomes from Mousavi et al. (2014). First, sequence annotation was carried out on the basis of unigenes from the merged group (Garcia-Seco et al., 2015). Then, the putative functions of all unigenes were analyzed based on GO and COG classifications (Young et al., 2010). In the present study, based on the merged group transcriptome data (98,676 unigenes), a basic sequence analysis was performed to understand the functions of the crayfish gill transcriptome before the analysis of DEGs related to WSSV infection. Among the predicted sequences, a total of 39,482 unigene sequences were annotated using a BLASTx alignment with an E-value ≤ 10-5. A total of 35,539 (90.01%), 14,931 (37.82%), 28,221 (71.48%), 25,290 (64.05%), 15,595 (39.50%), and 13,848 (35.07%) unigenes had significant matches with sequences in the Nr, Nt, Swiss-Prot, KEGG, COG and GO databases, respectively. In brief, a total of 35,539 transcripts (90.01% of all annotated transcripts) had significant hits in the Nr protein database. The gene names of the top BLAST hits were assigned to each transcript with significant hits. Among them, 2,682 (7.55%) transcripts were matched with genes from Paramecium tetraurelia, 2,362 (6.65%) transcripts were matched with genes from Daphnia pulex; 3,265 (9.19%) transcripts were matched with genes from Tetrahymena thermophila; 1,302 (3.66%) transcripts were matched with genes from Tribolium castaneum; 1,292 (3.64%) transcripts were matched with genes from Ichthyophthirius multifiliis; and 929 transcripts were matched with genes from Pediculus humanus.
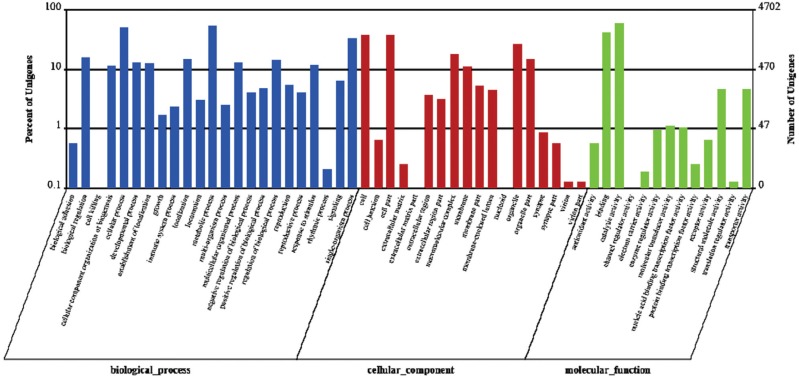
The GO classification is an international standardized gene function classification system that provides a dynamically updated controlled vocabulary and an exactly defined conception to describe genes' characteristics and their products in any organism (Di et al., 2015). The GO classification includes three ontologies: biological process, cellular component and molecular function (Chen et al., 2015). In this study, a GO analysis was carried out using Blast2GO software. A total of 13,848 transcripts annotated in the GO database were categorized into 58 functional groups, including the three main GO ontologies (Figure 1). Among these functional groups, “biological regulation”, “cellular process”, “metabolic process”, “cell”, “single-organism process”, “cell part”, “binding”, and “catalytic activity” terms were predominant.
Figure 1. Gene ontology (GO) classification of transcripts from the two gill samples (GN and GW). The three main GO categories include biological process (blue), cellular component (red), and molecular function (green).
COGs were delineated by comparing protein sequences encoded in complete genomes, representing major phylogenetic lineages (Tatusov et al., 2003). COG classification analysis is also an important method for unigene functional annotation and evolutionary research (Lai and Lin, 2013). We used the COG classification to further evaluate the completeness of the transcriptome library and the effectiveness of the annotation methods. A total of 15,959 unigenes were mapped to 25 different COG categories. In these 25 different categories, the largest COG group was category R, representing “general function prediction only” (6,435 unigenes, 41.26%), followed by category J, representing “translation, ribosomal structure and biogenesis” (3,347 unigenes, 21.46%); category K, representing “transcription” (3,044 unigenes, 19.52%); and category L, representing “replication, recombination and repair” (2,802 unigenes, 17.97%; Figure 2).
Figure 2. Cluster of orthologous groups (COG) classification of putative proteins.
KEGG is a bioinformatics resource for linking genomes to life and the environment (Mao et al., 2005). The KEGG pathway database records networks of molecular interactions in cells and variants specific to particular organisms (Chen et al., 2015). The genes from the merged groups (GN and GW) were categorized using the KEGG database to obtain more information to predict the unigene functions (Feng et al., 2015). A total of 25,290 unigenes were classified into 257 KEGG pathways. Among these KEGG pathways, the top 30 statistically significant KEGG classifications are shown in Table 3. Moreover, some important innate immunity-related pathways were predicted in this KEGG database, including Vibrio cholerae infection (1092 sequences, 4.32%), focal adhesion (910 sequences, 3.6%), Epstein-Barr virus infection (860 sequences, 3.40%), lysosome (610 sequences, 2.41%), HTLV-I infection (596 sequences, 2.36%), Herpes simplex infection (593 sequences, 2.34%), salmonella infection (576 sequences, 2.28%), MAPK signaling pathway (542 sequences, 2.14%), adherens junction (467 sequences, 1.85%), and so on (Table 3). Notably, the insulin signaling pathway, Wnt signaling pathway, mRNA surveillance pathway, endocytosis, phagosome, and tight junction functions were present in the top 40 statistically significant KEGG classifications.
Table 3. Top 30 statistically significant KEGG classifications.
| No. | Pathway | Pathway definition | Number of sequences |
|---|---|---|---|
| 1 | path: ko01100 | Metabolic pathways | 3371 (13.33%) |
| 2 | path: ko05146 | Amoebiasis | 1148 (4.54%) |
| 3 | path: ko05110 | Vibrio cholerae infection | 1092 (4.32%) |
| 4 | path: ko05016 | Huntington's disease | 973 (3.85%) |
| 5 | path: ko04810 | Regulation of actin cytoskeleton | 950 (3.76%) |
| 6 | path: ko04510 | Focal adhesion | 910 (3.6%) |
| 7 | path: ko03040 | Spliceosome | 894 (3.53%) |
| 8 | path: ko05200 | Pathways in cancer | 879 (3.48%) |
| 9 | path: ko05169 | Epstein-Barr virus infection | 860 (3.4%) |
| 10 | path: ko03013 | RNA transport | 847 (3.35%) |
| 11 | path: ko00230 | Purine metabolism | 781 (3.09%) |
| 12 | path: ko04145 | Phagosome | 643 (2.54%) |
| 13 | path: ko04144 | Endocytosis | 638 (2.52%) |
| 14 | path: ko04270 | Vascular smooth muscle contraction | 633 (2.5%) |
| 15 | path: ko03010 | Ribosome | 632 (2.5%) |
| 16 | path: ko04530 | Tight junction | 616 (2.44%) |
| 17 | path: ko00240 | Pyrimidine metabolism | 616 (2.44%) |
| 18 | path: ko04142 | Lysosome | 610 (2.41%) |
| 19 | path: ko04141 | Protein processing in endoplasmic reticulum | 607 (2.4%) |
| 20 | path: ko05166 | HTLV-I infection | 596 (2.36%) |
| 21 | path: ko05168 | Herpes simplex infection | 593 (2.34%) |
| 22 | path: ko05132 | Salmonella infection | 576 (2.28%) |
| 23 | path: ko03015 | mRNA surveillance pathway | 563 (2.23%) |
| 24 | path: ko04120 | Ubiquitin-mediated proteolysis | 557 (2.2%) |
| 25 | path: ko04010 | MAPK signaling pathway | 542 (2.14%) |
| 26 | path: ko04020 | Calcium signaling pathway | 538 (2.13%) |
| 27 | path: ko05414 | Dilated cardiomyopathy | 524 (2.07%) |
| 28 | path: ko05164 | Influenza A | 517 (2.04%) |
| 29 | path: ko05410 | Hypertrophic cardiomyopathy (HCM) | 495 (1.96%) |
| 30 | path: ko04970 | Salivary secretion | 493 (1.95%) |
Differentially expressed genes analysis of crayfish gills after WSSV infection
Previous sequence analysis and annotation for all unigenes in the merged group provided some useful information to analyze the crayfish gills transcriptome. Even more importantly, variation in the gene expression level of crayfish gills after WSSV infection was expected. In this study, an FDR ≤ 0.001 and an absolute value of log2Ratio ≥ 1 were used as the filtering threshold to determine up-regulated or down-regulated genes between normal and WSSV-challenged crayfish gills. As shown in Figure 3, a total of 22,062 DEGs with a > two-fold change were identified by the comparative analysis between the GN and GW samples, including 12,868 differentially up-regulated and 9,194 differentially down-regulated genes. In brief, among these 22,062 DEGs, 12,826 genes were expressed in both GN and GW, including 3,632 differentially up-regulated genes and 9,194 differently down-regulated genes. Moreover, 9,236 genes were only expressed in the GW sample.
Figure 3. Comparative analysis of the gene expression levels for two transcript libraries between normal (GN) and WSSV-challenged (GW) crayfish gills. Red dots represent transcripts significantly up-regulated in GW, while green dots represent significantly down-regulated transcripts. The parameters “FDR ≤ 0.001” and “| log2 Ratio| ≥ 1” were used as the threshold to judge the significance of gene expression differences.
To confirm the biological function of DEGs between GN and GW, GO classification and KEGG pathway analysis for the DEGs were carried out (Gupta et al., 2015). A GO classification analysis was conducted on the annotated transcripts using Blast2GO. As shown in Figure 4, a total of 4,702 DEGs were identified after a comparison between GN and GW. Significantly altered expression (up/down) was present for 45 GO terms (P < 0.05) and 32 GO terms, respectively (P < 0.01). Among the former group, there were 15, 8, and 22 GO terms that belonged to “cellular component (C)”, “molecular function (F)” and “biological process (P)”, respectively. Among the latter group, there were 14, 3, and 15 GO terms that belonged to “cellular component (C)”, “molecular function (F)” and “biological process (P)”, respectively. The GO analysis revealed that gene clusters with significant differential expression mainly concentrated in the aspect of biological process.
Figure 4. Gene ontology (GO) classification analysis of differentially expressed genes (DEGs) between GN and GW. The three main GO categories include biological process (blue), cellular component (red), and molecular function (green).
Next, all DEGs were mapped in the KEGG database to search for genes involved in innate immune response or signaling pathways. A total of 7,918 DEGs were assigned to 256 KEGG pathways. The KEGG pathway analysis identified 100 pathways that were significantly changed (P < 0.05),of which 80 reached a higher level of significance (P < 0.01) in the GW compared with the GN. Some of the significantly altered KEGG pathways were related to the innate immunity response, including apoptosis, Staphylococcus aureus infection, lysosome, melanogenesis, TGF-beta signaling pathway, NOD-like receptor signaling pathway, PPAR signaling pathway, focal adhesion, toll-like receptor signaling pathway, and bacterial invasion of epithelial cells (Table 4).
Table 4. Top 80 differentially expressed pathways between GW and GN.
| No. | Pathway | Number of DEGs | P-value | Pathway ID |
|---|---|---|---|---|
| 1 | Glycolysis / Gluconeogenesis | 99 (1.25%) | 3,78E-199 | ko00010 |
| 2 | RNA transport | 219 (2.77%) | 4.26E-92 | ko03013 |
| 3 | Epithelial cell signaling | 64 (0.81%) | 3.64E-19 | ko05120 |
| 4 | Apoptosis | 73 (0.92%) | 2.29E-17 | ko04210 |
| 5 | Allograft rejection | 3 (0.04%) | 7.15E-15 | ko05330 |
| 6 | Renin-angiotensin system | 31 (0.39%) | 2.42E-14 | ko04614 |
| 7 | p53 signaling pathway | 55 (0.69%) | 1.62E-12 | ko04115 |
| 8 | mTOR signaling pathway | 126 (1.59%) | 2.34E-12 | ko04150 |
| 9 | Endocytosis | 177 (2.24%) | 2.35E-11 | ko04144 |
| 10 | Staphylococcus aureus infection | 36 (0.45%) | 1.87E-10 | ko05150 |
| 11 | Leishmaniasis | 28 (0.35%) | 4.16E-10 | ko05140 |
| 12 | Protein processing in ER | 249 (3.14%) | 9.51E-10 | ko04141 |
| 13 | Synthesis and degradation of ketone bodies | 12 (0.15%) | 1.05E-09 | ko00072 |
| 14 | Metabolism of xenobiotics by P450 | 50 (0.63%) | 3.28E-09 | ko00980 |
| 15 | Phe, Tyr and Try biosynthesis | 13 (0.16%) | 4.22E-09 | ko00400 |
| 16 | Taurine and hypotaurine metabolism | 5 (0.06%) | 5.99E-09 | ko00430 |
| 17 | Collecting duct acid secretion | 44 (0.56%) | 9.75E-09 | ko04966 |
| 18 | Chronic myeloid leukemia | 43 (0.54%) | 2.35E-08 | ko05220 |
| 19 | Pentose phosphate pathway | 30 (0.38%) | 2.69E-08 | ko00030 |
| 20 | ABC transporters | 112 (1.41%) | 7.07E-08 | ko02010 |
| 21 | Thyroid cancer | 18 (0.23%) | 1.21E-07 | ko05216 |
| 22 | Lysosome | 268 (3.38%) | 2.02E-07 | ko04142 |
| 23 | Morphine addiction | 49 (0.62%) | 2.35E-07 | ko05032 |
| 24 | Valine, leucine and isoleucine biosynthesis | 12 (0.15%) | 2.72E-07 | ko00290 |
| 25 | Melanogenesis | 108 (1.36%) | 5.39E-07 | ko04916 |
| 26 | Butanoate metabolism | 40 (0.51%) | 5.58E-07 | ko00650 |
| 27 | Base excision repair | 45 (0.57%) | 6.18E-07 | ko03410 |
| 28 | Long-term depression | 41 (0.52%) | 6.57E-07 | ko04730 |
| 29 | Hepatitis C | 73 (0.92%) | 2.47E-06 | ko05160 |
| 30 | Mismatch repair | 23 (0.29%) | 3.24E-06 | ko03430 |
| 31 | Salivary secretion | 185 (2.34%) | 3.85E-06 | ko04970 |
| 32 | Histidine metabolism | 38 (0.48%) | 4.50E-06 | ko00340 |
| 33 | Bacterial invasion of epithelial cells | 102 (1.29%) | 1.05E-05 | ko05100 |
| 34 | Amoebiasis | 458 (5.78%) | 1.21E-05 | ko05146 |
| 35 | Propanoate metabolism | 72 (0.91%) | 1.25E-05 | ko00640 |
| 36 | Synaptic vesicle cycle | 85 (1.07%) | 1.39E-05 | ko04721 |
| 37 | Ubiquinone and terpenoid-quinone biosynthesis | 11 (0.14%) | 1.48E-05 | ko00130 |
| 38 | Influenza A | 201 (2.54%) | 1.78E-05 | ko05164 |
| 39 | Retrograde endocannabinoid signaling | 71 (0.9%) | 2.46E-05 | ko04723 |
| 40 | Prostate cancer | 113 (1.43%) | 3.11E-05 | ko05215 |
| 41 | Metabolic pathways | 1189 (15.02%) | 3.81E-05 | ko01100 |
| 42 | Starch and sucrose metabolism | 52 (0.66%) | 3.95E-05 | ko00500 |
| 43 | Glyoxylate and dicarboxylate metabolism | 50 (0.63%) | 7.25E-05 | ko00630 |
| 44 | Cocaine addiction | 39 (0.49%) | 0.000103 | ko05030 |
| 45 | Endocrine | 97 (1.23%) | 0.000119 | ko04961 |
| 46 | Phenylalanine metabolism | 33 (0.42%) | 0.000136 | ko00360 |
| 47 | HTLV-I infection | 198 (2.5%) | 0.000137 | ko05166 |
| 48 | Chemokine signaling pathway | 116 (1.47%) | 0.000142 | ko04062 |
| 49 | Vasopressin-regulated water reabsorption | 88 (1.11%) | 0.000148 | ko04962 |
| 50 | Endometrial cancer | 42 (0.53%) | 0.00015 | ko05213 |
| 51 | TGF-beta signaling pathway | 65 (0.82%) | 0.000158 | ko04350 |
| 52 | NOD-like receptor signaling pathway | 68 (0.86%) | 0.000224 | ko04621 |
| 53 | Regulation of actin cytoskeleton | 228 (2.88%) | 0.000291 | ko04810 |
| 54 | Transcriptional misregulation in cancer | 116 (1.47%) | 0.000355 | ko05202 |
| 55 | GnRH signaling pathway | 103 (1.3%) | 0.000478 | ko04912 |
| 56 | Glycine, serine and threonine metabolism | 68 (0.86%) | 0.000653 | ko00260 |
| 57 | Citrate cycle (TCA cycle) | 89 (1.12%) | 0.000814 | ko00020 |
| 58 | Pancreatic cancer | 67 (0.85%) | 0.000861 | ko05212 |
| 59 | Cardiac muscle contraction | 95 (1.2%) | 0.000893 | ko04260 |
| 60 | Tyrosine metabolism | 66 (0.83%) | 0.000972 | ko00350 |
| 61 | Sphingolipid metabolism | 31 (0.39%) | 0.00143 | ko00600 |
| 62 | Oocyte meiosis | 203 (2.56%) | 0.001598 | ko04114 |
| 63 | Adipocytokine signaling pathway | 68 (0.86%) | 0.001695 | ko04920 |
| 64 | Gastric acid secretion | 151 (1.91%) | 0.001769 | ko04971 |
| 65 | Peroxisome | 173 (2.18%) | 0.001809 | ko04146 |
| 66 | PPAR signaling pathway | 106 (1.34%) | 0.001873 | ko03320 |
| 67 | Drug metabolism - other enzymes | 44 (0.56%) | 0.001887 | ko00983 |
| 68 | Neurotrophin signaling pathway | 128 (1.62%) | 0.002169 | ko04722 |
| 69 | Tryptophan metabolism | 69 (0.87%) | 0.002208 | ko00380 |
| 70 | Cysteine and methionine metabolism | 45 (0.57%) | 0.002622 | ko00270 |
| 71 | Taste transduction | 26 (0.33%) | 0.003256 | ko04742 |
| 72 | Fatty acid biosynthesis | 2 (0.03%) | 0.003716 | ko00061 |
| 73 | Focal adhesion | 268 (3.38%) | 0.004214 | ko04510 |
| 74 | Glycosphingolipid biosynthesis - ganglio series | 13 (0.16%) | 0.004679 | ko00604 |
| 75 | Huntington's disease | 315 (3.98%) | 0.005037 | ko05016 |
| 76 | Toll-like receptor signaling pathway | 66 (0.83%) | 0.00549 | ko04620 |
| 77 | Insect hormone biosynthesis | 12 (0.15%) | 0.005932 | ko00981 |
| 78 | D-Arginine and D-ornithine metabolism | 13 (0.16%) | 0.007501 | ko00472 |
| 79 | Progesterone-mediated oocyte maturation | 151 (1.91%) | 0.007619 | ko04914 |
| 80 | Pancreatic secretion | 122 (1.54%) | 0.009396 | ko04972 |
Analysis of transcriptome data by qRT-PCR
We chose 15 genes related to the innate immunity response and evaluated their differential expression level between the GW and the GN, using qRT-PCR (Gao et al., 2015). For these candidate genes, the variation trends of the qRT-PCR expression profiles were found to be consistent with the RNA-seq data (Table 5). There were similar trends in the genes that were up-/down-regulated between the qRT-PCR and the transcriptome data, implying that the differential expression changes identified by the RNA-Seq data were reliable.
Table 5. Comparison of relative fold change of RNA-Seq and qRT-PCR results between GW and GN.
| Gene name | Protein identity | Fold variation (GW/GN) | |
|---|---|---|---|
| transcriptome | qRT-PCR | ||
| CL3739.Contig3_All | Lysozyme | 53.80 (up) | 20.51 (up) |
| Unigene68353_All | Integral membrane protein | 42.34 (up) | 25.63 (up) |
| Unigene56169_All | Apoptosis-regulated protein | 26.33 (up) | 16.77 (up) |
| CL88.Contig2_All | Serine protease inhibitor | 24.41 (up) | 18.52 (up) |
| CL4190.Contig1_All | Chitin binding-like protein | 7.29 (up) | 16.87 (up) |
| CL1412.Contig2_All | Interleukin enhancer binding factor | 4.83 (up) | 2.46 (up) |
| CL6575.Contig2_All | Anti-lipopolysaccharide factor | 4.11 (up) | 5.63 (up) |
| CL818.Contig4_All | Dicer 2 | 4.09 (up) | 3.48 (up) |
| CL1181.Contig4_All | Integrin | 3.59 (up) | 8.33 (up) |
| CL6415.Contig2_All | Cathepsin C | 3.49 (up) | 7.38 (up) |
| CL2737.Contig2_All | Ras | 3.16 (up) | 8.22 (up) |
| CL2514.Contig2_All | NF-kappa B inhibitor alpha | 0.46 (down) | 0.19 (down) |
| Unigene789_All | Clip domain serine proteinase 3 | 0.47 (down) | 0.66 (down) |
| Unigene31753_All | Tyrosine-protein kinase isoform | 0.36 (down) | 0.17 (down) |
| CL3161.Contig1_All | Phosphoinositide 3-kinase isoform | 0.49 (down) | 0.88 (down) |
Discussion
Transcriptome sequencing is an effective method for obtaining genomic information from non-model organism with no available reference genome. The genomic information is the important basis to discover molecular mechanisms of the organism's biological traits. Due to its superior antiviral immune characteristics, crayfish has become the economically most important aquacultured species in China. However, research on the antiviral molecular mechanisms is scarce. In the present study, de novo assembled transcriptomes of crayfish gills were analyzed, and a large number of sequences were obtained. DEGs that differed in expression between normal and WSSV-challenged crayfish gills were studied in detail. All these transcriptomes data are valuable to shed light on the antiviral immune mechanism of crayfish.
So far, several studies based on the NGS technology have reported transcriptome sequencing results for crayfish tissues, including hepatopancreas, muscle, ovary, testis, eyestalk, spermary, epidermis, branchia, intestines, and stomach. Those studies mainly focused on gonadal development (Jiang et al., 2014), neuroendocrinology (Manfrin et al., 2015), and genetic markers (Shen et al., 2014). Transcriptome sequencing results for crayfish antiviral immunity are very limited, and very few works about crayfish transcriptomes changing after virus challenge are reported. So, the aim of the present study was an in-depth analysis of DEGs by functional annotation, orthologous protein clustering, and annotation of signaling pathways related to the immune system to determine the underlying mechanisms involved in the crayfish anti-WSSV immune response.
Based on the KEGG pathway analysis for DEGs between the GN and GW, some signaling pathways related to the invertebrate innate immune system were identified. Apoptosis is an indispensable physiologic and biochemical process that evolved in eukaryotes to remove excessive and damaged cells to maintain organismal integrity. Apoptosis plays an important role in the processes of cell differentiation, development, and proliferation (Elmore, 2007). When organisms encounter a pathogen infection and other environment stresses, apoptosis can be mobilized to regulate cell metabolism (Igney and Krammer, 2002). Cell proliferation controlled by apoptosis has been reported to be involved in the antiviral immunity response (Du et al., 2013). In the present study, 73 representative innate immune-related genes were significantly differentially expressed in the apoptosis signaling pathway. Within this group, 52 DEGs were significantly up-regulated, and 21 DEGs were significantly down-regulated (Figure 5).
Figure 5. Significant differentially expressed genes (DEGs) identified by KEGG as involved in the apoptosis signaling pathway. Red boxes indicate significantly increased expression, green boxes indicate significantly decreased expression and blue boxes indicate unchanged expression.
Melanogenesis is an important biochemical pathway responsible for melanin synthesis that is controlled by complex regulatory mechanisms (Kim, 2015). Melanin is a vital intermediate in the arthropod humoral immunity response. In the present study, 108 innate immunity-related genes in the melanogenesis pathway were significantly differentially expressed, including 73 significantly up-regulated genes and 35 significantly down-regulated genes (Figure 6).
Figure 6. Significant differentially expressed genes (DEGs) identified by KEGG involved in the melanogenesis signaling pathway. Red boxes indicate significantly increased expression, green boxes indicate significantly decreased expression, and black boxes indicate unchanged expression.
Toll-like receptors (TLRs) are a group of highly conserved molecules that play a vital role in the recognition of pathogen-associated molecular patterns (PAMPs) and in the activation of innate immune responses to infectious agents (Zhang and Lu, 2015). Many studies have implied the TLRs signaling pathway in both antibacterial and antifungal defense (De Nardo, 2015). In this study, 66 innate immunity-related genes in the TLRs signaling pathway were significantly differentially expressed, including 49 significantly up-regulated genes and 17 significantly down-regulated genes (Figure 7).
Figure 7. Significant differentially expressed genes (DEGs) identified by KEGG involved in the Toll-like receptors (TLRs) signaling pathway. Red boxes indicate significantly increased expression, green boxes indicate significantly decreased expression, and black boxes indicate unchanged expression.
The three abovementioned signaling pathways were significantly changed (P < 0.01) between GN and GW, which suggests that they may play an important role in crayfish antiviral immunity responses. These results could provide new insights for future research on crayfish antiviral immunity.
In conclusion, many genes and pathways related to innate immunity were modified after WSSV infection of crayfish gills. Leveraging the gene expression changes into a model of altered network functions could provide new insights into the crayfish antiviral immunity mechanism and could also highlight candidate proteins that should be targeted to solve viral disease problems in the crayfish breeding process.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (Grant No. 31460698).
Footnotes
Associate Editor: Houtan Noushmehr
References
- Ali MY, Pavasovic A, Mather PB, Prentis PJ. Transcriptome analysis and characterisation of gill-expressed carbonic anhydrase and other key osmoregulatory genes in freshwater crayfish Cherax quadricarinatus . Data Brief. 2015;5:187–193. doi: 10.1016/j.dib.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani H, Marcet-Houben M, Ballester AR, Abbruscato P, González-Candelas L, Gabaldón T, Spadaro D. Genome sequencing and secondary metabolism of the postharvest pathogen Penicillium griseofulvum . BMC Genomics. 2016;17:e19. doi: 10.1186/s12864-015-2347-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Li E, Li T, Xu C, Wang X, Lin H, Qin JG, Chen L. Transcriptome and molecular pathway analysis of the hepatopancreas in the Pacific White Shrimp Litopenaeus vannamei under chronic low-salinity stress. PLoS One. 2015;7:e0131503. doi: 10.1371/journal.pone.0131503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Li E, Xu Z, Li T, Xu C, Qin JG, Chen L. Comparative transcriptome analysis in the hepatopancreas tissue of Pacific White Shrimp Litopenaeus vannamei fed different lipid sources at low salinity. PLoS One. 2015;12:e0144889. doi: 10.1371/journal.pone.0144889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie AE, Chi M, Lameyer TJ, Pascual MG, Shea DN, Stanhope ME, Schulz DJ, Dickinson PS. Neuropeptidergic signaling in the American Lobster Homarus americanus: New insights from high-throughput nucleotide sequencing. PLoS One. 2015;12:e0145964. doi: 10.1371/journal.pone.0145964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavero-Salas A, Sotelo-Mundo RR, Gollas-Galván T, Hernández-López J, Peregrino-Uriarte AB, Muhlia-Almazán A, Yepiz-Plascencia G. Transcriptome analysis of gills from the white shrimp Litopenaeus vannamei infected with White Spot Syndrome Virus. Fish Shellfish Immunol. 2007;2:459–472. doi: 10.1016/j.fsi.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;18:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- De Nardo D. Toll-like receptors: Activation, signalling and transcriptional modulation. Cytokine. 2015;2:181–189. doi: 10.1016/j.cyto.2015.02.025. [DOI] [PubMed] [Google Scholar]
- Dedeine F, Weinert LA, Bigot D, Josse T, Ballenghien M, Cahais V, Galtier N, Gayral P. Comparative analysis of transcriptomes from secondary reproductives of three Reticulitermes termitespecies. PLoS One. 2015;12:e0145596. doi: 10.1371/journal.pone.0145596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lena P, Domeniconi G, Margara L, Moro G. GOTA: GO term annotation of biomedical literature. BMC Bioinformatics. 2015;16:e346. doi: 10.1186/s12859-015-0777-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du ZQ, Jin YH. Molecular characterization and antibacterial activity analysis of two novel penaeidin isoforms from Pacific White Shrimp, Litopenaeus vannamei . Appl Biochem Biotechnol. 2015;8:1607–1620. doi: 10.1007/s12010-015-1840-7. [DOI] [PubMed] [Google Scholar]
- Du ZQ, Ren Q, Zhao XF, Wang JX. A double WAP domain (DWD)-containing protein with proteinase inhibitory activity in Chinese white shrimp, Fenneropenaeus chinensis . Comp Biochem Physiol B Biochem Mol Biol. 2009;2:203–210. doi: 10.1016/j.cbpb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Du ZQ, Li XC, Wang ZH, Zhao XF, Wang JX. A single WAP domain (SWD)-containing protein with antipathogenic relevance in red swamp crayfish, Procambarus clarkii . Fish Shellfish Immunol. 2010;1:134–142. doi: 10.1016/j.fsi.2009.10.009. [DOI] [PubMed] [Google Scholar]
- Du ZQ, Lan JF, Weng YD, Zhao XF, Wang JX. BAX inhibitor-1 silencing suppresses white spot syndrome virus replication in red swamp crayfish, Procambarus clarkii . Fish Shellfish Immunol. 2013;1:46–53. doi: 10.1016/j.fsi.2013.03.376. [DOI] [PubMed] [Google Scholar]
- Du ZQ, Yuan JJ, Ren DM. A novel single WAP domain-containing protein isoform with antibacterial relevance in Litopenaeus vannamei . Fish Shellfish Immunol. 2015;2:478–484. doi: 10.1016/j.fsi.2015.03.007. [DOI] [PubMed] [Google Scholar]
- Egas C, Pinheiro M, Gomes P, Barroso C, Bettencourt R. The transcriptome of Bathymodiolus azoricus gill reveals expression of genes from endosymbionts and free-living deep-sea bacteria. Mar Drugs. 2012;8:1765–1783. doi: 10.3390/md10081765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore S. Apoptosis: A review of programmed cell death. Toxicol Pathol. 2007;4:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolaeva MA, Schumacher B. Insights from the worm: The C elegans model for innate immunity. Semin Immunol. 2014;4:303–309. doi: 10.1016/j.smim.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Li Q, Yu H, Zhao X, Kong L. Comparative transcriptome analysis of the Pacific Oyster Crassostrea gigas characterized by shell colors: Identification of genetic bases potentially involved in pigmentation. PLoS One. 2015;12:e0145257. doi: 10.1371/journal.pone.0145257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Zhang X, Wei J, Sun X, Yuan J. Whole transcriptome analysis provides insights into molecular mechanisms for molting in Litopenaeus vannamei . PLoS One. 2015;12:e0144350. doi: 10.1371/journal.pone.0144350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Seco D, Zhang Y, Gutierrez-Mañero FJ, Martin C, Ramos-Solano B. RNA-Seq analysis and transcriptome assembly for blackberry (Rubus sp Var Lochness) fruit. BMC Genomics. 2015;16:e5. doi: 10.1186/s12864-014-1198-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P, Goel R, Agarwal AV, Asif MH, Sangwan NS, Sangwan RS, Trivedi PK. Comparative transcriptome analysis of different chemotypes elucidates withanolide biosynthesis pathway from medicinal plant Withania somnifera . Sci Rep. 2015;5:e18611. doi: 10.1038/srep18611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Zhuang H, Fu Y, Guo L, Guo B, Guo L, Zhang X, Wei Y. De novo transcriptome assembly of a Chinese Locoweed (Oxytropis ochrocephala) species provides insights into genes associated with drought, salinity, and cold tolerance. Front Plant Sci. 2015;6:e1086. doi: 10.3389/fpls.2015.01086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igney FH, Krammer PH. Death and anti-death: Tumour resistance to apoptosis. Nat Rev Cancer. 2002;4:277–288. doi: 10.1038/nrc776. [DOI] [PubMed] [Google Scholar]
- Jiang H, Xing Z, Lu W, Qian Z, Yu H, Li J. Transcriptome analysis of red swamp crawfish Procambarus clarkii reveals genes involved in gonadal development. PLoS One. 2014;8:e105122. doi: 10.1371/journal.pone.0105122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. Effect of ginseng and ginsenosides on melanogenesis and their mechanism of action. J Ginseng Res. 2015;1:1–6. doi: 10.1016/j.jgr.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai ZX, Lin YL. Analysis of the global transcriptome of longan (Dimocarpus longan Lour) embryogenic callus using Illumina paired-end sequencing. BMC Genomics. 2013;14:e561. doi: 10.1186/1471-2164-14-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;4:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng X, Jia H, Sun X, Shangguan L, Mu Q, Wang B, Fang J. Comparative transcriptome analysis of grapevine in response to copper stress. Sci Rep. 2015;5:e17749. doi: 10.1038/srep17749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Liang Y, Wang X, Wang L, Qi M, Yu Y, Luan Y. Transcriptomic analysis of Musca domestica to reveal key genes of the prophenoloxidase-activating system. G3 (Bethesda) 2015;9:1827–1841. doi: 10.1534/g3.115.016899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhang X, Sun Z, Li F, Xiang J. Transcriptome analysis on Chinese shrimp Fenneropenaeus chinensis during WSSV acute infection. PLoS One. 2013;3:e58627. doi: 10.1371/journal.pone.0058627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ST, Zhang P, Zhang M, Fu CH, Zhao CF, Dong YS, Guo AY, Yu LJ. Transcriptional profile of Taxus chinensis cells in response to methyl jasmonate. BMC Genomics. 2012a;13:295–295. doi: 10.1186/1471-2164-13-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Deng W, Yang K, Wang W. The expression of prophenoloxidase mRNA in red swamp crayfish, Procambarus clarkii, when it was challenged. Genomics. 2012b;6:355–360. doi: 10.1016/j.ygeno.2012.04.001. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Methods. 2001;4:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu X, Kim H, Zhong S, Chen H, Hu Z, Zhou B. De novo transcriptome assembly for rudimentary leaves in Litchi chinesis Sonn and identification of differentially expressed genes in response to reactive oxygen species. BMC Genomics. 2014;15:e805. doi: 10.1186/1471-2164-15-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfrin C, Tom M, De Moro G, Gerdol M, Giulianini PG, Pallavicini A. The eyestalk transcriptome of red swamp crayfish Procambarus clarkia . Gene. 2015;1:28–34. doi: 10.1016/j.gene.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Mao XZ, Cai T, Olyarchuk JG, Wei LP. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;19:3787–3793. doi: 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
- Martin JA, Wang Z. Next-generation transcriptome assembly. Nat Rev Genet. 2011;10:671–682. doi: 10.1038/nrg3068. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;7:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Mousavi S, Alisoltani A, Shiran B, Fallahi H, Ebrahimie E, Imani A, Houshmand S. De novo transcriptome assembly and comparative analysis of differentially expressed genes in Prunus dulcis Mill in response to freezing stress. PLoS One. 2014;8:e104541. doi: 10.1371/journal.pone.0104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos I, Fernandez-Sesma A. Modulating the innate immune response to influenza A virus: Potential therapeutic use of anti-inflammatory drugs. Front Immunol. 2015;6:e361. doi: 10.3389/fimmu.2015.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;1:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Hu Y, Ma Y, Zhou X, Xu Z, Shui Y, Li C, Xu P, Sun X. In-depth transcriptome analysis of the red swamp crayfish Procambarus clarkii . PLoS One. 2014;10:e110548. doi: 10.1371/journal.pone.0110548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taffoni C, Pujol N. Mechanisms of innate immunity in C elegans epidermis. Tissue Barriers. 2015;4:e1078432. doi: 10.1080/21688370.2015.1078432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: An updated version includes eukaryotes. BMC Bioinformatics. 2003;1:e41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Wang J, Su YQ, Mao Y, Zhang JS, Wu CW, Ke QZ, Han KH, Zheng WQ, Xu ND. Transcriptome analysis of the Larimichthys crocea liver in response to Cryptocaryon irritans . Fish Shellfish Immunol. 2015;48:1–11. doi: 10.1016/j.fsi.2015.11.011. [DOI] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010;2:R14–R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang E, Lu M. Toll-like receptor (TLR)-mediated innate immune responses in the control of hepatitis B virus (HBV) infection. Med Microbiol Immunol. 2015;1:11–20. doi: 10.1007/s00430-014-0370-1. [DOI] [PMC free article] [PubMed] [Google Scholar]