Abstract
P53 And DNA Damage-Regulated Gene 1 (PDRG1) is a novel gene which plays an important role in chaperone-mediated protein folding. In the present study, the full-length complementary DNA (cDNA) sequence of the PDRG1 gene from Penaeus monodon (PmPDRG1) was cloned by the rapid amplification of cDNA ends (RACE) method. The cDNA of PmPDRG1 spans 1,613 bp, interrupted by only one short intron, and encodes a protein of 136 amino acids with calculated molecular weight of 15.49 kDa. The temporal expression profile of PmPDRG1 in different tissues and in different developmental stages of the ovary was investigated by real-time quantitative PCR (RT-qPCR). An RNA interference (RNAi) experiment was performed to study the relationship between P. monodon p53 (Pmp53) and PmPDRG1, and the results showed that the relative expression level of PmPDRG1 mRNA was notably up-regulated from 12 h to 96 h after Pmp53 was silenced both in ovary and hepatopancreas. To further explore the role of PmPDRG1 in ovarian development, dopamine (DA) and 5-hydroxytryptamine (5-HT)-injected shrimps were analyzed by RT-qPCR, indicating that PmPDRG1 may be involved in the regulation of ovarian development of P. monodon.
Keywords: PDRG1, gene cloning, RT-qPCR, ovarian development, black tiger shrimp
Introduction
P53 And DNA Damage-Regulated Gene 1 (PDRG1, namely was first identified in 2003 by Luo et al. (2003). The human PDRG1 resides at the long arm of chromosome 20 and encodes a protein of 133 amino acids that is present within a distinct subcellular compartment of the cytoplasm (Deloukas et al., 2001; Luo et al., 2003). PDRG1 is usually strongly over-expressed in multiple human malignancies (Jiang et al., 2011; Wang et al., 2015a). PDRG1 protein was identified as a subunit of the R2TP/prefoldin-like complex, which is involved in the assembly of the RNA polymerase II complex (Pol II) in the cytoplasm of eukaryotic cells (Sardiu et al., 2008; Boulon et al., 2012; Mita et al., 2013). The tumor suppressor protein p53 can down-regulate the expression of PDRG1 mRNA, while ultraviolet (UV) radiation has the opposite effect (Luo et al., 2003). As certain interactions between PDRG1 and p53 exist, some scholars believe that PDRG1 has the potential to be a novel valuable tumor biomarker that could play a role in cancer development and/or progression or to be a DNA damage-associated maker (Jiang et al., 2011; Saigusa et al., 2012; Wang et al., 2015a). Furthermore, PDRG1 is proven to be involved in apoptosis and cell cycle regulation (Jiang et al., 2011; Wang et al., 2014). Although the roles of PDRG1 in DNA damage and tumor cell growth in vertebrates have been widely studied, the functions of PDRG1 in invertebrates, especially in crustaceans, are poorly understood.
The black tiger shrimp (P. monodon) is one of the most important aquatic commercial animals in Asia, especially in southern China. Because the eyestalk of P. monodon can secrete ovarian suppression hormones, unilateral eyestalk ablation is usually adopted to induce ovarian maturation of P. monodon, but this technique leads to the death of parent shrimps and lowers spawning quality (Benzie, 1998; Phinyo et al., 2013). Therefore, it is imperative to explore alternative technologies to eyestalk ablation and to understand the molecular mechanisms that control the development and maturation of ovaries/oocytes (Hiransuchalert et al., 2013; Phinyo et al., 2014). In our previous study, we found that Pmp53 plays an important role in the development and maturation of the ovaries in P. monodon (Dai et al., 2016). In the present study, we silenced Pmp53 to investigate its relationship with PmPDRG1 and the role that PmPDRG1 may play in the ovarian development of P. monodon. Biogenic amines such as dopamine (DA) and serotonin (5-hydroxytryptamine, 5-HT) are able to affect numerous physiological processes in crustaceans through their actions as neuroregulators. Both DA and 5-HT have been shown to be involved in the synthesis and release of neurohormones, such as crustacean hyperglycemic hormone (CHH), vitellogenesis-inhibiting hormone (VIH) and molt-inhibiting hormone (MIH) (Chen et al., 2003). It has been demonstrated that injected 5-HT can induce ovarian maturation in shrimp (Vaca and Alfaro, 2000), while dopamine depressed vitellogenin synthesis (Chen et al., 2003). The relationship between PDRG1 and ovarian maturation and the effects of DA and 5-HT on the expression levels of PmPDRG1 in ovaries and hepatopancreas of P. monodon are presented in this paper.
In this study, we cloned and characterized the full-length cDNA of P. monodon PmPDRG1 and assessed the distribution of PmPDRG1 transcripts in different tissues and ovary developmental stages. In addition, we investigated the expression profiles of PmPDRG1 mRNA in selected tissues after injection of Pmp53-dsRNA and exposure to 5-HT and DA. Results from this study will contribute to a better understanding of PDRG1 and its function in ovarian development of P. monodon.
Materials and Methods
Experimental animals and sample collection
Healthy black tiger shrimp, P. monodon (100 ± 18 g), cultivated in aerated seawater (salinity of 30 PSU) for three days at 25 ± 1 °C in the Shenzhen Base of South China Sea Fisheries Research Institute (Shenzhen, Guangdong province, China) were used as the material in the experiment. Various tissues (ovary, heart, intestine, brain, muscles, stomach and gills) from male and female individuals were dissected, snap frozen in liquid nitrogen, and stored at −80 °C. Five shrimp in each ovarian maturation stages were selected. The different ovarian stages used in this study were classified according to the morphology reported by Huang et al. (2006), as ovogonium stage (I), chromatin nucleolus stage (II), perinucleolus stage (III), yolky stage (IV), and cortical rod stage (V).
Total RNA extraction, first strand cDNA synthesis and DNA extraction
Total RNA was isolated from the examined tissue (about 100 mg) of the shrimp using TRIzol (Invitrogen, Shanghai, China) reagent following the manufacturer' protocol, resuspended in DEPC-treated water and stored at –80 °C (Jiang et al., 2009). The concentration of RNA was determined using a NanoDrop 2000 spectrometer (Thermo, USA), and RNA integrity was assessed by 1% agarose gel electrophoresis. The cDNA was synthesized from 1 μg of mRNA using a PrimeScript Reverse Transcriptase kit (TaKaRa, Dalian, China) following the manufacturer's protocol, as previously described (Wang et al., 2015b). The cDNA was used as the template for PCR reactions in gene cloning. The phenol-chloroform-isoamyl alcohol method was used to isolate total genomic DNA, which was then used as template to amplify introns.
Gene cloning and sequencing
A partial sequence of PDRG1 (970 bp) was isolated from the transcriptome database. Initially, PCR was carried out using the cDNA described above as template, using the primers PaF and PaR (Table 1) designed according to the partial sequence of PDRG1, for verification. Then, the 3′ end cDNA sequence of the PDRG1 gene was isolated using a SMARTTM RACE cDNA amplification kit (Clontech, Takara) (Jiang et al., 2009; Wang et al., 2015b). In the 3′RACE PCR, the touchdown PCR step was performed with the gene-specific primer pdrg-sp1 and a universal primer UPM (a mix of UPX-long and UPX-short, Table 1). The PCR cycling parameters were as follows: an initial denaturation at 94 °C for 3 min, followed by 30 cycles at 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 3 min, and the last cycle was followed by 10 min extension at 72 °C. Additionally, a nested PCR with pdrg-sp2 and NUP was carried out (PCR profile was as follows: 94 °C for 3 min; 94 °C for 30 s, 55 °C for 30 s, 72 °C for 3 min in 35 cycles; 72 °C for 10 min). The PCR products were purified using a PCR purification kit (Sangon, Shanghai, China) and cloned into the pMD18-T vector (TaKaRa). After transformtion into competent cells (E.coli DH5α), the positive clones were sequenced in both directions (Invitrogen, Guangzhou, China), and the resulting sequences were verified and subjected to cluster analysis.
Table 1. Primers used for gene cloning and expression analysis.
| Name | Primer sequence (5′ → 3′) | Application |
|---|---|---|
| 3′-CDS | AAGCAGTGGTATCAACGCAGAGTAC(T)30VN | reverse transcription |
| PaF | CTGGTGATGCAAGTGCAGTTCAG | PCR |
| PaR | TCCTCCTTACAATGAACTGTGCCA | PCR |
| pdrg-sp1 | GGAACGAACCTCAAGCCCTTATCACAA | 3′RACE |
| pdrg-sp2 | AAAATGTAGGGGGAGAAACTGTAGAAGC | 3′RACE |
| UPX-long | CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT | 3′RACE |
| UPX-short | CTAATACGACTCACTATAGGGC | 3′RACE |
| NUP | AAGCAGTGGTATCAACGCAGAGT | 3′RACE |
| qpdrg -F | TGCGGCAGAGGATGTTATATCA | Real time PCR |
| qpdrg-R | CCTGTGGACTGACTGGCTAAT | Real time PCR |
| rEF-F | AAGCCAGGTATGGTTGTCAACTTT | Real time PCR |
| rEF-R | CGTGGTGCATCTCCACAGACT | Real time PCR |
| pdrg B F | CGCGGATCCATGGCAGTGTCTCCAGAACGTATC | Prokaryotic expression |
| pdrg B R | CCCAAGCTTTCTACCAAGAACTTGCTTTACAGCT | Prokaryotic expression |
| pdrg NF1 | TGCGGCAGAGGATGTTATATCA | Intron amplification |
| pdrg NR1 | CCCAAGCTTTCTACCAAGAACTTGCTTTACAGCT | Intron amplification |
| pdrg NF2 | CGCGGATCCATGGCAGTGTCTCCAGAACGTATC | Intron amplification |
| pdrg NR2 | CCCAAGCTTTCTACCAAGAACTTGCTTTACAGCT | Intron amplification |
Sequence analysis, multiple sequence alignment, and phylogenetic analysis
The obtained PmPDRG1 cDNA sequence was compared with other known sequences in the NCBI database using the Blast algorithm (Altschul et al., 1997). The open reading frame (ORF) of PmPDRG1 cDNA was determined using ORF Finder software (http://www.ncbi.nlm.nih.gov/projects/gorf/). The molecular weight and pI of the deduced PmPDRG1 protein were examined using the Compute pI/Mw tool of the Expasy server (http://web.expasy.org/compute_pi/). N-glycosylation site prediction was done using NetNGlyc 1.0 software (http://www.cbs.dtu.dk/services/NetNGlyc/) and phosphorylation sites were predicted using NetPhos2.0 (http://www.cbs.dtu.dk/services/NetPhos/). Multiple sequence alignment was performed using the ClustalX 2.0.11 software. The signal peptide was predicted using the Signal P 4.1 program (http://www.cbs.dtu.dk/services/SignalP/) and the SMART 4.0 program (http://smart.embl-heidelberg.de/) was used to predict functional sites or domains in the amino acid sequence. A phylogenetic tree was constructed by the neighbor-joining (NJ) method and support of a bootstrap analysis with 1,000 replications implemented in the MEGA 5.0 package (Chen et al., 2013).
RNA interference
To investigate the relationship between PmPDRG1 and Pmp53 in ovarian development, an RNAi experiment was carried out using dsRNA specific for Pmp53. An in vitro expression system was adopted to obtain dsRNA. In brief, the recombined pD7 vector containing a bidirectional T7 RNA polymerase promoter was constructed using the pUC18 vector as described in previous studies (Robalino et al., 2007; Wang et al., 2010).
Subsequently, a recombinant plasmid (pD7-p53) containing a reverse complement of p53 was established and used as PCR template. Two separate PCR assays were set up with the primers pF/i53-R and pR/i53-F (Table 1). The PCR products of 763 bp and 590 bp were excised, gel-purified and used for in vitro transcription. Subsequently, dsRNA-p53 was synthesized using the in vitro Transcription T7 kit (TaKaRa) according to the manufacturer's instructions. The quality of dsRNAs was verified by 1.5% agarose gel electrophoresis and quantified using UV spectrophotometry. The dsRNA was stored at −80 °C until the experiment.
P. monodon shrimps (100 ± 2 g, 14 months old) were acclimatized for 2 days before dsRNA-p53 injection. The shrimps were injected with 300 μg of dsRNA-p53 dissolved in 40 μL of sterilized saline solution (10 mM Tris-HCl pH 7.5, 400 mM NaCl). Shrimps injected with sterilized saline solution were used as vehicle control (VC). Ovaries and hepatopancreas of the shrimp were randomly collected at 0, 12, 24, 48, 72, 96 h after injection, and frozen in liquid nitrogen for future RT-qPCR analysis.
5-HT and dopamine challenge
To examine the effects of 5-HT and DA on the expression levels of PmPDRG1, seven groups of female shrimp (100 ± 18 g, 14 months old) were injected intramuscularly into the first abdominal segment with 5-HT and DA (50 μg/g body weight). Shrimp injected with sterilized saline solution at 0 h were included as control. The ovaries and hepatopancreas were collected at 0, 6, 12, 24, 48, 72, 96 h post injection, and preserved in liquid nitrogen for future RT-qPCR analysis.
RT-qPCR for gene expression profile analysis
RT-qPCR was used to detect the temporal expression of the genes. cDNA was synthesized using the PrimeScriptTM RT reagent Kit with gDNA Eraser (Perfect Real Time) (TaKaRa) and used as templates for RT-qPCR assays. RT-qPCR was performed using SYBR® Premix Ex Taq TM II(TaKaRa) as described in our previous study (Jiang et al., 2009). The reference gene elongation factor-1alpha (EF-1α) (GenBank: DQ021452.1) was used as internal control for normalizing the cDNA template by menas of the primers rEF-F and rEF-R (Table 1). Each 25 μL reaction solution contained: 12.5 μL of 2SYBR® Premix Ex Taq II, 0.5 μL of forward primer (10 μM), 0.5 μL of reverse primer (10 μM), 2 μL of cDNA template equivalent to 70 ng total RNA, and 9.5 μL sterile distilled water. Each reaction was carried out simultaneously in three separate tubes and the test was repeated three times (Li et al., 2013). Thermal cycling conditions were 95 °C for 30 s, followed by 42 cycles of 95 °C for 5 s, 60 °C for 30 s. A melting curve analysis was added (95 °C for 1 s, 65 °C for 15 s, 95 °C for continuous acquisition) to demonstrate the specificity of the PCR products, as revealed by a single peak. The 2−ΔΔCT method was used to calculate relative gene expression levels (Livak and Schmittgen, 2001; Qiu et al., 2010).
Statistical analysis
Statistical analyses were carried out using SPSS software (SPSS Inc, USA). Data are reported as mean ± standard error (SE). Results obtained from qRT-PCR analysis were subjected to one-way analysis of variance (one-way ANOVA) followed by an unpaired, two-tailed t-test. Differences were considered significant at P < 0.05.
Results
Characterization of the PmPDRG1 full-length cDNA
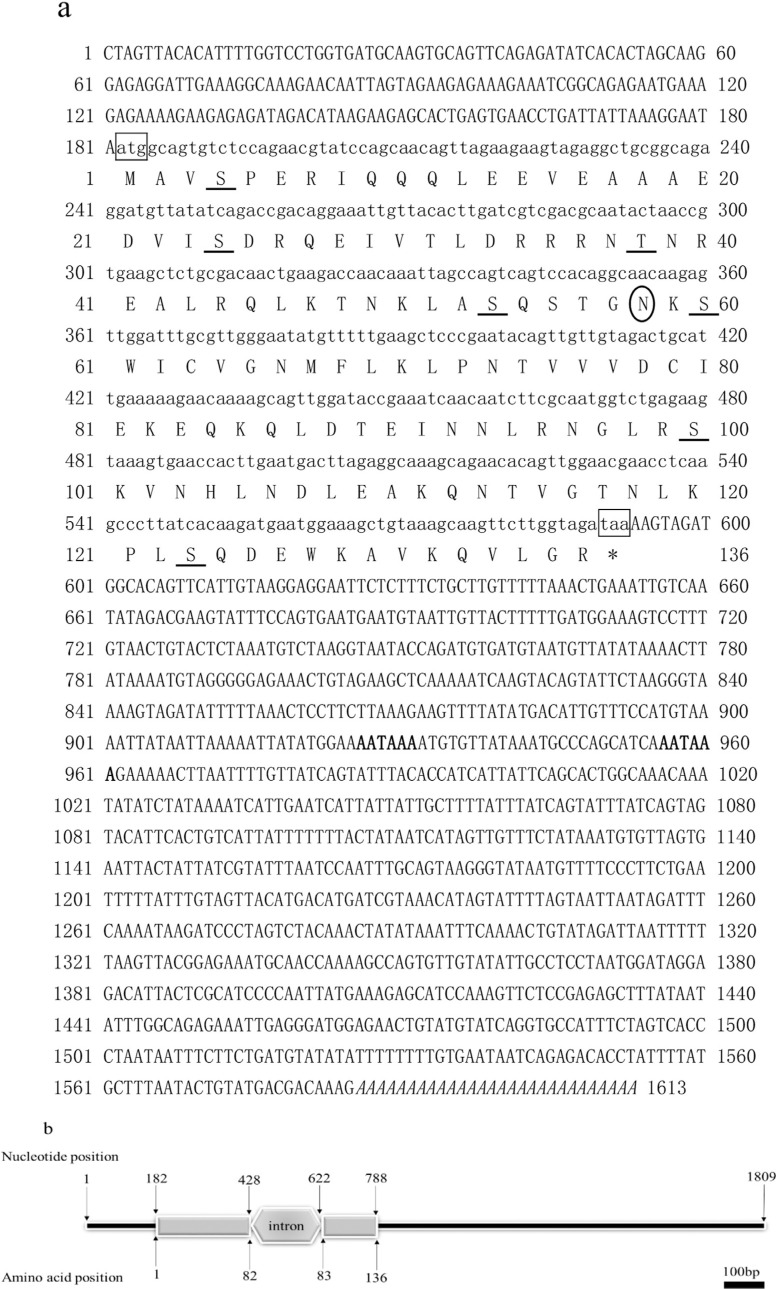
The full-length cDNA sequence of PmPDRG1 was obtained by RACE-PCR, and the complete nucleotide sequence and the deduced amino acid sequence are shown in Figure 1a (GenBank: KX156929). The cDNA sequence of PmPDRG1 is 1,613 bp in length, including an open reading frame of 411 bp (position 182-592 bp), a 5′UTR of 181 bp and a 3′UTR of 1,021 bp. Two eukaryotic polyadenylation signals AATAAA were located between nucleotides 926-931 bp and 956-961 bp and a poly (A) tail 28 bp downstream. The ORF sequence was predicted to encode a protein of 136 amino acids with a calculated molecular mass of about 15.49 kDa, and a theoretical isoelectric point of 8.62.
Figure 1. Deduced amino acid and sequences of PmPDRG1. (a) Numbers on the right and left of each row represent amino acid or nucleotide position; sequences in the boxes represent the start codon (ATG) and termination codon (TAA); eukaryotic polyadenylation signals AATAAA are highlighted in bold; the polyA signal sequence is italicized; the N-glycosylation site is marked by a circle; phosphorylation sites are underlined. (b) Schematic diagram representing the genomic DNA region of PmPDRG1.
The structure prediction results showed that PmPDRG1 contains 66.17% of α-helix, 5.15% of β-pleated sheet, and 28.68% of random coil. These conserved regions include seven phosphorylation sites and one N-glycosylation site (Figure 1a). Signal P 4.1 analysis revealed that PmPDRG1 does not contain a typical signal peptide sequence. PmPDRG1 is encoded in the nuclear genome, and has only one intron in the ORF (Figure 1b).
Phylogenetic analysis of PmPDRG1
The predicted amino acid sequence shared homology with previously published PDRGR1 sequences of other species in the GenBank database, detected using BLAST program. These include a 49% identity with Anoplopoma fimbria (ACQ58385.1), 48% identity with Oncorhynchus mykiss (NP_001154150.1), and 47% identity with Danio rerio (NP_001017757.1). The putative amino acid sequence of PmPDRG1 was aligned with other species in conserved regions. The deduced amino acid sequence QIVDLDTKRNQNREALRAL (30-48aa) of PmPDRG1 shared high homology with other species (Figure 2).
Figure 2. Multiple alignments of the deduced amino acid sequence of PmPDRG1 with other known PDRG1 aligned by Clustal X 2. 0. 11. Identical and similar sites are indicated with asterisks (*) and dots (. or :) , respectively. Black rectangles represent the phylogenetically conserved domain in different species. The species names and GenBank accession numbers are as follows: Homo sapiens (NP_110442.1); Mus musculus (NP_849270.1); Cricetulus griseus (ERE71423.1); Heterocephalus glaber (EHB11715.1); Ovis aries (XP_004014522.1); Bos taurus (NP_001071583.1); Gallus gallus (XP_015151961.1); Cuculus canorus (XP_009560934.1); Anolis carolinensis (XP_008119059.1); Alligator mississippiensis (KYO24169.1); Xenopus tropicalis (NP_001015688.1); Danio rerio (NP_001017757.1); Oncorhynchus mykiss (NP_001154150.1); Poecilia mexicana (XP_014829745.1); Cyprinodon variegatus (XP_015254726.1); Xiphophorus maculatus (XP_005810586.1); Lepisosteus oculatus (XP_006639564.1); Rousettus aegyptiacus (XP_016004320.1); Bombyx mori (XP_004928502.1); Penaeus monodon (KX156929).
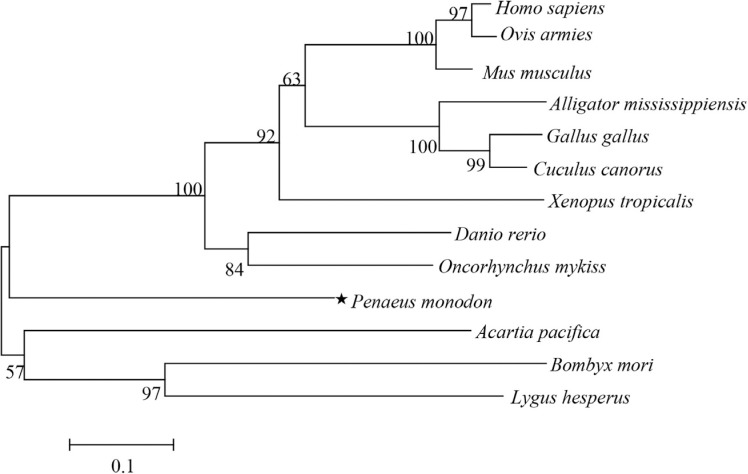
As shown in Figure 3, the PDRG1 phylogenetic tree comprised two main clusters: the upper cluster contained vertebrate PDRG1 sequences, while the second cluster contained invertebrate PDRG1 sequences. Vertebrate PDRG1 proteins appeared closely related to each other and converged into one subgroup, which include PmPDRG1. Although the shrimp PmPDRG1 was more similar to the vertebrate subgroup than the other main branch that includes the majority of the invertebrates, it still was an outlier in the main branch of the vertebrate subgroup.
Figure 3. NJ phylogenetic tree based on amino acid sequence encoded by PDRG1 as revealed by MEGA 5. 0 software. The numbers above the branches represent bootstrap values (1000 replicates). The following PDRG1 proteins family members were used in the phylogenetic analysis: Homo sapiens (NP_110442.1); Ovis aries (XP_004014522.1); Mus musculus (NP_849270.1); Alligator mississippiensis (KYO24169.1); Gallus gallus (XP_015151961.1); Cuculus canorus (XP_009560934.1); Xenopus tropicalis (NP_001015688.1); Danio rerio (NP_001017757.1); Oncorhynchus mykiss (NP_001154150.1); Penaeus monodon (KX156929); Acartia pacifica (ALS04393.1); Bombyx mori (XP_004928502.1); Lygus Hesperus (JAG33482.1).
Tissue expression analysis of PmPDRG1
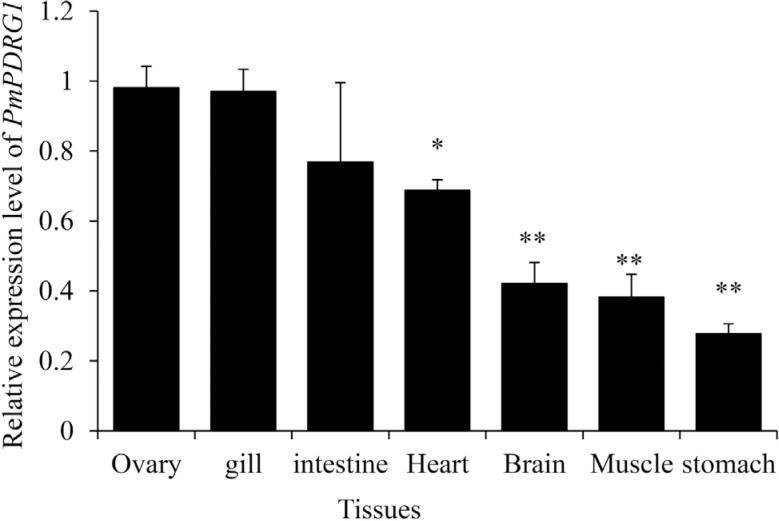
The tissue distribution pattern of PmPDRG1 mRNA is shown in Figure 4. The RT-qPCR results proved that the PmPDRG1 gene was expressed in all the examined tissues, with relatively high levels in the ovary, gill and intestine, moderate levels in the heart and brain, and low levels in th muscle and stomach (Figure 4).
Figure 4. Relative expression levels of PmPDRG1 mRNA in tissues. Tissue distribution of PDRG1 transcripts in the shrimp by RT-qPCR analysis using EF-1α as an internal reference. Vertical bars represented mean ± SD (n = 5). Significant differences between the experimental and the control group are indicated by asterisks * (P < 0.05); ** (P < 0.01).
Expression profiles of PmPDRG1 mRNA during ovarian maturation stages
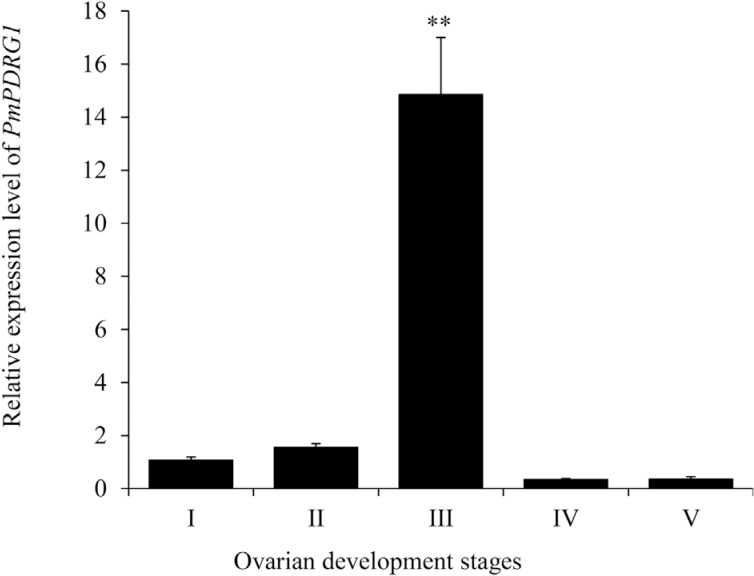
The relative expression levels of PmPDRG1 mRNA in different ovarian stages of P. monodon were investigated by RT-qPCR. The expression level in stage III ovaries was about 14.5-fold higher than in other stages (P < 0.05) as shown in Figure 5. The expression among stages I, II, IV, and V were not significantly different.
Figure 5. Relative expression levels of PmPDRG1 mRNA at different developmental stages of the ovaries. I, ovogonium stage; II, chromatin nucleolus stage; III, perinucleolus stage; IV, yolky stage; and V, cortical rod stage; Vertical bars represent the mean ± SD (n =5). ** P < 0.01.
PmPDRG1 mRNA expression profiles after Pmp53 gene silencing by Pmp53-dsRNA
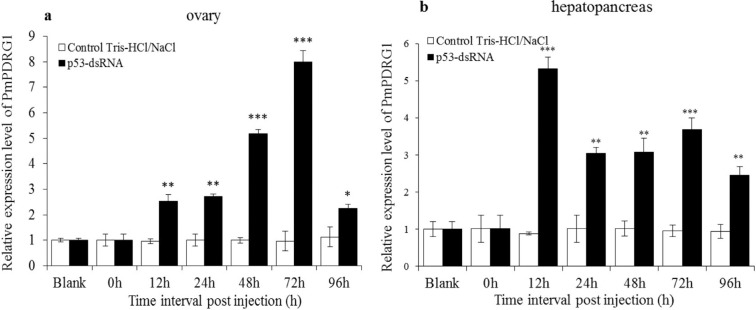
To investigate the relationship between Pmp53 and PmPDRG1, the Pmp53 gene was silenced by Pmp53-dsRNA. In ovary, the silencing efficiency of Pmp53 at 12, 24, 48 and 72 h of dsRNA-p53 post-injection were 65.86, 85.35, 64.06 and 25.45%, respectively, and in hepatopancreas, the silencing efficiency from 12, 24, 48, 72 and 96 h post injection were 37.70, 39.29, 64.24, 88.67 and 20.37%, respectively. Detailed data on Pmp53 gene silencing have been published in our previous study (Dai et al., 2016). After Pmp53 was successfully silenced, the PmPDRG1 mRNA expression pattern was analyzed by RT-qPCR assays in ovary and hepatopancreas. In ovary, the relative expression levels of PmPDRG1 mRNA were notably up-regulated at 12, 24, 48, 72 and 96 h post-injection of Pmp53-dsRNA compared to the control group. Additionally, its transcripts showed the lowest level at 96 h and the highest level at 72h post-injection (Figure 6a). In hepatopancreas of P. monodon, the relative expression levels of PmPDRG1 mRNA were notably up-regulated from 12 to 96 h post-injection of Pmp53-dsRNA compared to the control group (Figure 6b). The relative expression levels of PmPDRG1 mRNA were 5.3, 3.0, 3.1, 3.7 and 2.5-fold higher compared to the control group at 12, 24, 48, 72 and 96 h respectively.
Figure 6. PmPDRG1 mRNA expression profiles after silencing by Pmp53-dsRNA. (a) PmPDRG1 relative expression levels in ovary tissue post treatment with Pmp53-dsRNA; (b) PmPDRG1 relative expression levels in hepatopancreas tissue post treatment with Pmp53-dsRNA. Vertical bars represent the mean ± SD (n =3). Significant differences between the experimental and the control group are indicated by asterisks. * P < 0.05; ** P < 0.01; *** P < 0.001).
PmPDRG1 mRNA expression profiles after stimulation by 5-HT and DA
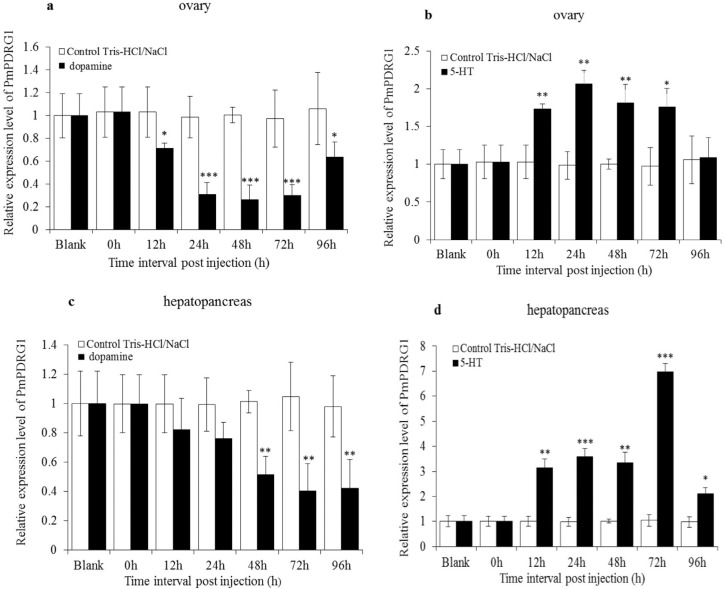
The expression levels of PmPDRG1 at 12–96 h post injection of Tris-HCl/NaCl were not significantly different from the untreated group. After the shrimp were injected with DA, the expression levels of PmPDRG1 were significantly reduced from 12–96 h in the ovary of P. monodon (Figure 7a) and at 48, 72 and 96 h in hepatopancreas of P. monodon (Figure 7c). In ovary, the expression levels of PmPDRG1 were significantly increased after injection of 5-HT at 12, 24, 48 and 72 h compared to the control group (Figure 7b), and in hepatopancreas, at 12–96h post injection. Specifically, after injection of 5-HT the expression level of PmPDRG1 was 6.98-fold higher compared with the control group at 72h (Figure 7d).
Figure 7. PmPDRG1 mRNA expression profiles after stimulation with 5-HT and DA. (a) PmPDRG1 relative expression levels in ovary tissue post treatment with DA. (b) PmPDRG1 relative expression levels in ovary tissue post treatment with 5-HT. (c) PmPDRG1 relative expression level in hepatopancreas tissue post treatment with DA. (d) PmPDRG1 relative expression levels in hepatopancreas tissue post treatment with 5-HT. Vertical bars represent the mean ± SD (n =3). Significant differences between the experimental and the control group are indicated by asterisks * P < 0.05; ** P < 0.01; *** P < 0.001).
Discussion
In the present study, the full-length cDNA sequence of the P. monodon PmPDRG1 gene was identified and characterized (Figure 1). Two potential polyadenylation signal sequences (AATAAA) were found in the 3′UTR of PmPDRG1, however, only one is found in PDRG1 of Apostichopus japonicus (Wang and Yang, 2013), and human (Luo et al., 2003). Although the deduced amino acid sequence QIVDLDTKRNQNREALRAL (30-48aa) of PmPDRG1 shares high homology with other species (Figure 2) (Wang and Yang, 2013), the total protein sequence of PmPDRG1 consists of 136 amino acids, compared to 133 amino acids deduced for PDRG1 of both human and mouse (Luo et al., 2003; Wang and Yang, 2013). Luo et al. (2003) reported that a helix-turn-helix motif (LNQDELKALKVILKG) exists at the C-terminal end of both human and mouse PDRG proteins, which is involved in protein-protein and protein-DNA interactions. However, we could not find such a motif in PmPDRG1, and further research is needed to study this difference. So far, the function of PDRG1 is still unclear because research about animal PDRG1 genes is relative rare and lacks thoroughness.
To study the evolutionary relationships of PmPDRG1 with other invertebrate and vertebrate PDRG1 family members, a phylogenetic analysis of the PDRG1 was performed. Vertebrate PDRG1 proteins are closely related to each other and converge into one subgroup, and even though PmPDRG1 was included in this vertebrate subgroup, the relationship was not very obvious. The results of the Blast and phylogenetic analysis suggested that PmPDRG1 iss a new member of the PDRG1 family. But the reason why PmPDRG1 was included in the vertebrate subgroup still needs further study.
The expression pattern in different tissues can indicate to some extent the main function(s) of the respective target gene. The results showed that PmPDRG1 is widely expressed in all the examined tissues, but especially high relative expression levels were detected in the ovary, gill and intestine (Figure 4). The results further indicate that the PmPDRG1 gene may play diverse roles in P. monodon, and that its main function sites may be the ovary, gill and intestine. The results for PmPDRG1 expression patterns during different maturation stages of the ovaries showed that PmPDRG1 mRNA increases sharply in stage III, and this is similar to previous reports in which the peak expression levels of PmCyclin A and PmCDK2, involved in ovarian development, were found at stage III (Visudtiphole et al., 2009; Dai et al., 2015). As stage III of ovary development is marked by massive cell proliferation and the presence of oocytes that have accumulated yolk substances in the cytoplasm (Huang et al., 2006), the results indicate that PmPDRG1 may be related to the oogenesis stage of ovarian development. In our previous study, we found that Pmp53 plays an important role in the development and maturation of the ovaries in P. monodon (Dai et al., 2016). To now study the relationship between Pmp53 and PmPDRG1 we successfully silenced the Pmp53 gene (Dai et al., 2016) causing an up-regulation of the relative expression of PmPDRG1 both in the ovary and hepatopancreas, indicating that Pmp53 could down-regulate PmPDRG1 transcript levels. The molecular regulatory mechanisms, however, still need to be further studied.
The study of molecular regulatory mechanisms related to promotion of reproductive development and maturation have begun to receive more attention,, especially in shrimp reproduction. Oocyte development includes a series of complex cellular events, in which differential genes express in a temporal and spatial fashion to guarantee the proper development of the oocytes or to store transcripts and proteins as maternal factors for early embryogenesis (Qiu et al., 2005). Vitellogenin (Vg) is synthesized in both the ovary and the hepatopancreas of P. monodon (Urtgam et al., 2015), and is a nutritive resource, playing an important role in embryonic growth and gonadal development (Bai et al., 2015). That is the reason why we selected ovary and hepatopancreas to perform the DA and 5-HT challenge assay. Molecular effects of DA and 5-HT on the relative expression levels of the PmpPDRG1 in ovaries and hepatopancreas are first reported in this study. The expression levels of PmPDRG1 mRNA were reduced after injection of DA, and increased after injection of 5-HT both in ovaries and hepatopancreas. Previous studies have shown that DA depresses vitellogenin synthesis and inhibits ovarian maturation. The expression level change of PmPDRG1 mRNA after DA or 5-HT injection may imply that PmPDRG1 is implicated in the regulation of ovarian maturation of P. monodon. However, knowledge on the detailed functional mechanisms of PmPDRG1 in ovarian maturation are still limited and require further research.
In summary, the complete cDNA sequence of PmPDRG1 was isolated and characterized in P. monodon. Subsequently, the mRNA distribution pattern of PmPDRG1 in different tissues and ovarian stages was studied to explore its role in the development and maturation of the ovaries. In addition, the expression pattern of PmPDRG1 post Pmp53-dsRNA was studied to explore the possible relationship between Pmp53 and PmPDRG1. Molecular effects of 5-HT and DA on the expression regulation of PmPDRG1 in ovaries and hepatopancreas are first reported in this study, which should help to improve our understanding of the molecular mechanisms of ovarian development in shrimp.
Acknowledgments
This work was supported by the special projects of the construction of fishing ports and the development of fisheries industry in Guangdong Province (A201601A08), the Special Scientific Research Funds for Central Non-profit Institutes (2015YD05), the Guangdong Provincial Science and Technology Program (2015A020209037) and the special projects of the marine fisheries science and technology and the industrial development in Guangdong Province (A201501A11).
Footnotes
Associate Editor: Luiz F. Z. Batista
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai H, Qiao H, Li F, Fu H, Sun S, Zhang W, Jin S, Gong Y, Jiang S, Xiong Y. Molecular characterization and developmental expression of vitellogenin in the oriental river prawn Macrobrachium nipponense and the effects of RNA interference and eyestalk ablation on ovarian maturation. Gene. 2015;562:22–31. doi: 10.1016/j.gene.2014.12.008. [DOI] [PubMed] [Google Scholar]
- Benzie JA. Penaeid genetics and biotechnology. Aquaculture. 1998;164:23–47. [Google Scholar]
- Boulon S, Bertrand E, Pradet-Balade B. HSP90 and the R2TP co-chaperone complex: Building multi-protein machineries essential for cell growth and gene expression. RNA Biol. 2012;9:148–154. doi: 10.4161/rna.18494. [DOI] [PubMed] [Google Scholar]
- Chen J, Liu P, Li Z, Chen Y, Qiu GF. The cloning of the cdk2 transcript and the localization of its expression during gametogenesis in the freshwater giant prawn, Macrobrachium rosenbergii . Mol Biol Rep. 2013;40:4781–4790. doi: 10.1007/s11033-013-2574-7. [DOI] [PubMed] [Google Scholar]
- Chen Y, Fan H, Hsieh S, Kuo C. Physiological involvement of DA in ovarian development of the freshwater giant prawn, Macrobrachium rosenbergii . Aquaculture. 2003;228:383–395. [Google Scholar]
- Dai W, Qiu L, Zhao C, Fu M, Ma Z, Zhou F, Yang Q. Characterization, expression and silencing by RNAi of p53 from Penaeus monodon. Mol Biol Rep. 2016;43:549–561. doi: 10.1007/s11033-016-3988-9. [DOI] [PubMed] [Google Scholar]
- Dai WT, Fu MJ, Zhao C, Zhou FL, Yang QB, Wang Y, Shi JX, Qiu LH. Molecular cloning and expression analysis of CDK2 gene from black tiger shrimps (Penaeus monodon) South China Fish Sci. 2015;11:1–11. [Google Scholar]
- Deloukas P, Matthews L, Ashurst J, Burton J, Gilbert J, Jones M, Stavrides G, Almeida J, Babbage A, Bagguley C. The DNA sequence and comparative analysis of human chromosome 20. Nature. 2001;414:865–871. doi: 10.1038/414865a. [DOI] [PubMed] [Google Scholar]
- Hiransuchalert R, Thamniemdee N, Khamnamtong B, Yamano K, Klinbunga S. Expression profiles and localization of vitellogenin mRNA and protein during ovarian development of the giant tiger shrimp Penaeus monodon . Aquaculture. 2013;412:193–201. [Google Scholar]
- Huang JH, Zhou FL, Ma ZM, Ye L, Jiang SG. Morphological and histological observation on ovary development of Penaeus monodon from northern South China Sea. J Trop Oceanogr. 2006;25:47–52. [Google Scholar]
- Jiang L, Luo X, Shi J, Sun H, Sun Q, Sheikh MS, Huang Y. PDRG1, a novel tumor marker for multiple malignancies that is selectively regulated by genotoxic stress. Cancer Biol Ther. 2011;11:567–573. doi: 10.4161/cbt.11.6.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Qiu L, Zhou F, Huang J, Guo Y, Yang K. Molecular cloning and expression analysis of a heat shock protein (Hsp90) gene from black tiger shrimp (Penaeus monodon) Mol Biol Rep. 2009;36:127–134. doi: 10.1007/s11033-007-9160-9. [DOI] [PubMed] [Google Scholar]
- Li WX, Huang HY, Huang JR, Yu JJ, Ma J, Ye HH. Molecular cloning, expression profiles and subcellular localization of cyclin B in ovary of the mud crab, Scylla paramamosain . Genes Genomics. 2013;35:185–195. [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luo X, Huang Y, Sheikh MS. Cloning and characterization of a novel gene PDRG that is differentially regulated by p53 and ultraviolet radiation. Oncogene. 2003;22:7247–7257. doi: 10.1038/sj.onc.1207010. [DOI] [PubMed] [Google Scholar]
- Mita P, Savas JN, Ha S, Djouder N, Yates JR, III, Logan SK. Analysis of URI nuclear interaction with RPB5 and components of the R2TP/prefoldin-like complex. PloS One. 2013;8:e63879. doi: 10.1371/journal.pone.0063879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phinyo M, Nounurai P, Hiransuchalert R, Jarayabhand P, Klinbunga S. Characterization and expression analysis of Cyclin-dependent kinase 7 gene and protein in ovaries of the giant tiger shrimp Penaeus monodon . Aquaculture. 2014;432:286–294. [Google Scholar]
- Phinyo M, Visudtiphole V, Roytrakul S, Phaonakrop N, Jarayabhand P, Klinbunga S. Characterization and expression of cell division cycle 2 (Cdc2) mRNA and protein during ovarian development of the giant tiger shrimp Penaeus monodon . Gen Comp Endocrinol. 2013;193:103–111. doi: 10.1016/j.ygcen.2013.07.012. [DOI] [PubMed] [Google Scholar]
- Qiu G, Yamano K, Unuma T. Cathepsin C transcripts are differentially expressed in the final stages of oocyte maturation in kuruma prawn Marsupenaeus japonicus . Comp Biochem Physiol B Biochem Mol Biol. 2005;140:171–181. doi: 10.1016/j.cbpc.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Qiu L, Ma Z, Jiang S, Wang W, Zhou F, Huang J, Li J, Yang Q. Molecular cloning and mRNA expression of peroxiredoxin gene in black tiger shrimp (Penaeus monodon) Mol Biol Rep. 2010;37:2821–2827. doi: 10.1007/s11033-009-9832-8. [DOI] [PubMed] [Google Scholar]
- Robalino J, Bartlett TC, Chapman RW, Gross PS, Browdy CL, Warr GW. Double-stranded RNA and antiviral immunity in marine shrimp: Inducible host mechanisms and evidence for the evolution of viral counter-responses. Dev Comp Immunol. 2007;31:539–547. doi: 10.1016/j.dci.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Saigusa S, Tanaka K, Toiyama Y, Matsushita K, Kawamura M, Okugawa Y, Hiro J, Inoue Y, Uchida K, Mohri Y. Gene expression profiles of tumor regression grade in locally advanced rectal cancer after neoadjuvant chemoradiotherapy. Oncol Rep. 2012;28:855–861. doi: 10.3892/or.2012.1863. [DOI] [PubMed] [Google Scholar]
- Sardiu ME, Cai Y, Jin J, Swanson SK, Conaway RC, Conaway JW, Florens L, Washburn MP. Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc Natl Acad Sci U S A. 2008;105:1454–1459. doi: 10.1073/pnas.0706983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urtgam S, Treerattrakool S, Roytrakul S, Wongtripop S, Prommoon J, Panyim S, Udomkit A. Correlation between gonad-inhibiting hormone and vitellogenin during ovarian maturation in the domesticated Penaeus monodon . Aquaculture. 2015;437:1–9. [Google Scholar]
- Vaca AA, Alfaro J. Ovarian maturation and spawning the white shrimp, Penaeus vannamei, by serotonin injection. Aquaculture. 2000;182:373–385. [Google Scholar]
- Visudtiphole V, Klinbunga S, Kirtikara K. Molecular characterization and expression profiles of cyclin A and cyclin B during ovarian development of the giant tiger shrimp Penaeus monodon . Comp Biochem Physiol A Mol Integr Physiol. 2009;152:535–543. doi: 10.1016/j.cbpa.2008.12.011. [DOI] [PubMed] [Google Scholar]
- Wang DQ, Wang K, Yan DW, Liu J, Wang B, Li MX, Wang XW, Liu J, Peng ZH, Li GX. Ciz1 is a novel predictor of survival in human colon cancer. Exp Bi Med. 2014;239:862–870. doi: 10.1177/1535370213520113. [DOI] [PubMed] [Google Scholar]
- Wang J, Zhang X, Wang L, Yang Y, Dong Z, Wang H, Du L, Wang C. MicroRNA-214 suppresses oncogenesis and exerts impact on prognosis by targeting PDRG1 in bladder cancer. PloS One. 2015a;10:e0118086. doi: 10.1371/journal.pone.0118086. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang KH-C, Tseng C-W, Lin H-Y, Chen I-T, Chen Y-H, Chen Y-M, Chen T-Y, Yang H-L. RNAi knock-down of the Litopenaeus vannamei Toll gene (LvToll) significantly increases mortality and reduces bacterial clearance after challenge with Vibrio harveyi . Dev Comp Immunol. 2010;34:49–58. doi: 10.1016/j.dci.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Wang T, Yang H. Cloning and characterization of PDRG gene from sea cucumber Apostichopus japonicus and the expression in intestine during aestivation. Marine Sci. 2013;37:1–9. [Google Scholar]
- Wang Y, Fu MJ, Zhao C, Zhou FL, Yang QB, Jiang SG, Qiu LH. Molecular cloning and expression analysis of MAT1 gene in black tiger shrimp (Penaeus monodon) Genet Mol Res. 2015b;5:1–13. doi: 10.4238/gmr.15017367. [DOI] [PubMed] [Google Scholar]