Abstract
Paradoxically, during early tumor development in many cancer types, TGF-β acts as a tumor suppressor, whereas in the advanced stages of these cancers, increased TGF-β expression is linked to high metastasis and poor prognosis. These findings suggest that unidentified mechanisms may function to rewire TGF-β signaling toward its prometastatic role in cancer cells. Our current study using non–small-cell lung carcinoma (NSCLC) cell lines, animal models, and clinical specimens demonstrates that suppression of SMAD2, with SMAD3 function intact, switches TGF-β–induced transcriptional responses to a prometastatic state. Importantly, we identified chaperonin containing TCP1 subunit 6A (CCT6A) as an inhibitor and direct binding protein of SMAD2 and found that CCT6A suppresses SMAD2 function in NSCLC cells and promotes metastasis. Furthermore, selective inhibition of SMAD3 or CCT6A efficiently suppresses TGF-β–mediated metastasis. Our findings provide a mechanism that directs TGF-β signaling toward its prometastatic arm and may contribute to the development of therapeutic strategies targeting TGF-β for NSCLC.
Introduction
Non–small-cell lung carcinoma (NSCLC) is among the most commonly diagnosed cancers worldwide (1, 2). The prognosis of NSCLC remains poor, and the overall 5-year relative survival rate, including all stages and subtypes, is less than 20% (1, 3). Like nearly all cancer types, metastasis represents the main cause of death in patients with NSCLC. Biologically, tumor metastasis is a multistep, complex process that is typically driven by aberrant activation or suppression of one or more signal transduction pathways (4). Notably, among the pathways frequently dysregulated in cancer metastasis, TGF-β signaling has been widely demonstrated as one of the most commonly activated and essential pathways for the metastasis of various cancer types (5, 6). Indeed, activation of TGF-β signaling is closely related to NSCLC progression and metastasis (7–9), whereas the mechanisms that activate and sustain prometastatic TGF-β signaling remain incompletely understood.
Activation of the TGF-β signaling cascade is typically initiated by binding of a TGF-β ligand with the TGF-β type II serine/threonine receptor (TGFBR2), followed by phosphorylation and oligomerization of TGFBR1/2, which causes phosphorylation of the cytoplasmic effectors SMAD2 and SMAD3. Phosphorylated SMAD2 or SMAD3 subsequently forms a heteromeric complex with SMAD4 and is transported to the nucleus, where it binds with other DNA-binding transcription factors and consequently regulates the transcription of TGF-β target genes (5, 6, 10, 11).
It has been noted that the biological and clinical outcomes of TGF-β signaling in cancer are far more complex than was previously understood, and these effects may be more cancer type and biological context dependent than expected. Alterations of pathway component proteins, binding partners of SMADs, and microenvironmental factors may lead to variable cellular responses to TGF-β stimulation (6, 12). For example, in benign epithelia and early tumor initiation, TGF-β inhibits epithelial growth and plays a tumor-suppressive role; in contrast, in advanced tumors, dysregulated TGF-β signaling promotes tumor progression and metastasis by enhancing the epithelial-mesenchymal transition (EMT) and cancer cell colonization in distant organs (6, 12–16). Moreover, additional studies have demonstrated that in several types of cancer, the tumor-suppressive arm of TGF-β signaling may be terminated (17–23). Notably, this complexity of TGF-β signaling has complicated the initially expected feasibility of targeting this pathway as an effective antimetastatic strategy. Both preclinically and clinically, the development of TGFBR inhibitors or ligand traps has not been successful (24, 25). Thus, while TGF-β is a central promoter of metastasis and may therefore represent a potentially promising antimetastatic target, a better understanding of the molecular mechanism that directs TGF-β signaling to promote metastasis will facilitate the development of effective TGF-β–targeting antimetastasis approaches.
In the context of distinguishing the antiproliferative and prometastatic arms of TGF-β signaling, it is particularly noteworthy that SMAD2 and SMAD3 comprise 2 major TGF-β receptor–regulated SMADs (R-SMADs) activated by TGF-β stimulation. Interestingly, SMAD2 and SMAD3 are structurally similar to each other and are functionally related in many physiological scenarios; however, abundant evidence suggests that these R-SMADs have different functions through the control of distinct transcriptional programs and play distinguishable roles during cancer progression (6). For example, depletion of SMAD2, but not SMAD3, results in enhanced cell invasion, metastasis (21, 26), and angiogenesis (26, 27) in skin squamous carcinoma and breast cancer. Notably, in specific types of cancer, such as colorectal and pancreatic cancers, genomic deletion or loss-of-function mutations of the SMAD2 gene are common, whereas in many other cancer types, including lung cancer and breast cancer, genomic alterations of SMAD2 are rarely present (5, 28, 29). Moreover, clinical data and specimens collected in The Cancer Genome Atlas (TCGA) public database indicate that less than 2% of NSCLC patients carry a genetic lesion in the SMAD2 gene. Thus, on the backdrop that overexpression of TGF-β ligand has been linked to metastasis and predicts poor prognosis in NSCLC patients (7, 8, 30), the mechanisms through which the TGF-β–induced metastasis-suppressive biological effects of SMAD2 are abrogated, such that TGF-β signaling becomes prometastatic in NSCLC, remain to be elucidated.
It is also noteworthy that SMAD2 and SMAD3 are responsible for the transactivation of cell-cycle inhibitory genes (31, 32) and the suppression of epithelial markers (33) following TGF-β stimulation; however, the overall transactivating effects of these 2 molecules at the genome-wide scale in cancer cells remain largely unknown. Furthermore, whether these molecules independently interact with different binding partners, such that the transcriptomes and the consequent biological effects resulting from SMAD2 and SMAD3 activation are distinct, remains to be resolved. In the present study, we sought to identify whether and how these 2 R-SMADs contribute to TGF-β–promoted cancer metastasis and aggressiveness. Our results demonstrate that SMAD2 and SMAD3 play distinct roles in the progression of NSCLC, with SMAD2 functioning as a mediator of the TGF-β–induced transcriptional program that suppresses metastasis, whereas SMAD3 transactivates a transcriptome that promotes metastasis following TGF-β stimulation. In additional mechanistic studies, we demonstrated that chaperonin containing TCP1 subunit 6A (CCT6A), a previously identified but functionally uncharacterized SMAD2-specific interactive protein (34), suppresses SMAD2 function and promotes TGF-β–induced metastasis in NSCLC. Our findings indicate that in NSCLC, suppression of the SMAD2-mediated transcription program may constitute a prominent mechanism that underlies the prometastatic effects of TGF-β signaling, whereas targeting SMAD3 or the specific SMAD2 inhibitor CCT6A may represent a novel, potentially promising antimetastasis strategy for the management of NSCLC.
Results
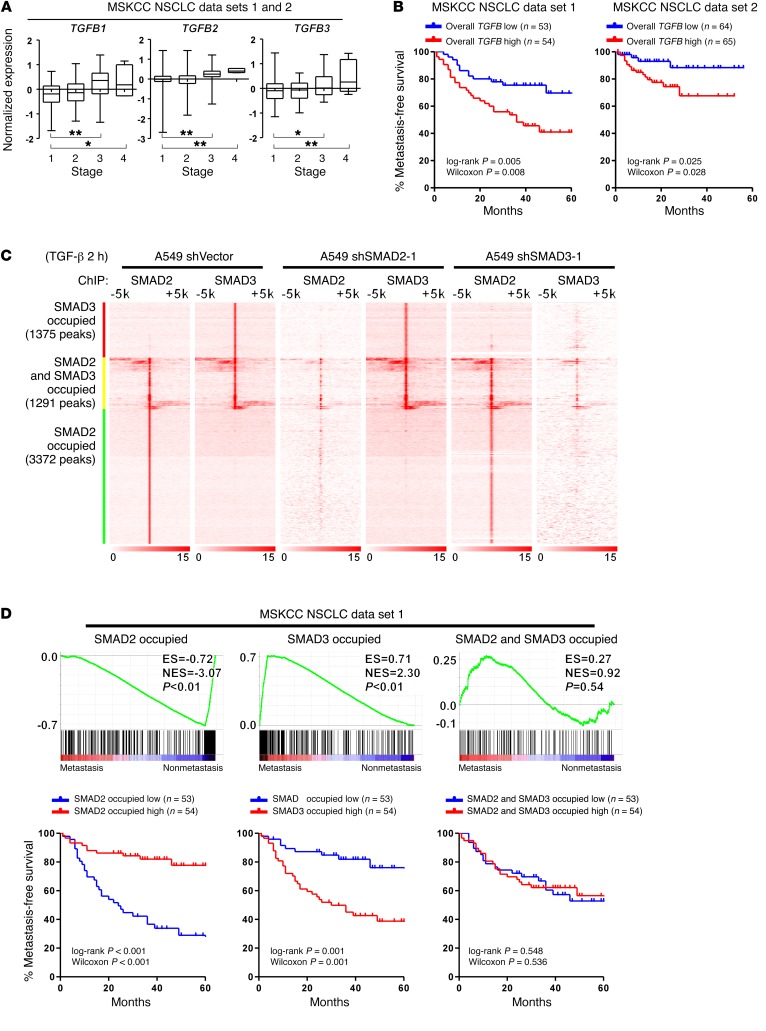
NSCLC metastasis is associated with inactivation of SMAD2-mediated and activation of SMAD3-mediated transcriptional programs.
To understand the correlation between TGF-β and NSCLC metastasis and progression, we initially analyzed a cohort of patients from the Memorial Sloan Kettering Cancer Center (MSKCC) NSCLC data set (35) and determined that TGF-β1, -β2, and -β3 (TGF-β1/2/3) levels were significantly increased in tumor tissues excised from patients with advanced-stage (stages 3 and 4) disease (Figure 1A). Moreover, a high overall TGF-β level (average of normalized TGF-β1/2/3 expression) (36) was associated with an increased metastatic potential in the same cohort of patients, which further supports the previously and widely recognized notion that TGF-β plays a promoting role in NSCLC metastasis (Figure 1B).
Figure 1. NSCLC metastasis is associated with inactivation of SMAD2-mediated and activation of SMAD3-mediated transcriptional programs.
(A) Analysis of publicly available MSKCC NSCLC data sets 1 and 2 indicates that the mRNA expression of TGFB1, TGFB2, and TGFB3 is significantly increased in patients with stage 3 or stage 4 NSCLC. n = 153 cases (stage 1); n = 35 cases (stage 2); n = 43 cases (stage 3); and n = 5 cases (stage 4). Box and whiskers plots represent the mRNA levels. *P < 0.05 and **P < 0.01, by 1-way ANOVA. (B) Kaplan-Meier analysis indicates that when stratifying patients by the median level of overall TGFB (average of TGFB1, TGFB2, and TGFB3), a high level of overall TGFB (average of TGFB1, TGFB2, and TGFB3) is associated with a shorter metastasis-free survival time in patients in the MSKCC NSCLC data sets 1 and 2. (C) Heatmap of the ChIP enrichment signal surrounding SMAD2 and SMAD3 peaks shows the chromosomal distribution of SMAD2 and SMAD3 in the indicated A549 cells. (D) GSEA and Kaplan-Meier analysis indicate that attenuated SMAD2-specific and enhanced SMAD3-specific transcripts are present in NSCLC patients with an increased metastatic potential in association with a shorter metastasis-free survival in the MSKCC NSCLC data set 1, whereas targeted genes shared by both SMAD2 and SMAD3 are not associated with metastasis status. The definition of “high” and “low” expression of each gene set was stratified by the median of the normalized expression levels of each gene in the set. ES, enrichment score; NES, normalized enrichment score.
To identify the downstream molecular events that mediate the prometastatic effect of TGF-β stimulation, we began by investigating whether SMAD2 and SMAD3 mediate the same or potentially different biological effects in TGF-β–promoted NSCLC metastasis. We initially analyzed the genome-wide distribution of SMAD2 and SMAD3 in response to TGF-β treatment in A549 and Calu3 NSCLC cells. Our ChIP-sequencing (ChIP-seq) results indicated that, following the addition of TGF-β to cultured A549 cells, SMAD2 and SMAD3 were recruited to 3,372 and 1,375 distinct enhancer elements, respectively; these 2 SMADs also simultaneously co-occupied 1,291 enhancer elements (Figure 1C and Supplemental Figure 1B; supplemental material available online with this article; https://doi.org/10.1172/JCI90439DS1). Similar recruitment patterns of SMAD2 and SMAD3 were also identified in Calu3 cells, and 87% of the SMAD-binding elements were common in A549 and Calu3 cells (Supplemental Figure 1, A and B). Previous reports have demonstrated competitive binding between SMAD2 and SMAD3 on individual promoters (21, 26). At the genome-wide level, to understand whether silencing of SMAD2 or SMAD3 would affect the distribution of the other factor, we performed ChIP-seq analysis of SMAD2 and SMAD3 in cells with 1 of the 2 R-SMADs silenced (Supplemental Figure 1C). Our results indicated that silencing SMAD2 only reduced the SMAD2 occupancy; it did not disrupt the SMAD3 distribution, and vice versa (Figure 1C and Supplemental Figure 1A). These findings suggest that SMAD2 and SMAD3 are not always recruited to the same sites in response to TGF-β stimulation and may be mutually and independently recruited to most of their corresponding enhancer sites. To further elucidate the positional relationship between the SMAD-binding sites and the transcription start sites (TSSs) on the genome, we compared the profiles of average binding densities between SMADs and H3K4me3 in TGF-β–treated A549 and Calu3 cells. We found that the binding of SMAD2 and SMAD3 on the genome was peaked upstream of the H3K4me3-enriched regions, indicating that SMAD2 and SMAD3 might primarily be recruited to the promoter/enhancer regions of their targeted genes and may be involved in the transcriptional regulation of their downstream genes (Supplemental Figure 1D). Analysis of mRNA expression profiles showed that silencing SMAD2 or SMAD3 significantly attenuated the expression of TGF-β–induced transcripts whose enhancers were distinctly occupied by SMAD2 or SMAD3, respectively, and to a lesser extent attenuated the expression of the target transcripts whose enhancers were shared by both R-SMADs (Supplemental Figure 1E). These findings suggest that SMAD2 or SMAD3 is required for increased transactivation of these distinctly or commonly bound enhancers in response to TGF-β stimulation, and the alteration of the transcriptional profile following silencing of SMAD2 or SMAD3 was mainly attributed to suppression of the respective set of target genes.
We subsequently investigated whether these SMAD target gene sets were associated with NSCLC metastasis. In both the MSKCC NSCLC data sets 1 and 2, we determined that decreases in SMAD2-specific and increases in SMAD3-specific transcripts were present in NSCLC patients with an increased metastatic potential and a shorter metastasis-free survival time (Figure 1D and Supplemental Figure 1F). Moreover, the expression levels of genes commonly transcribed by both SMAD2 and SMAD3 were not significantly correlated with the metastasis status (Figure 1D and Supplemental Figure 1F). These findings suggest that SMAD2-specific transcriptional targets may negatively contribute, whereas SMAD3-specific transcriptional targets positively contribute, to NSCLC metastasis.
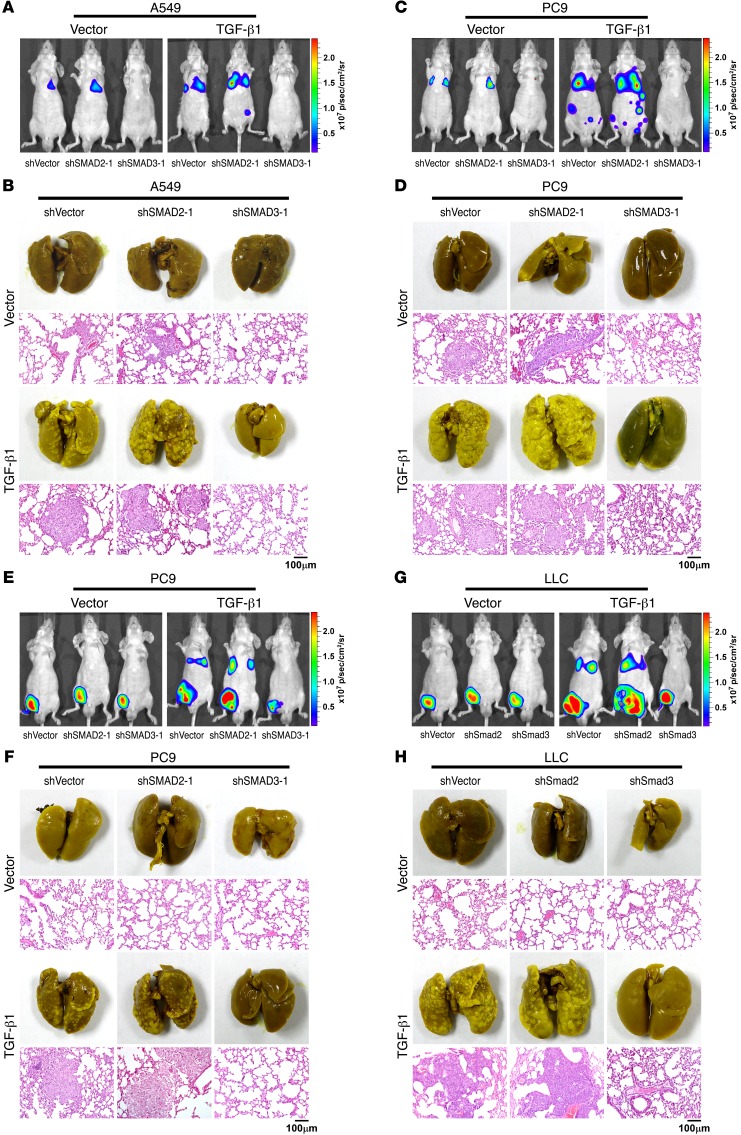
TGF-β–induced metastasis of NSCLC is suppressed by SMAD2 and promoted by SMAD3.
The findings that the suppression of SMAD2-mediated transcripts and the activation of SMAD3-dependent transcripts correlated with a high metastatic potential in NSCLC patients prompted us to investigate whether these 2 R-SMADs play different roles in TGF-β–induced metastasis in vivo. In our subsequent in vivo study, we determined that 30 days after tail-vein injection, the ectopic overexpression of TGF-β1 in A549 cells promoted metastasis, as shown by luciferase live-cell imaging, picric acid staining of metastatic lesions, H&E staining, and immunohistochemical staining, strongly indicating that TGF-β acts as a metastasis promoter in NSCLC cells (Figure 2, A and B; and Supplemental Figure 2, A, C, D, G, and I). Furthermore, silencing of SMAD2 further enhanced, whereas silencing of SMAD3 abrogated, TGF-β–induced metastasis (Figure 2, A and B; and Supplemental Figure 2, A, C, D, G, and I). Moreover, the survival time of A549 cell–injected mice was shortened when TGF-β1 was overexpressed, and the survival time was further shortened when SMAD2 was silenced; however, the survival time was prolonged when SMAD3 was silenced in TGF-β1–overexpressing A549 cells (Supplemental Figure 2, B and H). For PC9 cells, which have been demonstrated to be more metastatic than A549 cells, silencing of SMAD3 also significantly attenuated metastasis, whereas silencing of SMAD2 slightly enhanced TGF-β–induced metastasis and did not further reduce the survival time of the injected mice (Figure 2, C and D; and Supplemental Figure 2, A, B, and E–I). Furthermore, in a spontaneous metastasis model, using highly metastatic human PC9 NSCLC cells and murine Lewis lung carcinoma (LLC) cells, TGF-β promoted both primary tumor growth and the formation of metastases (Figure 2 , E–H, and Supplemental Figure 2, J–L). Consistent with the previously described results, silencing SMAD3 attenuated, whereas silencing SMAD2 slightly enhanced, metastasis and primary tumor growth (Figure 2, E–H, and Supplemental Figure 2, J–L).
Figure 2. TGF-β–induced metastasis of NSCLC is suppressed by SMAD2 and promoted by SMAD3.
(A–D) Mice were intravenously tail-vein injected with the indicated NSCLC cells. Luciferase live-cell imaging (A and C), picric acid staining of metastatic foci, and H&E staining of lung tissue (B and D) indicated that the knockdown of SMAD2 promoted, whereas knockdown of SMAD3 suppressed, the survival and metastasis of NSCLC cells expressing TGF-β. Luciferase-labeled images of live mice at different time points following intravenous tail-vein injection of the indicated NSCLC cells are presented in Supplemental Figure 2A. Representative images shown are from 2 independent experiments, with 6 mice per group in each independent experiment. Scale bars: 100 μm. (E–H) Mice were subcutaneously injected with the indicated NSCLC cells into inguinal folds. Luciferase live-cell imaging (E and G), picric acid staining of metastatic foci, and H&E staining of lung tissue (F and H) indicated that knockdown of SMAD2 promoted, whereas knockdown of SMAD3 suppressed, the metastasis potential of NSCLC cells. Luciferase-labeled images of live mice at different time points following subcutaneous injection of the indicated NSCLC cells into inguinal folds are presented in Supplemental Figure 2J. Representative images are from 2 independent experiments, with 6 mice per group in each independent experiment. Scale bars: 100 μm. p, photons; sr, steradian.
Furthermore, we used an immunocompetent mouse model (C57BL/6 mice), in which mice were tail-vein–injected with LLC cells. Our results showed that on day 50 after injection of shVector-, SMAD2-silenced, or SMAD3-silenced LLC cells, together with or without ectopic expression of TGF-β1, luciferase live imaging and picric acid staining of metastatic lesions showed that TGF-β moderately promoted metastasis and that silencing SMAD2 slightly enhanced the metastasis of LLC cells, whereas silencing SMAD3 diminished the metastasis of LLC cells induced by TGF-β (Supplemental Figure 2, M and N). Moreover, Kaplan-Meier plot analysis showed that the survival time of the mice was shortened when TGF-β was ectopically expressed, but silencing SMAD2 did not further reduce the survival time of the C57BL/6 mice (Supplemental Figure 2O). In contrast, the survival time was significantly (P < 0.01) prolonged when SMAD3 was silenced (Supplemental Figure 2O). The above results obtained from both the nude mice xenografts model and the immunocompetent C57BL/6 mice model further support the notion that SMAD2 suppresses, but SMAD3 enhances, the metastasis promoted by TGF-β in NSCLC.
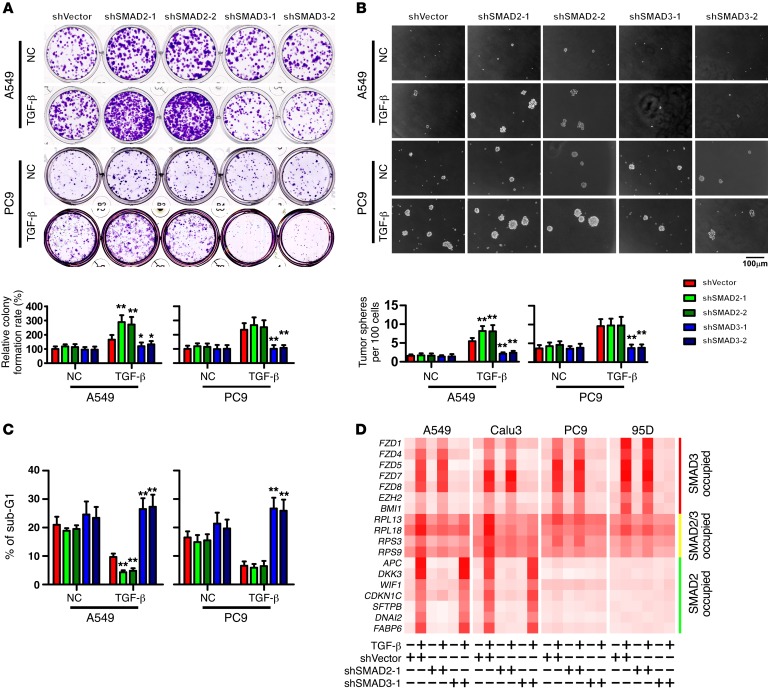
SMAD2 mediates a tumor-suppressive transcriptional program, whereas SMAD3 mediates a pro-cell survival transcriptional program, in NSCLC cells.
To gain a global view of the functions of genes distinctly governed by SMAD2 and SMAD3, a gene ontology (GO) enrichment analysis was performed. As shown in Supplemental Figure 3, A–C, SMAD3 mainly affected the expression of developmental regulators, particularly members of the Frizzled family. In contrast, SMAD2 tended to govern the expression of genes that induce apoptosis and differentiation, including negative regulators of Wnt signaling (Supplemental Figure 3, A–C). Interestingly, genes co-occupied by SMAD2 and SMAD3 were mainly involved in protein synthesis, particularly ribosome proteins and elongation factors (Supplemental Figure 3, A–C). These findings indicate that TGF-β signaling through SMAD2 or SMAD3 may attenuate or enhance multiple pro-cell survival pathways, respectively, including the Wnt pathway, which is closely correlated with NSCLC metastasis through the promotion of colony outgrowth (37).
To understand whether the cellular functions of SMAD2 and SMAD3 indicated by GO enrichment and in vivo assays are different, we analyzed the in vitro cellular effects of SMAD2 or SMAD3 depletion in both A549 and PC9 cells. We found that following TGF-β treatment, under standard and cell culture conditions, silencing SMAD3 suppressed colony and tumor sphere formation (Figure 3, A and B; and Supplemental Figure 3D) and reduced the proportion of side-population cells (Supplemental Figure 3E). Furthermore, when cells were detached from their substratum and cultured in suspension, silencing SMAD3 significantly increased the cellular sensitivity to anoikis (Figure 3C and Supplemental Figure 3F). These findings suggest that SMAD3 is a promoter of cell survival, particularly under stress conditions, such as the loss of cellular contact. Furthermore, following TGF-β stimulation, silencing of SMAD2 enhanced cell survival only in A549 cells and not in highly metastatic PC9 cells (Figure 3, A–C; and Supplemental Figure 3, D–F), which suggests that in highly metastatic NSCLC cells, SMAD2 may function differently compared with non- or low-metastatic cells.
Figure 3. SMAD2 mediates a tumor-suppressive transcriptional program, whereas SMAD3 mediates a pro-cell survival transcriptional program, in NSCLC cells.
(A–C) Analyses of colony formation in an adherent culture (A), tumor sphere formation (B), and sub-G1 DNA content of detached cell (C) were performed in the indicated cells. Representative images were derived from 3 independent experiments. Error bars represent the mean ± SD of 3 independent experiments. *P < 0.05 and **P < 0.01, by ANOVA with Dunnett’s t test. Scale bar: 100 μm. (D) qPCR analysis of the expression levels of target genes distinctly specific for, or commonly shared by, SMAD2 and SMAD3 in the indicated cells. Heatmaps represent the mRNA levels. NC, the vehicle control of TGF-β solution.
In support of this notion, following TGF-β stimulation, the expression levels of genes regulated by SMAD3 and co-occupied by SMAD2 and SMAD3 were upregulated, whereas silencing of SMAD3 reversed the alteration of these expression levels in both low-metastatic and high-metastatic NSCLC cells. However, treatment with TGF-β or silencing of SMAD2 expression in highly metastatic PC9 or 95D cells did not change the SMAD2-mediated transcriptional pattern, in contrast to the SMAD2-associated expression profile changes induced by TGF-β treatment and SMAD2 silencing in low-metastatic NSCLC cells (Figure 3D and Supplemental Figure 3G). Notably, in this context, further Western blot analysis indicated that the expression levels of SMAD2 and SMAD3 as well as their phosphorylation statuses following TGF-β stimulation were not significantly different among primary cultured lung epithelial cells (LE-1 and LE-2), immortalized bronchial epithelial cells (Beas2B), or low- (A549 and Calu3) or high-metastatic (PC9 and 95D) NSCLC cells (Supplemental Figure 3H). These findings suggest that in highly metastatic NSCLC cells with intact TGF-β signaling, unknown molecules yet to be identified may act to interfere with the function of SMAD2, particularly following its activation by phosphorylation, such that the suppressive effect of SMAD2 on cellular survival is inhibited.
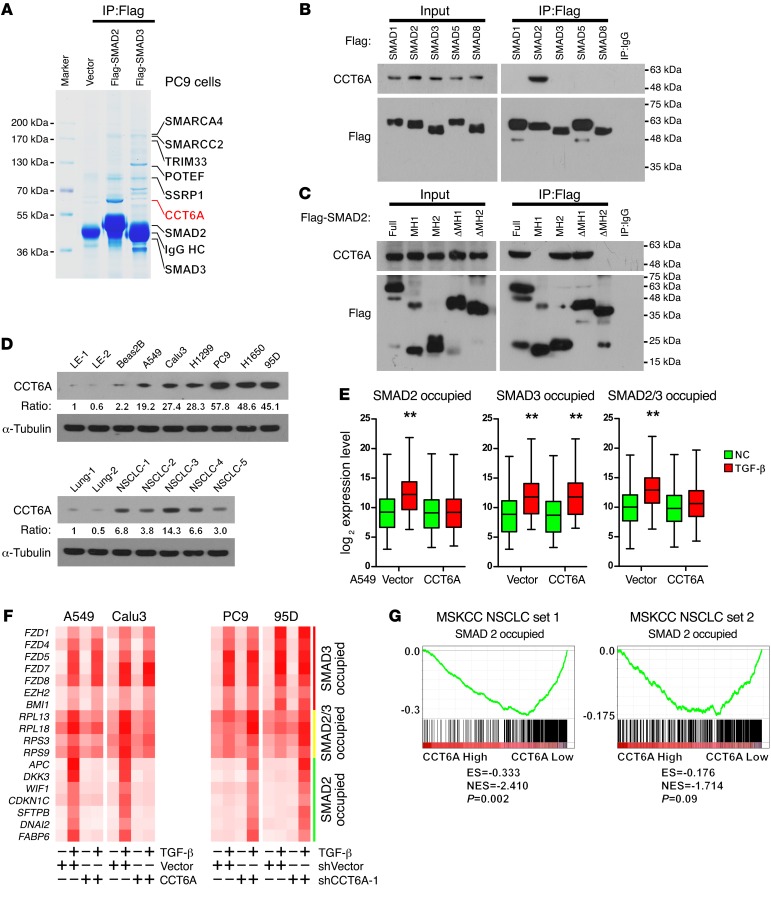
CCT6A specifically interacts with SMAD2 and suppresses the SMAD2-mediated transcriptional program.
The biological effects of R-SMADs may be modulated by their binding partners; thus, we were prompted to identify proteins that interact with SMAD2 in highly metastatic NSCLC cells. To this end, we performed an immunoprecipitation assay using ectopically expressed Flag-tagged SMAD2 and SMAD3 in PC9 cells. In the subsequent mass spectroscopic analysis, several previously reported and previously unknown R-SMAD–interactive partners were identified. Among these proteins, CCT6A, a previously reported but functionally uncharacterized SMAD2-interactive protein, was highly enriched in SMAD2 precipitates (Figure 4A and Supplemental Figure 4A). A further immunoprecipitation assay demonstrated that endogenous CCT6A interacted with SMAD2 specifically, but not with other R-SMADs, i.e., SMADs 1, 3, 5, and 8 (Figure 4B and Supplemental Figure 4B). Moreover, an immunoprecipitation assay using truncated SMAD2 fragments indicated that CCT6A specifically interacted with the MH2 domain of SMAD2 (Figure 4C). Western blot detection of CCT6A in a series of NSCLC cell lines, using LE-1, LE-2, and Beas2B cells as controls, indicated that CCT6A was significantly overexpressed in NSCLC cells, particularly in those with an increased metastatic ability, e.g., PC9, H1650, and 95D cells (Figure 4D, upper panel). Accordingly, Western blot analysis of CCT6A in NSCLC tissue specimens, using benign lung tissues as controls, also demonstrated that CCT6A was highly expressed in patient-derived NSCLC tissue (Figure 4D, lower panel).
Figure 4. CCT6A specifically interacts with SMAD2 and suppresses the SMAD2-mediated transcriptional program.
(A) Immunoprecipitation and subsequent mass spectroscopic analysis identified CCT6A as a highly specific SMAD2-interactive protein in PC9 cells. Representative image was derived from 3 independent experiments. (B) Immunoprecipitation in 293T cells that expressed Flag-tagged SMADs 1, 2, 3, 5, and 8 and HA-tagged CCT6A indicated that CCT6A specifically interacted with SMAD2. Representative blot was derived from 3 independent experiments. (C) Immunoprecipitation of truncated SMAD2 constructs indicated that CCT6A specifically interacted with the MH2 domain of SMAD2. Representative blot was derived from 3 independent experiments. (D) Western blot analysis of CCT6A in NSCLC cell lines using 2 primary lung epithelial cell lines (LE-1 and LE-2) and an immortalized lung epithelial cell line (Beas2B) as a control showed that CCT6A was highly expressed in NSCLC cell lines, particularly in highly metastatic lines (PC9, H1650, and 95D) (upper panel). Western blot analysis of CCT6A in NSCLC tissue using 2 normal lung tissue specimens as controls indicated that CCT6A was highly expressed in patient-derived NSCLC tumors (lower panel). Representative blots were derived from 3 independent experiments. (E) Microarray-based transcription profiling demonstrated that CCT6A overexpression significantly attenuated the expression of SMAD2-specific target genes, as well as target genes shared by SMAD2 and SMAD3 in A549 cells. Box and whiskers plots represent the mRNA levels. **P < 0.01, by Student’s t test. (F) qPCR analysis of the expression level of SMAD2- or SMAD3-specific and shared target genes in the indicated cells. Heatmaps represent the mRNA levels. (G) GSEA analysis indicated that the CCT6A levels were inversely associated with the expression levels of SMAD2-specific targets. The defined “high” and “low” expression levels of CCT6A were stratified by the median expression level.
To understand whether CCT6A affects the SMAD2-mediated transcriptional program, microarray assays were used to profile the mRNA expression pattern. In A549 cells, CCT6A overexpression significantly attenuated the transcription of SMAD2-specific targets; however, it did not affect the expression of SMAD3-specific targets following the induction of TGF-β (Figure 4E). Quantitative PCR (qPCR) assessment of representative SMAD2- or SMAD3-specific or shared target genes further confirmed that CCT6A overexpression in A549 and Calu3 cells abrogated the SMAD2-specific transcriptome in response to TGF-β stimulation, whereas in PC9 and 95D cells, silencing of CCT6A relieved the blockade of these genes (Figure 4F and Supplemental Figure 4C). These findings support the notion that CCT6A expression is negatively correlated with the expression of SMAD2-specific transcripts, as further demonstrated by data presented in the MSKCC NSCLC data sets (Figure 4G). Together, these findings indicate that CCT6A acts as a specific negative regulator of SMAD2 function in NSCLC patients.
CCT6A promotes NSCLC cell survival and blocks SMAD2-SMAD4 interaction.
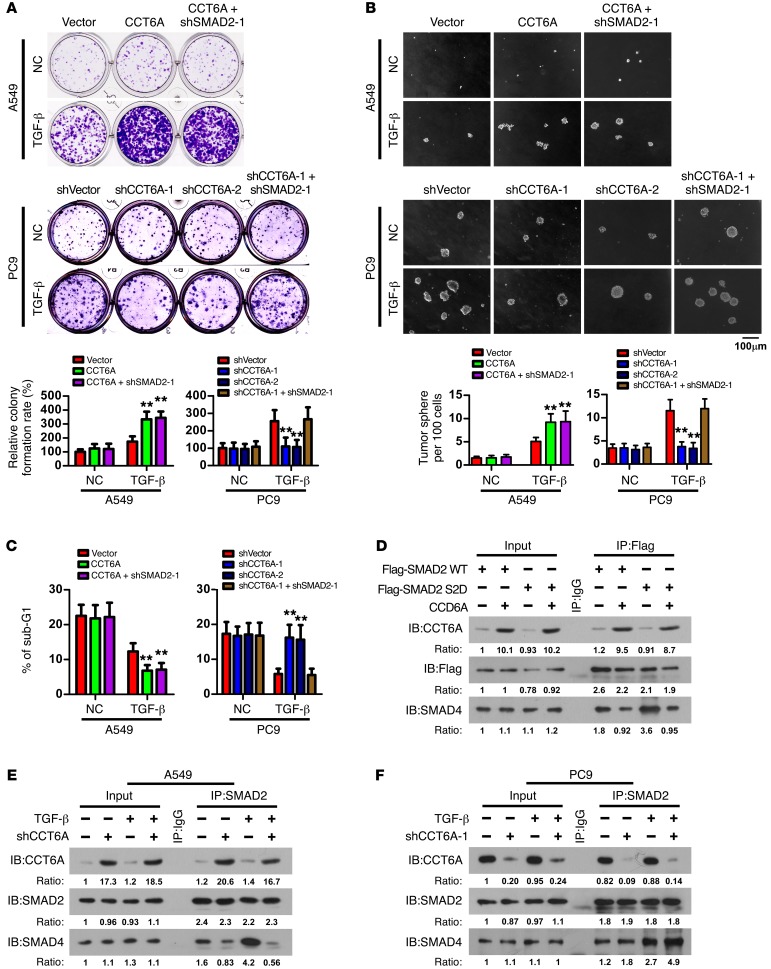
We subsequently aimed to study the functions of CCT6A in NSCLC cell survival. Further analyses indicated that, following TGF-β stimulation, CCT6A overexpression promoted colony and tumor sphere formation (Figure 5, A and B; and Supplemental Figure 5A), increased the side population (Supplemental Figure 5B), and reduced sensitivity to anoikis (Figure 5C and Supplemental Figure 5C) in A549 cells, whereas silencing CCT6A had opposite effects in PC9 cells (Figure 5, A–C, and Supplemental Figure 5, A–C). Furthermore, silencing SMAD2 in A549 NSCLC cells engineered to ectopically overexpress CCT6A did not further enhance CCT6A-promoted cell survival, and in PC9 cells, silencing SMAD2 markedly reversed the effects induced by CCT6A silencing (Figure 5, A–C, and Supplemental Figure 5, A–C). This line of evidence further supports the roles of CCT6A as an essential promoter of the TGF-β–induced aggressiveness of NSCLC cells and as a negative regulator of SMAD2 functions.
Figure 5. CCT6A promotes NSCLC cell survival and blocks SMAD2-SMAD4 interaction.
(A–C) Analyses of colony formation in an adherent culture (A), tumor sphere formation (B), and sub-G1 DNA content of detached cells (C) for the indicated cells. Representative images were derived from 3 independent experiments. Error bars represent the mean ± SD of 3 independent experiments. **P < 0.01, by ANOVA with Dunnett’s t test. Scale bar: 100 μm. (D) Immunoprecipitation analysis indicated that CCT6A attenuated the interaction of SMAD4 with both the WT and the phosphorylation mimicry mutant SMAD2 (S2D). Representative blots were derived from 3 independent experiments. (E and F) Immunoprecipitation analysis showed that in A549 cells, CCT6A overexpression attenuated SMAD2-SMAD4 interaction (E), whereas in PC9 cells, silencing of CCT6A enhanced SMAD2-SMAD4 interaction (F). Representative blots (IB) were derived from 3 independent experiments.
To further elucidate the mechanism by which CCT6A specifically blocks SMAD2 function, we noted that in Beas2B and A549 cells, the total protein and phosphorylation levels of SMAD2 and SMAD3 were barely changed after ectopic expression of CCT6A, with or without TGF-β treatment (Supplemental Figure 5D). Furthermore, an immunofluorescence assay showed that SMAD2 and CCT6A were always colocalized in the cytoplasm, even after treatment with TGF-β (Supplemental Figure 5E), indicating that CCT6A may block the nuclear localization of SMAD2 and further suppress the function of SMAD2 as a transcriptional regulator. In such a context, our data showed that CCT6A specifically interacted with the MH2 domain of SMAD2 (Figure 4C), which has been previously demonstrated to interact with SMAD4 and form a functional R-SMAD–SMAD4 complex in response to TGF-β stimulation (38, 39); thus, we aimed to determine whether CCT6A affected the interaction between SMAD4 and SMAD2. As shown in Figure 5D, CCT6A interacted with both WT and the phosphorylation mimicry mutant (40) of SMAD2 (S2D), which has high binding affinity for SMAD4. Importantly, when CCT6A was overexpressed, the interaction between SMAD2 (S2D) and SMAD4 was drastically abrogated (Figure 5D); this result was confirmed by the finding that in A549 cells (with a low endogenous CCT6A level), CCT6A overexpression markedly decreased SMAD2-associated SMAD4 following treatment with TGF-β (Figure 5E). In contrast, in PC9 cells, which express a high level of CCT6A, silencing of CCT6A increased SMAD2-associated SMAD4 in response to TGF-β stimulation (Figure 5F), which indicates that CCT6A may block SMAD2-induced transcription by blocking the formation of a functional SMAD2-SMAD4 complex.
CCT6A mediates TGF-β–promoted metastasis of NSCLC cells.
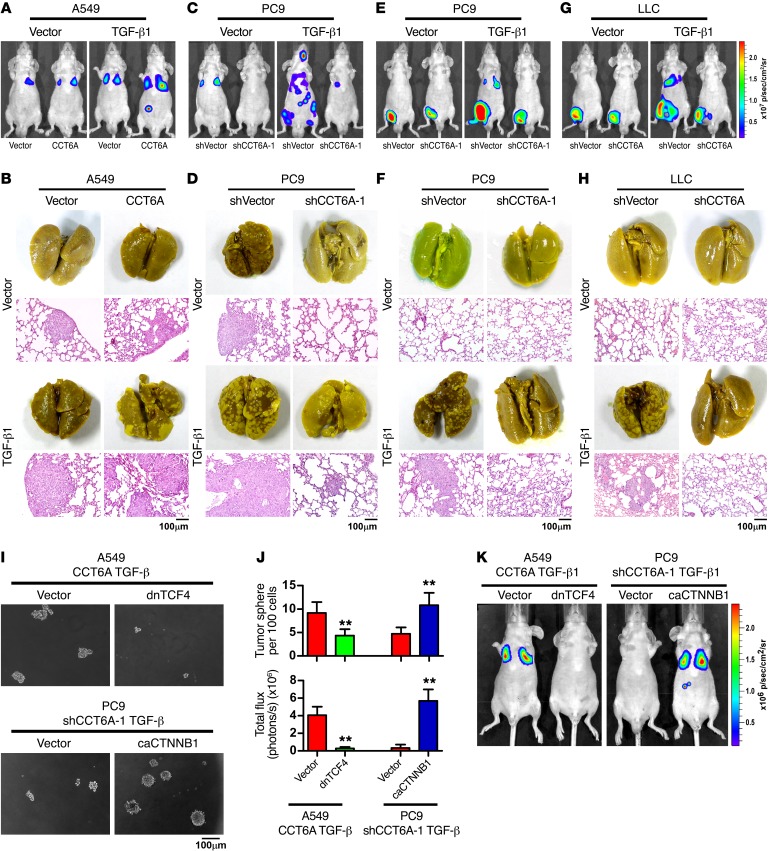
We subsequently investigated whether CCT6A is essential for TGF-β–induced metastasis in vivo. The animal studies showed that in A549 cells, CCT6A overexpression promoted, whereas silencing of CCT6A in PC9 cells inhibited, TGF-β–induced metastasis (Figure 6, A–D, and Supplemental Figure 6, A, E, G–I, and K). Accordingly, the survival time of mice injected with A549 cells overexpressing CCT6A was significantly (P < 0.01) shortened, whereas injection with CCT6A-silenced PC9 cells prolonged the survival of tumor-bearing mice (Supplemental Figure 6, B, F, and J). Similarly, in a spontaneous metastasis model, silencing of CCT6A also significantly inhibited the dissemination of tumor cells and improved animal survival (Figure 6, E–H, and Supplemental Figure 6, L–N). In addition, in an immunocompetent mouse model (C57BL/6 mice), on day 50 after tail-vein injection of shVector-, CCT6A silenced–, or CCT6A- and SMAD2-cosilenced LLC cells, together with or without ectopic expression of TGF-β1, luciferase live imaging, picric acid staining of metastatic lesions, and Kaplan-Meier plot analysis of mouse survival time demonstrated that silencing CCT6A inhibited TGF-β–induced metastasis and prolonged the survival of the tumor cell–injected mice, whereas silencing SMAD2 in CCT6A–knocked-down LLC cells could significantly (P < 0.01) reverse the metastasis-inhibiting effect of CCT6A silencing (Supplemental Figure 6, O–Q).
Figure 6. CCT6A mediates TGF-β–promoted metastasis of NSCLC cells.
(A–D) Mice were intravenously injected via tail vein with the indicated NSCLC cells. Luciferase live-cell imaging (A and C), picric acid staining of metastatic foci, and H&E staining of lung tissue (B and D) revealed that CCT6A promoted survival and metastasis of NSCLC cells with TGF-β expression. Luciferase-labeled images of live mice at different time points following intravenous tail-vein injection of the indicated NSCLC cells are presented in Supplemental Figure 6A. Representative images were from 2 independent experiments, with 6 mice per group in each independent experiment. Scale bars: 100 μm. (E–H) Mice were subcutaneously injected into inguinal folds with the indicated NSCLC cells. Luciferase live-cell imaging (E and G), picric acid staining of metastatic foci, and H&E staining of lung tissue (F and H) showed that silencing CCT6A diminished the metastasis potential of NSCLC cells. Luciferase-labeled images of live mice at different time points following subcutaneous injection into inguinal folds of the indicated NSCLC cells are presented in Supplemental Figure 6L. Representative images were from 2 independent experiments, with 6 mice per group in each independent experiment. Scale bars: 100 μm. (I–K) Tumor sphere formation assays with the indicated NSCLC cells (I and J) and luciferase live-cell images of mice intravenously injected via the tail vein with a low dose (5 × 104) of NSCLC cells (n = 6 per group) on day 30 (J and K) suggested that CCT6A promoted the survival and metastasis of NSCLC cells in a TGF-β signaling–dependent manner. Error bars represent the mean ± SD. **P < 0.01, by Student’s t test.
Furthermore, silencing SMAD2 in A549 cells ectopically overexpressing CCT6A could not further enhance CCT6A-promoted tumor cell metastasis, and in CCT6A-silenced PC9 cells, silencing SMAD2 markedly promoted TGF-β–induced cancer cell metastasis and shortened the survival time of mice transplanted with PC9 cells, with CCT6A and SMAD2 jointly silenced (Supplemental Figure 6, C–K). Meanwhile, comparison of the biological outcomes between knockdown of SMAD2 alone (Figures 2 and 3) and together with overexpression or knockdown of CCT6A (Figure 5 and 6) showed that when SMAD2 was silenced, ectopically overexpressing or silencing CCT6A had no effect on cell survival or metastasis of the NSCLC cells, further suggesting that CCT6A is a negative regulator of SMAD2 and that the metastasis-promoting function of CCT6A is SMAD2 dependent (Supplemental Figure 6, R and S).
The previously described results demonstrated that several key factors of Wnt/β-catenin signaling were transcriptionally regulated by SMAD2 and SMAD3 following TGF-β stimulation. Consistently, we found that blockade of the Wnt/β-catenin signaling pathway with dominant-negative transcription factor 4 (TCF4) attenuated CCT6A-enhanced cancer cell survival and metastasis, and the constitutively active form of β-catenin (33Y mutant) reversed the cell death phenotype induced by silencing of CCT6A in vitro and promoted TGF-β–stimulated metastasis in vivo (Figure 6, I–K, and Supplemental Figure 6, T and U). Taken together, these findings support the notion that CCT6A sustains the oncogenic arm of TGF-β signaling and acts as a strong promoter of TGF-β–induced metastasis in vivo.
High CCT6A levels are associated with inhibition of the SMAD2-mediated transcriptional program in clinical NSCLC specimens.
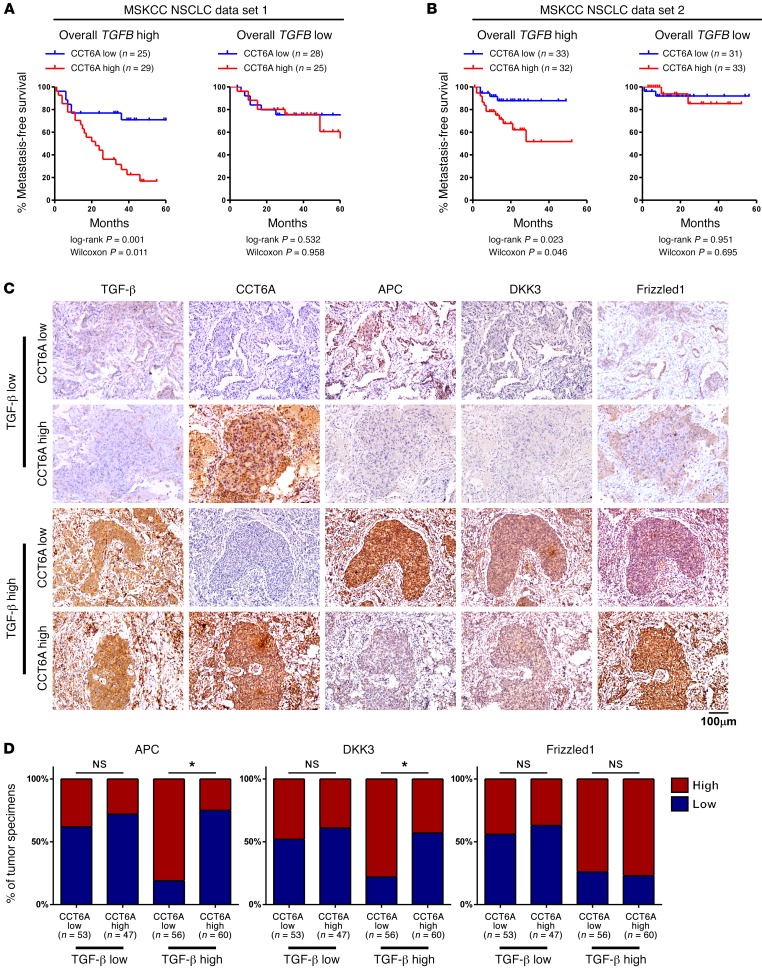
To understand whether CCT6A expression is associated with TGF-β–promoted metastasis in NSCLC patients, we analyzed CCT6A levels in the MSKCC NSCLC data sets 1 and 2. Consistent with the previously identified prometastatic function of CCT6A in TGF-β–stimulated tumor cells, we determined that CCT6A levels were correlated with a high metastasis potential in patients with high TGF-β levels, whereas in patients with low TGF-β levels, CCT6A expression was not associated with NSCLC metastasis (Figure 7, A and B). To further address whether CCT6A levels also correlate with the expression of TGF-β signaling transcriptional targets in NSCLC patients, we performed immunohistochemical staining of TGF-β, CCT6A, adenomatous polyposis coli (APC), dickkopf WNT signaling pathway inhibitor 3 (DKK3), and Frizzled1 in our collection of NSCLC specimens. In this cohort of patients, as demonstrated by imaging and statistical analyses across all 216 specimens, CCT6A expression was negatively associated with the expression of APC and DKK3, two transcriptional targets regulated by SMAD2, only in the specimens with high TGF-β levels. In contrast, the association between CCT6A levels and these TGF-β–regulated genes was not significant in the specimens with low TGF-β levels. Notably, Frizzled1 expression, which is regulated by SMAD3, did not correlate with CCT6A expression, regardless of TGF-β levels (Figure 7, C and D). This line of evidence supports the notion that high CCT6A expression levels are key to the metastasis-associated suppression of SMAD2 function and the high metastatic potential in NSCLC patients.
Figure 7. High CCT6A levels are associated with inhibition of the SMAD2-mediated transcriptional program in clinical NSCLC specimens.
(A and B) Kaplan-Meier survival analysis of data collected from MSKCC NSCLC data sets 1 and 2 indicated that CCT6A expression levels were positively correlated with metastasis in patients with high TGF-β expression levels. (C and D) Representative images (C) of immunohistochemical staining for TGF-β, CCT6A, APC, DKK3, and Frizzled1 and statistical analysis across 216 NSCLC specimens (D) show that CCT6A expression was negatively associated with expression of APC and DKK3 only in specimens with high TGF-β levels and that Frizzled1 expression was not correlated with CCT6A expression, regardless of TGF-β levels. “High” and “low” expression levels of each protein were stratified by the median optical density (OD) of staining in all specimens. Scale bar: 100 μm. *P < 0.05, by 1-way ANOVA.
Discussion
SMAD2 mediates a transcriptional program distinct from SMAD3 and plays a tumor-suppressive role in NSCLC.
Our current study using multiple NSCLC cell lines, animal models, and clinical specimens indicates that in NSCLC, the suppression of SMAD2, with the retention of SMAD3 function, switches TGF-β–induced transcriptional responses toward a prometastatic state. Notably, the activation of TGF-β signaling has been demonstrated in NSCLC tumors and cell lines, and a malignant phenotype of cancer cells, including cellular migration and invasion, angiogenesis, stem cell–like properties, and metastasis, has been linked to aberrant activation of TGF-β signaling in cancers (41, 42). Following stimulation with TGF-β, SMAD2 and SMAD3, two of the most important R-SMADs in the canonical TGF-β signaling pathway, are phosphorylated by TGF-β receptor 1 and interact with SMAD4 to form a molecular complex, which subsequently translocates to the nucleus and regulates the transcription of downstream target genes (5, 6, 10, 11). This cascade of TGF-β signaling has been identified in various types of cancers; however, the biological functions and target genes of SMAD2 and/or SMAD3 in NSCLC have remained largely unknown, and it is unclear whether SMAD2 and SMAD3 mediate similar or distinct biological functions in NSCLC cells. In the current study, the use of antibodies that specifically recognized SMAD2 or SMAD3 in ChIP-seq analyses led to the findings that substantial numbers of SMAD2- and SMAD3-binding sites are distinct in NSCLC cells and that the targeted genes of these 2 R-SMADs were different; however, a small number (~20%) of these binding sites are shared by SMAD2 and SMAD3. Our further analyses of target genes using whole-genome strategies indicated that SMAD3 mainly controls the transcription of developmental regulators, whereas SMAD2 target genes predominately govern apoptosis and differentiation; the genes jointly regulated by both SMAD2 and SMAD3 are mainly housekeeping genes involved in protein synthesis. In strong agreement with the results of previously described studies, further in vivo and in vitro experimental data demonstrated that silencing of SMAD2 promoted, while silencing of SMAD3 suppressed, the survival of NSCLC cells and metastasis following TGF-β stimulation. These findings suggest that SMAD2 and SMAD3 have opposite functions, with SMAD2 predominately acting as a tumor suppressor in the cancer cell. In this context, SMAD2 may be responsible, or in part responsible, for the tumor-suppressive effect of TGF-β signaling; however, the underlying mechanism has remained largely uncharacterized (21, 22, 27). Thus, the current study not only uncovered a novel mechanism that underlies the seemingly contradictory dual roles of TGF-β signaling as both an antitumorigenic and prometastatic pathway but also identified that 2 R-SMADs, SMAD2 and SMAD3, likely constitute a core switch that drives TGF-β signaling from its tumor-suppressive role toward its prometastatic function. Therefore, the practical development of antimetastatic strategies against NSCLC progression should be directed toward strategies that effectively inhibit SMAD3 and its governed transcriptional program and functionally maintain the transcriptional activity of SMAD2.
Abrogation of the SMAD2-mediated tumor-suppressive arm of TGF-β signaling during cancer progression.
In light of the finding that the SMAD2-SMAD3 switch plays a key role in the transition of TGF-β signaling from a tumor-suppressive to a metastasis-promoting factor, as previously discussed, it is of substantial interest to elucidate how the SMAD2-triggered tumor-suppressive transcriptional program is inhibited while the transactivating function of oncogenic SMAD3 remains active. Notably, in colon and pancreatic cancers, genetic alterations in SMAD2 and SMAD4 gene loci, such as genomic deletions, inactivating mutations, and heterozygosity, are frequently detected and have been demonstrated to abrogate SMAD2-dependent TGF-β signaling (28, 43, 44). However, in tumors with an intact TGF-β signaling pathway, including NSCLC and breast cancer, genomic alterations of the core components in the TGF-β signaling cascade have been detected in only 2% to 10% of patients (28), which strongly indicates that other endogenous factors highly expressed in cancer cells may contribute to inactivation of the SMAD2-mediated arm of TGF-β signaling. Moreover, how SMAD2-mediated transcription is overcome so that the tumor-suppressive role of TGF-β is blocked and the metastasis and progression of NSCLC may subsequently proceed following stimulation by TGF-β remains unknown. CCT6A is a subunit of chaperonin containing TCP1 complex, and it has been reported to be functionally related with immortalization and tumorigenicity of human mesenchymal stem cells and MMP3-dependent granule cell migration in neurite outgrowth and neuronal migration (45, 46). The expression levels of CCT6A had been found to be upregulated in various tumor types, including NSCLC, glioma, melanoma, colon cancer, and testicular cancer, and such an upregulation might be predominantly due to amplification of chromosomal region 7p11.2 (47–51). Nonetheless, thus far the functional significance of CCT6A in cancer development remains unclear. Our findings that CCT6A is overexpressed in NSCLC and that as a SMAD2-binding protein, CCT6A plays an essential role in diminishing the transactivating function of SMAD2, identify what we believe to be a novel mechanism to explain the abrogation of the SMAD2-mediated tumor-suppressive effect of TGF-β. Notably, we demonstrated that in NSCLC, CCT6A acts to inhibit SMAD2-SMAD4 interaction and interrupt the biological function of the SMAD2-SMAD4 complex in regulating the transcription of downstream target genes and that the suppression of CCT6A leads to diminished TGF-β–induced metastasis, which provides a mode of action to explain the cancellation of the tumor-suppressive function of SMAD2 in cancer cells, such as most NSCLC cells, that possess intact SMAD2-dependent TGF-β signaling. Interestingly, the amino acid sequence of the MH2 domain of SMAD2, which we demonstrated specifically interacts with CCT6A, is fairly similar to the MH2 domain of SMAD3. Therefore, as our data clearly demonstrate that CCT6A binds only with SMAD2 and not with SMAD3, it is of substantial interest to further elucidate whether the differential specificity of CCT6A-SMAD2 interaction is determined by the structural difference between SMAD2 and SMAD3 or, alternatively, by other protein-binding partners. Studies that aim to understand this question are currently underway in our laboratory.
CCT6A and SMAD3 may represent targets for the suppression of TGF-β–promoted metastasis in NSCLC.
Over decades, substantial efforts have been made to develop agents that target the TGF-β pathway as an antimetastasis strategy. Agents of this nature include anti–TGF-β–neutralizing antibodies, such as 1D11, which have been demonstrated to inhibit metastasis in breast cancer models derived from the 4T1 (52) and MDA-MB-231 breast cancer cell lines (53); the TGFBR1 inhibitors SB-431542, Ki26894, and LY2157299; and the TGFBR1/2 dual inhibitor LY2109761, which was shown to suppress metastasis in breast, colon, and pancreatic cancers in experimental animal models (36, 54–58). The antimetastatic properties of these agents have been demonstrated in various experimental settings; however, their effects on the overall survival of tumor-bearing animals are highly variable and appear to be context dependent. Highly variable responsiveness in a clinical trial with the TGF-β–neutralizing antibody GC-1008 further highlighted the challenges of a TGF-β–targeted strategy in humans (59). These studies warn that targeting the TGF-β pathway by inhibiting upstream signaling components, such as ligands and receptors, may not achieve effective blockade of the prometastatic arm of TGF-β signaling. Our current finding in NSCLC that TGF-β–mediated metastasis and tumor aggressiveness rely on the specific suppression of SMAD2, with SMAD3 remaining functionally intact, provides insight in this area. According to our data, silencing SMAD3 or the specific SMAD2 blocker CCT6A resulted in efficient suppression of metastasis in vivo and significantly prolonged the survival of tumor-bearing mice, which thus implicates a new potential strategy for specifically targeting the tumor-promoting branch of TGF-β signaling in cancer. In this context, it would be of substantial interest to determine whether optimal and potent antimetastatic efficacies may be achieved by combining anti-CCT6A or anti-SMAD3 agents with TGF-β inhibitors.
Methods
Tissue specimens and ethical approval.
A total of 216 paraffin-embedded, archived NSCLC specimens that had been clinically and histopathologically diagnosed at Sun Yat-Sen Memorial Hospital and Sun Yat-Sen University Cancer Center were subjected to immunostaining.
ChIP-seq analysis.
A total of 5 × 107 A549 cells were used for each ChIP assay. The ChIP procedure was performed according to a previously described protocol (60), using 10 μg anti-SMAD2, -SMAD3, and -H3K4me3 antibodies (Supplemental Table 2). Enriched DNA fragments were subjected to library preparation and next-generation sequencing with Annoroad Gene Technology using a HiSeq 2000 (Illumina). Short reads were mapped to the hg19 reference genome using Bowtie2 (61), and ChIP peaks were called using model-based analysis of ChIP-seq 14 (MACS 14), with the input sample as the control (62). Enrichment heatmaps that surrounded the ChIP peaks were generated using seqMINER (63), and signal plotting of individual genes was generated using the Integrated Genome Viewer (64). The raw sequencing data from the ChIP-seq analysis performed in the current study can be downloaded from the NCBI’s Sequence Read Archive (SRA) using accession numbers SR692803 and SRS976037.
Tumor xenografts and metastasis models.
A tail-vein injection model for metastasis was generated by injecting 2 × 106 A549 or 1 × 106 PC9 cells into the tail veins of nude mice. Spontaneous metastasis models were generated by subcutaneously injecting 1 × 106 PC9 or 5 × 105 LLC cells into the inguinal folds of nude mice. Mice that carried tumor xenografts were monitored using the IVIS Spectrum In Vivo Imaging System (PerkinElmer). Image calibration and visualization were performed using Living Image 4.2 software (PerkinElmer).
Immunoprecipitation and mass spectrometric analysis.
PC9 cells (3 × 107), transfected with Flag-tagged SMAD2, SMAD3, or pCDNA3 vector, were lysed with lysis buffer (150 mM NaCl, 10 mM HEPES, pH 7.4, and 1% Nonidet P-40) and incubated with anti-Flag affinity agarose (Sigma-Aldrich) overnight at 4°C. Beads that contained affinity-bound proteins were washed 6 times with immunoprecipitation wash buffer (150 mM NaCl, 10 mM HEPES, pH 7.4, and 0.1% Nonidet P-40). Following protein separation by SDS-PAGE and Coomassie blue staining, the SMAD2/3-interacting bands were subjected to mass spectrometric peptide sequence analysis.
Microarray assay and data analysis.
Total RNA isolated from A549 cells with SMAD2 or SMAD3 silenced, and the corresponding vector control cells were subjected to microarray analysis by the Shanghai Biochip Corporation using the Agilent Technologies 4 × 44k Human Genome Array Expression quantification data and normalized without median shift. The subsequent GO enrichment analysis was performed using GeneSpring GX (Agilent Technologies). Gene set enrichment analysis (GSEA) was performed using GSEA 2.09 (65). Expression profile data from microarray analysis performed in the current study can be downloaded from the NCBI’s Gene Expression Omnibus (GEO)database (GEO GSE61132).
The methods of cell line and primary cell culture, colony formation assay, three-dimension spheroid invasion assay, sphere formation assays, flow cytometric analysis, anoikis assay, RNA extraction, reverse transcription (RT), qPCR, plasmid construction and immunohistochemistry (IHC) are described in the Supplemental Methods, as well as Supplemental Table 1.
Statistics.
All statistical analyses except the microarray data were performed using the PASW Statistics 18 (SPSS Inc.) software package. Comparisons between any 2 sample groups were performed using a Student’s t test, while analyses comparing multiple treatments with a control group were performed using ANOVA with Dunnett’s t test. Data are presented as the mean ± SD. A P value of less than 0.05 was considered statistically significant.
Study approval.
All experimental procedures and use of NSCLC donors’ samples were approved by the IACUC of Sun Yat-sen University. Donors provided prior written informed consent.
Author contributions
ZY, HT, and ML designed the study. ZY, HT, YL, RL, WL, and SW performed the majority of experiments and analyzed data. HZZ provided patients’ samples and analyzed clinical data. JW, LL, JS, and XZ performed several experiments. HT and ZY wrote the manuscript. ML, JL, JC, and HG edited the manuscript.
Supplementary Material
Acknowledgments
This work was supported by the Natural Science Foundation of China (81330058, 81325013, 91529301, and 81490752); the Natural Science Foundation of Guangdong Province (2014A030313008 and 2016B030301002); and Guangdong Natural Science Funds for Distinguished Young Scholar (2014A030306023).
Footnotes
ZY’s present address is: Human Biology Division, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA.
YL’s present address is: Department of Immunobiology, Jinan University, Guangzhou, China.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2017;127(5):1725–1740.https://doi.org/10.1172/JCI90439.
Contributor Information
Zhe Ying, Email: zheying87@gmail.com.
Han Tian, Email: tianhan@mail2.sysu.edu.cn.
Yun Li, Email: yun.li2322@gmail.com.
Rong Lian, Email: lianrong@mail2.sysu.edu.cn.
Wei Li, Email: liwei72@mail2.sysu.edu.cn.
Shanshan Wu, Email: wushsh7@mail2.sysu.edu.cn.
Hui-Zhong Zhang, Email: zhhzhgd@163.com.
Jueheng Wu, Email: wujh@mail.sysu.edu.cn.
Lei Liu, Email: liulei3@mail2.sysu.edu.cn.
Junwei Song, Email: songjw5@mail2.sysu.edu.cn.
Hongyu Guan, Email: guanhongyu98@gmail.com.
Junchao Cai, Email: cjc_19860206@163.com.
Xun Zhu, Email: zhuxun8@mail.sysu.edu.cn.
Jun Li, Email: lijun37@mail.sysu.edu.cn.
Mengfeng Li, Email: limf@mail.sysu.edu.cn.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Global Burden of Disease Cancer Collaboration, et al. The Global Burden of Cancer 2013. JAMA Oncol. 2015;1(4):505–527. doi: 10.1001/jamaoncol.2015.0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. National Cancer Institute. Cancer Stat Fact: Lung and Bronchus Cancer. http://seer.cancer.gov/statfacts/html/lungb.html Accessed February 15, 2017.
- 4.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2(8):563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 5.Massagué J. TGFbeta in cancer. Cell. 2008;134(2):215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Massagué J. TGFβ signalling in context. Nat Rev Mol Cell Biol. 2012;13(10):616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hasegawa Y, Takanashi S, Kanehira Y, Tsushima T, Imai T, Okumura K. Transforming growth factor-beta1 level correlates with angiogenesis, tumor progression, and prognosis in patients with nonsmall cell lung carcinoma. Cancer. 2001;91(5):964–971. [PubMed] [Google Scholar]
- 8.Vicent S, et al. A novel lung cancer signature mediates metastatic bone colonization by a dual mechanism. Cancer Res. 2008;68(7):2275–2285. doi: 10.1158/0008-5472.CAN-07-6493. [DOI] [PubMed] [Google Scholar]
- 9.Kang HG, et al. Polymorphisms in TGF-beta1 gene and the risk of lung cancer. Lung Cancer. 2006;52(1):1–7. doi: 10.1016/j.lungcan.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 10.Moustakas A, Heldin CH. The regulation of TGFbeta signal transduction. Development. 2009;136(22):3699–3714. doi: 10.1242/dev.030338. [DOI] [PubMed] [Google Scholar]
- 11.Feng XH, Derynck R. Specificity and versatility in tgf-beta signaling through Smads. Annu Rev Cell Dev Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 12.Mullen AC, et al. Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell. 2011;147(3):565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blobe GC, Schiemann WP, Lodish HF. Role of transforming growth factor beta in human disease. N Engl J Med. 2000;342(18):1350–1358. doi: 10.1056/NEJM200005043421807. [DOI] [PubMed] [Google Scholar]
- 14.Yao Z, et al. TGF-beta IL-6 axis mediates selective and adaptive mechanisms of resistance to molecular targeted therapy in lung cancer. Proc Natl Acad Sci U S A. 2010;107(35):15535–15540. doi: 10.1073/pnas.1009472107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bruna A, et al. High TGFbeta-Smad activity confers poor prognosis in glioma patients and promotes cell proliferation depending on the methylation of the PDGF-B gene. Cancer Cell. 2007;11(2):147–160. doi: 10.1016/j.ccr.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 16.Jones E, Pu H, Kyprianou N. Targeting TGF-beta in prostate cancer: therapeutic possibilities during tumor progression. Expert Opin Ther Targets. 2009;13(2):227–234. doi: 10.1517/14728220802705696. [DOI] [PubMed] [Google Scholar]
- 17.Markowitz S, et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268(5215):1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 18.Inman GJ. Switching TGFβ from a tumor suppressor to a tumor promoter. Curr Opin Genet Dev. 2011;21(1):93–99. doi: 10.1016/j.gde.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 19.Wendt MK, Tian M, Schiemann WP. Deconstructing the mechanisms and consequences of TGF-β-induced EMT during cancer progression. Cell Tissue Res. 2012;347(1):85–101. doi: 10.1007/s00441-011-1199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui W, et al. TGFbeta1 inhibits the formation of benign skin tumors, but enhances progression to invasive spindle carcinomas in transgenic mice. Cell. 1996;86(4):531–542. doi: 10.1016/S0092-8674(00)80127-0. [DOI] [PubMed] [Google Scholar]
- 21.Hoot KE, et al. Keratinocyte-specific Smad2 ablation results in increased epithelial-mesenchymal transition during skin cancer formation and progression. J Clin Invest. 2008;118(8):2722–2732. doi: 10.1172/JCI33713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoot KE, Oka M, Han G, Bottinger E, Zhang Q, Wang XJ. HGF upregulation contributes to angiogenesis in mice with keratinocyte-specific Smad2 deletion. J Clin Invest. 2010;120(10):3606–3616. doi: 10.1172/JCI43304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xue J, et al. Sustained activation of SMAD3/SMAD4 by FOXM1 promotes TGF-β-dependent cancer metastasis. J Clin Invest. 2014;124(2):564–579. doi: 10.1172/JCI71104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yingling JM, Blanchard KL, Sawyer JS. Development of TGF-beta signalling inhibitors for cancer therapy. Nat Rev Drug Discov. 2004;3(12):1011–1022. doi: 10.1038/nrd1580. [DOI] [PubMed] [Google Scholar]
- 25.Connolly EC, Freimuth J, Akhurst RJ. Complexities of TGF-β targeted cancer therapy. Int J Biol Sci. 2012;8(7):964–978. doi: 10.7150/ijbs.4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoot KE, Oka M, Han G, Bottinger E, Zhang Q, Wang XJ. HGF upregulation contributes to angiogenesis in mice with keratinocyte-specific Smad2 deletion. J Clin Invest. 2010;120(10):3606–3616. doi: 10.1172/JCI43304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petersen M, et al. Smad2 and Smad3 have opposing roles in breast cancer bone metastasis by differentially affecting tumor angiogenesis. Oncogene. 2010;29(9):1351–1361. doi: 10.1038/onc.2009.426. [DOI] [PubMed] [Google Scholar]
- 28.Levy L, Hill CS. Alterations in components of the TGF-beta superfamily signaling pathways in human cancer. Cytokine Growth Factor Rev. 2006;17(1–2):41–58. doi: 10.1016/j.cytogfr.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 29.Jayaraman L, Massague J. Distinct oligomeric states of SMAD proteins in the transforming growth factor-beta pathway. J Biol Chem. 2000;275(52):40710–40717. doi: 10.1074/jbc.M005799200. [DOI] [PubMed] [Google Scholar]
- 30.Bennett WP, et al. p21waf1/cip1 and transforming growth factor beta 1 protein expression correlate with survival in non-small cell lung cancer. Clin Cancer Res. 1998;4(6):1499–1506. [PubMed] [Google Scholar]
- 31.Herrera RE, Mäkelä TP, Weinberg RA. TGF beta-induced growth inhibition in primary fibroblasts requires the retinoblastoma protein. Mol Biol Cell. 1996;7(9):1335–1342. doi: 10.1091/mbc.7.9.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seoane J, Le HV, Shen L, Anderson SA, Massagué J. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117(2):211–223. doi: 10.1016/S0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- 33.Vincent T, et al. A SNAIL1-SMAD3/4 transcriptional repressor complex promotes TGF-beta mediated epithelial-mesenchymal transition. Nat Cell Biol. 2009;11(8):943–950. doi: 10.1038/ncb1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown KA, et al. Identification of novel Smad2 and Smad3 associated proteins in response to TGF-beta1. J Cell Biochem. 2008;105(2):596–611. doi: 10.1002/jcb.21860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17(3):313–319. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- 36.Calon A, et al. Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation. Cancer Cell. 2012;22(5):571–584. doi: 10.1016/j.ccr.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen DX, et al. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell. 2009;138(1):51–62. doi: 10.1016/j.cell.2009.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Massagué J. TGFbeta in Cancer. Cell. 2008;134(2):215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Massagué J. TGFβ signalling in context. Nat Rev Mol Cell Biol. 2012;13(10):616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He W, Dorn DC, Erdjument-Bromage H, Tempst P, Moore MA, Massagué J. Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGFbeta pathway. Cell. 2006;125(5):929–941. doi: 10.1016/j.cell.2006.03.045. [DOI] [PubMed] [Google Scholar]
- 41.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 42.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 43.Tascilar M, et al. The SMAD4 protein and prognosis of pancreatic ductal adenocarcinoma. Clin Cancer Res. 2001;7(12):4115–4121. [PubMed] [Google Scholar]
- 44.Alazzouzi H, et al. SMAD4 as a prognostic marker in colorectal cancer. Clin Cancer Res. 2005;11(7):2606–2611. doi: 10.1158/1078-0432.CCR-04-1458. [DOI] [PubMed] [Google Scholar]
- 45.Huang GP, et al. Proteomic analysis of human bone marrow mesenchymal stem cells transduced with human telomerase reverse transcriptase gene during proliferation. Cell Prolif. 2008;41(4):625–644. doi: 10.1111/j.1365-2184.2008.00543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Hove I, Verslegers M, Hu TT, Carden M, Arckens L, Moons L. A proteomic approach to understand MMP-3-driven developmental processes in the postnatal cerebellum: Chaperonin CCT6A and MAP kinase as contributing factors. Dev Neurobiol. 2015;75(9):1033–1048. doi: 10.1002/dneu.22272. [DOI] [PubMed] [Google Scholar]
- 47.Xie M, et al. Integrative analysis of DNA copy number in metastatic NSCLC identifies drug sensitivity to Afatinib. J Clin Oncol. 2014;32(suppl):e19094. doi: 10.1111/resp.12416_10. [DOI] [Google Scholar]
- 48.Vogt N, Gibaud A, Almeida A, Ourliac-Garnier I, Debatisse M, Malfoy B. Relationships linking amplification level to gene over-expression in gliomas. PLoS One. 2010;5(12):e14249. doi: 10.1371/journal.pone.0014249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanic N, et al. Identification of differentially expressed mRNA transcripts in drug-resistant versus parental human melanoma cell lines. Anticancer Res. 2006;26(3A):2137–2142. [PubMed] [Google Scholar]
- 50.Alagaratnam S, Lind GE, Kraggerud SM, Lothe RA, Skotheim RI. The testicular germ cell tumour transcriptome. Int J Androl. 2011;34(4 Pt 2):e133–e150; discussion e150. doi: 10.1111/j.1365-2605.2011.01169.x. [DOI] [PubMed] [Google Scholar]
- 51.Yokota S, et al. Increased expression of cytosolic chaperonin CCT in human hepatocellular and colonic carcinoma. Cell Stress Chaperones. 2001;6(4):345–350. doi: 10.1379/1466-1268(2001)006<0345:IEOCCC>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nam JS, et al. An anti-transforming growth factor beta antibody suppresses metastasis via cooperative effects on multiple cell compartments. Cancer Res. 2008;68(10):3835–3843. doi: 10.1158/0008-5472.CAN-08-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Biswas S, et al. Anti-transforming growth factor β antibody treatment rescues bone loss and prevents breast cancer metastasis to bone. PLoS One. 2011;6(11):e27090. doi: 10.1371/journal.pone.0027090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ehata S, et al. Ki26894, a novel transforming growth factor-beta type I receptor kinase inhibitor, inhibits in vitro invasion and in vivo bone metastasis of a human breast cancer cell line. Cancer Sci. 2007;98(1):127–133. doi: 10.1111/j.1349-7006.2006.00357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Halder SK, Beauchamp RD, Datta PK. A specific inhibitor of TGF-beta receptor kinase, SB-431542, as a potent antitumor agent for human cancers. Neoplasia. 2005;7(5):509–521. doi: 10.1593/neo.04640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Korpal M, Yan J, Lu X, Xu S, Lerit DA, Kang Y. Imaging transforming growth factor-beta signaling dynamics and therapeutic response in breast cancer bone metastasis. Nat Med. 2009;15(8):960–966. doi: 10.1038/nm.1943. [DOI] [PubMed] [Google Scholar]
- 57.Zhang B, Halder SK, Zhang S, Datta PK. Targeting transforming growth factor-beta signaling in liver metastasis of colon cancer. Cancer Lett. 2009;277(1):114–120. doi: 10.1016/j.canlet.2008.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Melisi D, et al. LY2109761, a novel transforming growth factor beta receptor type I and type II dual inhibitor, as a therapeutic approach to suppressing pancreatic cancer metastasis. Mol Cancer Ther. 2008;7(4):829–840. doi: 10.1158/1535-7163.MCT-07-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lonning S, Mannick J, McPherson JM. Antibody targeting of TGF-β in cancer patients. Curr Pharm Biotechnol. 2011;12(12):2176–2189. doi: 10.2174/138920111798808392. [DOI] [PubMed] [Google Scholar]
- 60.Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1(2):729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ye T, et al. seqMINER: an integrated ChIP-seq data interpretation platform. Nucleic Acids Res. 2011;39(6):e35. doi: 10.1093/nar/gkq1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.