Abstract
Mutations in human Atrophin1, a transcriptional corepressor, cause dentatorubral-pallidoluysian atrophy, a neurodegenerative disease. Drosophila Atrophin (Atro) mutants display many phenotypes, including neurodegeneration, segmentation, patterning and planar polarity defects. Despite Atro’s critical role in development and disease, relatively little is known about Atro’s binding partners and downstream targets. We present the first genomic analysis of Atro using ChIP-seq against endogenous Atro. ChIP-seq identified 1300 potential direct targets of Atro including engrailed, and components of the Dpp and Notch signaling pathways. We show that Atro regulates Dpp and Notch signaling in larval imaginal discs, at least partially via regulation of thickveins and fringe. In addition, bioinformatics analyses, sequential ChIP and coimmunoprecipitation experiments reveal that Atro interacts with the Drosophila GAGA Factor, Trithorax-like (Trl), and they bind to the same loci simultaneously. Phenotypic analyses of Trl and Atro clones suggest that Atro is required to modulate the transcription activation by Trl in larval imaginal discs. Taken together, these data indicate that Atro is a major Trl cofactor that functions to moderate developmental gene transcription.
DOI: http://dx.doi.org/10.7554/eLife.23084.001
Research Organism: D. melanogaster
eLife digest
Cells with the same genetic information can look and behave differently to each other. This is because they can control the activity of their genes, changing the effects the genes have in the cell. Regulating genes in this way is important in allowing cells to adapt to their surroundings and to perform different tasks.
Proteins called transcription factors control the activity of genes through other proteins called transcriptional co-activators and co-repressors. Atrophins are a group of co-repressors found in many animals including humans and fruit flies. Atrophins suppress the activity of certain genes, reducing the effects that they have in the cell. Losing Atrophin from cells can lead to severe diseases, but how Atrophin causes these effects is currently not well understood.
Yeung et al. examined which genes Atrophin regulates in cells from the fruit fly Drosophila melanogaster. This investigation revealed that, amongst other genes, Atrophin controls several well-studied genes including engrailed and thickveins. These genes are important in allowing cells to communicate and co-ordinate before birth, ensuring cells work together to build complex tissues and organs. These results suggest Atrophin plays key roles in organising and shaping the body before birth.
Further examination revealed that Atrophin acts in partnership with another molecule called Trithorax-like. Inside the cell many genes are protected by structures called nucleosomes that make them difficult to access, and Trithorax-like helps Atrophin to gain access to these genes. Further work will examine whether Atrophin and Trithorax-like work directly together or if other molecules bring about their interaction. It will also be important to examine how Atrophins suppress the activity of the genes they control. Errors in Atrophin1 in humans result in a nerve-damaging disease known as DRPLA; this work could also help researchers to better understand this disorder.
Introduction
Atrophin family transcription factors are conserved transcriptional corepressors essential for development. Humans have two Atrophin genes; Atrophin 1 (ATN1) and Atrophin 2. A polyglutamine expansion in ATN1 is responsible for dentatorubral-pallidoluysian atrophy (DRPLA) a progressive disorder of ataxia, myoclonus, epilepsy, intellectual deterioration and dementia (Koide et al., 1994; Nagafuchi et al., 1994). Drosophila has only one Atrophin (Atro) gene, and expression of polyglutamine expansion Atro also lead to neurodegeneration (Napoletano et al., 2011). Loss of Atro results in defects in planar polarity, segmentation, and eye, wing and leg developmental defects (Erkner et al., 2002; Zhang et al., 2002; Fanto et al., 2003; Charroux et al., 2006; Napoletano et al., 2011; Saburi et al., 2012; Sharma and McNeill, 2013). Atro is required for proper expression of en in the embryo and represses the gap genes Krüppel and knirps (Erkner et al., 2002; Zhang et al., 2002; Haecker et al., 2007). Atro also affects epidermal growth factor receptor (EGFR), Decapentaplegic (Dpp) and Hedgehog signaling (Erkner et al., 2002; Charroux et al., 2006; Zhang et al., 2013). Despite regulating many pathways and processes, only three direct Atro targets have been defined (knirps, fat and dpp) (Wang et al., 2006; Napoletano et al., 2011; Zhang et al., 2013). These targets cannot adequately explain the diverse phenotypes exhibited by loss of Atro.
Atro physically interacts with histone deacetylase 1 (HDAC1) and the histone methyltransferase, G9a, through its SANT and ELM2 domains, respectively (Figure 1A) (Wang et al., 2006, 2008). These interactions are thought to contribute to Atro’s repressor activity. Atro also interacts with other cofactors such as the co-repressor, Scribbler (also called Brakeless) (Wang et al., 2006; Haecker et al., 2007). Atro does not have a DNA-binding sequence and has been shown to interact with DNA via interactions with nuclear receptors (Wang et al., 2006). We show here that a major means by which Atro regulates transcription is with the Drosophila GAGA factor, Trithorax-like (Trl, also called GAF). Trl is an essential transcription factor that directly binds to GA repeats (Biggin and Tjian, 1988; Soeller et al., 1988). Trl regulates many developmentally important genes including the segment polarity gene, engrailed (en) (Farkas et al., 1994; Bejarano and Busturia, 2004). Trl can interact with other transcription factors (e.g. Yorkie, Tramtrack) to regulate transcriptional targets in patterning and growth control (Pagans et al., 2002; Oh et al., 2013).
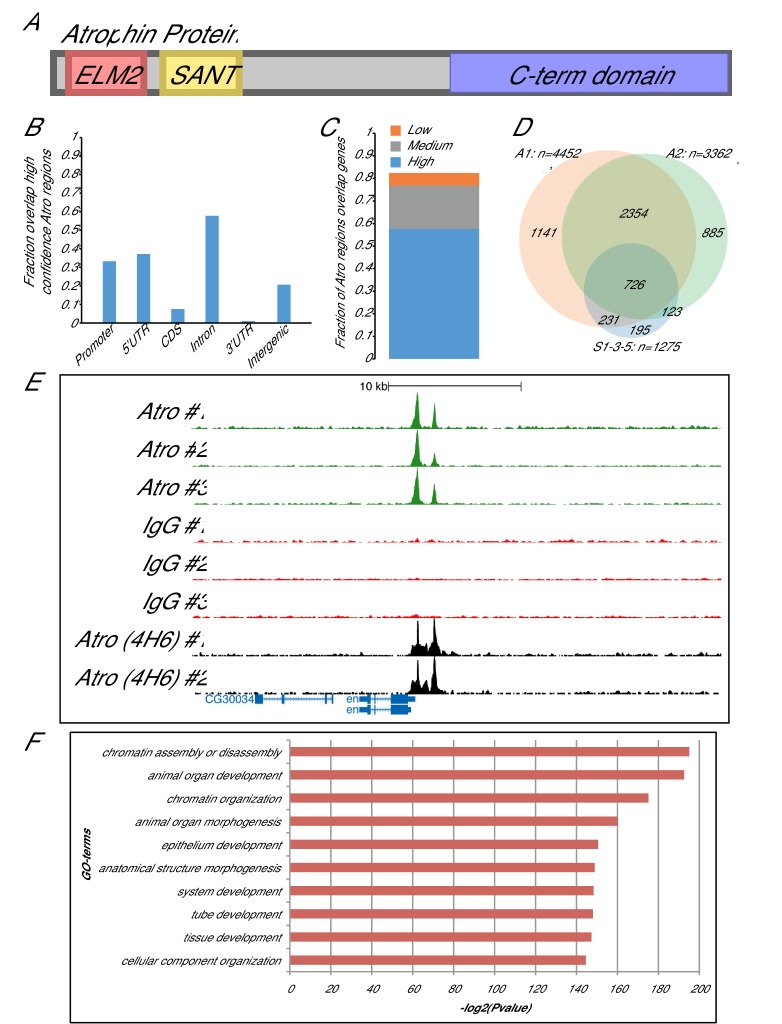
Figure 1. Atrophin ChIP-seq results.
(A) A schematic of Atro protein is shown. Atro’s N-terminal side has ELM2 and SANT domains to interact with HDAC1 and G9a, respectively. Atro’s C-terminal domain interacts with Tailless and Fat. (B) Fraction of the common Atro regions overlapping with various genomic features (genome release 5.57) (Attrill et al., 2016). (C) The fraction of common Atro regions overlapping genes (including 500 bp upstream of transcription start site) with high, medium or low expression in S2 cells (data from [Cherbas et al., 2011]). (D) Venn diagram showing overlap of the common regions from three biological replicates (S1-3-5) with two additional, independent biological replicates (A1 and A2) using a different Atro antibody (4H6). Only the peaks at the major chromosome arms (heterochromatin are excluded) are compared, and thus, only 1275 peaks are used in S1-3-5 for this analysis instead of 1377 peaks. (E) Example overlap of Atro ChIP-seq peaks at the engrailed (en) locus. Atro #1–3 are the triplicates Atro ChIP-seq data; IgG #1–3 are the corresponding IgG ChIP-seq controls for the Atro ChIP-seq data. Atro (4H6) #1–2 are the independent Atro ChIP replicates (shown in D). (F) Top 10 GO-term enrichment hits of the Atro ChIP-seq data. GO-term enrichments were done with PANTHER overrepresentation test with default parameters and Bonferroni correction (Mi et al., 2016).
DOI: http://dx.doi.org/10.7554/eLife.23084.003
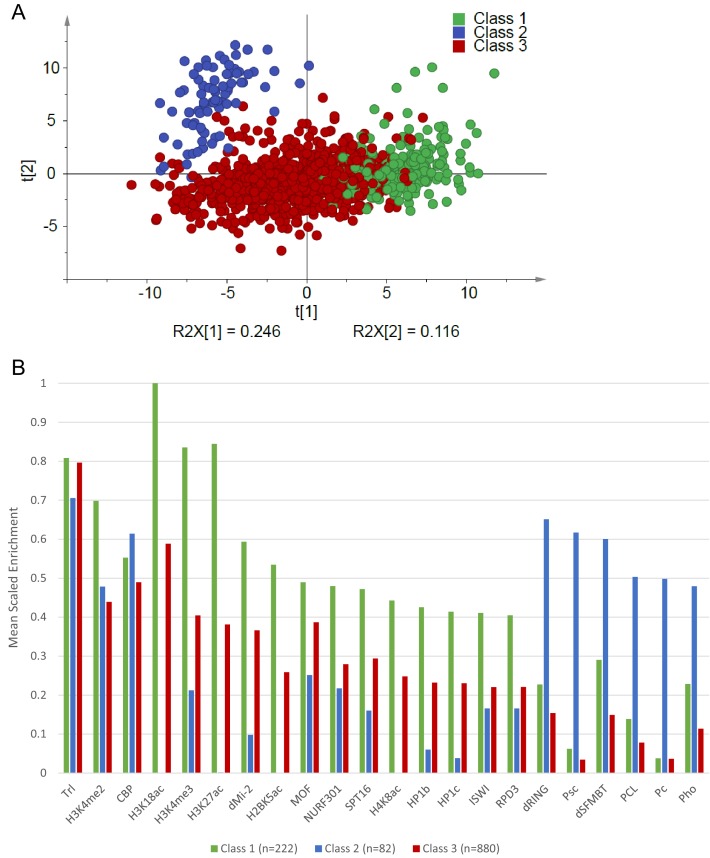
Figure 1—figure supplement 1. Principal component analysis of Atro peaks.
Trl was initially discovered to bind to the promoter of Ultrabithorax (Ubx) to activate transcription of Ubx in vitro (Biggin and Tjian, 1988). Trl is also required in transcriptional repression. This is supported by three pieces of evidence: First, Trl binds to Polycomb response elements, DNA sequences where Polycomb group proteins bind to repress transcription of target genes (Horard et al., 2000; Busturia et al., 2001). Second, Trl can physically associate with Polycomb Repressive Complex I (Poux et al., 2001). Third, Trl mutations enhance Polycomb group mutations, indicating that Trl is required for Polycomb repression (Mahmoudi et al., 2003). In addition to Trl’s ability to regulate transcription, Trl binding leads to open chromatin (Tsukiyama et al., 1994) and Trl maintains open chromatin (Fuda et al., 2015), at least in part by interacting with chromatin remodeling complexes such as NURF and FACT (Xiao et al., 2001; Shimojima et al., 2003). Trl is also required for transcriptional pausing (Lee et al., 2008; Fuda et al., 2015). A working model is that Trl first binds to DNA to maintain open chromatin and later associates with other proteins to activate or repress transcription (reviewed in [Granok et al., 1995; Lehmann, 2004]).
Here, we identify the genome-wide targets of Atro by chromatin-immunoprecipitation followed by sequencing (ChIP-seq). Our ChIP data show that Atro binds to regulatory regions of en, and in the putative regulatory regions of multiple Dpp and Notch signaling components. Our genetic and phenotypic analyses of Atro show that Atro regulates Dpp and Notch signaling, via transcriptional regulation of thickveins and likely fringe, respectively. We find that Atro negatively modulates En expression, while Trl promotes En expression. Bioinformatic analyses reveal that Atro and Trl ChIP-seq data strongly overlap and sequential ChIP and coimmunoprecipitation experiments confirm Atro and Trl bind to the same loci simultaneously and associate with one another. These data indicate that Atro modulates developmental gene expression via Trl binding. Our results suggest that Trl uses Atro as a cofactor to modulate transcription activation.
Results
Identification of genome-wide targets of Atro reveals enrichment in developmental patterning pathways
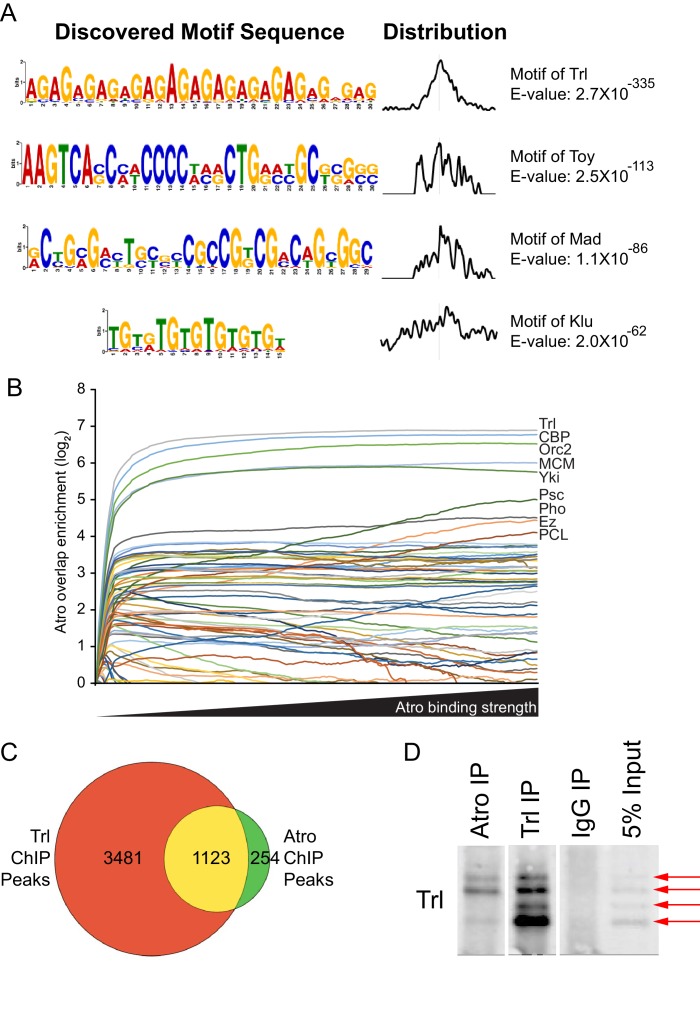
ChIP-seq against Atro was carried out in Drosophila Schneider 2 (S2) cells in three biological replicates (three sets of cells were grown and ChIP’ed on different days). ChIP peaks were called with MACS2 (Zhang et al., 2008) for each biological replicate using IgG ChIP-seq as the background model. The resulting three lists (containing 1757, 3064 and 3375 peaks) were intersected and peaks found in all three lists and with summits within 100 bp of each other were selected. The resulting list of 1377 peaks were associated with 1300 unique genes by proximity and treated as potential Atro targets (Supplementary file 1). We further mapped the 1377 Atro peaks relative to gene features and gene expression, which showed that the majority of peaks are located in actively transcribed genes, often close to the promoters or in introns (Figure 1B,C). In addition, we quantified the enrichment of all chromatin factors mapped by modENCODE (44 proteins and 23 histone modifications) at the Atro peaks and performed principal component analyses of our Atro peaks from major chromosome arms (with heterochromatin excluded, thus 1275 peaks were used). After hierarchical clustering of the significant principal components, we defined three classes (Figure 1—figure supplement 1A). We noticed that Class 2 (blue, Figure 1—figure supplement 1A and B) peaks are strongly enriched with Polycomb factors, indicating a potential connection between Atro and Polycomb factors.
To validate these peaks, we also performed independent ChIP-seq in two biological replicates with an antibody raised against a different part of Atro. Peaks were called with MACS2 using input as background model and intersected with the peaks identified above. We compared the peaks from major chromosome arms from each of the ChIP-seqs. A large fraction of the Atro-binding regions identified with the first antibody overlapped the ones found with this second antibody (Figure 1D,E).
Atro modulates developmental gene expression during larval development
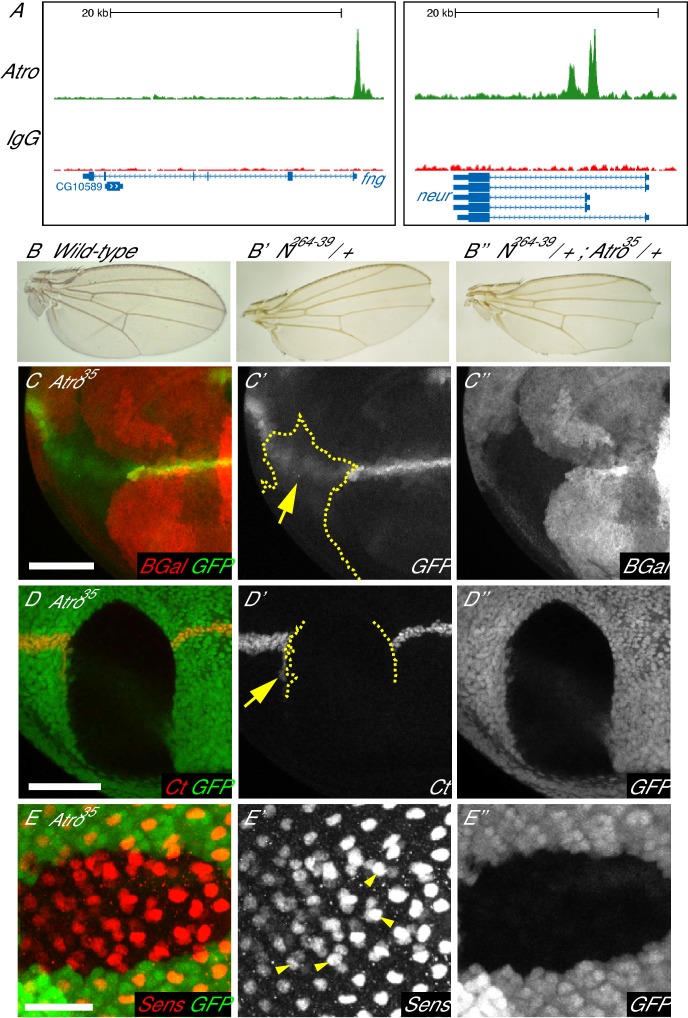
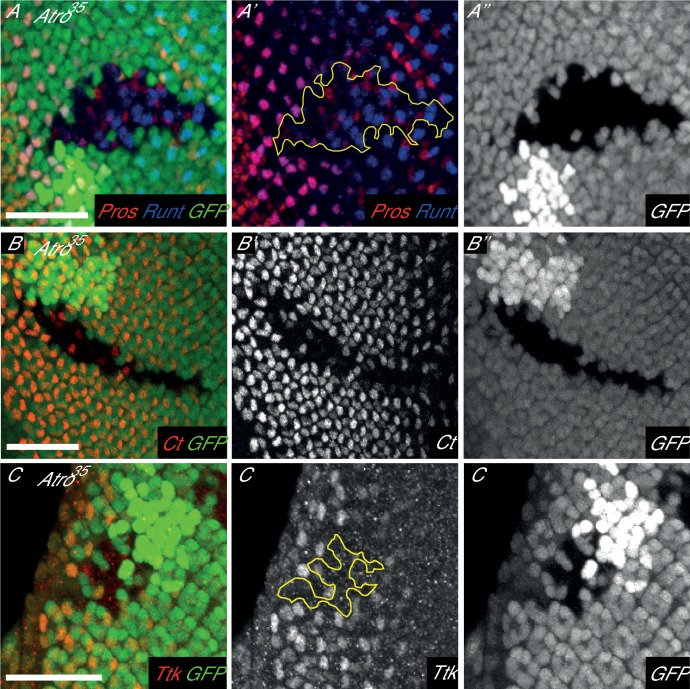
GO term enrichment analysis (PANTHER, [Mi et al., 2016]) of our ChIP-seq data revealed that Atro targets are enriched in chromatin organization, development and morphogenesis (Figure 1F). Interestingly, a potential direct target gene of Atro is engrailed (en, Figure 1E), a critical and conserved regulator of development. There are two strong Atro peaks in the en promoter (Figure 1E, within 2.4 kb upstream of the transcription start site [Kassis et al., 1992]). During embryogenesis, Atro is proposed to repress en expression with Even-skipped (Zhang et al., 2002). The regulatory region of en is complex (Cheng et al., 2014), and it is not known if Atro regulates en expression in larval stages. Therefore, we generated null mutant clones of Atro (Atro35) in larval imaginal discs and examined the expression of En by immunofluorescence.
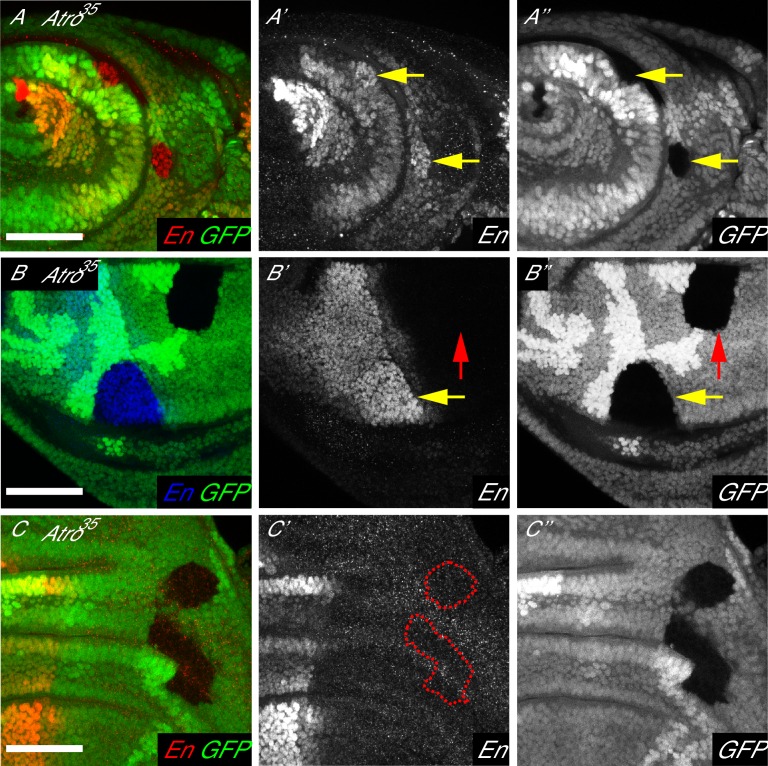
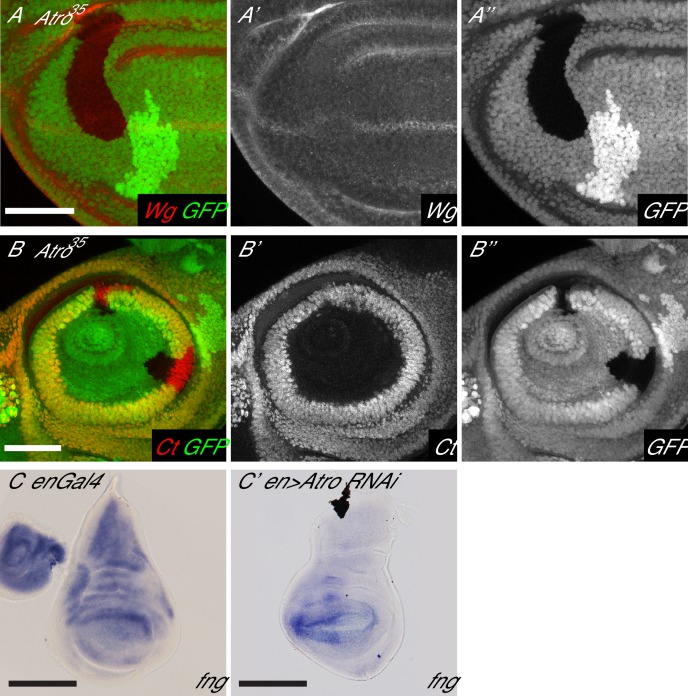
We found Atro35 clones have increased En levels in antennal, wing and leg imaginal discs (Figure 2A,B, and data not shown). Interestingly, Atro35 clones only show increased En levels in clones located in the posterior compartment, where En is endogenously expressed (Figure 2A–C). Notably, Atro35 clones in the anterior compartment cannot induce ectopic expression of En (Figure 2C). These data suggest that Atro negatively modulates En expression levels to maintain moderate expression in imaginal discs.
Figure 2. Atro is required for modulating En levels in larval imaginal discs.
(A) Antennal disc clones of Atro35 have increased En levels (arrows). (B) Atro35 wing disc clones in the posterior compartment have increased En levels (yellow arrow). But Atro35 clones cannot induce ectopic En expression in the anterior compartment (red arrow). (C) Atro35 clones cannot induce ectopic En expression (dotted lines mark clonal borders, wing disc is shown here). (A) has posterior to the right; (B–C) have posterior to the left, dorsal up. All scale bars are 50 μm.
Atro directly regulates Dpp signaling via thickveins
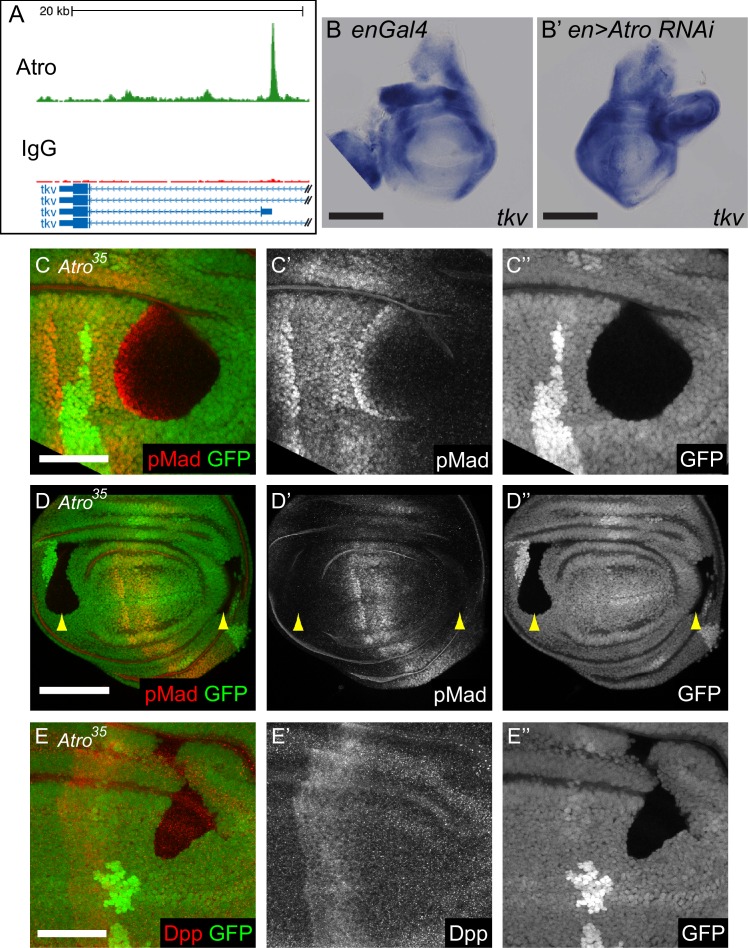
Atro mutant clones exhibit phenotypes similar to Dpp signaling defects such as leg patterning defects and expanded wing veins (Erkner et al., 2002; Zhang et al., 2002), suggesting Atro regulates Dpp signaling. Interestingly, our ChIP-seq data show that several Atro peaks occur in putative regulatory regions of several Dpp signaling pathway components (e.g. thickveins, Daughters against dpp and schnurri, Figure 3A and Supplementary file 1), suggesting Atro regulates Dpp signaling by directly regulating the expression of Dpp signaling pathway components. Thickveins (Tkv) is a critical Dpp receptor, and Atro35 clones show upregulation of the tkv-LacZ reporter in the wing (Wehn and Campbell, 2006). To confirm that Atro represses tkv expression, tkv transcript levels were assessed in Atro knocked down wing discs using in situ hybridization. Indeed, tkv expression is increased if Atro is knocked down (Figure 3B), confirming Atro represses tkv expression in the wing.
Figure 3. Atro regulates Dpp signaling.
(A) An Atro ChIP peak is found inside the tkv locus. This peak is directly upstream of the transcription start site of tkv isoform D. (B) In situ hybridization showing enGal4 control wing disc with normal tkv expression. (B’) shows in situ hybridization of Atro RNAi driven in the posterior half of the wing disc by enGal4. tkv expression is increased in the posterior half. (C) Atro35 wing disc clones that cross the endogenous pMad regions have increased pMad levels along the interior border of the clone that is closest to the middle of the disc (where the Dpp source is located). This indicates that Atro35 clones have increased Dpp signaling. (D) Atro35 wing disc clones cannot cause ectopic Mad phosphorylation (yellow arrow heads). (E) Atro35 wing disc clones cannot induce ectopic Dpp. All clones are marked by the absence of GFP; all figures have posterior to the left, dorsal up. Scale bars in A and C are 100 μm; in B and D are 50 μm.
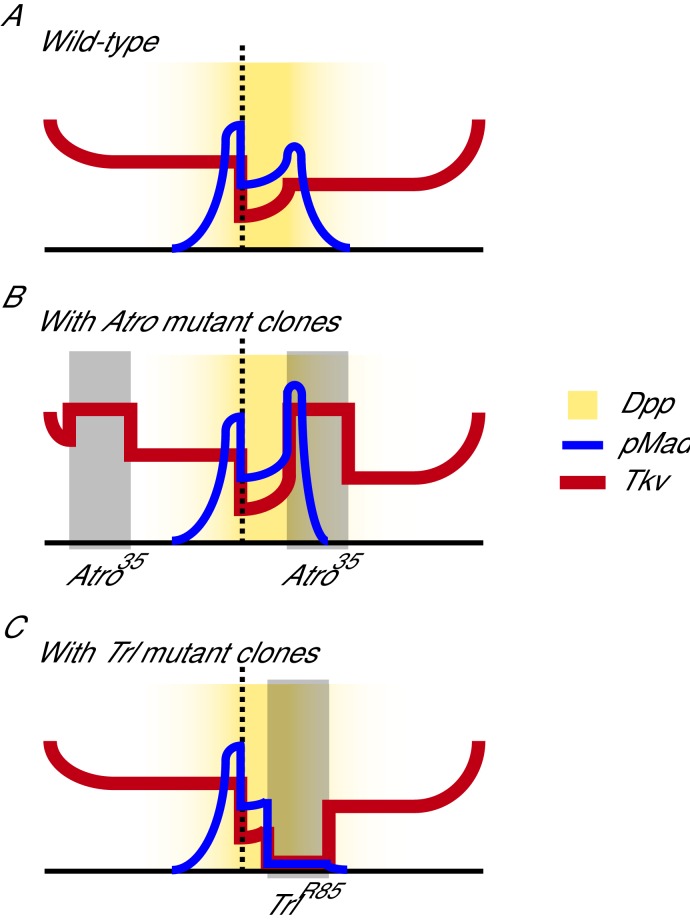
Figure 3—figure supplement 1. Model of how loss of Atro and Trl affect Dpp signaling in wing discs.
Atro binds within tkv’s gene locus and represses tkv transcription; however, it is not clear if this repression is important for the regulation of Dpp signaling. Mothers against dpp (Mad) is a downstream component of Dpp signaling, that is phosphorylated when the cell receives the Dpp signal (Newfeld et al., 1997). Phosphorylated Mad (pMad) is found in a broad stripe around the Dpp source and is used as a read-out for Dpp signaling. We stained Atro35 clones with anti-pMad antibodies to assess Dpp signaling. Atro35 clones that are found in the endogenous pMad regions have increased pMad levels along the interior border of the clones that is closest to the centre of the discs, where the endogenous Dpp source is located (Figure 3C). This pattern of increased pMad expression is expected if Tkv levels are increased in the clone, since increased Tkv levels in Atro35 clones would lead to an increase in Mad phosphorylation, as these cells will receive more Dpp signal compared to the adjacent non-clone cells (see Figure 3—figure supplement 1B). Atro35 clones also have increased Optomotor blind (Omb, a downstream target of Dpp signaling) expression (data not shown), indicating increased Dpp signaling in the clones (Grimm and Pflugfelder, 1996). However, Atro35 clones far from the Dpp source do not induce ectopic Mad phosphorylation, suggesting Dpp levels are not increased in Atro35 clones (Figure 3D, arrow heads). Consistent with this, Atro35 clones in the anterior or posterior compartments do not cause ectopic Dpp expression (Figure 3E). Our data indicate that Atro directly regulates tkv expression and thereby Dpp signaling.
Atro is required for Notch signaling and directly regulates Notch pathway gene expression
Atro ChIP data also revealed that Atro binds the putative regulatory regions of several Notch signaling components (mastermind (mam), Delta (Dl), neuralized (neur) and fringe (fng) (Figure 4A, Supplementary file 1), suggesting that Atro may regulate Notch signaling. Previous studies had not indicated that Atro loss of function affected Notch pathway activity. Therefore, to explore the biological relevance of the link with Notch, we first tested for genetic interactions between Atro and Notch (N). N is required for the development of the wing margin, and heterozygous N264-39 mutant (a null allele of N) flies have wing notches in the adult wing (Figure 4B’). We find that transheterozygous N264-39/+; Atro35 /+ mutant flies exhibit strikingly more severe wing notching than heterozygous N264-39 mutant alone (Figure 4B”). No notching is observed in Atro35 /+ mutant flies. Similar results were obtained with another independent Atro allele, Atroj5A3 (a P-element insertion allele, data not shown). Thus, Atro genetically interacts with N, suggesting Atro may play a role in N signaling.
Figure 4. Loss of Atro leads to loss of Notch signaling phenotypes.
(A) Atro ChIP peaks are found in the fng and neur loci, indicating that Atro may regulate these genes during development. (B) Wild-type wings have no notches at the wing margin. N264-39 heterozygous wings have wing notches (B’). Wing notch phenotype is enhanced in transheterozygous N264-39, Atro35 wings (B’’). (C) Atro35 clones cause the Notch signaling reporter (GFP) to express in a diffused pattern (arrow). Dotted line marks clonal borders. (D) Large Atro35 clones cause an autonomous loss of wing margin marker Cut as well as ectopic Cut expression on the posterior side of the clone (arrow). Clonal borders are marked with dotted lines. (E) Atro35 clones have extra cells expressing the early R8 marker, Sens. Extra Sens—positive cells are found in clusters of two to three cells but one cell within each cluster has more Sens staining than the rest (arrowheads). Clones are marked by absence of ß-Gal (red) in C and clones are marked by the absence of GFP in D and E. B have posterior to the bottom and C to E have posterior to the left. C and D have dorsal side up. Scale bars in C and D are 50 μm; in E is 25 μm.
Figure 4—figure supplement 1. Loss of Atro causes loss of wing margin markers via Notch signaling.
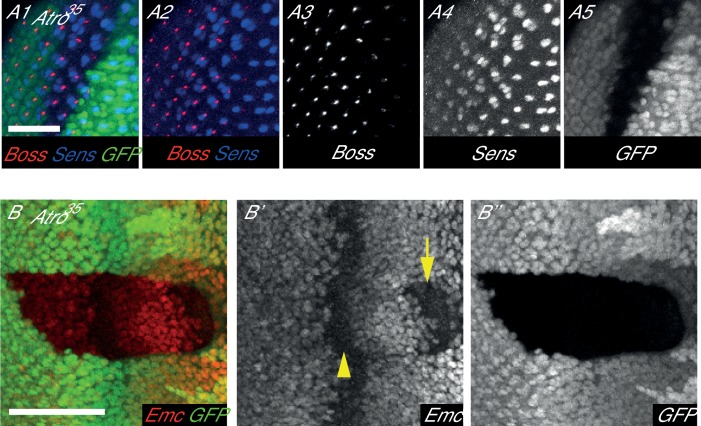
Figure 4—figure supplement 2. Atro35 clones have normal Boss expression but altered Emc levels.
Figure 4—figure supplement 3. Loss of Atro have eye phenotypes similar to loss of Notch signaling.
Since Atro and N genetically interact, we decided to test if loss of Atro affects wing margin development. In the wing, a line of N signaling induces the expression of wing margin markers Wingless (Wg) and Cut (Ct) (de Celis et al., 1996). N signaling in the wing can also be visualized with a N signaling reporter (NRE-GFP) (Housden et al., 2012), which expresses in a sharp line in the developing wing margin. Atro35 clones that cross the wing margin disrupt the expression pattern of the NRE-GFP reporter (Figure 4C), resulting in diffuse expression of the NRE-GFP reporter. In addition, Atro35 clones that cross the wing margin cause a loss of wing margin markers (Figure 4D and Figure 4—figure supplement 1A). In some cases, Atro35 clones induce ectopic Ct expression just outside of the posterior clonal border (Figure 4D, arrow). In contrast to the wing, Atro35 clones do not affect Ct expression in the antennal discs, where a requirement of N signaling for Ct expression has not been shown (Figure 4—figure supplement 1B). Atro binds to the fng promoter (Figure 4A, [Yang et al., 2013]), and in situ hybridization shows Atro knockdown causes increased fng transcription (Figure 4—figure supplement 1C). These observations suggest that Atro may be regulating fng and/or other factors to affect N signaling.
We also checked if Atro regulates N signaling in the eye. In larval eye discs, R8 photoreceptors differentiate from a three-cell equivalence group, which differentiates into R2, R5 and R8 photoreceptors (reviewed in [Frankfort and Mardon, 2002; Tsachaki and Sprecher, 2012]). N signaling is required for lateral inhibition in this equivalence group, such that one cell receives the least amount of N signaling and differentiates into R8. Thus, N signaling loss of function mutants have extra R8’s. To see if Atro affects R8 development, we stained Atro35 clones with Senseless (Sens), an early R8 marker. Atro35 clones have extra Sens positive cells (Figure 4E), clustered together in groups of two to three cells, resembling the number and shape of the R2/5/8 equivalence group. This suggests that lateral inhibition is defective in Atro35 clones. However, we noticed that there is one cell that has more Sens staining than the rest within each Sens-positive cell cluster (Figure 4E, arrowheads). Although Atro35 clones can induce extra Sens-positive cells, these clones do not have an excess of cells marked with a late R8 marker, Bride of sevenless (Boss) (Figure 4—figure supplement 2A). In addition, N signaling is required for the differentiation of R7 photoreceptors and cone cells, and loss of N signaling results in a lack of R7 and cone cells in the eye (Cooper and Bray, 2000; Flores et al., 2000; Tomlinson et al., 2011). We found that Atro35 clones also lose R7 and cone cell markers (Figure 4—figure supplement 3). Extra macrochaetae (emc), a downstream target of N signaling (Bhattacharya and Baker, 2009), is another potential direct target of Atro (Supplementary file 1). Indeed, Atro35 clones have reduced Emc protein levels anterior to the morphogenetic furrow (Figure 4—figure supplement 2B). Since Atro peaks are present in/very close to multiple N signaling components and downstream targets (fng, numb, Dl, neur, emc, Figure 4A, Supplementary file 1), Atro may directly regulate multiple targets to affect N signaling in the eye.
Atro is a cofactor of Trl
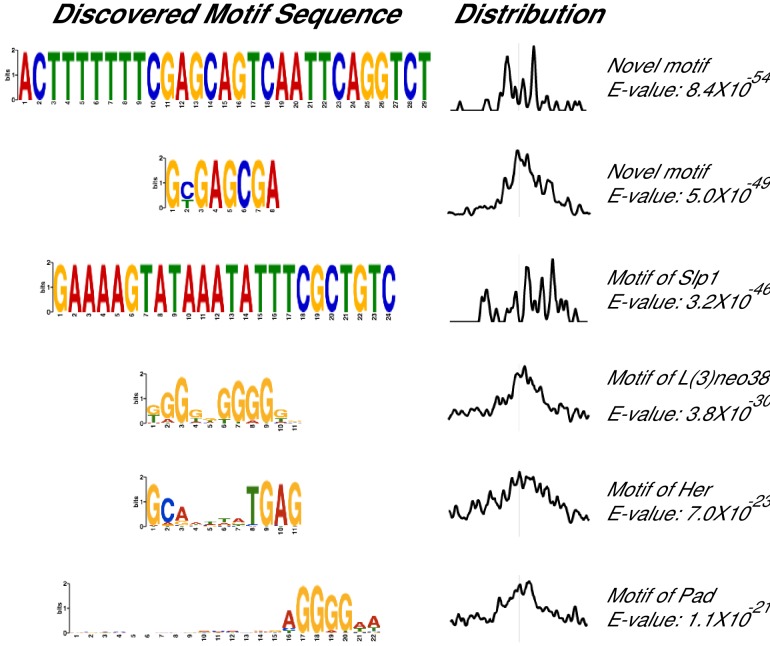
Although Atro does not bind to DNA directly, it binds with other cofactors to associate with DNA (e.g. Tailless [Wang et al., 2006]). We reasoned that analysis of the DNA sequences obtained from the Atro ChIP-seq could identify novel potential cofactors and binding motifs of Atro to gain insight into how Atro regulates developmental signaling. Therefore, we performed de novo motif analysis of our ChIP-seq data using MEME-ChIP (Bailey et al., 2009) (Figure 5A, Figure 5—figure supplement 1). The top motif discovered is a GA repeat, the binding motif of Trithorax-like (Trl, Figure 5A). The Trl binding motif is enriched within Atro ChIP peak summits, indicating strongest Atro binding. Other enriched motifs include Twin of eyeless (Toy) and Mad (Figure 5A). Next, we looked for overlap between the Atro ChIP-seq data with other ChIP datasets from S2 cells. We reasoned that we should see an increase of overlap between Atro and other ChIP data if we looked more specifically at genomic locations with higher Atro ChIP signal using Atro peaks from major chromosome arms (excluding heterochromatin). Therefore, we plotted the amount of Atro overlap with other transcription factors over that expected by chance against the number of genomic locations grouped according to the amount of Atro binding (using genomic data from modENCODE (downloaded from http://intermine.modencode.org/), CBP [Philip et al., 2015], and Yki [Oh et al., 2013]). In this analysis, several factors, including Trl, CBP (Nejire), the replication proteins Orc2 and MCM, Yorkie (Yki) and Polycomb proteins, showed increased overlap with increasing Atro binding (Figure 5B). Interestingly, the strongest overlap was found with Trl, confirming the MEME-ChIP analysis (Figure 5A). We further compared our list of Atro peaks with the published Trl ChIP-seq data (Fuda et al., 2015) and found striking overlap between the two data sets (1123/1377 Atro peaks overlap with Trl ChIP, Figure 5C). Visual inspection of Atro and Trl ChIP-seq peaks also shows binding at the same genomic regions (Figure 6C,E).
Figure 5. Atro and Trl bind to the same genomic locations.
(A) The top four hits from MEME-ChIP analysis using all Atro peaks. The top hit is a (GA) repeat, which is the binding motif of Trl. (B) Atro ChIP-seq data overlap with other factors in S2 cells. The y-axis represents the overlap over that expected by chance, where the fraction of the genome covered by each factor is the overlap expected by chance. Only positive values are shown. The Atro values are selected with increasing cut-off, so that fewer but stronger Atro-binding sites are shown along the x-axis. (C) shows the Venn diagram of the overlap of all Atro and Trl ChIP data. All 1377 Atro ChIP peaks were used and Trl ChIP peaks are from Fuda et al. (2015). Genomic coordinates from both data sets were used to construct this diagram.
DOI: http://dx.doi.org/10.7554/eLife.23084.014
Figure 5—figure supplement 1. Additional MEME ChIP hits of Atro ChIP-seq.
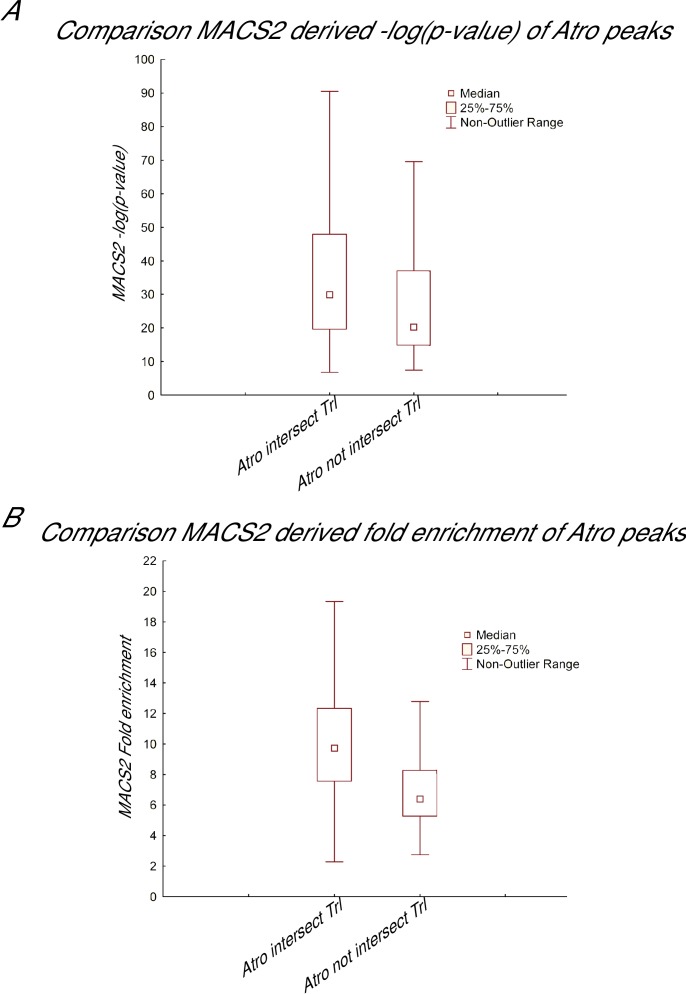
Figure 5—figure supplement 2. Comparison between Atro peaks that overlap Trl and those that do not.
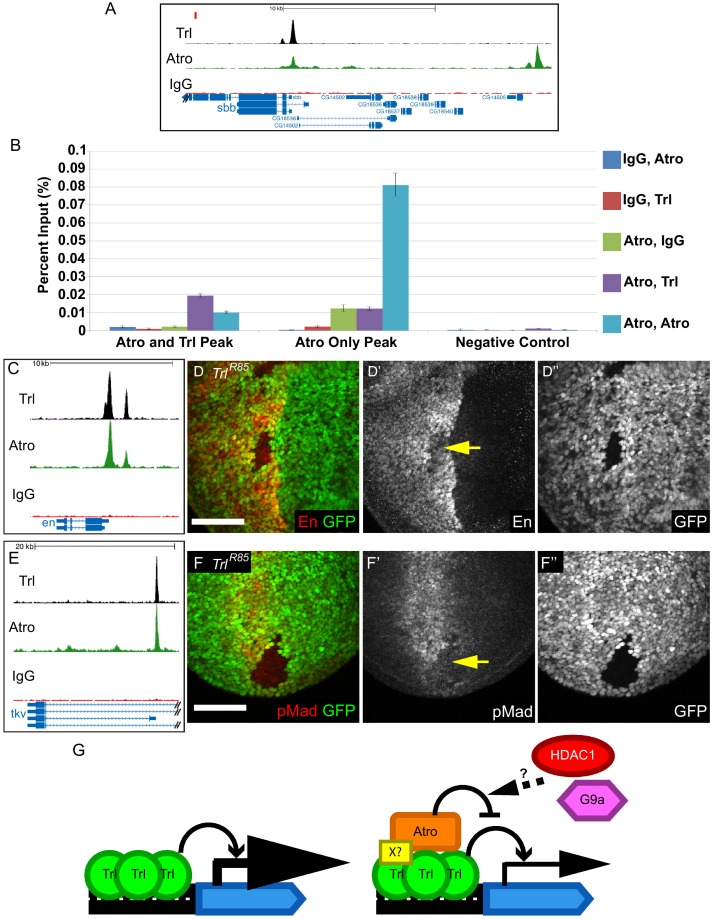
Figure 6. Trl and Atro bind to the same loci simultaneously and Trl is required for the expression of en and tkv.
(A) There is an Atro peak that overlaps with Trl peak within the sbb locus (left peak, Atro and Trl Peak in B). There is an Atro peak that does not overlap with Trl about 20 kb upstream of the sbb locus (right peak, Atro Only Peak in B). The red rectangle marks the region used as negative control in B. (B) ChIP-re-ChIP qPCR results. ChIP-re-ChIP samples are labeled as the sequence of antibody used (e.g. IgG, Atro means rabbit IgG ChIP followed by Atro re-ChIP). Negative controls for the ChIP-re-ChIP are any ChIPs with IgG. Atro, Trl ChIP-re-ChIP enriches the Atro and Trl Peak (purple bar) but it does not enrich the Atro only peak (same enrichment as Atro, IgG). Mean Ct value was used to calculate percent input and standard deviation of the Ct values was carried over in calculations and used as error bars. (C) Trl and Atro ChIP peaks coincide at the same loci upstream of en. (D) TrlR85 imaginal disc clones have decreased En levels (arrow, wing disc clone shown here). (E) Trl and Atro ChIP peaks coincide at the tkv locus. (F) TrlR85 wing disc clones have decreased pMad levels (arrow), possibly due to a decrease of tkv expression. All clones are marked by the absence of GFP; all figures have posterior to the left, dorsal up. Scale bars are 50 μm. (G) Model of how Atro and Trl function together to regulate expression of target genes. Trl is required to activate the transcription of its target genes. In the absence of Atro (Left), the target gene will express at a higher level than normal. Atro binds to the same site as Trl either directly or via some unknown cofactors (X?, Right). Atro modulates the expression of its target gene by counteracting Trl; potentially Atro is doing so by recruiting Histone deacetylase 1 (HDAC1) and G9a, a histone methyl transferase. Thus, target genes are expressed at the correct levels.
DOI: http://dx.doi.org/10.7554/eLife.23084.018
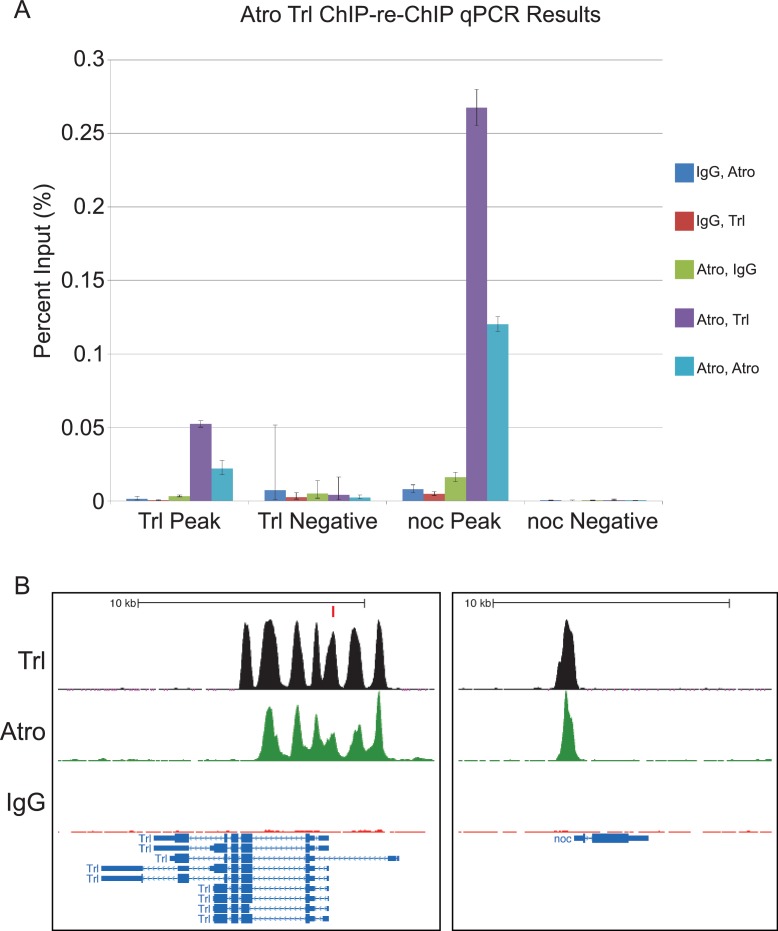
Figure 6—figure supplement 1. Additional ChIP-re-ChIP qPCR results of the Trl and no ocelli (noc) loci.
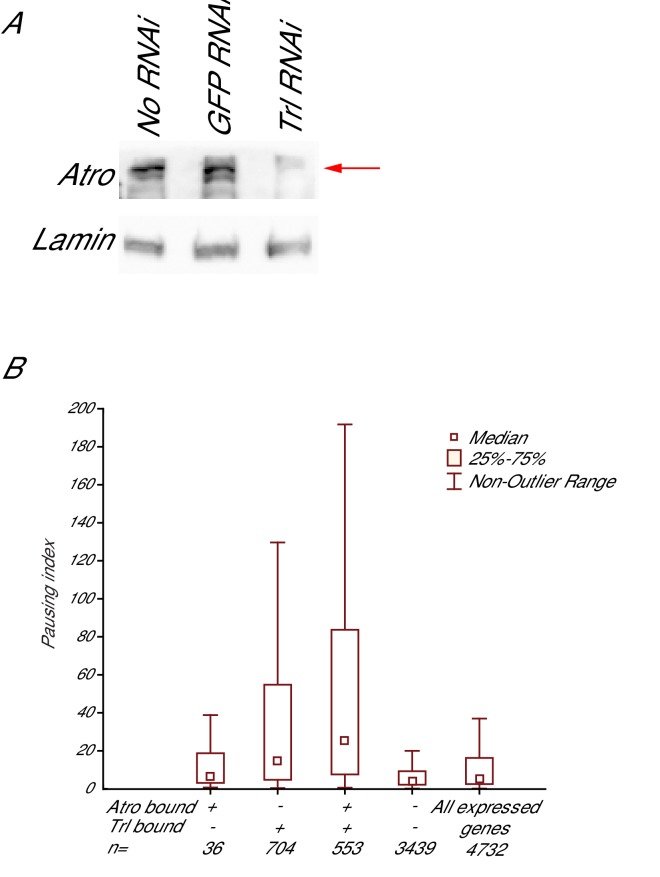
Figure 6—figure supplement 2. Trl knockdown decreases Atro protein levels, coimmunoprecipitation of Trl and Atro, and pausing indices of Trl and Atro bound genes.
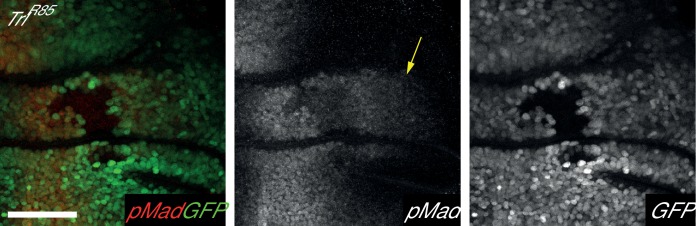
Figure 6—figure supplement 3. TrlR85 clones cause an autonomous decrease Mad phosphorylation but do not affect pMad levels outside clone (arrow).
Atro and Trl bind simultaneously and form a complex
The above analyses suggest that Atro and Trl bind to the same sites and may form a protein complex. To determine if Atro and Trl form a protein complex, we performed coimmunoprecipitation (coIP) of Atro and Trl in S2 cells. Although we did not detect an interaction using standard coIP conditions, pretreating lysates with micrococcal nuclease allowed us to coimmunoprecipitate endogenous Trl with endogenous Atro (Figure 5D).
To directly test if Atro and Trl bind to the same loci simultaneously, we performed sequential ChIP (ChIP-re-ChIP) on Atro followed by Trl in S2 cells. For this analysis, we selected Atro peaks at the scribbler (sbb) locus because of the presence of an Atro peak that overlaps with a Trl peak as well as a nearby Atro peak that does not overlap (Figure 6A). qPCR show that Atro-Trl ChIP-re-ChIP enriches for the locus with both Atro and Trl peaks, while it does not enrich for a locus with only Atro peak (enrichment is nearly equal to Atro-IgG ChIP-re-ChIP-negative control) (Figure 6B). Additional experiments showed that Atro and Trl co-occupy all the tested loci with both Atro and Trl peaks (Figure 6—figure supplement 1).
To see if Atro requires Trl to associate with DNA, we knocked down Trl using RNAi in S2 cells. Unfortunately, we could not directly test if Atro association with DNA requires Trl in cells, as knock down of Trl also leads to reduced abundance of Atro proteins (Figure 6—figure supplement 2A). These findings indicate that Atro and Trl bind to the same loci and to one another, forming a complex on chromatin to regulate transcription.
Trl is required for En and Tkv expression
Both ChIP-seq and ChIP-re-ChIP data show that Atro and Trl bind to the same loci simultaneously. Interestingly, there are strong overlaps of Atro and Trl ChIP-seq peaks at the en locus (Figure 6C). Since we found that Atro negatively modulates the expression of En during larval development, we wondered if Trl also regulates En expression. Therefore, we generated and analyzed Trl null mutant clones (TrlR85) in imaginal discs. In contrast to Atro35 clones, TrlR85 clones have reduced En expression (Figure 6D), indicating that Trl is required for the positive regulation of En expression in larval discs.
Atro and Trl ChIP-seq data also indicate that Atro and Trl bind to the same region of the tkv locus (Figure 6E), suggesting that Trl regulates tkv expression with Atro. Therefore, TrlR85 clones were generated to investigate the role of Trl in tkv regulation. pMad levels were used as a read out for Tkv levels. In contrast to Atro35 clones, TrlR85 clones have reduced pMad levels (Figure 6F, Figure 6—figure supplement 3). In addition, pMad staining can be found on the side of the TrlR85 clones further away from the middle of the wing (Figure 6—figure supplement 3, arrow), indicating Dpp expression was not disrupted. These changes can be explained by a decrease in Tkv levels in TrlR85 clones. In addition, we did not find any Trl or Atro peaks within 20 kb of the mad locus (data not shown). Thus, the simplest model to explain our data is that the reduced pMad in TrlR85 clones is caused by a downregulation of tkv expression. Therefore, similar to Atro and Trl’s regulation of en, Trl is required for tkv expression, while Atro is required for repression of tkv.
Discussion
Mutations in Atrophins lead to neurodegeneration, and loss of Atro leads to major defects in development. However, only a direct few targets of Atro are known and it is unclear how Atro regulates many genes. Here, using genome-wide ChIP-seq and in vivo analyses we show that Atro directly binds a number of developmentally critical genes; specifically regulating En expression and Dpp and Notch signaling. Trl has been proposed to activate transcription by opening and maintaining open chromatin to allow additional transcription factors to bind (reviewed in [Granok et al., 1995; Lehmann, 2004). We find that Atro and Trl participate in a complex and bind to the same loci simultaneously. Atro does not bind to DNA directly, while Trl has a DNA binding domain (Biggin and Tjian, 1988). Thus, our results suggest a model in which Atro binds to DNA via Trl, either directly or indirectly via someunknown cofactors, and Atro modulates transcriptional activation by Trl.
Our in vivo analyses of en and tkv show that Trl is required to promote their expression while Atro negatively modulates expression levels. To explain these phenotypes, we propose a model in which Trl binding leads to open chromatin to allow transcription, and Atro binding, either by directly interacting with Trl or through some unknown factors, negatively modulates Trl’s activity (Figure 6G). Atro binds to Histone deacetylase 1 (HDAC1) and a histone methyltransferase (G9a) to close chromatin and repress transcription (Wang et al., 2006, 2008), suggesting a mechanisms by which Atro is able to counteract Trl. Indeed, Atro- and Trl-binding sites at the en, tkv and fng loci overlap with HDAC1-binding sites (from modENCODE, RPD3-Q3451) (Celniker et al., 2009). Thus, loss of Trl leads to loss of expression of en and tkv where they are normally expressed. In the absence of Atro, Trl’s activity is no longer negatively modulated, and expression of target genes such as en and tkv are increased. However, in regions where en and tkv are not endogenously expressed, loss of Atro cannot induce ectopic expression as Atro’s function is to negatively modulate the transcription of its target genes.
Interestingly, Trl is enriched at paused promoters and is involved in transcription pausing (Lee et al., 2008; Fuda et al., 2015). Trl recruits the negative elongation factor NELF to paused genes (Li et al., 2013), but the precise mechanism of how Trl affects pausing is unknown. Our data suggest that Atro is present to modulate Trl’s transcription activation role. Perhaps, Atro is part of the transcriptional machinery used by Trl to pause transcription. In support of this, we noted that Atro preferentially binds to genes with higher expression (in S2 cells, genes with high expression are likely to be paused, Figure 1C) (Gilchrist et al., 2010). GO analysis of Atro targets shows enrichment of signaling and patterning genes, also consistent with a role in pausing (Figure 1F, data not shown). Notably, the pausing index for genes bound by both Atro and Trl is higher on average than for all expressed genes (Figure 6—figure supplement 2C, [Kwak et al., 2013]). This increase of pausing index is even higher than genes bound only by Trl (Figure 6—figure supplement 2B). These data suggest that Atro may be involved in transcriptional pausing with Trl.
We note that there are some Atro-binding sites that are not co-occupied by Trl. Atro also binds to other transcription factors (such as Tailless and Cubitus Interruptus [Wang et al., 2006; Zhang et al., 2013]) and Atro may interact with other factors to bind to sites not co-occupied by Trl. Visual inspection reveals strong Atro peaks that do overlap with Trl (Figure 6A); however, on a genome-wide basis, Atro peaks that do not overlap with Trl peaks are slightly weaker than the ones that do, as judged from the MACS2 derived significance (-log(p-value)) and fold enrichment values (Figure 5—figure supplement 2).
Atro maternal mutants have missing, malformed and/or expanded En stripes (Zhang et al., 2002), while in discs Atro modulates En expression. A possible explanation is that En regulation changes at different developmental times. During early embryogenesis, Pair-rule genes are required to first establish en expression (DiNardo et al., 1988). Atro is also required for the proper expression of Pair-rule genes such as fushi tarazu and even-skipped (Erkner et al., 2002; Zhang et al., 2002). This could explain why loss of Atro during early embryogenesis has more severe effects on en expression. However, from late embryogenesis on, En expression no longer requires Pair-rule genes but depends on Polycomb group genes and unidentified activators (Moazed and O'Farrell, 1992). We observed reduced En expression in Trl clones in antennal and wing imaginal discs, while Atro disc clones have increased expression. These results suggest that Trl and Atro are required for regulation of En expression from late embryogenesis through larval development.
Why is it important for Atro to moderate en expression? Intriguingly, expressing higher than normal levels of En in the posterior compartment leads to lethality and anterior-posterior patterning defects of the posterior wing, suggesting that moderating En levels is required for normal development, and high levels could be toxic (Guillén et al., 1995; Tabata et al., 1995).
Our Atro ChIP data show that Atro binds to the tkv locus and our in situ analysis reveals Atro represses tkv expression. We find that Atro35 clones have increased Dpp signaling, consistent with Atro’s role in tkv regulation. Conversely, TrlR85 clones have lower pMad levels. We reason that this is caused by decreased Tkv within the TrlR85 clones, consistent with a peak in the tkv locus. These observations mirror what we have seen with en, where Trl is required for activation and Atro is required for repression of transcription.
A model for Atro and Trl regulation of Dpp signaling is shown in Figure 3—figure supplement 1. In wild-type wings, the expression patterns of Dpp (yellow shading) and Tkv (red line) cause pMad to be found in a broad stripe (Figure 3—figure supplement 1A, blue line) (Tanimoto et al., 2000). Tkv levels are increased in Atro35 clones (Figure 3—figure supplement 1B, indicated by the shaded rectangle). Thus, pMad levels are increased along the interior border of the clone if the Atro35 clones are close enough to the Dpp source. Atro35 clones further away would not cause changes to pMad levels. Trl has the opposite effect where TrlR85 clones (Figure 3—figure supplement 1C, shaded rectangle) cause a decrease of Tkv and thus lower pMad levels. Interestingly, the Mad binding motif is enriched in the Atro ChIP, with a shift in distribution, suggesting Mad may bind adjacent to Atro, and that Atro-Mad interactions on chromatin may also affect Dpp signaling.
Although Atro35 clones can lead to ectopic dpp-LacZ expression (Erkner et al., 2002; Zhang et al., 2013), we did not see increased Dpp in Atro35 wing clones. Additionally, loss of Atro away from the endogenous Dpp stripe does not induce ectopic pMad. Therefore, the increase of pMad staining in Atro35 clones matches the pattern that is expected if Atro regulates Dpp signaling via tkv and not via dpp regulation. Thus, we suggest that Atro regulates Dpp signaling in the wing primarily by regulating tkv transcription.
To our knowledge, this report also provides the first evidence that Atro regulates N signaling. Our ChIP-seq analysis revealed Atro binding in multiple N pathway components, and genetic and molecular analysis reveals disruption of N signaling in Atro clones in eye and wing discs. Atro knockdown results in upregulation of fng transcripts in larval wings. Interestingly, patched-Gal4-driven overexpression of fng leads to an autonomous loss of wing margin marker expression as well as ectopic expression of wing margin marker on the posterior border of the Fng overexpressing region (Panin et al., 1997). All Atro35 clones that cross the wing margin cause an autonomous loss of wing margin markers and large Atro35 clones can induce some ectopic wing margin marker expression on the posterior edge of the clone, mimicking patched-Gal4-driven Fng overexpression. Atro35 clones also disrupt N signaling reporter expression. Atro35 clones cause the N signaling reporter to express diffusely instead of in a sharp line. This diffuse expression pattern may be an indication of a loss of precise N signaling at the wing margin. Interestingly, Fng is crucial for N signaling at the margin (de Celis and Bray, 1997; Panin et al., 1997). Thus, Atro represses fng expression, and loss of Atro and patched-Gal4-driven overexpression of Fng have similar N-related phenotype in the wing.
Loss of Atro also leads to Notch loss of function in the eye. Atro35 clones have extra cells with the early R8 marker, Sens, indicating a defect in lateral inhibition. Although there are extra Sens-positive cells in Atro clones, not all these cells express the late R8 marker, Boss, thus the extra Sens-positive cells do not differentiate into R8. Inspection of the Sens-positive cell clusters reveals there is one cell with more Sens than its neighbors in each cluster. We reason that the difference in Sens levels renders one cell with the most R8-like and this cell can express Boss. Atro35 clones also exhibit a loss of R7 and cone cell markers. Thus, Atro binds to the putative regulatory regions of genes that are connected to N signaling (such as emc, Dl, mam, fng, neur, numb, Supplementary file 1), genetically interacts with N and regulates N target expression.
While the strongest overlap in our ChIP-seq peaks was seen between Atro and Trl, we also observed significant overlap of our Atro ChIP-seq data with ChIP data sets of Yorkie (Yki), the key transcriptional co-activator of the Hippo pathway (Figure 5B). Interestingly, the atypical cadherin Fat regulates Yki activity via the Hippo pathway ([Bennett and Harvey, 2006; Cho et al., 2006; Silva et al., 2006; Willecke et al., 2006] and reviewed in Enderle and McNeill [2013] and planar polarity via Atro (Fanto et al., 2003; Saburi et al., 2012; Sharma and McNeill, 2013). Significantly, neurodegeneration by Atro has been shown to be mediated in part by Yki and the Hippo pathway (Napoletano et al., 2011). Thererfore, our finding that Yki and Atro are found at the same loci suggests a direct mechanism by which Atro may impact neurodegeneration, and suggests that Atro interactions with Yki may feed back into growth and patterning regulation by Fat cadherins.
To our knowledge, this is the first genome-wide analysis of Atrophin. Our genome-wide ChIP-seq and phenotypic analyses reveal many novel direct targets of Atro, and showed that Engrailed, Notch and Dpp signaling are directly regulated by Atro. Our analyses indicate that Atro preferentially binds to Trl binding sites. Interestingly, the fraction of paused genes is significantly more correlated with sites that bind both Atro and Trl, than just Trl alone, suggesting Atro may have a function in the regulation of pausing. Significantly, ChIP-re-ChIP experiments reveal that Atro and Trl bind to the same loci simultaneously, and phenotypic analyses indicate that Atro restricts expression of genes whose expression is promoted by Trl. Taken together, our data indicate that Atro is a critical component of developmental signaling and is an important general modulator of transcription activation by Trl.
Materials and methods
Double crosslink ChIP and subsequent ChIP-seq
Two step crosslinking ChIP was required in order for ChIP to enrich for positive controls when using the SG2524 anti-Atro antibody (see below). S2 cells (107 cells/mL, S2-DGRC, stock #6 from Drosophila Genomics Research Center) were washed three times in sterile PBS. Washed cells were first fixed with 2 mM ethylene glycol bis(succinimidyl succinate) (EGS) in PBS for 45 min at room temperature followed by three PBS washes. Washed cells were crosslinked in 1% formaldehyde in PBS for 15 min at room temperature and quenched in 125 mM glycine for 5 min on ice. Cells were then washed once in ChIP Wash Buffer A (10 mM Hepes pH7.6, 10 mM EDTA pH8.0, 0.5 mM EGTA pH8.0, 0.25% Triton X-100) and followed by a wash in ChIP Wash Buffer B (10 mM Hepes pH7.6, 100 mM NaCl, 1 mM EDTA pH8.0, 0.5 mM EGTA pH8.0, 0.01% Triton X-100) at 4°C, 5 min each. Washed cells were resuspended in Sonication buffer (50 mM Hepes pH7.6, 140 mM NaCl, 1 mM EDTA pH8.0, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, supplemented with proteinase inhibitors) at a concentration of 108 cells/mL. Cells were then sonicated using a Qsonica Q700 sonicator in an ice-water bath (until fragments were roughly 150 bp in length). 10 μL of 10% SDS, 100 μL of 1% sodium deoxycholate, 100 μL of 10% Triton X-100, 28 μL of 5M NaCl were added to each mL of sonicated chromatin and incubated at 4°C for 10 min. Sonicated chromatin was then spun down at ≥20,000g for 5 min to remove cellular debris and the supernatant was used for ChIP.
Protein G Dynabeads (Invitrogen, Lithuania) were blocked in 1 mg/mL BSA in sonication buffer for at least 2 hr at 4°C. Blocked beads were conjugated with the antibodies for at least 4 hr at 4°C. 5 μL anti-Atro sera (SG2524, raised in rabbits against Atro amino acids 121–134, KGIDKKWTEDETKK), and 5 μL normal rabbit IgG (Cell Signaling Technology, Danvers, MA) were used for ChIP. Of the sonicated chromatin, 300 μL were incubated with conjugated beads overnight on a rotating wheel at 4°C. Beads were washed for 5 min each in Sonication buffer, Wash A (50 mM Hepes pH7.6, 500 mM NaCl, 1 mM EDTA pH8.0, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, supplemented with proteinase inhibitors), Wash B (20 mM Tris pH8.0, 1 mM EDTA pH8.0, 250 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate), and TE buffer. Beads were then resuspended in Elution Buffer (50 mM Tris pH8.0, 50 mM NaCl, 2 mM EDTA, 0.75% SDS, 20 ug/mL RNase A) and incubated at 68°C overnight to remove crosslinks. Eluted chromatin was extracted by treating with Proteinase K followed by phenol chloroform DNA extraction (with 6 μg glycogen added during DNA precipitation [Thermo Scientific, Lithuania]). The extracted DNA was resuspended in 50 μL Tris pH8.0.
Three biological replicates were used for library construction (along with three corresponding IgG ChIP control). ChIP samples were treated with polynucleotide kinase and DNA polymerases for 30 min at room temperature (35 μL ChIP sample, 5 μL 10xNEB two buffer (New Englands Biolabs, Ipswich, MA), 2 μL of 25 mM ATP, 2 μL of 10 mM dNTP, 10U T4 polynucleotide kinase, 4.5U T4 DNA polymerase, 1U Klenow Large Fragment DNA Polymerase, water to a volume of 50 uL). Afterwards, DNA was purified with PEG bead slurry (1M NaCl, 23% PEG, Sera-Mag Speedbeads (Fisher Scientific, England) with final PEG concentration of 13.87%) and eluted with 35 uL Qiagen EB buffer (Qiagen, Valencia, CA). Single dA overhang was added to eluted ChIP samples by incubating samples in 35 μL ChIP samples, 5 μL 10X NEB two buffer, 1 μL 10 mM dATP, 5U Klenow Fragment (3’−5’ exo-) (New England Biolab), water to 50 μL for 30 min at 37°C. Samples were purified with PEG bead slurry and eluted with 35 μL Qiagen EB buffer. Short adapators for sequencing were ligated to samples by incubating samples at room temperature overnight in ligation buffer (35 μL DNA, 12 μL 5X quick ligation buffer, 2000U quick T4 DNA ligase, 2 μL 0.5 μM Illumina short sequencing adaptor, water to 60 μL). ChIP samples were purified two times with PEG bead slurry (started with 20% PEG instead of 23% PEG; final PEG of 8.89% and 10.91%, respectively) and eluted with 35 μL Qiagen EB buffer. Adaptor ligated libraries were PCR amplified (10 cycles), purified with PEG bead slurry (started with 20% PEG instead of 23%; final PEG concentration of 9.19%), and eluted with 35 μL Qiagen EB buffer. Libraries were then sequenced on Miseq (Illumina, San Diego, CA) using PE150V3 kit PE 85 bp. BWA program (v0.6.1; using default parameters) was used to align sequence reads to Drosophila genome release five (Attrill et al., 2016).
Single crosslink ChIP
A second, independent ChIP was performed with a monoclonal antibody (4H6) raised against Atro amino acids 1369–1378 (SRQSPLHPVP) using Drosophila S2 cells catalog #006 from the Drosophila Genomics Resource Center. This ChIP was performed using two biological replicates (cells grown and ChIP’ed at different times). The cells were grown to a density of 0.2–1 × 107 cells/mL and fixed in 1% formaldehyde for 15 min at ambient temperature. The reaction was quenched by 0.16 M glycine pH 7.0 for 5 min and washed in PBS. Cells were sequentially washed with ChIP Wash buffer A and ChIP Wash Buffer B for 10 min at 4°C followed by resuspension in Sonication buffer to a final concentration of 5–10 × 107 cells/mL. Nuclei were sonicated for 15 min using a Diagenode Bioruptor, rotated for 10 min followed by centrifugation for 10 min at 13,000 rpm at 4°C.
A mix of Protein A and G Dynabeads (Invitrogen, Lithuania) blocked with BSA (Sigma Aldrich, St Louis, MO) were mixed with the antibody. Beads and antibodies were incubated for at least 2 hr followed by the addition of 0.5–1 × 107 cells. Chromatin and antibody bead complexes were formed during at least 2 hr followed by 5 min washes with Sonication buffer, Wash A, Wash B and TE buffer. Beads were resuspended in Elution buffer (same as above but supplemented with 20 µg/mL glycogen) in a new tube. Cross-linking was reversed at 68°C for at least 4 hr and proteins removed by Proteinase K digestion. DNA was purified by phenol-chloroform extraction, ethanol precipitated and finally resuspended in 200 µl 0.1×TE.
The DNA was sequenced at the Uppsala Genome Center using SOLiD (TM) ChIP-Seq Library preparation, size selection (around 150 bp + adapters 95 bp) and sequenced using SOLiD4 75 bp fragment run. The number of mapped reads were 11270731 (Input 1), 13320338 (Atro ChIP 1), 7315911 (Input 2) and 6972016 (Atro ChIP 2).
ChIP-re-ChIP
S2 cells were double crosslinked, sonicated, and ChIP’ed as above, using 5 μL anti-Atro sera (SG2524), and 10 μL normal rabbit IgG (Cell Signaling Technology). After the first ChIP, beads were washed and eluted for re-ChIP as described in Truax and Greer (2012). Eluted chromatin was incubated in BSA-blocked Dynabeads for 2 hr to remove any leftover antibodies. The supernatant containing eluted chromatin was re-ChIP’ed by incubating in beads conjugated with the appropriate amount of antibodies overnight (5 μL anti-Atro sera (SG2524), 10 μL anti-Trl sera (gift from K. White), 10 μL normal rabbit IgG (Cell Signaling Technology)). After the re-ChIP, the beads were washed and eluted as normal double crosslink ChIP samples above. qPCRs were done in technical triplicates with SYBR green PCR Master Mix (Applied Biosystems, Canada, 20 μL reaction volume; qPCR was performed three times for each ChIP samples). Percent input and errors were calculated using standard percent input calculations. qPCR primer pairs are listed in Supplementary file 2.
Bioinformatics
For each biological replicate, peaks were called using MACS2 (version 2.1.0, FDR = 0.01, genome size 1.2 × 108). Each Atro ChIP-seq replicate was compared with its corresponding IgG ChIP-seq replicate control (or input control for the second independent Atro ChIP-seq replicates) in the MACS analysis. Peaks from the biological replicates were intersected and only peaks present across all replicates with summits within 100 bp were selected. Peaks were extended by 2 kb on both sides and then annotated by intersecting all Drosophila genes’ coordinates with the peaks coordinates using BEDTools (Quinlan and Hall, 2010). MEME-ChIP was carried out using first-order model, any number of repetitions, motif count of 10, score ≥5 and an E-value threshold of ≤10. Overlap of Atro ChIP with Trl ChIP data sets (Figure 5C) was done using BEDTools Intersect function. Atro-binding sites were compared to data from modENCODE (downloaded from http://intermine.modencode.org/), (Philip et al., 2015) for CBP, and (Oh et al., 2013) for Yki, and gene expression divided into three equally sized bins (low, medium and high expressions) from (Cherbas et al., 2011) using custom Perl scripts.
Principal component analyses
The enrichment values for each factor in the Atro binding regions were calculated by taking the mean of the top three consecutive enrichment values within each region (Philip et al., 2015). All factors were normalized so that 0 represents the genomic mean (background levels) and one represents the genomic maximum (mean of top 0.001 percentile) for each factor. Enriched Atro regions were used as observations and normalized enrichment values of each factor within the regions were used as variables as decribed in (Philip et al., 2015). Hierarchical clustering was done on all significant components of the analysis using Ward clustering to calculate tree distances. The three classes of Atro-bound regions were based on hierachical clustering.
Clones and immunofluorescence
Mitotic clones were generated in flies with the following genotypes:
hs-flp; ; Atro35 FRT80B / ubi-GFP FRT80B
hs-flp; ; TrlR85 FRT2A / hs-GFP, Minute, FRT2A
hs-flp; NRE-GFP / +; Atro35 FRT80B / arm-LacZ FRT80B
Mitotic clones were generated by heat shocking (at 37°C) larvae for 45 min at 48 hr and 72 hr after egg laying. TrlR85 clones were additionally heat shocked for 45 min about 1.5 hr prior to dissection to induce GFP expression. Larval tissues were dissected and prepared as in standard protocol. Sample sizes are listed in Supplementary file 3. The following antibodies were used for immunofluorescence: mouse antibodies against En (DSHB 4D9, 1/400), Pros (DSHB MR1A, 1/500), Ct (DSHB 2B10, 1/500), Wg (DSHB 4D4, 1/500), and ß-Gal (Promega [Madison, WI], 1/1000); rabbit antibodies against En (Santa Cruz (Santa Cruz, CA) d-300, 1/500), Omb (gift from G. Pflugfelder, 1/1000), Dpp (gift from M. Gibson, 1/100), Ttk (gift from W. Ge, 1/100), and pMad (Cell Signaling Technology (Danvers, MA) #9510, 1/500); guinea pig antibodies against Sens (gift from H. Bellen, 1/1000), and Runt (gift from C. Desplan, 1/500).
In situ hybridization
enGal4 / +; UAS Atro RNAi (Bloomington stock center, line 32961) / + larvae were used for in situ hybridization. Larval wing discs are prepared and stained as described in Morris et al. (2009), substituting wing discs for testes. Primers for probes are listed in Supplementary file 2.
S2 cells and knockdown
S2 cells were purchased from Drosophila Genomics Resource Center (S2-DGRC, stock #6). S2 cells were grown in S2 media + 10% fetal bovine serum in standard conditions, and cells were kept healthy for all experiments. Double stranded RNA was made using the Megascript T7 kit (Life Technologies, Canada). Primers used to make the dsRNA are listed in Supplementary file 2. S2 cells were dsRNA treatment using standard bathing S2 knockdown protocol. 10 ug/mL of dsRNA was used for each treatment.
Immunoprecipitation
S2 cells (107 cells per immunoprecipitation) were incubated in 0.5 mL LPC buffer (5% sucrose, 35 mM Hepes pH7.4, 80 mM KCl, 5 mM K2HPO4, 5 mM MgCl2, 5 mM CaCl2, 0.01% α-lysophospatidylcholine) for 3 min at room temperature. Cells were resuspended in 0.2 mL MNase buffer (5% Glycerol, 20 mM Tris pH7.4, 60 mM KCl, 15 mM NaCl, 5 mM CaCl2, 3 mM MgCl2, 0.5% NP-40, 1 mM DTT). 6 μL of micrococcal nuclease (40 U/μL, New England Biolabs) were added and incubated at 25°C for 5 min. 0.3 mL of MNase dilution buffer (3.6 mM Tris pH8.8, 12 mM EDTA, 225 mM NaCl, 60 mM KCl, 1.2% NP-40, supplemented with proteinase inhibitor) were added and incubated on shaker for 10 min at 4°C. MNase treated samples were centrifuged to remove cellular debris. Supernatant were used for immunoprecipitation using standard methods.
Accession number
ChIP-Sequencing data reported in this study are archived at the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) as accession numbers GSE87509 (ChIP using Atro antibody SG2524) and GSE87471 (second independent Atro ChIP with antibody 4H6).
Acknowledgements
We thank Hugo Bellen, Sarah Bray, Gerrard Campbell, Claude Desplan, Maxim Frolov, Wanzhong Ge, Matthew Gibson, Judy Kassis, Henry Krause, John Lis, Gert Pflugfelder, Kevin White and Jeff Wrana for antibody reagents and fly stocks; Tim Hughes, Hamed Shateri Najafabadi and Tim Westwood for ChIP analyses help. This work was supported by grants from the Candian Institutes of Health Research Foundation (FDN 143319) and a Terry Fox Research Institute Team Grant to HM. HM is a Tier 1 Canada Research Chair in Coordinating Growth and Polarity. This work was also partly supported by grants from the Swedish Research Council to MMannervik, the UK Medical Research Council (NIRG-G1002186) to MF, and Knut and Alice Wallenberg to EpiCoN, co-PI: PS.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Funding Information
This paper was supported by the following grants:
Canadian Institutes of Health Research FDN 143319 to Helen McNeill.
Medical Research Council NIRG-G1002186 to Manolis Fanto.
Knut och Alice Wallenbergs Stiftelse to Per Stenberg.
Terry Fox Research Institute team grant to Helen McNeill.
Additional information
Competing interests
HM: Reviewing editor, eLife.
The other authors declare that no competing interests exist.
Author contributions
KY, Conceptualization, Formal analysis, Validation, Investigation, Visualization, Methodology, Writing—original draft, Writing—review and editing.
AB, Data curation, Formal analysis, Investigation.
EK, Software, Formal analysis, Investigation, Methodology.
P-HH, Formal analysis, Investigation.
YT, Formal analysis, Investigation.
IN, Formal analysis, Investigation.
DY, Resources, Methodology.
AL, Resources, Data curation, Software, Methodology.
MMo, Resources, Data curation, Software, Methodology.
MH, Resources, Data curation, Software, Supervision, Writing—review and editing.
SA, Resources, Data curation, Supervision, Funding acquisition, Methodology.
MF, Formal analysis, Supervision, Funding acquisition, Investigation, Writing—review and editing.
PS, Software, Formal analysis, Supervision, Funding acquisition, Methodology, Writing—review and editing.
MMa, Resources, Formal analysis, Supervision, Investigation, Writing—review and editing.
HM, Conceptualization, Formal analysis, Supervision, Funding acquisition, Investigation, Visualization, Writing—review and editing.
Additional files
Perl script used to count the frequency of overlap between various data sets in gff3-format to a reference gff3 file. Each data set is divided into a user defined number of bins by equally distributed cut-offs by value or by rank. Alternatively, the bins can be created so that all data points below each cut-off are merged into one bin. The frequency of overlap between the reference file data to the data points in each bin is reported.
Perl script used to count the frequency of overlap between various data sets in sgr-format to a reference gff3 file. Each data set is divided into a user defined number of bins by equally distributed cut-offs by value or by rank. Alternatively, the bins can be created so that all data points below each cut-off are merged into one bin. The frequency of overlap between the reference file data to the data points in each bin is reported.
The user defines the number of data points within each region to include in the mean. The mean can be reported based on all values, the nth top values or the highest n consecutive values.
Major datasets
The following datasets were generated:
McNeill H,Yeung K,2017,ChIP-seq of Atrophin in Drosophila S2 cells,https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE87509,Publicly available at the NCBI Gene (accession no: GSE87509)
Boija A,Stenberg P,Mannervik M,2017,Atrophin (Atro) ChIP-seq data from Drosophila S2 cells,https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE87471,Publicly available at the NCBI Gene (accession no: GSE87471)
The following previously published datasets were used:
White K,2010,Chromatin Binding Site Mapping of Transcription Factors in D. melanogaster by ChIP-seq,http://intermine.modencode.org/query/experiment.do?experiment=Chromatin+Binding+Site+Mapping+of+Transcription+Factors+in+D.+melanogaster+by+ChIP-seq,Publicly available at Modencode (http://intermine.modencode.org)
White K,2009,Chromatin Binding Site Mapping,http://intermine.modencode.org/query/experiment.do?experiment=Chromatin+Binding+Site+Mapping,Publicly available at Modencode (http://intermine.modencode.org)
Fuda NJ,Guertin MJ,Sharma S,Danko CG,Martins AL,Siepel A,Lis JT,2013,The Genomic Binding Profile of GAGA Element Associated Factor (GAF) in Drosophila S2 cells,https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE40646,Publicly available at the NCBI Gene Expression Omnibus (accession no: GSE40646)
References
- Attrill H, Falls K, Goodman JL, Millburn GH, Antonazzo G, Rey AJ, Marygold SJ, FlyBase Consortium FlyBase: establishing a gene group resource for Drosophila melanogaster. Nucleic Acids Research. 2016;44:D786–D792. doi: 10.1093/nar/gkv1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Research. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejarano F, Busturia A. Function of the Trithorax-like gene during Drosophila development. Developmental Biology. 2004;268:327–341. doi: 10.1016/j.ydbio.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Bennett FC, Harvey KF. Fat cadherin modulates organ size in Drosophila via the salvador/Warts/Hippo signaling pathway. Current Biology. 2006;16:2101–2110. doi: 10.1016/j.cub.2006.09.045. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Baker NE. The HLH protein extramacrochaetae is required for R7 cell and cone cell fates in the Drosophila eye. Developmental Biology. 2009;327:288–300. doi: 10.1016/j.ydbio.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin MD, Tjian R. Transcription factors that activate the ultrabithorax promoter in developmentally staged extracts. Cell. 1988;53:699–711. doi: 10.1016/0092-8674(88)90088-8. [DOI] [PubMed] [Google Scholar]
- Busturia A, Lloyd A, Bejarano F, Zavortink M, Xin H, Sakonju S. The MCP silencer of the Drosophila Abd-B gene requires both pleiohomeotic and GAGA factor for the maintenance of repression. Development. 2001;128:2163–2173. doi: 10.1242/dev.128.11.2163. [DOI] [PubMed] [Google Scholar]
- Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, Micklem G, Piano F, Snyder M, Stein L, White KP, Waterston RH, modENCODE Consortium Unlocking the secrets of the genome. Nature. 2009;459:927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charroux B, Freeman M, Kerridge S, Baonza A. Atrophin contributes to the negative regulation of epidermal growth factor receptor signaling in Drosophila. Developmental Biology. 2006;291:278–290. doi: 10.1016/j.ydbio.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Brunner AL, Kremer S, DeVido SK, Stefaniuk CM, Kassis JA. Co-regulation of invected and engrailed by a complex array of regulatory sequences in Drosophila. Developmental Biology. 2014;395:131–143. doi: 10.1016/j.ydbio.2014.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherbas L, Willingham A, Zhang D, Yang L, Zou Y, Eads BD, Carlson JW, Landolin JM, Kapranov P, Dumais J, Samsonova A, Choi JH, Roberts J, Davis CA, Tang H, van Baren MJ, Ghosh S, Dobin A, Bell K, Lin W, Langton L, Duff MO, Tenney AE, Zaleski C, Brent MR, Hoskins RA, Kaufman TC, Andrews J, Graveley BR, Perrimon N, Celniker SE, Gingeras TR, Cherbas P. The transcriptional diversity of 25 Drosophila cell lines. Genome Research. 2011;21:301–314. doi: 10.1101/gr.112961.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a fat tumor suppressor pathway. Nature Genetics. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- Cooper MT, Bray SJ. R7 photoreceptor specification requires notch activity. Current Biology. 2000;10:1507–1510. doi: 10.1016/S0960-9822(00)00826-5. [DOI] [PubMed] [Google Scholar]
- de Celis JF, Bray S. Feed-back mechanisms affecting notch activation at the dorsoventral boundary in the Drosophila wing. Development. 1997;124:3241–3251. doi: 10.1242/dev.124.17.3241. [DOI] [PubMed] [Google Scholar]
- de Celis JF, Garcia-Bellido A, Bray SJ. Activation and function of notch at the dorsal-ventral boundary of the wing imaginal disc. Development. 1996;122:359–369. doi: 10.1242/dev.122.1.359. [DOI] [PubMed] [Google Scholar]
- DiNardo S, Sher E, Heemskerk-Jongens J, Kassis JA, O'Farrell PH. Two-tiered regulation of spatially patterned engrailed gene expression during Drosophila embryogenesis. Nature. 1988;332:604–609. doi: 10.1038/332604a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enderle L, McNeill H. Hippo gains weight: added insights and complexity to pathway control. Science Signaling. 2013;6:re7. doi: 10.1126/scisignal.2004208. [DOI] [PubMed] [Google Scholar]
- Erkner A, Roure A, Charroux B, Delaage M, Holway N, Coré N, Vola C, Angelats C, Pagès F, Fasano L, Kerridge S. Grunge, related to human Atrophin-like proteins, has multiple functions in Drosophila development. Development. 2002;129:1119–1129. doi: 10.1242/dev.129.5.1119. [DOI] [PubMed] [Google Scholar]
- Fanto M, Clayton L, Meredith J, Hardiman K, Charroux B, Kerridge S, McNeill H. The tumor-suppressor and cell adhesion molecule fat controls planar polarity via physical interactions with atrophin, a transcriptional co-repressor. Development. 2003;130:763–774. doi: 10.1242/dev.00304. [DOI] [PubMed] [Google Scholar]
- Farkas G, Gausz J, Galloni M, Reuter G, Gyurkovics H, Karch F. The Trithorax-like gene encodes the Drosophila GAGA factor. Nature. 1994;371:806–808. doi: 10.1038/371806a0. [DOI] [PubMed] [Google Scholar]
- Flores GV, Duan H, Yan H, Nagaraj R, Fu W, Zou Y, Noll M, Banerjee U. Combinatorial signaling in the specification of unique cell fates. Cell. 2000;103:75–85. doi: 10.1016/S0092-8674(00)00106-9. [DOI] [PubMed] [Google Scholar]
- Frankfort BJ, Mardon G. R8 development in the Drosophila eye: a paradigm for neural selection and differentiation. Development. 2002;129:1295–1306. doi: 10.1242/dev.129.6.1295. [DOI] [PubMed] [Google Scholar]
- Fuda NJ, Guertin MJ, Sharma S, Danko CG, Martins AL, Siepel A, Lis JT. GAGA factor maintains nucleosome-free regions and has a role in RNA polymerase II recruitment to promoters. PLoS Genetics. 2015;11:e1005108. doi: 10.1371/journal.pgen.1005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granok H, Leibovitch BA, Shaffer CD, Elgin SC. Chromatin. Ga-ga over GAGA factor. Current Biology. 1995;5:238–241. doi: 10.1016/S0960-9822(95)00048-0. [DOI] [PubMed] [Google Scholar]
- Grimm S, Pflugfelder GO. Control of the gene optomotor-blind in Drosophila wing development by decapentaplegic and wingless. Science. 1996;271:1601–1604. doi: 10.1126/science.271.5255.1601. [DOI] [PubMed] [Google Scholar]
- Guillén I, Mullor JL, Capdevila J, Sánchez-Herrero E, Morata G, Guerrero I. The function of engrailed and the specification of Drosophila wing pattern. Development. 1995;121:3447–3456. doi: 10.1242/dev.121.10.3447. [DOI] [PubMed] [Google Scholar]
- Haecker A, Qi D, Lilja T, Moussian B, Andrioli LP, Luschnig S, Mannervik M. Drosophila brakeless interacts with atrophin and is required for tailless-mediated transcriptional repression in early embryos. PLoS Biology. 2007;5:e145. doi: 10.1371/journal.pbio.0050145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horard B, Tatout C, Poux S, Pirrotta V. Structure of a polycomb response element and in vitro binding of polycomb group complexes containing GAGA factor. Molecular and Cellular Biology. 2000;20:3187–3197. doi: 10.1128/MCB.20.9.3187-3197.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housden BE, Millen K, Bray SJ. Drosophila reporter vectors compatible with φc31 integrase transgenesis techniques and their use to generate new notch reporter fly lines. G3: Genes|Genomes|Genetics. 2012;2:79–82. doi: 10.1534/g3.111.001321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassis JA, Noll E, VanSickle EP, Odenwald WF, Perrimon N. Altering the insertional specificity of a Drosophila transposable element. PNAS. 1992;89:1919–1923. doi: 10.1073/pnas.89.5.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide R, Ikeuchi T, Onodera O, Tanaka H, Igarashi S, Endo K, Takahashi H, Kondo R, Ishikawa A, Hayashi T. Unstable expansion of CAG repeat in hereditary dentatorubral-pallidoluysian atrophy (DRPLA) Nature Genetics. 1994;6:9–13. doi: 10.1038/ng0194-9. [DOI] [PubMed] [Google Scholar]
- Kwak H, Fuda NJ, Core LJ, Lis JT. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013;339:950–953. doi: 10.1126/science.1229386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Li X, Hechmer A, Eisen M, Biggin MD, Venters BJ, Jiang C, Li J, Pugh BF, Gilmour DS. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Molecular and Cellular Biology. 2008;28:3290–3300. doi: 10.1128/MCB.02224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M. Anything else but GAGA: a nonhistone protein complex reshapes chromatin structure. Trends in Genetics. 2004;20:15–22. doi: 10.1016/j.tig.2003.11.005. [DOI] [PubMed] [Google Scholar]
- Li J, Liu Y, Rhee HS, Ghosh SK, Bai L, Pugh BF, Gilmour DS. Kinetic competition between elongation rate and binding of NELF controls promoter-proximal pausing. Molecular Cell. 2013;50:711–722. doi: 10.1016/j.molcel.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi T, Zuijderduijn LM, Mohd-Sarip A, Verrijzer CP. GAGA facilitates binding of pleiohomeotic to a chromatinized polycomb response element. Nucleic Acids Research. 2003;31:4147–4156. doi: 10.1093/nar/gkg479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Research. 2016;44:D336–D342. doi: 10.1093/nar/gkv1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D, O'Farrell PH. Maintenance of the engrailed expression pattern by polycomb group genes in Drosophila. Development. 1992;116:805–810. doi: 10.1242/dev.116.3.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris CA, Benson E, White-Cooper H. Determination of gene expression patterns using in situ hybridization to Drosophila testes. Nature Protocols. 2009;4:1807–1819. doi: 10.1038/nprot.2009.192. [DOI] [PubMed] [Google Scholar]
- Nagafuchi S, Yanagisawa H, Ohsaki E, Shirayama T, Tadokoro K, Inoue T, Yamada M. Structure and expression of the gene responsible for the triplet repeat disorder, dentatorubral and pallidoluysian atrophy (DRPLA) Nature Genetics. 1994;8:177–182. doi: 10.1038/ng1094-177. [DOI] [PubMed] [Google Scholar]
- Napoletano F, Occhi S, Calamita P, Volpi V, Blanc E, Charroux B, Royet J, Fanto M. Polyglutamine atrophin provokes neurodegeneration in Drosophila by repressing fat. The EMBO Journal. 2011;30:945–958. doi: 10.1038/emboj.2011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newfeld SJ, Mehra A, Singer MA, Wrana JL, Attisano L, Gelbart WM. Mothers against dpp participates in a DDP/TGF-beta responsive serine-threonine kinase signal transduction cascade. Development. 1997;124:3167–3176. doi: 10.1242/dev.124.16.3167. [DOI] [PubMed] [Google Scholar]
- Oh H, Slattery M, Ma L, Crofts A, White KP, Mann RS, Irvine KD. Genome-wide association of yorkie with chromatin and chromatin-remodeling complexes. Cell Reports. 2013;3:309–318. doi: 10.1016/j.celrep.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagans S, Ortiz-Lombardía M, Espinás ML, Bernués J, Azorín F. The Drosophila transcription factor tramtrack (TTK) interacts with Trithorax-like (GAGA) and represses GAGA-mediated activation. Nucleic Acids Research. 2002;30:4406–4413. doi: 10.1093/nar/gkf570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panin VM, Papayannopoulos V, Wilson R, Irvine KD. Fringe modulates Notch-ligand interactions. Nature. 1997;387:908–912. doi: 10.1038/43191. [DOI] [PubMed] [Google Scholar]
- Philip P, Boija A, Vaid R, Churcher AM, Meyers DJ, Cole PA, Mannervik M, Stenberg P. CBP binding outside of promoters and enhancers in Drosophila melanogaster. Epigenetics & Chromatin. 2015;8:48. doi: 10.1186/s13072-015-0042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poux S, Melfi R, Pirrotta V. Establishment of polycomb silencing requires a transient interaction between PC and ESC. Genes & Development. 2001;15:2509–2514. doi: 10.1101/gad.208901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saburi S, Hester I, Goodrich L, McNeill H. Functional interactions between fat family cadherins in tissue morphogenesis and planar polarity. Development. 2012;139:1806–1820. doi: 10.1242/dev.077461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, McNeill H. Regulation of long-range planar cell polarity by Fat-Dachsous signaling. Development. 2013;140:3869–3881. doi: 10.1242/dev.094730. [DOI] [PubMed] [Google Scholar]
- Shimojima T, Okada M, Nakayama T, Ueda H, Okawa K, Iwamatsu A, Handa H, Hirose S. Drosophila FACT contributes to hox gene expression through physical and functional interactions with GAGA factor. Genes & Development. 2003;17:1605–1616. doi: 10.1101/gad.1086803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Current Biology. 2006;16:2081–2089. doi: 10.1016/j.cub.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Soeller WC, Poole SJ, Kornberg T. In vitro transcription of the Drosophila engrailed gene. Genes & Development. 1988;2:68–81. doi: 10.1101/gad.2.1.68. [DOI] [PubMed] [Google Scholar]
- Tabata T, Schwartz C, Gustavson E, Ali Z, Kornberg TB. Creating a Drosophila wing de novo, the role of engrailed, and the compartment border hypothesis. Development. 1995;121:3359–3369. doi: 10.1242/dev.121.10.3359. [DOI] [PubMed] [Google Scholar]
- Tanimoto H, Itoh S, ten Dijke P, Tabata T. Hedgehog creates a gradient of DPP activity in Drosophila wing imaginal discs. Molecular Cell. 2000;5:59–71. doi: 10.1016/S1097-2765(00)80403-7. [DOI] [PubMed] [Google Scholar]
- Tomlinson A, Mavromatakis YE, Struhl G. Three distinct roles for notch in Drosophila R7 photoreceptor specification. PLoS Biology. 2011;9:e1001132. doi: 10.1371/journal.pbio.1001132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truax AD, Greer SF. ChIP and Re-ChIP assays: investigating interactions between regulatory proteins, histone modifications, and the DNA sequences to which they bind. Methods in Molecular Biology. 2012;809:175–188. doi: 10.1007/978-1-61779-376-9_12. [DOI] [PubMed] [Google Scholar]
- Tsachaki M, Sprecher SG. Genetic and developmental mechanisms underlying the formation of the Drosophila compound eye. Developmental Dynamics. 2012;241:40–56. doi: 10.1002/dvdy.22738. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Becker PB, Wu C. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature. 1994;367:525–532. doi: 10.1038/367525a0. [DOI] [PubMed] [Google Scholar]
- Wang L, Charroux B, Kerridge S, Tsai CC. Atrophin recruits HDAC1/2 and G9a to modify histone H3K9 and to determine cell fates. EMBO Reports. 2008;9:555–562. doi: 10.1038/embor.2008.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Rajan H, Pitman JL, McKeown M, Tsai CC. Histone deacetylase-associating atrophin proteins are nuclear receptor corepressors. Genes & Development. 2006;20:525–530. doi: 10.1101/gad.1393506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehn A, Campbell G. Genetic interactions among scribbler, atrophin and groucho in Drosophila uncover links in transcriptional repression. Genetics. 2006;173:849–861. doi: 10.1534/genetics.105.055012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willecke M, Hamaratoglu F, Kango-Singh M, Udan R, Chen CL, Tao C, Zhang X, Halder G. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Current Biology. 2006;16:2090–2100. doi: 10.1016/j.cub.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Xiao H, Sandaltzopoulos R, Wang HM, Hamiche A, Ranallo R, Lee KM, Fu D, Wu C. Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Molecular Cell. 2001;8:531–543. doi: 10.1016/S1097-2765(01)00345-8. [DOI] [PubMed] [Google Scholar]
- Yang SA, Wang WD, Chen CT, Tseng CY, Chen YN, Hsu HJ. FOXO/Fringe is necessary for maintenance of the germline stem cell niche in response to insulin insufficiency. Developmental Biology. 2013;382:124–135. doi: 10.1016/j.ydbio.2013.07.018. [DOI] [PubMed] [Google Scholar]
- Zhang S, Xu L, Lee J, Xu T. Drosophila atrophin homolog functions as a transcriptional corepressor in multiple developmental processes. Cell. 2002;108:45–56. doi: 10.1016/S0092-8674(01)00630-4. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Feng J, Pan C, Lv X, Wu W, Zhou Z, Liu F, Zhang L, Zhao Y. Atrophin-Rpd3 complex represses hedgehog signaling by acting as a corepressor of CiR. The Journal of Cell Biology. 2013;203:575–583. doi: 10.1083/jcb.201306012. [DOI] [PMC free article] [PubMed] [Google Scholar]