Abstract
The genetic code, which defines the amino acid sequence of a protein, also contains information that influences the rate and efficiency of translation. Neither the mechanisms nor functions of codon-mediated regulation were understood. The prevailing model was that slow translation of codons decoded by rare tRNAs reduces efficiency. Recent genome-wide analyses have clarified three issues. Specific codons and codon combinations do modulate ribosome speed and facilitate protein folding. However, tRNA availability is not the sole determinant of rate, rather interactions between adjacent codons and wobble base pairing are key. One mechanism linking translation efficiency and codon use is that slower decoding is coupled to reduced mRNA stability. tRNA supply mediates biological regulation: changes in tRNA amounts facilitate cancer metastasis.
Keywords: Synonymous Codons, Codon Pair, Translation, mRNA decay, Protein Folding, Ribosome Profiling
Synonymous codon choice affects translation
Translation elongation is a major determinant of the composition of the proteome, affecting the amounts of each protein [1–3], the errors within each protein [4–10], and protein folding [11–16]. During translation elongation, each triplet nucleotide codon (see Glossary) in mRNA is decoded in the A-site of the ribosome by interactions with the anticodon of its cognate tRNA (aminoacyl or charged tRNA), resulting in insertion of an amino acid, followed by a precise three base translocation of the mRNA (and tRNA) to maintain the reading frame (illustrated in Figure 1). As elaborated below, translation elongation is influenced by the choice of synonymous codons, which specify insertion of the same amino acid, but differ in their decoding properties.
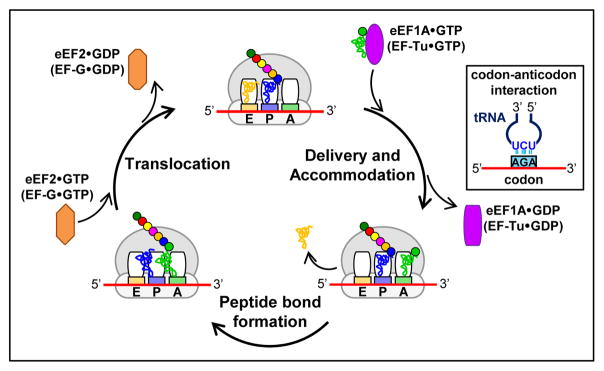
Figure 1. Schematic of translation elongation.
The top diagram shows an elongating ribosome, including the 3 sites for tRNA binding (A, P, and E) that span the large and small ribosomal subunits; the nascent polypeptide (which is attached to the P site tRNA and exits through the exit tunnel in the large subunit); and the mRNA (which enters and exits through the small subunit, moving 5’ to 3’). Bacteria specific components are shown in parentheses. First, addition of a new amino acid begins with the delivery, recognition and accommodation of an aminoacyl-tRNA into the A site of the ribosome. In this step, GTP-bound elongation factor (eEF1A GTP in eukaryotes or EF-Tu GTP in bacteria) delivers an aminoacyl-tRNA to the A site of the ribosome, where base pairing interactions between the anticodon of the tRNA and the 3 bases of the codon in the mRNA trigger hydrolysis of GTP, release of the deacylated tRNA from the E site of the ribosome, release of the GDP-bound elongation factor and further accommodation of the tRNA (a second proofreading step that depends upon codon-anticodon base pairing). Next, the addition of the amino acid to the growing polypeptide requires peptide bond formation, which involves close approximation of the 3’ ends of the aminoacyl and the peptidyl site tRNAs in the peptidyl transferase center of the large subunit, and large scale movement of the ribosomal subunits relative to each other to form a hybrid state. Finally, to complete the process, translocation of the mRNA (with its associated tRNAs) involves another GTP-bound elongation factor (eEF2 GTP in eukaryotes or EF-G GTP in bacteria), and large scale ribosome movement to restore the classical state. This process is repeated until the ribosome encounters a termination codon.
There is a compelling case that synonymous codon choice modulates translation elongation and translation efficiency [17–24]. Synonymous codons differ from each other in their relative use in the genome, in the abundance of tRNAs to decode them, and in the requirement of some codons for wobble interactions (non-Watson-Crick base pairing) between the third base of the codon and the first base of the tRNA anticodon [17, 24–27]. A subset of codons, called optimal codons, are decoded by abundant tRNAs, are efficiently translated, and are used nearly exclusively in many highly expressed genes in yeast and Escherichia coli [17, 18, 21, 23, 28]. Moreover, the influence of codon choice on translation efficiency is underscored by genome-wide correlations between codon use and translation efficiency [29–34], and by numerous examples in which recoding genes with optimal or suboptimal codons changes expression as predicted [32, 35–41].
However, despite more than 30 years of analysis, the mechanism(s) underpinning codon-mediated effects on translation are not well understood. The principal ideas have been that rate of translation elongation at different codons depends upon the supply of tRNA [18, 21, 34, 41–46], and that slower translation elongation rates reduce translation efficiency. Thus, it is generally thought that reduced translation rates and yield are due to an accumulation of small effects from individual codons decoded slowly by rare tRNAs. There were numerous indications that this model did not fit the facts including unpredictable effects of changing coding sequence on expression [37]. The rate-limiting factors in translation elongation were unknown, due in part to a lack of information about specific codons or codon combinations that either slow translation or reduce translation efficiency. Furthermore, it was not understood how differences in rates of translation elongation bring about reduced protein expression nor why suboptimal codon use is important for health of the organism. Recent work using newly developed high throughput methods has shed light on these questions.
Codon-dependent differences in elongation rate are due to wobble, tRNA and gene position
One impediment to understanding how codon choice affects translation has been the inability to distinguish effects of individual codons on the rate of translation elongation, an essential piece of information in the deduction of rate-limiting factors. Differences in the rates of decoding were detected in some early studies in E. coli in a variety of assays [47–50]. For instance, a three-fold difference in translation rates of Glu GAA and GAG codons, which are decoded by a single isoacceptor tRNA, was discerned by determining the in vivo rate of translation of lacZ with 30–60 codon inserts of GAA or GAG, (using a pulse-chase with [35S]methionine and examination of synthesis of radioactive β-galactosidase protein) [51]. However, these assays relied upon analysis of codons in a single position in a single gene, and upon non-natural constructs; thus, there was no general conclusion about differences in rates of decoding individual codons within native genes.
Ribosome profiling ushered in a new age by enabling genome-wide analysis of translation at single codon resolution [3]. In ribosome profiling, the sequence of a ribosome-protected mRNA fragment yields the identity of the codon in the ribosomal A and P sites, and the distribution of ribosome-protected fragments across a gene returns information on the relative time the ribosome spends at each codon in the gene (Box 1). By extension, a transcriptome-wide landscape of relative ribosome occupancy at each codon (which requires normalization by mRNA abundance) yields thousands of examples of decoding of each codon. Thus, ribosome profiling is theoretically a robust method to assess effects of individual codons on the rate of translation elongation.
Box 1. Ribosome profiling.
Ribosome profiling, which is based upon protection of mRNA sequences by the ribosome, provides nucleotide resolution of the locations of translating ribosomes, from which one may determine the ribosome occupancy of a specific sequence (relative to its occupancy of other sequences within the same transcript) and the identity of the codons in the A, P and E sites (Figure I). Ribosome profiling has been used to map the translated portions of the genome, to compare relative translation rates of different genes, to identify sites of ribosome pausing, and to identify genes translated in specific cellular locations, such as the endoplasmic reticulum [130].
This information can theoretically be used to determine the relative rate of translation of specific sequences or codons, but there have been several technical impediments to fulfilling this promise (discussed in [130]). Of particular note with respect to analysis of codon effects are the complications that arise from attempting to obtain instantaneous cessation of translation using antibiotics, which result in well-documented biases in the data [52, 53]. Furthermore, complications have arisen in aligning footprints to infer the codons in the A, P and E sites of the ribosome because of the non-uniform sizes of ribosome-protected fragments sizes (22–28 nucleotides in eukaryotes [54, 55] and 15–45 nucleotides in bacteria [53, 131]). Recent studies have addressed these issues [52–56].
Figure I. Schematic of the steps in ribosome profiling.
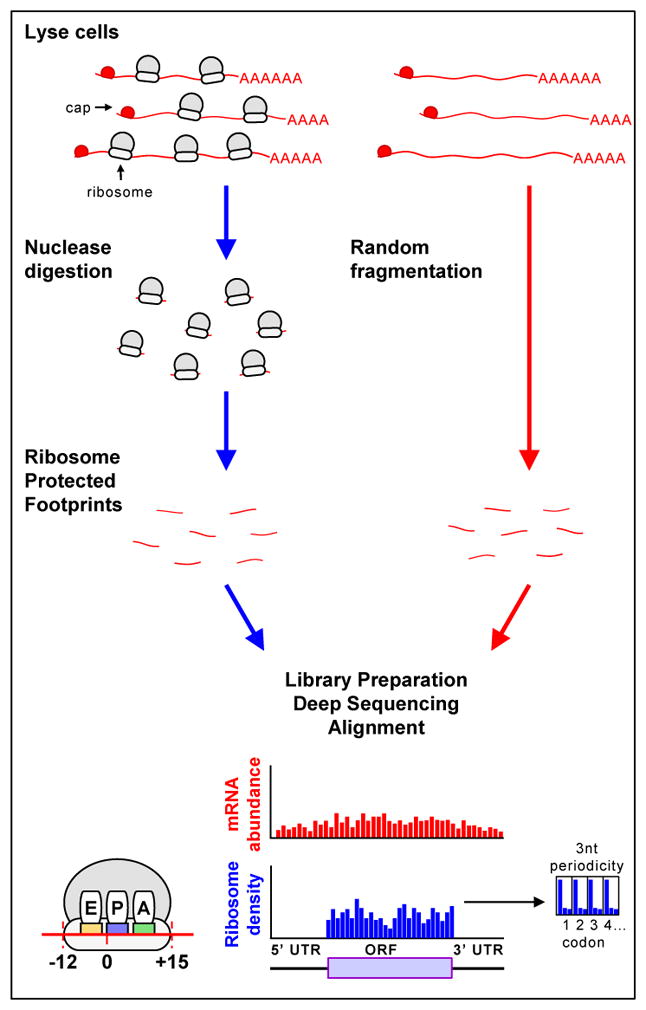
To obtain ribosome footprints, cell lysates, obtained from cells in which translation has been rapidly stopped, are treated with nucleases to digest unprotected RNAs. Ribosome-protected fragments of discrete sizes (22–28 nucleotides in eukaryotes and 15–45 nucleotides in bacteria) are isolated, converted to a library and subjected to deep sequencing. To obtain information on the identity and amounts of mRNA, mRNA is subjected to random fragmentation, isolated, converted to a similar library and sequenced. Sequences are aligned to the genome, such that codons in the A, P and E sites of the ribosome are distinguished. As expected, most ribosome footprints (blue) map to sequences that encode proteins (beginning with the AUG initiation codon and ending at the stop codon) and exhibit a 3 nucleotide periodicity (shown to right of ribosome reads); mRNA footprints (red) extend outside of the translated regions and do not exhibit this periodicity. UTR: untranslated region; ORF: open reading frame.
Early analysis of elongation rates at individual codons was complicated by at least two factors. First, rapid arrest of translation is essential to monitor in vivo rates, but the inclusion of antibiotics (to do this) distorts ribosome rates at specific codons. In yeast, addition of cycloheximide confounded analysis of individual codon rates, likely because elongation continues in the presence of the antibiotic at a slow rate with different individual rates for specific codons [52]. Similarly, arrest of translation in bacteria with chloramphenicol results in aberrant arrest with particular amino acids (Gly, Ser, Ala and Thr) in the E site [53]. To avoid biases that occur from antibiotic inclusion, current methods in yeast suggest flash freezing in liquid nitrogen [54–56]. Second, assessments of ribosome density at particular positions depend upon the size and alignment of the ribosome footprints (which vary in size from 15–45 nucleotides in bacteria and from 16 to 34 nucleotides for monosomes in yeast) [53–55]. In yeast, shorter ribosome footprints may correspond to ribosomes arrested at a different step in translation elongation than longer footprints [55] or on truncated mRNAs [54]. In E. coli, inclusion of all footprints revealed that pauses at Gly codons were responsible for a large portion of ribosome occupancy at Shine-Dalgarno motifs [53]. Nevertheless, several factors that influence the rates of elongation have been revealed in recent studies.
First, specific types of wobble decoding have been implicated in causing slow decoding in several different organisms. In both Caenorhabditis elegans and HeLa cells, decoding by G•U wobble interactions is slower than decoding by G-C Watson-Crick base pairing, based on comparisons among 16 NNU/NNC codon sets, each decoded by the same isoacceptor tRNA [58]. In yeast, decoding of two codons that use wobble interactions (I•A, Arg CGA and U•G, Pro CCG) was also particularly slow [55]. Decoding of the wobble codon is frequently facilitated by modifications of the interacting base in the anticodon of the tRNA (N34); tRNAs are highly post-transcriptionally modified with an average of 12.6 modifications per tRNA species in yeast [57]. Early studies demonstrated that failure to modify U34 in tRNAGlu resulted in differences in decoding rates of the two Glu codons in one context [49]. However, there has been no general examination of the effects of modification defects on decoding rates.
Second, tRNA amounts do correlate with codon-specific differences in ribosome occupancy, as expected based on the model in which translation elongation rates are limited by the supply of charged tRNA to fill the A site of the ribosome (see [59] for review). The expectations were that codons decoded by low abundance tRNAs are decoded slowly, which should result in higher ribosome occupancy at these codons. Indeed such inverse correlations between tRNA abundance (as assessed by one metric, tAI) and ribosome occupancy were seen in numerous studies in yeast, although the correlations were weak (see [52, 56]). Moreover, as we discuss below, changes in tRNA expression do affect biological regulation (see the discussion below on cell type specific changes in tRNA amounts [60–62]). Thus, there is a strong case that tRNA supply does regulate both translation elongation rate and translation efficiency. However, there is probably not a simple relationship between tRNA concentration and elongation rates. In one study, codon-specific rates (measured with ribosome profiling) were marginally affected by reducing the abundance of a particular tRNA to 30% and completely unaffected by overproducing a low copy tRNA [63]. Moreover, it remains unresolved precisely which properties actually limit tRNA supply: 1. the absolute concentration of tRNA, 2. the amount of tRNA relative to the number of codons that it decodes [64], or 3. the competition between the cognate tRNA and near cognate tRNAs [65–67].
Additional elements, from location within the gene to other local features, influence rates of decoding of individual codons [56]. Each of the 61 codons appears to be translated more slowly in the first 200 codons than in the remainder of the gene, based on an average 33% increase in ribosome protected fragments in this region [56]. In addition, codons are translated more rapidly if they are located within regions that form protein secondary structures, rather than inter-domain regions, and more slowly if they lie immediately downstream of polybasic regions [56]. These results underscore some of the complexities in understanding the regulation of translation by codon choice.
Specific codon pairs modulate the rate and efficiency of translation
Understanding how codons regulate translation efficiency has been challenging because it has been difficult to pinpoint specific codons or codon combinations that reduce translation efficiency. Changes in codon use affect mRNA structure, as well as both the rate and accuracy of translation elongation, thus complicating the analysis. For instance, the analysis of large libraries encoding multiple synonymous variants of a single assayable gene [68, 69] revealed that selection against strong mRNA secondary structures in the 5’ end is a major determinant of high expression, that is codon choices resulting in stronger structures inhibit expression. Indeed, genome scale analyses in both E. coli and yeast corroborate this point, since mRNA structure near the N-terminus is generally weak, which is likely to promote translation initiation [34]. However, a systematic analysis of effects of each codon on expression in the yeast Saccharomyces cerevisiae demonstrated that Arg CGA codons are very poorly decoded due to I·A wobble decoding, finding that adjacent CGA codons are much more potent inhibitors than separated CGA codons [70].
The effects of codons on translation are almost certainly dependent upon additional parameters, such as interactions with adjacent codons, or location in the gene [34, 64, 70–75] (reviewed in [76]). The idea that a codons effects were modulated by their adjacent nucleotides (or codons) was proposed coincidentally with the recognition that codon use might affect gene expression [17, 21, 77, 78], when it was noted that suppression of nonsense and missense codons is influenced by the surrounding sequences, a phenomenon called codon context [79, 80]. Soon thereafter, Gutman and Hatfield noted that codon pair use in E. coli is biased and correlates with expression levels [72]. Codon pair bias exists in many organisms in all three kingdoms [81, 82], but its role in expression was not clear [81], since some codon pairs are avoided in all three kingdoms (for instance nnUAnn codon pairs to decrease the likelihood of forming an out-of-frame stop codon) [82]. The observation that recoding viral genes with underrepresented codon pairs resulted in virus attenuation in mice [83] suggests that use of such pairs is deleterious to expression although the nature of their effects remains to be deciphered [84, 85]. In general, considerably less work has focused on the possibility that codon combinations or codon contexts are key to regulating translation elongation.
Effects of codons, codon context and codon combinations on the rate of translation elongation were directly examined using the his attenuator system in Salmonella enterica to measure relative translation speeds in vivo [86]. In this system, rapid translation of a leader polypeptide results in formation of a structure required for transcription termination (the attenuator) and a failure to translate the downstream genes, while slow translation of the leader polypeptide results in the absence of the attenuator and efficient translation of the downstream genes. Examination of the effects of all 64 codons upstream and downstream of a Ser UCA codon, indicated that the rate of translation depended upon both the identity and position of the neighboring codon [86]. Moreover, slow translation was also mediated by Pro-Pro dipeptides (known to mediate ribosome stalling) [87, 88], by codon combinations that base pair with the Shine-Dalgarno sequence, and by rare Arg-Arg codon pairs [86].
The idea that codon pairs exert effects on translation that are distinct from the effects of the individual codons in the pair was established with the identification of 17 inhibitory codon pairs in the yeast S. cerevisiae. These 17 inhibitory codon pairs were implicated in low expression in a high throughput screen of more than 35,000 GFP variants in which 3 adjacent codons were randomized [89]. The codon pair, rather than the six base sequence, the individual codons in the pair, or the dipeptide was responsible for low expression, since reduced GFP was only associated with variants in which both codons in the inhibitory pair were present, in-frame and adjacent. Inhibitory effects of these pairs were recapitulated by analysis of individual GFP variants, each compared to a synonymous variant with an optimal pair (a change of 2 nucleotides). The effects of these codon pairs were due to translation defects since, in 11/12 cases tested, their expression defects were suppressed either by overproduction of their native tRNA or by expression of a non-native tRNA engineered with an anticodon that forms W-C base pairs with all 3 bases of the codon and thus corrects normal wobble decoding of the codon [89].
Analysis of the properties of inhibitory pairs clarifies several aspects of the mechanisms underpinning codon-mediated effects on translation (Figure 2, Key Figure). First, many inhibitory pairs are translated very slowly in native yeast mRNAs [89], based on analysis of existing ribosome profiling data [90], confirming the idea that slow decoding is linked to inefficient translation. Second, codon-anticodon interactions in both positions are involved in inefficient translation, since 15 inhibitory pairs contain at least 1 codon that is decoded via I•A or U•G wobble interactions and inhibition was suppressed by correcting wobble interactions at either 5’ or 3’ codons (see above) [89]. Third, these pairs are likely to act as discrete regulatory signals, since the magnitude of their effects are large. Fourth, the order of codons in the pair is required for both inhibition and for slow decoding, implying that codon pair effects differ from effects of individual codons. These results challenge the notion that elongation is limited solely by availability of charged tRNA to fill the A site of the ribosome. Clearly, some interplay between adjacent sites in the ribosome is able to limit translation, but it is unknown at what step(s) in translation this interplay operates or how it works.
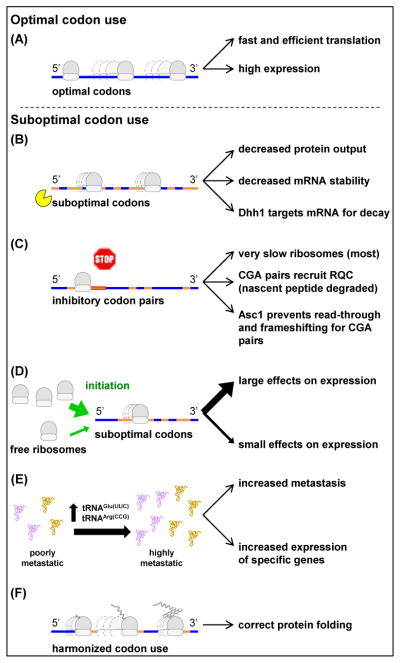
Figure 2. The consequences of codon choice.
Effects of different types of codon choice. (A) Genes encoded primarily with optimal codons (blue) are rapidly and efficiently translated. (B) Genes encoded primarily with suboptimal or slowly decoded codons (orange) are inefficiently translated; their mRNAs exhibit decreased stability, mediated in yeast by Dhh1 protein. (C) Inhibitory codon pairs (red box) are very slowly decoded, reduce protein output significantly, and sometimes recruit quality control machinery. (D) The efficiency of translation initiation influences the magnitude of the effects of suboptimal codon use on expression (ratio of expression of synonymous variants encoding the same protein). (E) A change in the amount of two specific tRNAs drives cancer metastasis and modulates expression of specific genes. (F) Harmonized codon use, a combination of rapidly and slowly decoded codons, can facilitate co-translational protein folding to obtain biologically active molecules.
Mechanisms coupling codon use to low expression
The mechanisms by which codon use affects the rate and output of translation are not completely understood, although information has emerged on three types of responses. First, suboptimal codon use drives mRNA decay [39]. Second, suboptimal codon use may feedback and limit initiation [36, 91]. Third, suboptimal codon use sometimes elicits a response from a quality control system that targets the nascent polypeptide and/or the mRNA for degradation [92, 93]. The evidence for each of these is discussed below, but much remains to be explained, including when each mechanism applies and how/if they impact each other.
Codon use modulates mRNA decay
The inference that codon usage and mRNA stability are linked was enunciated as early as a 1987 study of codon effects in the yeast PGK1 gene [20] and was reinforced in subsequent studies of the effects of rare codons on stability of MATα1 and PGK1 mRNAs [94, 95]. Indeed, there are numerous links between mRNA decay and translation; mRNA decay occurs in response to a variety of aberrant translation events (nonsense mediated decay, no-go decay and nonstop decay) [93]. However, since many factors regulate mRNA stability, from RNA binding proteins [96] to miRNAs [97] to RNA structure [98], it was unclear to what extent decay is generally dependent upon the translation rate and it was unknown how such decay might be executed.
Codon usage in yeast was recently implicated as the major determinant of mRNA stability, based upon a genome-wide analysis of mRNA stability which revealed a strong correlation between mRNA half-life and optimality of codons used to encode the gene (Figure 2) [39]. Moreover, synonymous substitutions in several genes resulted in the predicted change in mRNA stability, providing compelling evidence that the codon composition of genes dictate differences in their mRNA half-lives [39]. This correlation has been extended to bacteria [99] and to vertebrates [100, 101], including humans [102]. Recent analyses of the post-fertilization, maternal to zygotic transition in zebrafish implicated translation of specific codons and amino acids in the rapid degradation of maternal decay mRNAs [100, 101]. Strong evidence against specific sequence determinants of decay was provided by one study showing that insertion of a single nucleotide simultaneously alters codon optimality and decay rate of a gene [100]. While these findings establish a role for codon use in modulating mRNA decay, exactly how mRNA decay is triggered remained obscure.
The idea that some 5’ to 3’ mRNA decay closely tracks translation is strongly supported by a new method that monitors RNAs with a 5’ phosphate (expected of degradation products of the exonuclease Xrn1) [103]. These degradation intermediates exhibited the hallmarks of translation: a 3’ periodicity of products (which was used to estimate that ~34% of 5’ to 3’ degradation intermediates were co-translational) and an accumulation of intermediates at ribosome pause sites, such as stop codons, Arg CGA and Pro CCG [103]. Thus, decay intermediates are not only associated with ribosomes, but are limited by the speed of decoding, corroborating that decay and translation are intimately coupled.
Dhh1 protein was recently implicated in codon-mediated regulation as a link between mRNA metabolism and ribosome speed (Figure 2) [104]. Dhh1, a highly conserved DEAD-box helicase protein, was previously known to promote decapping, to repress translation of specific messages and to facilitate P-body formation [105]. Dhh1 was a good candidate to effect codon-mediated regulation, because it is highly abundant, acts during translation elongation and binds preferentially to slowly translating ribosomes [106]. Indeed, deletion of DHH1 results in the selective increase in the half-life of mRNAs with suboptimal codons, as evidenced by both analysis of individual genes and genome-wide correlations with codon content [104]. Dhh1 protein preferentially associates with mRNAs enriched in suboptimal codons, increasing the number of ribosomes on these mRNAs and on specific codons [104]. In addition, Dhh1 is more effective at promoting decay on mRNAs with a greater number of slow moving ribosomes, suggesting that Dhh1 both senses and affects the ribosome speed [104].
Codon use may modulate translation initiation
Codon effects on mRNA stability can only partly explain the connection between codon usage and protein output, since there are numerous examples in which translation efficiency (the protein output per mRNA) depends upon codon usage [20, 107]. Furthermore, differences in translation efficiency as a function of codon usage are recapitulated in a genome wide analysis in zebrafish in which translation efficiency is assessed by ribosome footprints per mRNA [100]. Similarly in yeast, suboptimal codon use results in lower protein output even after normalizing for loss of mRNA [39]. Furthermore, the location of suboptimal codons is a major factor in their effects on decay, but not protein output; mRNA decay preferentially affects mRNAs with suboptimal codons closer to the 3’ end [101, 104], but protein output is identical [104]. Thus, there must be additional mechanisms to couple elongation rate and protein output.
The long-standing idea has been that slow elongation is usually coupled to a reduced efficiency of translation initiation, consistent with evidence that free ribosomes are generally limiting [108–110] and thus, that initiation is frequently the rate-limiting step in translation. The mechanism by which such coupling works is unknown, although there is clear evidence of a link between initiation rates and elongation regulation. For instance, slow decoding caused by suboptimal codon use in yeast [36] or by polyproline motifs in Salmonella enterica [91] results in large effects on protein expression if and only if translation initiation is efficient (Figure 2). Translation elongation across slow codons could impair translation initiation, either because slow elongation limits clearance of the AUG start codon [36] or limits clearance through the stall [91]. Alternatively, slow elongation could reduce the availability of terminating ribosomes, which themselves may initiate translation more efficiently than the pool of free ribosomes [111]. Thus, elongation and initiation may be coupled, particularly for genes at which initiation is normally efficient, although there is much more to learn about this process.
Ribosomal protein RACK1/Asc1 and the Ribosome quality control complex mediate effects of some codon combinations
Cells contain a number of specific quality control systems to mitigate the downstream effects of translation of aberrant mRNAs, for instance those lacking a stop codon or containing a sufficiently strong structure to interfere with elongation [93]. In addition to the responses described above, there is evidence that translation of CGA codon repeats in yeast evokes responses from some cellular proteins that deal with aberrant translation (Figure 2) [92]. The yeast homolog of human RACK1, Asc1, is a small ribosomal subunit protein [112] located near the mRNA exit tunnel [113], is required for nascent polypeptide-dependent translation arrest [114] and is proposed to recruit the ribosome quality control (RQC) complex to stalled ribosomes in yeast by an unknown mechanism [115]. Asc1 is implicated in CGA-mediated inhibition of translation in S. cerevisiae: deletion of ASC1 results in both improved read-through of and frameshifting at CGA codon repeats [74]. Furthermore, Ltn1, an E3 ubiquitin ligase [116], which is a component of the RQC, targets the nascent polypeptide upstream of CGA codon repeats for degradation [92]. It is unknown if Asc1 and/or the RQC are generally involved in mediating effects of suboptimal codon use or of other strongly inhibitory codon pairs.
Genomic analyses of codon use reveal regulated changes for expression and accuracy
Most biological processes are facilitated by changes in gene expression, but it was unknown the extent to which changes in the efficacy of translation of synonymous codons contributes to such events. However, it was known that amounts of specific tRNAs vary in different human tissues, and that tRNA supply is correlated with codon use in those tissues, as expected if the supply of a tRNA modulates translation efficiency [60]. Furthermore, comparisons of tRNA amounts and codon use in 470 samples provided strong evidence of coordinated differences in tRNA supply and codon use between proliferating and differentiated cells (Figure 2) [61]. Two additional observations imply that these coordinated changes regulate important aspects of cell state. Regulatory sequences associated with the tRNA genes differ between proliferating and differentiated cells, suggesting that differences in tRNA amounts between these cell types are most likely due to differences in tRNA gene regulation [61]. The differences in codon usage between genes expressed in proliferating tissues and those expressed in differentiated tissues in humans are also observed in other vertebrates, and in the fruit fly Drosophila melanogaster [61]. Thus, tRNA supply and demand are coordinated to facilitate and perhaps reinforce the cell state [61].
Changes in amounts of specific tRNAs and translation of their codons drive the metastatic progression in specific breast cancer cells (Figure 2) [62]. Comparison of tRNA amounts between 2 highly metastatic breast cancer cell lines and their poorly metastatic parental lines revealed increased amounts of two tRNAs, tRNAArg(CCG) and tRNAGlu(UUC) [62]. A reduction in the amount of these two tRNAs in the metastatic cells reduced their metastatic phenotypes (lung colonization capacity and invasiveness), while overproduction of the same two tRNAs in their poorly metastatic parents dramatically increased the same metastatic phenotypes [62]. Thus, tRNA amounts are directly linked to regulation of a biological function (cancer metastasis). Moreover, these changes in tRNA amounts, particularly tRNAGlu(UUC), are responsible for increased amounts of one protein, EXOSC2, that is important for metastasis: not only is expression of this gene specifically increased by tRNA overproduction, but expression of a derivative gene in which all cognate Glu codons are mutated to a synonymous codon is not affected [62]. These observations provide unequivocal evidence of the biological importance of changes in tRNA supply and demand.
There has been extensive discussion about the idea that synonymous codons may be selected for specific translational properties (speed versus accuracy) [117]. An elegant analysis of selection across 12 drosophilid species began by distinguishing the set of gene positions at which amino acid identity is completely conserved across the 12 species, inferring that the accuracy of amino acid insertion is critical at these conserved residues [118]. If so, this subset of positions at which the amino acid is essential are likely to be encoded by codons selected for accurate insertion of the correct amino acid. Surprisingly, a switch in the relative translational accuracy of sets of codons occurred across the Drosophila/Sophophora genus between sets of codons decoded by the same tRNA [118]. This switch is correlated with changes in modification of their tRNA anticodons [118]. Similar changes are found in codon usage in genes expressed during different developmental stages in D. melanogaster and also correlate well with changes in modification of the tRNA anticodons [118]. Thus, not only are specific synonymous codons selected for specific translational properties (speed versus accuracy), but the optimal codon for a specific property can change in a manner coordinated with biological regulation.
One of the perplexing issues has been the overall biological role of suboptimal codons, although there are individual examples in which use of suboptimal codons is required for function [119, 120]. To assess the general importance of suboptimal codon choice, one study [121] attempted to replace the 13 most underused codons in E. coli in each of 42 genes. Although most substitutions were successful, most also resulted in reduced fitness. In a parallel approach, the 123 rare Arg codons (AGG and AGA) were replaced in all essential genes in E. coli [122]. Replacement of 110 rare Arg codons was straightforward, whereas the remaining 13 required alternative strategies which implicated mRNA secondary structure near the N-terminus and ribosome-binding sequences as determinants of codon choice. However, other parameters may constrain the choice of rare Arg codons at particular locations, since the resulting strain also exhibited slow growth and carried over 400 spontaneous non-synonymous mutations [122]. Thus, suboptimal codons can have important and substantive effects, but most of these remain to be discovered.
Synonymous codon choice regulates protein folding
The idea that synonymous codon choice influences protein folding was proposed as early as 1987 [123] and was supported by the observation that sequences encoding the inter-domain regions of 37 proteins were generally enriched in slowly translated codons [124]. Subsequent studies provided evidence that changing codon usage can predictably alter folding [12] and may affect protein activity [119, 120, 125]. However, we did not fully comprehend either the prevalence or functional consequences of codon constraints on protein folding.
The idea that has emerged from recent studies is that codon choice precisely modulates ribosome elongation rates to obtain biologically active molecules (Figure 2) [126–129]. The finding that sequences that interact with signal recognition particle (SRP) are followed by a region of low translation efficiency approximately 40 codons downstream (about the length of the ribosome protein exit tunnel) provided compelling evidence for this idea [126]. Moreover, this region of low translation efficiency is evolutionarily conserved among yeast, suggesting that these sequences serve functionally important roles, presumably in slowing translation to allow sufficient time for SRP binding [126].
The general link between translation rates and protein folding is underscored by studies showing that modulating local translation rates impacts protein homeostasis [127] as well as folding of specific proteins [128, 129]. For instance, inefficient decoding by a tRNA with an aberrant U34 modification reduces the rates of translation at specific codons, and results in protein misfolding and aggregation (proteotoxic stress) [127]. Conversely, increasing the speed of decoding by optimizing codon usage for Neurospera crassa in firefly luciferase results in protein with a lower specific activity and altered response to trypsin digestion [128]. Local translation rates within a 50 codon segment of luciferase [128] are crucial for correct folding of the native protein in vivo. Similarly, analysis of expression in E. coli of two synonymous variants of the mammalian eye-lens protein gamma-B crystalline demonstrated that the variant recoded to mimic local translation rates of the host organism (B. Taurus), yielded more protein that was correctly folded compared to the native B. Taurus sequence [129]. Moreover, the authors of this study directly demonstrated that folding differences occur co-translationally using real-time FRET and proposed that genes are encoded with a combination of fast and slow codons (harmonized codon use) to aid protein folding. Thus, modulating translation rates to obtain biologically active molecules appears to be a general function of codon choice and is prevalent at many sites in many proteins. It is still unknown if the sole or primary function of suboptimal codons is to regulate protein folding. It is also unclear whether or not inhibitory codon pairs are just stronger, more site-specific regulators of protein folding or have additional roles.
Concluding Remarks
The choice of synonymous codons used to encode a polypeptide affects the rate at which an mRNA is translated and the amount of protein produced per mRNA. Recent work has shed light on the mechanisms by which codon use modulates translation, specifically revealing that translation elongation rates are codon-dependent, that specific codon pairs are potent inhibitors of translation output and significantly slow ribosomes, as well as that codon use is tightly coupled to mRNA stability and decay. In addition, there is now compelling evidence that biological regulation is driven by changes in the efficacy of translating specific codons: increased expression of two tRNAs is necessary and sufficient to drive metastasis of two cancer cell lines [62]. Finally, based on evidence that codon choice affects nascent protein folding, the view emerges of genes encoded with codons that modulate local rates of elongation to obtain a correctly folded protein.
Much remains to be learned about specific mechanisms that slow the ribosome, about the coupling between elongation and output, as well as about the full spectrum of codon functions (see Outstanding Questions). What are the primary factors that limit the rate of translation elongation: the absolute concentration of an individual tRNA (or charged tRNA), the concentration of a tRNA relative to its use in translation, or the competition between cognate and near cognate tRNAs? To what extent do the interactions between codons (or adjacent nucleotides) modulate the rate of translation: are all or most codon pairs or triplets unique signals whose effects on translation elongation cannot be predicted based on their component codons? How do tRNA modifications and changes in these modifications affect translation speed, accuracy and output? How does a reduction in translation elongation result in decreased protein expression? What aspect of slow decoding is recognized? Why are some genes encoded with a large fraction of suboptimal codons: to maintain a constant supply of the mRNA with low protein output, to optimize production of a correctly folded protein, or to mediate protein-protein interactions? Are there a large number of biological regulatory events in which changes in tRNA supply or codon use are key to the transitions?
Outstanding Questions.
What limits the rate of translation elongation?
Is control of tRNA abundance or efficacy generally employed as a biological regulatory mechanism?
What are the functions of suboptimal synonymous codons and strongly inhibitory pairs? Are these functions different from each other?
How are translation elongation rates and translation efficiency connected? Is there a specific mechanism coupling translation elongation and initiation?
How many distinct systems mediate effects of suboptimal codons or codon pairs? Is there overlap or are they mutually exclusive?
At which steps of translation, does codon pair mediated inhibition occur? How does the interplay between adjacent sites in the ribosome occur?
How does Dhh1 sense codon use to preferentially degrade suboptimal codon transcripts?
Table I.
Comparison of different methods to prepare mRNA
| Method | Relative Amounts (r2) | Position dependence (3’ Bias) | ORF Length Bias |
|---|---|---|---|
| Dynabeads oligo(dT)25 (Life Technologies) | 0.85 | Strong | Strong |
| RiboMinus Yeast Transcriptome Isolation (Life Technologies) | 0.87 | Slight | Slight |
| Ribo-Zero Yeast Magnetic Gold (Epicenter) | 0.98 | Slight | None |
Box 2. High throughput measurements of mRNA abundance.
Accurate measurements of mRNA amounts are essential for many applications, including the precise determination of translation efficiencies and of mRNA half-lives. Two methods are commonly used to prepare libraries enriched for mRNA, the first is to select for polyA+ RNAs using oligo(dT) beads, and the second is to deplete rRNAs from the total RNA sample using hybridization. Poly(A) selection can lead to omission of mRNAs with short or no poly(A) tails, mRNAs that are endonucleolytically cleaved, or mRNAs that are stably deadenylated [55, 56]. Of course, rRNA depletion methods can lead to inclusion of the same types of mRNAs, and it is unclear a priori which of these approaches will provide more accurate information on the “active” pool of mRNAs. In one study, half-lives derived from oligo(dT) selected mRNAs were generally shorter and specifically skewed for some mRNAs compared to half-lives derived from rRNA-depleted mRNAs [39].
Three different methods of mRNA selection were evaluated by comparing RNA-seq results from mRNA prepared from identical samples (Table I) [56]. The Ribo-Zero mRNA displayed the closest equivalency to the total RNA sample, based on three criteria: relative amounts of individual messages, even distribution of reads across the coding sequence and equal representation of genes independent of length [56].
Trends.
Ribosome speed during elongation is modulated by codon choice, tRNA abundance and wobble decoding.
Codon pairs act as discrete signals that reduce expression and slow translation.
Codon usage modulates mRNA decay and Dhh1 preferentially targets mRNAs of low codon optimality for degradation.
Codon-mediated effects on translation rates facilitate co-translational protein folding.
Genome-wide and high throughput analyses of codon use and tRNAs have facilitated recent discoveries.
Glossary
- Aminoacyl-tRNA
a tRNA in which an amino acid is covalently attached to its 3’ end. Also referred to as charged tRNA.
- Anticodon
the three nucleotides in the anticodon loop of a tRNA that base pair with the codon in the mRNA. In the tRNA, these nucleotides are numbered 34, 35 and 36 in the 5’ to 3’ direction, with nucleotide 36 base pairing with nucleotide 1 of the codon. Thus, nucleotide 34 is responsible for wobble base pairing with nucleotide 3 of the codon.
- Codon
three adjacent nucleotides in an mRNA that specify insertion of a particular amino acid to the growing polypeptide chain.
- Cognate tRNA
the tRNA with an anticodon that forms complementary base pairs with the codon and is selected preferentially by the ribosome when decoding that codon.
- Isoacceptor tRNAs
the set of tRNAs that insert the same amino acid; they may interact with different codons.
- Near cognate tRNA
a tRNA that contains a single base mismatch between its anticodon and the codon in question.
- Peptidyl transferase center (PTC)
the location within the large ribosomal subunit at which a new peptide bond is formed, linking the nascent polypeptide to the next amino acid.
- Shine-Dalgarno
ribosomal binding site found in mRNAs in bacteria and archaea that directs translation initiation.
- Synonymous codons
different codons that specify insertion of the same amino acid to the growing polypeptide chain.
- Wobble decoding
interactions in the ribosome in which the tRNA anticodon forms Watson-Crick base pairs with the first two bases in the codon, but forms a non-Watson- Crick base pair between N34 of the tRNA and the third base in the codon. The wobble hypothesis was initially proposed by Francis Crick in 1966 [132].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Arava Y, et al. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2003;100:3889–94. doi: 10.1073/pnas.0635171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Godoy LM, et al. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–4. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- 3.Ingolia NT, et al. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–23. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–52. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kramer EB, Farabaugh PJ. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA. 2007;13:87–96. doi: 10.1261/rna.294907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kramer EB, et al. A comprehensive analysis of translational missense errors in the yeast Saccharomyces cerevisiae. RNA. 2010;16:1797–808. doi: 10.1261/rna.2201210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurland CG. Translational accuracy and the fitness of bacteria. Annu Rev Genet. 1992;26:29–50. doi: 10.1146/annurev.ge.26.120192.000333. [DOI] [PubMed] [Google Scholar]
- 8.Salas-Marco J, Bedwell DM. Discrimination between defects in elongation fidelity and termination efficiency provides mechanistic insights into translational readthrough. J Mol Biol. 2005;348:801–15. doi: 10.1016/j.jmb.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 9.Stansfield I, et al. Missense translation errors in Saccharomyces cerevisiae. J Mol Biol. 1998;282:13–24. doi: 10.1006/jmbi.1998.1976. [DOI] [PubMed] [Google Scholar]
- 10.Stoletzki N, Eyre-Walker A. Synonymous codon usage in Escherichia coli: selection for translational accuracy. Mol Biol Evol. 2007;24:374–81. doi: 10.1093/molbev/msl166. [DOI] [PubMed] [Google Scholar]
- 11.Kaiser CM, et al. The ribosome modulates nascent protein folding. Science. 2011;334:1723–7. doi: 10.1126/science.1209740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sander IM, et al. Expanding Anfinsen's principle: contributions of synonymous codon selection to rational protein design. J Am Chem Soc. 2014;136:858–61. doi: 10.1021/ja411302m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willmund F, et al. The cotranslational function of ribosome-associated Hsp70 in eukaryotic protein homeostasis. Cell. 2013;152:196–209. doi: 10.1016/j.cell.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang G, et al. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol. 2009;16:274–80. doi: 10.1038/nsmb.1554. [DOI] [PubMed] [Google Scholar]
- 15.Jacobson GN, Clark PL. Quality over quantity: optimizing co-translational protein folding with non-'optimal' synonymous codons. Curr Opin Struct Biol. 2016;38:102–10. doi: 10.1016/j.sbi.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodnina MV. The ribosome in action: Tuning of translational efficiency and protein folding. Protein Sci. 2016;25:1390–406. doi: 10.1002/pro.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bennetzen JL, Hall BD. Codon selection in yeast. J Biol Chem. 1982;257:3026–31. [PubMed] [Google Scholar]
- 18.dos Reis M, et al. Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res. 2004;32:5036–44. doi: 10.1093/nar/gkh834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duret L. tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes. Trends Genet. 2000;16:287–9. doi: 10.1016/s0168-9525(00)02041-2. [DOI] [PubMed] [Google Scholar]
- 20.Hoekema A, et al. Codon replacement in the PGK1 gene of Saccharomyces cerevisiae: experimental approach to study the role of biased codon usage in gene expression. Mol Cell Biol. 1987;7:2914–24. doi: 10.1128/mcb.7.8.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J Mol Biol. 1981;146:1–21. doi: 10.1016/0022-2836(81)90363-6. [DOI] [PubMed] [Google Scholar]
- 22.Ikemura T. Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting transfer RNAs. J Mol Biol. 1982;158:573–97. doi: 10.1016/0022-2836(82)90250-9. [DOI] [PubMed] [Google Scholar]
- 23.Man O, Pilpel Y. Differential translation efficiency of orthologous genes is involved in phenotypic divergence of yeast species. Nat Genet. 2007;39:415–421. doi: 10.1038/ng1967. [DOI] [PubMed] [Google Scholar]
- 24.Sharp PM, et al. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986;14:5125–43. doi: 10.1093/nar/14.13.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duret L, Mouchiroud D. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc Natl Acad Sci USA. 1999;96:4482–7. doi: 10.1073/pnas.96.8.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grantham R, et al. Codon catalog usage and the genome hypothesis. Nucleic Acids Res. 1980;8:r49–r62. doi: 10.1093/nar/8.1.197-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grantham R, et al. Codon catalog usage is a genome strategy modulated for gene expressivity. Nucleic Acids Res. 1981;9:r43–74. doi: 10.1093/nar/9.1.213-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharp PM, Li WH. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–95. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brockmann R, et al. Posttranscriptional expression regulation: what determines translation rates? PLoS Comput Biol. 2007;3:e57. doi: 10.1371/journal.pcbi.0030057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–41. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 31.Ishihama Y, et al. Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics. 2008;9:102. doi: 10.1186/1471-2164-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quax TE, et al. Differential translation tunes uneven production of operon-encoded proteins. Cell Rep. 2013;4:938–44. doi: 10.1016/j.celrep.2013.07.049. [DOI] [PubMed] [Google Scholar]
- 33.Tuller T, et al. Determinants of protein abundance and translation efficiency in S. cerevisiae. PLoS Comput Biol. 2007;3:e248. doi: 10.1371/journal.pcbi.0030248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tuller T, et al. Translation efficiency is determined by both codon bias and folding energy. Proc Natl Acad Sci USA. 2010;107:3645–50. doi: 10.1073/pnas.0909910107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burgess-Brown NA, et al. Codon optimization can improve expression of human genes in Escherichia coli: A multi-gene study. Protein Expr Purif. 2008;59:94–102. doi: 10.1016/j.pep.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 36.Chu D, et al. Translation elongation can control translation initiation on eukaryotic mRNAs. EMBO J. 2014;33:21–34. doi: 10.1002/embj.201385651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gustafsson C, et al. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22:346–53. doi: 10.1016/j.tibtech.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 38.Keppler-Ross S, et al. A new purple fluorescent color marker for genetic studies in Saccharomyces cerevisiae and Candida albicans. Genetics. 2008;179:705–10. doi: 10.1534/genetics.108.087080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Presnyak V, et al. Codon optimality is a major determinant of mRNA stability. Cell. 2015;160:1111–24. doi: 10.1016/j.cell.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quartley E, et al. Heterologous expression of L. major proteins in S. cerevisiae: a test of solubility, purity, and gene recoding. J Struct Funct Genomics. 2009;10:233–47. doi: 10.1007/s10969-009-9068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Welch M, et al. Design parameters to control synthetic gene expression in Escherichia coli. PLoS One. 2009;4:e7002. doi: 10.1371/journal.pone.0007002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elf J, et al. Selective charging of tRNA isoacceptors explains patterns of codon usage. Science. 2003;300:1718–22. doi: 10.1126/science.1083811. [DOI] [PubMed] [Google Scholar]
- 43.Novoa EM, et al. A role for tRNA modifications in genome structure and codon usage. Cell. 2012;149:202–13. doi: 10.1016/j.cell.2012.01.050. [DOI] [PubMed] [Google Scholar]
- 44.Parmley JL, Hurst LD. How do synonymous mutations affect fitness? Bioessays. 2007;29:515–9. doi: 10.1002/bies.20592. [DOI] [PubMed] [Google Scholar]
- 45.Rojiani MV, et al. Relationship between protein synthesis and concentrations of charged and uncharged tRNATrp in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:1511–5. doi: 10.1073/pnas.87.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharp PM, et al. Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens; a review of the considerable within-species diversity. Nucleic Acids Res. 1988;16:8207–11. doi: 10.1093/nar/16.17.8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bonekamp F, Jensen KF. The AGG codon is translated slowly in E. coli even at very low expression levels. Nucleic Acids Res. 1988;16:3013–24. doi: 10.1093/nar/16.7.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Curran JF, Yarus M. Rates of aminoacyl-tRNA selection at 29 sense codons in vivo. J Mol Biol. 1989;209:65–77. doi: 10.1016/0022-2836(89)90170-8. [DOI] [PubMed] [Google Scholar]
- 49.Kruger MK, et al. The modification of the wobble base of tRNAGlu modulates the translation rate of glutamic acid codons in vivo. J Mol Biol. 1998;284:621–31. doi: 10.1006/jmbi.1998.2196. [DOI] [PubMed] [Google Scholar]
- 50.Pedersen S. Escherichia coli ribosomes translate in vivo with variable rate. EMBO J. 1984;3:2895–8. doi: 10.1002/j.1460-2075.1984.tb02227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sorensen MA, Pedersen S. Absolute in vivo translation rates of individual codons in Escherichia coli The two glutamic acid codons GAA and GAG are translated with a threefold difference in rate. J Mol Biol. 1991;222:265–80. doi: 10.1016/0022-2836(91)90211-n. [DOI] [PubMed] [Google Scholar]
- 52.Hussmann JA, et al. Understanding biases in ribosome profiling experiments reveals signatures of translation dynamics in yeast. PLoS Genet. 2015;11:e1005732. doi: 10.1371/journal.pgen.1005732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mohammad F, et al. Clarifying the translational pausing landscape in bacteria by ribosome profiling. Cell Rep. 2016;14:686–94. doi: 10.1016/j.celrep.2015.12.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guydosh NR, Green R. Dom34 rescues ribosomes in 3' untranslated regions. Cell. 2014;156:950–62. doi: 10.1016/j.cell.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lareau LF, et al. Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. Elife. 2014;3:e01257. doi: 10.7554/eLife.01257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weinberg DE, et al. Improved ribosome-footprint and mRNA measurements provide insights into dynamics and regulation of yeast translation. Cell Rep. 2016;14:1787–99. doi: 10.1016/j.celrep.2016.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stadler M, Fire A. Wobble base-pairing slows in vivo translation elongation in metazoans. RNA. 2011;17:2063–73. doi: 10.1261/rna.02890211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24:1832–60. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Richter JD, Coller J. Pausing on polyribosomes: make way for elongation in translational control. Cell. 2015;163:292–300. doi: 10.1016/j.cell.2015.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dittmar KA, et al. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006;2:e221. doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gingold H, et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell. 2014;158:1281–92. doi: 10.1016/j.cell.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 62.Goodarzi H, et al. Modulated expression of specific tRNAs drives gene expression and cancer progression. Cell. 2016;165:1416–27. doi: 10.1016/j.cell.2016.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pop C, et al. Causal signals between codon bias, mRNA structure, and the efficiency of translation and elongation. Mol Syst Biol. 2014;10:770. doi: 10.15252/msb.20145524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pechmann S, Frydman J. Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat Struct Mol Biol. 2013;20:237–243. doi: 10.1038/nsmb.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chu D, et al. The role of tRNA and ribosome competition in coupling the expression of different mRNAs in Saccharomyces cerevisiae. Nucleic Acids Res. 2011;39:6705–14. doi: 10.1093/nar/gkr300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fluitt A, et al. Ribosome kinetics and aa-tRNA competition determine rate and fidelity of peptide synthesis. Comput Biol Chem. 2007;31:335–46. doi: 10.1016/j.compbiolchem.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zouridis H, Hatzimanikatis V. Effects of codon distributions and tRNA competition on protein translation. Biophys J. 2008;95:1018–33. doi: 10.1529/biophysj.107.126128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goodman DB, et al. Causes and effects of N-terminal codon bias in bacterial genes. Science. 2013;342:475–9. doi: 10.1126/science.1241934. [DOI] [PubMed] [Google Scholar]
- 69.Kudla G, et al. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–8. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Letzring DP, et al. Control of translation efficiency in yeast by codon-anticodon interactions. RNA. 2010;16:2516–28. doi: 10.1261/rna.2411710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boycheva S, et al. Codon pairs in the genome of Escherichia coli. Bioinformatics. 2003;19:987–98. doi: 10.1093/bioinformatics/btg082. [DOI] [PubMed] [Google Scholar]
- 72.Gutman GA, Hatfield GW. Nonrandom utilization of codon pairs in Escherichia coli. Proc Natl Acad Sci USA. 1989;86:3699–703. doi: 10.1073/pnas.86.10.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tuller T, et al. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141:344–54. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- 74.Wolf AS, Grayhack EJ. Asc1, homolog of human RACK1, prevents frameshifting in yeast by ribosomes stalled at CGA codon repeats. RNA. 2015;21:935–45. doi: 10.1261/rna.049080.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Irwin B, et al. Codon pair utilization biases influence translational elongation step times. J Biol Chem. 1995;270:22801–6. doi: 10.1074/jbc.270.39.22801. [DOI] [PubMed] [Google Scholar]
- 76.Brar GA. Beyond the triplet code: context cues transform translation. Cell. 2016;167:1681–1692. doi: 10.1016/j.cell.2016.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gouy M, Gautier C. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 1982;10:7055–74. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grosjean H, Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982;18:199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- 79.Bossi L, Ruth JR. The influence of codon context on genetic code translation. Nature. 1980;286:123–7. doi: 10.1038/286123a0. [DOI] [PubMed] [Google Scholar]
- 80.Yarus M, Folley LS. Sense codons are found in specific contexts. J Mol Biol. 1985;182:529–40. doi: 10.1016/0022-2836(85)90239-6. [DOI] [PubMed] [Google Scholar]
- 81.Buchan JR, et al. tRNA properties help shape codon pair preferences in open reading frames. Nucleic Acids Res. 2006;34:1015–27. doi: 10.1093/nar/gkj488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tats A, et al. Preferred and avoided codon pairs in three domains of life. BMC Genomics. 2008;9:463. doi: 10.1186/1471-2164-9-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Coleman JR, et al. Virus attenuation by genome-scale changes in codon pair bias. Science. 2008;320:1784–7. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Futcher B, et al. Reply to Simmonds et al: Codon pair and dinucleotide bias have not been functionally distinguished. Proc Natl Acad Sci USA. 2015;112:E3635–6. doi: 10.1073/pnas.1507710112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tulloch F, et al. RNA virus attenuation by codon pair deoptimisation is an artefact of increases in CpG/UpA dinucleotide frequencies. Elife. 2014;3:e04531. doi: 10.7554/eLife.04531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chevance FF, et al. The effects of codon context on in vivo translation speed. PLoS Genet. 2014;10:e1004392. doi: 10.1371/journal.pgen.1004392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Doerfel LK, et al. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science. 2013;339:85–8. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- 88.Ude S, et al. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science. 2013;339:82–5. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- 89.Gamble CE, et al. Adjacent codons act in concert to modulate translation efficiency in yeast. Cell. 2016;166:679–90. doi: 10.1016/j.cell.2016.05.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jan CH, et al. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science. 2014;346:1257521. doi: 10.1126/science.1257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hersch SJ, et al. Translation initiation rate determines the impact of ribosome stalling on bacterial protein synthesis. J Biol Chem. 2014;289:28160–71. doi: 10.1074/jbc.M114.593277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Letzring DP, et al. Translation of CGA codon repeats in yeast involves quality control components and ribosomal protein L1. RNA. 2013;19:1208–17. doi: 10.1261/rna.039446.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shoemaker CJ, Green R. Translation drives mRNA quality control. Nat Struct Mol Biol. 2012;19:594–601. doi: 10.1038/nsmb.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Caponigro G, et al. A small segment of the MATalpha1 transcript promotes mRNA decay in Saccharomyces cerevisiae: a stimulatory role for rare codons. Mol Cell Biol. 1993;13:5141–8. doi: 10.1128/mcb.13.9.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hu W, et al. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature. 2009;461:225–9. doi: 10.1038/nature08265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Singh G, et al. The clothes make the mRNA: past and present trends in mRNP fashion. Annual Review of Biochemistry. 2015;84:325–354. doi: 10.1146/annurev-biochem-080111-092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Iwakawa HO, Tomari Y. The functions of microRNAs: mRNA decay and translational repression. Trends Cell Biol. 2015;25:651–65. doi: 10.1016/j.tcb.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 98.Gu W, et al. A universal trend of reduced mRNA stability near the translation-initiation site in prokaryotes and eukaryotes. PLoS Comput Biol. 2010;6:e1000664. doi: 10.1371/journal.pcbi.1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Boel G, et al. Codon influence on protein expression in E. coli correlates with mRNA levels. Nature. 2016;529:358–63. doi: 10.1038/nature16509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bazzini AA, et al. Codon identity regulates mRNA stability and translation efficiency during the maternal-to-zygotic transition. EMBO J. 2016;35:2087–2103. doi: 10.15252/embj.201694699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mishima Y, Tomari Y. Codon usage and 3' UTR length determine maternal mRNA stability in zebrafish. Mol Cell. 2016;61:874–85. doi: 10.1016/j.molcel.2016.02.027. [DOI] [PubMed] [Google Scholar]
- 102.Radhakrishnan A, Green R. Connections underlying translation and mRNA stability. J Mol Biol. 2016;428:3558–64. doi: 10.1016/j.jmb.2016.05.025. [DOI] [PubMed] [Google Scholar]
- 103.Pelechano V, et al. Widespread co-translational RNA decay reveals ribosome dynamics. Cell. 2015;161:1400–12. doi: 10.1016/j.cell.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Radhakrishnan A, et al. The DEAD-Box protein Dhh1p couples mRNA decay and translation by monitoring codon optimality. Cell. 2016;167:122–132. e9. doi: 10.1016/j.cell.2016.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–86. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sweet T, et al. The DEAD-box protein Dhh1 promotes decapping by slowing ribosome movement. PLoS Biol. 2012;10:e1001342. doi: 10.1371/journal.pbio.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hense W, et al. Experimentally increased codon bias in the Drosophila Adh gene leads to an increase in larval, but not adult, alcohol dehydrogenase activity. Genetics. 2010;184:547–55. doi: 10.1534/genetics.109.111294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shah P, et al. Rate-limiting steps in yeast protein translation. Cell. 2013;153:1589–601. doi: 10.1016/j.cell.2013.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Subramaniam AR, et al. An Integrated Approach Reveals Regulatory Controls on Bacterial Translation Elongation. Cell. 2014;159:1200–1211. doi: 10.1016/j.cell.2014.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vind J, et al. Synthesis of proteins in Escherichia-Coli is limited by the concentration of free ribosomes - expression from reporter genes does not always reflect functional messenger-RNA levels. J Mol Biol. 1993;231:678–688. doi: 10.1006/jmbi.1993.1319. [DOI] [PubMed] [Google Scholar]
- 111.Merrick WC, Harris ME. Control not at initiation? Bah, humbug! EMBO J. 2014;33:3–4. doi: 10.1002/embj.201387388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gerbasi VR, et al. Yeast Asc1p and mammalian RACK1 are functionally orthologous core 40S ribosomal proteins that repress gene expression. Mol Cell Biol. 2004;24:8276–8287. doi: 10.1128/MCB.24.18.8276-8287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sengupta J, et al. Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM. Nat Struct Mol Biol. 2004;11:957–62. doi: 10.1038/nsmb822. [DOI] [PubMed] [Google Scholar]
- 114.Kuroha K, et al. Receptor for activated C kinase 1 stimulates nascent polypeptide-dependent translation arrest. EMBO Rep. 2010;11:956–61. doi: 10.1038/embor.2010.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Brandman O, et al. A ribosome–bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012;151:1042–54. doi: 10.1016/j.cell.2012.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bengtson MH, Joazeiro CAP. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature. 2010;467:470–473. doi: 10.1038/nature09371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Akashi H. Synonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy. Genetics. 1994;136:927–35. doi: 10.1093/genetics/136.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zaborske JM, et al. A nutrient-driven tRNA modification alters translational fidelity and genome-wide protein coding across an animal genus. PLoS Biol. 2014;12:e1002015. doi: 10.1371/journal.pbio.1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xu Y, et al. Non-optimal codon usage is a mechanism to achieve circadian clock conditionality. Nature. 2013;495:116–20. doi: 10.1038/nature11942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhou M, et al. Non-optimal codon usage affects expression, structure and function of clock protein FRQ. Nature. 2013;495:111–5. doi: 10.1038/nature11833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lajoie MJ, et al. Genomically recoded organisms expand biological functions. Science. 2013;342:357–60. doi: 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Napolitano MG, et al. Emergent rules for codon choice elucidated by editing rare arginine codons in Escherichia coli. Proc Natl Acad Sci USA. 2016;113:E5588–97. doi: 10.1073/pnas.1605856113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Purvis IJ, et al. The efficiency of folding of some proteins is increased by controlled rates of translation in vivo - a hypothesis. J Mol Biol. 1987;193:413–417. doi: 10.1016/0022-2836(87)90230-0. [DOI] [PubMed] [Google Scholar]
- 124.Thanaraj TA, Argos P. Ribosome-mediated translational pause and protein domain organization. Protein Sci. 1996;5:1594–612. doi: 10.1002/pro.5560050814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kimchi-Sarfaty C, et al. A "silent" polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–8. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 126.Pechmann S, et al. Local slowdown of translation by nonoptimal codons promotes nascent-chain recognition by SRP in vivo. Nat Struct Mol Biol. 2014;21:1100–5. doi: 10.1038/nsmb.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nedialkova DD, Leidel SA. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell. 2015;161:1606–18. doi: 10.1016/j.cell.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yu CH, et al. Codon usage influences the local rate of translation elongation to regulate co-translational protein folding. Mol Cell. 2015;59:744–54. doi: 10.1016/j.molcel.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Buhr F, et al. Synonymous codons direct cotranslational folding toward different protein conformations. Mol Cell. 2016;61:341–51. doi: 10.1016/j.molcel.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Brar GA, Weissman JS. Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat Rev Mol Cell Bio. 2015;16:651–664. doi: 10.1038/nrm4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Li GW, et al. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014;157:624–35. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Crick FH. Codon--anticodon pairing: the wobble hypothesis. J Mol Biol. 1966;19:548–55. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]