Abstract
Background
Intestinal failure-associated liver disease (IFALD) causes significant mortality in patients with short bowel syndrome (SBS). Steatosis, a major component of IFALD has been shown to persist even after weaning from parenteral nutrition. We sought to determine whether steatosis occurs in our murine model of SBS, and more importantly, whether steatosis was affected by manipulation of the intestinal microbiome.
Methods
Male C57BL6 mice underwent 50% small bowel resection (SBR) and orogastric gavage with vancomycin or vehicle for 10 weeks. DNA was extracted from stool samples then sequenced using 16s rRNA. Liver lipid content was analyzed. Bile acids were measured in liver and stool.
Results
Compared with unoperated mice, SBR resulted in significant changes in the fecal microbiome and was associated with > 25-fold increase in steatosis. Oral vancomycin profoundly altered the gut microbiome and was associated with a 15-fold reduction in hepatic lipid content after resection. There was a 17-fold reduction in fecal secondary bile acids after vancomycin treatment.
Conclusion
Massive SBR in mice is associated with development of steatosis, and prevented by oral vancomycin. These findings implicate a critical role for gut bacteria in IFALD pathogenesis and illuminate a novel surgical model for future investigation into this important morbidity.
Keywords: Small Bowel Resection, Short Bowel Syndrome, Farnesoid X Receptor, Steatosis, Vancomycin
Short bowel syndrome (SBS) results from a massive loss of small bowel associated with the treatment of several conditions in the pediatric population, which include necrotizing enterocolitis and midgut volvulus. The mortality of children with SBS ranges from 30–40% making SBS one of the most lethal diseases of infancy. 1 Intestinal failure associated liver disease (IFALD) is a major complication of SBS. IFALD affects 40%–60% of children with intestinal failure on parenteral nutrition and has been reported to account for up to 60% of the long term mortality from SBS.1,2 The economic impact of comprehensive care and complications arising from SBS is significant at over 6 billion US dollars annually. 3,4
The ability of the gut microbiome to modulate liver injury is well established in several experimental models of non-alcoholic fatty liver disease.5, 6 A central role for the intestinal microbiome has been further secured by experiments whereby lean and obese phenotypes of twin humans have been transferred to mice via fecal transplantation. 7 Increased abundance of Firmicutes (primarily Gram-positive bacteria) is one of the more prevalent findings involving an obese phenotype8. Similar to obesity, we have identified a remarkably similar pattern of increased Firmicutes abundance in the small intestine after SBR 9. Using our murine SBR model, we have established that mice exhibit a unique body composition profile in which fat is preferentially replaced over lean muscle during recovery.10 The initial purpose of this study was to test the hypothesis that modulating the microbiome with vancomycin to selectively eliminate Gram-positive microbiota would alter the resection-associated changes in body composition. Although vancomycin did not affect resection-associated body composition, significant differences in hepatic histology were observed, which led to further investigation.
1. Methods
1.1 Animals
C57BL6 mice were obtained from Jackson Laboratories (Bar Harbor, ME) at 7 weeks of age. Mice were housed on arrival in a facility with a 12-h light/dark cycle. Male mice were used exclusively in order to limit hormonal confounders when analyzing the enteric microbiome. This study was approved by the Washington University Animal Studies Committee (Protocol 20130308) in accordance with the National Institute of Health laboratory animal care and use guidelines.
1.1.1 Diets and Operation
Eight week-old male mice were placed on a standard liquid diet (LD; PMI Micro-Stabilized Rodent Liquid Diet LD 101,) for 24 hours prior to operation. The mice were randomized into 2 groups treated with either vancomycin (2 mg per mouse) or the equivalent volume of water (40 μL) by orogastric gavage 4 hours prior to the operation. The mice then underwent a 50% proximal SBR as we have previously described.11 Mice were maintained on standard LD for the remainder of the experiment. This SBR model does not involve parenteral nutrition, thereby eliminating multiple confounding variables including rates of glucose infusion, amino acid composition, different types of fat (Ω3 vs Ω6), and percent of enteral intake.
1.1.2 Experimental Design and Sample Collection
SBR mice were treated with vancomycin or water daily every 3 days for 10 weeks. Fecal samples were collected and sequenced prior to SBR and at 3 weeks following SBR. Body composition measurements were obtained every 3 days for the first 15 days and weekly after that for a total of 5 weeks. Food intake and weight were measured daily. An unoperated group of male C57BL6 mice were maintained on the same LD for 10 weeks without additional intervention to provide unoperated controls.
After 10 weeks, the small intestine was removed and prepared for isolation of enterocytes as previously described12. Blood was obtained via cardiac puncture. Liver was flash frozen and stored at −80 °C. Liver and small bowel 1cm distal to the anastomosis were collected and preserved in 10% buffered formalin.
1.2 RNA Isolation
RNA Isolation and real-time PCR was performed on isolated enterocytes and whole bowel from the terminal ileum and liver tissue as previously described.13 CYP7a1, Fgf15, and Nr0b2 (SHP) primers were obtained from Life Technologies (Carlsbad CA).
1.3 Lipid Quantification
Fecal fat quantification and fat absorption were measured using chloroform: methanol extraction protocol as previously described. 13 Liver sections were stained with Hematoxylin and eosin and analyzed using NIS Elements V4.3 software to obtain the percent lipid content using 4 random fields at 4x magnification per sample.
1.4 DNA Isolation from stool
DNA was extracted from frozen fecal samples per the manufacturer’s protocol for the QIAamp DNA stool mini kit (Qiagen, Germantown, MD). In addition, fecal pellets were placed in sterile tubes with buffer ASL, 425–600μm acid washed glass beads (Sigma-Aldrich, St. Louis, MO), 2.3mm zirconium/silica disruption beads (Research Products International Corp., Mt. Prospect, Il) and homogenized using a bead beater.
1.5 PCR Amplification and Sequencing of Bacterial 16S rRNA Genes
Fourteen PCR amplicons, representing all 9 16S variable regions, were constructed using the Fluidigm Access Array System. Five ng/ul of DNA were input into each reaction. The sample inlets consisted of 1X High Fidelity FastStart Reaction Buffer without MgCl2 (Roche), 4.5nM MgCl2 (Roche), 5% DMSO (Roche), 200uM PCR Grade Nucleotide Mix (Roche), 0.05 U/μL 5 U/μL FastStart High Fidelity Enzyme Blend (Roche), 1X Access Array Loading Reagent (Fluidigm), 1ul DNA, and water. Primers were added to the assay inlets at 200nM forward and reverse primers with 1X Access Array Loading Reagent. PCR amplification was performed on the BioMark HD system from Fluidigm. Each sample was harvested and indexed using unique 10 base pair sequences with 14 rounds of PCR to incorporate each index sequence. Samples were pooled into 48 sample libraries and cleaned using bead purification, then loaded on Miseq instruments and sequenced.
1.6 Sequencing Data Analysis
Of the 14 PCR amplicons, we sequenced only reads from one amplicon, which covers the 16S V4. Analysis of the V4 region reads was performed using the QIIME pipeline 14. Open-reference operational taxonomic unit (OTU) were called using the May, 2013 release of the Greengenes Databas. Reads were clustered into OTUs by QIIME using UCLUST 15 at a threshold of 97% similarity. Representative sequences for each OTU were classified taxonomically using the Ribosomal Database Project (RDP) Naïve Bayesian Classifier (training set 6) 16.
1.7 Bile Acid Analysis
Bile acid concentrations in stool and liver samples were measured using a high-performance liquid chromatography-mass spectroscopy (LCMS) system (4000 Q Trap, Applied Biosystems, Waltham, MA). Bile acids were extracted using a previously published protocol.17 Briefly, samples were collected, stored at −80°C and then lyophilized prior to extraction. Bile acids were extracted from dried samples (10mg) with NaOH (0.1mol/L) and incubated for 1 hour at 60°C. Samples were diluted with water and homogenized by sonication. Resultant samples were centrifuged at 25000 × g for 20 mins. The supernatant was extracted using solid phase extraction (SPE) cartridges (Waters Corporation, Milford, MA). Quantification was done using bile acid standards and labeled internal standards.
1.8 Statistical Analysis
Student’s t-test was used to compare the same OTU’s between untreated mice before and after SBR or untreated mice and treated mice at the same time point. Student’s t-test was also used to compare phyla before and after SBR or with and without vancomycin. Measurements of food intake, fecal fat, fat absorption, liver lipid content, bile acids, and mRNA expression were compared between groups before and after surgery or with and without vancomycin at the same time point using Student’s t-test. Significance was assigned to p-values of < 0.05.
2. Results
2.1 Effect of SBR and oral vancomycin on the fecal microbiome
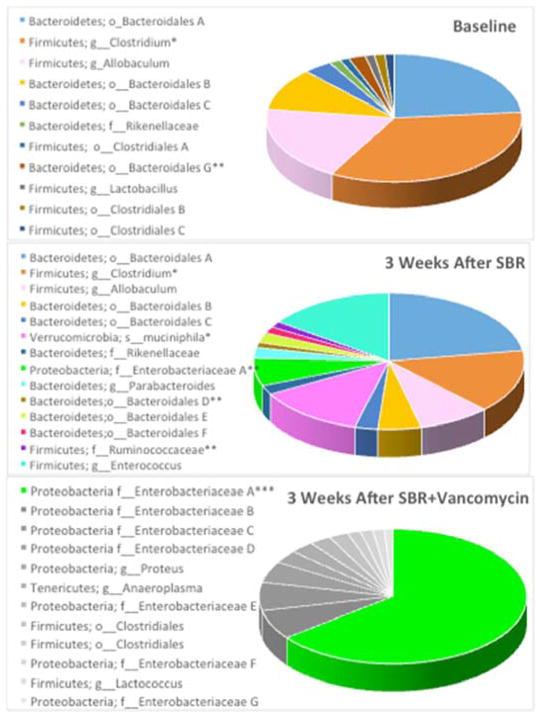
We elected to use vancomycin as a single agent due to its Gram-positive selectivity (primarily Firmicutes) and poor intestinal absorption. All fecal samples were collected at 3 weeks from mice that underwent SBR. Significance was assigned to any microbe contributing >1% of the total microbiome while major constituents composed >5% of the total. We identified dominant major phyla within our baseline samples; Firmicutes accounting for 41% and Bacteroidetes 35%. We identified 3 new significant phyla postoperatively that included Proteobacteria, Tenericutes, and Verrucomicrobia in addition to the preexisting significant Firmicutes and Bacteroidetes. (Figures 1A, 1B, and 1D).
Figure 1.
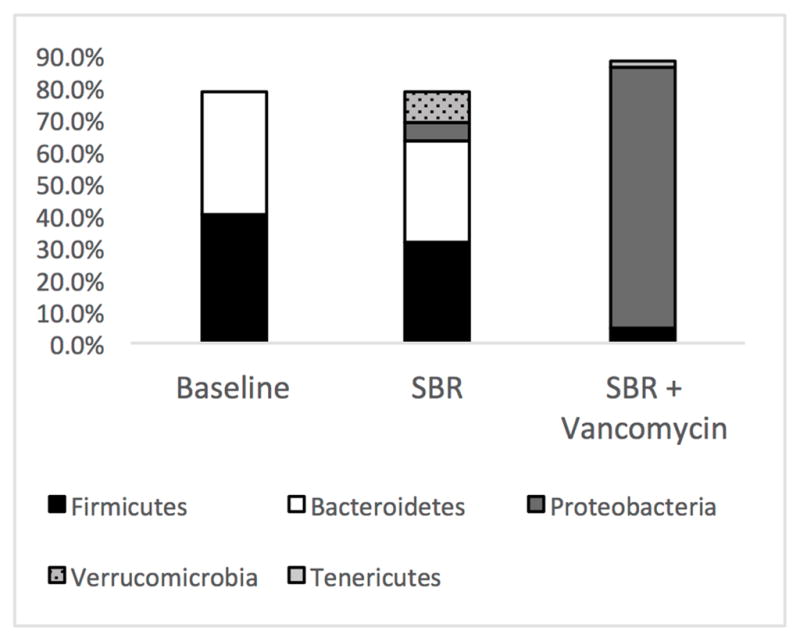
Massive small bowel resection (SBR) and vancomycin treatment alter the fecal microbiome. (A) 16s rRNA sequencing of feces prior to small bowel resection (baseline). (B) Fecal microbiome three weeks after resection (n=5). (C) Fecal microbiome three weeks after resection and oral vancomycin treatment (n=7). *p<0.05 when comparing baseline to 3 weeks after SBR, ** p<0.01 when comparing baseline to 3 weeks after SBR, *** The only common major constituent when comparing 3 weeks after SBR to 3 weeks after SBR + vancomycin. (p<0.001) All other major constituents of the SBR+vancomycin microbiome were not significant constituents in the mice 3 weeks after SBR alone. (D) Microbial constituents of the fecal microbiome at the phylum level from mice at baseline 3 weeks after massive SBR, and 3 weeks after massive SBR plus oral vancomycin treatment. *p<0.01 when comparing baseline to control for Proteobacteria, # p<0.05 when comparing baseline to control for Verrucomicrobia, ** p<0.01 when comparing control and vancomycin treated at 3 weeks for all 5 phyla.
SBR resulted in significant changes within 6 significant constituents of the gut microbiome at three weeks following surgery. The most substantial changes after SBR include a 55% reduction in a group from the Clostridium genus (27% to 12%, p=0.02), a 10% increase in Akkermansia muciniphila making it a significant species to evolve after SBR (0% to 10%, p=0.05), and a 5% increase in an unspecified group of members from the Enterobacteriaceae family (0% to 5%, p=0.003). Changes to the minor significant constituents consisted of an 86% decrease in a group of members from the Clostridiales order (1% to 0.1%, p=0.02) and a 43% decrease in a group of members from the Bacteriodales order (1.7% to 0.95%, p=0.002). Fecal microbiome alpha diversity was not significantly altered after SBR alone.
Oral vancomycin resulted in drastic alterations in the microbial community with a single group of members from the Escherichia/Shigella genus being the only significant constituents shared with controls at three weeks following SBR (Figure 1C). There were significant increases in several other members of the Proteobacteria phylum after vancomycin treatment and a loss of all significant members the Bacteroidetes phylum. Firmicutes demonstrated an 88% decrease over all. Alpha diversity decreased significantly as calculated by both Shannon (3.95 ± 0.42 versus 2.75 ± 0.28; p=0.00004) and Simpson (0.86 ± 0.03 versus 0.66 ± 0.07; p=0.00003) indices.
2.2 Body composition
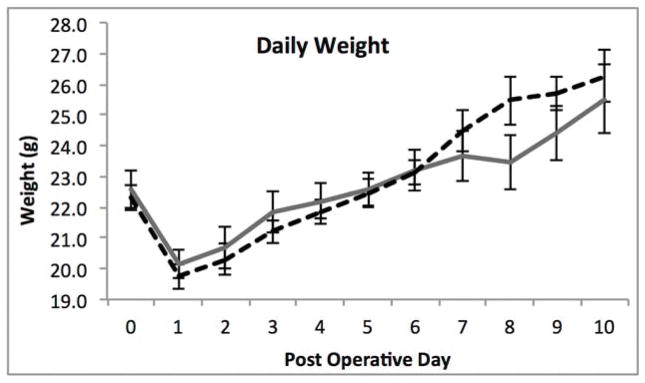
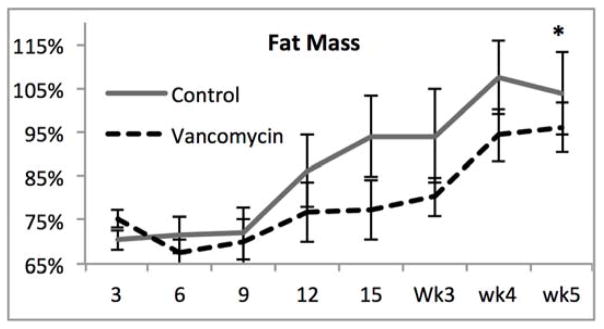
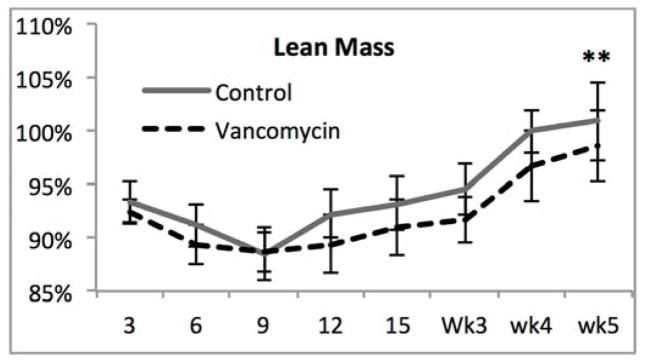
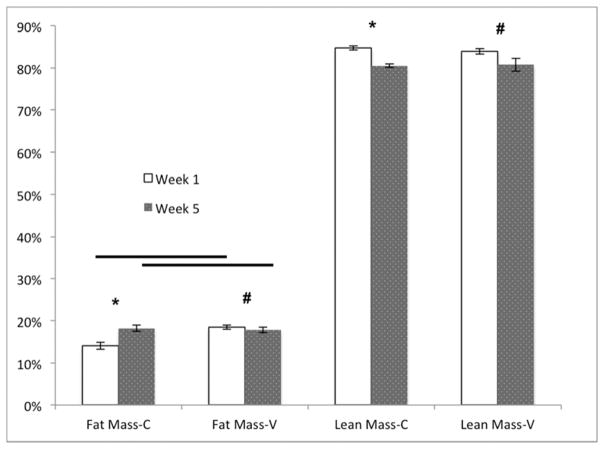
With significant alterations in the microbiome after SBR and profound differences after vancomycin treatment, we sought to determine whether these factors affected weight gain, body composition or the magnitude of intestinal adaptation after SBR. There were no significant differences in weight with vancomycin treatment (Figure 2A). Both groups exhibited the previously demonstrated resection-associated body composition alterations consisting of maintained or increased percent fat mass in the face of a decreasing percent lean mass (Figure 2B–C).10 The control mice demonstrated a significant increase in percent body fat from one week to five weeks postoperatively (4.2% ± 0.6%, p=0.006) while the Vancomycin treated animals demonstrated a very slight decrease (0.7% ± 0.6%, p=0.02). Although the change in % fat mass was significant in both controls and vancomycin-treated, there were no differences when comparing fat mass between groups at either time point. These results further supporting the notion that SBR rather than vancomycin is responsible for this phenotype. There was a more noticeable decrease in lean body mass from one week post operatively to 5 weeks post operatively with the control group demonstrating a decrease of 4.5% ± 0.4% (p=0.005), and the vancomycin group demonstrating a decrease of 3.2% ± 1%, (p=0.02). (Figure 2D).
Figure 2.
(A–C) Oral vancomycin does not affect weight gain, % body fat, or % lean body mass or body composition after 50% small bowel resection(SBR). *p<0.01, **p=0.03 (D) Mice demonstrate a significant decrease in percent lean mass after SBR regardless of vancomycin treatment. Mice treated with vancomycin demonstrated a marginally significant 1.2% decrease in fat at 5 weeks after SBR, while the control mice demonstrated a significant increase in fat at 5 weeks post operatively. There was no significant difference between percent fat or lean mass between the control groups at either time point. #p<0.01, *p<0.05, bar= not significant
There was no significant difference in food intake averaging 9.9ml/day and 10.1ml/day for control and vancomycin treated groups, respectively. All mice had histological evidence of normal adaptation with an average increase in villus height of 56% ± 12% in control mice versus 41% ± 12% in the vancomycin-treated animals. Similarly, the increase in crypt depth was 65% ± 14% in controls versus 48 % ± 5% in the vancomycin group. There were no significant differences in villus height or crypt depth measurements between the two groups following SBR.
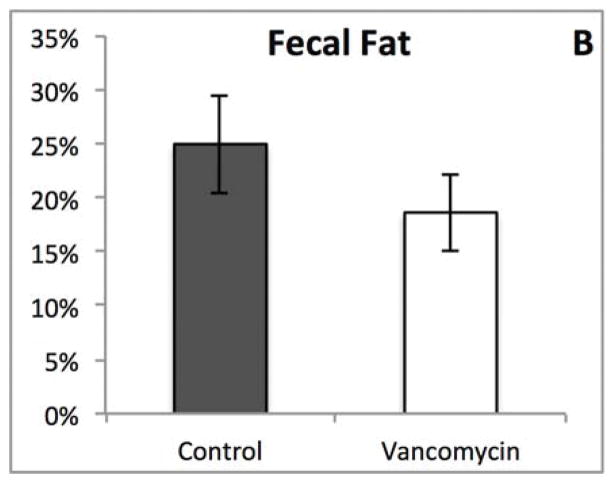
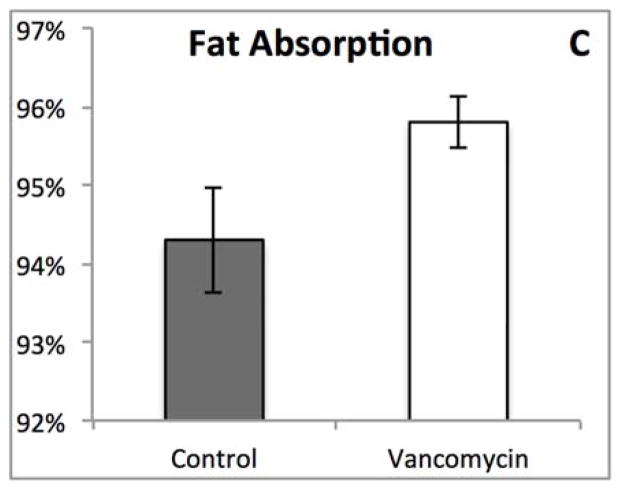
Although most of the physical characteristics were identical, we noted striking difference in the appearance of the feces between the groups (Figure 3A), which prompted us to measure fecal fat and determine fat absorption. There were no significant differences between the two groups (Figure 3B and C). The pale appearance of the feces from vancomycin treated animals combined with the results of the fecal fat analysis therefore directed us to explore the hepatobiliary system as the possible etiology for the appearance of the feces.
Figure 3.

(A) Oral vancomycin causes a distinct fecal phenotype. (B) % Fecal fat revealed no significant difference between groups. (p=0.314) (C) % Fat absorption shows no significant difference between groups. (p=0.094)
2.3 Hepatic Steatosis
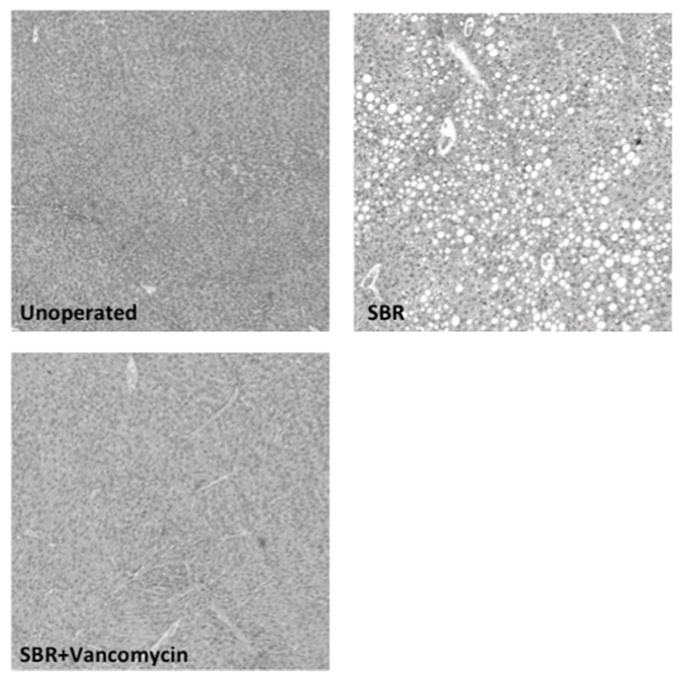
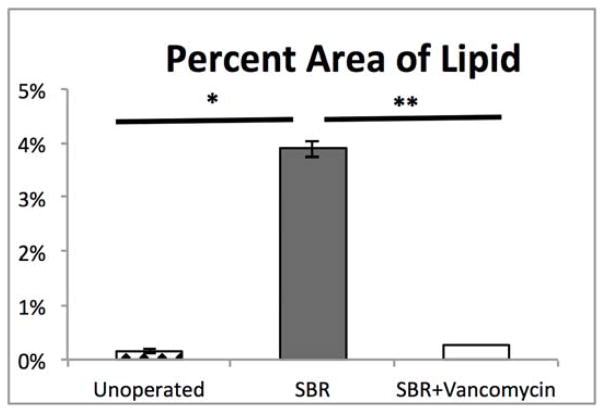
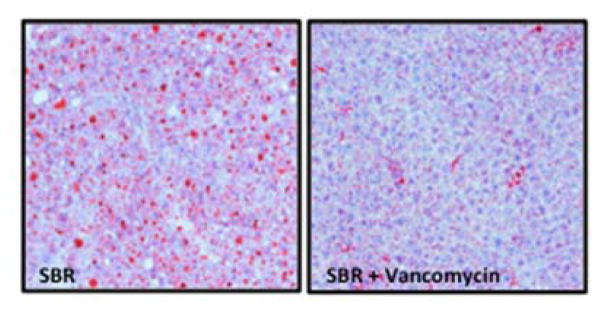
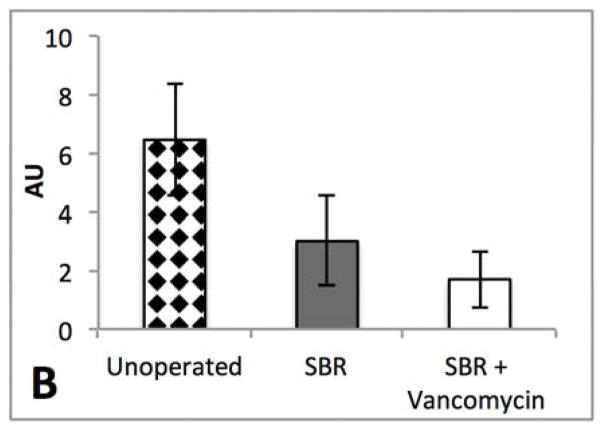
Hepatic histology revealed that SBR (N=8) was associated with greater than 25-fold increase in hepatic steatosis compared to unoperated mice (N=9) (3.89% vs. 0.14%, p=0.009). Strikingly, oral vancomycin treatment (N=7) resulted in a 15-fold reduction in hepatic lipid content (3.89% vs. 0.26%, p=0.02) in SBR mice (Figure 4). Liver sections obtained from treated and control mice 10 weeks after SBR were stained with oil-red-O in order to verify lipid. (Figure 5)
Figure 4.
SBR accelerates liver steatosis and is prevented by oral vancomycin (A) Hematoxylin and eosin stained liver sections at 10 weeks after liquid diet, SBR, or SBR + oral vancomycin treatment (H&E staining, 4x magnification). (B) Lipid content of liver samples was measured with NIS elements V4.3 software as percent area of 4 fields at 4x magnification. Liver samples were collected at 10 weeks after intervention *p<0.01, ** p<0.05
Figure 5.
Oil Red-O staining confirms diminished steatosis in mouse liver after oral vancomycin treatment in mice after 50% small bowel resection.
2.4 Bile Acid Data
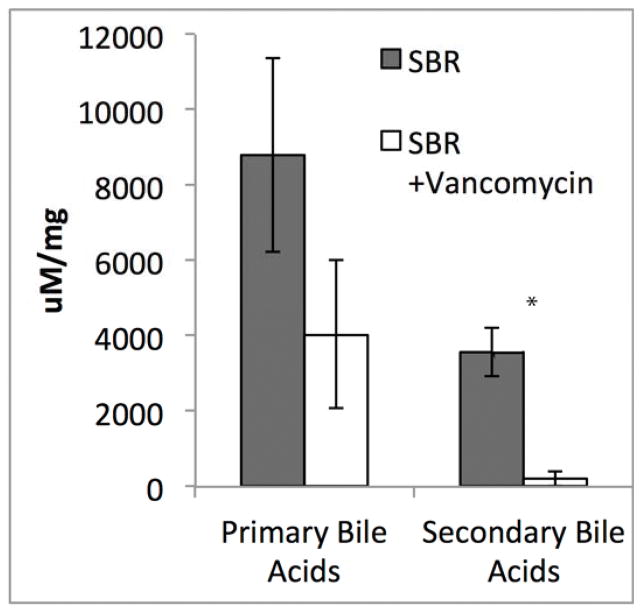
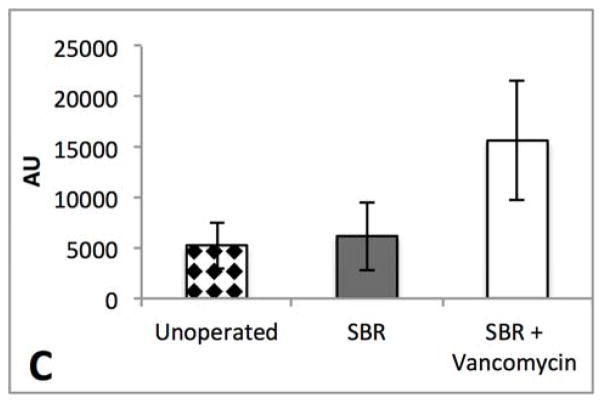
The striking liver histology, the fecal phenotype, and the profound changes in the vancomycin treated microbiome lead us to suspect that the fecal samples would differ with respect to bile acids. Although total bile acids were not significantly different between the groups, there was a 18-fold reduction in fecal secondary bile acids after vancomycin treatment (p=0.002)(Figure 6). Hepatic bile acids were also analyzed to in order to help delineate whether the significant differences in the fecal microbiome were due to a difference in synthesis or bacterial transformation. In the liver, there were no significant differences in the total bile acids, primary, or secondary bile acids.
Figure 6.
Secondary bile acids in the stool are reduced by >18-fold after oral vancomycin treatment. Average total is plotted ± SEM. *P= 0.002
2.5 mRNA Expression
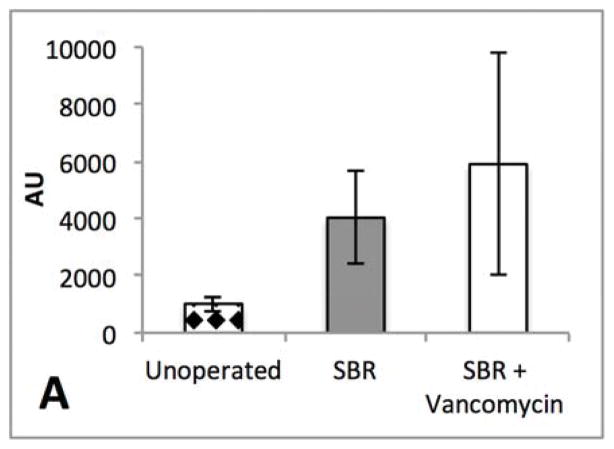
In addition to the hepatic bile acid content we also measured the relative expression of bile acid synthesis rate-limiting enzyme CYP7a1 and the FXR derived repressor, SHP in liver tissue. We did not demonstrate a significant difference between the groups with respect to expression levels of either mRNA. (Figure 7A–B) In previous studies, antibiotic treatment has been shown to alter downstream FXR signaling. 6 However, relative expression of FGF-15 in the terminal ileum after SBR did not demonstrate a significant difference between the control and vancomycin-treated animals. (Figure 7C) There was marked variability within each group.
Figure 7.
(A) Liver SHP expression is not significantly different in liver tissue after 10 weeks of liquid diet in unoperated, 50% SBR, and SBR + vancomycin treated mice. (B) FGF15 expression in intestinal tissue is not significantly different after 50% SBR or oral vancomycin treatment. (C) CYP7a1 expression in liver tissue is not significantly different after 50% SBR or oral vancomycin treatment. Averages are plotted ± SEM (N=4 per group) All expression levels were normalized to whole bowel. All units are arbitrary.
3. Discussion
Mice undergoing SBR and treated with oral vancomycin demonstrated profound changes in the gut microbiome. Despite these profound changes, gut adaptation, fat absorption, postoperative weight gain, and body composition were not affected. On the other hand, we have demonstrated for the first time that our murine SBR model is associated with the development of significant hepatic steatosis, an effect which is largely mitigated by oral vancomycin. Taken together, these data suggest that the gut microbiome is likely to be a key contributor to resection-associated liver injury. Administration of oral antibiotics, prebiotics, probiotics, or fecal transplantation may therefore be beneficial in the prevention of this important morbidity in patients with short bowel syndrome.
The role that massive SBR plays in contributing to resection associated liver disease has only recently been questioned in a pig model.18 The mechanism for resection causing liver disease is poorly understood and complex. The steatosis that we observed after SBR is similar to studies in pigs and therefore offers a much more cost efficient model to further explore the etiology behind resection-associated liver disease. One of the limitations to our murine model may be its dependence male gender. Previous work has demonstrated that estrogen confers protection against steatosis in non-alcoholic fatty liver disease (NAFLD). 19 The effects of estrogen on the presence and timing of development in resection associated liver disease is currently unknown and is worthy of further investigation.
The decrease in diversity and increase in Proteobacteria that we measured in our vancomycin-treated SBR mice is in line with what is seen in human patients after vancomycin treatment.20 Previous studies suggest that a predominance of Proteobacteria is associated with prolonged parenteral nutrition, intestinal inflammation, as well as hepatic fibrosis, steatosis and portal inflammation.21, 22 In contrast with these findings, a much greater abundance of Proteobacteria was present after vancomycin treatment and yet steatosis was prevented in the treated animals. This finding suggests that the presence of Proteobacteria alone is not likely the cause of the steatosis seen in SBS related hepatic disease. We also saw a near complete loss of the Bacteroidetes phyla in the treated mice similar to what was seen in obese mice after vancomycin treatment.23 However, there was no difference in the weight or percent body fat between the groups suggesting that a shift in Bacteroidetes alone is not responsible for the lean phenotype observed in previous studies. 7, 8 In contrast, vancomycin treatment did not induce an increase in Actinobacteria as seen in previous experiments with vancomycin.23 Our results in control SBR animals more closely resembled the short bowel microbiome of humans after SBR, with a scarcity of Actinobacteria.24 The combination of surgical resection and antibiotic treatment therefore appears to have created a unique microbiome, which shows evidence of each intervention. It must also be considered that vancomycin treatment might have resulted in bacterial resistance of some organisms. Antibiotic resistance is likely to play an important role clinically as many SBS patients are subjected to multiple courses of antibiotics during the adaptation period.
After SBR, the Bacteroidetes and Firmicutes remain dominant and maintain their proportionality. Vancomycin treatment abolishes this and it might be considered that Bacteroidetes and Firmicutes have a symbiotic relationship that contributes to the steatosis associated with SBR. However, the extent to which Proteobacteria is present in the vancomycin treated/non-steatotoic mice raises the possibility that the vancomycin treated SBR mice either lost a harmful microbiome constituent in Bacteroidetes/Firmicutes or alternatively, gained a protective one in Proteobacteria. It is also possible that in the absence of a significant number of gram positive organisms, the Proteobacteria had better survival and less cytokine-inducing liberation of proinflammatory bacteria products such as endotoxin.
In this experiment, we demonstrated significant alterations in the fecal microbiome after SBR. This contrasts with an earlier study in which we revealed significant alterations in the microbiota of the small bowel, but not in the feces after SBR9. The variations between current and prior studies might be due to altered time of fecal collection (7 and 90 days versus 21 days), small bowel lumen versus passed stool, or different housing conditions from one time of year to another. Consistent with this study, the fecal microbiome was found to be significantly altered in pig SBS models as well as in human infants and adults with SBS. 18, 24
Both groups of animals displayed a resection-associated metabolic phenotype of maintained fat mass in face of decreasing percent lean mass despite antibiotic intervention. Absolute, relative, and percent lean and fat mass were measured and not found to be significantly different after vancomycin treatment despite the profound changes seen in the microbiome. From this data, we can conclude that the microbiome does not play a major role in the propensity to increase percent fat mass after SBR. This would contrast with the postulated role for the gut microbiome contributing to an obese phenotype.7, 8 The types microbiome changes associated with obesity include an increase in Firmicutes and a decrease in Bacteroidetes.8 In our study, neither of the groups demonstrates an increase in Firmicutes and while both groups demonstrate a decrease in Bacteroidetes, the decrease does not correlate with percent body fat. Despite the drastic differences in the magnitude by which Bacteroidetes decreased, there was no difference in body composition between groups. Because of this, our data would not support a role for the intestinal microbiota in contributing to an obese phenotype associated with intestinal resection.
β-Murocholic acid (β –MCA), the most prominent primary bile acid in mice antagonizes farnesoid X receptor (FXR) and its downstream inhibitory effects on bile acid synthesis. 25 The major focus has been on FXR signaling and its ability to be regulated by gut microbiome-induced changes in bile acids, specifically taurine-conjugated-β–MCA. Previous studies have looked at antibiotic treatment in unoperated mice using broad-spectrum antibiotics and have found decreased FXR signaling from the intestine6. However, we did not see significant differences in intestinal FXR signaling between vancomycin treated and control groups. Studies in an SBS piglet model of resection-induced liver disease also demonstrated no changes in FXR or its major target gene fibroblast growth factor 19 (FGF15 homolog) in piglets. They concluded that piglets fail to have an appropriate FXR-FGF19 response to the change in bile acid composition found after SBR.18 It appears that the resection-induced dysfunction of FXR-FGF15/19 prevents the expected antibiotic induced up regulation of FGF15/19, and therefore is unlikely to play a major role in vancomycin’s ability to prevent resection-associated steatosis.
The major constituents in the gut microbiome that affect bile acids are those bacteria with bile salt hydrolase (BSH) and 7α-dehydroxylase-enzymatic activity. BSH deconjugates bile acids, a step required for transformation from primary to secondary bile acids by 7α-dehydroxylase. Studies in an SBS piglet model of resection induced liver disease demonstrated decreased BSH and 7α-dehydroxylase containing bacteria after resection and a predictable shift to increased primary bile acids. 18 Our data also demonstrates a significant decrease in transforming bacteria, mainly members of the Clostridium and Lactobacillus genuses in the vancomycin treated group. This is reflected in the decreased secondary bile acids found in the fecal samples of this group. Although there were significant differences in fecal bile acid excretion, these differences were not observed in the liver. Given the minimal changes of bile acids present in the liver after vancomycin treatment, it is unlikely that the hepatotoxicity of bile acids is responsible for the steatosis seen after resection.
The specific mechanism for how the gut microbiome induces hepatic steatosis in the context of massive SBR remains unknown. It is possible that toll-like receptor signaling in the intestine is stimulated, which then leads to steatosis. This has recently been identified to be a potential factor in patients with non-alcoholic fatty liver disease.26
Acknowledgments
Grants
This work was supported by the NIH National Institute of Diabetes and Digestive and Kidney Diseases (F32DK103490 – Dr. Barron), The Genome Technology Access Center at Washington University School of Medicine (NIH #P30 CA91842), ICTS/CTSA Grant# UL1TR000448 from the National Center for Research Resources, The March of Dimes, The St. Louis Children’s Hospital Foundation Children’s Surgical Sciences Research Institute, and The Digestive Disease Research Core Center (NIH# P30DK052574)
Footnotes
Conflicts of Interest: Dr. Warner is on the Data Safety Monitoring Board of Shire Pharmaceutical Company.
This work was presented at The 11th annual Academic Surgical Congress February 2–4; Jacksonville FL Revised 6/14/2016
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Citations
- 1.Wales PW, Christison-Lagay ER. Short bowel syndrome: epidemiology and etiology. Seminars in Pediatric Surgery. 2010;19:3–9. doi: 10.1053/j.sempedsurg.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Lauriti G, Zani A, Aufieri R, Cananzi M, Chiesa PL, Eaton S, et al. Incidence, prevention, and treatment of parenteral nutrition-associated cholestasis and intestinal failure-associated liver disease in infants and children: a systematic review. JPEN Journal of parenteral and enteral nutrition. 2014;38:70–85. doi: 10.1177/0148607113496280. [DOI] [PubMed] [Google Scholar]
- 3.Hamilton Brady E, PDJAM, M.P.H, Osterman Michelle JK, M.H.S, Curtin Sally C, M.A, Mathews TJ, M.S Division of Vital Statistics. National Vital Statistics Reports. Hyattsville, MD: National Center for Health Statistics; 2014. Births: Final Data for 2014. [Google Scholar]
- 4.Spencer AU, Kovacevich D, McKinney-Barnett M, Hair D, Canham J, Maksym C, et al. Pediatric short-bowel syndrome: the cost of comprehensive care. Am J Clin Nutr. 2008;88:1552–9. doi: 10.3945/ajcn.2008.26007. [DOI] [PubMed] [Google Scholar]
- 5.Le Roy T, Llopis M, Lepage P, Bruneau A, Rabot S, Bevilacqua C, et al. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut. 2013;62:1787–94. doi: 10.1136/gutjnl-2012-303816. [DOI] [PubMed] [Google Scholar]
- 6.Jiang C, Xie C, Li F, Zhang L, Nichols RG, Krausz KW, et al. Intestinal farnesoid X receptor signaling promotes nonalcoholic fatty liver disease. The Journal of clinical investigation. 2015;125:386–402. doi: 10.1172/JCI76738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11070–5. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sommovilla J, Zhou Y, Sun RC, Choi PM, Diaz-Miron J, Shaikh N, et al. Small bowel resection induces long-term changes in the enteric microbiota of mice. Journal of gastrointestinal surgery: official journal of the Society for Surgery of the Alimentary Tract. 2015;19:56–64. doi: 10.1007/s11605-014-2631-0. discussion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tantemsapya N, Meinzner-Derr J, Erwin CR, Warner BW. Body composition and metabolic changes associated with massive intestinal resection in mice. Journal of pediatric surgery. 2008;43:14–9. doi: 10.1016/j.jpedsurg.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 11.Helmrath MA, VanderKolk WE, Can G, Erwin CR, Warner BW. Intestinal adaptation following massive small bowel resection in the mouse. Journal of the American College of Surgeons. 1996;183:441–9. [PubMed] [Google Scholar]
- 12.Guo J, Longshore S, Nair R, Warner BW. Retinoblastoma Protein (pRb), but Not p107 or p130, Is Required for. The Journal of Biological Chemistry. 2009;284:134–40. doi: 10.1074/jbc.M806133200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi PM, Guo J, Erwin CR, Wandu WS, Leinicke JA, Xie Y, et al. Disruption of retinoblastoma protein expression in the intestinal epithelium impairs lipid absorption. American journal of physiology Gastrointestinal and liver physiology. 2014;306:G909–15. doi: 10.1152/ajpgi.00067.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics (Oxford, England) 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 16.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Applied and Environmental Microbiology. 2007;73:5261–7. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Humbert L, Maubert MA, Wolf C, Duboc H, Mahe M, Farabos D, et al. Bile acid profiling in human biological samples: comparison of extraction procedures and application to normal and cholestatic patients. J Chromatogr B Analyt Technol Biomed Life Sci. 2012;899:135–45. doi: 10.1016/j.jchromb.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 18.Pereira-Fantini PM, Lapthorne S, Joyce SA, Dellios NL, Wilson G, Fouhy F, et al. Altered FXR signalling is associated with bile acid dysmetabolism in short bowel syndrome-associated liver disease. Journal of hepatology. 2014;61:1115–25. doi: 10.1016/j.jhep.2014.06.025. [DOI] [PubMed] [Google Scholar]
- 19.Hewitt KN, Pratis K, Jones ME, Simpson ER. Estrogen replacement reverses the hepatic steatosis phenotype in the male aromatase knockout mouse. Endocrinology. 2004;145:1842–8. doi: 10.1210/en.2003-1369. [DOI] [PubMed] [Google Scholar]
- 20.Vrieze A, Out C, Fuentes S, Jonker L, Reuling I, Kootte RS, et al. Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. Journal of hepatology. 2014;60:824–31. doi: 10.1016/j.jhep.2013.11.034. [DOI] [PubMed] [Google Scholar]
- 21.Korpela K, Mutanen A, Salonen A, Savilahti E, de Vos WM, Pakarinen MP. Intestinal Microbiota Signatures Associated With Histological Liver Steatosis in Pediatric-Onset Intestinal Failure. JPEN Journal of parenteral and enteral nutrition. 2015 doi: 10.1177/0148607115584388. [DOI] [PubMed] [Google Scholar]
- 22.Michail S, Lin M, Frey MR, Fanter R, Paliy O, Hilbush B, et al. Altered gut microbial energy and metabolism in children with non-alcoholic fatty liver disease. FEMS Microbiol Ecol. 2015;91:1–9. doi: 10.1093/femsec/fiu002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy EF, Cotter PD, Hogan A, O’Sullivan O, Joyce A, Fouhy F, et al. Divergent metabolic outcomes arising from targeted manipulation of the gut microbiota in diet-induced obesity. Gut. 2013;62:220–6. doi: 10.1136/gutjnl-2011-300705. [DOI] [PubMed] [Google Scholar]
- 24.Davidovics ZH, Carter BA, Luna RA, Hollister EB, Shulman RJ, Versalovic J. The Fecal Microbiome in Pediatric Patients With Short Bowel Syndrome. JPEN Journal of parenteral and enteral nutrition. 2015 doi: 10.1177/0148607115591216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sayin SI, Wahlstrom A, Felin J, Jantti S, Marschall HU, Bamberg K, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell metabolism. 2013;17:225–35. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Kapil S, Duseja A, Sharma BK, Singla B, Chakraborti A, Das A, et al. Small intestinal bacterial overgrowth and toll-like receptor signaling in patients with non-alcoholic fatty liver disease. J Gastroenterol Hepatol. 2016;31:213–21. doi: 10.1111/jgh.13058. [DOI] [PubMed] [Google Scholar]