Abstract
Infectious bronchitis is a highly contagious respiratory disease of poultry caused by the coronavirus infectious bronchitis virus (IBV). It was thought that coronavirus virions were composed of three major viral structural proteins until investigations of other coronaviruses showed that the virions also include viral non-structural and genus-specific accessory proteins as well as host-cell proteins. To study the proteome of IBV virions, virus was grown in embryonated chicken eggs, purified by sucrose-gradient ultracentrifugation and analysed by mass spectrometry. Analysis of three preparations of purified IBV yielded the three expected structural proteins plus 35 additional virion-associated host proteins. The virion-associated host proteins had a diverse range of functional attributions, being involved in cytoskeleton formation, RNA binding and protein folding pathways. Some of these proteins were unique to this study, while others were found to be orthologous to proteins identified in severe acute respiratory syndrome coronavirus virions and also virions from a number of other RNA and DNA viruses.
Introduction
Infectious bronchitis is a highly contagious respiratory disease of chickens that causes substantial economic losses to the poultry industry worldwide (Cavanagh, 2005). The causative agent, infectious bronchitis virus (IBV), belongs to the family Coronaviridae and is composed of an enveloped virion encapsulating a 27.6 kb positive-sense ssRNA genome. The genome encodes four structural proteins, 15 non-structural proteins (Boursnell et al., 1987; Brierley et al., 1987; Lim & Liu, 1998; Lu et al., 1996) and at least five lineage-specific accessory proteins found only in other coronaviruses in the genus Gammacoronavirus (Bentley et al., 2013; Casais et al., 2005; Hodgson et al., 2006). The virion is composed of four structural proteins, the spike (S), envelope (E), membrane (M) and nucleocapsid (N). S, M and E are all found integral to the virion membrane, while N complexes with the RNA genome, packaged inside the virion. The 15 non-structural proteins (nsps) are expressed as two replicase polyproteins that are post-translationally cleaved. The nsps make up the replicase complex, responsible for replicating the genomic RNA and transcribing viral mRNA within an infected cell (Ziebuhr et al., 2000). Little is known about the structure or function of the other virally encoded proteins such as the lineage-specific accessory proteins of IBV – 3a, 3b, 5a, 5b and the putative ORF4 (Bentley et al., 2013; Casais et al., 2005; Hodgson et al., 2006). Previous studies of purified coronavirus and related nidovirus virions have shown that a number of nsps such as the viral RNA-dependent RNA polymerase nsp12 (Nogales et al., 2012) and genus-specific accessory proteins (Neuman et al., 2008) are associated with the virus particle, along with a number of host proteins (Zhang et al., 2010), but it is still unclear why these proteins associate with the virion. There is relatively little crossover between the severe acute respiratory syndrome coronavirus (SARS-CoV) proteome and host proteins that are upregulated during infection (Vogels et al., 2011). At present, only the known structural proteins and some host proteins have been identified in association with the IBV virion (Kong et al., 2010), with no nsps or lineage-specific accessory proteins identified. Studies with a variety of other DNA and RNA viruses also indicate that many host proteins are found in association with virions (Bechtel et al., 2005; Chertova et al., 2006; Chung et al., 2006; Kattenhorn et al., 2004; Segura et al., 2008; Shaw et al., 2008; Varnum et al., 2004), suggesting far more complexity in the virion proteome than previously thought.
In this study, the proteome of the IBV strain Beau-R virion was investigated using gel-free liquid chromatography-coupled mass spectrometry (MS) methods. The Beau-R strain of IBV is a molecular clone of the Beaudette strain, which grows to high titres in embryonated chicken eggs, making it ideal for this study. Using MS analysis, three of the viral structural proteins were identified in association with the IBV virion, in addition to 35 host-cell proteins, which could be split into 17 groups based on their biological function.
Results
Analysis of IBV-infected allantoic fluid
IBV Beau-R was grown in the allantoic membrane of 10-day-old specific-pathogen-free, Rhode Island Red embryonated chicken eggs. Virus or 1 × BES medium (mock) was inoculated into the allantoic cavity and grown for 24 h, before the embryos were sacrificed and the allantoic fluid harvested. To determine the complexity of the starting material, Coomassie blue-stained SDS-PAGE gels were used to determine the protein content of fluid harvested from uninfected eggs, mock-infected eggs and IBV Beau-R-infected eggs (Fig. 1a). The low number of proteins identified in uninfected eggs demonstrated that the proteome of allantoic fluid is dominated by a few key proteins. Upon inoculation with either 1 × BES IBV growth medium or IBV Beau-R, the proteome changed, resulting in the presence of more low-abundance protein bands and a reduction in the abundance of prominent proteins from uninfected allantoic fluid. Numerous low-intensity high-molecular-mass bands in the 110 kDa and larger molecular mass range were present in mock-infected or IBV-infected allantoic fluid compared with uninfected allantoic fluid. However, several of the prominent bands found in untreated allantoic fluid were less prominent or absent after mock treatment or infection, including bands present between 110 and 260 kDa, and at 38 kDa and 58 kDa. This was taken to indicate that the process of inoculating eggs, which includes piercing the chorioallantoic membrane, was associated with a major change in the allantoic proteome.
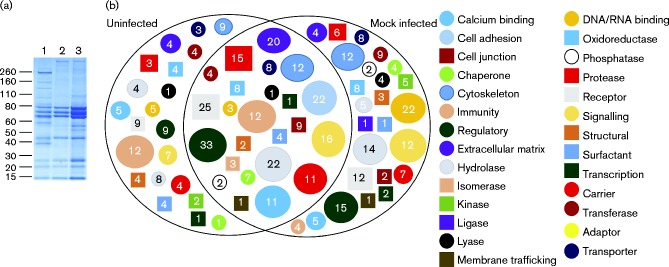
Fig. 1.
Analysis of the proteome of uninfected and mock infected allantoic fluid. (a) Allantoic fluid (100 μg protein) from 10-day-old embryonated chicken eggs which were uninfected (lane 1), mock infected with cell-culture medium without virus (lane 2) or infected with IBV Beau-R in cell culture medium (lane 3) were analysed by SDS-PAGE using 4–20 % Tris/glycine/SDS gels and stained by colloidal Coomassie blue. Molecular mass markers are indicated (kDa). (b) Proteins identified in the proteomes of uninfected and mock-infected allantoic fluid were analysed by the panther classification system (Mi et al., 2013) to identify classes of protein function, using the default settings. Groups that were identified were compared between uninfected and mock-infected allantoic fluid. The numbers of proteins classified in each category are indicated.
In comparison with a previous gel-free proteomic study of the egg-white proteome (Mann & Mann, 2011), common egg proteins such as ovalbumin, ovotransferrin, lysozyme C, ovomucoid, ovoinhibitor, ovalbumin-related proteins, cystatin, gallinacin proteins and apolipoproteins could be identified in mock-infected and uninfected allantoic fluid. This is particularly relevant given that the egg white is what becomes of allantoic fluid in an unfertilized egg, although the compositions would not be expected to be identical given the very different physiological states between a fertilized and unfertilized egg.
Proteins within uninfected and mock-infected allantoic fluid were identified using MS. A total of 354 proteins were detected in uninfected allantoic fluid, while 420 proteins were detected in mock-infected allantoic fluid. To compare whether there were any changes in the groups of proteins that could be identified between uninfected and mock-infected allantoic fluid, lists of proteins identified in each sample were analysed using the panther classification system (Mi et al., 2013) to identify classes of protein function, using the default settings. The lists of protein classes present in each sample were then compared with each other to generate a Venn diagram (Fig. 1b), which showed that 252 of the identified proteins were present in both uninfected and mock-infected allantoic fluid. Comparison of the two samples demonstrated that the majority of the overall protein classes identified were shared between both uninfected and mock-infected allantoic fluid. Despite this, the individual proteins identified within a specific protein class varied between the two samples, resulting in overlap of proteins as shown in Fig. 1(b).
Analysis of purified virus
IBV was purified by a mixture of polyethylene glycol (PEG) precipitation and sucrose-gradient ultracentrifugation. First, the virions were concentrated, by precipitation from allantoic fluid, by the addition of PEG-8000 to a final concentration of 10 %. The precipitate was reconstituted and loaded onto a sucrose gradient for purification. IBV virions formed a band between the 30 and 50 % sucrose cushions and were collected by side puncture. Mock-infected allantoic fluid was also purified in the same way and the interface between the 30 and 50 % sucrose cushions was collected. Purified virions were analysed by negative-stain electron microscopy to confirm the presence of virus particles (Fig. 2a). Intact virus particles, some with spikes decorating the lipid envelope, could be identified in each preparation. However, some damaged virions were also present. In addition, purified IBV and mock samples were resolved by SDS-PAGE followed by Coomassie blue staining to determine the protein complexity of each of the samples. While many bands were present in the purified IBV sample (Fig. 2b), no bands were seen in the purified mock sample by Coomassie blue-stained SDS-PAGE (data not shown). When the presence of IBV proteins in purified virions and mock samples was analysed by Western blotting using an anti-IBV antibody, viral proteins could be detected in the virion sample but not in the mock sample (data not shown).
Fig. 2.
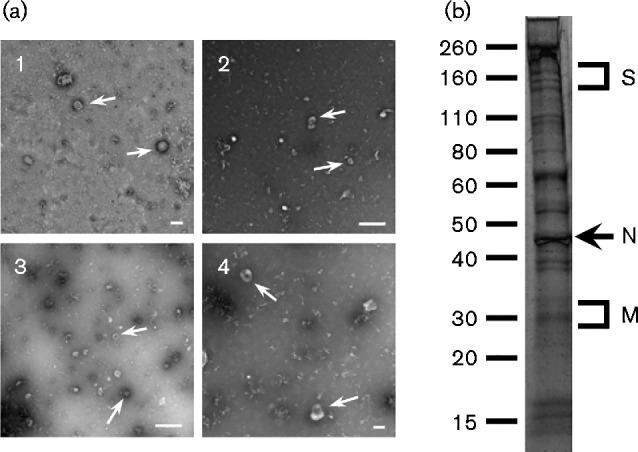
Electron microscopy and SDS-PAGE analysis of purified IBV. (a) Purified virions from preparations 1, 2 and 3 were negatively stained using methylamine vanadate and imaged using an electron microscope. An enlarged image showing virus particles with visible spike protein is shown in (4). Virus particles are indicated with arrows. Bars, 100 nm (1, 4); 500 nm (2, 3). (b) Representative banding profile of purified IBV on a 4–20 % Tris/glycine SDS-PAGE gel, stained with colloidal Coomassie blue. Molecular mass markers (kDa) and the positions of viral proteins are indicated.
IBV virion-associated proteins
MS analysis was performed on three preparations of purified IBV to identify the proteins repeatedly associated with the IBV virion. Only proteins that had at least two unique peptide matches to the identified protein and were present in all three preparations were classified as associated with the IBV virion. Three IBV structural proteins N, S and M were detected in each of the experiments with multiple unique peptide matches identified for each protein in each replicate (N – 144, 81 and 50; S – 21, 14 and 12; M – 11, 7 and 7). The fourth viral structural protein, E, the replicase proteins and the lineage-specific accessory proteins could not be identified. The total number of proteins identified in each preparation was 399, 289 and 273, respectively. To identify host proteins more likely to be selectively incorporated into the IBV virion, the proteins identified in each biological replicate were compared and those present in each replicate were determined. This identified 35 host proteins that were classified as associated with the IBV virion (Table 1). The list of 35 host proteins in common between the three experiments was submitted to PANTHER.db to determine the biological functions and process that these proteins belonged to. Of the avian proteins that were annotated in PANTHER.db, 17 diverse classes of proteins could be defined in the host proteins that were identified as being associated with the IBV virion (Fig. 3a). These proteins belonged to groups responsible for making up the cytoskeletal system and transport around it, proteins involved in metabolic processes and their modulators, and proteins involved in cellular signalling.
Table 1. Avian proteins identified in association with the IBV virion.
| UniProt accession no. | Gene name | Protein name | Previously detected in IBV virion* | Detected in SARS-CoV virion† | No. unique peptide matches‡ |
|---|---|---|---|---|---|
| O73885 | HSPA8 | Heat-shock cognate 71 kDa protein | ✓ | 8, 5, 5 | |
| P00356 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | ✓ | 11, 76, 36 | |
| P01994 | HBAA | Haemoglobin subunit αA | ✓ | 6, 4, 5 | |
| P02001 | HBAD | Haemoglobin subunit αD | 4, 2, 5 | ||
| P02457 | COL1A1 | Collagen α1(I) chain | 3, 19, 19 | ||
| P02552 | Tubulin α1 chain | 10, 18, 8 | |||
| P08251 | ATP1B1 | Sodium/potassium-transporting ATPase subunit β1 | 3, 5, 5 | ||
| P08629 | TXN | Thioredoxin | 4, 4, 4 | ||
| P08636 | RPS17 | 40S ribosomal protein S17 | ✓ | 5, 6, 2 | |
| P09206 | Tubulin β3 chain | ✓ | ✓ | 14, 3, 2 | |
| P09244 | Tubulin β7 chain | 16, 8, 3 | |||
| P09652 | Tubulin β4 chain | ✓ | 11, 1, 1 | ||
| P09654 | VIM | Vimentin | 6, 32, 12 | ||
| P0C1H5 | H2B-VII | Histone H2B 7 | ✓ | 12, 2, 2 | |
| P0CB50 | PRDX1 | Peroxiredoxin-1 | ✓ | 3, 16, 9 | |
| P11501 | HSP90AA1 | Heat-shock protein HSP 90α | ✓ | 13, 11, 4 | |
| P14105 | MYH9 | Myosin-9 | 14, 312, 191 | ||
| P17785 | ANXA2 | Annexin A2 | ✓ | ✓ | 32, 27, 21 |
| P18359 | DSTN | Destrin | ✓ | 3, 21, 12 | |
| P24479 | S100A11 | Protein S100-A11 | ✓ | 5, 3, 4 | |
| P24797 | ATP1A2 | Sodium/potassium-transporting ATPase subunit α2 | 2, 5, 2 | ||
| P41125 | RPL13 | 60S ribosomal protein L13 | 3, 5, 3 | ||
| P47836 | RPS4 | 40S ribosomal protein S4 | 5, 20, 7 | ||
| P51913 | ENO1 | α-Enolase | ✓ | 7, 12, 7 | |
| P68034 | ACTC1 | Actin, α cardiac muscle 1 | 14, 2, 9 | ||
| P84247 | H3-IX | Histone H3.3 | 8, 3, 4 | ||
| Q5ZJ54 | CCT6 | T-complex protein 1 subunit ζ; | ✓ | 11, 26, 8 | |
| Q5ZLC5 | ATP5B | ATP synthase subunit β, mitochondrial | 2, 5, 7 | ||
| Q5ZMD1 | YWHAQ | 14-3-3 protein θ | 6, 15, 7 | ||
| Q5ZMQ2 | ACTG1 | Actin, cytoplasmic 2 | ✓ | 27, 2, 1 | |
| Q5ZMT0 | YWHAE | 14-3-3 protein θ | ✓ | 6, 1, 2 | |
| Q8JFP1 | EIF4A2 | Eukaryotic initiation factor 4A-II | ✓ | 4, 15, 5 | |
| Q90705 | EEF2 | Elongation factor 2 | ✓ | 5, 41, 10 | |
| Q90835 | EEF1A | Elongation factor 1α1 | ✓ | 8, 44, 18 | |
| Q98953 | S100A6 | Protein S100-A6 | ✓ | 4, 3, 3 |
* Proteins previously reported to be associated with the IBV virion by Kong et al. (2010).
† Proteins orthologous to those reported to be associated with the SARS virion by Neuman et al. (2008).
‡ The number of peptide matches in each of the three preparations is shown.
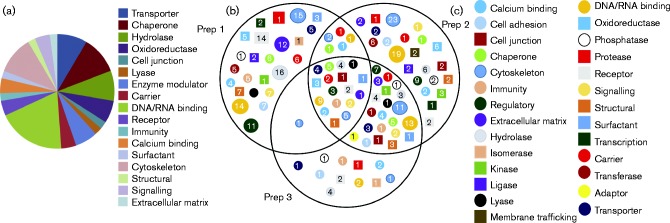
Fig. 3.
Analysis of classes of proteins associated with the IBV virion. (a) Proteins identified in association with the IBV virion described in Table 1 were analysed by panther (Mi et al., 2013) to identify classes of protein function, using the default settings. (b) Following panther analysis, groups of proteins that were identified were compared between the three IBV preparations. The numbers of proteins classified in each category are indicated.
To compare whether there were any differences in the groups of proteins that could be identified among the three preparations of purified IBV, the complete lists of proteins identified in each preparation were analysed by the PANTHER classification system (Mi et al., 2013) to identify classes of protein function, using the default settings. The lists of groups present in each preparation were then compared with each other to generate a Venn diagram (Fig. 3b). Similar to observations made when comparing uninfected and mock-infected allantoic fluids, the protein classes identified for the three preparations of IBV were largely the same. However, specific proteins within each protein class varied among the three preparations, resulting in the overlap shown in Fig. 3(b). Interestingly, preparations 2 and 3 showed more similarity to one another in the proteins identified than with preparation 1.
Discussion
In this study, 35 host proteins were observed to be repeatedly associated with egg-grown IBV Beau-R virions. Three viral proteins were also detected; S, M and N. Growth of IBV in embryonated chicken eggs is an established technique to produce large quantities of high-titre virus (Guy, 2015). Repeated passaging of virus through embryonated eggs is routinely used to attenuate viruses to create live-attenuated vaccine strains. In addition, virus is grown in embryonated eggs for the production of inactivated virus vaccines. This analysis showed that the proteome of allantoic fluid is relatively simple but that inoculation of the egg alters the proteome. The reason for this is unknown; however, it is probably due to embryonic stress caused by puncturing the shell and allantoic membrane and injecting foreign material into the egg. Comparison of the groups of proteins identified between uninfected and mock-infected allantoic fluid showed that the differences were not in the functional types of protein that were identified but were in the individual proteins that fell into these groups. At present, it is unknown why this might be the case. However, one explanation is that, because the analysis does not only represent complete proteins present within the sample but also identifies individual subunits of proteins, this is likely to affect the observed overlap.
IBV Beau-R is able to grow in embryonated eggs, primary chicken cells and cell lines, and in Vero cells (Casais et al., 2001), all potential sources of virions. However, the simple proteome of allantoic fluid demonstrated here offers the advantage that there are fewer potential contaminating host proteins in a virus preparation grown in eggs. In addition to this, higher titres of infectious virus are generally achieved through cultivation of IBV Beau-R in ovo when compared with cell culture. Consistent with the egg-white proteome (Mann & Mann, 2011), all of the major components of the egg such as ovalbumin, ovotransferrin, lysozyme C, ovomucoid and others were present in the uninfected and mock-infected allantoic fluids. However, none of these proteins was found in the purified IBV preparation, indicating that the virus purification method used was effective at removing them. This was further evidenced by the lack of proteins present in mock-infected preparations of allantoic fluid, purified in the same way as IBV.
Following MS analysis, three viral proteins could be identified that were associated with the IBV virion. In comparison with a previous study of purified IBV virions (Kong et al., 2010), it was also possible to identify the IBV M protein. The M protein is the most abundant protein in the IBV virion (Stern et al., 1982), and the enhanced sensitivity of gel-free methods used here in comparison with difference gel elecrophoresis used by Kong et al. (2010) may explain why this protein was detected in this study. The E protein was not detected in either study. E is a small membrane protein involved in virus budding. It is known to be a very minor component of the viral particle, and the majority of mouse hepatitis virus (MHV) E remains within the cell at the site of virus budding (Venkatagopalan et al., 2015). This may explain why E could not be detected in either study.
Thirty-five host-cell proteins were identified in association with the IBV Beau-R virion. When the number of proteins identified in the individual preparations was compared, this number was very small, approximately 10 % of the total proteins identified. This indicated high batch-to-batch variability in the purification of virions by sucrose-density gradients but, interestingly, suggested that the virion itself is likely to be a carrier for many transiently associated proteins. Comparison of the groups of proteins identified among three preparations of purified IBV showed that the main differences were not in the functional types of protein that were identified but were in the individual proteins that fell into these groups. The number of groups that were represented in all three preparations was much smaller than the number of groups present in the individual preparations. This might be expected as the virus may target specific proteins for incorporation. Another consideration is that some of the proteins identified were subunits of whole proteins, resulting in reduced overlap between individual preparations. By comparing three separate preparations from allantoic fluid, proteins systematically associated with the virion were identified. Of the 35 proteins that were identified, 13 were identical to proteins identified in a previous proteomic study of the IBV virion (Kong et al., 2010). Eight proteins were orthologous to proteins identified in the related SARS-CoV virion (Neuman et al., 2008). As these studies were all performed using different approaches but these proteins were consistently identified, this suggests that these proteins are likely to be specifically incorporated into IBV, or more generally coronavirus, virions and may confer an advantage to the virus. However, the function they perform remains to be identified. In addition, 15 proteins were found to be unique to this study.
The host proteins identified could be split into 17 functional groups. Cytoskeletal proteins are a common group of proteins identified with virions, probably due to their abundance within the cell and the hijacking of the cytoskeleton to aid in assembly and transportation of virion components. β-Tubulin has previously been shown to associate with IBV (Kong et al., 2010). Vimentin, another cytoskeletal protein, was found here to be associated with the IBV virion. Interestingly, a proteomic study of subcellular fractions of IBV-infected Vero cells showed an increased abundance in the cytoplasmic fraction of vimentin, accumulating at virus-induced syncytia (Emmott et al., 2010). It has also been shown that expression of the SARS-CoV papain-like protease increased the levels of cellular vimentin due to activation of TGF-1 (Li et al., 2012). The presence of actin is also interesting, given that the IBV M protein is known to interact with actin (Wang et al., 2009). A number of binding proteins were identified such as annexin A2, destrin, S100 proteins and T-complex protein 1. All are known to interact with cytoskeletal proteins, and their presence may well be coincidental, as they were associated with the cytoskeletal protein at the time it was packaged with the virion. Annexin A2 binds to cytoplasmic actin (Gerke et al., 2005) and also binds tightly to S100 proteins to promote membrane fusion events and is involved in exocytosis (Lewit-Bentley et al., 2000).
Nucleic acid-binding proteins such as EIF4A, proteins of the 40S ribosomal subunit, EEF2 and proteins of the 60S ribosomal subunit were detected. Identification of ribosomal protein L13 is also interesting, indicating some interaction with the 60S ribosomal subunit and a component of the virus. A study of endoplasmic reticulum/Golgi-enriched fractions of MHV-infected cells identified other subunits of the 60S ribosome, which could not be identified in mock-infected cells (Vogels et al., 2011). It is not surprising to find ribosomal subunits associated with the virion, given that they are present at the endoplasmic reticulum where virion components are transported for virus assembly. Ribosomes have previously been found associated with SARS-CoV double-membrane vesicles (Knoops et al., 2008) but were not found in those of IBV (Maier et al., 2013).
A number of other interesting proteins were also identified associated with the IBV virion but have not been reported previously in association with other nidovirus virions. Chaperone proteins HSP8A and HSP90 were also identified, which are known to be involved in viral replication and assembly (Chromy et al., 2003; Momose et al., 2002; Hu & Anselmo, 2000; Swameye & Schaller, 1997). Energy generation and cellular metabolism proteins such as GAPDH were also found, which have previously been shown to interact with viral proteins (Kishimoto et al., 2012; Yi et al., 2000; Zang et al., 1998). Some of the proteins, however, may actually be present due to the growth of the virus in embryonated eggs and their abundance, rather than any kind of selective mechanism to include them. The egg contains a number of membranous structures and has numerous blood vessels running through it, which could account for the presence of proteins such as collagen, myosin and haemoglobin.
Further studies to investigate host-cell protein incorporation into nidovirus virions are warranted. The advantage of using IBV, an avian coronavirus, such as Beau-R, is that studies can take place in an in ovo system that is far less complex proteomically than cell-culture preparations. This system also allows studies to investigate the cellular localization of these proteins and how they may be redistributed during infection, providing important clues into both IBV replication and pathogenesis.
Methods
Viruses and cells
IBV Beau-R was propagated in 10-day-old, specific-pathogen-free Rhode Island Red chicken eggs. Eggs were inoculated with IBV into the allantoic cavity. At 24 h post-infection, eggs were sacrificed by refrigeration and allantoic fluid was harvested. Viruses were diluted for inoculation into 1 × BES IBV growth medium [1 × modified Eagle's medium (Sigma), 0.3 % tryptose phosphate broth (BDH), 0.2 % BSA (Sigma), 20 mM BES (N,N-bis(2-hydroxyethyl)-2-aminoethanesulphonic acid; Sigma), 0.21 % sodium bicarbonate, 2 mM l-glutamine, 250 U nystatin ml− 1, and 100 U penicillin and streptomycin ml− 1]. This was also used for mock infections by inoculation with 1 × BES only.
IBV purification
Purification was performed using the method of Dent & Neuman (2015). Briefly, harvested allantoic fluid was centrifuged at 1000 g for 10 min to pellet particulate material. IBV in allantoic fluid was concentrated by PEG precipitation. PEG-8000 and sodium chloride were added to the allantoic fluid to final concentrations of 10 and 2.2 %, respectively. The solution was stirred constantly on ice for 30 min once the PEG and sodium chloride had dissolved to allow precipitation of IBV. The suspension was then centrifuged at 10 000 g to pellet the precipitate. The precipitate was resuspended in HEPES saline [10 mM HEPES (pH 7.0), 0.9 % NaCl in water] and then layered onto sucrose. The virus formed a band between the 30 and 50 % sucrose cushions after centrifugation at 100 000 g for 90 min. Purifications of mock-infected allantoic fluid from eggs inoculated with 1 × BES IBV growth medium were also performed in the same way. Although it was possible to collect the interface between the two sucrose cushions, unlike IBV-infected allantoic fluid there was no cloudy white band present.
Analysis of purified virus by electron microscopy
Purified IBV virions were negatively stained using methylamine vanadate (NanoVan; Nanoprobes) before imaging on a Philips CM-20 with images recorded on a 2k × 2k charge coupled device.
Analysis of purified virus by SDS-PAGE
Purified virus was mixed with sample buffer [200 mM Tris/HCl (pH 6.8), 400 mM DTT, 8 % SDS, 0.4 % bromophenol blue, 40 % glycerol] and heated. Ten microlitres was loaded on a 4–20 % Tris/glycine SDS-PAGE gel. The gels were stained with colloidal Coomassie blue (Neuhoff et al., 1988). For mock-infected material purified by sucrose gradient, no proteins were found to be present in the sample.
MS analysis
Purified IBV was resuspended in 0.1 % RapiGest (Waters) diluted in 50 mM ammonium bicarbonate (pH 7.8). To lyse virions, the suspension was sonicated for 15 min and then heated to 80 °C for 10 min before a further 15 min sonication. Total protein concentration was determined by a BCA assay (Pierce) using the microplate method. Briefly, 25 μl protein sample was incubated with 200 μl BCA assay reagent for 30 min at 37 °C. Absorbance was measured at 562 nm and protein concentrations were determined from a BSA standard curve. Samples were reduced by the addition of DTT to a final concentration of 3 mM and heating at 60 °C for 10 min. Samples were cooled to room temperature and then alkylated by the addition of iodoacetamide to a final concentration of 9 mM and incubation at 20 °C for 10 min in the dark. Trypsin (Sigma-Aldrich) was reconstituted in 50 mM acetic acid to a concentration of 0.2 μg μl− 1. Samples were digested by the addition of 10 μl trypsin to the sample, followed by incubation at 37 °C overnight. The RapiGest was removed from the sample by acidification (addition of 1 μl trifluoroacetic acid and incubation at 37 °C for 45 min) and centrifugation (15 000 g for 15 min). The peptide mixtures were analysed by online nanoflow liquid chromatography using the nanoACQUITY-nLC system (Waters MS Technologies) coupled to an LTQ-Orbitrap Velos (ThermoFisher Scientic) mass spectrometer equipped with the manufacturer's nanospray ion source. The analytical column (nanoAcquity UPLC TM BEH130 C18, 15 cm × 75 μm, 1.7 μm capillary column) was maintained at 35 °C and a flow rate of 300 nl min− 1. The gradient consisted of 3–40 % acetonitrile in 0.1 % formic acid for 90 min, followed by a ramp of 40–85 % acetonitrile in 0.1 % formic acid for 3 min. Full-scan MS spectra (m/z range 300–2000) were acquired by the Orbitrap at a resolution of 30 000. Analysis was performed in data-dependent mode. The top 20 most intense ions from the MS1 scan (full MS) were selected for tandem MS by collision-induced dissociation and all product spectra were acquired in the LTQ ion trap. Ion trap and Orbitrap maximal injection times were set to 50 ms and 500 ms, respectively. Analysis of purified IBV was performed on three biological replicates, with one technical replicate of each.
Data analysis
Thermo RAW files were imported into Progenesis LC MS (version 4.1; Non-linear Dynamics). Replicate runs were time aligned using default settings. Peaks were picked by the software using default settings and filtered to include only peaks with a charge state of between +2 and +6. Peptide intensities of replicates were normalized against the reference run by Progenesis LC-MS. Spectral data were exported for peptide identification using Mascot version 2.3 (Matrix Science). Tandem MS data were searched against a custom database that contained the common contamination and internal standards such as keratins and trypsin, and the chicken database (UniProt, reviewed October 2013) concatenated to a custom database of IBV ORFs. The custom IBV database consisted of all known IBV ORFs and any other potential ORFs (an AUG start codon to a STOP codon) outside of the known reading frames greater than 50 aa, due to the recent discovery of the potential for IBV to encode extra proteins (Bentley et al., 2013). Only proteins that had at least two unique peptide matches to the identified protein and were present in all three preparations were classified as being associated with the IBV virion. Lists of all identified proteins were submitted to PANTHER.db to determine the functional classification of those proteins (Mi et al., 2013). The MS proteomics data have been deposited in the ProteomeXchange Consortium (Vizcaíno et al., 2014) via the PRIDE partner repository with the dataset identifier PXD002936.
Acknowledgements
This work was supported by a Biotechnology and Biological Sciences Research Council (BBSRC) studentship to SDD. The authors would like to thank Julian Hiscox, University of Liverpool, UK, for helpful discussions.
References
- Bechtel J. T., Winant R. C., Ganem D. (2005). Host and viral proteins in the virion of Kaposi's sarcoma-associated herpesvirus J Virol 794952–4964 10.1128/JVI.79.8.4952-4964.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley K., Keep S. M., Armesto M., Britton P. (2013). Identification of a noncanonically transcribed subgenomic mRNA of infectious bronchitis virus and other gammacoronaviruses J Virol 872128–2136 10.1128/JVI.02967-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursnell M. E., Brown T. D., Foulds I. J., Green P. F., Tomley F. M., Binns M. M. (1987). Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus J Gen Virol 6857–77 10.1099/0022-1317-68-1-57. [DOI] [PubMed] [Google Scholar]
- Brierley I., Boursnell M. E., Binns M. M., Bilimoria B., Blok V. C., Brown T. D., Inglis S. C. (1987). An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV EMBO J 63779–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casais R., Thiel V., Siddell S. G., Cavanagh D., Britton P. (2001). Reverse genetics system for the avian coronavirus infectious bronchitis virus J Virol 7512359–12369 10.1128/JVI.75.24.12359-12369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casais R., Davies M., Cavanagh D., Britton P. (2005). Gene 5 of the avian coronavirus infectious bronchitis virus is not essential for replication J Virol 798065–8078 10.1128/JVI.79.13.8065-8078.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. (2005). Coronaviruses in poultry and other birds Avian Pathol 34439–448 10.1080/03079450500367682. [DOI] [PubMed] [Google Scholar]
- Chertova E., Chertov O., Coren L. V., Roser J. D., Trubey C. M., Bess J. W., Jr, Sowder R. C., II, Barsov E., Hood B. L., other authors (2006). Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages J Virol 809039–9052 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chromy L. R., Pipas J. M., Garcea R. L. (2003). Chaperone-mediated in vitro assembly of polyomavirus capsids Proc Natl Acad Sci U S A 10010477–10482 10.1073/pnas.1832245100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C. S., Chen C. H., Ho M. Y., Huang C. Y., Liao C. L., Chang W. (2006). Vaccinia virus proteome: identification of proteins in vaccinia virus intracellular mature virion particles J Virol 802127–2140 10.1128/JVI.80.5.2127-2140.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent S., Neuman B. W. (2015). Purification of coronavirus virions for Cryo-EM and proteomic analysis Methods Mol Biol 128299–108 10.1007/978-1-4939-2438-7_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmott E., Rodgers M. A., Macdonald A., McCrory S., Ajuh P., Hiscox J. A. (2010). Quantitative proteomics using stable isotope labeling with amino acids in cell culture reveals changes in the cytoplasmic, nuclear, and nucleolar proteomes in Vero cells infected with the coronavirus infectious bronchitis virus Mol Cell Proteomics 91920–1936 10.1074/mcp.M900345-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke V., Creutz C. E., Moss S. E. (2005). Annexins: linking Ca2+ signalling to membrane dynamics Nat Rev Mol Cell Biol 6449–461 10.1038/nrm1661. [DOI] [PubMed] [Google Scholar]
- Guy J. S. (2015). Isolation and propagation of coronaviruses in embryonated eggs Methods Mol Biol 128263–71 10.1007/978-1-4939-2438-7_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson T., Britton P., Cavanagh D. (2006). Neither the RNA nor the proteins of open reading frames 3a and 3b of the coronavirus infectious bronchitis virus are essential for replication J Virol 80296–305 10.1128/JVI.80.1.296-305.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Anselmo D. (2000). In vitro reconstitution of a functional duck hepatitis B virus reverse transcriptase: posttranslational activation by Hsp90 J Virol 7411447–11455 10.1128/JVI.74.24.11447-11455.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattenhorn L. M., Mills R., Wagner M., Lomsadze A., Makeev V., Borodovsky M., Ploegh H. L., Kessler B. M. (2004). Identification of proteins associated with murine cytomegalovirus virions J Virol 7811187–11197 10.1128/JVI.78.20.11187-11197.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto N., Onitsuka A., Kido K., Takamune N., Shoji S., Misumi S. (2012). Glyceraldehyde 3-phosphate dehydrogenase negatively regulates human immunodeficiency virus type 1 infection Retrovirology 9107. 10.1186/1742-4690-9-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Kikkert M., Worm S. H., Zevenhoven-Dobbe J. C., van der Meer Y., Koster A. J., Mommaas A. M., Snijder E. J. (2008). SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum PLoS Biol 6e226. 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q., Xue C., Ren X., Zhang C., Li L., Shu D., Bi Y., Cao Y. (2010). Proteomic analysis of purified coronavirus infectious bronchitis virus particles Proteome Sci 829. 10.1186/1477-5956-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewit-Bentley A., Réty S., Sopkova-de Oliveira Santos J., Gerke V. (2000). S100-annexin complexes: some insights from structural studies Cell Biol Int 24799–802 10.1006/cbir.2000.0629. [DOI] [PubMed] [Google Scholar]
- Li S. W., Yang T. C., Wan L., Lin Y. J., Tsai F. J., Lai C. C., Lin C. W. (2012). Correlation between TGF-β1 expression and proteomic profiling induced by severe acute respiratory syndrome coronavirus papain-like protease Proteomics 123193–3205 10.1002/pmic.201200225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K. P., Liu D. X. (1998). Characterization of the two overlapping papain-like proteinase domains encoded in gene 1 of the coronavirus infectious bronchitis virus and determination of the C-terminal cleavage site of an 87-kDa protein Virology 245303–312 10.1006/viro.1998.9164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Lu Y., Denison M. R. (1996). Intracellular and in vitro-translated 27-kDa proteins contain the 3C-like proteinase activity of the coronavirus MHV-A59 Virology 222375–382 10.1006/viro.1996.0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier H. J., Hawes P. C., Cottam E. M., Mantell J., Verkade P., Monaghan P., Wileman T., Britton P. (2013). Infectious bronchitis virus generates spherules from zippered endoplasmic reticulum membranes MBio 4e00801–e00813 10.1128/mBio.00801-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann K., Mann M. (2011). In-depth analysis of the chicken egg white proteome using an LTQ Orbitrap Velos Proteome Sci 97. 10.1186/1477-5956-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., Casagrande J. T., Thomas P. D. (2013). Large-scale gene function analysis with the PANTHER classification system Nat Protoc 81551–1566 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momose F., Naito T., Yano K., Sugimoto S., Morikawa Y., Nagata K. (2002). Identification of Hsp90 as a stimulatory host factor involved in influenza virus RNA synthesis J Biol Chem 27745306–45314 10.1074/jbc.M206822200. [DOI] [PubMed] [Google Scholar]
- Neuman B. W., Joseph J. S., Saikatendu K. S., Serrano P., Chatterjee A., Johnson M. A., Liao L., Klaus J. P., Yates J. R., III, other authors (2008). Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3 J Virol 825279–5294 10.1128/JVI.02631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhoff V., Arold N., Taube D., Ehrhardt W. (1988). Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250 Electrophoresis 9255–262. [DOI] [PubMed] [Google Scholar]
- Nogales A., Márquez-Jurado S., Galán C., Enjuanes L., Almazán F. (2012). Transmissible gastroenteritis coronavirus RNA-dependent RNA polymerase and nonstructural proteins 2, 3, and 8 are incorporated into viral particles J Virol 861261–1266 10.1128/JVI.06428-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura M. M., Garnier A., Di Falco M. R., Whissell G., Meneses-Acosta A., Arcand N., Kamen A. (2008). Identification of host proteins associated with retroviral vector particles by proteomic analysis of highly purified vector preparations J Virol 821107–1117 10.1128/JVI.01909-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw M. L., Stone K. L., Colangelo C. M., Gulcicek E. E., Palese P. (2008). Cellular proteins in influenza virus particles PLoS Pathog 4e1000085. 10.1371/journal.ppat.1000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. F., Burgess L., Sefton B. M. (1982). Structural analysis of virion proteins of the avian coronavirus infectious bronchitis virus J Virol 42208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swameye I., Schaller H. (1997). Dual topology of the large envelope protein of duck hepatitis B virus: determinants preventing pre-S translocation and glycosylation J Virol 719434–9441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnum S. M., Streblow D. N., Monroe M. E., Smith P., Auberry K. J., Paša-Tolic L., Wang D., Camp D. G., II, Rodland K., other authors (2004). Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome J Virol 7810960–10966 10.1128/JVI.78.20.10960-10966.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatagopalan P., Daskalova S. M., Lopez L. A., Dolezal K. A., Hogue B. G. (2015). Coronavirus envelope (E) protein remains at the site of assembly Virology 47875–85 10.1016/j.virol.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Ríos D., Dianes J. A., Sun Z., Farrah T., other authors (2014). ProteomeXchange provides globally coordinated proteomics data submission and dissemination Nat Biotechnol 32223–226 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogels M. W., van Balkom B. W., Kaloyanova D. V., Batenburg J. J., Heck A. J., Helms J. B., Rottier P. J., de Haan C. A. (2011). Identification of host factors involved in coronavirus replication by quantitative proteomics analysis Proteomics 1164–80 10.1002/pmic.201000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Fang S., Xiao H., Chen B., Tam J. P., Liu D. X. (2009). Interaction of the coronavirus infectious bronchitis virus membrane protein with β-actin and its implication in virion assembly and budding PLoS One 4e4908. 10.1371/journal.pone.0004908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi M., Schultz D. E., Lemon S. M. (2000). Functional significance of the interaction of hepatitis A virus RNA with glyceraldehyde 3-phosphate dehydrogenase (GAPDH): opposing effects of GAPDH and polypyrimidine tract binding protein on internal ribosome entry site function J Virol 746459–6468 10.1128/JVI.74.14.6459-6468.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang W. Q., Fieno A. M., Grant R. A., Yen T. S. (1998). Identification of glyceraldehyde-3-phosphate dehydrogenase as a cellular protein that binds to the hepatitis B virus posttranscriptional regulatory element Virology 24846–52 10.1006/viro.1998.9255. [DOI] [PubMed] [Google Scholar]
- Zhang C., Xue C., Li Y., Kong Q., Ren X., Li X., Shu D., Bi Y., Cao Y. (2010). Profiling of cellular proteins in porcine reproductive and respiratory syndrome virus virions by proteomics analysis J Virol 7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., Snijder E. J., Gorbalenya A. E. (2000). Virus-encoded proteinases and proteolytic processing in the Nidovirales J Gen Virol 81853–879 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]