Abstract
Human members of the solute carrier 1 (SLC1) family of transporters take up excitatory neurotransmitters in the brain and amino acids in peripheral organs. Dysregulation of their functions is associated to neurodegenerative disorders and cancer. Here we present the first crystal structures of a thermostabilized human SLC1 transporter, the excitatory amino acid transporter 1 (EAAT1), with and without allosteric and competitive inhibitors bound. The structures show novel architectural features of the human transporters, including intra- and extracellular domains with potential roles in transport function, as well as regulation by lipids and post-translational modifications. The coordination of the inhibitor in the structures and the change in the transporter dynamics measured by hydrogen-deuterium exchange mass spectrometry, reveal an allosteric mechanism of inhibition, whereby the transporter is locked in the outward-facing states of the transport cycle. Our results provide unprecedented insights into the molecular mechanisms of function and pharmacology of human SLC1 transporters.
SLC1 transporters constitute a large family of ion-coupled transporters present in all kingdoms of life1. There are seven human SLC1 transporters (Extended Data Fig. 1) that evolved to serve two specialized functions2: in the central nervous system, SLC1 excitatory amino acid transporters (EAAT1-5) take up the neurotransmitter glutamate into the cell. In peripheral organs, EAATs take up glutamate and aspartate, while SLC1 neutral amino acid transporters (ASCT1-2) exchange small amino acids between the extra- and intracellular compartments, contributing to the cellular solute homeostasis.
Glutamate is the most important excitatory transmitter in the mammalian brain and it has to be continuously pumped into the cytoplasm to allow for rounds of transmission and prevent cytotoxicity. This essential neurological function is done by EAAT1-5 expressed at the plasma membrane of astrocytes and neurons3. In particular, astroglial EAAT1 and EAAT2 orthologs are highly expressed in the hind- and forebrain, respectively, and are responsible for most of the glutamate uptake in the rodent brain4. EAATs are powerful molecular pumps capable of maintaining up to 104-fold glutamate gradients by using energy stored in sodium, proton and potassium gradients5. Remarkably, their dysregulation has been associated with several neurological diseases, including amyotrophic lateral sclerosis6, ataxia7,8, stroke9, depression10 and glioma11, making them important drug targets.
ASCTs are structurally related to EAATs, and function as sodium-dependent neutral amino acid exchangers at the plasma membrane12. Importantly, ASCT2 is up regulated in several forms of cancer, including melanoma13, lung14, prostate15 and breast cancer16, and it is a key drug target in cancer therapy.
Despite the need for small compounds that selectively and allosterically modulate SLC1 human transporters, most of their pharmacology is based on substrate-analogs that inhibit transport competitively17,18. The only known selective allosteric modulators of SLC1 transporters are a series of non-competitive EAAT1-selective inhibitors, of which 2-Amino-4-(4-methoxyphenyl)-7-(naphthalen-1-yl)-5-oxo-5,6,7,8-tetrahydro-4H-chromene-3-carbonitrile (UCPH101) is the best studied19,20. However, its mechanism of action is still poorly understood at the molecular level.
In structural terms, most our knowledge on the transport mechanism and pharmacology of SLC1 transporters comes from the prokaryotic homolog GltPh that has been crystallized in the main conformational states of the transport cycle, outward-21 and inward-facing states22,23, as well as in complex with a non-selective and competitive inhibitor of the EAATs24, DL-threo-β-benzyloxyaspartic acid (TBOA). However, the presence of amino acid insertions and deletions, as well as important differences in the transport function and pharmacology of GltPh, make this homolog a limited structural model to understand the molecular mechanism of the human SLC1 proteins.
Here we present 3.1-3.3 Å X-ray crystal structures of thermostable EAAT1 variants in complex with a substrate (L-aspartate), and the allosteric inhibitor UCPH101. The structures, and supporting functional data, show new architectural features of the EAATs and ASCTs, and unravel the allosteric mechanism of UCPH101-like inhibitors in atomic detail. Taken together, these structural data can prove useful for the design of novel allosteric compounds with improved selectivity for both EAATs and ASCTs.
EAAT1 engineering and crystallization
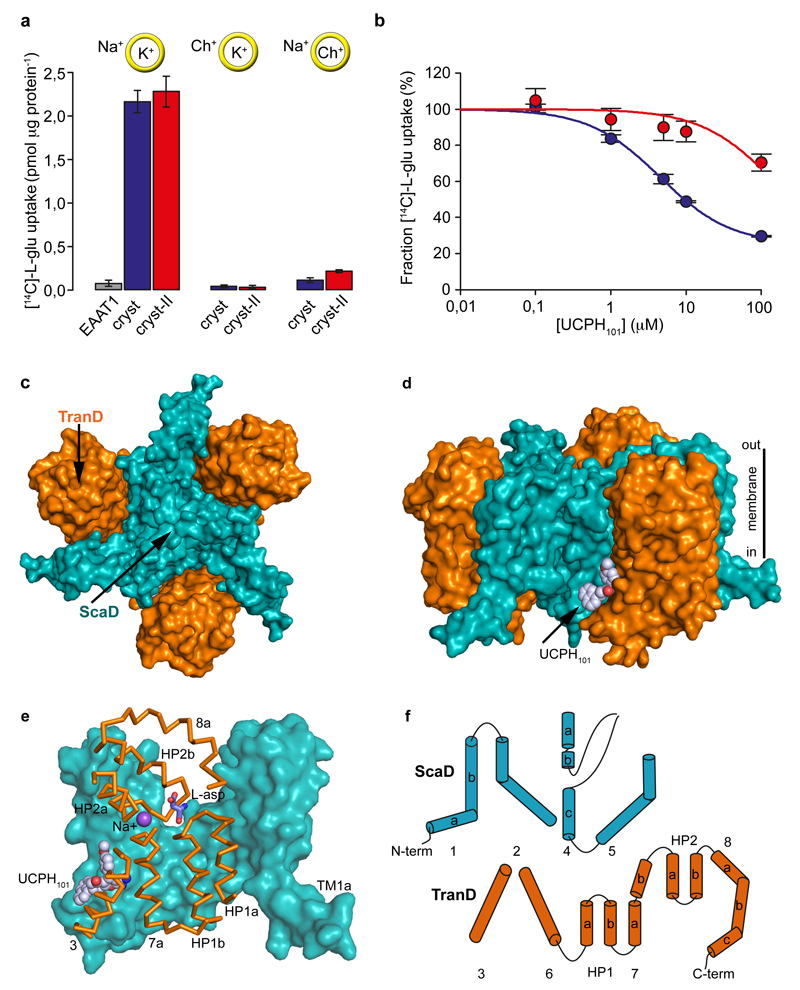
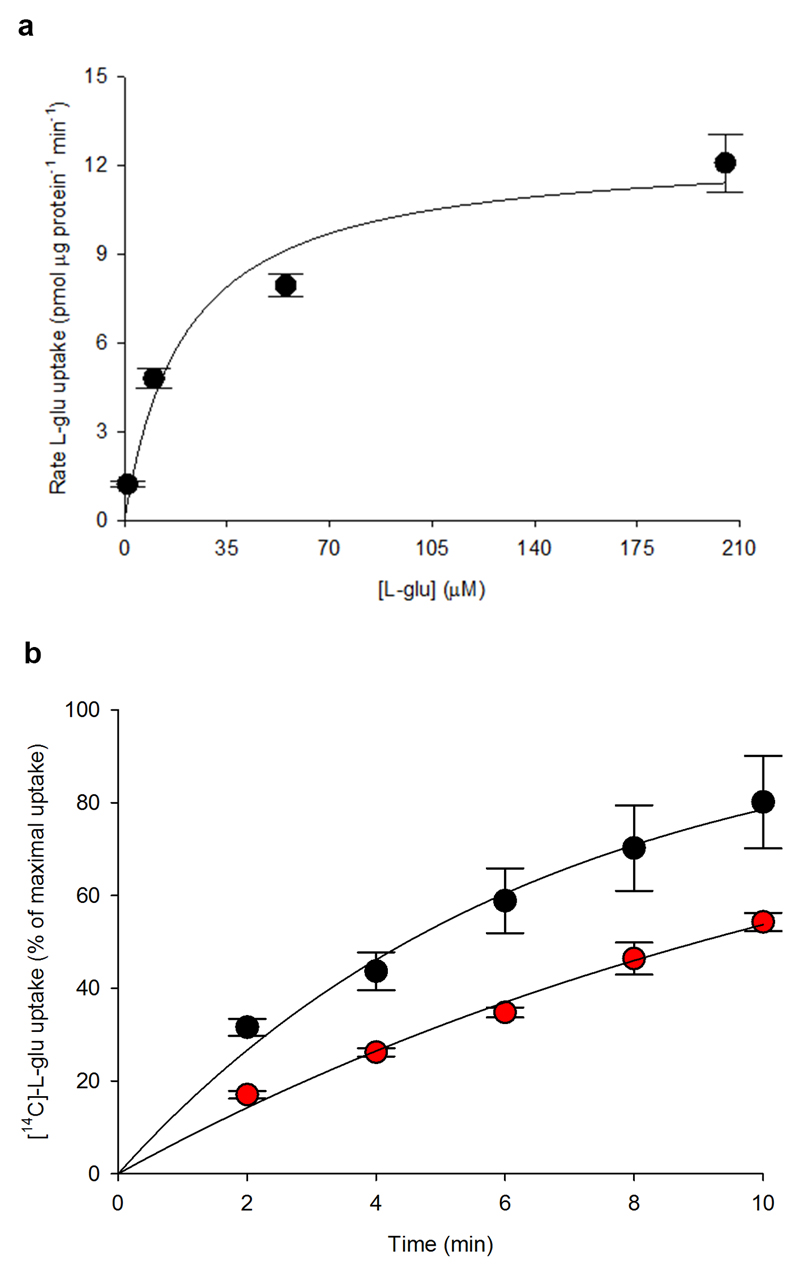
Purified wild-type EAAT1 lacks transport activity upon reconstitution in synthetic liposomes (Fig. 1a), and was recalcitrant to crystallization. To obtain functional protein suitable for crystallographic studies, we engineered a thermostable EAAT1, called EAATcryst that shares an overall ~75% sequence identity with the wild type, and up to ~90% identity at the C-terminal core of the protein (Extended Data Fig. 1; Methods), where the transported substrate and coupled ions are expected to bind24–29. Indeed, purified EAAT1cryst reconstituted in liposomes showed robust glutamate uptake that depends on opposite gradients of sodium and potassium ions across the bilayer (Fig. 1a and Extended Data Fig. 2a), and was inhibited by the EAAT1-selective compound UCPH101 (IC50 of 4.5±0.3 μM, Hill coefficient 0.92±0.07) (Fig. 1b). These data show that the transport mechanism and pharmacological selectivity are conserved in EAAT1cryst.
Figure 1. Function and architecture of EAAT1cryst.
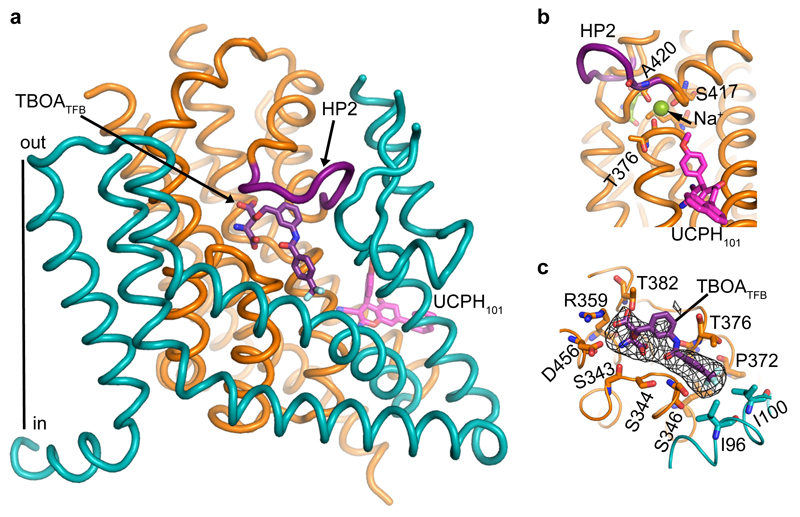
a-b, Uptake of radioactive L-glutamate by purified EAAT1 (grey), EAAT1cryst (blue), and EAAT1cryst-II (red) reconstituted in liposomes. Transport was abolished when choline (Ch+) was used in the extra- or intra-liposomal solutions (yellow circles) (a). UCPH101 inhibits glutamate transport in a concentration dependent manner (b). Plots depict an average of three independent experiments performed with duplicate measurements, and error bars represent s.e.m. c-d, Structure of EAAT1cryst trimer viewed from the extracellular solution (c) and from the membrane (d) highlighting the ScaD (teal) and TranD (orange). e, EAAT1cryst monomer viewed parallel to the membrane. The ScaD domain is represented as surface (teal), and several helices and loops in the TranD (orange) have been removed. f, Domain organization diagram of EAAT1cryst monomer.
Notably, EAAT1cryst crystallized in the presence of UCPH101, and we solved its inhibitor-bound structure (see Methods and Extended Data Table 1), but it was refractory to crystallization in the absence of UCPH101. To overcome this, we introduced M231I and F235I mutations in the inhibitor-binding pocket (Extended Data Fig. 1), and solved the crystal structures of the so-called EAAT1cryst-II both in the presence and absence of the inhibitor (Extended Data Table 1). Purified EAAT1cryst-II also showed robust sodium- and potassium-dependent glutamate uptake, while the UCPH101 IC50 increased >30-fold (>131±38 μM, Hill coefficient 0.92±0.0; Fig. 1a,b), as expected due to the mutations in the inhibitor binding pocket (see below).
Domain organization
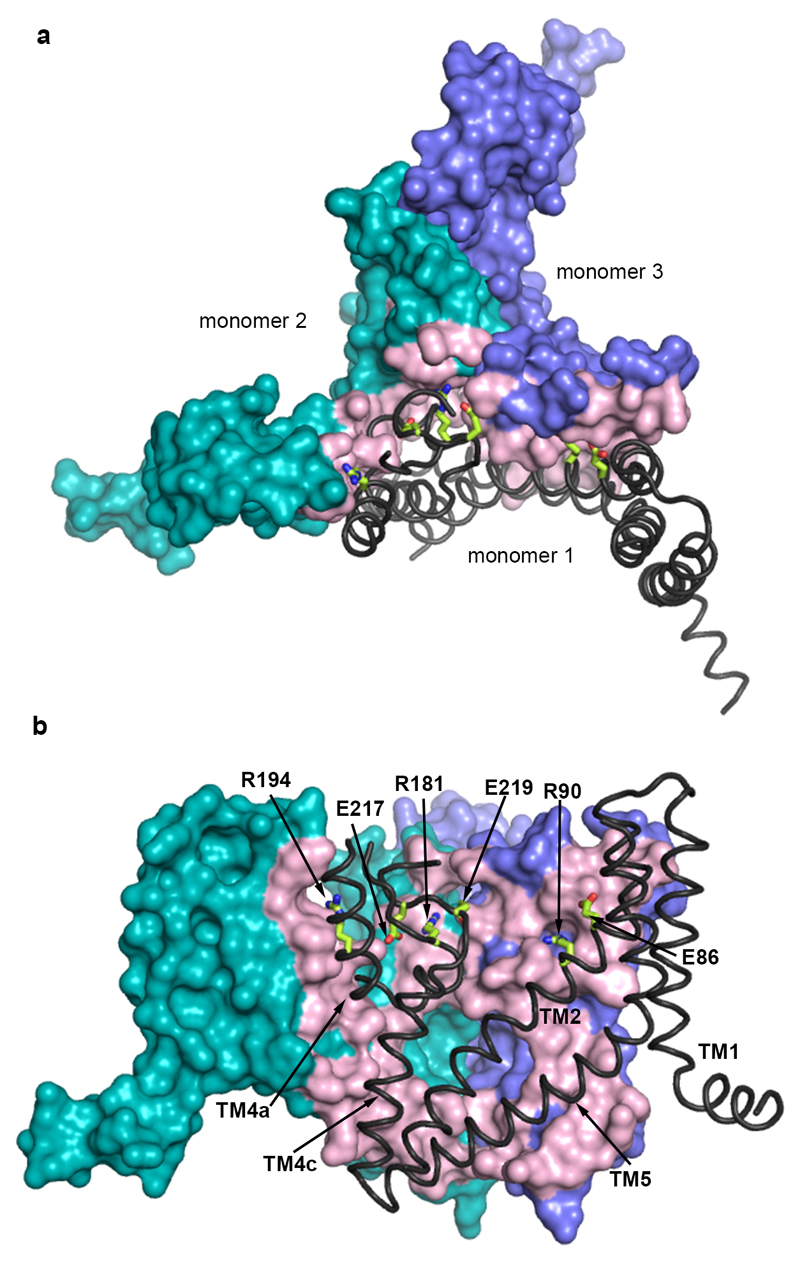
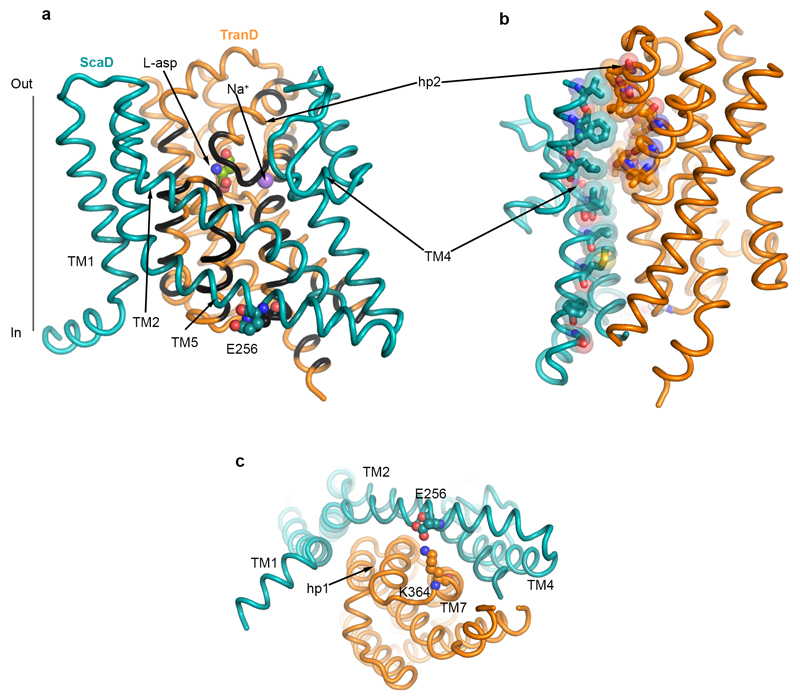
The structure of EAAT1cryst shows a symmetric homotrimer in a substrate- and UCPH101-bound outward-facing conformation (Fig. 1c-e) with an overall GltPh-like fold21,22 (Extended Data Fig. 3). Each monomer is composed of two domains: a scaffold domain (ScaD), including transmembrane helices TM1-2 and TM4-5; and a transport domain (TranD), including TM3, TM6-8 and re-entrant helical loops 1-2 (HP1-2; Fig. 1f). The three ScaDs form a compact central structure with a propeller-like shape that ensures the trimeric form of the transporter and anchors it to the membrane (Extended Data Fig. 4). The three TranDs are more peripheral and localize between the blades of the propeller, making protein contacts exclusively with the ScaDs of their own monomer. The TranD-ScaD interface buries ~3,500 Å2, including a conserved salt bridge between E256 and K364, through interactions of residues in the cytoplasmic parts of HP1, TM7, and TM3 (TranD), and TM2, TM4c, and TM5 (ScaD). On the extracellular side, additional contacts occurred between residues in HP2 and TM4 that are well conserved among human transporters (Extended Data Fig. 5).
Substrate and ion translocation in SLC1 transporters is thought to occur through large rigid-body movements of the TranD, relative to the static ScaD, that move the cargo in an elevator-like fashion across the membrane22,30. Thus, during the isomerization to the inward-facing state the TranD-ScaD interface changes drastically on the TranD side, and the novel features observed at this interface in EAAT1cryst might influence the distinct TranD dynamics in human SLC1 proteins.
Transport domain
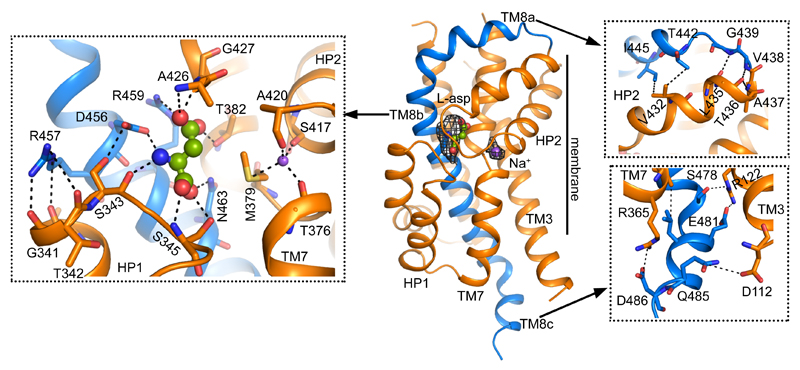
One of the most remarkable architectural features of the EAAT1cryst TranD is in TM8, where there are deletions and insertions compared to prokaryotic homologs (Fig. 2 and Extended Data Fig. 3). In EAAT1cryst, TM8 can be divided into extracellular (TM8a), transmembrane (TM8b), and cytoplasmic (TM8c) helices. The loop connecting TM8a and HP2 is six residues shorter in human SLC1 transporters, and brings their extracellular ends in close contact through hydrogen bonding and hydrophobic interactions (Fig. 2). HP2 is a dynamic element that controls the access of substrate and ions to their binding sites in the TranD24,31–33, and its interactions with TM8b likely play an important role in determining HP2 movements. Consistently, single-cysteine mutations at positions along TM8a in EAAT134, and in a rodent EAAT2 ortholog35 impaired glutamate transport, highlighting the significance of this extracellular region for function.
Figure 2. Transport domain.
EAAT1cryst TranD with substrate (spheres) and one sodium ion (purple spheres) bound. Omit Fo-Fc densities contoured at 2.3 σ (black mesh) show the position of the ligands. TM8a-c (blue) interacts with other regions on the TranD (orange), including HP1 (left inset), HP2 (upper right inset), and TM3 and TM7 (lower right inset).
In TM8b, we found strong electron density for the substrate (L-aspartate) and one of the sodium ions (Na2) in similar sites than in the structures of prokaryotic homologs24,29 (Fig. 2). Remarkably, the carboxylate group of D456 (TM8b), which coordinates the α-amino group of the substrate, is also at hydrogen bond distance with the hydroxyl group of S343 (HP1). Moreover, the guanidinium group of R457 (TM8b) engages in hydrogen bonding with HP1 residue G341, and possibly L340 and T342 that point their backbone carbonyl oxygen atoms towards TM8b. Residues S343 and R457 are well conserved in human SLC1 transporters, and substitutions at equivalent positions in EAAT1 (S363 and R477)36, and EAAT3 (R445)37 inactivate transport. In addition, the loss-of-function mutation R445W in EAAT3, equivalent to R457 in EAAT1cryst, causes human dicarboxylic aminoaciduria38. Overall, the functional studies and our structural data converge to suggest that interactions between conserved human residues at HP1 and TM8b are important to the correct folding and function of the transporters.
On the cytoplasmic side, TM8 extends beyond the membrane through a hydrophilic helix (TM8c) that makes contact with TM3 and TM7a (Fig. 2). EAAT2 deletion mutants in this region have a deleterious effect on transport function and membrane trafficking39. Accordingly, an EAAT1 deletion in TM8c decreased glutamate uptake rate by ~2-fold (Extended Data Fig. 2b). Indeed, the functional data and the TM8c amino acid conservation among EAATs underscore the pivotal role of this novel structural motif in protein folding and transport kinetics.
Scaffold domain
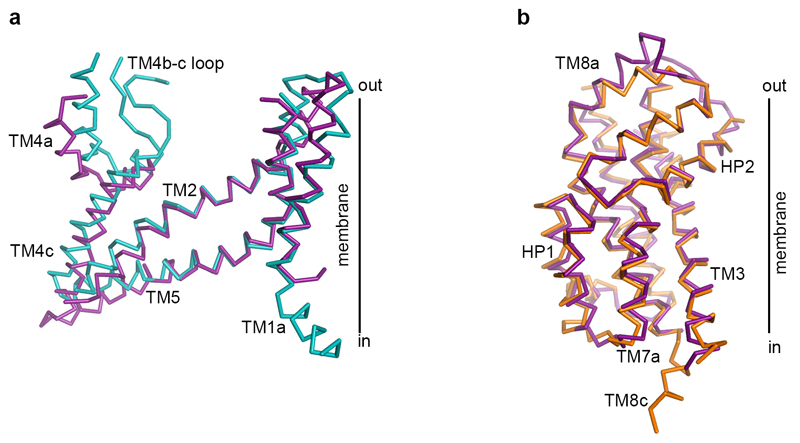
The ScaD contains highly divergent regions, especially in TM4 (Extended Data Fig. 1), where EAAT1cryst shows several unique architectural features. On the extracellular side, TM4a forms inter- and intra-monomeric contacts with TM2 and HP2, respectively. Moreover, an amino acid insertion between TM4b and TM4c (TM4b-c loop) that appeared during the evolution of eukaryotic transporters protrudes into the central vestibule of the EAAT1cryst trimer (Fig. 3a and Extended Data Fig. 4). The TM4b-c loop forms the center of the propeller, and makes extensive contacts within and between protomers. Although, we could not model the outermost residues in the TM4b-c loop (Y200-V210), they are expected to each out to the bulk solvent exposing an N-glycosylation site (N204). Notably, all human SLC1 transporters contain predicted N-glycosylation sites in the TM4b-c loop, suggesting a conserved role of this loop in the posttranslational processing of these proteins.
Figure 3. Scaffold domain.
a, TM4 (purple) makes extensive contacts with the ScaD of the neighboring monomer (monomer 2, teal surface), and the TranD (orange) of the same subunit (monomer 1). The TranD of monomer 2, and several TMs of monomer 1 were removed for clarity of display. Fo-Fc density in the HP2-TM4 crevice is contoured at 2.7σ (red mesh). b, Two monomers of EAATcryst show TM1a laying parallel to the membrane. The third monomer is not shown. TM1 (teal) and HP1a (dark blue) form a hydrophobic crevice containing non-protein Fo-Fc density (purple mesh, contour 2.7σ).
On the cytoplasmic side, TM1 bends and re-orients its N-terminal part (TM1a) nearly parallel to the membrane plane, which forms the tips of the propeller’s blades (Fig. 1c and Fig. 3b). The position and amphipathic nature of TM1a suggests that it somehow interacts with the inner leaflet of the membrane. Consistently, in the hydrophobic crevice between TM1a and HP1a, we observe strong non-protein electron density that likely corresponds to bound detergent or lipid molecules (Fig. 3b). Interestingly, a second hydrophobic crevice between the extracellular part of TM4 and HP2, also shows strong non-protein electron density (Fig. 3a). A similar crevice was also noted in a substrate-bound structure of GltPh21.
Lipids strongly regulate SLC1 transport function40–44, and those hydrophobic crevices at the interface between the TranD and ScaD, where large conformational changes occur during substrate translocation, might constitute sites for lipid regulation of transport function.
UCPH101 binding site
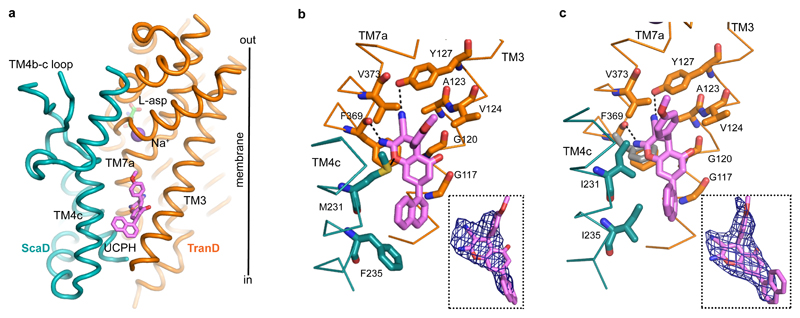
The structure of EAAT1cryst showed strong electron density for UCPH101 in a hydrophobic pocket between TM3, TM7 and TM4c at the TranD-ScaD interface, where the compound extends the interface by ~ 500 Å2 (Fig. 1d,e and Fig. 4a,b). The chromene skeleton, the parental group of the UCPH series of compounds, is buried deeply in the domains interface, and coordinated by a direct ring-stacking interaction with F369 (TM7a), as well as hydrophobic interactions with G120 (TM3), V373 (TM7a) and M231 (TM4c) (Fig. 4b). In addition, the amine group of UCPH101 forms a hydrogen bond with the main-chain carbonyl of F369, while its carbonitrile group interacts with Y127 (TM3). The methoxy-phenyl and naphthalene groups appear partially facing the hydrocarbon core of the membrane. Yet, the former establishes hydrophobic interactions with V124 (TM3), V373 and M231, while the latter is mainly coordinated by F235 (TM4c). The majority of the above-mentioned residues are important for UCPH101 transport inhibition of an EAAT1 rodent ortholog20. Furthermore, the M231I-F235I double mutant (EAAT1cryst-II) showed >30-fold increase in UCPH101 IC50 compared to EAAT1cryst in proteo-liposomes (Fig. 1b). Hence, there is an excellent agreement between the crystallographic and functional data.
Figure 4. UCPH101 binding site.
a, Lateral view of EAAT1cryst monomer showing UCPH101 bound (pink) between the TranD (orange) and ScaD (teal). b-c, UCPH101 coordination and Fo-Fc densities contoured at 2.0σ (blue mesh) in EAAT1cryst (b) and EAAT1cryst-II (c), respectively. Side chains of residues in TM3, TM4c, and TM7 involved in coordination are shown. F369 side chain moves outward in the EAAT1cryst-II unbound state (grey) (c).
Several mechanistically-relevant observations can be made regarding the UCPH101 binding pocket in EAAT1cryst: i) it is over 15 Å away from the substrate and sodium binding sites, suggesting that UCPH101 does not preclude extracellular substrate binding; ii) it faces the inner leaflet of the membrane, implying that UCPH101 accesses its binding site from the lipidic, and not the aqueous phase, when applied extracellularly; iii) it is fully contained in a single subunit, in agreement with the lack of cooperativity observed in proteo-liposome (Fig. 1b), and cell assays20; iv) a comparison of the EAAT1-5 sequences suggests that the main determinants of UCPH101 selectivity for EAAT1 are in TM4c, where M231 and F235 are the only coordinating residues that differ between EAAT1 and all other EAATs (Extended Data Fig. 1). Consistently, the double mutant EAAT1cryst-II containing EAAT2 residues equivalent to M231-F235 shows a large IC50 increase.
UCPH101-unbound state
To better understand the conformational changes of the transporter induced by UCPH101, we set out to determine the structure of the EAAT1cryst UCPH101-unbound state, but it was refractory to crystallization. Instead, we solved the structure of the EAAT1cryst-II UCPH101-unbound state and for comparison, the EAAT1cryst-II UCPH101-bound state, using an excess of the compound in the crystallization conditions (see Methods).
The EAAT1cryst-II UCPH101-bound state is nearly identical to that of the EAAT1cryst with the exception of the I231 and I235 side chains, and a ~2 Å movement of UCPH101 methoxy-phenyl and naphthalene groups away from them (Fig. 4c and Extended Data Fig. 6a). However, the EAAT1cryst-II UCPH101-unbound structure shows notable differences with bound state. First, there is no excess electron density for UCPH101, and the side chain of F369 moves outward by as much as 1.9 Å, partly occupying the volume of the UCPH101 chromene group (Fig. 4c). Second, there is a small rigid-body movement of the entire TranD that shifts by as much as 0.7 Å, compared to the UCPH101-bound structures (Extended Data Fig. 6b). This conformational change shows the EAAT1cryst TranD is able to move as rigid bodies relative to the ScaD, and highlights the importance of such movements for the function of the human transporters, as it has been shown for GltPh22,23.
The structural changes observed in the UCPH101-unbound structure unambiguously demonstrate that the assigned binding pocket of UCPH101 is correct, and that within the restricted environment of the crystal lattice, UCPH101 induces both local and global conformational changes of the transporter that optimize its coordination in an outward-facing state.
Transport domain dynamics
The coordination of UCPH101 in the crystal structures, wedged in the TranD-ScaD interface, as well as the effect of the M231I-F235I mutations on its potency strongly suggest that UCPH101 inhibits transport by trapping the transporter in an outward-facing state. Consistently, the rigid-body movements of the TranD to isomerize the transporter into the inward-facing state would separate the coordinating residues in the TranD from those in the ScaD, and disrupt the UCPH101 coordination. Hence, under equilibrium conditions where the transporters are sampling outward- and inward-facing states, the expected effect of UCPH101 binding is to shift the equilibrium in favor of the outward-facing state.
To gain insights into the effects of UCPH101 binding to the transporters at equilibrium, we probed the detergent solubilized EAAT1cryst by hydrogen-deuterium exchange mass spectrometry (HDX-MS). HDX-MS measures the rate of exchange of backbone amide hydrogen atoms that depends on solvent accessibility and hydrogen bonding, and provides valuable information on the dynamics and conformational changes of proteins45,46.
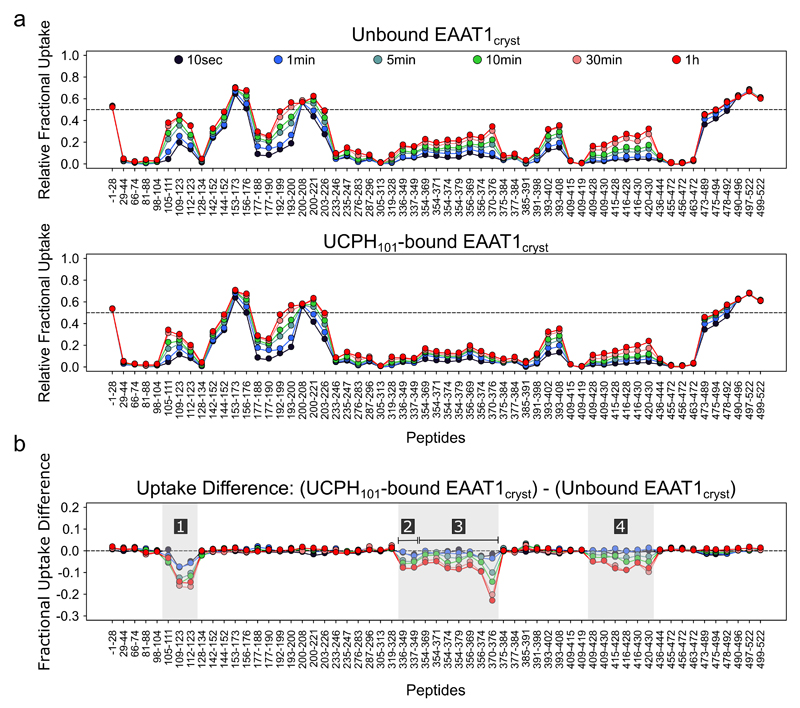
We compared the HDX behavior of the EAAT1cryst in the presence and absence of UCPH101. Overall, the deuterium uptake pattern of EAAT1cryst shows dynamic structural elements in both the TranD and ScaD (Extended Data Fig. 7-8), and reveals the unstructured and solvent-exposed nature of several regions that were not resolved in the crystal structures, including the TM3-TM4a (peptide 153-173) and TM4b-c (peptide 200-208) loops, as well as the N- (peptide 1-28) and C-termini (peptides 490-522) (Extended Data Fig. 9).
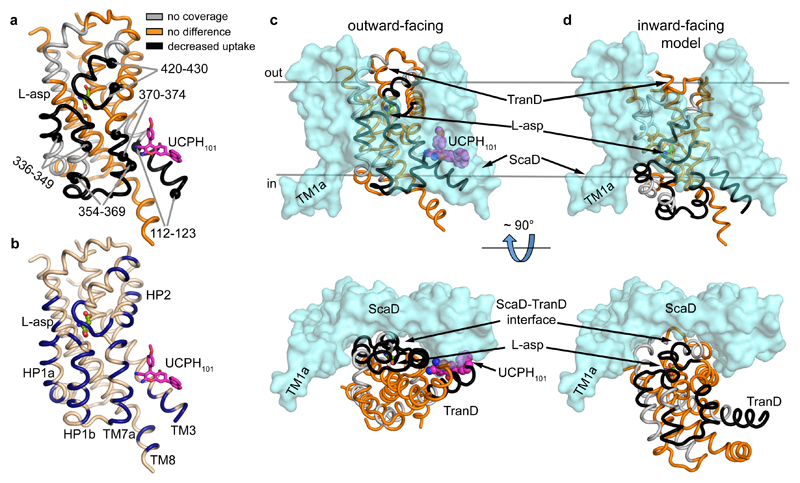
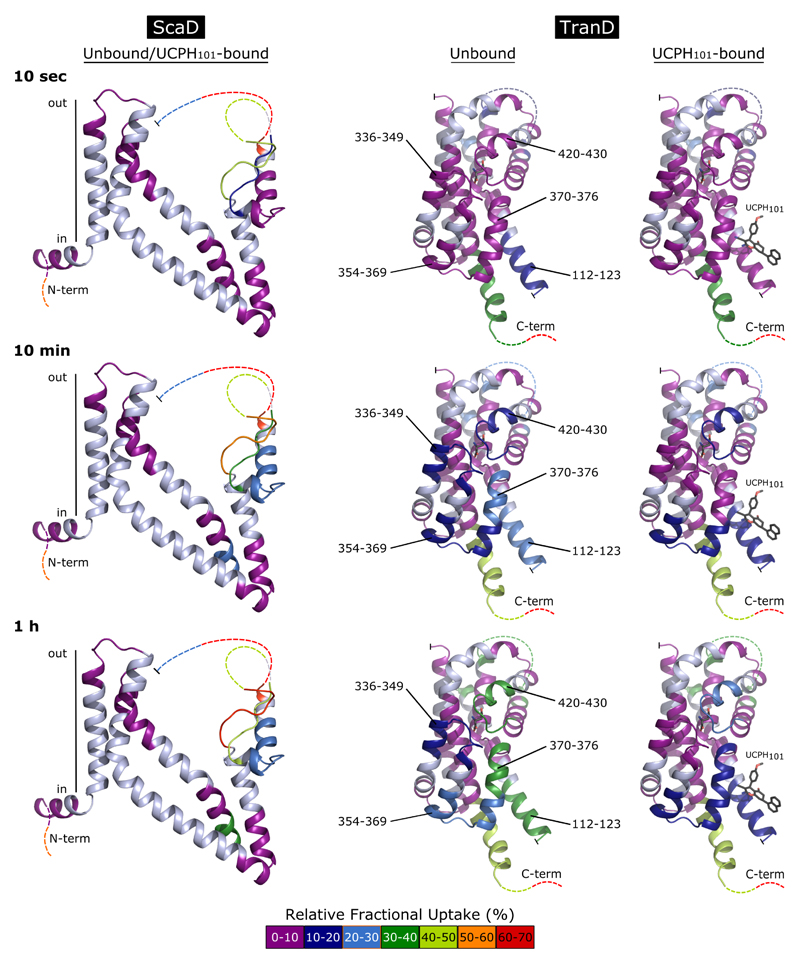
Binding of UCPH101 significantly decreased deuterium uptake in several areas of the TranD including its binding pocket (residues 112-123 and 370-374), and the surrounding area (residues 354-369), while it left the uptake in the ScaD unchanged (Fig. 5a). It also decreased deuterium uptake in distant residues (336-349 and 420-430) at the tips of HP1 and HP2 involved in substrate coordination and occlusion, suggesting that UCPH101 induces conformational changes in the transporter upon binding. To gain insights into the nature of those conformational changes, we first compared the TranD areas in which UCPH101 decreased uptake with those buried at the interface with ScaD in the EAAT1cryst structure, and found that they correlate remarkably well (Fig. 5a-c). Second, we built a model of the EAAT1cryst inward-facing state, based on a recently solved structure of GltPh23, to assess the changes in solvent accessibility in a possible transition between inward- and outward-facing states (Fig. 5c,d). Indeed, the comparison between the structure and the model shows that the UCPH101-modified areas detected by HDX-MS transit as rigid bodies from being solvent-exposed, in the inward-facing state, to buried at the TranD-ScaD interface, in the outward-facing state. Such conformational change is expected to decrease the dynamics of alpha helices and/or the solvent accessibility of the loops in the UCPH101-modified areas and thus, is consistent with the observed decrease in deuterium uptake. Overall, the HDX-MS and structural analysis support the stabilization of the outward-facing state, at the expense of the inward-facing state(s), induced by UCPH101.
Figure 5. Transport domain dynamics.
a, UCPH101 decreased deuterium uptake in several regions (black) of the TranD at the interface with the ScaD. The alpha carbon atoms delimiting such regions are shown as black spheres. Unaffected regions of the TranD (orange) or those not detected during HDX-MS analysis (grey) are also highlighted (sequence coverage ~77%). b, TranD residues buried at the interface with the ScaD, in the UPCH101-bound outward-facing state, are colored dark blue for comparison with (a). c-d, Comparison between EAAT1cryst structure in the UPCH101-bound outward-facing state (c) and a model of an inward-facing state based on the GltPh “unlocked” state (PDB 4X2S) (d), in which the TranD (color code as in a) undergoes a large rigid body movement towards the cytoplasm.
UCPH101- and TBOATFB-bound state
The distant position of UCPH101 from the substrate and HP2, a structural element that controls extracellular access to the substrate binding site24, suggests that the UCPH101-bound transporters could undergo the conformational changes required to exchange the substrate with the extracellular solution. To test this, we solved the crystal structure of EAAT1cryst in complex with both UCPH101 and (2S,3S)-3-[3-[4-(trifluoromethyl)benzoylamino]benzyloxy]aspartate (TBOATFB), a potent and non-selective TBOA derivative47, at 3.7 Å resolution (Extended Data Table 1).
Overall, the UCPH101-TBOATFB-bound structure is similar to that of the UCPH101-bound state, with the exception of HP2 that adopts an “open” conformation and packs against the TM4b-c loop, disrupting the coordination of the Na2 (Fig. 6a,b). These conformational changes resemble those previously observed in the structure of the GltPh-TBOA complex, and are in excellent agreement with the proposed competitive inhibitory mechanism of TBOA-like compounds24.
Figure 6. UCPH101-TBOATFB bound EAAT1cryst structure.
a, The movement of HP2 (purple) partly exposes the substrate-binding pocket to the solvent and shows a molecule of TBOATFB (purple sticks) bound to it. UCPH101 is also observed in this structure (pink sticks). b, The tip of HP2 moves as much as 9.5 Å in the UCPH101-TBOATFB- (purple), compared to the UCPH101-substrate bound (orange) structures, placing the carbonyl oxygen of A420 away from Na2 (green sphere). c, Omit map Fo-Fc density for the TBOATFB molecule is contoured at 2.3σ (black mesh), and some of the residues at Van deer Waals or H-bond distance from the compound are represented as sticks.
In the substrate-binding site, we observed excess electron density for the bulky TBOATFB (Fig. 6c), but due to lack of resolution, we were not able to unambiguously orient the compound. Hence, we initially positioned the TBOA moiety of TBOATFB using the TBOA-bound GltPh structure as a guide. After crystallographic refinement, the additional benzoylamino and trifluoromethyl groups of TBOATFB in EAAT1cryst localized in a hydrophobic cavity mainly formed by residues in HP1b and TM7a, and possibly by residues in TM2 and TM4c, likely explaining the observed ~1500-fold increase in EAAT1 inhibitory potency over TBOA47.
Indeed, in the TBOATFB-bound structure UCPH101 is bound with an identical coordination than in the substrate-bound state. Therefore, the EAAT1cryst structures show that UCPH101 binding at its allosteric site does not preclude the movements of HP2 involved in substrate and sodium binding from the extracellular solution.
Inhibitory mechanisms of EAAT1
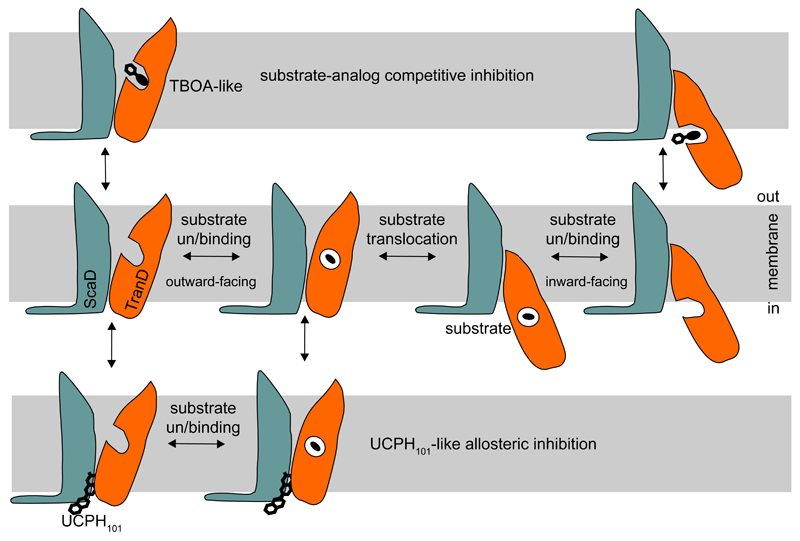
The structural and functional EAAT1cryst data reveal new architectural features of human SLC1 transporters, and their first molecular mechanism of allosteric inhibition. UCPH101 binding “glues” the TranD to the ScaD in the outward-facing states, and precludes substrate translocation, but not binding from the extracellular solution (Fig. 7). Non-conserved residues in the ScaD determined UCPH101 selectivity for EAAT1. This inhibitory mechanism contrasts with that of substrate-analog inhibitors like TBOA. The binding pocket of TBOA-like compounds overlaps with the conserved substrate binding site24, explaining their lack of selectivity among glutamate transporters and the inhibition of substrate binding observed on either side of the membrane48,49.
Figure 7. EAAT1 inhibitory mechanisms.
Schematic representation of TBOA-like competitive and UCPH101-like allosteric inhibition. Competitive inhibitors, occupy the substrate-binding pocket and preclude substrate binding. UCPH101-like compounds bind at the TranD-ScaD interface and block the movement of the TranD (orange) relative to the ScaD (teal).
The unique inhibitory mechanism of UCPH101 makes it an extremely valuable pharmacological tool to study the conformational changes that EAAT1 undergoes upon substrate and ion binding. Remarkably, the UCPH101 allosteric binding site highlights a cavity that can facilitate the design of selective compounds for other human SLC1 transporters, and possibly the long-sought positive modulators of glutamate uptake.
Methods
Construct optimization
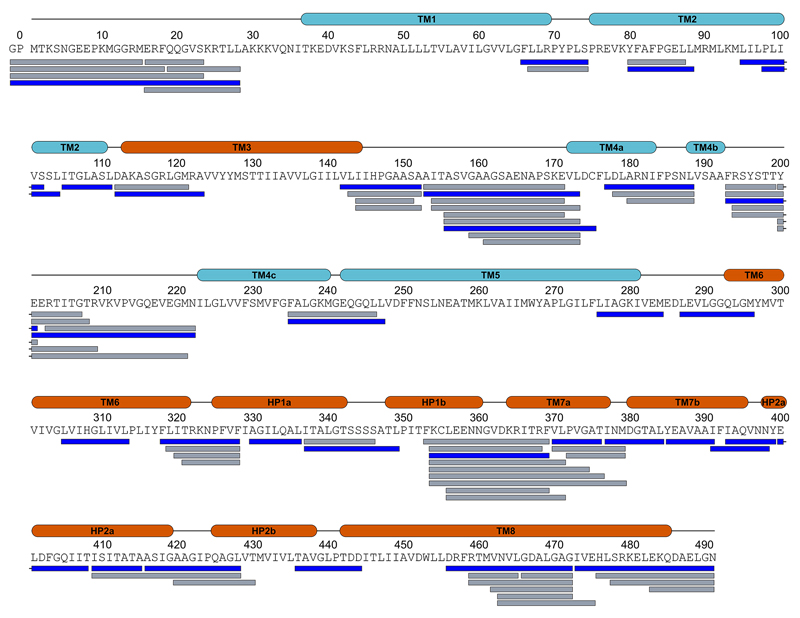
We used fluorescence-detection size-exclusion chromatography (FSEC)50 to screen solubilization conditions and EAAT1 variants fused to enhanced green fluorescent protein (eGFP). EAAT1 N-terminal fusions solubilized in dodecanoyl sucrose (DDS, Anatrace) were found to have good solubility and mono-dispersity by FSEC in clear lysates. However, EAAT1 looses its transport activity and chromatographic monodispersity upon purification. To increase its stability, we used consensus mutagenesis51, and screened EAAT1 variants with different consensus mutations in the predicted transmembrane helices by FSEC. The apparent melting temperature (Tm) of the most stable EAAT1 construct was >20 °C over that of the wild-type EAAT1, but the mutated transporter was still refractory to crystallization. We reasoned that the divergent extracellular region between TM3-4c could interfere with crystallization, and changed it for the equivalent region in ASCT2, the shortest one among human SLC1 members (Extended Data Fig.1). In addition, we mutated the two predicted N-glycosylation sites of the transporter (N155T and N204T mutations) to further improve crystallizability.
Expression and purification
All constructs were introduced into pcDNA3.1(+) (Invitrogen) with N-terminal Strep-tag II affinity tag followed by eGFP and PreScission protease cleavage site, and expressed in HEK293F cells (ATCC, mycoplasma test negative) grown in Excell293 medium (Sigma) and supplemented with 4mM L-glutamine (Sigma) and 5µg/ml Phenol red (Sigma-Aldrich) to densities of 2.5 x 106 cells ml-1. Cells were transiently transfected in Freestyle293 medium (Invitrogen) using poly-ethylenimine (PEI) (Polysciences) at a density of 2.5 x 106 cells ml-1, diluted with an equivalent volume of Excell293 6 hours post-transfection, and treated with 2.2 mM valproic acid (Sigma) 12 hours after dilution of the cultures. Cells were collected at ~48 h post-transfection.
Initial screens of constructs and detergent solubilization buffers were done in small-scale (5-10 ml), and cells were collected, mechanically disrupted with a douncer and solubilized in 50 mM HEPES/Tris-base, pH 7.4, 200 mM NaCl buffer supplemented with 1 mM L-asp, 1 mM EDTA, 1 mM Phenylmethylsulfonyl fluoride (PMSF), 1 mM Tris(2-carboxyethyl)phosphine (TCEP), 1:200 (v/v) dilution of mammalian protease inhibitor cocktail (Sigma), 10 % glycerol, 2% detergent and 0.4% cholesterol hemisuccinate (CHS) (Anatrace). After 1-hour incubation at 4 °C, clear lysates were obtained by ultracentrifugation (247,000 g for 45 min). A high-throughput auto-sampler was used to inject the lysates in a SRT SEC-500 column (Sepax Technologies) equilibrated in 50 mM HEPES/Tris-base, pH 7.4, 200 mM NaCl buffer supplemented with 1 mM L-asp, 1 mM (TCEP), 5 % glycerol, 3 x CMC detergent, and ~0.01% CHS, in line with fluorescence detection (Photon technology international) for FSEC analysis.
Large-scale expression was done in 2-4 l cultures with cells collected in 50 mM HEPES/Tris-base, pH 7.4, 50 mM NaCl buffer supplemented with 1 mM L-asp, 1 mM EDTA, 1 mM PMSF, 1 mM TCEP, and 1:200 (v/v) dilution of mammalian protease inhibitor cocktail (Sigma), and disrupted in an cell homogenizer (EmulsiFlex-C5, Avestin) after 3 runs at 15,000 Psi. The resulting homogenate was clarified by centrifugation (4,500 g, 0.5 h) and the crude membranes were collected by ultracentrifugation (186,000 g for 1.5 h). Membranes were washed once with the above-mentioned buffer and finally homogenized with a douncer in a buffer containing 50 mM HEPES/Tris-base, pH 7.4, 200 mM NaCl, 1 mM L-asp, 1 mM EDTA, 1 mM TCEP, and 10% Glycerol, snap-frozen in liquid N2 and stored at -80°C at 0.5 g of membranes ml-1.
Membrane solubilization was done by thawing out and supplementing the membrane homogenate with 2% DDS, 0.4% CHS, and 25 μM UCPH101 (Abcam). After 1-hour incubation, the insoluble material was removed by ultracentrifugation (186,000g for 1 h), and Strep-Tactin sepharose resin (GE Healthcare) was added to the supernatant and rotated for 2 h. Resin was washed with 25 column volumes of 50 mM HEPES/Tris-base, pH 7.4, 200 mM NaCl, 1 mM L-asp, 1 mM TCEP, 5% Glycerol, 0.05% DDS, 0.01% CHS and 25 μM UCPH101, and the protein was eluted with the same buffer supplemented with 2.5 mM D-desthiobiotin.
The eluted eGFP-transporter fusion was concentrated to 1-2 mg ml-1 using 100-kDa cutoff membranes (Millipore), and digested with His-tagged PreScission protease overnight at 4 °C. The protease was removed by reverse Ni-NTA (Qiagen) affinity chromatography, and the flow through containing the transporter was concentrated to 500 μl, ultra-centrifuged (86,900 g, 20 min), and applied to a Superose 6 10/300 gel filtration column (GE Healthcare) equilibrated with 50 mM HEPES/Tris-base, pH 7.4, 200 mM NaCl, 1 mM L-asp, 1 mM TCEP, 5% Glycerol, 0.25% decanoyl sucrose (DS, Sigma), 0.05% CHS and 100 μM UCPH101. To obtain the UCPH101 unbound structure, the protocol was identical, but the allosteric inhibitor was omitted from all buffers. To obtain the UCPH101-TBOATFB bound structure, the protein sample was supplemented with 3 mM TBOATFB (Tocris) before the injection in the gel filtration column equilibrated with 50 mM HEPES/Tris-base, pH 7.4, 200 mM NaCl, 1 mM TCEP, 5% Glycerol, 0.25% decanoyl sucrose (DS, Sigma), 0.05% CHS, 300 μM TBOATFB and 100 μM UCPH101.
Protein samples after the solubilization step were kept on ice or at 4 °C at all times.
Crystallization and structure determination
Purified protein was concentrated to 3.5-4.0 mg ml-1 and 1 mM UCPH101 was added in experiments with the inhibitor-bound transporters. Initial vapor diffusion crystallization screens were done by mixing 300 nl of protein and reservoir solution in sitting drops, dispensed by a Mosquito robot (TTP labtech) in 96-well Greiner plates. The purified transporters form three-dimensional crystals in several conditions containing low molecular weight polyethylene glycols. The best-diffracting crystals were obtained after manual optimization using 1.6 μl hanging drops at 4 °C, obtained by mixing equal volumes of protein supplemented with 0.2% n-Octyl-b-D-glucopyranoside (BOG, Anatrace) and 0.04% CHS, and reservoir solutions containing 100 mM Tris, pH 8.2, 50 mM CaCl2, 50 mM BaCl2, and 28-30% PEG 400. Crystals appeared after 24-48 h and reached their maximum size after a week. Crystals were flash-frozen in liquid nitrogen before X-ray diffraction data collection without any further cryo protection.
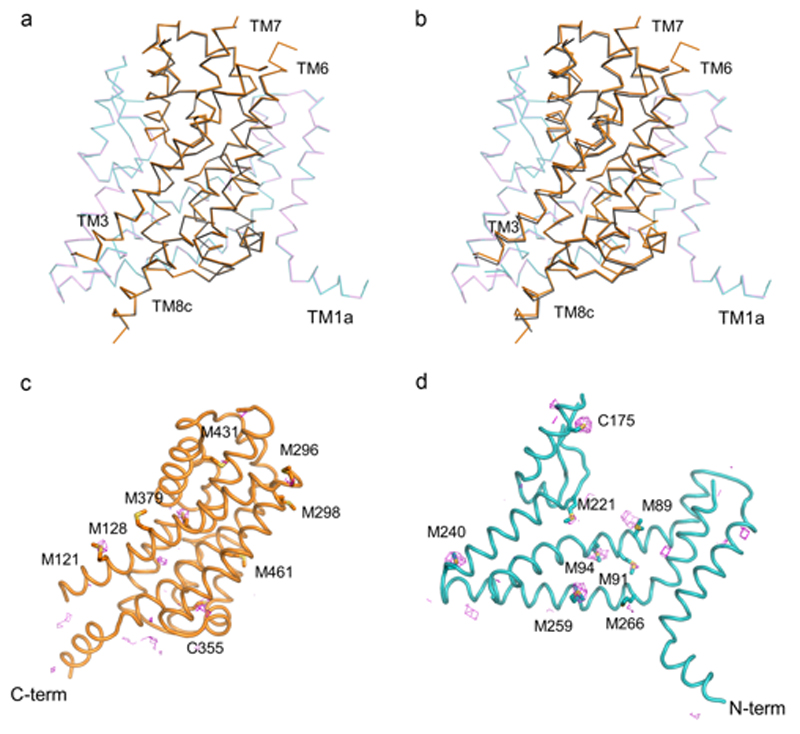
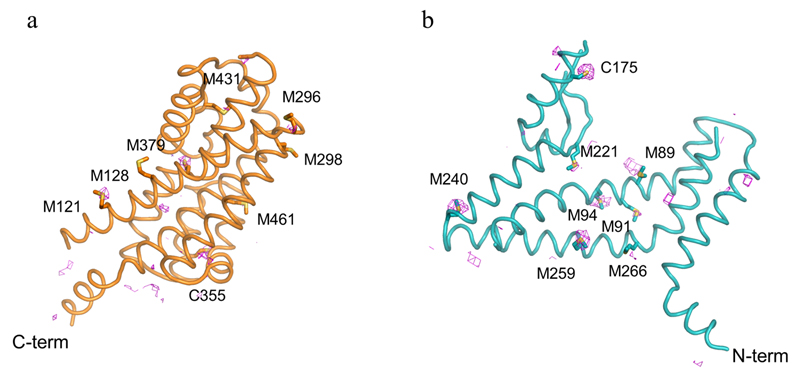
X-ray diffraction data were collected at beamlines PROXIMA-1 at the SOLEIL synchrotron (St Aubin, France) and at beamlines at the European Synchrotron Radiation Facility (Grenoble, France). In general, 2-3 data sets from single crystals were collected, and indexed, integrated, scaled and merged using XDS package52. Due to the anisotropic nature of the diffraction data, the DEBYE and STARANISO programs were applied to scale it using the STARANISO server (http://staraniso.globalphasing.org/). The software performs an anisotropic cut-off of merged intensity data with a Bayesian estimation of the structure amplitudes, and applies an anisotropic correction to the data. Table 1 shows the refinement statistics for the full sets of reflections truncated at the best high-resolution along h, k or l axis, values given by AIMLESS53, before the anisotropic corrections computed by the STARANISO software. The corrected anisotropic amplitudes were then used for molecular replacement in PHASER54, using the scaffold and transport domains of GltPh (PDB code 2NWL) as independent search models. The initial electron density maps were clearly interpretable, and the final model was obtained through rounds of manual building in COOT55 and refinement in Buster56, until reaching good crystallographic statistics and stereochemistry (Extended Data Table 1). The model contains one EAAT1cryst monomer per asymmetric unit and most of the EAAT1cryst polypeptide (residues 37-487), with the exception of some residues in the extracellular loops between TM3-4a, TM4b-4c, TM5-6 and TM7b-HP2a. Sequence assignment was aided by anomalous difference Fourier maps from diffraction data collected with low energy X-rays (1.77 Å) to highlight the sulfur atoms of methionine and cysteine residues (Extended Data Fig. 6c,d). EAAT1cryst-II, as well as the EAAT1cryst UCPH101-TBOATFB bound structures were solved by the same approach above mentioned, but using the EAAT1cryst TranD and ScaD as independent search models for molecular replacement.
The stereochemical properties of the final models were analyzed with the Molprobity server (http://molprobity.biochem.duke.edu/). At least 95% of the residues in all models are in the Ramachandran favored region. Protein interfaces were analyzed with the PISA server (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html). Structural alignments were done with Superpose in the CCP4 suite. All structural figures were prepared with PyMOL Molecualr Graphics System, Schrodinger, LLC.
Radioactive substrate transport assays
Unilamellar liposomes were made at 9:1 molar ratio of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids) and CHS, in a buffer containing 50 mM HEPES/Tris-base, pH 7.4, 200 mM NaCl and 1 mM L-asp. The transporters were purified as described above, but excluding the reverse chromatography step after protease cleavage, and using a Superose 6 10/300 column equilibrated with 50 mM HEPES/Tris-base, pH 7.4, 200 mM NaCl, 1 mM L-asp, 0.5 mM TCEP, 0.0632% DDS, 0.01264% CHS, and 5% glycerol.
To reconstitute the protein, liposomes were first mixed with DDS at a 1:2 (w/w) lipid-to-detergent ratio for 1 h, and then the purified transporters were added at a 1:40 (w/w) protein-to-lipid ratio. Detergent removal was done at 4° C using SM-2 biobeads (BioRad) at 100 mg ml-1. The internal solution of the liposomes was exchanged using 10 freeze-thaw cycles in the appropriate buffer. After extrusion through 400-nm polycarbonate membranes (Avanti Polar Lipids), the proteoliposomes were concentrated by ultracentrifugation (150,000 g for 30 min at 4°C) and resuspended at 20 mg of lipids ml-1, for immediate use.
Substrate transport was assayed at 37°C. The uptake reaction was initiated by diluting the proteo-liposomes 10-fold into a buffer containing 50 mM HEPES/Tris-base, pH 7.4, 200 mM NaCl, 50 µM L-glutamate, and 5 µM [14C]-L-glutamate (PerkinElmer), and 2.5% glycerol. After 30 min, 200-µl aliquots were diluted 5-fold into ice-cold quench buffer (50 mM HEPES/Tris-base, pH 7.4, 200 mM ChCl, and 2.5% glycerol), followed by immediate filtration and wash on nitrocellulose 0.22-µm filters (Millipore). Radioactivity was quantified by liquid scintillation using a Tri-Carb 3110TR counter (PerkinElmer). For the UCPH101 titrations, proteo-liposomes were both pre-incubated for 20 min at room temperature, and assayed in the presence of UCPH101. Background radioactivity was estimated from protein-free liposomes, and subtracted from the uptake data. Data was fitted to a Hill equation of the form:
F=F∞ + ΔF∞/(1+(IC50/[UCPH101])n)
Where F∞ is the final level of inhibition, ΔF∞ is the final amplitude of the UCPH101 effect, and n is the Hill coefficient.
To titrate the rate of L-glutamate transport by EAAT1cryst, proteo-liposomes were assayed in the presence of 0, 5, 50 or 200 μM L-glutamate supplemented with 1, 5, 5, or 5 μM [14C]-L-glutamate, respectively. At each substrate concentration, the initial rate of transport was calculated by a linear fit to 120 s and 180 s uptake measurements with origin fixed at zero. Background radioactivity was estimated from protein-free liposomes, and subtracted from the uptake data.
For the cell-based transport uptake, cells were collected 36 h post-transfection, and washed three times and resuspended at a density of 50 x 106 cells ml-1 in 11 mM HEPES/Tris-base, pH 7.4, 140 mM ChCl, 4.7 mM KCl, 2.5 mM CaCl2, 1.2 mM MgCl2, and 10 mM D-glucose, for immediate use. The uptake assay was performed similarly to the one described for the proteo-liposomes, but using a reaction buffer containing 11 mM HEPES/Tris-base, pH 7.4, 140 mM NaCl, 4.7 mM KCl, 2.5 mM CaCl2, 1.2 mM MgCl2, 10 mM D-glucose, 50 µM L-glutamate, and 5 µM [14C]-L-glutamate, and 0.8-µm nitrocellulose filters. Background radioactivity was estimated from cells transfected with empty vector, and subtracted from the uptake data.
Hydrogen-deuterium exchange mass spectrometry
HDX-MS experiments were performed with transporters purified as described in the proteo-liposome section, and using a Superose 6 5/150 gel filtration column equilibrated with 50 mM HEPES/Tris-base, 200 mM NaCl, pH7.4, 1 mM L-asp, 0.5 mM TCEP, 0.0632% DDS, 0.01264% CHS, and 5% glycerol.
The purified EAAT1cryst was incubated in ice for 30 min with 2.2% DMSO at a monomer concentration of 5.2 µM, in the presence and absence of 102 µM UCPH101, respectively. Prior to labeling, 10 µL of the unbound and UCPH101-bound EAAT1cryst solution was equilibrated for 10 min at room temperature. Deuterium exchange was initiated by adding 40 µL of D2O buffer (50 mM HEPES, pD 7.3, 200 mM NaCl, 1 mM L-asp, 5% glycerol, 0.0632% DDS, 0.01264% CHS, 0.5 mM TCEP) supplemented or not with 101.2 µM UCPH101. Assuming that the KD and IC50 values of UCPH101 are similar, ~96% of the transporters would remain bound during deuterium labeling. Aliquots of 10.4 pmols of protein were removed at defined deuterium exchange time points (from 10 sec to 60 min) and quenched upon mixing with an ice-cold acidic solution (0.75% formic acid, 5% glycerol) to decrease the pH to 2.6. Quenched samples were immediately snap-frozen in liquid N2 and stored at -80°C until analysis.
Prior to mass analysis, quenched samples were rapidly thawed and immediately injected into a cooled nanoACQUITY UPLC HDX system (Waters corp.) maintained at 0°C. 8.6-pmol protein samples were on-line digested for 2 min at 20 °C using an in-house packed immobilized pepsin cartridge (2.0 x 20 mm, 66 µL bed volume). The resulting peptides were trapped and desalted onto a C18 Trap column (VanGuard BEH 1.7 µm, 2.1 x 5 mm, Waters corp.) at a flow rate of 100 µL/min of 0.15% formic acid, and then separated in 10 min by a linear gradient of acetonitrile from 5 to 40% at 40 µL/min using an ACQUITY UPLC™ BEH C18 analytical column (1.7 µm, 1 x 100 mm, Waters corp.). After each run, the pepsin cartridge was manually cleaned with two consecutive washes of 1% formic acid, 5% acetonitrile, 1.5 M guanidinium chloride, pH 2.5. Blank injections were performed between each run to confirm the absence of carry-over.
Mass spectra were acquired in resolution and positive mode on a Synapt G2-Si HDMS mass spectrometer (Waters corp.) equipped with a standard electrospray ionization source, as described previously57. Peptides were identified from undeuterated protein samples acquired in MSE mode by database searching in ProteinLynX Global Server 3.0 (Waters corp.). Each fragmentation spectrum was manually inspected for assignment validation. Deuterium uptake values were calculated for each peptide using DynamX 3.0 (Waters corp.). Only one unique charge state was considered per peptide and no adjustment was made for back-exchange. HDX-MS results are reported as relative deuterium uptake values expressed in mass unit or fractional exchange57. A statistical analysis was performed with MEMHDX58 using a False Discovery Rate of 1%.
Extended Data
Extended Data Figure 1. Alignment of human SLC1 transporters.
Amino acid sequences of EAAT1-5, ASCT1-2 and EAAT1cryst are compared. The boundaries of the α-helices (cylinders) in the TranD (orange) and the ScaD (teal) seen in the EAAT1cryst structure are shown. In order to confer crystallizability, the region between TM3 and TM4c (arrows) from ASCT2 was transferred to a thermally stabilized EAAT1. To further improve crystal formation in the absence of UPCH101, mutations M231I and F235I (circles) were introduced to generate EAAT1cryst-II. These substitutions are found in EAAT2. Other residues involved in UPCH101 coordination are more conserved (triangles). Sequences were aligned with Jalview59.
Extended Data Figure 2. EAAT1cryst and EAAT1 glutamate uptake.
a, Initial rates of L-glutamate uptake from purified EAAT1cryst reconstituted in liposomes. The solid line is the fit of a Michaelis-Menten equation to the data with KM=21 ± 10 μM and Vmax= 13 ± 1 pmol μg-1 protein min-1. The graph is the mean of three independent experiments, and error bars represent s.e.m.. b, L-glutamate uptake was measured in HEK293 cells expressing WT EAAT1 (black circles) and a truncated mutant beyond E501 (red symbols). The initial rate of uptake decreased by ~2-fold in the EAAT1 truncated mutant. Data were normalized to the asymptotic level of glutamate uptake based on a monoexponential function. The rates obtained from the fits were 0.16 ± 0.03 and 0.08 ± 0.03 min-1 for EAAT1 and the truncated mutant, respectively. The graphs are means of 4 independent experiments done in duplicates. Error bars represent the s.e.m..
Extended Data Figure 3. EAAT1cryst and GltPh structural comparison.
a-b EAAT1cryst aligns to a monomer of GltPh (PDB 2NWL) with α-carbon r.m.s.d. of 1.4 Å. The ScaDs (EAAT1cryst teal, and GltPh purple, a), and TranDs (EAAT1cryst orange and GltPh, purple, b) are shown separately for clarity of display.
Extended Data Figure 4. EAAT1cryst trimeric interface.
a-b, Interface of three ScaDs of the EAAT1cryst UCPH101-bound structure viewed from the extracellular side (a) and from the membrane (b). The TranDs are not shown. The ScaD of one monomer (black) buries 3,000 Å2 in the trimerization interface through extensive contacts with the two other subunits (teal and purple surfaces), including six intermolecular salt bridges (shown as green sticks for the monomer in black). The surface area in the other two monomers is colored in light pink. Only residues that contribute ≥ 10 Å2 of buried surface area are highlighted.
Extended Data Figure 5. TranD-ScaD interface.
a-b, EAAT1cryst monomer viewed from the membrane (solid black line). Residues in the TranD (colored black) bury 1,760 Å2 at the interface with the ScaD (a). This interface extends to the extracellular side of the transporter through interactions between HP2-TM4 (sticks and pseudo-transparent spheres) (b). c, Cytoplasmic view of the monomer displaying the salt bridge between TM7 and TM5.
Extended Data Figure 6. Superposition of EAAT1cryst and EAAT1cryst-II structures.
a-b, The transport domains of EAAT1cryst (teal) and EAAT1cryst-II (pink) UCPH101-bound structures superimpose accurately after aligning their scaffold domains (a). The overall α-carbon r.m.s.d. was 0.3 Å. However, the same alignment done with EAAT1cryst-II UCPH101-bound and -unbound structures shows a small but global movement of the transport domain (b), with a small increase in the overall α-carbon r.m.s.d. of 0.1 Å. c,d Anomalous difference Fourier maps contoured at the 2.8σ (pink mesh), from data collected at low energy X-rays (1.77 Å), show the correct sequence registry in both the TranD (orange, a) and the ScaD (teal, b).
Extended Data Figure 7. Peptide coverage map of EAAT1cryst.
A total of 111 peptides covering 76.3% of the EAAT1cryst sequence were identified by data independent MS/MS acquisition after 2 min digestion with immobilized pepsin. Each bar below the EAAT1cryst sequence corresponds to a unique peptide. The 57 peptides colored in blue were further selected for HDX-MS data extraction and analysis. The two additional N-terminal residues (i.e., GP) that remain after protein purification are also shown. The transmembrane helices (TM) of the TranD (orange) and the ScaD (cyan) are indicated above the sequence.
Extended Data Figure 8. UCPH101 effect on the local hydrogen exchange behavior of EAAT1cryst.
a, HDX profiles of EAAT1cryst (see Methods) in the apo (upper panel) and UCPH101-bound state (middle panel). The relative fractional uptake determined for each peptide and at each time point is plotted as a function of peptide position. The black to red lines correspond to data acquired from 10 sec up to 1h, respectively. b, The fractional uptake difference plot was generated by subtracting the deuterium uptake values in the UCPH101-unbound from those in the bound state. Negative uptake difference indicates UCPH101-induced decrease in amide hydrogen exchange. Each dot corresponds to an average of three independent HDX-MS experiments. The four regions (labeled 1 to 4) showing a statistically significant modification (p <0.01) of deuterium uptake upon binding of UCPH101 are highlighted in grey.
Extended Data Figure 9. HDX-MS results mapped on the crystal structure of ScaD and TranD of EAAT1cryst in the unbound and UCPH101-bound state.
The color code at the bottom shows the average relative fractional uptake measured in both domains after 10 sec (upper panel), 10 min (middle panel) and 1h (lower panel) labeling. Missing regions in the crystal structure are represented by dashed lines. Peptides showing a statistically significant (p <0.01) modification of deuterium uptake upon UCPH101 binding are labeled. Uncovered regions are colored in light blue.
Extended Data Figure 10.
Extended Data Table 1. Data collection and refinem ent statistics.
| EAATlcryst UCPH101 bound |
EAATIcryst-II UCPH101 bound |
EAATIcryst-II | EAATlcryst UCPH101 and TBOATFB bound |
|
|---|---|---|---|---|
| Data collection * | ||||
| Space group | P63 | P63 | P63 | P63 |
| Cell dimensions | ||||
| a=b, c (Å) | 123.27, 89.87 | 123.11, 89.62 | 123.32, 89.57 | 124.33, 90.81 |
| α=β, γ (°) | 90.0, 120.0 | 90.0, 120.0 | 90.0, 120.0 | 90.0, 120.0 |
| Wavelength | 0.979 | 0.976 | 1.009 | 0.977 |
| Resolution (Å) | 34.50-3.25 (3.34-3.25) | 45.82 -3.1 (3.18-3.1) | 45.87-3.32 (3.41-3.32) | 46.31-3.71 (3.81-3.71) |
| Anisotropy direction$ Resolution where CC1/2 >0.3 |
||||
| Overall (Å) | 3.37 | 3.1 | 3.32 | 3.71 |
| along h, k axis (Å) | 3.75 | 3.68 | 3.85 | 4.35 |
| along l axis (Å) | 3.25 | 3.1 | 3.32 | 3.71 |
| Measured reflections | 333978 (24261) | 290672 (21906) | 326273 (21829) | 141904 (11176) |
| Unique reflections | 12338 (902) | 14115 (1 032) | 11556 (834) | 8570 (628) |
| Completeness (%) | 100 (100) | 99.9 (99.9) | 100 (100) | 99.9 (100.0) |
| Mn (I) half-set correlation | 0.99 (0.22) | 1 (0.43) | 0.99 (0.31) | 0.99 (0.373) |
| I/σ(I) | 11.7 (0.7) | 15.8 (0.8) | 13 (0.7) | 12.1 (0.9) |
| R merge | 0.19 (6.8) | 0.1 (6.71) | 0.15 (8.03) | 0.14 (3.71) |
| Redundancy | 27.1 (26.9) | 20.6 (21.2) | 28.2 (26.2) | 16.3 (14.3) |
|
Structure
determination Refinement |
||||
| Resolution cut-off (Å) | 34.50-3.25 | 45.80-3.10 | 20.00-3.32 | 25.00-3.71 |
| No. of Work / Test reflections | 9888/474 | 10725/528 | 9251/445 | 6860/684 |
| Rcrvst (%) / Rfree (%) | 216/23.5 | 21.7/25.9 | 20.9/25.3 | 22.7/25.4 |
| No. of protein atoms | 3008 | 2960 | 2995 | 3008 |
| No. of
heteroatoms B factors (Å)2 |
42 | 42 | 10 | 62 |
| Protein | 126.2 | 111.8 | 137.0 | 135.5 |
| Heteroatoms | 123.5 | 99.5 | 125.6 | 132.7 |
| R.m.s. deviations from ideal | ||||
| Bond lengths (Å) | 0.009 | 0.01 | 0.009 | 0.009 |
| Bond angles (°) | 1.06 | 1.12 | 1.05 | 1.03 |
One crystal was used to collect diffraction datasets for each structure, except in the EAAT1cryst UCPH101 and TFB-TBOA bound structure, where datasets from three crystals were merged.
5% of reflections were used for calculations of Rfree, except for the EAAT1cryst UCPH101-TBOATFB bound structure where 10% was used due to the lower number of total reflections.
Values in parentheses are for the highest-resolution shell.
The anisotropy directions where computed with AIMLESS.
Acknowledgements
We thank Olga Boudker for comments on the manuscript and discussion on consensus mutagenesis; Petya V. Krasteva for comments on the manuscript; Ahmed Haouz and the staff at the crystallogenesis core facility of the Institut Pasteur for assistance with crystallization screens; Staff at Synchrotron Soleil and the European Synchrotron Radiation Facility for assistance with data collection; Darragh O’Brien for discussion of HDX results. The work was funded by the ERC Starting grant 309657 (N.R.). Further support from G5 Institut Pasteur funds (N.R.), CACSICE grant (ANR-11-EQPX-008), and CNRS UMR3528 (N.R, J.C.-R.) is acknowledged.
Footnotes
Author contribution
J.C.C.-T., and R.A. optimized and performed protein expression, purification and crystallization, and R.A. performed molecular biology; J.C.C.-T., R.A., and N.R. collected crystallographic data, and J.C.C.-T., P.L., and N.R. analyzed diffraction data and structures; E.C. and R.A performed and analyzed uptake experiments; E.C. prepared protein samples for HDX-MS; S.B. collected and analyzed HDX-MS data with help from E.C.; All authors contributed to the experimental design of the project and manuscript preparation. N.R. conceived and supervised the project.
Author Information The authors declare no competing financial interests.
Data availability
Coordinates and structure factors for the EAAT1cryst-UCPH101, EAAT1cryst-UCPH101-TBOATFB, EAAT1cryst-II-UCPH101, and EAAT1cryst-II crystal structures have been deposited in the Protein Data Bank with accession codes 5LLM, 5MJU, 5LM4, 5LLU, respectively. All other data are available from the corresponding author upon reasonable request.
References
- 1.Slotboom DJ, Konings WN, Lolkema JS. Structural features of the glutamate transporter family. Microbiol Mol Biol Rev. 1999;63:293–307. doi: 10.1128/mmbr.63.2.293-307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanai Y, Hediger MA. The glutamate and neutral amino acid transporter family: physiological and pharmacological implications. Eur J Pharmacol. 2003;479:237–247. doi: 10.1016/j.ejphar.2003.08.073. [DOI] [PubMed] [Google Scholar]
- 3.Danbolt NC. Glutamate uptake. Prog Neurobiol. 2001;65:1–105. doi: 10.1016/s0301-0082(00)00067-8. [DOI] [PubMed] [Google Scholar]
- 4.Lehre KP, Danbolt NC. The number of glutamate transporter subtype molecules at glutamatergic synapses: chemical and stereological quantification in young adult rat brain. J Neurosci. 1998;18:8751–8757. doi: 10.1523/JNEUROSCI.18-21-08751.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zerangue N, Kavanaugh MP. Flux coupling in a neuronal glutamate transporter. Nature. 1996;383:634–637. doi: 10.1038/383634a0. [DOI] [PubMed] [Google Scholar]
- 6.Rothstein JD, Van Kammen M, Levey AI, Martin LJ, Kuncl RW. Selective loss of glial glutamate transporter GLT-1 in amyotrophic lateral sclerosis. Ann Neurol. 1995;38:73–84. doi: 10.1002/ana.410380114. [DOI] [PubMed] [Google Scholar]
- 7.Winter N, Kovermann P, Fahlke C. A point mutation associated with episodic ataxia 6 increases glutamate transporter anion currents. Brain. 2012;135:3416–3425. doi: 10.1093/brain/aws255. [DOI] [PubMed] [Google Scholar]
- 8.Choi KD, et al. Late-onset episodic ataxia associated with SLC1A3 mutation. J Hum Genet. 2016 doi: 10.1038/jhg.2016.137. [DOI] [PubMed] [Google Scholar]
- 9.Chao XD, Fei F, Fei Z. The role of excitatory amino acid transporters in cerebral ischemia. Neurochem Res. 2010;35:1224–1230. doi: 10.1007/s11064-010-0178-3. [DOI] [PubMed] [Google Scholar]
- 10.Pilc A, Wieronska JM, Skolnick P. Glutamate-based antidepressants: preclinical psychopharmacology. Biol Psychiatry. 2013;73:1125–1132. doi: 10.1016/j.biopsych.2013.01.021. [DOI] [PubMed] [Google Scholar]
- 11.Robert SM, Sontheimer H. Glutamate transporters in the biology of malignant gliomas. Cell Mol Life Sci. 2014;71:1839–1854. doi: 10.1007/s00018-013-1521-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zerangue N, Kavanaugh MP. ASCT-1 is a neutral amino acid exchanger with chloride channel activity. J Biol Chem. 1996;271:27991–27994. doi: 10.1074/jbc.271.45.27991. [DOI] [PubMed] [Google Scholar]
- 13.Wang Q, et al. Targeting glutamine transport to suppress melanoma cell growth. Int J Cancer. 2014;135:1060–1071. doi: 10.1002/ijc.28749. [DOI] [PubMed] [Google Scholar]
- 14.Shimizu K, et al. ASC amino-acid transporter 2 (ASCT2) as a novel prognostic marker in non-small cell lung cancer. Br J Cancer. 2014;110:2030–2039. doi: 10.1038/bjc.2014.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Q, et al. Targeting ASCT2-mediated glutamine uptake blocks prostate cancer growth and tumour development. J Pathol. 2015;236:278–289. doi: 10.1002/path.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Geldermalsen M, et al. ASCT2/SLC1A5 controls glutamine uptake and tumour growth in triple-negative basal-like breast cancer. Oncogene. 2016;35:3201–3208. doi: 10.1038/onc.2015.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimamoto K. Glutamate transporter blockers for elucidation of the function of excitatory neurotransmission systems. Chem Rec. 2008;8:182–199. doi: 10.1002/tcr.20145. [DOI] [PubMed] [Google Scholar]
- 18.Grewer C, Grabsch E. New inhibitors for the neutral amino acid transporter ASCT2 reveal its Na+-dependent anion leak. J Physiol. 2004;557:747–759. doi: 10.1113/jphysiol.2004.062521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jensen AA, et al. Discovery of the first selective inhibitor of excitatory amino acid transporter subtype 1. J Med Chem. 2009;52:912–915. doi: 10.1021/jm8013458. [DOI] [PubMed] [Google Scholar]
- 20.Abrahamsen B, et al. Allosteric modulation of an excitatory amino acid transporter: the subtype-selective inhibitor UCPH-101 exerts sustained inhibition of EAAT1 through an intramonomeric site in the trimerization domain. J Neurosci. 2013;33:1068–1087. doi: 10.1523/JNEUROSCI.3396-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yernool D, Boudker O, Jin Y, Gouaux E. Structure of a glutamate transporter homologue from Pyrococcus horikoshii. Nature. 2004;431:811–818. doi: 10.1038/nature03018. [DOI] [PubMed] [Google Scholar]
- 22.Reyes N, Ginter C, Boudker O. Transport mechanism of a bacterial homologue of glutamate transporters. Nature. 2009;462:880–885. doi: 10.1038/nature08616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akyuz N, et al. Transport domain unlocking sets the uptake rate of an aspartate transporter. Nature. 2015;518:68–73. doi: 10.1038/nature14158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boudker O, Ryan RM, Yernool D, Shimamoto K, Gouaux E. Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter. Nature. 2007;445:387–393. doi: 10.1038/nature05455. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Bendahan A, Zarbiv R, Kavanaugh MP, Kanner BI. Molecular determinant of ion selectivity of a (Na+ + K+)-coupled rat brain glutamate transporter. Proc Natl Acad Sci U S A. 1998;95:751–755. doi: 10.1073/pnas.95.2.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seal RP, Amara SG. A reentrant loop domain in the glutamate carrier EAAT1 participates in substrate binding and translocation. Neuron. 1998;21:1487–1498. doi: 10.1016/s0896-6273(00)80666-2. [DOI] [PubMed] [Google Scholar]
- 27.Tao Z, et al. Mechanism of cation binding to the glutamate transporter EAAC1 probed with mutation of the conserved amino acid residue Thr101. J Biol Chem. 2010;285:17725–17733. doi: 10.1074/jbc.M110.121798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larsson HP, et al. Evidence for a third sodium-binding site in glutamate transporters suggests an ion/substrate coupling model. Proc Natl Acad Sci U S A. 2010;107:13912–13917. doi: 10.1073/pnas.1006289107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guskov A, Jensen S, Faustino I, Marrink SJ, Slotboom DJ. Coupled binding mechanism of three sodium ions and aspartate in the glutamate transporter homologue GltTk. Nat Commun. 2016;7:13420. doi: 10.1038/ncomms13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crisman TJ, Qu S, Kanner BI, Forrest LR. Inward-facing conformation of glutamate transporters as revealed by their inverted-topology structural repeats. Proc Natl Acad Sci U S A. 2009;106:20752–20757. doi: 10.1073/pnas.0908570106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Focke PJ, Moenne-Loccoz P, Larsson HP. Opposite movement of the external gate of a glutamate transporter homolog upon binding cotransported sodium compared with substrate. J Neurosci. 2011;31:6255–6262. doi: 10.1523/JNEUROSCI.6096-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brocke L, Bendahan A, Grunewald M, Kanner BI. Proximity of two oppositely oriented reentrant loops in the glutamate transporter GLT-1 identified by paired cysteine mutagenesis. J Biol Chem. 2002;277:3985–3992. doi: 10.1074/jbc.M107735200. [DOI] [PubMed] [Google Scholar]
- 33.Qu S, Kanner BI. Substrates and non-transportable analogues induce structural rearrangements at the extracellular entrance of the glial glutamate transporter GLT-1/EAAT2. J Biol Chem. 2008;283:26391–26400. doi: 10.1074/jbc.M802401200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seal RP, Leighton BH, Amara SG. A model for the topology of excitatory amino acid transporters determined by the extracellular accessibility of substituted cysteines. Neuron. 2000;25:695–706. doi: 10.1016/s0896-6273(00)81071-5. [DOI] [PubMed] [Google Scholar]
- 35.Grunewald M, Bendahan A, Kanner BI. Biotinylation of single cysteine mutants of the glutamate transporter GLT-1 from rat brain reveals its unusual topology. Neuron. 1998;21:623–632. doi: 10.1016/s0896-6273(00)80572-3. [DOI] [PubMed] [Google Scholar]
- 36.Ryan RM, Kortt NC, Sirivanta T, Vandenberg RJ. The position of an arginine residue influences substrate affinity and K+ coupling in the human glutamate transporter, EAAT1. J Neurochem. 2010;114:565–575. doi: 10.1111/j.1471-4159.2010.06796.x. [DOI] [PubMed] [Google Scholar]
- 37.Borre L, Kanner BI. Arginine 445 controls the coupling between glutamate and cations in the neuronal transporter EAAC-1. J Biol Chem. 2004;279:2513–2519. doi: 10.1074/jbc.M311446200. [DOI] [PubMed] [Google Scholar]
- 38.Bailey CG, et al. Loss-of-function mutations in the glutamate transporter SLC1A1 cause human dicarboxylic aminoaciduria. J Clin Invest. 2011;121:446–453. doi: 10.1172/JCI44474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leinenweber A, Machtens JP, Begemann B, Fahlke C. Regulation of glial glutamate transporters by C-terminal domains. J Biol Chem. 2011;286:1927–1937. doi: 10.1074/jbc.M110.153486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shouffani A, Kanner BI. Cholesterol is required for the reconstruction of the sodium- and chloride-coupled, gamma-aminobutyric acid transporter from rat brain. J Biol Chem. 1990;265:6002–6008. [PubMed] [Google Scholar]
- 41.Butchbach ME, Tian G, Guo H, Lin CL. Association of excitatory amino acid transporters, especially EAAT2, with cholesterol-rich lipid raft microdomains: importance for excitatory amino acid transporter localization and function. J Biol Chem. 2004;279:34388–34396. doi: 10.1074/jbc.M403938200. [DOI] [PubMed] [Google Scholar]
- 42.McIlwain BC, Vandenberg RJ, Ryan RM. Transport rates of a glutamate transporter homologue are influenced by the lipid bilayer. J Biol Chem. 2015;290:9780–9788. doi: 10.1074/jbc.M114.630590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fairman WA, Sonders MS, Murdoch GH, Amara SG. Arachidonic acid elicits a substrate-gated proton current associated with the glutamate transporter EAAT4. Nat Neurosci. 1998;1:105–113. doi: 10.1038/355. [DOI] [PubMed] [Google Scholar]
- 44.Raunser S, et al. Heterologously expressed GLT-1 associates in approximately 200-nm protein-lipid islands. Biophys J. 2006;91:3718–3726. doi: 10.1529/biophysj.106.086900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom Rev. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 46.Konermann L, Pan J, Liu YH. Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem Soc Rev. 2011;40:1224–1234. doi: 10.1039/c0cs00113a. [DOI] [PubMed] [Google Scholar]
- 47.Shimamoto K, et al. Characterization of novel L-threo-beta-benzyloxyaspartate derivatives, potent blockers of the glutamate transporters. Mol Pharmacol. 2004;65:1008–1015. doi: 10.1124/mol.65.4.1008. [DOI] [PubMed] [Google Scholar]
- 48.Watzke N, Grewer C. The anion conductance of the glutamate transporter EAAC1 depends on the direction of glutamate transport. FEBS Lett. 2001;503:121–125. doi: 10.1016/s0014-5793(01)02715-6. [DOI] [PubMed] [Google Scholar]
- 49.Reyes N, Oh S, Boudker O. Binding thermodynamics of a glutamate transporter homolog. Nat Struct Mol Biol. 2013;20:634–640. doi: 10.1038/nsmb.2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14:673–681. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 51.Steipe B, Schiller B, Pluckthun A, Steinbacher S. Sequence statistics reliably predict stabilizing mutations in a protein domain. J Mol Biol. 1994;240:188–192. doi: 10.1006/jmbi.1994.1434. [DOI] [PubMed] [Google Scholar]
- 52.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blanc E, et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr. 2004;60:2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 57.O'Brien DP, et al. Structural models of intrinsically disordered and calcium-bound folded states of a protein adapted for secretion. Sci Rep. 2015;5:14223. doi: 10.1038/srep14223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hourdel V, et al. MEMHDX: An interactive tool to expedite the statistical validation and visualization of large HDX-MS datasets. Bioinformatics. 2016 doi: 10.1093/bioinformatics/btw420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]