Abstract
The central roles of luteinizing hormone (LH), progestin and their receptors for initiating ovulation have been well established. However, signaling pathways and downstream targets such as proteases that are essential for the rupture of follicular cells are still unclear. Recently, we found anovulation in nuclear progestin receptor (Pgr) knockout (Pgr-KO) zebrafish, which offers a new model for examining genes and pathways that are important for ovulation and fertility. In this study, we examined expression of all transcripts using RNA-Seq in preovulatory follicular cells collected following the final oocyte maturation, but prior to ovulation, from wild-type (WT) or Pgr-KO fish. Differential expression analysis revealed 3,567 genes significantly differentially expressed between WT and Pgr-KO fish (fold change ≥2, p<0.05). Among those, 1,543 gene transcripts were significantly more expressed, while 2,024 genes were significantly less expressed, in WT than those in Pgr-KO. We then retrieved and compared transcriptional data from online databases and further identified 661 conserved genes in fish, mice, and humans that showed similar levels of high (283 genes) or low (387) expression in animals that were ovulating compared to those with no ovulation. For the first time, ovulatory genes and their involved biological processes and pathways were also visualized using Enrichment Map and Cytoscape. Intriguingly, enrichment analysis indicated that the genes with higher expression were involved in multiple ovulatory pathways and processes such as inflammatory response, angiogenesis, cytokine production, cell migration, chemotaxis, MAPK, focal adhesion, and cytoskeleton reorganization. In contrast, the genes with lower expression were mainly involved in DNA replication, DNA repair, DNA methylation, RNA processing, telomere maintenance, spindle assembling, nuclear acid transport, catabolic processes, and nuclear and cell division. Our results indicate that a large set of genes (>3,000) is differentially regulated in the follicular cells in zebrafish prior to ovulation, terminating programs such as growth and proliferation, and beginning processes including the inflammatory response and apoptosis. Further studies are required to establish relationships among these genes and an ovulatory circuit in the zebrafish model.
Keywords: ovulation, transcriptomics, Pgr, knockout, TALENs, zebrafish
1. Introduction
Ovulation is a physiological process that releases a fertilizable oocyte from follicular cells and is an essential reproductive event for the preservation of a species. It is well established that luteinizing hormone (LH) initiates a cascade of signaling, including up-regulation of progestin and its nuclear progestin receptor (PGR) which activates various downstream targets and signaling pathways, eventually leading to follicular rupture. However, our understanding of the molecular mechanisms that control ovulation is far from complete. For example, there is limited evidence of downstream targets and signaling pathways that PGR regulates. A few genome-wide transcriptome analyses of differentially expressed genes in the follicular cells of preovulatory oocytes suggest conserved mechanisms in the regulation of gene expression in humans, macaques, and mice (Hernandez-Gonzalez et al., 2006; Wissing et al., 2014; Xu et al., 2011). It is still unknown whether these “ovulatory genes and pathways” are also presented and conserved in other vertebrates. To our knowledge, there are no published transcriptomic analyses of gene expression focusing on the follicular cells of preovulatory oocytes in non-mammalian vertebrates. All studies conducted so far have not separated follicular cells from the oocytes or used mixed stages of oocytes that preclude detailed comparisons between different species (Chapman et al., 2014; Gohin et al., 2010; Reading et al., 2012).
Zebrafish is an alternative vertebrate model for studying gene function, signaling pathways in development, and various physiological processes because of their low cost, rapid development, and relative simplicity. Unlike mammalian models, zebrafish release and fertilize mature oocytes outside the body, developing their embryos externally. Because embryos develop externally, females do not undergo cumulus-oocytes complex (COC) expansion, luteinization, or implantation processes that happen concurrently or subsequently with ovulation, making it relatively easy to distinguish genes exclusively involved in ovulation (Hagiwara et al., 2014). In addition, follicular cell layers can be collected separately from preovulatory oocytes, which are relative large (>650 μm), for biochemical and molecular analyses (Hanna and Zhu, 2011). Our recent study has also shown that Pgr is an important transcription factor induced by luteinizing hormone (LH) and is essential for ovulation in zebrafish (Zhu et al., 2015). In Pgr knockout (Pgr-KO) female zebrafish mature oocytes were trapped within follicular cells unable to ovulate, leading to infertility. Our results are consistent with the complete anovulatory and infertile phenotype reported in PGR-KO mice and rats (Lydon et al., 1995; Kubota et al., 2016). These results prompted us to hypothesize that ovulation is controlled by conserved genes and signaling pathways in vertebrates.
In this study, we first conducted a genome-wide differential gene expression analysis in the follicular cells of preovulatory oocytes from wildtype (WT) in comparison to Pgr-KO zebrafish using RNA-Seq and bioinformatics tools. We hypothesize that changes in gene expression in WT would be important for ovulation, while lack of changes in gene expression in Pgr-KO due to anovulation could serve as reference. We then conducted a comparison analysis of genome-wide differentially regulated genes in the follicular cells of preovulatory oocytes of three key vertebrate species, i.e., zebrafish, mouse, and human. We found that Pgr regulates a network of conserved signaling pathways, biological processes, and genes that control ovulation. Various genes with dramatic differences in the expression between wt and Pgr-KO include ptgs2 (prostaglandin-endoperoxide synthase 2a, 2b), runx1 (runt-related transcription factor 1), ptger4b (prostaglandin E receptor 4b), rgs2 (regulator of G-protein signaling 2), and adamts9 (a disintegrin-like and metalloproteinase with thrombospondin motifs). This varied expression across species provides a list of candidate genes to study the molecular mechanisms underlying ovulation.
2. Materials and Methods
2.1. Zebrafish husbandry
Generation and characterization of Pgr mutant lines have been described previously (Zhu et al., 2015). The WT zebrafish used in this study was a Tübingen strain initially obtained from the Zebrafish International Resource Center and propagated in our lab according to the following procedure. Fish were kept at constant water temperature (280C), a photoperiod of 14 hrs of light with 10 hrs of dark (lights on 9:00, lights off at 23:00), pH at 7.2, and salinity conductivity from 500–1200 μS in automatically controlled zebrafish rearing systems (Aquatic Habitats Z-Hab Duo systems, Florida, USA). Fish were fed three times daily to satiation with a commercial food (Otohime B2, Reed Mariculture, CA, USA) containing high protein content and supplemented with newly hatched artemia (Brine Shrimp Direct, Utah, USA). The Institutional Animal Care and Use Committee (IACUC) at East Carolina University approved experimental protocols.
2.2. Collection of preovulatory follicular cell layers
Follicular cells of preovulatory oocytes were collected from three WT or three Pgr-KO female zebrafish at the same developmental stage, i.e., immediately following oocyte maturation but prior to ovulation (See Fig.1 for detail). We limited our sample size to n=3 for each group, to balance the high cost of RNA-seq and minimum requirement of statistical analyses. Follicular cells are two thin layers of cells (~20 μm in thickness, panels D1 and D2 in Fig.1) containing theca and granulosa cells, surrounding a gigantic oocyte in zebrafish (Ø>650 μm for preovulatory oocyte, Fig.1). Follicular cells could be physically separated from oocytes (Hanna and Zhu, 2011), which typically have an approximate 1000 fold higher amount of total RNAs than those in surrounding follicular cells. However, physical separation of granulosa cells from theca cells is impractical due to small cell size and no distinguishable physical properties of these two cell types.
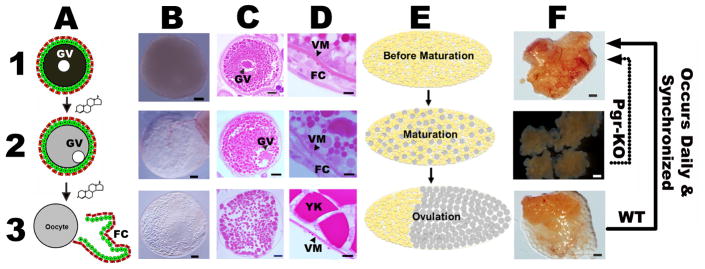
Figure 1. Final oocyte maturation and ovulation are easily observable and distinguishable with naked eyes in zebrafish.
Schematic drawing or microscopic images of representative stage IV fully-grown immature oocytes (A1, B1, and C1); stage IV oocytes that undergo final maturation, germinal vesicle migration and breakdown (A2, B2, and C2); stage V ovulated oocytes (A3, B3, and C3); representative ovaries prior to final oocyte maturation (E1 and F1), during final oocyte maturation (E2 and F2), and ovulated ovaries (E3 and F3). High magnification images of vitelline membrane (VM) and follicular cells (FC) are also shown in panel D (D1-D3). Final oocyte maturation occurs daily prior to ovulation, and synchronized by the lights in zebrafish. The maturation process includes germinal vesicle migration (GM) and breakdown (GVBD), chromosome condensation, assembly of meiotic spindle, and formation of the first polar body. Change in cytoplasm appearance of stage IV oocytes from opaque to transparent is the most reliable indication of final oocyte maturation (panel 2). Once ovulation is complete, these mature and transparent oocytes migrate to the posterior of the animal body (E3), ready to be released and fertilized outside. These recurring tissue remodeling processes, including maturation and ovulation, happened almost daily in wildtype (WT) zebrafish. In contrast, oocyte growth and maturation occur normally but ovulation was completely blocked in Pgr knockout (Pgr-KO). Follicular cells used in this study were all collected from the same stage of oocytes, i.e., prior to ovulation but after occurrence of oocyte maturation (panels 2) from WT or Pgr-KO fish. Scale bar: 100 μm (panel B and C); 10 μm (panel D); 1 mm (panel F). FC: follicular cells. GV: germinal vesicle. VM: vitelline membrane. YK: yolk.
All the fish used in the experiment were approximately four months old. Individual, well-fed, mature, and healthy female zebrafish were housed separately from male fish by a middle divider in a spawning tank the night before sampling. Oocyte maturation and ovulation were synchronized between different individuals, and spawning typically happened within 30 minutes once the lights switched on and the middle divider was removed from these well-fed and individually housed fish. To obtain preovulatory oocytes, ovaries were removed within an hour before the lights turned on in the morning following an appropriate anesthetic overdose (MS-222: 300 mg/L in buffered solution). Each excised individual ovary was then placed in a zebrafish Ringer’s solution (116mM NaCl, 2.9mM KCl, 1.8mM CaCl2, 5mM HEPES, pH 7.2) and examined under a dissecting microscope. We selected majority of ovaries containing healthy preovulatory follicles with a translucent appearance, indicating the occurrence of oocyte maturation and proximal to ovulation (panel F2 in Fig.1). A few ovaries had no preovulatory oocytes, incomplete absorption of previous left-over matured oocytes, or unhealthy oocytes were discarded. Individual, follicle-enclosed, healthy mature oocytes were teased away from immature oocytes by gently pipetting in and out several times using a Pasteur pipette. To determine cell specific changes of transcripts regulated by Pgr in the follicular cells, we manually peeled follicular cells off preovulatory oocytes in zebrafish Ringers’ solution under a dissecting microscope using a pair of fine, clean glass needles as described previously (Hanna and Zhu, 2011). Follicular cells were collected into a 1.7ml microcentrifuge tube and homogenized immediately in a 300 μl TRIzol solution by a sonicator (Sonic Dismembrator, Fisher Scientific, Pittsburgh, PA, USA). Samples were stored in a −80°C freezer until RNA extraction. Each sample contained follicular cell layers separated carefully from 45 to 130 preovulatory oocytes of one fish. The process for collecting one sample was limited to less than an hour, avoiding significant degradation or changes of the transcripts (unpublished data). We collected six samples in six different days, in order to sample at the same time point and same developmental stages.
2.3. RNA isolation
Total RNA was extracted using TRIzol and a Qiagen RNeasy kit according to a modified protocol. An equal volume of cold 100% ethanol was added into the aqueous phase of the solution following phase separation of TRIzol. Samples were then loaded onto an RNeasy spin column, centrifuged (8,000g, 30 seconds), washed once with 700 μl RW1, twice with 500 μl RPE, and eluted in 25 μl of RNase free water according to Qiagen’s instructions. The approximate concentration and purity of samples were examined using a Nanodrop 2000 Spectrophotometer. An aliquot of RNA sample with OD 260/280 > 1.8 and OD 260/230 > 1.6 was used for RNA-Seq analysis and the remainder was used for quantitative real-time PCR (qPCR) assay.
2.4. Library construction and Illumina sequencing
RNA-Seq library preparation and high-throughput NGS sequencing were carried out at HudsonAlpha Genomic Services Lab (Huntsville, AL). Qubit® 3.0 Fluorometer and Agilent 2100 Bioanalyzer further examined the concentration and integrity of total RNA samples prior to library construction. About 800ng of total RNA was used to construct a cDNA library according to a protocol of Illumina TruSeq RNA Sample Preparation Kit (Illumina). Ribosomal reduction was used to remove non-coding rRNA. The library was then PCR amplified with 15 cycles using TruSeq indexes adaptor primers, submitted for Kapa quantification and dilution, and sequenced with a single end read (50bp) on an Illumina HiSeq 2000 instrument.
2.5. Genome mapping and differential expression analysis
The quality control and adaptor trimming of raw FastQ files were performed using Trim_Galore. Trimmed raw files were inspected using FastQC. The entire zebrafish genome sequence (version GRCz10) was downloaded from Ensembl, and the alignment of sequence reads to the zebrafish genome was carried out using STAR aligner (Dobin et al., 2013). Binning of sequencing reads to genes/exons was accomplished by HTseq-count, where reads with an alignment score of less than 10 were skipped (Anders et al., 2014). DESeq2 was chosen to normalize the raw counts with respect to the gene length and sequencing depth as well as to identify differentially expressed genes (Anders and Huber, 2010).
2.6. Validation of differential expression by quantitative real-time PCR (qPCR)
Twenty-two of the differentially expressed genes that have potential roles in ovulation were selected, and their expression levels were further validated using traditional qPCR. Briefly, 250 ng of total RNA from a subset of samples that were used for transcriptomic analysis were reverse transcribed using SuperScript III Reverse Transcriptase in a 10 μl reaction volume following the manufacturer’s instructions (Invitrogen, Carlsbad, CA). Specific PCR primer pairs (supplemental Table S1) for target genes were designed to span at least two adjacent exons to avoid genomic DNA interference. Glyceraldehyde-3-phosphate dehydrogenase (gapdhs) was chosen as an internal control for qPCR because gapdhs was expressed evenly among all samples in RNA-Seq analysis. Absolute copy numbers of each transcript were calculated from a standard curve generated from a serial dilution of plasmid DNA with known concentrations (Hanna and Zhu, 2011). Then, the expression of each transcript was normalized with gapdhs and expressed as fold change by comparing WT to knockout (log2 (WT/Pgr-KO)), since genes important for ovulation would change significantly in WT, but were not expected to do so in Pgr-KO.
2.7. Comparison of genes that are important for ovulation in human, mouse and zebrafish
Human (E-MTAB-2203) and mouse (GSE4260) (Hernandez-Gonzalez et al., 2006; Wissing et al., 2014) transcriptomic data sets that were focused on differentially regulated genes in the follicular cells of preovulatory oocytes were downloaded from EMBL and GEO databases, respectively. Two important marker genes, LHCGR and PGR, were found in these two data sets. Therefore, both human and mouse samples contain granulosa cells, and are comparable to the follicular cells of zebrafish.
Multiple CEL data files containing human transcripts were first imported into Expression Console software (Affymetrix) for data normalization, then the robust Multi-array Average (RMA) normalized our data and they were transferred into Transcriptome Analysis Console program (v3.0, Affymetrix) for differential expression analysis using One-Way Repeated Measure ANOVA (paired). We used the R packages Affy (Gautier et al., 2004) and limma (Ritchie et al., 2015) for differential expression analysis, due to only two biological replicates in mouse samples.
Differentially expressed genes were defined as having a minimal 2 fold difference in the expression of the transcript observed in treated or mutant samples, compared to controls (absolute log2FoldChange > 1) with a corrected FDR p-value < 0.05. Ensembl gene IDs of human and zebrafish were converted to the mouse version to determine ovulatory genes that are conserved between humans, mice, and zebrafish.
2.8. Enrichment analysis
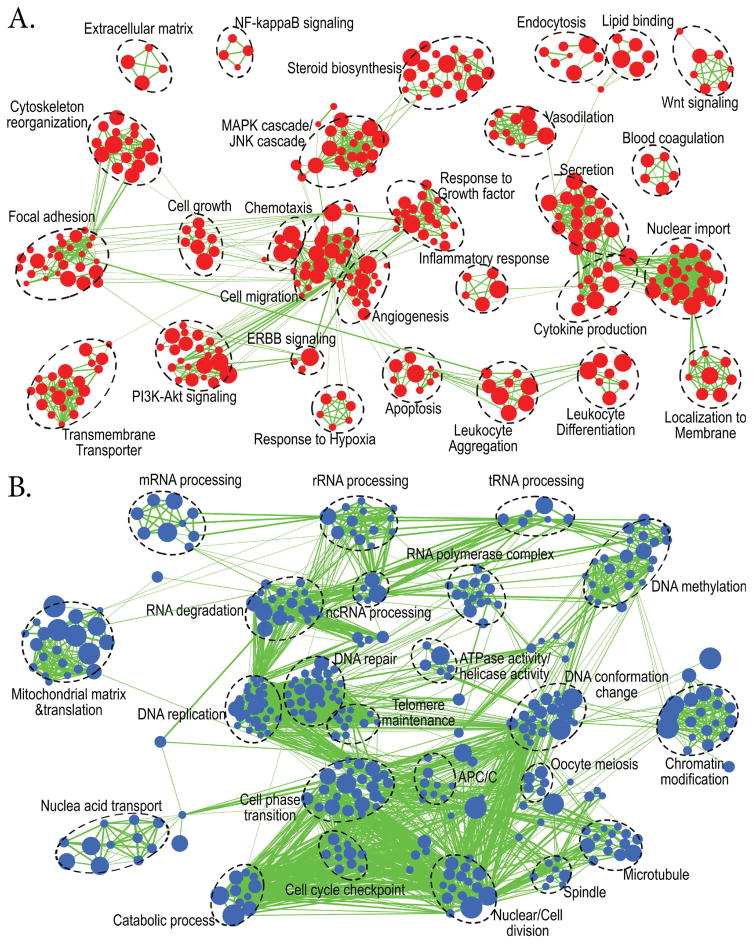
Both up-regulated and down-regulated genes were first imported into the online g:Profiler (http://biit.cs.ut.ee/gprofiler/). We selected the “no filtering” option and kept significant gene sets (FDR p≤0.05) that had three or more differentially expressed genes (DEGs) for follow-up analyses. These gene sets were compiled from KEGG, Reactome, and GO databases. To visualize the results of enriched gene sets and pathways for WT and Pgr-KO samples, the Enrichment Map plugin for Cytoscape (Merico et al., 2010) was used. Enriched gene sets were first loaded into Enrichment Map plugin for Cytoscape using a p-value cutoff at 0.0005 to achieve an operable clustered network. The resulting network map was curated manually by removing uninformative gene sets and grouping functionally related gene sets together. We then labeled these functional groups to highlight prevalent biological functions that were enriched.
3. Results
3.1. A large set of genes (>10%) are differentially regulated prior to ovulation
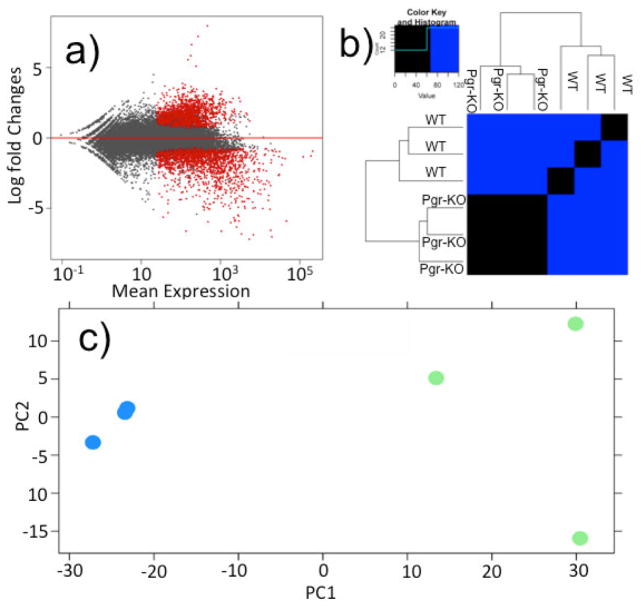
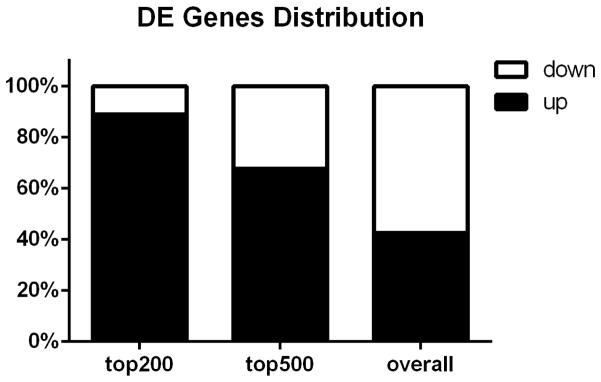
In total, we found that 3,567 zebrafish genes were significantly differentially expressed in WT compared to those in Pgr-KO, using a cutoff of fold change ≥ 2 and FDR adjusted p-value ≤ 0.05 (Fig.2a; supplementary Table S2). The principal component and heat-map analyses showed clear difference in gene expression between WT and Pgr-KO (Fig.2b and 2c), further confirming the role of Pgr as a top regulator/mediator of ovulation. We retained 2,888 significantly regulated zebrafish genes that have mouse homologs for subsequent analyses and removed duplicates with less significance (supplementary Table S3). We did this to reduce the complexity caused by teleost specific gene duplication and to search for conserved genes in vertebrates. Among these differentially regulated genes, 1,230 genes had higher expression and 1,658 genes had lower expression in WT than in Pgr-KO. Intriguingly, among the top 200 differentially expressed genes ranked by their adjusted p-value, the majority of these genes (178 genes, 89%) had higher levels of expression in WT than those in Pgr-KO, while a small number of these genes (22 genes, 11%) had lower levels of expression in WT than those in Pgr-KO (Fig.3). Among these differentially expressed genes, 154 genes had 8- to 147-fold increase, whereas only 16 genes had an 8-fold decrease in WT than those in Pgr-KO (Supplementary Table S4).
Figure 2. A large set of genes expressed differentially in preovulatory follicular cells in wildtype (WT) in comparison to those in Pgr-KO.
a) Volcano plot shows the global transcriptional changes in WT vs Pgr-KO. All genes present on the RNA-seq analysis were plotted. Each circle represents one gene. Genes with significant difference (FDR corrected p<0.05, fold change> 2) are indicated by red circles, while grey circles indicate those genes that did not show significant differences in their expression between WT and Pgr-KO. b) Heat map comparison of top 2000 differentially expressed genes ranked by FDR-corrected p-value. Each column represents an independent sample collected from follicular cells of stage IV oocytes that had completed final oocyte maturation, but prior to ovulation. c) Principal component analysis (PCA) of top 2000 significant differentially expressed genes from preovulatory follicular cells of WT and Pgr-KO. The blue circles represent transcriptomic data from three preovulatory follicular samples collected from three Pgr-KO fish, and the green circles represent the data from three samples of three WT fish.
Figure 3. Distribution of up- or down-regulated genes in wildtype (WT) compared to Pgr knockout (Pgr-KO).
The majority of the top 500 genes were up-regulated in WT compared to Pgr-KO, indicating that an increase of gene expression is important for ovulation.
3.2. RNA-Seq results were confirmed by qRT-PCR
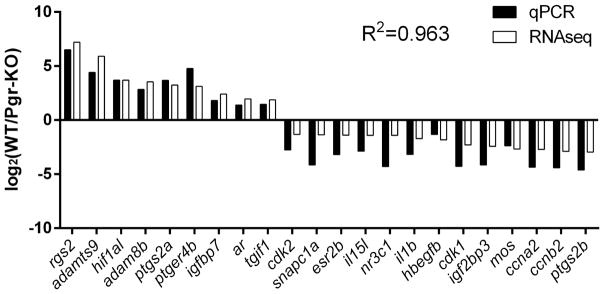
The specificities of each PCR primer set used in qPCR was confirmed using a melting curve analysis and by sequencing of PCR products. Similar significant differences in gene expression among selected genes were observed in the follicular cells of WT compared to those in Pgr-KO by qPCR analysis, which validated the results of RNA-seq (Fig.4).
Figure 4. Validation of RNA-Seq results using real-time quantitative PCR (qPCR) based on relative fold changes of 22 selected genes in WT compared to Pgr-KO.
Relative fold changes were expressed as log 2 of normalized average count (RNA-seq), or mean copy number (qPCR) of each gene in WT normalized by those respective numbers in Pgr-KO (n=3). Correlation coefficient (R2) of gene expression values from qPCR compared to RNA-Seq was 0.963. FDR-adjusted p-values from RNA-Seq analysis for these selected genes ranged from 4.08E-86 to 0.0193, while student t-test p-values from qPCR analysis ranged from 0.05 to 1.02E-05.
3.3. Enrichment analysis shows clear distinction of biological processes and pathways between WT and Pgr-KO fish prior to ovulation
In total, 1,962 gene sets were enriched significantly (FDR p≤0.05) when all up-regulated genes, i.e., 1,230 corresponding mouse homolog IDs, in WT fish were input into g:Profiler. A small 943 gene sets were enriched when all down-regulated genes, i.e., 1,658 mouse homolog IDs, were input into the same program. We retained 649 up-regulated gene sets and 469 down-regulated gene sets, with a more stringent cutoff at p≤0.0005, to draw better and less congested enrichment maps. Interestingly, multiple sets of biological processes, molecular functions, and cellular components important for ovulation were significantly enriched in WT but not in Pgr-KO, explaining why Pgr-KO fish fail to ovulate (Fig.5). These enriched biological processes in WT include: angiogenesis, cell migration, chemotaxis, focal adhesion, response to growth factor, vasodilation, blood coagulation, cytokine production, inflammatory response, leukocyte aggregation and differentiation, cytoskeleton reorganization, extracellular matrix organization, response to hypoxia, and apoptosis (Fig.5a). In contrast, biological processes enriched in the Pgr-KO were mainly related to cell growth, proliferation, and cell cycle instead of ovulation (Wissing et al., 2014). These processes include: DNA replication, DNA repair, DNA methylation, cell phase transition, and cell division (Fig.5b).
Figure 5. Enrichment map analysis showed two distinct groups of biological processes and pathways which were significantly enhanced or suppressed in wildtype (WT) compared to Pgr-KO (p<0.0005).
Nodes represent enriched gene sets, clustered automatically by the Enrichment Map plugin for Cytoscape program (Merico et al., 2010) according to the number of genes shared within sets. Node size is proportional to the total number of genes within each gene set. The proportion of shared genes among gene sets is represented by the thickness of the line between the nodes. Functionally related gene sets were manually grouped, encircled by a dashed black line, and labeled. A. Enrichment map for 1230 up-regulated genes in WT compared to Pgr-KO. B. Enrichment map for 1658 down-regulated genes in WT compared to those in Pgr-KO.
Among the entire enriched gene sets, we found 104 and 191 KEGG or Reactome pathways respectively, enriched in WT and Pgr-KO. We listed the top 20 significant pathways as representatives for each genotype (supplemental Table S5). Interestingly, representative pathways were also quite distinct between WT and Pgr-KO. Multiple signaling and metabolic pathways that seem to be important for ovulation were enriched in WT. These signaling pathways included Ras, Rap1, FoxO, cGMP-PKG, PI3K-Akt, Hippo, and oxytocin, as well as pathways in cancer, transport of inorganic cations/anions and amino acids/oligopeptides, and neurophilin interactions with VEGF and VEGFR. In contrast, few signaling pathways enriched in Pgr-KO were related to ovulation. Enriched pathways in Pgr-KO included cell cycle related pathways such as mitochondrial translation, DNA strand elongation, activation of ATR in response to replication stress, and extension of telomeres, which were very similar to those in GO annotations.
The clear distinction between up- and down-regulated genes in WT compared to Pgr-KO demonstrates that Pgr is essential for determining the cell fate and transition of preovulatory follicular cells. A lack of Pgr would inhibit the biological processes and signaling pathways required for ovulation.
3.4. Comparison with human and mouse data shows conserved biological processes, pathways, and genes that are important for ovulation in human, mouse, and zebrafish
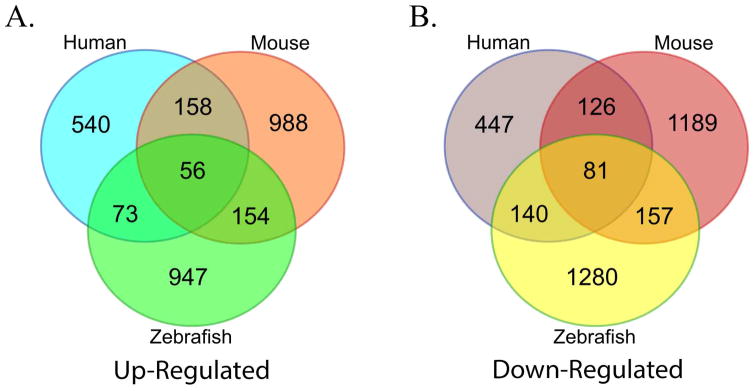
In order to examine conserved genes and biological processes important for ovulation, we compared our RNA-Seq data with transcriptomic data obtained from HCG induced ovulation samples in humans (Wissing et al., 2014) and mice (Hernandez-Gonzalez et al., 2006). In follicular cell samples of human treated with HCG, 852 genes were significantly up-regulated and 884 were down-regulated when compared to those without HCG treatment; whereas, in mouse follicular cell samples treated with HCG, 1,356 genes were significantly up-regulated and 1,553 were significantly down-regulated compared to those without HCG exposure (Table 1). We focused on analyzing genes that showed similar increases or decreases between zebrafish and mammals (human and/or mouse) and found 283 up-regulated and 378 down-regulated conserved ovulatory genes (Fig.6). Similarly, enrichment analysis of these conserved genes also showed distinct pathways between up-regulated and down-regulated genes, covering those main GO terms that distinguished HCG-treated or WT from non-treated or Pgr-KO group (Fig.7). Based on their reported molecular functions, we categorized and highlighted 64 genes that showed similar expression (increase or decrease) among these three model vertebrates (Table 2). Conserved genes showing high expression in ovulating animal could be categorized into those related to inflammatory response or apoptosis (e.g. runx1, ptgs2a, zbtb16a, tnfrsf21, adam8b, ptger4b and furin), vascularization (e.g. rgs2, f3a, f5, nrp1a, tfpi2 and serpine1), and cell-matrix adhesion and extracellular matrix remodeling (e.g. tnfaip6, adamts9, ptx3b, timp2a, cd151, mkln1 and cldn11a); whereas, conserved genes showing low expression in ovulating animals were those related to cell cycle (e.g. ccnb2, ccna2, ccnb1, cdk1, cdk2, mki67, pcna, mcm10, cdc20 and mos). We were also particularly interested in the changes of proteinase expression and their roles during ovulation. Intriguingly, we found that several metalloproteinases including adamts9, adam8b, and mmp9 had dramatically changed their expressions during ovulation, which were also confirmed by qPCR and a series of experiments (Fig.4; unpublished).
Table 1.
Comparison of three transcriptomic experiments for analyzing differentially regulated genes during ovulation in the follicular cells of zebrafish, human and mouse.
| Species | Human | Mouse | Zebrafish |
|---|---|---|---|
| GEO or EMBL accession | E-MTAB-2203 | GSE4260 | current study |
| Platform | Microarray | Microarray | NGS |
| Genotype | WT | WT | Pgr-KO & WT |
| Treatment | HCG 36h | HCG 8h | no |
| Biological replicates | 9 paired | 2 repeats | 3 Pgr-KO or 3 WT |
| Cutoff | |lfc| > 1 ; padj < 0.05 a | |lfc| > 1 ; padj < 0.05 | |lfc| > 1 ; padj < 0.05 |
| Comparison | 36h vs 0h | 8h vs 0h | WT vs KO |
| Up-regulated | 852 | 1356 | 1544 |
| Down-regulated | 884 | 1553 | 2025 |
Lfc, log2 fold change. padj, FDR-corrected p-value.
Figure 6. Comparison of significantly regulated genes in preovulatory follicular cells of zebrafish, human, and mouse during ovulation.
A. a total of 283 genes (73 + 56 + 154) were up-regulated in zebrafish, humans and/or mice prior to ovulation. B. A total of 378 genes (140 + 81 +157) were down-regulated in zebrafish, humans and/or mice during ovulation.
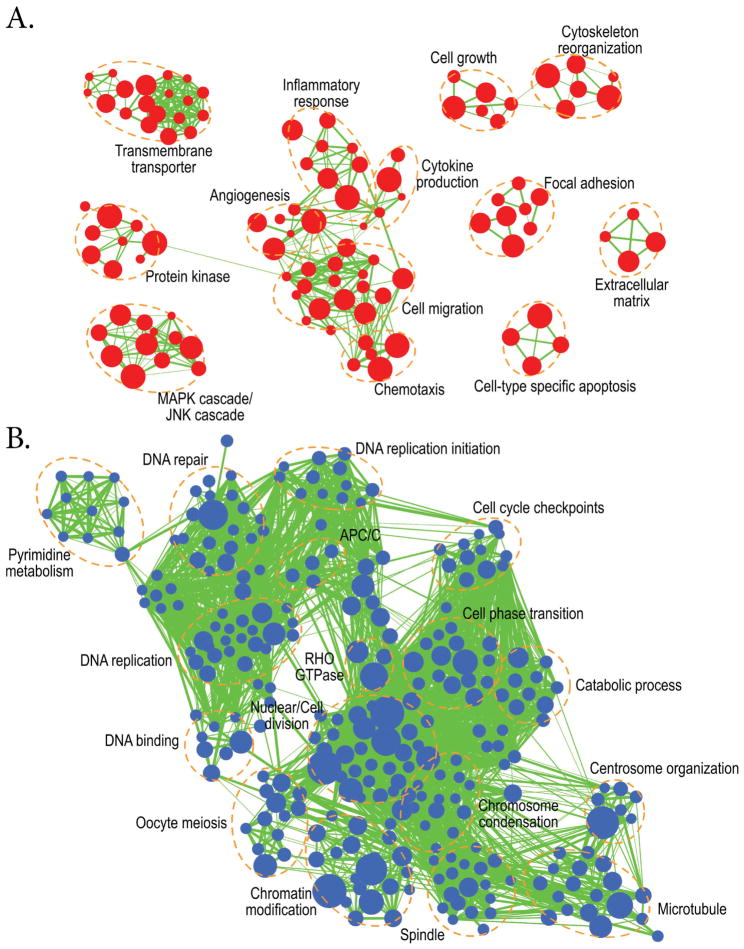
Figure 7. Enrichment map analysis using conserved 283 up-regulated and 378 down- regulated genes shared between zebrafish and mammals (human and/or mice) during ovulation (p<0.0005).
A. Up-regulated biological processes during ovulation which include: transmembrane transportation, protein kinases, MAPK/JNK cascades, angiogenesis, the inflammatory response, cytokine production, cell migration, chemotaxis, extracellular matrix organization, cell growth, and cytoskeleton reorganization. B. Down-regulated biological processes during ovulation include: Pyrimidine metabolism, DNA repair, DNA methylation, DNA binding, DNA replication, cell cycle checkpoints, oocyte meiosis, chromatin modification, and cell phase transition.
Table 2.
Representative differentially regulated genes in zebrafish during ovulation that are conserved between zebrafish, human and mouse.
| gene symbol (mouse) | gene title | gene symbol (zebrafish) | shared type a | log2 Fold Changeb | FDR p-value |
|---|---|---|---|---|---|
| Inflammatory response and apoptosis | |||||
| Maf | Avian musculoaponeurotic fibrosarcoma (v-maf) AS42oncogene homolog | MAF (2 OF 2) | T1 | 3.65 | 1.52E-14 |
| Pla2g4a | Phospholipase A2, group IVA | cpla2 | T1 | 3.46 | 2.43E-17 |
| Azin1 | Antizyme inhibitor 1 | azin1a | T1 | 3.38 | 2.77E-14 |
| Runx1 | Runt related transcription factor 1 | runx1 | T1 | 3.27 | 2.39E-20 |
| Ptgs2 | Prostaglandin endoperoxide synthase 2 | ptgs2a | T1 | 3.25 | 4.30E-08 |
| Zbtb16 | Zinc finger and BTB domain containing 16 | zbtb16a | T1 | 3.11 | 9.48E-18 |
| Tnfrsf21 | Tumor necrosis factor receptor superfamily, member 21 | tnfrsf21 | T1 | 2.66 | 1.47E-08 |
| Gadd45a | Growth arrest and DNA-damage-inducible 45 alpha | gadd45ab | T1 | 2.45 | 7.78E-11 |
| Mcl1 | myeloid cell leukemia sequence 1 | bcl2l10 | T1 | 2.32 | 2.35E-08 |
| Litaf | LPS-induced TN factor | litaf | T1 | 1.63 | 3.44E-05 |
| Slco2a1 | Solute carrier organic anion transporter family, member 2a1 | slco2a1 | T1 | 1.62 | 7.43E-04 |
| Hipk2 | Homeodomain interacting protein kinase 2 | hipk2 | T1 | 1.28 | 4.11E-03 |
| Nr1d2 | Nuclear receptor subfamily 1, group D, member 2 | nr1d2a | T2 | 4.11 | 9.24E-26 |
| Snai2 | Snail family zinc finger 2 | snai2 | T2 | 3.43 | 8.90E-19 |
| Clu | Clusterin | clu | T2 | 2.90 | 9.23E-08 |
| Socs3 | Suppressor of cytokine signaling 3 | socs3a | T2 | 1.42 | 1.96E-04 |
| Adam8 | A disintegrin and metallopeptidase domain 8 | adam8b | T3 | 3.55 | 7.95E-25 |
| Ptger4 | Prostaglandin E receptor 4 | ptger4b | T3 | 3.13 | 5.77E-11 |
| Csf1r | Colony stimulating factor 1 receptor | csf1ra | T3 | 2.92 | 4.13E-08 |
| Nt5e | ecto-5'-nucleotidase | nt5e | T3 | 2.37 | 1.89E-05 |
| Furin | Furin | furina | T3 | 1.84 | 1.32E-04 |
| Vascularization | |||||
| Rgs2 | Regulator of G-protein signaling 2 | rgs2 | T1 | 7.21 | 4.08E-86 |
| F3 | Coagulation factor III | f3a | T1 | 4.86 | 7.20E-30 |
| Zfand5 | Zinc finger, AN1-type domain 5 | zfand5a | T1 | 3.12 | 3.93E-16 |
| Nrp1 | Neuropilin 1 | nrp1a | T1 | 1.98 | 9.89E-05 |
| Tfpi2 | Tissue factor pathway inhibitor 2 | tfpi2 | T1 | 1.71 | 8.49E-04 |
| Sema3a | Semaphorin 3A | sema3ab | T1 | 1.15 | 7.00E-03 |
| F5 | Coagulation factor V | f5 | T2 | 6.56 | 4.92E-54 |
| Serpine1 | Serine (or cysteine) peptidase inhibitor, clade E, member 1 | serpine1 | T2 | 1.84 | 1.87E-05 |
| Apoe | Apolipoprotein E | apoea | T2 | 1.67 | 9.51E-05 |
| Ppap2b | Phosphatidic acid phosphatase type 2B | ppap2b | T3 | 3.24 | 2.22E-15 |
| Adipor2 | Adiponectin receptor 2 | adipor2 | T3 | 2.07 | 1.37E-06 |
| Cell-matrix adhesion and extracellular matrix remodeling | |||||
| Tnfaip6 | Tumor necrosis factor alpha induced protein 6 | tnfaip6 | T1 | 2.25 | 1.26E-07 |
| Adamts9 | A disintegrin-like and metallopeptidase with thrombospondin type 1 motif, 9 | adamts9 | T1 | 5.91 | 8.44E-41 |
| Ptx3 | Pentraxin 3 | ptx3b | T1 | 4.68 | 3.45E-15 |
| Timp2 | Tissue inhibitor of metalloproteinase 2 | timp2a | T1 | 3.43 | 2.34E-21 |
| Rnd3 | Rho family GTPase 3 | rnd3b | T1 | 2.33 | 1.68E-07 |
| Ezr | Ezrin | ezra | T1 | 1.84 | 1.91E-04 |
| Cd151 | CD151 antigen | cd151 | T1 | 1.76 | 5.99E-05 |
| Mkln1 | Muskelin 1 | mkln1 | T2 | 3.51 | 7.11E-18 |
| Cldn11 | Claudin 11 | cldn11a | T2 | 2.19 | 4.65E-08 |
| Palld | Palladin | palld | T2 | 2.19 | 9.19E-07 |
| Thbs4 | Thrombospondin 4 | thbs4a | T2 | 2.05 | 3.09E-04 |
| Ctnnd1 | Catenin delta 1 | ctnnd1 | T3 | 4.20 | 2.41E-12 |
| Net1 | Neuroepithelial cell transforming gene 1 | net1 | T3 | 2.79 | 2.84E-12 |
| Dapk3 | Death-associated protein kinase 3 | dapk3 | T3 | 2.09 | 1.49E-09 |
| Lox | Lysyl oxidase | lox | T3 | 1.92 | 1.83E-04 |
| Cell cycle | |||||
| Rgcc | Regulator of cell cycle | rgcc | T1 | 1.44 | 1.21E-03 |
| Ccnb2 | Cyclin B2 | ccnb2 | T1 | −2.89 | 4.82E-08 |
| Ccna2 | Cyclin A2 | ccna2 | T1 | −2.70 | 1.00E-07 |
| Mki67 | antigen identified by monoclonal antibody Ki 67 | mki67 | T1 | −2.68 | 9.26E-13 |
| Mcm3 | Minichromosome maintenance deficient 3 | mcm3l | T1 | −2.16 | 1.45E-05 |
| Cdc6 | Cell division cycle 6 | cdc6 | T1 | −1.86 | 1.06E-05 |
| Mcm10 | Minichromosome maintenance deficient 10 | mcm10 | T1 | −1.77 | 5.31E-06 |
| Chek1 | Checkpoint kinase 1 | chek1 | T1 | −1.70 | 2.67E-04 |
| Cdc20 | Cell division cycle 20 | cdc20 | T2 | −2.88 | 2.49E-10 |
| Ccnb1 | Cyclin B1 | ccnb1 | T2 | −2.72 | 5.35E-06 |
| Chek2 | Checkpoint kinase 2 | chek2 | T2 | −2.31 | 2.65E-08 |
| Cdk1 | Cyclin-dependent kinase 1 | cdk1 | T2 | −2.29 | 4.47E-07 |
| Pcna | Proliferating cell nuclear antigen | pcna | T2 | −2.12 | 2.32E-05 |
| Cdk2 | Cyclin-dependent kinase 2 | cdk2 | T2 | −1.33 | 3.34E-03 |
| Mos | Moloney sarcoma oncogene | mos | T3 | −2.67 | 2.48E-08 |
| Hormone receptor | |||||
| Lhcgr | Luteinizing hormone/choriogonadotropin receptor | lhcgr | T3 | 1.74 | 1.19E-04 |
| Esr2 | Estrogen receptor beta | esr2b | T1 | −1.39 | 1.29E-03 |
T1, genes that were regulated similarly among all three species; T2, genes that were regulated similarly only between human and zebrafish; T3, genes that were regulated similarly only between mice and zebrafish.
Changes in gene expression were based on log2 fold of the expression in wildtype relative to those in Pgr-KO. Genes were listed by fold change within each shared type in each category.
3.5. Novel genes that may be important for ovulation
Approximately 2/3 of DEGs (77%) found during ovulation in zebrafish did not show a significant difference in two selected studies of human and mice (Fig.6). It is likely some of these DEGs have important roles for ovulation in vertebrates, but were missed in these two studies. Therefore, we focused on the analyses of the top 200 significant DEGs in zebrafish (Table S4), specially the top 54 genes that showed large differences in zebrafish but were missed in human and mice studies, using a much more stringent cutoff (base mean count>1000, fold change>8; Table S6). Among the top 54 genes, only 33 genes had relative good annotations, which allowed to be grouped into several categories using PANTHER functional classification (http://www.pantherdb.org/). We identified two cell adhesion molecules, six ion channels or small molecule transporters, eight transcription factors or DNA binding proteins, seven kinases or kinase activators, and four G-protein modulators (Table 3). These genes dramatically up-regulated in WT compared to those in Pgr-KO. High expressions of two zebrafish genes, hypoxia-inducible factor 1 alpha subunit like (hif1al) and platelet-derived growth factor receptor beta polypeptide (pdgfrb), partially validated our hypothesis that important ovulation genes were missed in those two mammalian studies, since homologs of these two zebraifsh genes have been suggested to play important roles in ovulation in mouse and rat, respectively (Kim J et al., 2009a; Sleer and Taylor, 2007). However, the majority of these genes had not been linked to ovulation previously. Periostin (postnb, 60-fold increase, p=3.77E-72) and Pendrin (slc26a4, 128-fold increase, p=1.07E-53) were two typical examples. Periostin has been shown to increase its expression at sites of injury or inflammation and in tumors within adult organisms (Liu AY et al., 2014). Pendrin is a sodium independent anion channel that functions as a coupled iodide/chloride, iodide/bicarbonate and chloride/bicarbonate exchanger. Functions of Pendrin have been associated with the activities of inflammatory mediators in the airways (Bizhanova and Kopp, 2011; Scanlon et al., 2014); however, its functions in the zebrafish ovary may be related to ion exchanges and water absorption that are necessary for ovulation in fish in addition to inflammation. We also found that two genes important for methylation/demethylation, i.e., lysine (K)-specific demethylase 6B, a (kdm6ba) and spermne oxidase (smox), increased significantly during ovulation. Interestingly, high expressions of five tumor suppressor genes including forkhead box O3b (foxo3b), inhibin beta B (inhbb), DLC1 Rho GTPase activating protein (dlc1), DAB2 interacting protein b (dab2ipb), and cyclin G2 (ccng2) were also found in the follicular cells of preovulatory oocytes from WT female fish (labeled in bold font in Table 3).
Table 3.
Representative transcripts with most significant changes during ovulation in zebrafish, but were missed in two mammalian studies
| Ensembl(DR) | Gene Symbol (DR) | Base Mean | Mean counts (Mut) | Mean counts (WT) | log2 Fold Changea | FDR p-value |
|---|---|---|---|---|---|---|
| cell adhesion molecule | ||||||
| ENSDARG00000061219 | postnb | 2300.96 | 63.30 | 4538.63 | 5.91 | 3.77E-72 |
| ENSDARG00000038446 | nrp2b | 1332.76 | 148.54 | 2516.99 | 3.87 | 6.09E-25 |
| membrane ion channels and transporters | ||||||
| ENSDARG00000069431 | slc26a4 | 19657.61 | 192.42 | 39122.79 | 7.03 | 1.07E-53 |
| ENSDARG00000051914 | slc14a2 | 2860.52 | 83.73 | 5637.32 | 5.55 | 1.71E-32 |
| ENSDARG00000041107 | cftr | 17651.10 | 814.65 | 34487.56 | 5.07 | 3.67E-37 |
| ENSDARG00000034473 | ttyh3a | 2623.56 | 174.79 | 5072.33 | 4.27 | 1.68E-14 |
| ENSDARG00000007449 | slc25a39 | 1828.73 | 198.12 | 3459.35 | 3.90 | 1.45E-24 |
| ENSDARG00000073952 | slc4a7 | 1808.63 | 337.88 | 3279.39 | 3.05 | 4.82E-12 |
| transcription factors/DNA binding protein | ||||||
| ENSDARG00000036967 | smox | 5293.05 | 371.84 | 10214.26 | 4.32 | 5.87E-18 |
| ENSDARG00000031420 | wt1a | 3444.20 | 275.33 | 6613.08 | 4.17 | 1.01E-17 |
| ENSDARG00000017121 | mafba | 10269.01 | 1024.45 | 19513.58 | 3.99 | 1.96E-22 |
| ENSDARG00000042904 | foxo3bb | 1418.13 | 152.05 | 2684.22 | 3.74 | 2.72E-13 |
| ENSDARG00000036965 | rnf24 | 2446.91 | 301.67 | 4592.16 | 3.73 | 7.83E-24 |
| ENSDARG00000041169 | hif1al | 9990.20 | 1099.61 | 18880.78 | 3.69 | 1.43E-12 |
| ENSDARG00000063417 | erf | 2276.04 | 384.57 | 4167.52 | 3.20 | 3.01E-13 |
| ENSDARG00000088087 | kdm6ba | 16785.29 | 2950.99 | 30619.58 | 3.16 | 4.02E-14 |
| kinase/kinase activatiors/receptor kinase | ||||||
| ENSDARG00000057345 | pdgfrb | 11660.91 | 466.15 | 22855.68 | 5.02 | 4.38E-22 |
| ENSDARG00000027552 | mapk1 | 1885.05 | 109.33 | 3660.78 | 4.62 | 4.77E-22 |
| ENSDARG00000088967 | si:dkeyp-61g11.1 | 2423.68 | 148.23 | 4699.13 | 4.60 | 4.2E-25 |
| ENSDARG00000013841 | abl2 | 3639.77 | 271.60 | 7007.95 | 4.51 | 6.82E-46 |
| ENSDARG00000032103 | mapk6 | 6851.22 | 803.15 | 12899.29 | 3.77 | 6.57E-21 |
| ENSDARG00000017602 | ccng2 | 1693.47 | 301.35 | 3085.58 | 3.18 | 6.69E-17 |
| ENSDARG00000019420 | etnk1 | 10622.72 | 2022.70 | 19222.75 | 3.05 | 9.77E-14 |
| G-protein modulator | ||||||
| ENSDARG00000014577 | rhpn2 | 4165.14 | 214.52 | 8115.77 | 5.00 | 1.91E-46 |
| ENSDARG00000040177 | rgs16 | 2312.65 | 214.29 | 4411.01 | 3.91 | 6.19E-14 |
| ENSDARG00000051836 | si:dkeyp-19e1.3 | 1044.75 | 135.45 | 1954.04 | 3.62 | 1.36E-19 |
| ENSDARG00000075110 | dab2ipb | 2919.94 | 368.84 | 5471.04 | 3.58 | 1.46E-14 |
| others | ||||||
| ENSDARG00000074652 | smtna | 3356.04 | 39.86 | 6672.21 | 6.82 | 1.93E-55 |
| ENSDARG00000077688 | cpo | 1050.21 | 11.49 | 2088.93 | 6.62 | 4.41E-38 |
| ENSDARG00000016868 | rhobtb2a | 2929.81 | 60.08 | 5799.54 | 5.90 | 4.28E-31 |
| ENSDARG00000040777 | inhbb | 1056.45 | 70.63 | 2042.27 | 4.61 | 1.52E-38 |
| ENSDARG00000074414 | sema4ba | 3626.31 | 466.64 | 6785.98 | 3.65 | 6.91E-21 |
| ENSDARG00000079317 | Dlc1 | 13590.55 | 2280.00 | 24901.11 | 3.28 | 6.85E-19 |
Changes in gene expression were based on log2 fold of the expression in wildtype relative to those in Pgr-KO. Genes were ranked in the order of fold change and classified into functional group based on PANTHER functional classification (http://www.pantherdb.org/).
Bold indicates tumor suppressor.
4. Discussion
We have performed the first genome-wide differential gene expression analysis that is specifically designed for the follicular cells of preovulatory oocytes in the zebrafish and conducted the first comparison of differentially regulated genes in the follicular cells of preovulatory oocytes among zebrafish, mice, and humans. Our analysis indicates that ovulation is involved in a large network of approximately 3,000 genes, which is 11.5% of 26,000 available genes in zebrafish, all working in concert in the follicular cells of preovulatory zebrafish oocytes. The number of ovulation-related genes found in the zebrafish is about three times more than those reported in humans, but only slightly more than those reported in mice (Table.1). One likely reason is that RNA-Seq is much more sensitive than microarrays used in previous studies. Or, it is possible that we overestimate the ovulation-related genes in zebrafish due to differences in gene expression that may be present before ovulation occurs in Pgr knockout. It is impossible to predict which WT fish will undergo ovulation unless they have completed final oocyte maturation (see Fig.1 for detail). Not all fish ovulate daily, so we did not collect follicular cells prior to maturation in the current study due to the lack of a reliable marker to identify which fish will ovulate before the completion of oocyte maturation in vivo. Therefore, it will be necessary to establish an ovulation assay in vitro and to study gene expression during ovulation. Nevertheless, by comparing our data sets with mammalian data sets we are able to show that many conserved genes are related to ovulation and that associated signaling pathways are very similar whether we use the entire set of differentially regulated zebrafish genes or conserved “ovulatory” genes of fish, mice, and humans. These differentially expressed genes activate signaling pathways and biological processes that are important for ovulation to proceed while also down regulating pathways involved in growth and proliferation prior to ovulation. The switch from a period of growth and proliferation to ovulation is important for follicular cell rupture and for the process to proceed appropriately. These ovulatory genes and signaling pathways are highly conserved among fish, mice, and humans. Therefore, as demonstrated herein and elsewhere, zebrafish offers an alternative model for studying molecular mechanisms and biological processes that control ovulation.
We also found a large number of genes (approximately 2/3) that changed significantly in their expression in zebrafish prior to ovulation, but did not show significant difference in two data sets from mice and humans. The difference could be due to different experimental treatments (hormonal exposure vs. knockouts), different sensitivities of assay methods (RNA-seq vs microarray), or differences in species (e.g. external vs. internal fertilization/implantation/development). For an accurate comparison, we would have to generate transcriptomic data sets using the same sensitive sequencing technique, and the same genotype (e.g. non-functional PGR) in different model species. We will focus our discussion on similarities and conserved genes and pathways potentially important for ovulation, since additional information and experiments are required for defining unique mechanisms in different species.
4.1. Inflammatory response
Substantial evidence from studies in mammals has shown that biological events occurring in an ovulating follicle are similar to those in an acute inflammatory response (Kim J et al., 2009b; Wissing et al., 2014; Xu et al., 2011). One major indicator is the increase of prostaglandins (PGs) resulting from the up-regulation of prostaglandin reductase 2 (Ptgs2), a rate-limiting step in prostaglandin biosynthesis. It is still unclear whether Pgr and Ptgs are two separate downstream pathways of the LH signal, as there is no evidence for Pgr binding sites on ptgs2 promoter. Some had suggested that several genes including Pparg and Il6 might act as mediators between Pgr and Ptgs2, as demonstrated by conditional PPARG knockout mice (Harris et al., 2011; Kim SO et al., 2014). However, the transcripts of these mediators were not significantly regulated in the follicular cells of preovulatory oocytes in zebrafish, implying the Ppar pathway might not be an important mediator in zebrafish ovulation. Another transcriptional factor, Runx1, was a candidate mediator since up-regulation of Ptgs2 by Runx1 could be inhibited by a Pgr antagonist, and direct binding of Runx1 to two Runx-binding motifs in the Ptgs2 promoter region had also been confirmed by CHIP and EMSA assays (Jo and Curry, 2006; Liu J et al., 2009). Significant up-regulation of runx1 (fold change>10, p=2.40E-20) in the follicular cells of preovulatory oocytes supports Runx1 is a mediator regulating ptgs2. On the other hand, the present study showed one of the PGs receptors, Ptger4, was markedly up-regulated prior to ovulation in both mouse and zebrafish data sets. It has already been shown that Pgr directly regulates ptger4 in the preovulatory follicles of medaka (Fujimori et al., 2012; Hagiwara et al., 2014). In contrast, the role of PTGER4 in ovulation has been overlooked in mammals as studies have focused more on the role of PTGER2 in COC expansion and ovulation (Harris et al., 2011; Kim SO et al., 2014; Trau et al., 2016). Our study provides possible candidates that are involved in Pgr signaling pathway and ovulation. Clearly, more studies are needed to understand the molecular mechanisms that regulate members of Ptgs and Ptger and their functions in ovulation.
4.2. Vascularization
Vasodilation induced by LH in the preovulatory ovary is required for increased vascular permeability. This drives leukocytes to migrate from the blood vessels to the interior of the preovulatory follicles to release multiple cytokines and elicit inflammatory reactions leading to follicle wall breakdown (Kim J et al., 2009b). The expression of rgs2, which showed the greatest increase during ovulation in our RNA-Seq data (>100 fold), had been suggested to be essential for stabilizing blood pressure via inhibition of Gq/11-mediated signaling in human cardiovascular system (Doggrell, 2004; Zhang et al., 2014). Evidence also suggested that rgs2 expression could be stimulated by LH, but was attenuated by PGR antagonist or PTGS2 inhibitor in the preovulatory follicles of mouse and bovine (Ujioka et al., 2000; Sayasith et al., 2014). It remains to be elucidated whether or not RGS2 can play a similar role in follicle rupture as it does in the cardiovascular system, increasing vascular permeability.
NRP1, a well-known membrane bound co-receptor involved in VEGF signaling and vascularization, showed increased expression in preovulatory follicles of zebrafish, mouse, and human. NRP1 up-regulated KDR downstream signaling in response to VEGF in angiogenic modulation (Kofler and Simons, 2015). We also found a significant increase of kdr expression in the follicular cells of preovulatory oocytes in zebrafish. To prevent microbleeding during ovulation, multiple coagulation factors such as f3, f5, and tissue factor pathway inhibitor 2 (tfpi2) were concomitantly up-regulated in zebrafish, consistent with previous findings in humans (Wissing et al., 2014). The up-regulation of serpine 1(pai1), a blood clotting promoting factor, before ovulation in zebrafish is consistent with the recent finding in medaka. Serpine1 had been suggested to be a limiting regulator controlling plasmin hydrolyzing laminin, a major basement membrane component situated between the granulosa and theca cells of the follicle in medaka (Ogiwara et al., 2015). However, since PAI1-deficient mice were viable, the role of PAI1 in vascularization during ovulation needs to be explored further (Carmeliet et al., 1993; Binder et al., 2002).
4.3. Extracellular matrix remodeling and follicle wall breakdown
Proteases are required for extracellular matrix degradation and remodeling and are essential for follicular cell rupture and the release of mature oocytes. The involvement of several metalloproteinases, including members of MMP (matrix metalloproteinase), ADAM, and ADAMTS families, has been examined (Sriraman et al., 2008; Brown et al., 2010; Peluffo et al., 2011; Robker et al., 2000). However, evidence of these enzymes being involved in ovulation is limited, partly due to a lack of obvious effects on ovulation or embryonic lethality in knockouts (Enomoto et al., 2010). Interestingly, expression and knockout studies of Adamts1 have suggested the involvement of ADAMTS1 in ovulation and fertility in mammals. Adamts1 was up-regulated in the preovulatory follicles of several mammalian species. This up-regulation was partly dependent on the expression of Pgr in the granulosa cells (Robker et al., 2000). Knockout of Adamts1 in mice suggests an important role of this protease as a downstream effector of PGR in ovulation. Homozygous Adamts1 knockouts were subfertile, producing litters 4- to 5-fold less than control littermates, partially due to failed rupture of some large follicles (Shozu et al., 2005). Adamts1 knockout mice were subfertile and had less severe phenotypes than Pgr knockout mice, which could not ovulate and therefore had no litters. This indicates that other PGR regulated proteases may also contribute to the ovulatory mechanism. ADAMTS1 had been suggested to cleave versican in COC matrix (Brunet et al., 2015; Russell et al., 2003), so the functions of this gene in non-mammalian vertebrates may be different since no COC is necessary.
Intriguingly, up-regulation of adamts9 in mammals and zebrafish during ovulation suggests that this enzyme is most likely involved in ovulation and its function conserved across vertebrate species. Adamts9 was found to be up-regulated in preovulatory follicles or GCs following HCG treatment in macaque and human (Dehner et al., 2008; Peluffo et al., 2011; Wissing et al., 2014; Xu et al., 2011). GON-1, an ortholog of ADAMTS9, was involved in the degradation of extracellular matrix (ECM) and was essential for gonadal morphogenesis in C. elegans (Blelloch and Kimble, 1999; Blelloch et al., 1999). GON-1 helped migration of distal tip cells by degrading extracellular matrix components. In GON-1 mutants, the adult gonad was severely disorganized with no arm extension and no recognizable somatic structure. The developmental defects in gon-1 mutants were limited to the gonad. Other cells, tissues, and organs developed normally in C. elegans (Blelloch and Kimble, 1999; Blelloch et al., 1999). However, functions of ADAMTS9 in vertebrates have not been established, partly due to Adamts9 knockout mice dying before gastrulation (Enomoto et al., 2010). Thus, using alternative models or establishing conditional knockouts is required for examining the function of ADAMTS9 in vertebrates.
4.4. Missing information was compensated by our fish data
Some important ovulatory genes and signaling pathways such as ERBB, PI3K, and WNT signaling were unexpectedly missing in the shared gene lists and conserved enrichment maps, but significantly enriched in the zebrafish data set (Fig.5). ERBB signaling (epidermal growth factor receptor signaling) was induced by the LH surge through the activation of EGF-like factors (Hsieh et al., 2007). The increased expression of lhcgr, egfr, and mapk1 along with downstream genes (e.g. ptgs2, tnfaip6) in the follicular cells of WT preovulatory zebrafish oocytes suggests that ERBB signaling is conserved and involved in the ovulation of vertebrates. Knockouts of PTEN activated PI3K signaling and decreased susceptibility to apoptosis and enhanced ovulation in mice (Fan et al., 2008). Interestingly, we found that pten (a tumor suppressor) was highly expressed in the follicular cells of WT and that PI3K/Akt signaling was also enriched in up-regulated genes (Fig.5). Similarly, tumor suppressor genes dab2ip and foxo3 were expressed much higher in WT, which correlated with enriched WNT signaling in zebrafish. FOXO3 had been shown to inhibit WNT signaling and cancer development (Dehner et al., 2008; Liu H et al., 2015). Notably, we found at least 232 human tumor suppressor (Zhao et al., 2016) homolog genes that were differentially expressed in the follicular cells of zebrafish preovulatory oocytes. Molecular mechanisms and functions of these tumor-related genes during vertebrate ovulation are still unclear.
Hypoxia typically occurs at the time of ovulation (Kim J et al., 2009a). This was supported by high expression levels of hif1al (hypoxia-inducible factor 1, alpha subunit like) in the follicular cells of WT preovulatory oocytes. Hypoxia response also activated angiogenesis, consistent with the high expression of vascular endothelial growth factor receptors flt1 and kdr in WT fish. Activation of NF-kB (nuclear factor kappaB) was a critical part of transcriptional response to hypoxia and the local inflammatory response (Culver et al., 2010; Pahl, 1999). High expression of rela/p65, a component of NF-kB, increased expression of nfkbia (NF-KappaB inhibitor, alpha), encoding the cognate binding inhibitor of Rela, and several ubiquitin related genes (ubb, ubc, uba52, ube2a,Table S2). This suggested possible activation of NF-kB signaling in preovulatory follicular cells in response to hypoxia-like conditions of ovulation (Verma et al., 1995). The high expression of tnfrsf21 (tumor necrosis factor receptor superfamily, member 21 death receptor 6) during ovulation suggested that activation of NF-kB signaling was also involved in local inflammation (Pan et al., 1998). Intriguingly, il1b showed low expression in WT fish, although IL1B could induce direct binding of NF-kB to the promoter sequence of ADAMTS9 in human chondrocytes (Altuntas et al., 2015). More studies are required to determine the activation and involvement of NF-kB in response to hypoxia, and its regulation of inflammation and ADAMTS9 during ovulation.
In summary, for the first time we successfully identified genes and signaling pathways that were potentially important for ovulation in zebrafish, a non-mammalian vertebrate model, using high-throughput sequencing and Pgr-KO. The comparison of differentially regulated genes among human, mouse, and zebrafish data sets further confirmed that genes and signaling pathways important for ovulation were conserved among vertebrates. Zebrafish should serve as an excellent model for studying the function of genes and signaling pathways that are important for ovulation.
Supplementary Material
Highlights.
A large set of genes (>10%) differentially expressed during ovulation.
Over 600 genes showed similar increase or decrease in fish, mice, and humans.
Signaling pathways for ovulation are highly conserved in vertebrates.
New candidates for the regulation of ovulation have been identified.
Acknowledgments
Funding
This work was supported by NIH 1R15GM100461-01A1 (Y.Z.), North Carolina Biotechnology Center Biotechnology Research Grant #2012-BRG-1210 (Y.Z.), National Natural Science Foundation of China No. 31201977 (S. C), and No. 41276129 (W. H).
We would like to express our great appreciation to an anonymous reviewer, and to Ms. Joyce J. Newman for proof reading and improvement of language.
Footnotes
Author Contributions
DL performed experiments, conducted bioinformatics analyses, and wrote the paper. MB initiated bioinformatics analyses. SC and WH supervised the project and discussed the results. YZ conceived the idea, supervised the project, performed experiments, and wrote the paper. All the experiments and analyses were conducted at East Carolina University.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:1–12. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2014;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altuntas A, Halacli SO, Cakmak O, Erden G, Akyol S, Ugurcu V, Hirohata S, Demircan K. Interleukin-1beta induced nuclear factor-kappaB binds to a disintegrin-like and metalloproteinase with thrombospondin type 1 motif 9 promoter in human chondrosarcoma cells. Mol Med Rep. 2015;12:595–600. doi: 10.3892/mmr.2015.3444. [DOI] [PubMed] [Google Scholar]
- Binder BR, Christ G, Gruber F, Grubic N, Hufnagl P, Krebs M, Mihaly J, Prager GW. Plasminogen Activator Inhibitor 1: Physiological and Pathophysiological Roles. Physiology. 2002;17:56–61. doi: 10.1152/nips.01369.2001. [DOI] [PubMed] [Google Scholar]
- Bizhanova A, Kopp P. Controversies concerning the role of pendrin as an apical iodide transporter in thyroid follicular cells. Cell Physiol Biochem. 2011;28:485–490. doi: 10.1159/000335103. [DOI] [PubMed] [Google Scholar]
- Blelloch R, Kimble J. Control of organ shape by a secreted metalloprotease in the nematode Caenorhabditis elegans. Nature. 1999;399:586–90. doi: 10.1038/21196. [DOI] [PubMed] [Google Scholar]
- Blelloch R, Anna-Arriola SS, Gao D, Li Y, Hodgkin J, Kimble J. The gon-1 gene is required for gonadal morphogenesis in Caenorhabditis elegans. Dev Biol. 1999;216:382–93. doi: 10.1006/dbio.1999.9491. [DOI] [PubMed] [Google Scholar]
- Brown HM, Dunning KR, Robker RL, Boerboom D, Pritchard M, Lane M, Russell DL. ADAMTS1 cleavage of versican mediates essential structural remodeling of the ovarian follicle and cumulus-oocyte matrix during ovulation in mice. Biol Reprod. 2010;83:549–557. doi: 10.1095/biolreprod.110.084434. [DOI] [PubMed] [Google Scholar]
- Brunet FG, Fraser FW, Binder MJ, Smith AD, Kintakas C, Dancevic CM, Ward AC, McCulloch DR. The evolutionary conservation of the A Disintegrin-like and Metalloproteinase domain with Thrombospondin-1 motif metzincins across vertebrate species and their expression in teleost zebrafish. BMC Evol Biol. 2015;15:1–15. doi: 10.1186/s12862-015-0281-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P, Kieckens L, Schoonjans L, Ream B, van Nuffelen A, Prendergast G, Cole M, Bronson R, Collen D, Mulligan RC. Plasminogen activator inhibitor-1 gene-deficient mice. I. Generation by homologous recombination and characterization. J Clin Invest. 1993;92:2746–2755. doi: 10.1172/JCI116892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman RW, Reading BJ, Sullivan CV. Ovary transcriptome profiling via artificial intelligence reveals a transcriptomic fingerprint predicting egg quality in striped bass, Morone saxatilis. PLoS One. 2014;9:e96818. doi: 10.1371/journal.pone.0096818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver C, Sundqvist A, Mudie S, Melvin A, Xirodimas D, Rocha S. Mechanism of Hypoxia-Induced NF-κB. Mol Cell Biol. 2010;30:4901–4921. doi: 10.1128/MCB.00409-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehner M, Hadjihannas M, Weiske J, Huber O, Behrens J. Wnt signaling inhibits Forkhead box O3a-induced transcription and apoptosis through up-regulation of serum- and glucocorticoid-inducible kinase 1. J Biol Chem. 2008;283:19201–19210. doi: 10.1074/jbc.M710366200. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doggrell SA. Is RGS-2 a new drug development target in cardiovascular disease? Expert Opin Ther Targets. 2004;8:355–358. doi: 10.1517/14728222.8.4.355. [DOI] [PubMed] [Google Scholar]
- Enomoto H, Nelson CM, Somerville RP, Mielke K, Dixon LJ, Powell K, Apte SS. Cooperation of two ADAMTS metalloproteases in closure of the mouse palate identifies a requirement for versican proteolysis in regulating palatal mesenchyme proliferation. Development. 2010;137:4029–4038. doi: 10.1242/dev.050591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H-Y, Liu Z, Cahill N, Richards JS. Targeted Disruption of Pten in Ovarian Granulosa Cells Enhances Ovulation and Extends the Life Span of Luteal Cells. Mol Endocrinol. 2008;22:2128–2140. doi: 10.1210/me.2008-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori C, Ogiwara K, Hagiwara A, Takahashi T. New evidence for the involvement of prostaglandin receptor EP4b in ovulation of the medaka, Oryzias latipes. Mol Cell Endocrinol. 2012;362:76–84. doi: 10.1016/j.mce.2012.05.013. [DOI] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- Gohin M, Bobe J, Chesnel F. Comparative transcriptomic analysis of follicle-enclosed oocyte maturational and developmental competence acquisition in two non-mammalian vertebrates. BMC Genomics. 2010;11:18. doi: 10.1186/1471-2164-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara A, Ogiwara K, Katsu Y, Takahashi T. Luteinizing Hormone-Induced Expression of Ptger4b, a Prostaglandin E2 Receptor Indispensable for Ovulation of the Medaka Oryzias latipes, Is Regulated by a Genomic Mechanism Involving Nuclear Progestin Receptor. Biol Reprod. 2014;90:126–121. 114. doi: 10.1095/biolreprod.113.115485. [DOI] [PubMed] [Google Scholar]
- Hanna RN, Zhu Y. Controls of meiotic signaling by membrane or nuclear progestin receptor in zebrafish follicle-enclosed oocytes. Mol Cell Endocrinol. 2011;337:80–88. doi: 10.1016/j.mce.2011.02.004. [DOI] [PubMed] [Google Scholar]
- Harris SM, Aschenbach LC, Skinner SM, Dozier BL, Duffy DM. Prostaglandin E2 Receptors Are Differentially Expressed in Subpopulations of Granulosa Cells from Primate Periovulatory Follicles. Biol Reprod. 2011;85:916–923. doi: 10.1095/biolreprod.111.091306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Gonzalez I, Gonzalez-Robayna I, Shimada M, Wayne CM, Ochsner SA, White L, Richards JS. Gene Expression Profiles of Cumulus Cell Oocyte Complexes during Ovulation Reveal Cumulus Cells Express Neuronal and Immune-Related Genes: Does this Expand Their Role in the Ovulation Process? Mol Endocrinol. 2006;20:1300–1321. doi: 10.1210/me.2005-0420. [DOI] [PubMed] [Google Scholar]
- Hsieh M, Lee D, Panigone S, Horner K, Chen R, Theologis A, Lee DC, Threadgill DW, Conti M. Luteinizing hormone-dependent activation of the epidermal growth factor network is essential for ovulation. Mol Cell Biol. 2007;27:1914–1924. doi: 10.1128/MCB.01919-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydon JP, DeMayo FJ, Funk CR, Mani SK, Hughes AR, Montgomery CA, Jr, Shyamala G, Conneely OM, O'Malley BW. Mice lacking progesterone receptor exhibit pleiotropic reproductive abnormalities. Genes Dev. 1995;9:2266–2278. doi: 10.1101/gad.9.18.2266. [DOI] [PubMed] [Google Scholar]
- Jo M, Curry TE. Luteinizing Hormone-Induced RUNX1 Regulates the Expression of Genes in Granulosa Cells of Rat Periovulatory Follicles. Mol Endocrinol. 2006;20:2156–2172. doi: 10.1210/me.2005-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Bagchi IC, Bagchi MK. Signaling by Hypoxia-Inducible Factors Is Critical for Ovulation In Mice. Endocrinology. 2009a;150:3392–3400. doi: 10.1210/en.2008-0948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Bagchi IC, Bagchi MK. Control of ovulation in mice by progesterone receptor-regulated gene networks. Mol Hum Reprod. 2009b;15:821–828. doi: 10.1093/molehr/gap082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SO, Harris SM, Duffy DM. Prostaglandin E2 (EP) Receptors Mediate PGE2-Specific Events in Ovulation and Luteinization Within Primate Ovarian Follicles. Endocrinology. 2014;155:1466–1475. doi: 10.1210/en.2013-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler NM, Simons M. Angiogenesis versus arteriogenesis: neuropilin 1 modulation of VEGF signaling. F1000Prime Rep. 2015;2015(7):26. doi: 10.12703/P7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota K, Cui W, Dhakal P, Wolfe MW, Rumi MA, Vivian JL, Roby KF, Soares MJ. Rethinking progesterone regulation of female reproductive cyclicity. Proc Natl Acad Sci U S A. 2016;113:4212–7. doi: 10.1073/pnas.1601825113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AY, Zheng H, Ouyang G. Periostin, a multifunctional matricellular protein in inflammatory and tumor microenvironments. Matrix Biol. 2014;37:150–156. doi: 10.1016/j.matbio.2014.04.007. [DOI] [PubMed] [Google Scholar]
- Liu H, Yin J, Wang H, Jiang G, Deng M, Zhang G, Bu X, Cai S, Du J, He Z. FOXO3a modulates WNT/beta-catenin signaling and suppresses epithelial-to-mesenchymal transition in prostate cancer cells. Cell Signal. 2015;27:510–518. doi: 10.1016/j.cellsig.2015.01.001. [DOI] [PubMed] [Google Scholar]
- Liu J, Park E-S, Jo M. Runt-Related Transcription Factor 1 Regulates Luteinized Hormone-Induced Prostaglandin-Endoperoxide Synthase 2 Expression in Rat Periovulatory Granulosa Cells. Endocrinology. 2009;150:3291–3300. doi: 10.1210/en.2008-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiwara K, Hagiwara A, Rajapakse S, Takahashi T. The role of urokinase plasminogen activator and plasminogen activator inhibitor-1 in follicle rupture during ovulation in the teleost medaka. Biol Reprod. 2015;92:10. doi: 10.1095/biolreprod.114.121442. [DOI] [PubMed] [Google Scholar]
- Pahl HL. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene. 1999;18:6853–6866. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- Pan G, Bauer JH, Haridas V, Wang S, Liu D, Yu G, Vincenz C, Aggarwal BB, Ni J, Dixit VM. Identification and functional characterization of DR6, a novel death domain-containing TNF receptor. FEBS Lett. 1998;431:351–356. doi: 10.1016/s0014-5793(98)00791-1. [DOI] [PubMed] [Google Scholar]
- Peluffo MC, Murphy MJ, Baughman ST, Stouffer RL, Hennebold JD. Systematic analysis of protease gene expression in the rhesus macaque ovulatory follicle: metalloproteinase involvement in follicle rupture. Endocrinology. 2011;152:3963–3974. doi: 10.1210/en.2011-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reading BJ, Chapman RW, Schaff JE, Scholl EH, Opperman CH, Sullivan CV. An ovary transcriptome for all maturational stages of the striped bass (Morone saxatilis), a highly advanced perciform fish. BMC Res Notes. 2012;5:111. doi: 10.1186/1756-0500-5-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robker RL, Russell DL, Espey LL, Lydon JP, O'Malley BW, Richards JS. Progesterone-regulated genes in the ovulation process: ADAMTS-1 and cathepsin L proteases. Proc Natl Acad Sci U S A. 2000;97:4689–4694. doi: 10.1073/pnas.080073497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DL, Doyle KM, Ochsner SA, Sandy JD, Richards JS. Processing and localization of ADAMTS-1 and proteolytic cleavage of versican during cumulus matrix expansion and ovulation. J Biol Chem. 2003;278:42330–42339. doi: 10.1074/jbc.M300519200. [DOI] [PubMed] [Google Scholar]
- Sayasith K, Sirois J, Lussier JG. Expression and regulation of regulator of G-protein signaling protein-2 (RGS2) in equine and bovine follicles prior to ovulation: molecular characterization of RGS2 transactivation in bovine granulosa cells. Biol Reprod. 2014;91:1–12. doi: 10.1095/biolreprod.114.121186. [DOI] [PubMed] [Google Scholar]
- Scanlon KM, Gau Y, Zhu J, Skerry C, Wall SM, Soleimani M, Carbonetti NH. Epithelial anion transporter pendrin contributes to inflammatory lung pathology in mouse models of Bordetella pertussis infection. Infect Immun. 2014;82:4212–4221. doi: 10.1128/IAI.02222-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriraman V, Eichenlaub-Ritter U, Bartsch JW, Rittger A, Mulders SM, Richards JS. Regulated expression of ADAM8 (a disintegrin and metalloprotease domain 8) in the mouse ovary: evidence for a regulatory role of luteinizing hormone, progesterone receptor, and epidermal growth factor-like growth factors. Biol Reprod. 2008;78:1038–1048. doi: 10.1095/biolreprod.107.066340. [DOI] [PubMed] [Google Scholar]
- Shozu M, Minami N, Yokoyama H, Inoue M, Kurihara H, Matsushima K, Kuno K. ADAMTS-1 is involved in normal follicular development, ovulatory process and organization of the medullary vascular network in the ovary. J Mol Endocrinol. 2005;35:343–55. doi: 10.1677/jme.1.01735. [DOI] [PubMed] [Google Scholar]
- Sleer LS, Taylor CC. Platelet-derived growth factors and receptors in the rat corpus luteum: localization and identification of an effect on luteogenesis. Biol Reprod. 2007;76:391–400. doi: 10.1095/biolreprod.106.053934. [DOI] [PubMed] [Google Scholar]
- Trau HA, Brannstrom M, Curry TE, Jr, Duffy DM. Prostaglandin E2 and vascular endothelial growth factor A mediate angiogenesis of human ovarian follicular endothelial cells. Hum Reprod. 2016;31:436–444. doi: 10.1093/humrep/dev320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujioka T, Russell DL, Okamura H, Richards JS, Espey LL. Expression of regulator of G-protein signaling protein-2 gene in the rat ovary at the time of ovulation. Biol Reprod. 2000;63:1513–1517. doi: 10.1095/biolreprod63.5.1513. [DOI] [PubMed] [Google Scholar]
- Verma IM, Stevenson JK, Schwarz EM, Van Antwerp D, Miyamoto S. Rel/NF-kappa B/I kappa B family: intimate tales of association and dissociation. Genes Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- Wissing ML, Kristensen SG, Andersen CY, Mikkelsen AL, Host T, Borup R, Grondahl ML. Identification of new ovulation-related genes in humans by comparing the transcriptome of granulosa cells before and after ovulation triggering in the same controlled ovarian stimulation cycle. Hum Reprod. 2014;29:997–1010. doi: 10.1093/humrep/deu008. [DOI] [PubMed] [Google Scholar]
- Xu F, Stouffer RL, Muller J, Hennebold JD, Wright JW, Bahar A, Leder G, Peters M, Thorne M, Sims M, Wintermantel T, Lindenthal B. Dynamics of the transcriptome in the primate ovulatory follicle. Mol Hum Reprod. 2011;17:152–165. doi: 10.1093/molehr/gaq089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Mende U. Functional role, mechanisms of regulation, and therapeutic potential of regulator of G protein signaling 2 in the heart. Trends Cardiovasc Med. 2014;24:85–93. doi: 10.1016/j.tcm.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Kim P, Mitra R, Zhao J, Zhao Z. TSGene 2.0: an updated literature-based knowledgebase for tumor suppressor genes. Nucleic Acids Res. 2016;44:D1023–1031. doi: 10.1093/nar/gkv1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Liu D, Shaner ZC, Chen S, Hong W, Stellwag EJ. Nuclear progestin receptor (pgr) knockouts in zebrafish demonstrate role for pgr in ovulation but not in rapid non-genomic steroid mediated meiosis resumption. Front Endocrinol (Lausanne) 2015;6:37. doi: 10.3389/fendo.2015.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.