Abstract
The human malaria parasite, Plasmodium falciparum, depends on a coordinated regulation of gene expression for development and propagation within the human host. Recent developments suggest that gene regulation in the parasite is largely controlled by epigenetic mechanisms. Here, we discuss recent advancements contributing to our understanding of the mechanisms controlling gene regulation in the parasite, including nucleosome landscape, histone modifications, and nuclear architecture. In addition, various processes involved in regulation of parasite-specific genes and gene families are examined. Finally, we address the use of epigenetic processes as targets for novel antimalarial therapies. Collectively, these topics highlight the unique biology of P. falciparum, and contribute to our understanding of mechanisms regulating gene expression in this deadly parasite.
Keywords: epigenetics, gene regulation, chromatin, nucleosome, Plasmodium, malaria
The Malaria Parasite
The human malaria parasite remains one of the deadliest infectious agents worldwide. In 2015, an estimated 214 million cases of infection and 438 000 malaria-related deaths were reported [1]. Most malaria infections occur in sub-Saharan Africa; however, developing countries in South East Asia and South America are also affected. Children under the age of five and pregnant women are most susceptible to the disease, and in 2015 children under the age of five accounted for approximately 70% of all malaria-related deaths.
P. falciparum, one of five Plasmodium species that can infect humans, is responsible for the most severe disease symptoms and the highest mortality rate in humans. The parasite develops through a complex life cycle that involves two hosts: the Anopheles mosquito and the human host (Figure 1). The parasite’s life cycle begins as an infected Anopheles mosquito takes a blood meal from a human and in the process injects sporozoites into the host's bloodstream. The sporozoites translocate to the liver, invade liver cells (hepatocytes), and replicate multiple times for a 2-week period, producing thousands of merozoites that leave the liver and invade red blood cells (erythrocytes) [2,3].
Figure 1.
Schematic Representation of the Life Cycle of the Malaria Parasite. In the human host, the parasite first develops through the liver stage, followed by subsequent 48-h replication cycles inside red blood cells, which is the stage responsible for symptomatic disease. During this replication cycle, a small proportion of parasites will commit to sexual differentiation into male and female gametocytes that can be taken up by a mosquito. Sexual reproduction takes place inside the mosquito midgut and ultimately results in the formation of sporozoites that can be transmitted to a new human host.
During the intraerythrocytic developmental cycle (IDC), the parasite develops asexually through ring, trophozoite, and schizont stages and multiplies by a process of replication termed schizogony. As the parasite progresses through the three distinct developmental stages, it undergoes multiple rounds of nuclear replication and cytokinesis to produce 16–32 daughter cells at the end of each IDC. The daughter merozoites then burst out of the host red blood cell and invade new healthy red blood cells. During the IDC, environmental stress can trigger the parasites into committing to sexual development, resulting in differentiation into male and female gametocytes. The mature gametocytes can be ingested by a feeding mosquito, undergo sexual replication in the mosquito midgut, and develop further into salivary gland sporozoites to be transmitted to a new human host as the mosquito takes the next blood meal. This multistage life cycle of the parasite is tightly regulated, most likely by strict control of stage-specific gene expression. In eukaryotes, stage-specific regulation of gene expression can be a combined effect of transcriptional, post-transcriptional and translational control. In P. falciparum, the nature and the contribution of mechanisms regulating gene expression at the transcriptional level, including the role of chromatin structure, are starting to emerge. In this review, we summarize the current knowledge on the role of chromatin structure and epigenetics in gene regulation of the human malaria parasite and its potential to identify much-needed new therapies.
P. falciparum Genome
The human malaria parasite P. falciparum has a relatively compact genome of twenty three million base pairs that is organized into 14 chromosomes (per haploid genome) [4]. The P. falciparum genome is the most AT-rich eukaryotic genome sequenced to date, with an overall AT composition of ~80%, rising to 90–95% in introns and intergenic regions. The distinct developmental stages of the P. falciparum life cycle (Figure 1) are characterized by coordinated changes in gene expression [5–10]. In eukaryotes, gene expression is partly controlled by transcription factors that bind to cell- or tissue-specific promoters to regulate transcription [11]. However, a surprisingly low number of specific transcription factors have been identified in the parasite's genome [12,13] and, in particular, only a few stage-specific transcription factors have been validated [14–20]. Therefore, the coordinated cascade of transcripts observed throughout the parasite life cycle is unlikely to be regulated only by this limited collection of specific transcription factors, and suggests that additional components and mechanisms, such as post-transcriptional [21–25], translational, and post-translational regulation [21,26,27], as well as change of chromatin structure, may control the expression of the predicted 6372 genes in the malaria parasite.
How DNA is packaged inside the nucleus greatly influences gene expression. After a general overview of what is known in higher eukaryotes, we describe important features of the P. falciparum nuclear and chromatin landscape and how these features, including histone modifications, nucleosome occupancy, and the three-dimensional (3D) nuclear organization, may affect gene expression.
Eukaryotic Chromatin Structure
In a eukaryotic cell, genomic DNA is tightly wrapped around histone proteins and assembled as nucleosomes. These nucleosomes are then coiled and packaged together, resulting in a fiber also known as chromatin. Interactions between chromatin and protein complexes as well as the dynamics of nucleosome positioning and post-translational modifications (PTMs) of histone core proteins are of vital importance to the usage of DNA.
The major step in gene transcription initiation is the recruitment of RNA polymerase II, along with other general transcription factors (TFIIs), to promoter regions to form the basal preinitiation complex (PIC). Recent genome-wide nucleosome mapping studies in model organisms, such as yeast and human, have revealed consensus patterns in nucleosome organization, including lower nucleosome density at intergenic regions as compared to genic regions, a strong nucleosome-depleted region (NDR) near the promoter, and well-positioned nucleosomes (i.e., −1 and +1 nucleosomes) containing variant histone H2A.Z around the transcription start site (TSS) [28–31]. These findings suggest that specific positioning of nucleosomes, especially at promoter and transcription start or stop regions, largely contributes to transcriptional control by governing the access of components of the transcription initiation machinery to their binding sites. Furthermore, to ensure nucleosome dynamics and gene expression regulation, nucleosome components or the entire nucleosome may be repositioned, removed, or replaced through the action of ATP-dependent chromatin remodeling enzymes. In addition, post-translational modifications of histone proteins can have large effects on chromatin structure and gene activity. For instance, acetylation of histone H3 at lysine 9 or 14 of their N-terminal tail (H3K9ac and H3K14ac) often alters the physical and chemical stability of nucleosomes, resulting in an open chromatin structure and a transcriptionally permissive state [32]. On the other hand, trimethylation of histone H3 at lysine 9 and 27 (H3K9me3 and H3K27me3) is often associated with a heterochromatin state and repression of gene expression [33].
Besides precise positioning of nucleosomes, gene expression requires physical interaction between promoter regions and their distal regulatory elements, yet promoters and their regulatory elements are often linearly separated along the chromosome. To overcome this spatial constraint, chromatin loops are formed to bring together the regulatory elements and their promoters for gene activation. For example, the distal enhancer known as locus control region (LCR) of beta-globin (β-globin) genes makes contact with globin gene promoters through chromatin looping and dynamically changes its interactions with the promoters of embryonic, fetal, and adult β-globin genes to ensure expression of the correct set of β-globin genes at the proper developmental stage [34–38]. Chromatin loops can also play a role in gene silencing. For example, the maternal copy of the insulin-like growth factor 2 (Igf2) gene is silenced by placement in an inactive chromatin loop that prevents enhancer–promoter interaction and allows the exclusive expression of the paternal allele [39,40].
Over the past decade, a series of molecular and genomic approaches have been developed (3C, 4C, 5C, Hi-C, etc.) to study the higher order organization of chromosomes by mapping interactions between genomic loci (Box 1) [41]. Hi-C analyses of mouse and human chromosome structures have revealed that eukaryotic genomes are organized into large blocks that show high levels of chromatin interactions within that region, but not with other loci in the genome. These regions, called topologically associated domains (TADs) [42,43], are well defined by insulator proteins [44,45] and are composed of many chromatin loops that have important functional roles in regulating gene expression. On a higher dimension, individual chromosomes organize and occupy distinct territories within the nucleus, and such organization is highly associated with gene density; gene-dense chromatin is usually enriched in the internal part of the nucleus, while gene-poor regions tend to locate toward the nuclear periphery [46–53].
Box 1. Selected Methods for Assaying 3D Chromatin Structure.
Chromosome Conformation Capture (3C) method identifies pairwise interactions between two selected fragments in a one-to-one approach. The major steps of the 3C protocol include cross-linking cells with formaldehyde, digesting chromatin with a restriction enzyme, ligating chromatin fragments that are in close proximity, reversal of cross-links, and identifying the interaction frequency of the targeted regions using primers specific to those regions in a quantitative PCR. For 3C experiments, prior knowledge of which genomic loci are likely to interact is required, and the technique is best suited to study small numbers of loci or relatively small genomic regions [124].
Circularized Chromosome Conformation Capture/Chromosome Conformation Capture-on-Chip (4C) method uses high-throughput sequencing to identify all contacts for a selected locus in a one-versus-all approach. The 4C methodology is similar to 3C up to the reverse cross-linking step. Following an additional round of digestion, DNA fragments are self-ligated to create circular DNA containing both the locus of interest and its interacting partner. Using inverse PCR, the interacting partner sequence is amplified and sequenced, generating a genome-wide interaction profile for the region of interest. The 4C technique is suitable for studying both inter and intrachromosomal contacts of a specific region at high resolution [125].
Chromosome Conformation Capture Carbon Copy (5C) method is a many-to-many approach capturing chromosome interactions for multiple loci in one experiment. This methodology uses a pool of primers that are hybridized to different restriction sites in the genome. During the ligation step, primer pairs on interacting fragments can be ligated together. Each primer contains a universal sequence at its 5′ end to allow the amplification of these ligation products, thus detecting the interaction frequencies for multiple loci or genomic regions. One major bottleneck for 5C is the requirement for the design of individual primers for each restriction site [126].
Chromosome Conformation Capture Coupled with Next-Generation Sequencing (Hi-C) method provides true genome-wide capture of chromosome interactions, also described as an all-to-all approach. Hi-C follows the basic steps of 3C procedures with a modified ligation step: after restriction digestion of cross-linked DNA, biotin-labeled nucleotides are incorporated into the overhangs followed by blunt-end ligation. The ligated fragments are then isolated using streptavidin-coated magnetic beads and sequenced on a next-generation sequencing platform. Hi-C provides a complete genome-wide interaction map, including both local and long-range intrachromosomal contacts as well as interchromosomal contacts; however, large amounts of sequencing data and complex computational analyses are required [114].
Emerging evidence shows that long noncoding RNAs (lncRNAs) are also involved in the control of transcription and genome activity by affecting chromatin-remodeling events, including nucleosome positioning and chromatin looping [54]. A well known example is the lncRNA known as Xist, which mediates X-chromosome inactivation during zygotic development [55]. Deposition of Xist on the X chromosome recruits histone-modifying enzymes that place repressive histone marks, such as H3K9 and H3K27 methylation, leading to gene silencing and the formation of heterochromatin. How many of these typical chromatin features are maintained in Plasmodium, as well as the areas where additional research is necessary, will be highlighted in the upcoming topics.
Chromatin Structure and Gene Regulation in P. falciparum
Transcriptional Machinery in Plasmodium
Since the publication of the P. falciparum genome in 2002 [4], researchers have attempted to explore the transcriptional machinery of the parasite in detail. The basal transcriptional machinery, RNA polymerase II and all its subunits, have been identified in the parasite [13,56]. Additionally, a total of 23 TFII components have been found. Although four TATA-binding protein (TBP)-associated factors (TAFs) have been discovered in P. falciparum, the parasite seems to lack the classical TFIID subunits with a histone fold domain. The histone fold domain allows the TAFs to assemble into heterodimers [56]. In yeast, the TFIID complex contains the TATA-binding protein (TBP) and TBP-associated proteins (TAFs) and more than half of these proteins contain a histone fold motif [57]. The fact that the TAFs in P. falciparum do not contain this motif suggests that the parasite TFIID complex is divergent from other eukaryotes. As so many TAFs are missing in P. falciparum, compared to other eukaryotes, alternative mechanisms may be more important for transcriptional regulation in the parasite.
In eukaryotes, specific transcription factors (TFs) recruit and activate the transcription preinitiation complex. Remarkably, in P. falciparum, only about 30 specific transcription factors have been identified [12,13]. Twenty-seven of these TFs belong to the apicomplexan-specific family of transcription factors and contain a modified form of the AP2 domain found in plant TFs (ApiAP2) [12]. These TFs are present throughout the parasite's life cycle and are believed to control the transition between specific developmental stages. Some examples include AP2-G for the development of gametocytes [17,18], AP2-Sp for sporozoite development [19], AP2-L for the development of liver-stage parasites [16], and AP2-O for the development of ookinetes in the mosquito [20]. Another member of the ApiAP2 family, PfSIP2, is shown to bind to heterochromatic regions of the genome and act as a transcriptional repressor [58]. Despite continued experimentation, it remains to be determined how ApiAP2 transcription factors recruit RNA polymerase II to sites of transcription. Similar to the lack of TAFs mentioned above, the small number of specific TFs identified in P. falciparum highlights the role of alternative mechanisms regulating gene expression.
It is now evident that most of the P. falciparum genome is maintained in a decondensed chromatin environment called euchromatin, while only a small subset, including subtelomeric regions and a few internal loci, are contained within highly condensed heterochromatin cluster(s) [59,60]. The heterochromatin cluster(s) of the parasite's genome are marked by H3K9me3 modifications and heterochromatin protein 1 (PfHP1), and harbor gene families encoding clonally variant antigens (var, rifin, stevor, and pfmc-2tm), invasion gene families (eba and clag), and a few other loci such as the gametocyte-specific transcription factor pfap2-g during the IDC [61–65]. The presence of these repressive marks on parasite stage-specific gene families suggests that mechanisms regulating transcription of these genes may be more conserved with higher eukaryotes, than the rest of the genes in the Plasmodium genome.
Epigenetic Regulation of Clonally Variant Gene Families
P. falciparum Virulence Genes
Disease pathogenesis in malaria is the result of the parasite’s ability to escape host immune responses. The var gene family, the best characterized multigene family in P. falciparum, encodes erythrocyte membrane proteins 1 (PfEMP1s) that are expressed at the surface of the infected erythrocyte and play a key role in cytoadherence and antigenic variation [66]. Approximately 60 var genes are present in a haploid genome of P. falciparum, but only one var gene is expressed at any given time [67]. By switching var gene expression, the parasite is able to avoid host immune responses. The mechanisms regulating var gene expression in P. falciparum in vitro have emerged recently and are discussed below.
Histone Post-Translational Modifications Affecting Virulence Gene Expression
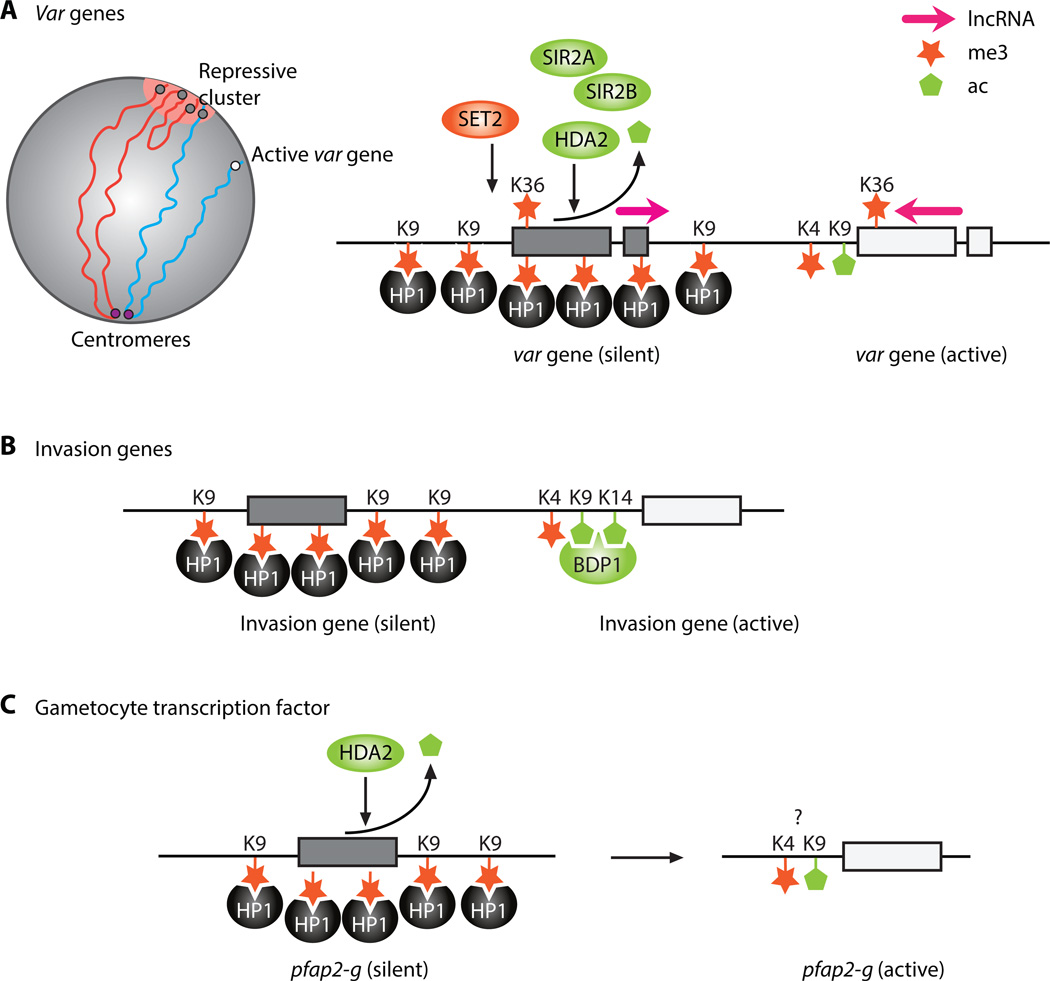
Silent var genes are clustered to one or more repressive regions at the nuclear periphery, marked by H3K9me3 and PfHP1 (Figure 2A) [59–63,65,68,69]. Histone deacetylases (HDACs), in particular NAD+-dependent class III HDAC proteins PfSIR2A and PfSIR2B and class II HDAC protein PfHDA2, play a role in regulating the repressive clusters containing silent var genes, as manipulated parasite lines lacking these proteins show loss of monoallelic var gene expression [65,70–72]. PfSET2 is a histone lysine methyltransferase (HKMT) that specifically marks var genes, and the disruption of PfSET2 results in the derepression of the silenced var gene cluster(s) [73,74]. Absence of PfHP1 in the parasite has also been shown to result in loss of monoallelic var gene expression as well as result in parasite growth arrest [75], which indicates that PfHP1 plays an essential role in maintaining repressive heterochromatin.
Figure 2.
Epigenetic Regulation of Specific Genes and Gene Families in Plasmodium falciparum. (A) The family of var genes is controlled by clustering of silent var genes at the nuclear periphery and the deposition of repressive H3K9me3 marks, which recruits PfHP1 and results in the formation of heterochromatin. The single active var gene is isolated from all other var genes, marked by H3K4me3 and H3K9ac, and localized in a euchromatic environment. LncRNAs transcribed from a bidirectional promoter in the var introns also contribute to regulation of var gene expression. (B) Several families of invasion genes are epigenetically regulated through repressive and active histone marks that recruit heterochromatin marker PfHP1 and gene activator PfBDP1, respectively. (C) During the IDC, gametocyte-specific TF pfap2-g localizes to the nuclear periphery and is silenced by repressive histone marks, including H3K9me3 and PfHP1. Abbreviations: IDC, intraerythrocytic developmental cycle; HDA2, histone deacetylase 2; H3K9me3, trimethylation of histone H3 at lysine 9; H3K9ac, acetylation of histone H3 at lysine 9; LncRNAs, long noncoding RNAs; PfBDP1, bromodomain protein; PfHP1, heterochromatin protein 1; SET2, histone lysine methyltransferase that specifically marks var genes; SIR2A, silent information regulator 2A; SIR2B, silent information regulator 2B; TF, transcription factor; pfap2-g, gametocyte-specific transcription factor.
The active var gene, transcribed at the ring stage, is distinguished by the presence of H3K4me3 and H3K9ac marks and resides in a region of the nucleus away from the repressive heterochromatin cluster(s) [65,70,76]. At the later trophozoite and schizont stages, the active var gene is controlled by the HKMT PfSET10, which is suggested to play a role in maintaining epigenetic memory of var gene expression [77]. Collectively, these results highlight the relationship between proper chromatin assembly and regulation of antigenic variation in the parasite.
Long Noncoding RNAs
Another mechanism regulating the monoallelic expression of the var gene family is the transcription of lncRNAs. Two lncRNAs transcribed from a bidirectional promoter within the var intron have been identified [78,79]. Both of these lncRNAs are incorporated into chromatin after being capped but not polyadenylated. Transcription of the sense lncRNA may play a role in positing the repressive histone mark H3K36me3 at the var gene loci, thereby functioning as a silencer of var gene expression, while the antisense lncRNA is proposed to be required for expression of the single active var gene [79].
Recently, a novel family of 22 lncRNAs transcribed from the telomere-associated repetitive elements (TAREs) was identified [80–82]. The exact role of these lncRNAs is yet to be determined. However, these lncRNA TARE loci are enriched with ApiAP2 transcription factor PfSIP2 binding sites. PfSIP2 has been implicated in heterochromatin formation around subtelomeric var gene regions [58]. Therefore, lncRNA-TAREs may, directly or indirectly, help regulate var gene expression. The TARE-lncRNAs show functional similarities to the eukaryotic family of noncoding RNAs involved in telomere and heterochromatin maintenance [83], which further validates the role for lncRNA-TAREs regulating heterochromatin and repressive centers in the parasite genome.
In vivo Regulation of Virulence Gene Expression
While in vivo studies of var gene regulation are challenging, results from human infections suggest that infection-induced stress responses in the host, such as fever and changes in bloodstream metabolites, can modify expression of PfEMP1 via changes in histone modifications [84], confirming the importance of this epigenetic mechanism for var gene regulation. Another in vivo study using patient samples found that not only do the parasites switch between different PfEMP1 variants, but they also vary the expression level of the PfEMP1 variants, most likely in response to host immune responses [85]. With these studies it is becoming clearer that the selection pressures in vitro are different from those in vivo, and epigenetic regulation of the clonally variant gene families may contribute to the differences observed in vivo and in vitro.
P. falciparum Invasion Genes
Invasion of a new erythrocyte by the malaria parasite involves binding of parasite ligands to specific recognition surface receptors on the red blood cell [86]. Eba, rhoph1/clag, acbp, and PfRH are among some of the gene families involved in the invasion process, but are not essential for parasite survival. The genes in these families are thought to be partially regulated through epigenetic mechanisms and show differential expression patters in different parasite lines, as they can be in either active or inactive states [87] (Figure 2B). According to a more recent study exploring the parasite-specific bromodomain protein PfBDP1 using in vitro culture, invasion genes are regulated in a more 'classical' manner by transcription factors interacting with specific promoters [88]. In schizonts, an enrichment of PfBDP1 was observed at the transcription start sites of invasion genes. PfBDP1 was shown to positively regulate transcription of invasion genes by binding to acetylated histone H3. Additionally, conditional knockdown of PfBDP1 resulted in erythrocyte invasion defects and parasite growth inhibition, further confirming the essentiality of this bromodomain protein for the coordinated expression of invasion genes in P. falciparum.
Epigenetic Regulation of Gametocytogenesis
As the malaria parasites continue asexual replication, a small fraction of parasites will commit to sexual differentiation and form gametocytes with every replication cycle. It is believed that this commitment is made during the schizont stage; however, what prompts the asexual stage parasites to commit to sexual differentiation is not well understood. The AP2 transcription factor, pfap2-g, located on chromosome 12, is one of the master regulators of gametocyte differentiation [17,18]. In asexual parasites, the locus containing pfap2-g is localized to the nuclear periphery and silenced by H3K9me3 and PfHP1 (Figure 2C) [65,69]. In vitro studies show that downregulation of PfHDA2 activates pfap2-g and induces the formation of gametocytes [72]. Similarly, depletion of PfHP1 activates pfap2-g and increases the rate of gametocyte production [75]. However, these observations have not been confirmed in vivo.
Histones and Nucleosome Landscape of P. falciparum
Recently, high-sensitivity mass spectrometry experiments have identified a total of 232 different histone PTMs during the P. falciparum intraerythrocytic stages, including acetylations, methylations, phosphorylations, ubiquitylations, and sumoylations [89]. Many of these histone PTMs had never been detected in Plasmodium or in other organisms, and their exact function remains to be determined. It is, however, important to mention that a majority of the parasite genome carries a large proportion of activating histone marks (H3K9ac and H3K4me3) compared to silencing marks (H3K9me3 and H3K36me3). This contrasts with what has been identified in multicellular eukaryotes [90], but validates further the transcriptionally permissive euchromatic state of the parasite genome. In mammalian genomes, H3K9ac and H3K4me3 strictly localize to active promoters [91–95], while in P. falciparum these modifications not only mark promoters and 5′ coding regions of genes that are highly transcribed [96,97], but are also found in intergenic regions and ‘silenced’ promoters [61,96,98].
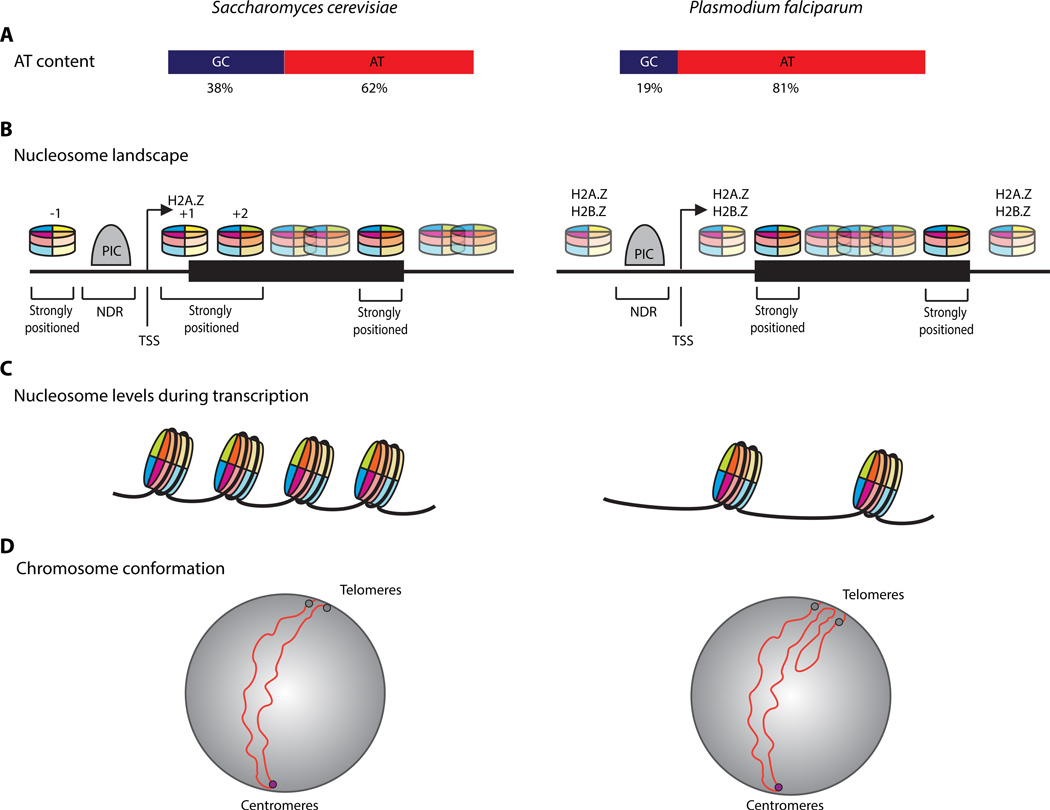
The nucleosome landscape of P. falciparum is similar to that of other eukaryotes in some aspects. First, the parasite's genome exhibits a depletion of nucleosome coverage in the promoter regions of genes [99–101], most likely to allow binding of transcription factors and other components of the transcription machinery. Second, lower levels of nucleosomes are observed in intergenic regions compared to coding regions [100–102]. Some studies dispute this finding and attribute the difference in nucleosome levels to sequencing biases introduced by the high AT content of the intergenic regions [103]. In these studies, the authors normalize the nucleosome coverage data using sonicated genomic DNA, but because sonication tends to degrade AT-rich sequences faster than GC-rich sequences, normalization using sonicated genomic DNA may over-correct for AT-rich sequencing biases. Alternative methodologies that enrich for nucleosome-depleted regions such as FAIRE-Seq [102] or ATAC-Seq [104] should be considered to better evaluate sequencing biases of nucleosome positioning in an AT-rich genome. Indeed, lower nucleosome levels are observed in intergenic regions of all other eukaryotes [105–108], including Tetrahymena thermophila, another organism with an AT-rich genome [109]. Third, genes with higher transcription levels exhibit a more open chromatin structure at their core promoter region than do silenced genes [99,100,103]. However, the nucleosome landscape of P. falciparum also displays some unique characteristics (Figure 3). First, the parasite's genome lacks a strongly positioned +1 nucleosome that marks the TSS [99,100] and instead, the most strongly positioned nucleosomes are found at the start and stop of the coding region. Second, the high AT content of the P. falciparum genome is likely to be the cause for the parasite-specific nucleosome landscape as AT-rich DNA is relatively inflexible and thus does not easily wrap around the nucleosome core. To solve this issue, the parasite has evolved its histone variants H2A.Z and H2B.Z that bind weakly but more effectively to AT-rich DNA [110,111]. H2A.Z and H2B.Z are found throughout intergenic regions of the parasite genome, instead of being present only at the +1 nucleosome in other eukaryotes.
Figure 3.
Differences in Chromatin and Genome Organization between Saccharomyces cerevisiae and Plasmodium falciparum. (A) The genome of P. falciparum is more AT-rich compared to the similarly sized budding yeast. (B) In yeast, the promoter region is flanked by strongly positioned nucleosomes (–1 and +1). These nucleosomes often contain the histone variant H2A.Z, which destabilizes the nucleosome and promotes gene activation. The nucleosome landscape at the end of the gene mirrors the landscape at the gene start. The nucleosome landscape around P. falciparum genes shows strongly positioned nucleosomes at the start and stop of the coding region, and histone variants H2A.Z and H2B.Z throughout the intergenic regions. This parasite-specific nucleosome landscape is most likely the result of the high AT-content of the parasite genome. As AT-rich DNA is relatively rigid, and does not easily wrap around a nucleosome core, the parasite may have partially solved this issue by evolving its histone variants H2A.Z and H2B.Z to become more efficient at binding AT-rich DNA. (C) In yeast, the process of transcription involves the local displacement of nucleosomes to allow the transcription machinery to progress along the gene. In the malaria parasite, nucleosome levels decrease dramatically during the trophozoite stage to facilitate massive transcription. This global nucleosome eviction may contribute to a general transcriptionally permissive state of the genome, allowing the efficient transcription of 70–80% of all P. falciparum genes at this stage of its life cycle. (D) S. cerevisiae chromosomes have a folded architecture anchored at the centromere, with clustering of the centromeres and telomeres on opposite sites of the nucleus. In P. falciparum, clusters of var genes that are located internally on chromosomes colocalize with subtelomeric var genes, resulting in additional looping structures in 5 out of 14 chromosomes. This contributes to a more complex genome organization than what is observed in yeast. Abbreviations: NDR, nucleosome-depleted region; PIC, preinitiation complex; TSS, transcription start site.
Another unique feature of the P. falciparum nucleosome landscape, that is also an area of debate, is that nucleosome levels change as the parasite develops through the asexual replication cycle. Other studies propose a more sequence-driven and transcription-independent nucleosome positioning in the parasite [103]. These controversies seem to be stemming from discrepancies in data normalization. When nucleosome occupancy profiles are normalized by input parasite content or by the number of nuclei, these opposing datasets display similar nucleosome dynamics throughout the parasite's life cycle. Additionally, changes in nucleosome occupancy during parasite development have also been confirmed by alternative approaches. Experiments, including western blots [112], mass spectrometry [89,100,113], MNase-Seq, FAIRE-Seq [102], and ChIP-seq [100], showed that histone levels are lower during the transcriptionally active trophozoite stage. This observation has prompted a model for gene regulation in the parasite genome where nucleosome eviction drives the massive transcriptional event observed at the trophozoite stage, which is followed by the schizont stage where the genome repacks in preparation for reinvasion. At the schizont stage, nucleosomes are reassembled and global histone levels are restored to the levels observed before the transcriptionally active trophozoite stage. At this later stage, regulation of transcription at the initiation level may be controlled by more classical mechanisms of regulation such as stage-specific transcription factors (AP2) and histone PTMs [88].
P. falciparum Nuclear Architecture
Much like in complex metazoans, the 3D genome structure of P. falciparum plays important roles in regulating gene expression. Initial observations of global chromatin arrangement within the parasite's nucleus were studied using immunofluorescence microscopy and fluorescence in situ hybridization (FISH) experiments [65,114,115]. Earlier FISH experiments revealed that var genes localize to two to five clusters around the parasite's nucleus [60,65]. More recent studies showed a single locus for the var gene-associated repressive cluster marked by H3K9me3 and PfHP1 [116], as well as H3K36me3, which is a mark of both active and silent var genes [74]. These observations were confirmed with the recent advancement of chromosome conformation capture techniques such as Hi-C that capture genome-wide intra- and interchromosomal interactions. However, because Hi-C captures the nuclear architecture at the population level, unlike FISH that is performed at the single-cell level, Hi-C data cannot be used to distinguish between multiple var gene clusters if the genes are randomly distributed from cell to cell. In addition, technical challenges, such as cell cycle timing as well as cross-linking strategies, may contribute to the discrepancies between these methodologies. Since 3C-based and imaging methods inherently measure different aspects of the 3D genome, there will be instances where the two datasets appear to be inconsistent [117]. Nevertheless, Hi-C and FISH experiments, together with chromatin immunoprecipitation followed by microarray or next-generation sequencing technologies (ChIP-on-Chip or ChIP-Seq), have contributed to the discovery of atypical chromatin features in the parasite's genome [59].
P. falciparum chromosomes are arranged into folded structures, which are attached at the centromere with the two chromosomal arms folding over each other in a parallel orientation [59]. The clustering of the centromeres and telomeres on opposite regions of the nucleus is comparable to the 3D genome structure observed in the similarly sized budding and fission yeast [118,119]. Unlike the yeast genome, P. falciparum has several additional complex structures mostly generated by var genes located internally on 5 out of 14 chromosomes [59]. These internal var genes colocalize with the subtelomeric var genes at the nuclear periphery by creating additional loops in the chromosomes.
Hi-C experiments [59] as well as advanced microscopy examination [120] throughout the IDC reveal that the nuclear organization undergoes several distinct changes during its developmental progression, most likely in order to accommodate the high level of transcriptional activity necessary during these stages of the parasite life cycle. Together with the expansion of the nucleus, which reaches the maximum size and volume at the trophozoite stage, the number of nuclear pores increases from 3 to 7 pores at the ring stage to 12–58 pores at the trophozoite stage [120]. Nuclear pores are located next to euchromatic areas, and the increased number of pores suggests a transcriptionally active trophozoite stage with the need to facilitate messenger RNA transport into the cytoplasm. As the parasite undergoes schizogony, the contents of the nucleus together with the pores are distributed among the resulting daughter nuclei. Increased nuclear pores at the trophozoite stage correlate with partial loss of chromosomal territories for more chromosome intermingling [59]. Additional observations from Hi-C experiments include similar expression profiles for genes that are located close to each other with colocalization of genes that are silenced during the IDC but expressed at other stages [59]. Taken together, these observations highlight the importance of understanding mechanisms regulating the dynamic nuclear organization and their role in controlling gene expression.
Using Chromatin Structure and Epigenetic Regulation as Drug Targets
As mentioned previously, the maintenance and regulation of the repressive heterochromatin environment within the parasite nucleus is essential for parasite survival. In particular, conditional deletions of PfHP1 or PfHDA2 have been shown to cause developmental arrest of blood-stage parasites [72,75]. Profound transcriptional changes have been observed for parasites treated with the drug apicidin, a potent inhibitor of HDACs [121]. Apicidin does so by causing hypermethylation of H3K9 and H4K8 residues, which leads to deregulation of the global transcriptional cascade. Disruption of histone acetylation and methylation levels, and in particular HKMTs, have also been shown to interfere with parasite growth and survival, although so far only a few small molecules potent enough to inhibit HKMT activity in P. falciparum have been identified [122,123]. Targeting HDAC and HKMT classes of enzymes as antimalarial therapies is a promising strategy. However, many eukaryotic histone-modifying enzymes share conserved catalytic domains and when targeted for antimalarial therapies could be toxic to the human host as well. Therefore, potential drug compounds should be studied extensively before they can be considered as novel antimalarial therapies.
Concluding Remarks and Future Perspectives
P. falciparum uses a combination of different epigenetic mechanisms to regulate its gene expression. However, our understanding of the parasite epigenome is far from complete (see Outstanding Questions). Although most chromatin modifications used by the parasite are also common to other eukaryotes, several features of chromatin regulation are unique to P. falciparum. Exploring the underlying regulatory mechanisms of how the repressive cluster(s) are established and maintained could lead to identification of specific proteins important for chromatin regulation in the malaria parasite. At the epigenetic level, as outlined above, the Plasmodium genome architecture points towards a binary structure, with the majority of the genome existing as transcriptionally permissive euchromatin and a small subset of genes present in a transcriptionally silent heterochromatin state. This heterochromatin cluster is localized at the periphery of the nucleus and is characterized by high levels of H3K9me3 and H3K36me3 histone marks, PfHP1 and high nucleosome density. The euchromatin environment harbors active genes, including the single active var gene at the nuclear periphery, and is characterized by high levels of H3K4me3 and H3K9ac histone modifications. During the asexual cycle, the parasite's nucleus and chromatin undergo drastic remodeling to accommodate the high transcriptional activity at the trophozoite stage. Chromatin structure remains relatively compact during the ring and schizont stages, but opens substantially during the trophozoite stage. This open-and-close chromatin structure is also reflected at the nucleosome landscape and global histone levels. As the nucleus expands in size, which can also be visualized using immunofluorescence and Giemsa staining, the number of nuclear pores increases greatly and distribute around the nucleus. As the parasite transitions from the trophozoite stage to the schizont stage, the changes in nuclear architecture are reversed by reassembling nucleosomes, increasing global histone levels and compacting the genome. In the transition from trophozoite to schizont stage, the DNA is replicated, and the nucleus is divided into multiple daughter nuclei, each with a small number of the nuclear pores previously present in the nucleus. Collectively, these observations suggest that the majority of the parasite genome is regulated via genome-wide changes in chromatin structure, while a small subset of genes are regulated by classical transcriptional regulation mechanisms, such as changes in local chromatin structure and specific transcription factors. Exploration of regulatory mechanisms that regulate large chromatin rearrangements throughout the parasite's life cycle could enable the discovery of molecules that can target parasite development with high specificity.
Outstanding Questions.
Does the nucleus of P. falciparum harbor one or more repressive heterochromatin clusters at the nuclear periphery?
Are the epigenetic control mechanisms observed throughout the asexual life cycle similar in other stages?
Through what mechanisms do stage-specific transcription factors control the transition between specific developmental stages?
Which chromatin-associated proteins are vital for maintaining the epigenetics features and the 3D structure of the parasite nucleus and can these key proteins be targeted for development of antimalarial drug therapies?
What is the role of epigenetic control in gene regulation in other Plasmodium species?
Trends.
Chromatin organization within the parasite nucleus plays a role in gene regulation.
Parasite-specific genes involved in pathogenesis, immune evasion, and host cell invasion are regulated at the epigenetic level.
Histone variants and the nucleosome landscape of the parasite genome are associated with gene expression.
Most of the parasite genome is maintained as euchromatin, while only a small subset of genes are maintained in heterochromatin clusters.
Mediators of epigenetic control and nuclear remodeling could be promising targets for antimalarial drugs.
Acknowledgments
This work was financially supported by the National Institutes of Health (grants R01 AI85077-01A1 and R01 AI06775-01 to KGLR), the National Science Foundation (grant IIS-1302134 to KGLR), the University of California, Riverside (USDA-NIFA-Hatch-225935 to KGLR), and The University of Texas Health Science Center at San Antonio (EMB). The funding bodies had no role in the design of the study, in collection, analysis, and interpretation of data, or in writing the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.WHO. World Malaria Report, 2015. 2015 http://www.who.int/malaria/publications/world-malaria-report-2015/report/en/
- 2.Rosenberg R, et al. An estimation of the number of malaria sporozoites ejected by a feeding mosquito. Trans. Roy. Soc. Trop. Med. Hyg. 1990:209–212. doi: 10.1016/0035-9203(90)90258-g. [DOI] [PubMed] [Google Scholar]
- 3.Yuda M, Ishino T. Liver invasion by malarial parasites – how do malarial parasites break through the host barrier? Cell. Microbiol. 2004;6:1119–1125. doi: 10.1111/j.1462-5822.2004.00474.x. [DOI] [PubMed] [Google Scholar]
- 4.Gardner MJ, et al. Genome sequence of the human malaria parasite Plasmodium falciparum . Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Roch KG, et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- 6.Bozdech Z, et al. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum . PLoS Biol. 2003;1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bunnik EM, et al. Polysome profiling reveals translational control of gene expression in the human malaria parasite Plasmodium falciparum . Genome Biol. 2013;14:R128. doi: 10.1186/gb-2013-14-11-r128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Otto TD, et al. New insights into the blood-stage transcriptome of f Plasmodium falciparum using RNA-Seq. Mol. Microbiol. 2010;76:12–24. doi: 10.1111/j.1365-2958.2009.07026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopez-Barragan MJ, et al. Directional gene expression and antisense transcripts in sexual and asexual stages of Plasmodium falciparum . BMC Genom. 2011;12:587. doi: 10.1186/1471-2164-12-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rovira-Graells N, et al. Transcriptional variation in the malaria parasite Plasmodium falciparum . Genome Res. 2012;22:925–938. doi: 10.1101/gr.129692.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balaji S, et al. Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Res. 2005;33:3994–4006. doi: 10.1093/nar/gki709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coulson RM, et al. Comparative genomics of transcriptional control in the human malaria parasite Plasmodium falciparum . Genome Res. 2004;14:1548–1554. doi: 10.1101/gr.2218604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young JA, et al. In silico discovery of transcription regulatory elements in Plasmodium falciparum . BMC Genom. 2008;9:70. doi: 10.1186/1471-2164-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell TL, et al. Identification and genome-wide prediction of DNA binding specificities for the ApiAP2 family of regulators from the malaria parasite. PLoS Pathog. 2010;6:e1001165. doi: 10.1371/journal.ppat.1001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwanaga S, et al. Identification of an AP2-family protein that is critical for malaria liver stage development. PLoS One. 2012;7:e47557. doi: 10.1371/journal.pone.0047557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kafsack BF, et al. A transcriptional switch underlies commitment to sexual development in malaria parasites. Nature. 2014;507:248–252. doi: 10.1038/nature12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sinha A, et al. A cascade of DNA-binding proteins for sexual commitment and development in Plasmodium . Nature. 2014;507:253–257. doi: 10.1038/nature12970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuda M, et al. Transcription factor AP2-Sp and its target genes in malarial sporozoites. Mol. Microbiol. 2010;75:854–863. doi: 10.1111/j.1365-2958.2009.07005.x. [DOI] [PubMed] [Google Scholar]
- 20.Yuda M, et al. Identification of a transcription factor in the mosquito-invasive stage of malaria parasites. Mol. Microbiol. 2009;71:1402–1414. doi: 10.1111/j.1365-2958.2009.06609.x. [DOI] [PubMed] [Google Scholar]
- 21.Kirchner S, et al. Recent advances in malaria genomics and epigenomics. Genome Med. 2016;8:92. doi: 10.1186/s13073-016-0343-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balu B, et al. CCR4-associated factor 1 coordinates the expression of Plasmodium falciparum egress and invasion proteins. Eukaryot. Cell. 2011;10:1257–1263. doi: 10.1128/EC.05099-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bunnik EM, et al. The mRNA-bound proteome of the human malaria parasite Plasmodium falciparum . Genome Biol. 2016;17:147. doi: 10.1186/s13059-016-1014-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eshar S, et al. PfSR1 controls alternative splicing and steady-state RNA levels in Plasmodium falciparum through preferential recognition of specific RNA motifs. Mol. Microbiol. 2015;96:1283–1297. doi: 10.1111/mmi.13007. [DOI] [PubMed] [Google Scholar]
- 25.Vembar SS, et al. The PfAlba1 RNA-binding protein is an important regulator of translational timing in Plasmodium falciparum blood stages. Genome Biol. 2015;16:212. doi: 10.1186/s13059-015-0771-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caro F, et al. Genome-wide regulatory dynamics of translation in the Plasmodium falciparum asexual blood stages. eLife. 2014;3:e04106. doi: 10.7554/eLife.04106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foth BJ, et al. Quantitative protein expression profiling reveals extensive post-transcriptional regulation and post-translational modifications in schizont-stage malaria parasites. Genome Biol. 2008;9:R177. doi: 10.1186/gb-2008-9-12-r177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang C, Pugh BF. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol. 2009;10:R109. doi: 10.1186/gb-2009-10-10-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tolstorukov MY, et al. Comparative analysis of H2A.Z nucleosome organization in the human and yeast genomes. Genome Res. 2009;19:967–977. doi: 10.1101/gr.084830.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raisner RM, et al. Histone variant H2A.Z marks the 5' ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guillemette B, et al. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karmodiya K, et al. H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genom. 2012;13:424. doi: 10.1186/1471-2164-13-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 34.Ragoczy T, et al. The locus control region is required for association of the murine beta-globin locus with engaged transcription factories during erythroid maturation. Genes Dev. 2006;20:1447–1457. doi: 10.1101/gad.1419506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tolhuis B, et al. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 36.Carter D, et al. Long-range chromatin regulatory interactions in vivo. Nature Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- 37.Deng W, et al. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell. 2014;158:849–860. doi: 10.1016/j.cell.2014.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palstra RJ, et al. The beta-globin nuclear compartment in development and erythroid differentiation. Nature Genet. 2003;35:190–194. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- 39.Murrell A, et al. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nature Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 40.Dean A. In the loop: long range chromatin interactions and gene regulation. Brief Funct. Genom. 2011;10:3–10. doi: 10.1093/bfgp/elq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Wit E, de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 2012;26:11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nora EP, et al. Segmental folding of chromosomes: a basis for structural and regulatory chromosomal neighborhoods? Bioessays. 2013;35:818–828. doi: 10.1002/bies.201300040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hou C, et al. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol. Cell. 2012;48:471–484. doi: 10.1016/j.molcel.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li L, et al. Widespread rearrangement of 3D chromatin organization underlies polycomb-mediated stress-induced silencing. Mol. Cell. 2015;58:216–231. doi: 10.1016/j.molcel.2015.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bolzer A, et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 2005;3:e157. doi: 10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boyle S, et al. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum. Mol. Genet. 2001;10:211–219. doi: 10.1093/hmg/10.3.211. [DOI] [PubMed] [Google Scholar]
- 48.Cremer T, et al. Chromosome territories – a functional nuclear landscape. Curr. Opin. Cell Biol. 2006;18:307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 49.Croft JA, et al. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Federico C, et al. Gene-rich and gene-poor chromosomal regions have different locations in the interphase nuclei of cold-blooded vertebrates. Chromosoma. 2006;115:123–128. doi: 10.1007/s00412-005-0039-z. [DOI] [PubMed] [Google Scholar]
- 51.Foster HA, Bridger JM. The genome and the nucleus: a marriage made by evolution. Genome organisation and nuclear architecture. Chromosoma. 2005;114:212–229. doi: 10.1007/s00412-005-0016-6. [DOI] [PubMed] [Google Scholar]
- 52.Zink D, et al. Structure and dynamics of human interphase chromosome territories in vivo. Hum. Genet. 1998;102:241–251. doi: 10.1007/s004390050686. [DOI] [PubMed] [Google Scholar]
- 53.Lanctot C, et al. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat. Rev. Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- 54.Bohmdorfer G, Wierzbicki AT. Control of chromatin structure by long noncoding RNA. Trends Cell Biol. 2015;25:623–632. doi: 10.1016/j.tcb.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maclary E, et al. Long nonoding RNAs in the X-inactivation center. Chromosome Res. 2013;21:601–614. doi: 10.1007/s10577-013-9396-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Callebaut I, et al. Prediction of the general transcription factors associated with RNA polymerase II in Plasmodium falciparum: conserved features and differences relative to other eukaryotes. BMC Genom. 2005;6:100. doi: 10.1186/1471-2164-6-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gangloff YG, et al. The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem. Sci. 2001;26:250–257. [Google Scholar]
- 58.Flueck C, et al. A major role for the Plasmodium falciparum ApiAP2 protein PfSIP2 in chromosome end biology. PLoS Pathog. 2010;6:e1000784. doi: 10.1371/journal.ppat.1000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ay F, et al. Three-dimensional modeling of the P. falciparum genome during the erythrocytic cycle reveals a strong connection between genome architecture and gene expression. Genome Res. 2014;24:974–988. doi: 10.1101/gr.169417.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Freitas-Junior LH, et al. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum . Nature. 2000;407:1018–1022. doi: 10.1038/35039531. [DOI] [PubMed] [Google Scholar]
- 61.Salcedo-Amaya AM, et al. Dynamic histone H3 epigenome marking during the intraerythrocytic cycle of Plasmodium falciparum . Proc. Natl. Acad. SciU.SA. 2009;106:9655–9660. doi: 10.1073/pnas.0902515106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Crowley VM, et al. Heterochromatin formation in bistable chromatin domains controls the epigenetic repression of clonally variant Plasmodium falciparum genes linked to erythrocyte invasion. Mol. Microbiol. 2011;80:391–406. doi: 10.1111/j.1365-2958.2011.07574.x. [DOI] [PubMed] [Google Scholar]
- 63.Freitas-Junior LH, et al. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell. 2005;121:25–36. doi: 10.1016/j.cell.2005.01.037. [DOI] [PubMed] [Google Scholar]
- 64.Chookajorn T, et al. Epigenetic memory at malaria virulence genes. Proc. Natl. Acad. SciU.SA. 2007;104:899–902. doi: 10.1073/pnas.0609084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lopez-Rubio JJ, et al. Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell Host Microbe. 2009;5:179–190. doi: 10.1016/j.chom.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 66.Miller LH, et al. Malaria pathogenesis. Science. 1994;264:1878–1883. doi: 10.1126/science.8009217. [DOI] [PubMed] [Google Scholar]
- 67.Scherf A, et al. Antigenic variation in Plasmodium falciparum . Annu. Rev. Microbiol. 2008;62:445–470. doi: 10.1146/annurev.micro.61.080706.093134. [DOI] [PubMed] [Google Scholar]
- 68.Perez-Toledo K, et al. Plasmodium falciparum heterochromatin protein 1 binds to tri-methylated histone 3 lysine 9 and is linked to mutually exclusive expression of var genes. Nucleic Acids Res. 2009;37:2596–2606. doi: 10.1093/nar/gkp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Flueck C, et al. Plasmodium falciparum heterochromatin protein 1 marks genomic loci linked to phenotypic variation of exported virulence factors. PLoS Pathog. 2009;5:e1000569. doi: 10.1371/journal.ppat.1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Duraisingh MT, et al. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum . Cell. 2005;121:13–24. doi: 10.1016/j.cell.2005.01.036. [DOI] [PubMed] [Google Scholar]
- 71.Tonkin CJ, et al. Sir2 paralogues cooperate to regulate virulence genes and antigenic variation in Plasmodium falciparum . PLoS Biol. 2009;7:e84. doi: 10.1371/journal.pbio.1000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Coleman BI, et al. A Plasmodium falciparum histone deacetylase regulates antigenic variation and gametocyte conversion. Cell Host Microbe. 2014;16:177–186. doi: 10.1016/j.chom.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jiang L, et al. PfSETvs methylation of histone H3K36 represses virulence genes in Plasmodium falciparum . Nature. 2013;499:223–227. doi: 10.1038/nature12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ukaegbu UE, et al. Recruitment of PfSET2 by RNA polymerase II to variant antigen encoding loci contributes to antigenic variation in P. falciparum . PLoS Pathog. 2014;10:e1003854. doi: 10.1371/journal.ppat.1003854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brancucci NM, et al. Heterochromatin protein 1 secures survival and transmission of malaria parasites. Cell Host Microbe. 2014;16:165–176. doi: 10.1016/j.chom.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 76.Lopez-Rubio JJ, et al. 5' flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites. Mol. Microbiol. 2007;66:1296–1305. doi: 10.1111/j.1365-2958.2007.06009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Volz JC, et al. PfSET10, a Plasmodium falciparum methyltransferase, maintains the active var gene in a poised state during parasite division. Cell Host Microbe. 2012;11:7–18. doi: 10.1016/j.chom.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 78.Epp C, et al. Chromatin associated sense and antisense noncoding RNAs are transcribed from the var gene family of virulence genes of the malaria parasite Plasmodium falciparum . RNA. 2009;15:116–127. doi: 10.1261/rna.1080109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Amit-Avraham I, et al. Antisense long noncoding RNAs regulate var gene activation in the malaria parasite Plasmodium falciparum . Proc. Natl. Acad. SciU.SA. 2015;112:E982–E991. doi: 10.1073/pnas.1420855112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Broadbent KM, et al. A global transcriptional analysis of Plasmodium falciparum malaria reveals a novel family of telomere-associated lncRNAs. Genome Biol. 2011;12:R56. doi: 10.1186/gb-2011-12-6-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Raabe CA, et al. A global view of the nonprotein-coding transcriptome in Plasmodium falciparum . Nucleic Acids Res. 2010;38:608–617. doi: 10.1093/nar/gkp895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sierra-Miranda M, et al. Two long non-coding RNAs generated from subtelomeric regions accumulate in a novel perinuclear compartment in Plasmodium falciparum . Mol. Biochem Parasitol. 2012;185:36–47. doi: 10.1016/j.molbiopara.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Luke B, Lingner J. TERRA: telomeric repeat-containing RNA. EMBO J. 2009;28:2503–2510. doi: 10.1038/emboj.2009.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Merrick CJ, et al. Epigenetic dysregulation of virulence gene expression in severe Plasmodium falciparum malaria. J. Infect. Dis. 2012;205:1593–1600. doi: 10.1093/infdis/jis239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Abdi AI, et al. Global selection of Plasmodium falciparum virulence antigen expression by host antibodies. Sci. Rep. 2016;6:19882. doi: 10.1038/srep19882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cowman AF, Crabb BS. Invasion of red blood cells by malaria parasites. Cell. 2006;124:755–766. doi: 10.1016/j.cell.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 87.Cortes A, et al. Epigenetic silencing of Plasmodium falciparum genes linked to erythrocyte invasion. PLoS Pathog. 2007;3:e107. doi: 10.1371/journal.ppat.0030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Josling GA, et al. A Plasmodium falciparum bromodomain protein regulates invasion gene expression. Cell Host Microbe. 2015;17:741–751. doi: 10.1016/j.chom.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 89.Saraf A, et al. Dynamic and combinatorial landscape of histone modifications during the intraerythrocytic developmental cycle of the malaria parasite. J. Proteome Res. 2016;15:2787–2801. doi: 10.1021/acs.jproteome.6b00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Garcia BA, et al. Organismal differences in post-translational modifications in histones H3 and H4. J. Biol. Chem. 2007;282:7641–7655. doi: 10.1074/jbc.M607900200. [DOI] [PubMed] [Google Scholar]
- 91.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 92.Bernstein BE, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 93.Kim TH, et al. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nishida H, et al. Histone H3 acetylated at lysine 9 in promoter is associated with low nucleosome density in the vicinity of transcription start site in human cell. Chromosome Res. 2006;14:203–211. doi: 10.1007/s10577-006-1036-7. [DOI] [PubMed] [Google Scholar]
- 95.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nature Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bartfai R, et al. H2A.Z demarcates intergenic regions of the plasmodium falciparum epigenome that are dynamically marked by H3K9ac and H3K4me3. PLoS Pathog. 2010;6:e1001223. doi: 10.1371/journal.ppat.1001223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cui L, et al. PfGCN5-mediated histone H3 acetylation plays a key role in gene expression in Plasmodium falciparum . Eukaryot. Cell. 2007;6:1219–1227. doi: 10.1128/EC.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Trelle MB, et al. Global histone analysis by mass spectrometry reveals a high content of acetylated lysine residues in the malaria parasite Plasmodium falciparum . J. Proteome Res. 2009;8:3439–3450. doi: 10.1021/pr9000898. [DOI] [PubMed] [Google Scholar]
- 99.Ponts N, et al. Nucleosome occupancy at transcription start sites in the human malaria parasite: a hard-wired evolution of virulence? Infect. Genet. Evol. 2011;11:716–724. doi: 10.1016/j.meegid.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bunnik EM, et al. DNA-encoded nucleosome occupancy is associated with transcription levels in the human malaria parasite Plasmodium falciparum . BMC Genom. 2014;15:347. doi: 10.1186/1471-2164-15-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Westenberger SJ, et al. Genome-wide nucleosome mapping of Plasmodium falciparum reveals histone-rich coding and histone-poor intergenic regions and chromatin remodeling of core and subtelomeric genes. BMC Genom. 2009;10:610. doi: 10.1186/1471-2164-10-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ponts N, et al. Nucleosome landscape and control of transcription in the human malaria parasite. Genome Res. 2010;20:228–238. doi: 10.1101/gr.101063.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kensche PR, et al. The nucleosome landscape of Plasmodium falciparum reveals chromatin architecture and dynamics of regulatory sequences. Nucleic Acids Res. 2016;44:2110–2124. doi: 10.1093/nar/gkv1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schep AN, et al. Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions. Genome Res. 2015;25:1757–1770. doi: 10.1101/gr.192294.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lee CK, et al. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nature Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- 106.Pokholok DK, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 107.Mavrich TN, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Valouev A, et al. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 2008;18:1051–1063. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Beh LY, et al. DNA-guided establishment of nucleosome patterns within coding regions of a eukaryotic genome. Genome Res. 2015;25:1727–1738. doi: 10.1101/gr.188516.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hoeijmakers WA, et al. H2A.Z/H2B.Z double-variant nucleosomes inhabit the AT-rich promoter regions of the Plasmodium falciparum genome. Mol. Microbiol. 2013;87:1061–1073. doi: 10.1111/mmi.12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Petter M, et al. H2A.Z and H2B.Z double-variant nucleosomes define intergenic regions and dynamically occupy var gene promoters in the malaria parasite Plasmodium falciparum . Mol. Microbiol. 2013;87:1167–1182. doi: 10.1111/mmi.12154. [DOI] [PubMed] [Google Scholar]
- 112.Le Roch KG, et al. Global analysis of transcript and protein levels across the Plasmodium falciparum life cycle. Genome Res. 2004;14:2308–2318. doi: 10.1101/gr.2523904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Oehring SC, et al. Organellar proteomics reveals hundreds of novel nuclear proteins in the malaria parasite Plasmodium falciparum . Genome Biol. 2012;13:R108. doi: 10.1186/gb-2012-13-11-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ralph SA, et al. Antigenic variation in Plasmodium falciparum is associated with movement of var loci between subnuclear locations. Proc. Natl. Acad. SciU.SA. 2005;102:5414–5419. doi: 10.1073/pnas.0408883102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dahan-Pasternak N, et al. PfSec13 is an unusual chromatin-associated nucleoporin of Plasmodium falciparum that is essential for parasite proliferation in human erythrocytes. J. Cell Sci. 2013;126:3055–3069. doi: 10.1242/jcs.122119. [DOI] [PubMed] [Google Scholar]
- 117.Dekker J. Mapping the 3D genome: Aiming for consilience. Nature Rev. Mol. Cell Biol. 2016;17:741–742. doi: 10.1038/nrm.2016.151. [DOI] [PubMed] [Google Scholar]
- 118.Duan Z, et al. A three-dimensional model of the yeast genome. Nature. 2010;465:363–367. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tanizawa H, et al. Mapping of long-range associations throughout the fission yeast genome reveals global genome organization linked to transcriptional regulation. Nucleic Acids Res. 2010;38:8164–8177. doi: 10.1093/nar/gkq955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Weiner A, et al. 3D nuclear architecture reveals coupled cell cycle dynamics of chromatin and nuclear pores in the malaria parasite Plasmodium falciparum . Cell. Microbiol. 2011;13:967–977. [Google Scholar]
- 121.Chaal BK, et al. Histone deacetylases play a major role in the transcriptional regulation of the Plasmodium falciparum life cycle. PLoS Pathog. 2010;6:e1000737. doi: 10.1371/journal.ppat.1000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Malmquist NA, et al. Small-molecule histone methyltransferase inhibitors display rapid antimalarial activity against all blood stage forms in Plasmodium falciparum . Proc. Natl. Acad. SciU.SA. 2012;109:16708–16713. doi: 10.1073/pnas.1205414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Malmquist NA, et al. Histone methyltransferase inhibitors are orally bioavailable, fast-acting molecules with activity against different species causing malaria in humans. Antimicrob. Agents Chemother. 2015;59:950–959. doi: 10.1128/AAC.04419-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dekker J, et al. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 125.Gondor A, et al. High-resolution circular chromosome conformation capture assay. Nature Protocols. 2008;3:303–313. doi: 10.1038/nprot.2007.540. [DOI] [PubMed] [Google Scholar]
- 126.Dostie J, et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]