Abstract
Skeletal muscle is a heterogeneous tissue that represents between 30 and 38% of the human body mass and has important functions in the organism, such as maintaining posture, locomotor impulse, or pulmonary ventilation. The genesis of skeletal muscle during embryonic development is a process controlled by an elaborate regulatory network combining the interplay of extrinsic and intrinsic regulatory mechanisms that transform myogenic precursor cells into functional muscle fibers through a finely tuned differentiation program. However, the capacity of generating muscle still remains once these fibers have matured. Adult myogenesis resembles many of the embryonic morphogenetic episodes and depends on the activation of satellite cells that have the potential to differentiate into new muscle fibers. Pitx2 is a member of the bicoid family of homeodomain transcription factors that play an important role in morphogenesis. In the last decade, Pitx2 has emerged as a key element involved in the fine-tuning mechanism that regulates skeletal-muscle development as well as the differentiation and cell fate of satellite cells in adult muscle. Here we present an integrative view of all aspects of embryonic and adult myogenesis in which Pitx2 is involved, from embryonic development to satellite-cell proliferation, fate specification, and differentiation. Those new Pitx2 functions on satellite-cell biology might open new perspectives to develop therapeutic strategies for muscular disorders.
Keywords: Pitx2, myogenic precursor cells, embryonic myogenesis, adult myogenesis, satellite cell and regeneration
Introduction
Skeletal muscle is a heterogeneous tissue that represents between 30 and 38% of the human body mass (Janssen et al., 2000). It is composed of individual muscle fibers, diversified in size, shape, and contractile protein content, to fulfill the different functional needs of the vertebrate body such as maintaining body posture, locomotor impulse, or pulmonary ventilation. The genesis of skeletal muscle during embryonic development and postnatal life is a process controlled by an extremely elaborate regulatory network that combines the interplay of extrinsic (e.g., morphogens, neurohormonal input, muscle damage, etc.) and intrinsic elements (gene regulatory elements). The intrinsic elements form hierarchical interactions between transcriptional regulators, regulatory RNAs, and chromatin-remodeling factors. In this sense, during embryogenesis, muscle progenitors are specified by the sequential expression of a network of transcription factors composed of PAX3 and PAX7, and the basic helix-loop-helix (bHLH) myogenic regulatory factors (MRFs) MYOD, MYF5, MYF6 (also called MFR4), and MYOG (Bentzinger et al., 2012; Moncaut et al., 2013). In addition, during adult life the skeletal muscle has the ability to resume developmental mechanisms that compensate for the physiological turnover and damage in order to maintain tissue homeostasis (Schmalbruch and Lewis, 2000; Pellettieri and Alvarado, 2007). This adult myogenesis depends on the activation of satellite cells (SCs), that have the potential to proliferate, differentiate, and generate new fibers, or repair existing ones (Chargé and Rudnicki, 2004). It has been well-established that SCs are closely related to progenitors of embryonic origin (Gros et al., 2005; Relaix et al., 2005; Schienda et al., 2006; Hutcheson et al., 2009; Lepper and Fan, 2010). Thus, many similarities have been discovered between prenatal myogenesis and regeneration in the mature skeletal musculature, such as common transcription factors and signaling molecules (Tajbakhsh, 2009).
During the last two decades the homeobox transcription factor Pitx2 has emerged as a key element in the fine-tuning mechanism that regulates skeletal-muscle development. Concurrently, several recent experimental pieces of evidence point to the role of Pitx2 in SC biology. Here, we present an integrative view of the role of Pitx2 in prenatal and adult myogenesis (from embryonic development to SC proliferation), fate specification, and differentiation. Finally we discuss the potential therapeutic use of Pitx2 in the future.
Prenatal and adult myogenesis
In vertebrates, skeletal-muscle development is a biphasic process. A primary (embryonic) myogenesis takes place to generate primary muscle fibers, between embryonic day (E) 9.5 and E14.5 in the mouse. This is followed during fetal stages by a secondary myogenesis which gives rise to the bulk of skeletal-muscle fibers present at birth (Kelly and Zacks, 1969; Biressi et al., 2007; Tajbakhsh, 2009; Deries and Thorsteinsdóttir, 2016). All skeletal-muscle cells have the same underlying functions, although their progenitors within the paraxial mesoderm are spread throughout the embryo at the onset of myogenesis. This bears emphasizing since the genetic networks that control myogenesis present differences depending on the location of those myogenic precursors in the embryo.
Embryonic myogenesis: the trunk and limb muscles
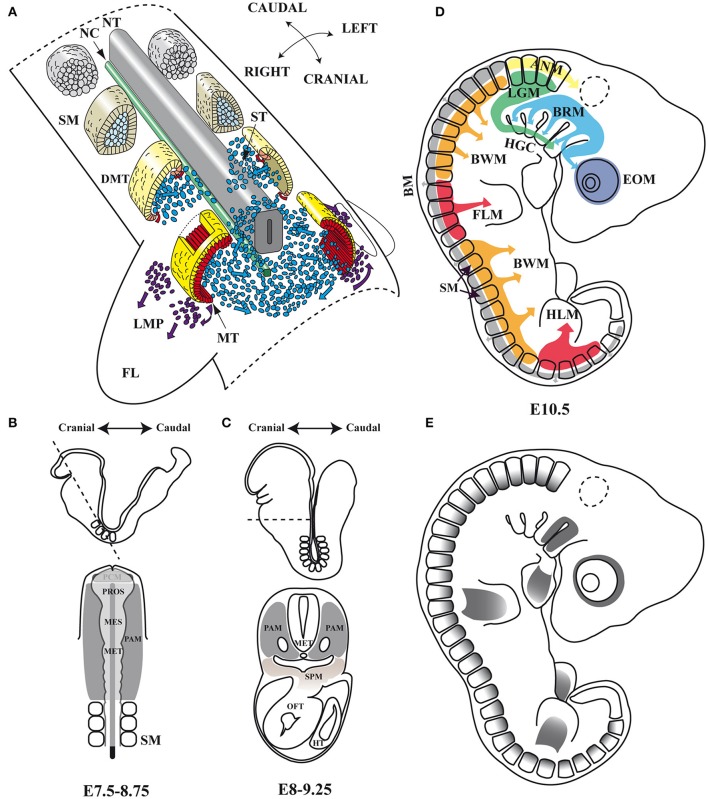
The muscles of the trunk and limbs derive from somites (Figure 1A), which are transient paraxial mesodermal structures that form pairwise on either side of the neural tube, following an anterior-posterior developmental gradient. The somite is initially a spherical unit of polarized epitheloid cells that soon after subdivides into two compartments, the ventral mesenchymal sclerotome and the dorsal epithelial dermomyotome. Shortly afterwards, myogenic precursor cells from the epaxial and hypaxial lips of the dermomyotome undergo an epithelial-mesenchymal transition (EMT) and accumulate underneath, where they differentiate and elongate to form the myocytes of the myotome, the first myogenic structure to develop in the body (Buckingham and Relaix, 2015; Deries and Thorsteinsdóttir, 2016). The epaxial region of the myotome gives rise to the deep back muscles, whereas the hypaxial myotome is the source of body wall muscles and most other trunk muscles (Buckingham and Relaix, 2015; Deries and Thorsteinsdóttir, 2016). In segments adjacent to the limb-region cells of the hypaxial dermomyotome undergo an EMT, leave the epithelial structure, and migrate toward the fore and hind limbs to form dorsal and ventral muscle masses in the limb-bud mesenchyme, where they begin to differentiate and express muscle-specific genes (Biressi et al., 2007; Deries and Thorsteinsdóttir, 2016).
Figure 1.
Embryonic myogenesis (A) Schematic representation of somite maturation. Somites mature following an anterior to posterior developmental gradient (Modified from Gray's Anatomy. The Anatomical Basis of Clinical Practice, 40th Edition Standring, 2008): myogenic precursor cells arise from the epaxial and hypaxial lips of the dermomyotome after archive epithelial-mesenchymal transition (EMT) and migrate toward the limbs to form dorsal and ventral muscle masses where they begin to differentiate. (B,C) Head frontal and transverse planes of a mouse embryo between stages of development E7.5–8.75 and E8–9.25 in mouse. At an open neural plate stage, head mesoderm in a frontal plane includes the prechordal mesoderm and the paraxial mesoderm. When the neural tube closes dorsally and the endoderm ventrally, the prechordal mesoderm is integrated within the remaining paraxial mesoderm, which is located anterior to the somites. Dashed line illustrates the cutting plane. (D) Origins of skeletal muscles: Myogenic precursors arise from different paraxial mesoderm compartments. (E) Pitx2 expression domains at the E10.5 stage of development in mouse. NT, neural tube; NC, notochord; SM, somites; DMT, dermomyotome; ST, sclerotome; MT, myotome; LMP, limb muscle precursors; FL, forelimb; PAM, head paraxial mesoderm; PCM, prechordal mesoderm; PROS, prosencephalon; MES, mesencephalon; MET, metencephalon; SPM, splanchnic mesoderm; OFT, outflow tract of heart; HT, heart tube; EOM, extra-ocular muscles; BRM, branchial muscles; LGM, laryngoglossal muscles; HGC, hypoglossal cord; ANM, axial neck muscles; BM, back muscles; BWM, body wall muscles; FLM, forelimbs muscles; HLM, hind limbs muscles.
Cell commitment in the somite is highly dependent on a number of transcription factors which act in a hierarchical molecular cascade to orchestrate the specification, determination, and differentiation of myogenic precursors. In the genetic hierarchy that regulates the onset of trunk myogenesis, Pax3 and Myf5 play a dominant role (Buckingham and Relaix, 2015). Pax3 is already transcribed in pre-somitic mesoderm adjacent to the first somite (Schubert et al., 2001) and then throughout the newly formed somites. As somites mature Pax3 expression becomes confined to the dermomyotome (Goulding et al., 1991) and persists in myogenic progenitor cells that delaminate and migrate from the somite to more distant sites of myogenesis such as the limb (Buckingham and Relaix, 2015). Myogenic cells that have activated the myogenic determination genes Myf5/Myf6 and MyoD downregulate Pax3 and delaminate from the edges of the dermomyotome (Buckingham and Relaix, 2015). The epaxial myotome then start to form. This depends on the early epaxial activation of Myf5, which is driven by Wnt and Shh signaling, without any Pax3 and/or Pax7 requirement (Borello et al., 2006; Buckingham and Relaix, 2015). These cells do not activate MyoD but rather Myog and differentiate (Kablar et al., 2003). On the other hand, the activation of Myf5 in the hypaxial somite as well as in the limb depends on PAX3 (Bajard et al., 2006; Buckingham and Relaix, 2015). At this stage MYF6 also acts as a myogenic determination factor (Kassar-Duchossoy et al., 2004). The Myod gene is activated after the onset of Myf5 expression in the rest of the dermomyotome and limbs (Hu et al., 2008). Finally, the transcription factor MYOG is required for the onset of the expression of terminal differentiation genes needed for the fusion of myocytes and the formation of myotubes (Bentzinger et al., 2012).
Embryonic myogenesis: the head muscles
Although, all skeletal muscle throughout the body originates within paraxial mesoderm, in the head, identifiable compartments such as the somites in the trunk are not evident histologically or by most molecular criteria. This unsegmented head mesoderm is remodeled at the early stages of embryonic development (Figures 1B,C). The unsegmented head mesoderm gives rise to all craniofacial skeletal muscles, which can be cataloged as four distinct populations: extra-ocular (EOMs), branchial, laryngoglossal, and axial neck muscles (Noden and Francis-West, 2006; Tzahor, 2015). EOMs are formed by cells from the cranial paraxial mesoderm that migrate through the first branchial arch (FBA) as well as from the prechordal mesoderm (Jacob et al., 1984; Evans and Noden, 2006; Tzahor, 2015; Figure 1D). Branchial arch muscles are formed mainly by migrating cells from the cranial paraxial mesoderm and the lateral splanchnic mesoderm (Harel et al., 2009; Sambasivan et al., 2009; Tzahor, 2015). Laryngoglossal muscles develop from migratory myoblasts arising from occipital somites that form a condensed mesenchymal band, the hypoglossal cord, which elongates and similarly brings myoblasts ventral to pharynx (Hammond, 1965; Hazelton, 1970; Tzahor, 2015). Finally, in the transition zone between the head and the trunk are the axial neck muscles. They arise from medio-dorsal and latero-ventral domains of occipital and cervical somites (Noden, 1983; Couly et al., 1992; Matsuoka et al., 2005).
The genetic hierarchy governing primary myogenesis in the trunk does not appear to operate for head-muscle formation. Activation of the myogenic program in the head therefore depends on different upstream factors, responds differently to signaling pathways and also displays site-dependent regulation. Branchial-arch-derived muscles depend on Myf5/Myf6/Myod, whereas extra-ocular muscle formation is initiated by Myf5/Myf6 and in their absence cannot be restored by Myod (Tajbakhsh and Buckingham, 1999).
Fetal myogenesis
During fetal myogenesis, secondary fibers in trunk, limbs, and head are generated by the fusion of fetal myoblasts. Secondary fibers form initially at the site of innervation of the primary fiber and are surrounded by the same basal lamina as the primary fiber on which they lie (Duxson et al., 1989). The secondary myotubes remain attached for a short period to primary fibers and subsequently elongate and become independent fibers, which can be distinguished from primary fibers by their relative small size (Kelly and Zacks, 1969). Although, the genetic networks that rule this second stage of prenatal myogenesis is less understood, it is known that the MRFs MYF5, MYOD, and MYOG are also crucial, since in Myog−/− as well as Myf5−/−:MyoD−/− double-mutant secondary myogenesis is completely inhibited (Venuti et al., 1995; Kassar-Duchossoy et al., 2004).
Adult myogenesis
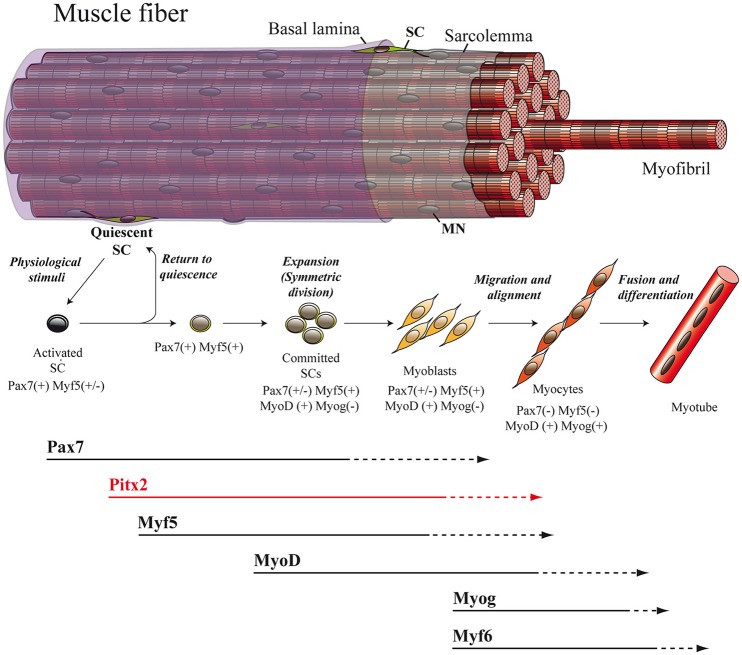
The regulatory inputs that orchestrate myogenesis during prenatal myogenesis are partially reactivated in adult muscle repair. In adulthood, the maintenance as well as the repair of muscle tissue are both directed mainly by SCs. These cells, originally identified via electron microscopy in 1961 by Alexander Mauro, are located underneath the basal lamina and adjacent to the plasma membrane of the skeletal-muscle myofiber (Mauro, 1961; Figure 2). In their quiescent state, SC express the transcription factor Pax7 and represent a genuine stem-cell population indispensable for skeletal-muscle repair (Lepper et al., 2011; Murphy et al., 2011; Sambasivan et al., 2011; Miersch et al., 2017; Stuelsatz et al., 2017). It has been established that SCs in adult muscle represent a lineage continuum of the embryonic myogenic progenitor cells. Thus, while SCs of the body and limbs arise from somites, in common with the muscle that they are associated with (Armand et al., 1983; Gros et al., 2005; Relaix et al., 2005; Schienda et al., 2006), the SCs located in head muscles also originate from the cranial mesoderm (Harel et al., 2009). Within a context of physiological stimuli (physical exercise or pathological conditions) SCs become activated, proliferate, differentiate and fuse to form multinucleated myofibers in order to undergo proper myogenesis (Lepper et al., 2011; Murphy et al., 2011; Sambasivan et al., 2011; Miersch et al., 2017; Stuelsatz et al., 2017; Figure 2). In this regard, numerous studies have revealed a striking similarity between adult and embryonic myogenesis, where the core regulatory network composed of the MRFs MYF5, MYOD, MYOG, and MYF6 is mainly required (Bentzinger et al., 2012; Segalés et al., 2016; Figure 2).
Figure 2.
Adult myogenesis. The overall myogenic differentiation pathway includes the activation of quiescent SCs, commitment to differentiation, proliferation, fusion to form myotubes and ultimately maturation into myofibers. SC, satellite cell; MN, myonucleus.
The Pitx2 gene
The Pitx gene family includes three vertebrate paralogues, Pitx1, Pitx2, and Pitx3, which have been cloned in multiple organisms (Gage et al., 1999b; Knopp et al., 2013). These three genes encode transcription factors that belong to the bicoid-related subclass of homeodomain proteins (Gage et al., 1999b) The members of this family share an almost identical protein sequence within their homeodomains, varying mainly in the N-terminal region (Gage et al., 1999b; Knopp et al., 2013). Mutations or misregulation of Pitx1, Pitx2, and Pitx3 result in developmental disorders in humans, such as Facioscapulohumeral Muscular Dystrophy (FSHD; Dixit et al., 2007), Axenfeld-Rieger syndrome (Semina et al., 1996), and Anterior Segment Mesenchymal Dysgenesis (ASMD; Semina et al., 1998), respectively. Muscle expression of these genes during development has been systematically studied. Thus, Pitx1 is highly expressed in developing hind-limb-bud mesenchyme and is shown to determine hind-limb identity in mice (Lanctôt et al., 1999; Szeto et al., 1999), chicks (Logan and Tabin, 1999), and fish (Shapiro et al., 2004). On the other hand, Pitx3 is widely expressed in all skeletal muscles of the head, trunk and limbs (Semina et al., 1998; L'honoré et al., 2007). Curiously, despite its apparent importance in muscle development, the investigation of Pitx3−/− mice indicates that Pitx3 on its own is not required for myogenesis (L'honoré et al., 2007). In this scenario Pitx2, the third Pitx family member is strongly upregulated and appears to fully compensate for the loss of Pitx3 during muscle formation (L'honoré et al., 2007). Pitx2 is also able to control the growth ability of hind-limb mesenchyme together with Pitx1 (Marcil et al., 2003), indicating the importance of Pitx2 in the control of skeletal myogenesis during development.
In mice, the Pitx2 (Pituitary homeobox 2 or Paired-like homeodomain transcription factor 2) gene is mapped on chromosome 3 (3G3; 3 57.84 cM) (Gage and Camper, 1997) and is transcribed into three distinct isoforms: Pitx2a, Pitx2b, and Pitx2c. Pitx2a and Pitx2b share the same promoter while Pitx2c uses an alternative one upstream of exon 4 (Schweickert et al., 2000). In human, PITX2 is mapped on chromosome 4 (4q25) and maintains a similar genetic structure, but presents a fourth isoform (Arakawa et al., 1998; Cox et al., 2002). This fourth isoform is generated by the PITX2C alternative promoter and differential splicing, being able to suppress the transcriptional activity of the other PITX2 isoforms (Cox et al., 2002). All Pitx2 isoforms share a K50 DNA-binding homeodomain which binds to the consensus sequence TAATCC (Amendt et al., 1998; Chaney et al., 2005), thus being able to induce a transcriptional activation of Prl (Amendt et al., 1998) or Anf (Ganga et al., 2003) promoters. The Pitx2 gene was isolated independently by several research groups and designated as Otlx2 (Muccielli et al., 1996), Rieg (Semina et al., 1996), Ptx2 (Gage and Camper, 1997), Brx1 (Kitamura et al., 1997), and Arp1 (Arakawa et al., 1998). Although, most of these works focused on the role of this gene in the development of brain structures, the authors reported the expression of Pitx2 in the mesenchyme of the eye, the first and second branchial arches, the fore and hind limbs as well as the dermomyotome at somite stages E8.5 and 10.5 in mouse, and its equivalent stages in chicken (Figure 1E). Soon afterwards, a role for Pitx2 was also described in left-right asymmetry, being proposed as the molecular transducer of embryonic left-right signaling during early developmental stages at the level of organs such as heart, gut, and/or stomach (Logan et al., 1998; Ryan et al., 1998; Yoshioka et al., 1998; Campione et al., 1999).
Pitx2 within the genetic hierarchies that control muscle development
Pitx2 function during embryonic myogenesis
Pitx2 in trunk and limb muscle development
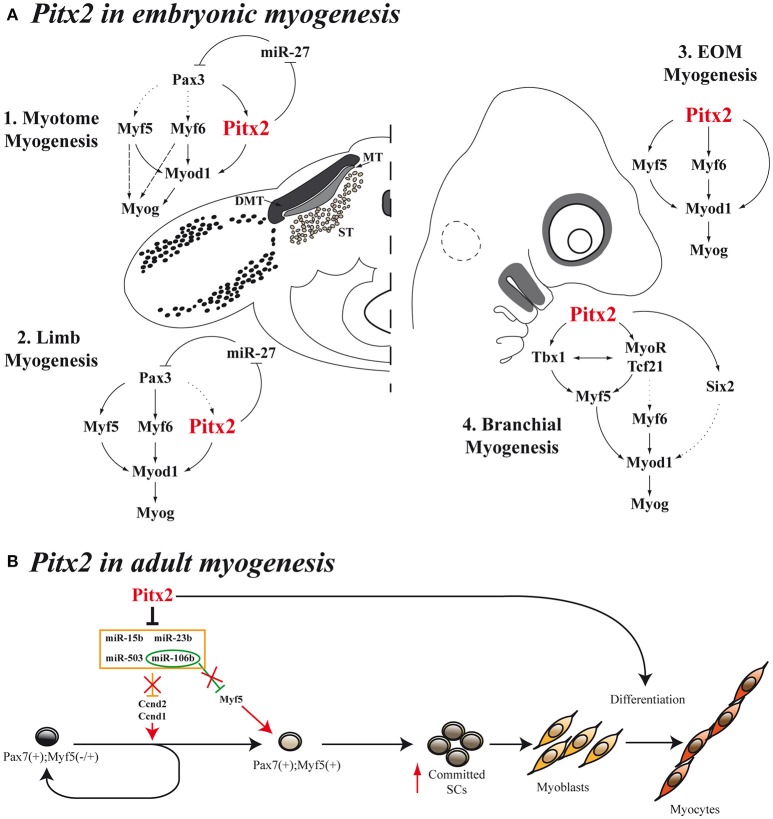
The first evidence involving Pitx2 in the molecular process controlling myogenesis was provided by Kitamura et al. (1999). These authors reported Pitx2 expression co-localizing in dermomyotomes with Pax3, a muscle specification marker playing a key role in delamination and migration of the somitic muscle progenitor cells to the limb buds (Goulding et al., 1994; Tajbakhsh et al., 1997). Later, Marcil et al. demonstrated the presence of PITX2 protein in the myoblasts of the limb bud, displaying an expression pattern similar to that of PAX3 and MYOD (Marcil et al., 2003). All these data suggested that PITX2 was involved in muscle patterning. A more detailed temporal and spatial analysis during initial muscle-cell-cluster formation, by using lacZ expression from a Pitx2 gene insertion, revealed the presence of a Pitx2-expressing cell cluster lateral to the dermomyotome (Shih et al., 2007b). This cluster first appeared at the forelimb level at E10.25. After E10.5, Pitx2(+/LacZ)-expressing cells were then also detected on sections of the limbs. Curiously, Pax3 and the muscle-regulatory factors (MRFs) stained only subsets of Pitx2+ cells within these areas, and virtually all Pitx2+ cells in these areas express at least one of these known myogenic markers (Shih et al., 2007b). These observations led the authors to conclude that Pitx2 marks the muscle lineage more completely than any of the known markers does. In agreement with the interpretation that muscle progenitors express Pitx2, L'Honoré et al. (2007) found extensive co-labeling of myotome- and dermomyotome-proliferating cells with PITX2, PAX3, and with PAX7. Notably, they also observed PAX3-positive cells that have completed migration at the proximal limb bud also express PITX2 while not all PITX2-positive cells expressed PAX3. All these data suggest that Pitx2 might be a player within the molecular pathways controlling muscle-progenitor fate.
Sometime afterwards, additional information regarding the hierarchical position occupied by Pitx2 within the genetic cascade that control somite-derived myogenesis was inferred by using Pitx2−/−, Myf5nlacZ/nlacZ and Pitx2−/−;Myf5nlacZ/nlacZ double-mutant mice (L'honoré et al., 2010). In this work, the authors showed that PITX2 protein directly regulates Myod expression through binding to its core enhancer in wild-type limbs. In agreement, the authors described a delayed myogenic differentiation and a Myod down regulation in Pitx2−/− limb buds and proposed that this phenotype appears to be due to the failure to activate the Myod core enhancer. However, although the inactivation of Myf5 and Myf6 in Myf5nlacZ/nlacZ mutant embryos (Myf6 is inactivated in cis in this mutant; Tajbakhsh et al., 1997) did not affect Myod expression in limb buds, this inactivation in a Pitx2−/− background (Pitx2−/−;Myf5nlacZ/nlacZ) induced a synergic effect that resulted not in a downregulation but in almost a complete loss of Myod expression compared with Pitx2−/−;Myf5nlacZ/+ embryos, where the presence of one active Myf5 allele prevented Myod loss in about 60% of myogenic precursors cells. These results imply that Myf5 and/or Myf6 cooperate with Pitx2 to control Myod expression during early limb-bud myogenesis (Figure 3A2). In contrast to limb-muscle cells, myotome expression of Myod was not delayed in Pitx2−/− embryos. Nevertheless, in Myf5nlacZ/nlacZ mutant embryos, Myod expression was delayed by ~2 days. Therefore, the onset of Myod expression in the myotome does not appear to depend on PITX2 but mostly on MYF5/MYF6. Nonetheless the inactivation of Myf5 and/or Myf6 in a Pitx2−/− background (Pitx2−/−;Myf5nlacZ/nlacZ) led to an almost complete loss of Myod expression in myotome, as happened in limbs (L'honoré et al., 2010). These results indicate that MYF5 and/or MYF6 also cooperate with PITX2 to control Myod expression during myotome development (Figure 3A1).
Figure 3.
Models for Pitx2 functions in myogenesis. (A) During embryonic stages, Pitx2 contribution is different depending on the initial muscle-cell clusters [myotome myogenesis (A1), limb myogenesis (A2), EOM myogenesis (A3), or branchial myogenesis (A4)]. First myocytes of the myotome differentiate through Myf5 and/or Myf6 directly to Myog without turning on MyoD. This is represented by dashed arrows. Dotted arrows represent direct molecular relationships that still remain elusive (B) Proposed model for Pitx2 in adult myogenesis promoting activation and commitment of SCs.
In addition, it should be stressed that the analysis of Pax3 mutant Splotch mice revealed a deficit of Pitx2 expression restricted to the myotome (L'honoré et al., 2010). This deficit is not observed in neighboring mesenchyme, indicating that Pitx2 is downstream of Pax3 during myotome myogenesis. This is also supported by transcriptome analyses of Pax3GFP/+ and Pax3GFP/PAX3−FKHR transgenic mice carried out by Lagha et al. (2010) since, in gain-of-function screens for PAX3 targets, they found an up-regulation of Pitx2 in somites but not in limb buds. Although, all these seminal works suggest that Pitx2 could be acting downstream of Pax3 and in parallel with Myf5, at least in the myotome, as noted above, not all PITX2-expressing cells were positive for PAX3, and limb expression of Pitx2 precedes Myf5 (L'honoré et al., 2010). Therefore, additional studies using conditional Pitx2 inactivation in specific myogenic cell populations would help to elucidate the function of Pitx2 in embryonic myogenesis.
Scientific evidence also relates Pitx2 to cell proliferation in myogenic cells and somite derivates. Notably, Pitx2 has been reported to be a target gene in the Wnt/Dvl2/beta-catenin pathway and operates in specific cell types to control proliferation by regulating expression of the growth-control genes Ccnd1, Ccnd2, and c-Myc (Kioussi et al., 2002; Baek et al., 2003). These authors established that the PITX2 N-terminal domain is required for its effects on proliferation in a myoblast cell line. We have previously demonstrated that Pitx2c is the main Pitx2-isoform expressed in Sol8 myoblasts and that overexpression of Pitx2c in Sol8 cells led to an increase in proliferative capacity and completely blocked terminal differentiation, mainly because high levels of Pax3 expression were maintained (Martínez-Fernández et al., 2006). Additional data in vivo have supported the role of Pitx2 in cell proliferation during myogenesis. In this sense, Abu-Elmagd et al. (2010) showed that Pitx2 loss of function in chicken embryos decreased the number of differentiated myocytes/myofibers in the somites, whereas Pitx2 overexpression increased myocyte/myofiber numbers, particularly in the epaxial region of the myotome. In agreement with Abu-Elmagd et al. and by using Pitx2c−/− mutant embryos, we have reported that Pitx2c plays a pivotal role in the control of the subtle equilibrium between proliferation and differentiation during trunk and limb myogenesis. This control is exercised by balancing Pax3+/Pax7+ myogenic population in vivo as well as regulating key myogenic transcription factors such as Pax3 through the repression of miR-27 (Lozano-Velasco et al., 2011; Figures 3A1,A2). This new function of Pitx2c mediated by miRNAs introduces a new level of complexity in the intricate regulatory network that governs myogenesis in the embryo.
Pitx2 during head-muscle development
As mentioned above, Pax3 controls the myogenic specification of muscle embryonic progenitors in trunk and limbs (Tajbakhsh et al., 1997). However, it has been proposed that, instead of Pax3, Pitx2 plays a major role as an upstream regulator of craniofacial myogenesis (Zacharias et al., 2011; Buckingham and Rigby, 2014). This is supported by the fact that EOM development is impaired in Pitx2 null mice (Gage et al., 1999a; Kitamura et al., 1999). However, in the early studies it was not evident whether this muscle dysgenesis in Pitx2 mutant mice resulted from an intrinsic defect in the developing myoblasts or was secondary to the loss of Pitx2 expression in the periocular mesenchyme. Other authors have subsequently suggested that this phenotype could be due to the Pitx2 effect on proliferation rate of myogenic precursors (Noden and Francis-West, 2006), in agreement with previously reported data (Kioussi et al., 2002; Martínez-Fernández et al., 2006). The hypothesis that Pitx2 plays a part in controlling cell proliferation in myogenic cells in this context is also supported by the fact that conditional inactivation of Pitx2 in neural-crest-derived cells does not affect the early differentiation of eye muscles (Evans and Gage, 2005), while conditional Pitx2 deletion in the mesoderm induces a down-regulation of Myf5, Myf6, Myod1, and Myog expression and, therefore, blocks the onset of myogenesis of EOM (Zacharias et al., 2011). In this regard, in 2009, Sambasivan et al. by analyzing double defective mutant mouse embryos Myf5(Myf6) (Myf5nlacZ/+, Myf5nlacZ/nlacZ) and Myf4−/− mutants, showed that Pitx2 cannot ensure survival and activation of Myod expression in EOM in the absence of both Myf5 and Myf6 (Sambasivan et al., 2009). Shortly afterwards, Zacharias et al. were able to inactivate the expression of Pitx2 in mesodermal EOM precursors by using a tamoxifen inducible UBC- CreERT2 promoter (Zacharias et al., 2011). This inactivation clearly showed that Pitx2 is required for EOM precursor specification and survival, acting as an anti-apoptotic factor in the pre-myogenic mesoderm and subsequently activating the myogenic program in these cells through direct binding to Myf5 and Myod promoters (Zacharias et al., 2011). Taken together, all these data clearly suggest that Pitx2 is an upstream regulator of Myf5, Myf6, and Myod in EOM embryonic myogenesis (Figure 3A3).
Pitx2 is also expressed in the myogenic precursors of the FBA. Tbx1 expression on FBA premyoblast is required for specification leading to Myf5 and Myod1 activation in those cells (Kelly et al., 2004). Notably, systemic Pitx2 mutants, whether Pitx2−/− (Dong et al., 2006) or Pitx2LacZ/LacZ (Shih et al., 2007a), display a down regulation of Tbx1 expression in this structure, although Pitx2 expression is unaffected in Tbx1 null mutants (Dong et al., 2006). These data, together with the fact that Pitx2 directly interacts with Tbx1 regulatory elements (Shih et al., 2007a) suggest that Pitx2 is an upstream activator of Tbx1 in FBA. A fuller analysis of both systemic Pitx2 mutants reveals that the inactivation of Pitx2 in FBA results in increased cell death in the mesodermal core and loss of early premyoblast specification markers such as Six2, Tcf21, and MyoR (Dong et al., 2006; Shih et al., 2007a). Although, the role that Six2 could play in the myogenesis of the FBA remains elusive, Tcf21 and MyoR are known to be upstream effectors of Myf5, Myod, and Myog in these initial muscle-cell clusters (Lu et al., 2002). Jointly, these results indicate that Pitx2 controls the expression of Myod1 and the onset of myogenesis in FBA through Tbx1, Tcf21, and MyoR (Figure 3A4).
Pitx2 during fetal myogenesis
Most of what is known about Pitx2 concerns early (embryonic) myogenesis. However, a new role for Pitx2 has recently been unraveled during fetal myogenesis. L'Honoré et al. by using Pitx2:Pitx3 double conditional mutants, have shown that Pitx2/3 control the expression of the antioxidant system through the regulation of Nrf1 and antioxidant enzymes during muscle differentiation (L'honoré et al., 2014a). Thus, Pitx2/3 depletion at the onset of differentiation induces an abnormal increase of reactive oxygen species (ROS) levels in differentiating myoblasts and leads to impaired myogenesis due to apoptosis of these cells. These results emphasize the role of Pitx2 controlling redox conditions during fetal myogenesis.
Pitx2 is emerging as a key transcription factor that modulates adult myogenesis
During adult life the maintenance and repair of skeletal-muscle tissue is directed by SCs. The regulation of SC function in adults requires the redeployment of many of the regulatory networks fundamental for developmental myogenesis. Although, several efforts have been made during the last few years to disentangle the role of Pitx2 in embryonic and fetal stages of myogenesis, studies linking Pitx2 to adult myogenesis have only recently emerged and are still controversial.
The first evidence regarding Pitx2 expression in SCs was reported by Ono et al. (2010). These authors showed that all Pitx2 isoforms are expressed in proliferating SC-derived myoblasts. They analyzed SCs with a different ontology, comparing those of the extensor digitorum longus (EDL) of the limb with SCs from the masseter of the head (MAS). They found that Pitx2b and Pitx2c levels were higher in cells from the EDL than from the MAS, with Pitx2c being the main Pitx2 isoform expressed in proliferating limb SCs (Ono et al., 2010). Based on these distinct gene-expression profiles, the authors suggest that, even after activation and entry into the cell cycle, SCs retain an identity consistent with their ontogeny underlying their distinct properties. Subsequent studies have pointed out that Pitx2a, Pitx2b, and Pitx2c were expressed at very low levels in proliferating SCs, but increased during the early stages of myogenic differentiation. Meanwhile the constitutive expression of any Pitx2 isoform suppressed SC proliferation, with the cells undergoing greater myogenic differentiation (Knopp et al., 2013). However, additional evidence underlying the functional relevance of Pitx2 on SC proliferation has been reported. For example, Herbet et al. demonstrated that Pitx2 is crucial in maintaining the phenotype of myogenic precursor cells in the extraocular muscles (EOM; Hebert et al., 2013). In this analysis, the authors found that the higher levels of Pitx2 expression in EOM in comparison with limb muscles were concomitant with longer proliferative state in EOM-derived SCs as compared with limb cells. In addition, the knockdown of Pitx2 in SCs isolated from EOM slowed their proliferation rate, and a similar trend was seen for SCs isolated from tibialis anterioris muscle. These data led to the conclusion that Pitx2 helps maintain a proliferating pool of myogenic precursor cells. Finally, the authors highlight that this greater proliferative capacity may facilitate the repair of damaged EOM tissue, thereby contributing to the sparing of EOM in muscular dystrophies (Hebert et al., 2013).
More recently, a study conducted in our laboratory has provided additional information about the molecular mechanisms by which the Pitx2 transcription factor regulates cell proliferation in SCs (Lozano-Velasco et al., 2015). We have reported that Pitx2c expression is higher in early-activated SCs than in long-term activated ones, and our in vitro Pitx2c gain-of-function experiments have revealed that Pitx2c stimulates Ccnd1 and Ccnd2 expression, accelerating cell proliferation during early satellite-cell activation. Moreover, we have demonstrated that such Pitx2c effect on SCs proliferation is due to Pitx2c-mediated downregulation of the miRNAs miR-15b, miR-106b, miR-23b, and miR-503 (Figure 3B). The existence of the Pitx2-miRNA pathway controlling the expression of key regulatory cell-cycle genes in early-activated SCs revealed a role of Pitx2 in satellite-cell activation. Although, muscle SCs are promising targets for cell therapies, the paucity of SCs that can be isolated or expanded from adult muscle tissue is limiting; thus these findings provide new molecular tools to overcome such a bottleneck. It bears noting that our analyses also showed that Pitx2c can increase Myf5 expression by down-regulating miR-106b (Figure 3B), thus expanding the Myf5+ satellite-cell population and revealing a role for Pitx2c in promoting satellite-cell populations more primed for myogenic commitment (Lozano-Velasco et al., 2015). In this context it should be highlighted that in several muscular disorders such as muscular dystrophies, the progressive muscle wasting and weakness is often associated with exhaustion of muscle-regeneration potential. Therefore, the progressive loss of muscle mass has been attributed, at least partly, to the inability of muscle stem cells to efficiently regenerate tissue loss as the result of the disease (Berardi et al., 2014). Thus, critical for the development of effective strategies to treat muscle disorders is the optimization of approaches targeting muscle stem cells and capable of regenerating tissue loss as the result of the disease or as the result of normal muscle turnover (Bertoni, 2014). Notably, very recent reports have been pointed out that muscle stem cells should be considered as a therapeutic target for restoring muscle function in individuals with DMD (Chal et al., 2015; Dumont et al., 2015). Therefore, identification of new Pitx2 functions in the context of SC biology may significantly contribute to the clarification of the molecular and cellular mechanisms of skeletal-muscle regeneration and may help to develop therapeutic strategies for muscular disorders.
Notably, the analysis of adult single and double Pitx2:Pitx3 conditional mutant mouse lines targeted to the muscle stem-cell compartment revealed that double mutant SCs undergo senescence with impaired regeneration after injury, suggesting that Pitx2-mediated changes in ROS levels are required for differentiation of SCs (L'honoré et al., 2014b).
All these data provide new insight into the function of Pitx2 in the molecular mechanisms that control SC behavior and might thus have future application to enhance the regenerative capacity of these myogenic precursor cells. Further analysis using in vivo models could aid in understanding how the Pitx2-mediated effects on SCs can influence the kinetics of muscle regeneration.
Conclusions and future challenges
The data reviewed above show that Pitx2 is a comprehensive marker for cells undergoing myogenic progression, more so than any of the MRFs. This supports models that include a Pitx2-dependent pathway in virtually all skeletal muscles. Many pieces of experimental evidence have pointed out that Pitx2 is the first molecular signal specifying all myogenic precursors in the head muscles. However, although several works have characterized Pitx2 as a key transcription factor in the molecular cascade regulating trunk- and limb-muscle progenitors, additional work is needed to elucidate the function of Pitx2 in specification vs. determination during trunk and limb myogenesis. In addition, since seminal works have revealed that Pitx2 functions on myogenic cells may be due to Pitx2-mediated regulation of miRNAs, the role of Pitx2 in the post-transcriptional control of myogenesis should be further explored.
In parallel, the role of Pitx2 during adult myogenesis is beginning to be explored. Skeletal muscle has the ability to repair and regenerate due to the presence of resident SCs. SC function in adults requires redeployment of many of the regulatory networks fundamental to developmental myogenesis. Currently, SCs are considered potential therapeutic targets for restoring muscle function in muscle degenerative disorders such as muscular dystrophies. Recent works indicate that Pitx2 is expressed in proliferating SCs and can promote differentiation of satellite-cell-derived myoblasts. Moreover, the identification of Pitx2-miRNA pathways that regulate satellite-cell behavior as well as the impact of Pitx2 on redox condition during satellite-cell differentiation may open insights toward future applications to modulate satellite-cell fate during muscle regeneration. Therefore, these findings propose Pitx2 as a new player on skeletal-muscle satellite-cell biology and may help to develop therapeutic strategies for muscular disorder.
Author contributions
FH, LR, and AA conceived of the structure and content. FH wrote the first draft document. FH and LR designed and produced the figures. LR and DF critically revised the manuscript for intellectual content. AA corrected, edited, and approved the final version of the document to be published.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was partially supported by grants BFU2012-38111, BFU2015-67131 (Ministerio de Economía y Competitividad, Gobierno de España), CTS-1614, P08-CTS-03878, BIO-302 (Junta de Andalucia), and AFM2012-16074 (AFM).
References
- Abu-Elmagd M., Robson L., Sweetman D., Hadley J., Francis-West P., Münsterberg A. (2010). Wnt/Lef1 signaling acts via Pitx2 to regulate somite myogenesis. Dev. Biol. 337, 211–219. 10.1016/j.ydbio.2009.10.023 [DOI] [PubMed] [Google Scholar]
- Amendt B. A., Sutherland L. B., Semina E. V., Russo A. F. (1998). The molecular basis of Rieger syndrome. Analysis of Pitx2 homeodomain protein activities. J. Biol. Chem. 273, 20066–20072. 10.1074/jbc.273.32.20066 [DOI] [PubMed] [Google Scholar]
- Arakawa H., Nakamura T., Zhadanov A. B., Fidanza V., Yano T., Bullrich F., et al. (1998). Identification and characterization of the ARP1 gene, a target for the human acute leukemia ALL1 gene. Proc. Natl. Acad. Sci. U.S.A. 95, 4573–4578. 10.1073/pnas.95.8.4573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armand O., Boutineau A. M., Mauger A., Pautou M. P., Kieny M. (1983). Origin of satellite cells in avian skeletal muscles. Arch. Anat. Microsc. Morphol. Exp. 72, 163–181. [PubMed] [Google Scholar]
- Baek S. H., Kioussi C., Briata P., Wang D., Nguyen H. D., Ohgi K. A., et al. (2003). Regulated subset of G1 growth-control genes in response to derepression by the Wnt pathway. Proc. Natl. Acad. Sci. U.S.A. 100, 3245–3250. 10.1073/pnas.0330217100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajard L., Relaix F., Lagha M. (2006). A novel genetic hierarchy functions during hypaxial myogenesis: Pax3 directly activates Myf5 in muscle progenitor cells in the limb. Genes Dev. 20, 2450–2464. 10.1101/gad.382806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentzinger C. F., Wang Y. X., Rudnicki M. A. (2012). Building muscle: molecular regulation of myogenesis. Cold Spring Harb. Perspect. Biol. 4:a008342. 10.1101/cshperspect.a008342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardi E., Annibali D., Cassano M., Crippa S., Sampaolesi M. (2014). Molecular and cell-based therapies for muscle degenerations: a road under construction. Front. Physiol. 5:119. 10.3389/fphys.2014.00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoni C. (2014). Emerging gene editing strategies for Duchenne muscular dystrophy targeting stem cells. Front. Physiol. 5:148. 10.3389/fphys.2014.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biressi S., Molinaro M., Cossu G. (2007). Cellular heterogeneity during vertebrate skeletal muscle development. Dev. Biol. 308, 281–293. 10.1016/j.ydbio.2007.06.006 [DOI] [PubMed] [Google Scholar]
- Borello U., Berarducci B., Murphy P., Bajard L., Buffa V., Piccolo S., et al. (2006). The Wnt/beta-catenin pathway regulates Gli-mediated Myf5 expression during somitogenesis. Development 133, 3723–3732. 10.1242/dev.02517 [DOI] [PubMed] [Google Scholar]
- Buckingham M., Relaix F. (2015). PAX3 and PAX7 as upstream regulators of myogenesis. Semin. Cell Dev. Biol. 44, 115–125. 10.1016/j.semcdb.2015.09.017 [DOI] [PubMed] [Google Scholar]
- Buckingham M., Rigby P. W. J. (2014). Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev. Cell 28, 225–238. 10.1016/j.devcel.2013.12.020 [DOI] [PubMed] [Google Scholar]
- Campione M., Steinbeisser H., Schweickert A., Deissler K., van Bebber F., Lowe L. A., et al. (1999). The homeobox gene Pitx2: mediator of asymmetric left-right signaling in vertebrate heart and gut looping. Development 126, 1225–1234. [DOI] [PubMed] [Google Scholar]
- Chal J., Oginuma M., Al Tanoury Z., Gobert B., Sumara O., Hick A., et al. (2015). Differentiation of pluripotent stem cells to muscle fiber to model Duchenne muscular dystrophy. Nat. Biotechnol. 33, 962–969. 10.1038/nbt.3297 [DOI] [PubMed] [Google Scholar]
- Chaney B. A., Clark-Baldwin K., Dave V., Ma J., Rance M. (2005). Solution structure of the K50 class homeodomain PITX2 bound to DNA and implications for mutations that cause Rieger syndrome. Biochemistry 44, 7497–7511. 10.1021/bi0473253 [DOI] [PubMed] [Google Scholar]
- Chargé S. B. P., Rudnicki M. A. (2004). Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 84, 209–238. 10.1152/physrev.00019.2003 [DOI] [PubMed] [Google Scholar]
- Couly G. F., Coltey P. M., Le Douarin N. M. (1992). The developmental fate of the cephalic mesoderm in quail-chick chimeras. Development 114, 1–15. [DOI] [PubMed] [Google Scholar]
- Cox C. J., Espinoza H. M., McWilliams B., Chappell K., Morton L., Hjalt T. A., et al. (2002). Differential regulation of gene expression by PITX2 isoforms. J. Biol. Chem. 277, 25001–25010. 10.1074/jbc.M201737200 [DOI] [PubMed] [Google Scholar]
- Deries M., Thorsteinsdóttir S. (2016). Axial and limb muscle development: dialogue with the neighbourhood. Cell. Mol. Life Sci. 73, 4415–4431. 10.1007/s00018-016-2298-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit M., Ansseau E., Tassin A., Winokur S., Shi R., Qian H., et al. (2007). DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc. Natl. Acad. Sci. U.S.A. 104, 18157–18162. 10.1073/pnas.0708659104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F., Sun X., Liu W., Ai D., Klysik E., Lu M. F., et al. (2006). Pitx2 promotes development of splanchnic mesoderm-derived branchiomeric muscle. Development 133, 4891–4899. 10.1242/dev.02693 [DOI] [PubMed] [Google Scholar]
- Dumont N. A., Wang Y. X., von Maltzahn J., Pasut A., Bentzinger C. F., Brun C. E., et al. (2015). Dystrophin expression in muscle stem cells regulates their polarity and asymmetric division. Nat. Med. 21, 1455–1463. 10.1038/nm.3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duxson M. J., Usson Y., Harris A. J. (1989). The origin of secondary myotubes in mammalian skeletal muscles: ultrastructural studies. Development 107, 743–750. [DOI] [PubMed] [Google Scholar]
- Evans A. L., Gage P. J. (2005). Expression of the homeobox gene Pitx2 in neural crest is required for optic stalk and ocular anterior segment development. Hum. Mol. Genet. 14, 3347–3359. 10.1093/hmg/ddi365 [DOI] [PubMed] [Google Scholar]
- Evans D. J. R., Noden D. M. (2006). Spatial relations between avian craniofacial neural crest and paraxial mesoderm cells. Dev. Dyn. 235, 1310–1325. 10.1002/dvdy.20663 [DOI] [PubMed] [Google Scholar]
- Gage P. J., Camper S. A. (1997). Pituitary homeobox 2, a novel member of the bicoid-related family of homeobox genes, is a potential regulator of anterior structure formation. Hum. Mol. Genet. 6, 457–464. 10.1093/hmg/6.3.457 [DOI] [PubMed] [Google Scholar]
- Gage P. J., Suh H., Camper S. A. (1999a). Dosage requirement of Pitx2 for development of multiple organs. Development 126, 4643–4651. [DOI] [PubMed] [Google Scholar]
- Gage P. J., Suh H., Camper S. A. (1999b). The bicoid-related Pitx gene family in development. Mamm. Genome 10, 197–200. 10.1007/s003359900970 [DOI] [PubMed] [Google Scholar]
- Ganga M., Espinoza H. M., Cox C. J., Morton L., Hjalt T. A., Lee Y., et al. (2003). PITX2 isoform-specific regulation of atrial natriuretic factor expression: synergism and repression with Nkx2.5. J. Biol. Chem. 278, 22437–22445. 10.1074/jbc.M210163200 [DOI] [PubMed] [Google Scholar]
- Goulding M., Lumsden A., Paquette A. J. (1994). Regulation of Pax-3 expression in the dermomyotome and its role in muscle development. Development 120, 957–971. [DOI] [PubMed] [Google Scholar]
- Goulding M. D., Chalepakis G., Deutsch U., Erselius J. R., Gruss P. (1991). Pax-3, a novel murine DNA binding protein expressed during early neurogenesis. EMBO J. 10, 1135–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros J., Manceau M., Thomé V., Marcelle C. (2005). A common somitic origin for embryonic muscle progenitors and satellite cells. Nature 435, 954–958. 10.1038/nature03572 [DOI] [PubMed] [Google Scholar]
- Hammond W. S. (1965). Origin of hypoglossal muscles in the chick embryo. Anat. Rec. 151, 547–557. 10.1002/ar.1091510405 [DOI] [PubMed] [Google Scholar]
- Harel I., Nathan E., Tirosh-Finkel L., Zigdon H., Guimarães-Camboa N., Evans S. M., et al. (2009). Distinct origins and genetic programs of head muscle satellite cells. Dev. Cell 16, 822–832. 10.1016/j.devcel.2009.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelton R. D. (1970). A radioautographic analysis of the migration and fate of cells derived from the occipital somites in the chick embryo with specific reference to the development of the hypoglossal musculature. J. Embryol. Exp. Morphol. 24, 455–466. [PubMed] [Google Scholar]
- Hebert S. L., Daniel M. L., McLoon L. K. (2013). The role of Pitx2 in maintaining the phenotype of myogenic precursor cells in the extraocular muscles. PLoS ONE 8:e58405. 10.1371/journal.pone.0058405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P., Geles K. G., Paik J.-H., DePinho R. A., Tjian R. (2008). Codependent activators direct myoblast-specific MyoD transcription. Dev. Cell 15, 534–546. 10.1016/j.devcel.2008.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutcheson D. A., Zhao J., Merrell A., Haldar M., Kardon G. (2009). Embryonic and fetal limb myogenic cells are derived from developmentally distinct progenitors and have different requirements for beta-catenin. Genes Dev. 23, 997–1013. 10.1101/gad.1769009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob M., Jacob H. J., Wachtler F., Christ B. (1984). Ontogeny of avian extrinsic ocular muscles. I. A light- and electron-microscopic study. Cell Tissue Res. 237, 549–557. 10.1007/BF00228439 [DOI] [PubMed] [Google Scholar]
- Janssen I., Heymsfield S. B., Wang Z. M., Ross R. (2000). Skeletal muscle mass and distribution in 468 men and women aged 18-88 yr. J. Appl. Physiol. 89, 81–88. [DOI] [PubMed] [Google Scholar]
- Kablar B., Krastel K., Tajbakhsh S., Rudnicki M. A. (2003). Myf5 and MyoD activation define independent myogenic compartments during embryonic development. Dev. Biol. 258, 307–318. 10.1016/S0012-1606(03)00139-8 [DOI] [PubMed] [Google Scholar]
- Kassar-Duchossoy L., Gayraud-Morel B., Gomès D., Rocancourt D., Buckingham M., Shinin V., et al. (2004). Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature 431, 466–471. 10.1038/nature02876 [DOI] [PubMed] [Google Scholar]
- Kelly A. M., Zacks S. I. (1969). The histogenesis of rat intercostal muscle. J. Cell Biol. 42, 135–153. 10.1083/jcb.42.1.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly R. G., Jerome-Majewska L. A., Papaioannou V. E. (2004). The del22q11.2 candidate gene Tbx1 regulates branchiomeric myogenesis. Hum. Mol. Genet. 13, 2829–2840. 10.1093/hmg/ddh304 [DOI] [PubMed] [Google Scholar]
- Kioussi C., Briata P., Baek S. H., Rose D. W., Hamblet N. S., Herman T., et al. (2002). Identification of a Wnt/Dvl/beta-Catenin –> Pitx2 pathway mediating cell-type-specific proliferation during development. Cell 111, 673–685. 10.1016/S0092-8674(02)01084-X [DOI] [PubMed] [Google Scholar]
- Kitamura K., Miura H., Miyagawa-Tomita S., Yanazawa M., Katoh-Fukui Y., Suzuki R., et al. (1999). Mouse Pitx2 deficiency leads to anomalies of the ventral body wall, heart, extra- and periocular mesoderm and right pulmonary isomerism. Development 126, 5749–5758. [DOI] [PubMed] [Google Scholar]
- Kitamura K., Miura H., Yanazawa M., Miyashita T., Kato K. (1997). Expression patterns of Brx1 (Rieg gene), Sonic hedgehog, Nkx2.2, Dlx1 and Arx during zona limitans intrathalamica and embryonic ventral lateral geniculate nuclear formation. Mech. Dev. 67, 83–96. 10.1016/S0925-4773(97)00110-X [DOI] [PubMed] [Google Scholar]
- Knopp P., Figeac N., Fortier M., Moyle L., Zammit P. S. (2013). Pitx genes are redeployed in adult myogenesis where they can act to promote myogenic differentiation in muscle satellite cells. Dev. Biol. 377, 293–304. 10.1016/j.ydbio.2013.02.011 [DOI] [PubMed] [Google Scholar]
- Lagha M., Sato T., Regnault B., Cumano A., Zuniga A., Licht J., et al. (2010). Transcriptome analyses based on genetic screens for Pax3 myogenic targets in the mouse embryo. BMC Genomics 11:696. 10.1186/1471-2164-11-696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanctôt C., Moreau A., Chamberland M., Tremblay M. L., Drouin J. (1999). Hindlimb patterning and mandible development require the Ptx1 gene. Development 126, 1805–1810. [DOI] [PubMed] [Google Scholar]
- Lepper C., Fan C.-M. (2010). Inducible lineage tracing of Pax7-descendant cells reveals embryonic origin of adult satellite cells. Genesis 48, 424–436. 10.1002/dvg.20630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C., Partridge T. A., Fan C.-M. (2011). An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development 138, 3639–3646. 10.1242/dev.067595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'honoré A., Commère P.-H., Ouimette J.-F., Montarras D., Drouin J., Buckingham M. (2014a). Redox regulation by Pitx2 and Pitx3 is critical for fetal myogenesis. Dev. Cell 29, 392–405. 10.1016/j.devcel.2014.04.006 [DOI] [PubMed] [Google Scholar]
- L'honoré A., Coulon V., Marcil A., Lebel M., Lafrance-Vanasse J., Gage P., et al. (2007). Sequential expression and redundancy of Pitx2 and Pitx3 genes during muscle development. Dev. Biol. 307, 421–433. 10.1016/j.ydbio.2007.04.034 [DOI] [PubMed] [Google Scholar]
- L'honoré A., Drouin J., Buckingham M., Montarras D. (2014b). Pitx2 and Pitx3 transcription factors: two key regulators of the redox state in adult skeletal muscle stem cells and muscle regeneration. Free Radic. Biol. Med. 75, S37. 10.1016/j.freeradbiomed.2014.10.781 [DOI] [PubMed] [Google Scholar]
- L'honoré A., Ouimette J.-F., Lavertu-Jolin M., Drouin J. (2010). Pitx2 defines alternate pathways acting through MyoD during limb and somitic myogenesis. Development 137, 3847–3856. 10.1242/dev.053421 [DOI] [PubMed] [Google Scholar]
- Logan M., Pagán-Westphal S. M., Smith D. M., Paganessi L., Tabin C. J. (1998). The transcription factor Pitx2 mediates situs-specific morphogenesis in response to left-right asymmetric signals. Cell 94, 307–317. 10.1016/S0092-8674(00)81474-9 [DOI] [PubMed] [Google Scholar]
- Logan M., Tabin C. J. (1999). Role of Pitx1 upstream of Tbx4 in specification of hindlimb identity. Science 283, 1736–1739. 10.1126/science.283.5408.1736 [DOI] [PubMed] [Google Scholar]
- Lozano-Velasco E., Contreras A., Crist C., Hernandez-Torres F., Franco D., Aránega A. E. (2011). Pitx2c modulates Pax3+/Pax7+ cell populations and regulates Pax3 expression by repressing miR27 expression during myogenesis. Dev. Biol. 357, 165–178. 10.1016/j.ydbio.2011.06.039 [DOI] [PubMed] [Google Scholar]
- Lozano-Velasco E., Vallejo D., Esteban F. J., Doherty C., Hernandez-Torres F., Franco D., et al. (2015). A Pitx2-miRNA pathway modulates cell proliferation in myoblasts and skeletal-muscle satellite cells and promotes their commitment to a myogenic cell fate. Mol. Cell. Biol. 35, 2892–2909. 10.1128/MCB.00536-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J.-R., Bassel-Duby R., Hawkins A., Chang P., Valdez R., Wu H., et al. (2002). Control of facial muscle development by MyoR and capsulin. Science 298, 2378–2381. 10.1126/science.1078273 [DOI] [PubMed] [Google Scholar]
- Marcil A., Dumontier E., Chamberland M., Camper S. A., Drouin J. (2003). Pitx1 and Pitx2 are required for development of hindlimb buds. Development 130, 45–55. 10.1242/dev.00192 [DOI] [PubMed] [Google Scholar]
- Martínez-Fernández S., Hernandez-Torres F., Franco D., Lyons G. E., Navarro F., Aránega A. E. (2006). Pitx2c overexpression promotes cell proliferation and arrests differentiation in myoblasts. Dev. Dyn. 235, 2930–2939. 10.1002/dvdy.20924 [DOI] [PubMed] [Google Scholar]
- Matsuoka T., Ahlberg P. E., Kessaris N., Iannarelli P., Dennehy U., Richardson W. D., et al. (2005). Neural crest origins of the neck and shoulder. Nature 436, 347–355. 10.1038/nature03837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro A. (1961). Satellite cell of skeletal muscle fibers. J. Biophys. Biochem. Cytol. 9, 493–495. 10.1083/jcb.9.2.493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miersch C., Stange K., Hering S., Kolisek M., Viergutz T., Röntgen M. (2017). Molecular and functional heterogeneity of early postnatal porcine satellite cell populations is associated with bioenergetic profile. Sci. Rep. 7:45052. 10.1038/srep45052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncaut N., Rigby P. W. J., Carvajal J. J. (2013). Dial M(RF) for myogenesis. FEBS J. 280, 3980–3990. 10.1111/febs.12379 [DOI] [PubMed] [Google Scholar]
- Muccielli M. L., Martinez S., Pattyn A., Goridis C., Brunet J. F. (1996). Otlx2, an Otx-related homeobox gene expressed in the pituitary gland and in a restricted pattern in the forebrain. Mol. Cell. Neurosci. 8, 258–271. 10.1006/mcne.1996.0062 [DOI] [PubMed] [Google Scholar]
- Murphy M. M., Lawson J. A., Mathew S. J., Hutcheson D. A., Kardon G. (2011). Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development 138, 3625–3637. 10.1242/dev.064162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noden D. M. (1983). The embryonic origins of avian cephalic and cervical muscles and associated connective tissues. Am. J. Anat. 168, 257–276. 10.1002/aja.1001680302 [DOI] [PubMed] [Google Scholar]
- Noden D. M., Francis-West P. (2006). The differentiation and morphogenesis of craniofacial muscles. Dev. Dyn. 235, 1194–1218. 10.1002/dvdy.20697 [DOI] [PubMed] [Google Scholar]
- Ono Y., Boldrin L., Knopp P., Morgan J. E., Zammit P. S. (2010). Muscle satellite cells are a functionally heterogeneous population in both somite-derived and branchiomeric muscles. Dev. Biol. 337, 29–41. 10.1016/j.ydbio.2009.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellettieri J., Alvarado A. S. (2007). Cell turnover and adult tissue homeostasis: from humans to planarians. Annu. Rev. Genet. 41, 83–105. 10.1146/annurev.genet.41.110306.130244 [DOI] [PubMed] [Google Scholar]
- Relaix F., Rocancourt D., Mansouri A., Buckingham M. (2005). A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature 435, 948–953. 10.1038/nature03594 [DOI] [PubMed] [Google Scholar]
- Ryan A. K., Blumberg B., Rodriguez-Esteban C., Yonei-Tamura S., Tamura K., Tsukui T., et al. (1998). Pitx2 determines left-right asymmetry of internal organs in vertebrates. Nature 394, 545–551. 10.1038/29004 [DOI] [PubMed] [Google Scholar]
- Sambasivan R., Gayraud-Morel B., Dumas G., Cimper C., Paisant S., Kelly R. G., et al. (2009). Distinct regulatory cascades govern extraocular and pharyngeal arch muscle progenitor cell fates. Dev. Cell 16, 810–821. 10.1016/j.devcel.2009.05.008 [DOI] [PubMed] [Google Scholar]
- Sambasivan R., Yao R., Kissenpfennig A., Van Wittenberghe L., Paldi A., Gayraud-Morel B., et al. (2011). Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development 138, 3647–3656. 10.1242/dev.067587 [DOI] [PubMed] [Google Scholar]
- Schienda J., Engleka K. A., Jun S., Hansen M. S., Epstein J. A., Tabin C. J., et al. (2006). Somitic origin of limb muscle satellite and side population cells. Proc. Natl. Acad. Sci. U.S.A. 103, 945–950. 10.1073/pnas.0510164103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalbruch H., Lewis D. M. (2000). Dynamics of nuclei of muscle fibers and connective tissue cells in normal and denervated rat muscles. Muscle Nerve 23, 617–626. [DOI] [PubMed] [Google Scholar]
- Schubert F. R., Tremblay P., Mansouri A., Faisst A. M., Kammandel B., Lumsden A., et al. (2001). Early mesodermal phenotypes in splotch suggest a role for Pax3 in the formation of epithelial somites. Dev. Dyn. 222, 506–521. 10.1002/dvdy.1211 [DOI] [PubMed] [Google Scholar]
- Schweickert A., Campione M., Steinbeisser H., Blum M. (2000). Pitx2 isoforms: involvement of Pitx2c but not Pitx2a or Pitx2b in vertebrate left-right asymmetry. Mech. Dev. 90, 41–51. 10.1016/S0925-4773(99)00227-0 [DOI] [PubMed] [Google Scholar]
- Segalés J., Perdiguero E., Muñoz-Cánoves P. (2016). Regulation of muscle stem cell functions: a focus on the p38 MAPK signaling pathway. Front. Cell Dev. Biol. 4:91. 10.3389/fcell.2016.00091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semina E. V., Ferrell R. E., Mintz-Hittner H. A., Bitoun P., Alward W. L., Reiter R. S., et al. (1998). A novel homeobox gene PITX3 is mutated in families with autosomal-dominant cataracts and ASMD. Nat. Genet. 19, 167–170. 10.1038/527 [DOI] [PubMed] [Google Scholar]
- Semina E. V., Reiter R., Leysens N. J., Alward W. L., Small K. W., Datson N. A., et al. (1996). Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat. Genet. 14, 392–399. 10.1038/ng1296-392 [DOI] [PubMed] [Google Scholar]
- Shapiro M. D., Marks M. E., Peichel C. L., Blackman B. K., Nereng K. S., Jónsson B., et al. (2004). Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature 428, 717–723. 10.1038/nature02415 [DOI] [PubMed] [Google Scholar]
- Shih H. P., Gross M. K., Kioussi C. (2007a). Cranial muscle defects of Pitx2 mutants result from specification defects in the first branchial arch. Proc. Natl. Acad. Sci. U.S.A. 104, 5907–5912. 10.1073/pnas.0701122104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih H. P., Gross M. K., Kioussi C. (2007b). Expression pattern of the homeodomain transcription factor Pitx2 during muscle development. Gene Expr. Patterns 7, 441–451. 10.1016/j.modgep.2006.11.004 [DOI] [PubMed] [Google Scholar]
- Standring S. (2008). Gray's Anatomy. The Anatomical Basis of Clinical Practice. London: Churchill Livingstone Elsevier. [Google Scholar]
- Stuelsatz P., Keire P., Yablonka-Reuveni Z. (2017). Isolation, culture, and immunostaining of skeletal muscle myofibers from wildtype and nestin-GFP mice as a means to analyze satellite cell. Methods Mol. Biol. 1556, 51–102. 10.1007/978-1-4939-6771-1_4 [DOI] [PubMed] [Google Scholar]
- Szeto D. P., Rodriguez-Esteban C., Ryan A. K., O'Connell S. M., Liu F., Kioussi C., et al. (1999). Role of the Bicoid-related homeodomain factor Pitx1 in specifying hindlimb morphogenesis and pituitary development. Genes Dev. 13, 484–494. 10.1101/gad.13.4.484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajbakhsh S. (2009). Skeletal muscle stem cells in developmental versus regenerative myogenesis. J. Intern. Med. 266, 372–389. 10.1111/j.1365-2796.2009.02158.x [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S., Buckingham M. (1999). The birth of muscle progenitor cells in the mouse: spatiotemporal considerations. Curr. Top. Dev. Biol. 48, 225–268. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S., Rocancourt D., Cossu G., Buckingham M. (1997). Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell 89, 127–138. 10.1016/S0092-8674(00)80189-0 [DOI] [PubMed] [Google Scholar]
- Tzahor E. (2015). Head muscle development. Results Probl. Cell Differ. 56, 123–142. 10.1007/978-3-662-44608-9_6 [DOI] [PubMed] [Google Scholar]
- Venuti J. M., Morris J. H., Vivian J. L., Olson E. N., Klein W. H. (1995). Myogenin is required for late but not early aspects of myogenesis during mouse development. J. Cell Biol. 128, 563–576. 10.1083/jcb.128.4.563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka H., Meno C., Koshiba K., Sugihara M., Itoh H., Ishimaru Y., et al. (1998). Pitx2, a bicoid-type homeobox gene, is involved in a lefty-signaling pathway in determination of left-right asymmetry. Cell 94, 299–305. 10.1016/S0092-8674(00)81473-7 [DOI] [PubMed] [Google Scholar]
- Zacharias A. L., Lewandoski M., Rudnicki M. A., Gage P. J. (2011). Pitx2 is an upstream activator of extraocular myogenesis and survival. Dev. Biol. 349, 395–405. 10.1016/j.ydbio.2010.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]