Abstract
Microbial oils, which are mainly extracted from yeasts, molds, and algae, have been of considerable interest as food additives and biofuel resources due to their high lipid content. While these oleaginous microorganisms generally produce only small amounts of lipids under optimal growth conditions, their lipid accumulation machinery can be induced by environmental stresses, such as nutrient limitation and an inhospitable physical environmental. As common second messengers of many stress factors, reactive oxygen species (ROS) may act as a regulator of cellular responses to extracellular environmental signaling. Furthermore, increasing evidence indicates that ROS may act as a mediator of lipid accumulation, which is associated with dramatic changes in the transcriptome, proteome, and metabolome. However, the specific mechanisms of ROS involvement in the crosstalk between extracellular stress signaling and intracellular lipid synthesis require further investigation. Here, we summarize current knowledge on stress-induced lipid biosynthesis and the putative role of ROS in the control of lipid accumulation in oleaginous microorganisms. Understanding such links may provide guidance for the development of stress-based strategies to enhance microbial lipid production.
Keywords: reactive oxygen species, oleaginous microorganisms, stress response, signaling molecules, lipid accumulation
Introduction
Lipids can be produced by practically all living organisms, and they play pivotal structural and functional roles, including the formation of cell membranes, as well as carbon and energy storage. However, only a few microorganisms, including yeasts, molds, and algae, can accumulate microbial lipids to more than 20% of their dry cell weight (DCW), and thus be defined as oleaginous microorganisms (Donot et al., 2014). Such oleaginous microorganisms are currently emerging as production strains for a variety of applications, and are already significant sources of fatty acid- and lipid derivatives in various industrial fields (Laoteng et al., 2011). The production of microbial lipids is not restricted by season, climate, and location, and can be realized utilizing a variety of inexpensive substrates, such as waste streams from the food industry (Ochsenreitheri et al., 2016). Considering the threat of global warming, the increasing demand for energy, and the foreseeable depletion of easily extracted crude oil, microbial lipids are receiving worldwide attention as a cleaner alternative to fossil fuels. Also, from a dietary point of view, microbial lipids can be a safe, edible substitute for animal fats, which is a traditional source of polyunsaturated fatty acids (PUFAs) (Ratledge, 2004; Ryan et al., 2010; Bellou et al., 2016a). This is particularly significant since PUFAs, such as those belonging to the omega-3 and omega-6 series, are well known for their benefits to human health (Ji et al., 2014, 2015; Xie et al., 2015).
Oleaginous microorganisms are able to accumulate cellular lipids from 25% to an astonishing 70% of their DCW, and this accumulation is normally induced under various environmental stresses, such as high light stress, salt stress, and deprivation of nutrients (Wang et al., 2004; Jeennor et al., 2006; Donot et al., 2014; Fan et al., 2014). Consequently, stress-based strategies are widely used as environmentally friendly approaches to induce lipid overproduction in cultured microorganisms (Sharma et al., 2012), and a wide range of studies were carried out to identify and develop efficient induction techniques for lipid accumulation. Advances provided by the development of “-omics” analysis methods have illuminated the global reorganization of metabolic and transcriptional states and their integration between different stress regimens (Morano et al., 2012; Yang Z.K. et al., 2014; Yu et al., 2017).
On the other hand, oxidative metabolism is the major form of energy production in most living organisms, and reactive oxygen species (ROS) are generated as by-products of various metabolic pathways, including aerobic respiration. It is well established that ROS can inhibit photosynthesis and cause damage to DNA, proteins, lipids. However, over the past two decades, there has been a growing appreciation for the role of ROS as “second messengers” of various signal transduction mechanisms (Schieber and Chandel, 2014; Zhang J. et al., 2016). For example, nicotinamide adenine dinucleotide phosphate (NADPH) oxidases can produce ROS to regulate different cellular processes, such as defense, growth, and acclimation (Lara-Ortiz et al., 2003; Aguirre et al., 2005; Sandalio et al., 2013).
Here we review recent work on stress-induced lipid biosynthesis and the putative role of ROS in controlling lipid accumulation in oleaginous microorganisms. Furthermore, future perspectives on emerging analysis methods and new research directions in the field of ROS signaling are discussed.
Lipid Accumulation in Oleaginous Microorganisms Under Different Types of Stress
Oleaginous microorganisms produce only small amounts of lipids under favorable environmental conditions, and their lipid biosynthesis machinery can be induced by stress conditions such as nutrient limitation and exposure to damaging physical factors (Chisti, 2007; Georgianna and Mayfield, 2012; Ji et al., 2015).
Nutrient Limitation
Oleaginous microorganisms can accumulate considerable amounts of lipids under various nutrient limitation conditions, among which nitrogen limitation is recognized as the most successful induction strategy, and is most widely used. Large amounts of omics data had shown some clues of the regulation of carbon flux redistribution to lipids. For example, proteomic analysis demonstrated that nitrogen depletion can upregulate the glycolytic pathway, while the activity of TCA cycle was retarded, thus, leading more carbon flux to fatty acid biosynthesis in Mucor circinelloides (Tang et al., 2016). Other types of nutrient starvation that can enhance lipid accumulation include phosphate, silicon, and sulfate limitation (Wu et al., 2011; Ren et al., 2013; Bellou et al., 2016b; Ota et al., 2016).
Generally, two different pathways are involved in lipid synthesis: de novo lipid synthesis and ex novo lipid synthesis. Fundamental differences on the biochemical level exist between de novo lipid synthesis from hydrophilic substrates and ex novo lipid synthesis from hydrophobic substrates (Donot et al., 2014). Some studies have reported that nutrient limitation is only effective in the induction of de novo lipid synthesis, when growth is carried out on various hydrophilic substances (Papanikolaou and Aggelis, 2011a,b; Donot et al., 2014). Furthermore, nutrient limitation will induce lipid biosynthesis while concomitantly causing growth inhibition (Sajbidor et al., 1990; Nie et al., 2014; Zienkiewicz et al., 2016). However, it is a significant goal to obtain both a great quantity of biomass and a high lipid content for improved lipid productivity. Thus, in practice, oleaginous microorganisms are first cultured in nutrient-rich media for rapid propagation in early process stages, while nutrient limitation is introduced in later stages to induce the overproduction of lipids (Lin et al., 2011; Zhu et al., 2016). For instance, Nie et al. (2014) reported a three-stage fermentation strategy that changes the nutrient components of the medium, which was designed for the efficient production of oil rich in arachidonic acid (ARA) using Mortierella alpina.
Physical Environmental Stresses
The composition and quantity of microbial lipids is species-dependent and can be affected by external environmental factors, such as light intensity, temperature, salinity, dissolved oxygen, etc. (Vitova et al., 2015). Furthermore, in autotrophic microalgae, light capture for photosynthesis is crucial for growth and accumulation of lipid reserves. Thus, adequate light intensity favors the overproduction of microalgal lipids. For example, high light conditions trigger the formation of neutral lipids in the microalgae Scenedesmus abundans and Nannochloropsis sp., as well as Botryococcus braunii and a further Botryococcus species (Kojima and Zhang, 1999; Pal et al., 2011; Yeesang and Cheirsilp, 2011; Mandotra et al., 2016).
The incubation temperature has been reported as an important abiotic factor affecting growth and microbial oil accumulation (Laoteng et al., 2011). In Nannochloropsis oculata, heat stress rapidly stimulates neutral lipid formation, while raising the temperature up to 25°C causes an elevation of total lipids (Converti et al., 2009). On the other hand, the incubation temperature of oleaginous microorganisms influences their fatty acid composition by changing the desaturase enzyme activity (Amaretti et al., 2010; Xu et al., 2012). James et al. (2013) found that a switch to temperatures lower than 25°C decreased the content of saturated fatty acids in the microalga Chlamydomonas reinhardtii. Likewise, the concentration of ARA in M. alpina changed from 7.3 to 9.2 g/L, when the temperature changed from 25 to 20°C (Peng et al., 2010).
In some species of microalgae, an increase of intracellular lipid content was observed as a response to osmotic pressure due to salinity stress (Takagi et al., 2006; Yang H. et al., 2014). For example, Duan et al. (2012) reported that Chlorella vulgaris experienced a 21.1% increase of lipid yield when exposed to salt pressure. Biosynthesis of lipids is also induced by other physical environmental stresses, such as low oxygen in the microalga Aurantiochytrium sp., and dehydration in the green alga Chlorella kessleri (Jakobsen et al., 2008; Shiratake et al., 2013).
Stress-Induced ROS Generation and Its Potential Role in Lipid Accumulation
Since stress-based strategies are widely used as an environmentally friendly approach to microbial lipid overproduction, understanding the relationship between various stress factors and lipid accumulation is of industrial and biotechnological importance (Breuer et al., 2013). With advances in biotechnology and bioinformatics, such as proteomic and genomic analysis, many studies revealed metabolic network shifts toward lipid accumulation under different kinds of stress (Lei et al., 2012; Yang Z.K. et al., 2014; Tang et al., 2016). However, little is known about the link between extracellular stress signals and intracellular lipid synthesis. There may exist potential signal transduction mechanisms that trigger carbon partitioning and lipid accumulation in response to different environmental stresses, which serve to control homeostasis at the cellular level. In recent years, some research results indicated that ROS may be important mediators in this process (Urbano et al., 2014; Yilancioglu et al., 2014).
Redox Homeostasis and Oxidative Stress
Reactive oxygen species, such as superoxide (O2-), hydroxyl radical (OH•), and hydrogen peroxide (H2O2) are formed by the partial reduction of oxygen, which is an inevitable aspect of life under aerobic conditions (Montibus et al., 2015). Cellular ROS metabolism is tightly regulated by a battery of biological redox mechanisms, including both antioxidant enzymes and non-enzymatic antioxidant molecules. In a normal physiological state, the level of cellular ROS is in a stable dynamic equilibrium (Zhang J. et al., 2016). However, under specific adverse environmental stimuli, the balance between cellular ROS production and elimination can be disturbed, leading to a locally increased concentration of ROS – i.e., “oxidative stress” (Lushchak, 2011).
Similar to other forms of environmental stress, oxidative stress results in damage of nucleic acids, proteins, and lipids. However, indeed in many eukaryotes, there are well-described mechanisms in which ROS play a key role in the response and adaptation to environmental changes (Bhattacharjee, 2012; Hideg et al., 2013). According to recent reports, ROS may act as decisive signaling molecules of the cellular responses to environmental stresses in oleaginous microorganisms. The relationship between ROS levels and a number of different kinds of stress has been reported, including adverse light conditions, nitrogen depletion, unfavorable salinity, as well as high or low temperature (Shih et al., 2014; Chokshi et al., 2015; Garcia-Rios et al., 2016). In C. vulgaris, the specific intracellular ROS level can serve as a general quantitative marker for stress, irrespective of the type of stress induced (Menon et al., 2013).
Stress Sensing and Putative Concomitant ROS Generation
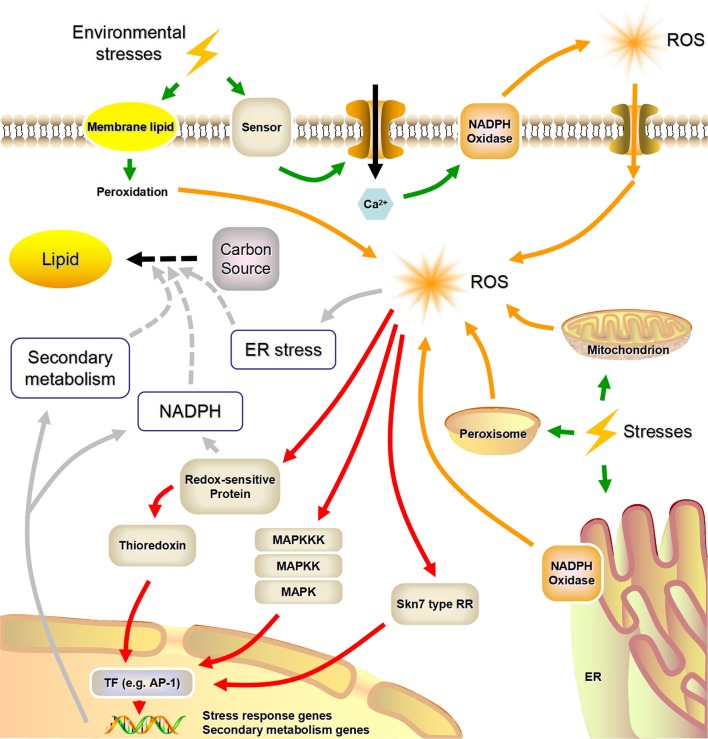
Eukaryotic microorganisms (e.g., yeasts, molds, and microalgae) possess highly conserved molecular mechanisms that enable them to survive in adverse environments. The machinery encompassing stress sensors, signal transduction and response elements is common to these mechanisms (Lushchak, 2011). In Figure 1, we summarized what is known about stress sensing and the putative concomitant ROS generation.
FIGURE 1.
A possible link between environmental stress factors and lipid biosynthesis mediated by reactive oxygen species (NADPH, nicotinamide adenine dinucleotide phosphate; ROS, reactive oxygen species; MAPK, mitogen-activated protein kinase; MAPKK, MAPK kinase; MAPKKK, MAPKK kinase; TF, transcription factor; RR, response regulator; ER, endoplasmic reticulum; Green arrows, stress sensing; Orange arrows, stress-induced ROS generation; Red arrows, ROS signal transduction; Gray arrows, ROS-induced lipid accumulation). This figure was modified from Hagiwara et al. (2016) and Zhu (2016).
Nicotinamide adenine dinucleotide phosphate oxidases are highly conserved plasma- and endo-membrane enzymes present in animals, plants (including microalgae), and fungi. Some researchers also summarized the functions of certain macromolecules as possible sensors for environmental changes in plants and mammals (Hernandez-Onate and Herrera-Estrella, 2015; Zhu, 2016). Specific signals (e.g., Ca2+) that are generated by these stress sensors can lead to the phosphorylation and activation of NADPH oxidases, and activated NADPH oxidases can in turn utilize cytosolic NADPH as the electron donor to reduce extracellular O2 to O2-, causing the subsequent formation of H2O2 (Herve et al., 2006; Mignolet-Spruyt et al., 2016).
The electron transfer chains (ETC), which exist in mitochondria, endoplasmic reticulum, and peroxisomes, are regarded as the main sources of intracellular ROS (Starkov, 2008; Sandalio et al., 2013). Under certain stimuli, growing ROS concentrations can overwhelm the cellular antioxidant defense systems, causing ROS to be released into the matrix via osmosis (Manoli et al., 2007; Mignolet-Spruyt et al., 2016). Using TEM, Yu et al. (2016) observed mitochondria in M. alpina that seemed to be enlarged and less compact than normal with carbon limitation, which in turn may generate ROS. Furthermore, abiotic stresses such as UV and heat instantaneously lead to modifications of the membranes, such as membrane-lipid peroxidation, which are potential sources of ROS. Unlike NADPH oxidase-generated ROS, these membrane-lipid peroxidation products induce non-specific responses to various kinds of environmental stress (Bhattacharjee, 2012).
Transduction of Intracellular ROS Signals
Compared with other types of ROS, such as O2- and OH•, H2O2 is much more stable and can pass membranes through aquaporins, which is an advantage with regard to its signaling capacity (Reczek and Chandel, 2015). The transduction of H2O2-based signals is mainly centered on sulfur chemistry, with the main player being the reversible oxidative modification of cellular sulfur-containing groups (e.g., cysteine residues and thioredoxin), which in turn results in disturbances of cellular metabolism and signaling pathways in eukaryotic microorganisms (Veal et al., 2007; Scott and Eaton, 2008).
As shown in Figure 1, the key part of the adaptive response to H2O2 is the transcriptional reprogramming of gene expression. The AP-1 family transcription factors are highly conserved among eukaryotes. They are involved in genetic responses to H2O2 signals associated with the Skn7-type response regulators (Carmel-Harel et al., 2001; Hagiwara et al., 2016). H2O2 can also oxidize critical cysteine thiol groups of phosphatases in mitogen-activated protein kinase (MAPK) pathways, which appears to be a conserved signaling mechanism for diverse environmental stimuli (Ikner and Shiozaki, 2005; Lushchak, 2011). For instance, the unicellular yeast Schizosaccharomyces pombe utilizes a two-component signaling system coupled to a MAPK pathway to perceive and respond to local ROS accumulation (Nguyen et al., 2000; Lara-Rojas et al., 2011). Finally, a number of recent studies have shown that post-translational changes induced by ROS also play an important role in the rapid response to oxidative stress, in addition to the altered global gene expression patterns (Ralser et al., 2009; Morano et al., 2012).
Possible Links between ROS and Lipid Accumulation
Oleaginous microorganisms mainly accumulate neutral lipids, which account for nearly 90% of their lipid storage (Juneja et al., 2013; Donot et al., 2014). In addition to their traditional and most-studied function in carbon and energy storage, these lipids, and especially PUFAs, may act as antioxidants or otherwise protective defense molecules in the stress response (Hu et al., 2008). For example, the increase of fatty acid unsaturation under low temperature stress has been generally linked to an adaptation mechanism which the lipid-producing fungi use to maintain membrane fluidity (Certik and Shimizu, 1999; Ji et al., 2014). The overproduction of lipids presents an indispensable buffer against stress conditions.
As shown in Table 1, improving experimental evidence seems to point in the direction that intracellular ROS may in fact be mediators of lipid accumulation in oleaginous microorganisms (Yilancioglu et al., 2014; Chokshi et al., 2015). A number of studies showed that stress-induced lipid accumulation always goes along with increasing antioxidant defenses (e.g., oxidative-stress response proteins) or increasing intracellular ROS levels (Yang et al., 2013; Yilancioglu et al., 2014). As stress markers, the specific intracellular ROS levels have been found to be linked to the specific intracellular neutral lipid levels in an inverse and direct power law fashion in C. vulgaris (Menon et al., 2013). In fact, the proper exposure to exogenous ROS, such as H2O2, but also certain nanomaterials, can trigger neutral lipid formation (Yu et al., 2015).
Table 1.
Stress-induced reactive oxygen species (ROS) generation in different oleaginous microorganisms.
| Stress factor | Oleaginous microorganism | Lipid production | ROS generation | Reference |
|---|---|---|---|---|
| Salinity | Scenedesmus sp. | 33.13% of lipids accumulated under salinity stress (400 mM NaCl). | Higher H2O2, MDA, APX, and proline contents were observed with an increase of NaCl. | Pancha et al., 2015 |
| High light intensity | Chlorella sp. and Monoraphidium sp. | Carbon allocation converted from protein and carbohydrate to lipid under high light. Neutral lipid productivity was significantly promoted. | Along with increasing lipid accumulation, ROS-scavenging enzymes also increased. | He et al., 2015 |
| Low temperature | Scenedesmus sp. | At low temperatures, the lipid content per microalgal biomass increased. | The ROS levels at 10°C and 20°C were higher than that under higher temperature. | Li et al., 2011 |
| Nitrogen limitation | Dunaliella salina | Nitrogen limitation increased cellular lipid content up to 35% under 0.05 mM nitrogen concentration. | Under nitrogen depleted cultivation conditions, higher MDA, CAT, APX, and SOD were observed. | Yilancioglu et al., 2014 |
| Nitzschia closterium f. minutissima | Neutral lipid increased 48, 111, 171, and 216% compared to the control, with nitrogen concentrations of 712, 491, 270, and 159 μM. | Intercellular ROS was enhanced under low nitrogen stress. | Liu et al., 2012 | |
| Acutodesmus dimorphus | Nitrogen limitation is effective to produce biomass containing 29.92% of lipid (comprising about 75% of neutral lipid). | Nitrogen limitation resulted in the accumulation of ROS. | Chokshi et al., 2017 | |
| Mucor circinelloides | The rate of lipid production goes up after nitrogen exhaustion. | Some proteins involved in signal transduction and redox homeostasis were upregulated upon nitrogen depletion. | Tang et al., 2016 | |
| Chlorella pyrenoidosa | The total lipid and neutral lipid contents exhibit the most marked increment under nitrogen deficiency, achieving 50.32 and 34.29% of DCW, respectively. | Nitrogen limitation resulted in the co-occurrence of ROS and lipid accumulation. | Fan et al., 2014 | |
However, only a few possible mechanisms of ROS-mediated lipid accumulation have been illuminated, and direct experimental evidence is still absent. Some hypotheses surrounding the mechanisms of ROS-dependent lipid accumulation are shown in Figure 1. As lipids are highly reduced molecular species, neutral lipid overproduction requires large quantities of NADPH, whose primary sources are the oxidative pentose phosphate pathway and malic enzyme (Wasylenko et al., 2015). In what appears to be a common mechanism, within seconds of oxidative stress the carbon metabolic flux changes from glycolysis to the oxidative pentose phosphate pathway by post-translational modification of glycolytic enzymes (Ralser et al., 2009). On the other hand, the stored neutral lipids in oleaginous microorganisms are deposited within lipid droplets, and there is evidence that endoplasmic reticulum stress is an activator of lipid droplets formation (Jacquier et al., 2011). Since ROS are a well-known trigger of endoplasmic reticulum stress, this is another possible mechanism by which ROS stress enhances the formation of lipid droplets (Yu et al., 2015). Moreover, previous studies have demonstrated that some transcription factors, such as AP-1, which are associated with the oxidative-stress response, also participate in a regulatory network that induces secondary metabolism (Montibus et al., 2015). Considering the crosstalk between lipid biosynthesis and secondary metabolism, this appears to be another potential link between intracellular ROS signaling and lipid metabolism (Reyes et al., 2014; Zhang Z. et al., 2016).
Furthermore, the overall level of intracellular ROS cannot be ignored. The generation of low levels of ROS by environmental stresses initiates adaptive responses, including lipid biosynthesis and accumulation. By contrast, high levels of ROS may cause cellular damage, leading to the consumption of stored lipids as energy source in order to maintain cellular homeostasis. It has also been reported that the external addition of antioxidants, such as sesamol, ginsenosides, and ascorbic acid, can scavenge intracellular ROS and enhance PUFA content in oleaginous microorganisms (Liu et al., 2015; Ren et al., 2017).
Conclusion and Perspectives
Microbial lipid accumulation can be regarded as a buffer against environmental stresses, such as nutrient limitation and adverse physical environments. Stress-based lipid production strategies have been widely used in almost all oleaginous microorganisms due to their significant effectiveness. However, the signal transduction mechanisms that trigger metabolic changes in response to different stress factors are very poorly understood. ROS signaling may act as a mediator in cellular responses to extracellular environmental stresses. As a quantitative marker of cellular stress, increased ROS levels have been frequently reported in association with lipid overproduction. Here, we list the possible links among environmental stress factors, ROS signaling, and lipid metabolism, which may provide guidance for the development of stress-based strategies to enhance microbial lipid production.
The research on ROS signaling was often hindered by the lack of quantitative, dynamic, and specific techniques to monitor different activities of ROS in vivo (Marschall and Tudzynski, 2014). However, a variety of specific imaging tools, including small molecule fluorescent probes and genetically encoded redox probes, have been developed in recent years, and these new methods provide improved selectivity, making them able to characterize the subcellular localization and flux of ROS (Meyer and Dick, 2010; Dikalov and Harrison, 2014). Moreover, advanced proteomics techniques, such as the “redox proteome,” have paved the way for the study of cellular redox metabolism and its tight connection with biological structure and function (Go and Jones, 2013). Recent work showed importance of Ca2+ signal transduction in lipid accumulation in response to nitrogen starvation, and there may exist potential cross-talking between these two signaling pathways (Chen et al., 2014). Unraveling these complex signaling networks may provide guidance for the development of new stress-based strategies for enhanced microbial lipid overproduction.
Author Contributions
KS and XJ-J wrote the manuscript. ZG, T-QS, PS, L-JR, and HH revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (Nos. 21376002 and 21476111), Jiangsu Province Natural Science Foundation (No. BK20131405), the National High-Tech R&D Program of China (No. 2014AA021703), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and the Program for Innovative Research Team in University of Jiangsu Province.
References
- Aguirre J., Rios-Momberg M., Hewitt D., Hansberg W. (2005). Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol. 13 111–118. 10.1016/j.tim.2005.01.007 [DOI] [PubMed] [Google Scholar]
- Amaretti A., Raimondi S., Sala M., Roncaglia L., De Lucia M., Leonardi A., et al. (2010). Single cell oils of the cold-adapted oleaginous yeast Rhodotorula glacialis DBVPG 4785. Microb. Cell Fact. 9:73 10.1186/1475-2859-9-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellou S., Triantaphyllidou I. E., Aggeli D., Elazzazy A. M., Baeshen M. N., Aggelis G. (2016a). Microbial lipids as food additives: recent approaches for improving microbial oil production and its polyunsaturated fatty acid content. Curr. Opin. Biotechnol. 37 24–35. 10.1016/j.copbio.2015.09.005 [DOI] [PubMed] [Google Scholar]
- Bellou S., Triantaphyllidou I. E., Mizerakis P., Aggelis G. (2016b). High lipid accumulation in Yarrowia lipolytica cultivated under double limitation of nitrogen and magnesium. J. Biotechnol. 234 116–126. 10.1016/j.jbiotec.2016.08.001 [DOI] [PubMed] [Google Scholar]
- Bhattacharjee S. (2012). The language of reactive oxygen species signaling in plants. J. Bot. 2012:985298 10.1155/2012/985298 [DOI] [Google Scholar]
- Breuer G., Lamers P. P., Martens D. E., Draaisma R. B., Wijffels R. H. (2013). Effect of light intensity, pH, and temperature on triacylglycerol (TAG) accumulation induced by nitrogen starvation in Scenedesmus obliquus. Bioresour. Technol. 143 1–9. 10.1016/j.biortech.2013.05.105 [DOI] [PubMed] [Google Scholar]
- Carmel-Harel O., Stearman R., Gasch A. P., Botstein D., Brown P. O., Storz G. (2001). Role of thioredoxin reductase in the Yap1p-dependent response to oxidative stress in Saccharomyces cerevisiae. Mol. Microbiol. 39 595–605. 10.1046/j.1365-2958.2001.02255.x [DOI] [PubMed] [Google Scholar]
- Certik M., Shimizu S. (1999). Biosynthesis and regulation of microbial polyunsaturated fatty acid production. J. Biosci. Bioeng. 87 1–14. 10.1016/S1389-1723(99)80001-2 [DOI] [PubMed] [Google Scholar]
- Chen H., Zhang Y. M., He C. L., Wang Q. (2014). Ca2+ signal transduction related to neutral lipid synthesis in an oil-producing green alga Chlorella sp. C2. Plant Cell Physiol. 55 634–644. 10.1093/pcp/pcu015 [DOI] [PubMed] [Google Scholar]
- Chisti Y. (2007). Biodiesel from microalgae. Biotechnol. Adv. 25 294–306. 10.1016/j.biotechadv.2007.02.001 [DOI] [PubMed] [Google Scholar]
- Chokshi K., Pancha I., Ghosh A., Mishra S. (2017). Nitrogen starvation-induced cellular crosstalk of ros-scavenging antioxidants and phytohormone enhanced the biofuel potential of green microalga Acutodesmus dimorphus. Biotechnol. Biofuels 10:60 10.1186/s13068-017-0747-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chokshi K., Pancha I., Trivedi K., George B., Maurya R., Ghosh A., et al. (2015). Biofuel potential of the newly isolated microalgae Acutodesmus dimorphus under temperature induced oxidative stress conditions. Bioresour. Technol. 180 162–171. 10.1016/j.biortech.2014.12.102 [DOI] [PubMed] [Google Scholar]
- Converti A., Casazza A. A., Ortiz E. Y., Perego P., Del Borghi M. (2009). Effect of temperature and nitrogen concentration on the growth and lipid content of Nannochloropsis oculata and Chlorella vulgaris for biodiesel production. Chem. Process. Eng. 48 1146–1151. 10.1016/j.cep.2009.03.006 [DOI] [Google Scholar]
- Dikalov S. I., Harrison D. G. (2014). Methods for detection of mitochondrial and cellular reactive oxygen species. Antioxid. Redox Signal. 20 372–382. 10.1089/ars.2012.4886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donot F., Fontana A., Baccou J. C., Strub C., Schorr-Galindo S. (2014). Single cell oils (SCOs) from oleaginous yeasts and moulds: production and genetics. Biomass Bioenerg. 68 135–150. 10.1016/j.biombioe.2014.06.016 [DOI] [Google Scholar]
- Duan X., Ren G. Y., Liu L. L., Zhu W. X. (2012). Salt-induced osmotic stress for lipid overproduction in batch culture of Chlorella vulgaris. Afr. J. Biotechnol. 11 7072–7078. [Google Scholar]
- Fan J. H., Cui Y. B., Wan M. X., Wang W. L., Li Y. G. (2014). Lipid accumulation and biosynthesis genes response of the oleaginous Chlorella pyrenoidosa under three nutrition stressors. Biotechnol. Biofuels 7:17 10.1186/1754-6834-7-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rios E., Ramos-Alonso L., Guillamon J. M. (2016). Correlation between low temperature adaptation and oxidative stress in Saccharomyces cerevisiae. Front. Microbiol. 7:1199 10.3389/fmicb.2016.01199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgianna D. R., Mayfield S. P. (2012). Exploiting diversity and synthetic biology for the production of algal biofuels. Nature 488 329–335. 10.1038/nature11479 [DOI] [PubMed] [Google Scholar]
- Go Y. M., Jones D. P. (2013). The redox proteome. J. Biol. Chem. 288 26512–26520. 10.1074/jbc.R113.464131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara D., Sakamoto K., Abe K., Gomi K. (2016). Signaling pathways for stress responses and adaptation in Aspergillus species: stress biology in the post-genomic era. Biosci. Biotech. Biochem. 80 1667–1680. 10.1080/09168451.2016.1162085 [DOI] [PubMed] [Google Scholar]
- He Q. N., Yang H. J., Wu L., Hu C. X. (2015). Effect of light intensity on physiological changes, carbon allocation and neutral lipid accumulation in oleaginous microalgae. Bioresour. Technol. 191 219–228. 10.1016/j.biortech.2015.05.021 [DOI] [PubMed] [Google Scholar]
- Hernandez-Onate M. A., Herrera-Estrella A. (2015). Damage response involves mechanisms conserved across plants, animals and fungi. Curr. Genet. 61 359–372. 10.1007/s00294-014-0467-5 [DOI] [PubMed] [Google Scholar]
- Herve C., Tonon T., Collen J., Corre E., Boyen C. (2006). NADPH oxidases in Eukaryotes: red algae provide new hints! Curr. Genet. 49 190–204. 10.1007/s00294-005-0044-z [DOI] [PubMed] [Google Scholar]
- Hideg E., Jansen M. A. K., Strid A. (2013). UV-B exposure, ROS, and stress: inseparable companions or loosely linked associates? Trends Plant Sci. 18 107–115. 10.1016/j.tplants.2012.09.003 [DOI] [PubMed] [Google Scholar]
- Hu Q., Sommerfeld M., Jarvis E., Ghirardi M., Posewitz M., Seibert M., et al. (2008). Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J. 54 621–639. 10.1111/j.1365-313X.2008.03492.x [DOI] [PubMed] [Google Scholar]
- Ikner A., Shiozaki K. (2005). Yeast signaling pathways in the oxidative stress response. Mutat. Res. 569 13–27. 10.1016/j.mfrmmm.2004.09.006 [DOI] [PubMed] [Google Scholar]
- Jacquier N., Choudhary V., Mari M., Toulmay A., Reggiori F., Schneiter R. (2011). Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Sci. 124 2424–2437. 10.1242/jcs.076836 [DOI] [PubMed] [Google Scholar]
- Jakobsen A. N., Aasen I. M., Josefsen K. D., Strom A. R. (2008). Accumulation of docosahexaenoic acid-rich lipid in thraustochytrid Aurantiochytrium sp. strain T66: effects of N and P starvation and O2 limitation. Appl. Microbiol. Biotechnol. 80 297–306. 10.1007/s00253-008-1537-8 [DOI] [PubMed] [Google Scholar]
- James G. O., Hocart C. H., Hillier W., Price G. D., Djordjevic M. A. (2013). Temperature modulation of fatty acid profiles for biofuel production in nitrogen deprived Chlamydomonas reinhardtii. Bioresour. Technol. 127 441–447. 10.1016/j.biortech.2012.09.090 [DOI] [PubMed] [Google Scholar]
- Jeennor S., Laoteng K., Tanticharoen M., Cheevadhanarak S. (2006). Comparative fatty acid profiling of Mucor rouxii under different stress conditions. FEMS Microbiol. Lett. 259 60–66. 10.1111/j.1574-6968.2006.00242.x [DOI] [PubMed] [Google Scholar]
- Ji X. J., Ren L. J., Huang H. (2015). Omega-3 biotechnology: a green and sustainable process for omega-3 fatty acids production. Front. Bioeng. Biotechnol. 3:158 10.3389/fbioe.2015.00158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X. J., Ren L. J., Nie Z. K., Huang H., Ouyang P. K. (2014). Fungal arachidonic acid-rich oil: research, development and industrialization. Crit. Rev. Biotechnol. 34 197–214. 10.3109/07388551.2013.778229 [DOI] [PubMed] [Google Scholar]
- Juneja A., Ceballos R. M., Murthy G. S. (2013). Effects of environmental factors and nutrient availability on the biochemical composition of algae for biofuels production: a review. Energies 6 4607–4638. 10.3390/en6094607 [DOI] [Google Scholar]
- Kojima E., Zhang K. (1999). Growth and hydrocarbon production of microalga Botryococcus braunii in bubble column photobioreactors. J. Biosci. Bioeng. 87 811–815. 10.1016/S1389-1723(99)80158-3 [DOI] [PubMed] [Google Scholar]
- Laoteng K., Certik M., Cheevadhanark S. (2011). Mechanisms controlling lipid accumulation and polyunsaturated fatty acid synthesis in oleaginous fungi. Chem. Pap. 65 97–103. 10.2478/s11696-010-0097-4 [DOI] [Google Scholar]
- Lara-Ortiz T., Riveros-Rosas H., Aguirre J. (2003). Reactive oxygen species generated by microbial NADPH oxidase NoxA regulate sexual development in Aspergillus nidulans. Mol. Microbiol. 50 1241–1255. 10.1046/j.1365-2958.2003.03800.x [DOI] [PubMed] [Google Scholar]
- Lara-Rojas F., Sanchez O., Kawasaki L., Aguirre J. (2011). Aspergillus nidulans transcription factor AtfA interacts with the MAPK SakA to regulate general stress responses, development and spore functions. Mol. Microbiol. 80 436–454. 10.1111/j.1365-2958.2011.07581.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei A. P., Chen H., Shen G. M., Hu Z. L., Chen L., Wang J. X. (2012). Expression of fatty acid synthesis genes and fatty acid accumulation in Haematococcus pluvialis under different stressors. Biotechnol. Biofuels 5:18 10.1186/1754-6834-5-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Hu H. Y., Zhang Y. P. (2011). Growth and lipid accumulation properties of a freshwater microalga Scenedesmus sp under different cultivation temperature. Bioresour. Technol. 102 3098–3102. 10.1016/j.biortech.2010.10.055 [DOI] [PubMed] [Google Scholar]
- Lin J., Shen H., Tan H., Zhao X., Wu S., Hu C., et al. (2011). Lipid production by Lipomyces starkeyi, cells in glucose solution without auxiliary nutrients. J. Biotechnol. 152 184–188. 10.1016/j.jbiotec.2011.02.010 [DOI] [PubMed] [Google Scholar]
- Liu B., Liu J., Sun P. P., Ma X. N., Jiang Y., Chen F. (2015). Sesamol enhances cell growth and the biosynthesis and accumulation of docosahexaenoic acid in the microalga Crypthecodinium cohnii. J. Agric. Food Chem. 63 5640–5645. 10.1021/acs.jafc.5b01441 [DOI] [PubMed] [Google Scholar]
- Liu W. H., Huang Z. W., Li P., Xia J. F., Chen B. (2012). Formation of triacylglycerol in Nitzschia closterium f. minutissima under nitrogen limitation and possible physiological and biochemical mechanisms. J. Exp. Mar. Biol. Ecol. 418 24–29. 10.1016/j.jembe.2012.03.005 [DOI] [Google Scholar]
- Lushchak V. I. (2011). Adaptive response to oxidative stress: bacteria, fungi, plants and animals. Comp. Biochem. Physiol. C 153 175–190. 10.1016/j.cbpc.2010.10.004 [DOI] [PubMed] [Google Scholar]
- Mandotra S. K., Kumar P., Suseela M. R., Nayaka S., Ramteke P. W. (2016). Evaluation of fatty acid profile and biodiesel properties of microalga Scenedesmus abundans under the influence of phosphorus, pH and light intensities. Bioresour. Technol. 201 222–229. 10.1016/j.biortech.2015.11.042 [DOI] [PubMed] [Google Scholar]
- Manoli I., Alesci S., Blackman M. R., Su Y. A., Rennert O. M., Chrousos G. P. (2007). Mitochondria as key components of the stress response. Trends Endocrinol. Metab. 18 190–198. 10.1016/j.tem.2007.04.004 [DOI] [PubMed] [Google Scholar]
- Marschall R., Tudzynski P. (2014). A new and reliable method for live imaging and quantification of reactive oxygen species in Botrytis cinerea technological advancement. Fungal Genet. Biol. 71 68–75. 10.1016/j.fgb.2014.08.009 [DOI] [PubMed] [Google Scholar]
- Menon K. R., Balan R., Suraishkumar G. K. (2013). Stress induced lipid production in Chlorella vulgaris: relationship with specific intracellular reactive species levels. Biotechnol. Bioeng. 110 1627–1636. 10.1002/bit.24835 [DOI] [PubMed] [Google Scholar]
- Meyer A. J., Dick T. P. (2010). Fluorescent protein-based redox probes. Antioxid. Redox Signal. 13 621–650. 10.1089/ars.2009.2948 [DOI] [PubMed] [Google Scholar]
- Mignolet-Spruyt L., Xu E. J., Idanheimo N., Hoeberichts F. A., Muhlenbock P., Brosche M., et al. (2016). Spreading the news: subcellular and organellar reactive oxygen species production and signalling. J. Exp. Bot. 67 3831–3844. 10.1093/jxb/erw080 [DOI] [PubMed] [Google Scholar]
- Montibus M., Pinson-Gadais L., Richard-Forget F., Barreau C., Ponts N. (2015). Coupling of transcriptional response to oxidative stress and secondary metabolism regulation in filamentous fungi. Crit. Rev. Microbiol. 41 295–308. 10.3109/1040841X.2013.829416 [DOI] [PubMed] [Google Scholar]
- Morano K. A., Grant C. M., Moye-Rowley W. S. (2012). The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics 190 1157–1195. 10.1534/genetics.111.128033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A. N., Lee A., Place W., Shiozaki K. (2000). Multistep phosphorelay proteins transmit oxidative stress signals to the fission yeast stress-activated protein kinase. Mol. Biol. Cell 11 1169–1181. 10.1091/mbc.11.4.1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z. K., Deng Z. T., Zhang A. H., Ji X. J., Huang H. (2014). Efficient arachidonic acid-rich oil production by Mortierella alpina through a three-stage fermentation strategy. Bioprocess. Biosyst. Eng. 37 505–511. 10.1007/s00449-013-1015-2 [DOI] [PubMed] [Google Scholar]
- Ochsenreitheri K., Gluck C., Stressler T., Fischer L., Syldatk C. (2016). Production strategies and applications of microbial single cell oils. Front. Microbiol. 7:1539 10.3389/Fmicb.2016.01539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota S., Oshima K., Yamazaki T., Kim S., Yu Z., Yoshihara M., et al. (2016). Highly efficient lipid production in the green alga Parachlorella kessleri: draft genome and transcriptome endorsed by whole-cell 3D ultrastructure. Biotechnol. Biofuels 9:13 10.1186/s13068-016-0424-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal D., Khozin-Goldberg I., Cohen Z., Boussiba S. (2011). The effect of light, salinity, and nitrogen availability on lipid production by Nannochloropsis sp. Appl. Microbiol. Biot. 90 1429–1441. 10.1007/s00253-011-3170-1 [DOI] [PubMed] [Google Scholar]
- Pancha I., Chokshi K., Maurya R., Trivedi K., Patidar S. K., Ghosh A., et al. (2015). Salinity induced oxidative stress enhanced biofuel production potential of microalgae Scenedesmus sp. CCNM 1077. Bioresour. Technol. 189 341–348. 10.1016/j.biortech.2015.04.017 [DOI] [PubMed] [Google Scholar]
- Papanikolaou S., Aggelis G. (2011a). Lipids of oleaginous yeasts. Part I: biochemistry of single cell oil production. Eur. J. Lipid Sci. Technol. 113 1031–1051. 10.1002/ejlt.201100014 [DOI] [Google Scholar]
- Papanikolaou S., Aggelis G. (2011b). Lipids of oleaginous yeasts. Part II: technology and potential applications. Eur. J. Lipid Sci. Technol. 113 1052–1073. 10.1002/ejlt.201100015 [DOI] [Google Scholar]
- Peng C., Huang H., Ji X. J., Liu X., You J. Y., Lu J. M., et al. (2010). A temperature-shift strategy for efficient arachidonic acid fermentation by Mortierella alpina in batch culture. Biochem. Eng. J. 53 92–96. 10.1016/j.bej.2010.09.014 [DOI] [Google Scholar]
- Ralser M., Wamelink M. M. C., Latkolik S., Jansen E. E. W., Lehrach H., Jakobs C. (2009). Metabolic reconfiguration precedes transcriptional regulation in the antioxidant response. Nat. Biotechnol. 27 604–605. 10.1038/nbt0709-604 [DOI] [PubMed] [Google Scholar]
- Ratledge C. (2004). Fatty acid biosynthesis in microorganisms being used for Single Cell Oil production. Biochimie 86 807–815. 10.1016/j.biochi.2004.09.017 [DOI] [PubMed] [Google Scholar]
- Reczek C. R., Chandel N. S. (2015). ROS-dependent signal transduction. Curr. Opin. Cell Biol. 33 8–13. 10.1016/j.ceb.2014.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L. J., Feng Y., Li J., Qu L., Huang H. (2013). Impact of phosphate concentration on docosahexaenoic acid production and related enzyme activities in fermentation of Schizochytrium sp. Bioprocess. Biosyst. Eng. 36 1177–1183. 10.1007/s00449-012-0844-8 [DOI] [PubMed] [Google Scholar]
- Ren L. J., Sun X. M., Ji X. J., Chen S. L., Guo D. S., Huang H. (2017). Enhancement of docosahexaenoic acid synthesis by manipulation of antioxidant capacity and prevention of oxidative damage in Schizochytrium sp. Bioresour. Technol. 223 141–148. 10.1016/j.biortech.2016.10.040 [DOI] [PubMed] [Google Scholar]
- Reyes L. H., Gomez J. M., Kao K. C. (2014). Improving carotenoids production in yeast via adaptive laboratory evolution. Metab. Eng. 21 26–33. 10.1016/j.ymben.2013.11.002 [DOI] [PubMed] [Google Scholar]
- Ryan A. S., Zeller S., Nelson E. B., Cohen Z., Ratledge C. (2010). “Safety evaluation of single cell oils and the regulatory requirements for use as food ingredients,” in Single Cell Oils. Microbial and Algal Oil, 2nd Edn, eds Cohen Z., Ratledge C. (Champaign, IL: AOCS Press; ), 317–350. [Google Scholar]
- Sajbidor J., Dobronova S., Certik M. (1990). Arachidonic acid production by Mortierella Sp S-17 influence Of C/N ratio. Biotechnol. Lett. 12 455–456. 10.1007/Bf01024404 [DOI] [Google Scholar]
- Sandalio L. M., Rodríguez-Serrano M., Romero-Puertas M. C., Luis A. (2013). “Role of peroxisomes as a source of reactive oxygen species (ROS) signaling molecules,” in Peroxisomes and Their Key Role in Cellular Signaling and Metabolism, ed. del Río L. A. (Dordrecht: Springer; ), 231–255. 10.1007/978-94-007-6889-5_13 [DOI] [PubMed] [Google Scholar]
- Schieber M., Chandel N. S. (2014). ROS function in redox signaling and oxidative stress. Curr. Biol. 24 R453–R462. 10.1016/j.cub.2014.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott B., Eaton C. J. (2008). Role of reactive oxygen species in fungal cellular differentiations. Curr. Opin. Microbiol. 11 488–493. 10.1016/j.mib.2008.10.008 [DOI] [PubMed] [Google Scholar]
- Sharma K. K., Schuhmann H., Schenk P. M. (2012). High lipid induction in microalgae for biodiesel production. Energies 5 1532–1553. 10.3390/en5051532 [DOI] [Google Scholar]
- Shih S. C. C., Mufti N. S., Chamberlain M. D., Kim J., Wheeler A. R. (2014). A droplet-based screen for wavelength-dependent lipid production in algae. Energ. Environ. Sci. 7 2366–2375. 10.1039/c4ee01123f [DOI] [Google Scholar]
- Shiratake T., Sato A., Minoda A., Tsuzuki M., Sato N. (2013). Air-drying of cells, the novel conditions for stimulated synthesis of triacylglycerol in a green alga, Chlorella kessleri. PLoS ONE 8:e79630 10.1371/journal.pone.0079630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkov A. A. (2008). The role of mitochondria in reactive oxygen species metabolism and signaling. Ann. N. Y. Acad. Sci. 1147 37–52. 10.1196/annals.1427.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi M., Karseno K., Yoshida T. (2006). Effect of salt concentration on intracellular accumulation of lipids and triacylglyceride in marine microalgae Dunaliella cells. J. Biosci. Bioeng. 101 223–226. 10.1263/jbb.101.223 [DOI] [PubMed] [Google Scholar]
- Tang X., Zan X. Y., Zhao L. N., Chen H. Q., Chen Y. Q., Chen W., et al. (2016). Proteomics analysis of high lipid-producing strain Mucor circinelloides WJ11: an explanation for the mechanism of lipid accumulation at the proteomic level. Microb. Cell Fact. 15:35 10.1186/s12934-016-0428-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbano S. B., Di Capua C., Cortez N., Farias M. E., Alvarez H. M. (2014). Triacylglycerol accumulation and oxidative stress in Rhodococcus species: differential effects of pro-oxidants on lipid metabolism. Extremophiles 18 375–384. 10.1007/s00792-013-0623-8 [DOI] [PubMed] [Google Scholar]
- Veal E. A., Day A. M., Morgan B. A. (2007). Hydrogen peroxide sensing and signaling. Mol. Cell 26 1–14. 10.1016/j.molcel.2007.03.016 [DOI] [PubMed] [Google Scholar]
- Vitova M., Bisova K., Kawano S., Zachleder V. (2015). Accumulation of energy reserves in algae: from cell cycles to biotechnological applications. Biotechnol. Adv. 33 1204–1218. 10.1016/j.biotechadv.2015.04.012 [DOI] [PubMed] [Google Scholar]
- Wang S. B., Chen F., Sommerfeld M., Hu Q. (2004). Proteomic analysis of molecular response to oxidative stress by the green alga Haematococcus pluvialis (Chlorophyceae). Planta 220 17–29. 10.1007/s00425-004-1323-5 [DOI] [PubMed] [Google Scholar]
- Wasylenko T. M., Ahn W. S., Stephanopoulos G. (2015). The oxidative pentose phosphate pathway is the primary source of NADPH for lipid overproduction from glucose in Yarrowia lipolytica. Metab. Eng. 30 27–39. 10.1016/j.ymben.2015.02.007 [DOI] [PubMed] [Google Scholar]
- Wu S. G., Zhao X., Shen H. W., Wang Q. A., Zhao Z. B. K. (2011). Microbial lipid production by Rhodosporidium toruloides under sulfate-limited conditions. Bioresour. Technol. 102 1803–1807. 10.1016/j.biortech.2010.09.033 [DOI] [PubMed] [Google Scholar]
- Xie D. M., Jackson E. N., Zhu Q. (2015). Sustainable source of omega-3 eicosapentaenoic acid from metabolically engineered Yarrowia lipolytica: from fundamental research to commercial production. Appl. Microbiol. Biot. 99 1599–1610. 10.1007/s00253-014-6318-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J. Y., Zhao X. B., Wang W. C., Du W., Liu D. H. (2012). Microbial conversion of biodiesel byproduct glycerol to triacylglycerols by oleaginous yeast Rhodosporidium toruloides and the individual effect of some impurities on lipid production. Biochem. Eng. J. 65 30–36. 10.1016/j.bej.2012.04.003 [DOI] [Google Scholar]
- Yang H., He Q. N., Rong J. F., Xia L., Hu C. X. (2014). Rapid neutral lipid accumulation of the alkali-resistant oleaginous Monoraphidium dybowskii LB50 by NaCl induction. Bioresour. Technol. 172 131–137. 10.1016/j.biortech.2014.08.066 [DOI] [PubMed] [Google Scholar]
- Yang Z. K., Ma Y. H., Zheng J. W., Yang W. D., Liu J. S., Li H. Y. (2014). Proteomics to reveal metabolic network shifts towards lipid accumulation following nitrogen deprivation in the diatom Phaeodactylum tricornutum. J. Appl. Phycol. 26 73–82. 10.1007/s10811-013-0050-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. K., Niu Y. F., Ma Y. H., Xue J., Zhang M. H., Yang W. D., et al. (2013). Molecular and cellular mechanisms of neutral lipid accumulation in diatom following nitrogen deprivation. Biotechnol. Biofuels 6:67 10.1186/1754-6834-6-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeesang C., Cheirsilp B. (2011). Effect of nitrogen, salt, and iron content in the growth medium and light intensity on lipid production by microalgae isolated from freshwater sources in Thailand. Bioresour. Technol. 102 3034–3040. 10.1016/j.biortech.2010.10.013 [DOI] [PubMed] [Google Scholar]
- Yilancioglu K., Cokol M., Pastirmaci I., Erman B., Cetiner S. (2014). Oxidative stress is a mediator for increased lipid accumulation in a newly isolated Dunaliella salina strain. PLoS ONE 9:e91957 10.1371/journal.pone.0091957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q. L., Liu Z., Xu H. M., Zhang B., Zhang M., Li M. C. (2015). TiO2 nanoparticles promote the production of unsaturated fatty acids (UFAs) fighting against oxidative stress in Pichia pastoris. RSC Adv. 5 41033–41040. 10.1039/c5ra02366a [DOI] [Google Scholar]
- Yu Y., Li T., Wu N., Jiang L., Ji X., Huang H. (2017). The role of lipid droplets in Mortierella alpina aging revealed by integrative subcellular and whole-cell proteome analysis. Sci. Rep. 7:43896 10.1038/srep43896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y. D., Li T., Wu N., Ren L. J., Jiang L., Ji X. J., et al. (2016). Mechanism of arachidonic acid accumulation during aging in Mortierella alpina: a large-scale label-free comparative proteomics study. J. Agric. Food Chem. 64 9124–9134. 10.1021/acs.jafc.6b03284 [DOI] [PubMed] [Google Scholar]
- Zhang J., Wang X. L., Vikash V., Ye Q., Wu D. D., Liu Y. L., et al. (2016). ROS and ROS-mediated cellular signaling. Oxid. Med. Cell. Longe. 2016:4350965 10.1155/2016/4350965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Sun D. Z., Mao X. M., Liu J., Chen F. (2016). The crosstalk between astaxanthin, fatty acids and reactive oxygen species in heterotrophic Chlorella zofingiensis. Algal Res. 19 178–183. 10.1016/j.algal.2016.08.015 [DOI] [Google Scholar]
- Zhu J. K. (2016). Abiotic stress signaling and responses in plants. Cell 167 313–324. 10.1016/j.cell.2016.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L. D., Li Z. H., Hiltunen E. (2016). Strategies for lipid production improvement in microalgae as a biodiesel feedstock. Biomed. Res. Int. 2016:8792548 10.1155/2016/8792548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zienkiewicz K., Du Z. Y., Ma W., Vollheyde K., Benning C. (2016). Stress-induced neutral lipid biosynthesis in microalgae – molecular, cellular and physiological insights. Biochim. Biophys. Acta 1861 1269–1281. 10.1016/j.bbalip.2016.02.008 [DOI] [PubMed] [Google Scholar]