Abstract
Levodopa (L-DOPA) remains the most effective pharmacological treatment for Parkinson’s Disease (PD) but its use is limited by the development of debilitating drug-related side effects, particularly L-DOPA induced dyskinesia (LID). LID is a consequence of long-term L-DOPA use, and in model systems is characterized by a “priming effect”, whereby initial administrations of L-DOPA trigger a sensitized biochemical and transcriptional response upon subsequent dopaminergic stimulation. Preliminary studies into the mechanisms underlying this cellular memory have indicated an important role for epigenetic change but many of the downstream mechanisms remain unknown. The family of bromodomain and extraterminal (BET) proteins, which bind acetylated histones, play a critical effector role in the regulation of transcription. BET proteins have been implicated in several forms of neural plasticity, but their potential relevance to LID remains unexplored. Using the 6-OHDA rodent model of LID, we show that dyskinesia development induces alterations in BET protein expression along with enhanced occupation of sites at the promoter and enhancer regions of genes dysregulated during dyskinesia development. When BET function was blocked using the pharmacologic inhibitor JQ1, LID was prevented. In addition, we found that JQ1 treatment blocked the transcriptional upregulation of several immediate-early genes known to participate in the pathogenesis of dyskinesia. Together, these results demonstrate an essential role for BET protein activity as an epigenetic “reader” of the altered histone acetylation required for LID development and suggest that modulation of BET protein function is a potential therapeutic avenue for the treatment prevention or reversal of LID in PD.
Keywords: BET proteins, JQ1, dyskinesia, L-DOPA
Introduction
While levodopa (L-DOPA) is currently the best available treatment for Parkinson disease (PD), its therapeutic potential is limited due to a progressive narrowing of the therapeutic window and the development of motor complications from L-DOPA therapy. Levodopa-induced dyskinesia (LID) is often the most troubling of these complications, producing involuntary, choreiform movements that can be extremely disabling (Grandas et al., 1999; Fahn, 2000). Dyskinetic behaviors develop following repeated L-DOPA treatment and are usually seen during the peak of plasma L-DOPA concentrations. In clinical populations the prevalence of LID in L-DOPA-treated PD patients is nearly 50% at 5 years, and is estimated as high as 90% at 10 years (Rascol et al., 2000; Poewe, 2009). Once dyskinesias have been established they are usually persistent and recur with every L-DOPA dose, suggesting the pathophysiological release of unintended motor programs stored in the basal ganglia (Pisani et al., 2005; Belujon et al., 2010).
Biochemical and physiologic investigations have indicated that dysregulation of corticostriatal plasticity is an important mechanism underlying LID development. In animal models, there is a loss of bidirectional synaptic plasticity and sensitization in striatal MAPK/Erk signaling that causes downstream changes to CREB and AP-1 dependent transcription (Picconi et al., 2003; Santini et al., 2009; Heiman et al., 2014; Charbonnier-Beaupel et al., 2015). Examples of these alterations in cellular behavior have been found in rodent and non-human primate models following LID development, and are known to be very long-lasting, quickly reemerging following periods of prolonged treatment interruption (Bezard et al., 2001; Fahn et al., 2004; Brotchie, 2005). Although sensitization of striatal signaling is able to explain the expression of dyskinetic behaviors following L-DOPA administration, the mechanisms behind the long-term maintenance of these changes in cellular responsiveness remain unknown.
Following LID development, dysregulated transcription factor activity leads to the sustained over expression of several activity regulated immediate-early genes (IEG) essential to the formation of long-term neuronal memory (Nestler et al., 2001; West et al., 2002; Figge et al., 2013). Studies in LID animal models have indicated that the aberrant gene expression is due to sustained alterations in epigenetic modifications that control chromatin accessibility. Recent work has indicated an essential role for dynamic striatal DNA methylation in the maintenance of LID, identifying locus-specific methylation changes and the bidirectional modification of dyskinetic behaviors following global manipulations of DNA methylation (Figge et al., 2016). Alterations in histone post-translational modifications have also been observed following dyskinesia development, with animal models displaying enhancements in histone phosphorylation and acetylation in D1 neurons; however, the genes specifically effected by this chromatin remodeling remain unknown (Nicholas et al., 2008; Darmopil et al., 2009; Santini et al., 2009). Although enhancements in histone acetylation correlate to dyskinetic behaviors, attempts to modify LID development through histone deacetylase inhibitors counter-intuitively decreased dyskinetic behaviors, indicating there remains an additional layer of regulation involved in the translation of the pathologic epigenetic code (Johnston et al., 2013). While these data indicate histone acetylation as being pivotally involved in LID development, the specific loci effected and the proteins involved in its functional output in the form of pathologic gene transcription remain a mystery.
The interpretation of epigenetic marks requires a family of “reader” proteins that act as functional scaffolding for additional effector complexes to directly integrate the epigenetic code into transcriptional behavior. Members of the bromodomain and extra-terminal domain (BET) protein family, including Brd2, Brd3, Brd4, and BrdT, serve as known readers of histone acetylation, an epigenetic mark dysregulated in LID (Filippakopoulos and Knapp, 2014; Shi and Vakoc, 2014). BET proteins act to bind acetylated lysines to facilitate cellular transcription by assisting in transcription complex assembly, recruiting P-TEFb to initiate transcriptional elongation, and facilitate productive transcriptional elongation (Jang et al., 2005; Filippakopoulos and Knapp, 2014). Recent work on the mechanisms of activity-dependent neuronal plasticity has demonstrated an essential role for BET proteins as a link between the mechanisms of epigenetic regulation and stimulus-dependent neuronal transcription (West et al., 2002; Korb et al., 2015). Inhibition of BET activity has also been found sufficient to inhibit the development of hippocampal dependent memory and prevented behavioral sensitization following repeated cocaine administration (Korb et al., 2015; Sartor et al., 2015). Due to its importance in neuronal transcriptional regulation, we hypothesized that the aberrant activity of BET proteins directly contributes to the development of LID through the binding of L-DOPA dependent histone acetylation changes to contribute to the dysregulation of striatal gene expression. Here we show that the aberrant activity of BET proteins in the dorsal striatum is necessary for the development of dyskinetic behaviors and provide a critical link between the long-term transcriptional sensitization and epigenetic dysregulation underlying LID.
Methods
Animals
Male Sprague-Dawley rats, approximately 60–90 d old and weighing 180–200 g, were housed in pairs in plastic cages in an AAALAC-approved animal care facility on a 12 h light/dark cycle with food and water available ad libitum. All procedures were performed in accordance with the University of Alabama at Birmingham Institutional Animal Care and Use Committee.
Surgical procedures
One week after arrival, rats were given a unilateral dopamine lesion to the left medial forebrain bundle. Prior to surgery, rats were given injections of desipramine (25 mg/kg, ip) to protect norepinephrine neurons, and buprenorphine (0.03 mg/kg, ip) as pre-emptive analgesia. Animals were then anaesthetized with 1–2% isoflurane (Baxter Healthcare Corp., Deerfield, IL, USA) mixed with oxygen (2 L/min) and placed in a stereotaxic apparatus (Kopf Instruments). Under aseptic conditions, rats were rendered hemi-parkinsonian using unilateral injections of 6-OHDA (12 ug in 4 uL, Sigma) into the medial forebrain bundle (stereotaxic coordinates: anteroposterior, +1.8 mm from bregma, ±2 mm lateral from midline, and −8.6 mm from the dura). Animals were given at least 7 d of recovery, during which they received buprenorphine and wound care for pain management.
Pharmacology
Daily injections commenced three weeks after lesioning with either vehicle or L-DOPA (6 mg/kg + benserazide, 15 mg/kg, sc) for 7 consecutive days. This 7-day paradigm ensures that dyskinesia is displayed on the first day of treatment in most rats and that stable dyskinesia is developed by day 7. Animals were sacrificed after completion of the day 7 behavioral sessions and 3 hours following the last daily injection by rapid decapitation. Brains were immediately removed, the dorsal striatum dissected, flash frozen on dry ice and stored at −80°C.
For JQ1 (ApexBio) experiments, animals were injected intraperitoneal with 25 mg/kg JQ1 dissolved in 10% DMSO and 10% 2-hydroxypropyl-β-cyclodextrin in sterile PBS daily for 14 d (Korb et al., 2015; Sartor et al., 2015). This treatment paradigm included 7 days prior to and concurrently with L-DOPA (8 mg/kg, a higher dose which provides a better test of blockade by JQ1) treatment, with JQ1 administration immediately prior to L-DOPA injection. Animals were euthanized 30 mins following the 8th day of L-DOPA administration in order to assess JQ1’s effect on immediate-early gene expression. Brains were immediately removed, the dorsal striatum dissected, flash frozen on dry ice and stored at −80°C.
Behavioral Testing
Two weeks after lesion, the forepaw adjusting steps test was performed for use as a behavioral correlate of unilateral dopamine depletion (Chang et al., 1999). Rats were held such that they had only one free forelimb; for each trial, rats were moved laterally across a table at a steady rate of 90 cm/10 s. Each stepping test consisted of 6 trials for each forepaw, alternating between forehand and backhand. To create a “percent intact” stepping score, the total number of steps with the lesioned forelimb were divided them by total number of steps with the intact forelimb and multiplied by 100. Lower scores indicate greater parkinsonian impairment. Percent intact scores were used to rank-order the rats according to degree of impairment and assign them to 2 equivalently-impaired treatment groups for all behavioral experiments.
The abnormal involuntary movements (AIMs) test is a metric of dyskinesia. Rats were assessed for AIMs using a procedure as previously described (Figge et al., 2016). Following treatment with vehicle or L-DOPA, rats were placed in clear, plastic cages and rated by a trained observer blind to drug treatment for 1 min every 10 min over a 180 min period. Individual dyskinesia severity scores ranging from 0 (not present) to 4 (severe and not interruptible) were given for axial, limb, orolingual, and rotational dyskinesias. The AIMs subscores were summed to create a single AIMs score for data analysis. Rotational behavior was assessed by the number of rotations made contralateral to the lesioned side during the 1 min observation period.
RNA Isolation and qRT-PCR
For all experiments, flash frozen samples were then homogenized in RNA Stat 60 (Amsbio, UK) and processed according to manufacturer’s instructions. RNA was purified with RNAeasy Mini columns (Qiagen, CA, USA) and reverse transcribed using an iScript Kit (BioRad, CA, USA). Specific primers were designed to amplify appropriate cDNA regions depending upon the target of interest (primer sequences available upon request). q-PCR amplifications were performed in triplicate using an CFX96 real-time PCR system (Bio-Rad) at 95°C for 5 min, followed by 40 cycles of 95°C for 15 s and 60°C for 45 s, and then incubation at 72°C for 10 min followed by real-time melt analysis to verify product specificity. Hprt was used as an internal control for normalization using the ΔΔCt method (Livak and Schmittgen, 2001).
Chromatin Immunoprecipitation
For ChIP experiments striatal tissue was minced and then cross-linked in 1% formaldehyde for 10 min. The reaction was quenched for 5 minutes with 1.25 m glycine and then washed three times in cold PBS with protease inhibitors. Samples were homogenized in hypotonic lysis buffer (250mm Sucrose, 50mm Tris-HCL, 25mm Kcl, 1x protease inhibitors) using a BeadRuptor5 on power 5 for 30 seconds. The samples were pelleted, and the supernatant was transferred to new tubes; 500 μl of nuclear lysis buffer (50mm Tris, pH 8.0, 10mm 500mm EDTA, 1% SDS, 1× protease inhibitor mixture) was added, and 100 uL of each chromatin sample was sheared using the Bioruptor Pico (Diagenode) 6 times (30s ON, 30s off) to obtain 300- to 600-bp fragments. The quality of DNA shearing was assessed by running 600 ng of sheared DNA in a 1% agarose gel. Cellular debris were pelleted, and the supernatants were transferred to new tubes; 5 μl of each sample was saved to run as an input control. Immunoprecipitation was performed using 5 μg of ChIP validated rabbit antibody bound to 20 μL of protein A–coated Dynabeads (Invitrogen) in ChIP dilution buffer (1% Triton X- 100, 2mM EDTA, 20mm Tris-HCl, pH 8.1, 150mm NaCl). Antibodies used were Brd2 (Bethyl Labs, A302-583A), Brd4 (Bethyl Labs, A301-985A), H3K14ac (Active motif, 39697), pS10AcH3K14 (Millipore, 07-081), and rabbit IgG (Bethyl Labs, P120-101). Antibody bead mixtures were incubated with the sheared chromatin overnight (~16 h) at 4°C on rotation and then washed once with 800 μl of the following solutions (in order): low salt (0.1% SDS, 1% Triton X-100, 2mM EDTA, 20mM Tris-HCl, pH 8.1, 150mM NaCl), high salt (0.1% SDS, 1% Triton X-100, 2mM EDTA, 20mM Tris-HCl, pH 8.1, 500mM NaCl.), LiCl wash buffer (0.25 m LiCl, 1% NP-40, 1% sodium deoxycholate, 1mm EDTA, 10mm Tris-HCl, pH 8.1), and TE buffer (10mm Tris-HCl, pH 7.5, 1mm EDTA). The beads were then incubated for 2 h at 60°C with proteinase K in TE buffer with 1% SDS and then incubated at 95°C for 10 min to deactivate all enzymes. DNA was then extracted using the QIAquick PCR Purification Kit (QIAGEN) according to the manufacturer’s instructions.
For ChIP samples, previously validated primers at regions showing dynamic DNA methylation were used to amplify the promoter regions of Dab1, Ntrk2, Fos, and Esr1, and enhancer regions of Arc, Nedd4l, and FosB (Massart et al., 2015; Figge et al., 2016). DNA regions was assayed via qPCR using SYBR Green as described earlier to assess for enrichment of the specfied histone marks. Ct values for IP samples were normalized to unprocessed (input) DNA.
Statistical analysis
Data analysis was performed and graphed in GraphPad Prism for Mac OS X (version 6.00; GraphPad Sofware). Statistical significance was designated at p<0.05 for all analyses. Statistical significance was measured using two-way ANOVA or Student’s unpaired t-test as indicated for all biochemical results. For ANOVAs, Tukey’s post hoc comparisons were performed when appropriate. Repeated-measures two-way ANOVAs were used for all behavioral analysis and followed by Bonferonni’s post-hoc test when indicated. Data are presented as mean ± SEM.
Results
Chronic L-DOPA Treatment Leads to the Aberrant Transcriptional Expression of BET Domain Containing Proteins
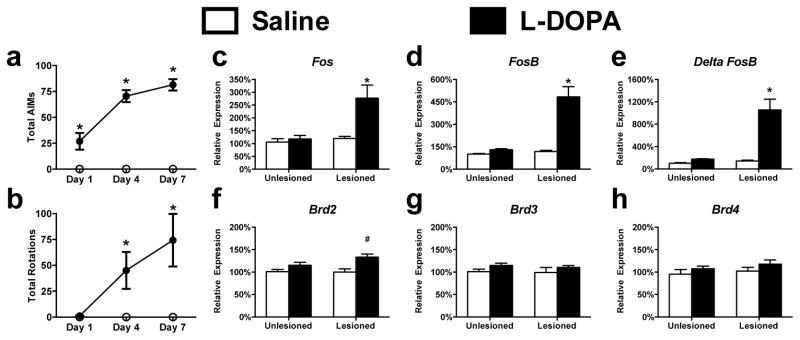
To examine the role of BET containing proteins in LID, we unilaterally lesioned the nigrostriatal dopaminergic pathway by the injection of 6-OHDA into the medial forebrain bundle to render rats hemi-parkinsonian. Animals (n=7–8 per group) were allowed to recover for three weeks and then treated with L-DOPA for 7 consecutive days leading to a robust behavioral response with the induction of abnormal involuntary movements (assessed by AIMs score) as well as a progressive sensitization of rotational behavior (Fig 1A/B) (Time x Treatment interaction, 1A: F(2, 26) = 56.78, p < 0.0001; 1B: F (2, 26) = 9.271, p < 0.0009; Bonferroni’s post hoc tests). Following the conclusion of behavioral testing on the seventh day and three hours post L-DOPA administration, RNA was collected from the dorsal striatum and we confirmed the transcriptional sensitization of Fos, FosB, and ΔFosB in the dopamine-depleted striatum following repeated L-DOPA treatment (Fig 1C–E) (Lesion x Treatment interaction, Fos: F(1, 25) = 7.114, p = 0.0132; FosB: F(1, 25) = 24.92, p < 0.0001; ΔFosB: F(1, 25) = 20.78, p = 0.0001; Tukey’s post hoc tests). To determine if there were alterations in the transcriptional expression of BET containing proteins following dyskinesia development, we measured the mRNA levels of several BET proteins: Brd2, Brd3, and Brd4 (Fig 1F–H). We found significant enhancements in Brd2 (L-DOPA dependent effect, Brd2: F(1, 25) = 12.41, p = 0.0017, Tukey’s Post-Hoc) a result consistent with previous studies using genome-wide approaches (Heiman et al., 2014). Along with the known alterations in histone acetylation, the aberrant expression of a BET protein in the context of dyskinesia indicates a potential role for this epigenetic reader in the mechanisms associated with the transcriptional sensitization underlying LID development.
Figure 1. Aberrant expression of BET Protein isoforms following LID development.
(a/b) Repeated L-DOPA administration induces a sensitized dyskinetic response as measured by ALO score (a) or rotational behavior (b) (Repeated-measures 2-way ANOVA; Bonferroni’s post hoc tests, *p<0.05 vs. Saline treatment; n=7–8 per group). (c–e) L-DOPA induces the sensitized transcriptional expression of immediate-early genes 3 hours following treatment. (f–h) mRNA expressional profile for three BET proteins 3 hrs following L-DOPA administration in dyskinetic animals (2-way ANOVA; Tukey’s post hoc tests; n=6–7 per group). Error bars represent SEM. *p<0.05 for interaction between lesion and treatment, #p<0.05 vs both Saline treated groups
LID Development Leads to Aberrant Function of BET Proteins on a Locus-Specific Level
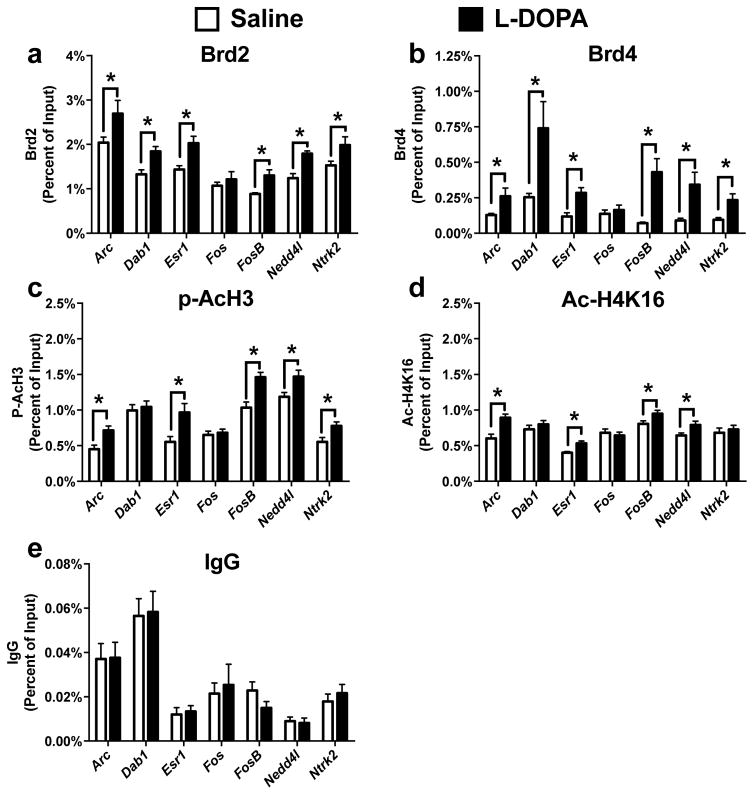
To directly determine if there are alterations in dorsal striatal BET function, we used ChIP to measure the levels of Brd2 and Brd4 bound to specific regions of DNA following the development of dyskinesia. We chose to target the promoter and enhancer regions of genes known to be aberrantly expressed following LID development and which have also been previously shown to exhibit dynamic DNA methylation (Heiman et al., 2014; Charbonnier-Beaupel et al., 2015; Massart et al., 2015; Figge et al., 2016). We found that he promoter regions of Dab1, Esr1, and Ntrk2 exhibited enhancements in both Brd2 and Brd4 binding in an L-DOPA dependent manner (Fig 2A, Unpaired t-test, Dab1: p = 0.007; Esr1: p = 0.004; Ntrk2: p = 0.034) (Fig 2B, Unpaired t-test, Dab1: p = 0.012; Esr1: p = 0.004; Ntrk2: p = 0.006). Similarly, predicted enhancer regions of Arc, FosB, and Nedd4l also showed increases in the association of Brd2 and Brd4 following repeated L-DOPA administration (Fig 2A, Unpaired t-test, Arc: p = 0.038; FosB: p = 0.002; Nedd4l: p = 0.002) (Fig 2B, Arc: p = 0.022; FosB: p = 0.001; Nedd4l: p = 0.008). Together, these data show that locus-specific changes to BET protein DNA binding are dependent upon repeated L-DOPA administration in the dopamine deficient striatum and implicate alterations in the mechanisms of epigenetic “readers” as contributing to the development of LID.
Figure 2.
BET protein function is aberrantly regulated following LID development. (a/b) Using ChIP for Brd2 (a) and Brd4 (b) finds increased binding 1 hr following L-DOPA administration at regions near genes aberrantly transcribed following dyskinesia development (c/d) L-DOPA treatment increases histone marks associated with active transcription in a locus dependent manner at genes aberrantly transcribed following L-DOPA administration in dyskinetic animals.. (e) L-DOPA treatment had no effect on IgG enrichment at multiple regions. (Unpaired t-test; n=6–7 per group) Error bars represent SEM. *p<0.05 for treatment effect
As BET proteins are known to read acetylation patterns on histones, we next sought to investigate whether any of the regions displaying dynamic changes to BET binding also displayed associative changes in histone acetylation. LID is known to induce the phospho (Ser10)-acetylation (K16) of histone 3 (pAcH3), a mark of active transcription, in D1-receptor expressing neurons following acute L-DOPA administration in dyskinetic animals; the specific loci effected, however, have been unknown up to this point (Darmopil et al., 2009; Santini et al., 2009). We found that L-DOPA administration increased pAcH3 at the majority of regions showing altered Brd2 and Brd4 occupancy (Fig 2C, Unpaired t-test, Arc: p = 0.008; Esr1: p = 0.016; FosB: p = 0.002; Nedd4l: p = 0.021; Ntrk2: p = 0.024). Acetylated H4K16 (AcH4K16) is known to be a more stable histone acetylation mark that is also associated with actively transcribed genes and BET protein binding in neurons (Korb et al., 2015). Interestingly, we also found several enhancements in AcH4K16 at regions displaying increased BET protein binding (Fig 2D, Unpaired t-test, Arc: p = 0.006; Esr1: p = 0.003; FosB: p = 0.044; Nedd4l: p = 0.036). These findings indicate that the enhanced BET protein occupancy is directly associated with LID dependent alterations in histone acetylation.
Inhibition of BET Protein Function Modulates LID Expression and Development
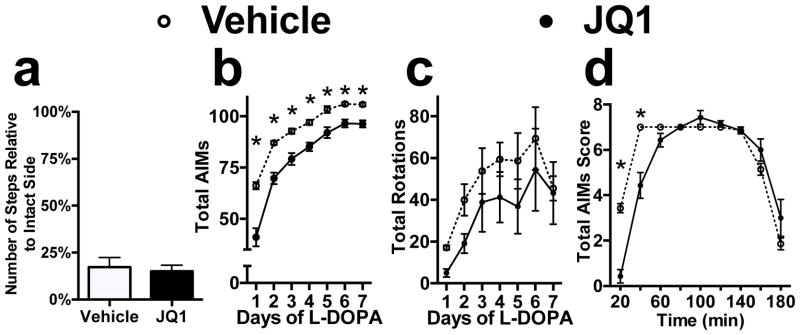
Collectively, these data support the idea that the dynamic changes in histone acetylation following LID development lead to alterations in BET protein genomic association that are necessary for the transcriptional dysfunction underlying dyskinesia expression. To determine if BET protein function is required for LID development and expression, we administered JQ1, a selective competitive inhibitor of BET protein’s histone acetylation binding domain, one week prior to and concurrently with L-DOPA treatment in a new cohort of animals. This treatment paradigm was based on prior studies in which JQ1 was shown to effect long-term memory formation (Filippakopoulos et al., 2010; Shi and Vakoc, 2014). We found that chronic JQ1 administration using this strategy suppressed the induction of LID and was able to decrease the maximum level of stable dyskinesia following repeated L-DOPA treatment (Time x Treatment interaction, Fig 3B, F(6, 72) = 8.897, p < 0.0001, Bonferroni’s post hoc). This behavioral effect was not due to differences in the level of dopaminergic denervation or alterations in L-DOPA efficacy, as each group had an equal level of motor impairment prior to treatment and JQ1 was found to have no effect on the L-DOPA dependent sensitization of rotational behavior (Fig 3A, unpaired t-test, p = 0.7272, Fig 3C, drug-dependent effect, F(1, 12) = 1.223, p = 0.2904).
Figure 3.
Chronic Inhibition of BET protein function inhibits the development of LID. (a) 6-OHDA lesion led to equivalent behavioral deficits in both treatment groups as measured by the forelimb adjusting steps test. (b/c) Treatment with JQ1 for one week prior and concurrently with L-DOPA prevents sensitization of AIM scores (b) with no effect on rotational behavior (c). (d) Time profile of AIM scores on day 7 of L-DOPA treatment when measured every twenty minutes (Repeated-measures 2-way ANOVA; Bonferroni’s post hoc tests, *p<0.05 vs. Vehicle; n=7 per group) Error bars represent SEM.
Inhibition of BET Protein Function Inhibits BET Protein Genomic Occupation and LID-related Transcriptonal Sensitization
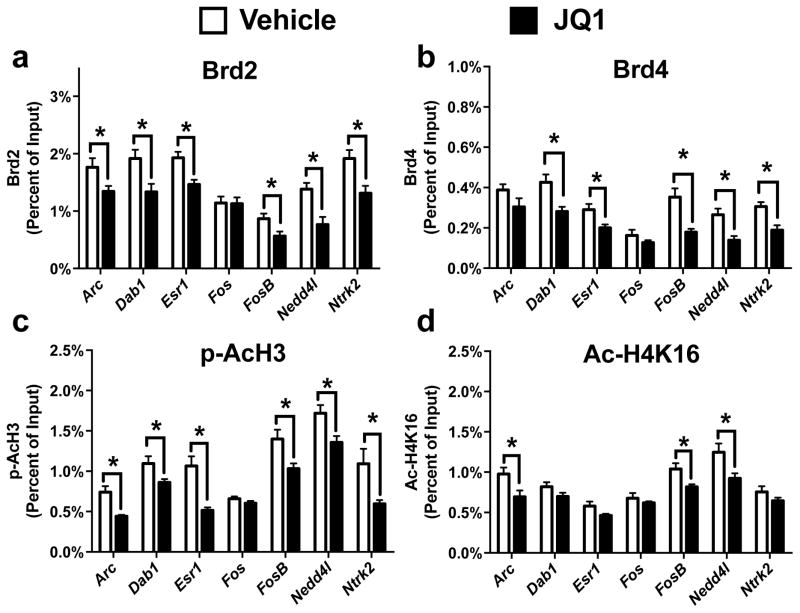
To evaluate JQ1’s effect on BET protein function, we then used ChIP to determine if our previously identified LID dependent changes in Brd2 and Brd4 DNA binding were inhibited by the JQ1 treatment paradigm. We found that JQ1 treatment occluded the Brd2 and Brd4 DNA binding enhancements at all of the locations we previously found to display dynamic changes following LID development (Fig 4a, Unpaired t-test, Arc: p = 0.048; Dab1: p = 0.018; Esr1: p = 0.006, FosB: p = 0.031; Nedd4l: p = 0.005; Ntrk2: p = 0.012) (Fig 4b, Unpaired t-test, Dab1: p = 0.011; Esr1: p = 0.026, FosB: p = 0.004; Nedd4l: p = 0.007; Ntrk2: p = 0.005). Interestingly, the inhibition of BET protein function also prevented several of the increases in pAcH3 and AcH4K16 we previously identified (Fig 4c, Unpaired t-test, Arc: p = 0.002; Dab1: p = 0.034; Esr1: p = 0.0006, FosB: p = 0.015; Nedd4l: p = 0.016; Ntrk2: p = 0.017) (Fig 4d, Unpaired t-test, Arc: p = 0.028; FosB: p = 0.019; Nedd4l: p = 0.029). Although JQ1 selectively inhibits bromodomain activity, in the context of repeated L-DOPA exposure and JQ1 treatment, our data indicates that BET protein activity is necessary for the alterations in cellular signaling leading to enhanced phosphoacetylation.
Figure 4.
Chronic Inhibition of BET protein function prevented enhancements in BET protein genomic occupation. (a/b) ChIP revealed that treatment with JQ1 blocked Brd2 (a) and Brd4 (b) enhancements 30 mins following L-DOPA treatment (c/d) Repeated JQ1 treatment blocked the LID dependent effects on histone H3 phosphoacetylation (c) and histone H4K16 acetylation (Unpaired t-test; n=6–7 per group). Error bars represent SEM. *p<0.05 for treatment effect.
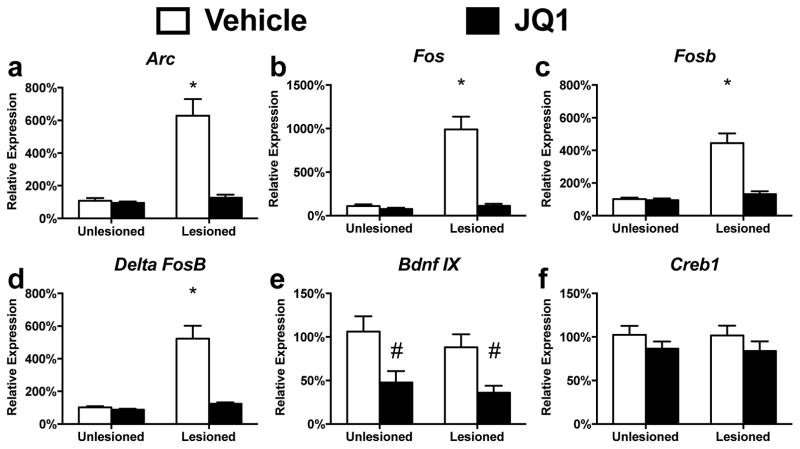
To determine if the alterations in BET protein genomic occupancy had an effect on the transcriptional sensitization central to LID, we measured the expression of several genes known to be transcriptionally sensitized following LID development (Bezard et al., 2001; Corvol et al., 2004; Crittenden et al., 2009; Nadjar et al., 2009; Heiman et al., 2014). Chronic administration of JQ1 prevented the enhanced expression of Arc, Fos, FosB and ΔFosB observed in dyskinetic animals following L-DOPA adminstration. JQ1s effects were found to selectively prevent the L-DOPA dependent transcriptional sensitization, with no effect on their baseline expression patterns (Fig 5, Lesion x Treatment effect, Arc: F(1, 20) = 21.96, p = 0.000; Fos: F(1, 20) = 31.59, p = 0.0132; FosB: F(1, 20) = 23.19, p < 0.0001; ΔFosB: F(1, 20) = 23.31, p = 0.0001; Tukey’s post hoc tests). Similar to previously published reports on BET protein inhibition in the striatum, we also found a JQ1 dependent decrease in BDNF IX expression, another pathway linked to corticostriatal plasticity and LID (JQ1 Treatment effect, BDNFIX: F(1, 16) = 16.189, p = 0.001) (Foltynie et al., 2009; Sartor et al., 2015).
Figure 5.
Chronic Inhibition of BET protein function prevents the transcriptional sensitization of immediate early genes following LID development (a–d) Thirty minutes following the last L-DOPA treatment, JQ1 is shown to inhibit the transcriptional sensitization of immediate early genes aberrantly transcribed following LID development (e) Repeated JQ1 treatment decreased the expression of BDNF exon 9 as previously reported (Sartor et al., 2015) (f) Chronic JQ1 treatment had no effect on Creb expression. Error bars represent SEM. (2-way ANOVA; Tukey’s post hoc tests; n=6–7 per group). Error bars represent SEM. *p<0.05 for interaction between lesion and treatment, #p<0.05 vs both Vehicle treated groups
Discussion
Our results demonstrate an essential role for BET proteins in the development and expression of LID. We observed an enhancement in the transcriptional expression of Brd2 following the development of dyskinetic behaviors that correlated with increases in locus-specific Brd2 and Brd4 binding in the dorsal striatum. The changes in BET protein chromatin association were found at both the promoter and enhancer regions of Arc, Fos, and FosB, genes known to be aberrantly transcribed following L-DOPA administration, and were associated with enhancements in histone acetylation at many of these same regions. We found that using JQ1 to block the histone acetylation binding domain of BET proteins hindered dyskinesia development, reduced BET protein chromatin binding, prevented L-DOPA-induced histone acetylation changes, and reversed the aberrant expression of several genes dysregulated in LID. Collectively, these data indicate that the reader functions of BET proteins have a critical role in the development and maintenance of dyskinesia.
During LID development, the dysregulation of L-DOPA dependent gene expression is known to underlie the aberrant corticostriatal plasticity necessary for the expression of dyskinetic behaviors. Although several pieces of evidence have indicated that epigenetic remodeling directly contributes to the pathologic transcription in LID, the cellular mechanisms responsible for actually translating this molecular code into aberrant transcriptional behavior have been unclear (Nicholas et al., 2008; Darmopil et al., 2009; Santini et al., 2009; Johnston et al., 2013). The BET family of proteins is known to regulate transcriptional initiation by binding the acetylated N-terminal tails of histones to assist in transcription complex assembly and initiation (West et al., 2002; Jang et al., 2005; Yang et al., 2005; Wu et al., 2013; Korb et al., 2015). Our data indicates that the dysregulation of BET protein function is required for LID induction and that this happens in a stimulus dependent manner selectively following striatal dopamine denervation (Fig 2/5). This change in BET protein function causes locus-specific alterations in BET protein binding essential to the transcriptional dysfunction observed in LID, and intricately linked to the dynamic regulation of histone acetylation necessary for LID development (Fig 2/4). BET proteins do not, however, appear to participate directly in regulating motor behavior, as we did not observe any direct effect of JQ1 on the motor efficacy of L-DOPA as measured by rotational behavior. This is consistent with several previous studies, which have shown JQ1 to have no effect on a variety of motor measures including open-field activity and cocaine induced locomotor sensitization, however, future investigations ensuring BET inhibition has no effect on L-DOPA efficacy will be necessary to therapeutic development (Korb et al., 2015; Sartor et al., 2015).
BET proteins are known to interact with multiple regions of the genome. For example, we observed that JQ1 modified Fos transcriptional expression, but ChIP targeted to it’s known promoter regions showed that BET protein binding at these sites was not altered (Fig 4/5). These results most likely reflect BET protein modulating genomic enhancer regions, rather than promoters, and is consistent with the locus-specific changes in DNA methylation at both promoter and enhancer regions which we have described previously (Figge et al., 2016).
Although the aberrant expression of Brd2 provides a potential explanation for the enhancements in BET protein chromatin associations, the post-translational regulation of BET proteins has been shown necessary for their appropriate genomic localization and effect on transcription (Wu et al., 2013). In other models of long-term neuronal memory, BET protein function is controlled by the activity-dependent regulation of casein kinase 2 (CK2), a protein whose dysregulation has been previously implicated in LID development (Svenningsson et al., 2004; Santini et al., 2007; Korb et al., 2015). As our data indicates that the BET proteins may be a critical link between the epigenetic dysregulation and abnormal synaptic plasticity found in LID, future studies determining the post-translational regulators of BET protein function during dyskinesia development could lead to a deeper understanding of the molecular mechanisms behind the aberrant transcription necessary for dyskinesia expression opening new avenues for therapeutic intervention.
The striatum consists of two parallel motor circuits that have antagonistic effects on motor behavior, direct pathway neurons activated through D1 receptors that facilitate motor behaviors and indirect pathway neurons inhibited by D2 receptor activity that prevent motor cortex activity (Albin et al., 1989; Gradinaru et al., 2009). The dorsal striatum is a heterogenous cellular structure that contains both of these neuronal populations, yet previous work on LID has identified D1-receptor expressing neurons as the primary site of biochemical sensitization, altered gene expression, and modified histone acetylation following LID (Darmopil et al., 2009; Fieblinger et al., 2014; Heiman et al., 2014). Although our data is derived from dorsal striatal samples containing both D1 and D2 neurons, we found that many of the regions of the genome displaying differential histone acetylation and BET protein binding were near genes known to be aberrantly transcribed exclusively in D1-expressing neurons. Additionally, studies conducting transcriptional profiling following LID development have also shown enhancements in Brd2 expression selectively in D1-expressing neurons (Heiman et al., 2014). The dorsal striatum does contain several different populations of glial cells, but recent studies in multiple models of experience dependent plasticity have observed minimal epigenetic change in non-neuronal cell populations (Halder et al., 2016). Together, these data suggest that the alterations in BET protein function and the consequence of JQ1 treatment observed in this study are likely due to effects on D1 expressing direct pathway dorsal striatal neurons. Future experiments using cellular profiling techniques will help to identify the contribution of different striatal cellular populations to the epigenetic changes necessary for dyskinesia development and clarify their role in the aberrant corticostriatal plasticity.
Investigations of the molecular mechanisms behind LID have found several alterations in histone post-translational modifications, including enhanced histone phosphorylation and acetylation. However, administration of a histone deacetylase inhibitor, a treatment that should enhance histone acetylation and therefore increase dyskinesia, counter-intuitively blocked dyskinetic behaviors in non-human primates suggesting direct alterations to the epigenetic code could lead to unpredicted cellular effects (Johnston et al., 2013). The ability to target the downstream mechanisms of chromatin reorganization would create an opportunity for interventions directly blocking the interpretation of the pre-existing epigenetic modifications that enhance transcription. Our data suggests that interventions inhibiting the effector proteins binding the altered histone acetylation pattern following LID development are sufficient to prevent the induction of dyskinetic behaviors (Fig 3).
Although JQ1 itself is unlikely to be clinically useful due to off-target effects at the tachykinin and adenosine A3 receptor as well as alterations to other forms of behavioral memory, the further study of this molecular mechanism could lead to targeted therapies for LID. Currently, BET protein inhibitors with enhanced specificity are in human clinical trials for the treatment of multiple diseases. Future studies attempting to determine if the inhibition of these mechanisms are capable of preventing the sensitization of dyskinetic behaviors or reversing LID in the context of stable dyskinesia will provide valuable insight into the therapeutic potential of this class of molecules. Additionally, the recent advent of locus-specific chromatin remodeling with the Crispr-Cas9 system would allow for targeted approaches to reverse the dysregulated epigenetic mechanisms underlying LID in the future (Gilbert et al., 2013; Zalatan et al., 2015).
In summary, our results reveal an essential role for BET proteins in the establishment of LID and demonstrate that manipulation of their function can modify the development and expression of dyskinetic behaviors. These findings provide further evidence that the transcriptional dysfunction necessary for the long-term maintenance of LID is strongly associated with the reorganization of dorsal striatal chromatin. These results suggest modulation of epigenetic readers, either globally or selectively, could be a useful therapeutic approach for the improvement of L-DOPA therapy in PD patients that requires further investigation.
Highlights.
BET proteins, which read epigenetic marks, are key regulators of neural plasticity
L-DOPA treatment in a 6-OHDA model alters BET gene expression and chromatin binding
Pharmacologic inhibition of BET proteins prevents L-DOPA induced dyskinetic behaviors
Acknowledgments
We thank K. Eskow Jaunarajs and J. Randolph for guidance and technical assistance. This work was supported by the Alacare Mary Sue Beard Pre-doctoral Fellowship, the American Parkinson Disease Association, and NINDS grant F31NS090641 (D.A.F.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albin RL, Young AB, Penney JB. The functional anatomy of basal ganglia disorders. Trends Neurosci. 1989;12:366–375. doi: 10.1016/0166-2236(89)90074-x. [DOI] [PubMed] [Google Scholar]
- Belujon P, Lodge DJ, Grace AA. Aberrant striatal plasticity is specifically associated with dyskinesia following levodopa treatment. Mov Disord. 2010;25:1568–1576. doi: 10.1002/mds.23245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezard E, Brotchie JM, Gross CE. Pathophysiology of levodopa-induced dyskinesia: potential for new therapies. Nat Rev Neurosci. 2001;2:577–588. doi: 10.1038/35086062. [DOI] [PubMed] [Google Scholar]
- Brotchie JM. Nondopaminergic mechanisms in levodopa-induced dyskinesia. Mov Disord. 2005;20:919–931. doi: 10.1002/mds.20612. [DOI] [PubMed] [Google Scholar]
- Chang JW, Wachtel SR, Young D, Kang UJ. Biochemical and anatomical characterization of forepaw adjusting steps in rat models of Parkinson’s disease: studies on medial forebrain bundle and striatal lesions. Neuroscience. 1999;88:617–628. doi: 10.1016/s0306-4522(98)00217-6. [DOI] [PubMed] [Google Scholar]
- Charbonnier-Beaupel F, Malerbi M, Alcacer C, Tahiri K, Carpentier W, Wang C, During M, Xu D, Worley PF, Girault JA, Herve D, Corvol JC. Gene expression analyses identify Narp contribution in the development of L-DOPA-induced dyskinesia. J Neurosci. 2015;35:96–111. doi: 10.1523/JNEUROSCI.5231-13.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corvol JC, Muriel MP, Valjent E, Feger J, Hanoun N, Girault JA, Hirsch EC, Herve D. Persistent increase in olfactory type G-protein alpha subunit levels may underlie D1 receptor functional hypersensitivity in Parkinson disease. J Neurosci. 2004;24:7007–7014. doi: 10.1523/JNEUROSCI.0676-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden JR, Cantuti-Castelvetri I, Saka E, Keller-McGandy CE, Hernandez LF, Kett LR, Young AB, Standaert DG, Graybiel AM. Dysregulation of CalDAG-GEFI and CalDAG-GEFII predicts the severity of motor side-effects induced by anti-parkinsonian therapy. Proc Natl Acad Sci U S A. 2009;106:2892–2896. doi: 10.1073/pnas.0812822106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmopil S, Martin AB, De Diego IR, Ares S, Moratalla R. Genetic inactivation of dopamine D1 but not D2 receptors inhibits L-DOPA-induced dyskinesia and histone activation. Biol Psychiatry. 2009;66:603–613. doi: 10.1016/j.biopsych.2009.04.025. [DOI] [PubMed] [Google Scholar]
- Fahn S. The spectrum of levodopa-induced dyskinesias. Ann Neurol. 2000;47:S2–9. discussion S9–11. [PubMed] [Google Scholar]
- Fahn S, Oakes D, Shoulson I, Kieburtz K, Rudolph A, Lang A, Olanow CW, Tanner C, Marek K Parkinson Study G. Levodopa and the progression of Parkinson’s disease. N Engl J Med. 2004;351:2498–2508. doi: 10.1056/NEJMoa033447. [DOI] [PubMed] [Google Scholar]
- Fieblinger T, Graves SM, Sebel LE, Alcacer C, Plotkin JL, Gertler TS, Chan CS, Heiman M, Greengard P, Cenci MA, Surmeier DJ. Cell type-specific plasticity of striatal projection neurons in parkinsonism and L-DOPA-induced dyskinesia. Nat Commun. 2014;5:5316. doi: 10.1038/ncomms6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figge DA, Eskow Jaunarajs KL, Standaert DG. Dynamic DNA Methylation Regulates Levodopa-Induced Dyskinesia. J Neurosci. 2016;36:6514–6524. doi: 10.1523/JNEUROSCI.0683-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figge DA, Rahman I, Dougherty PJ, Rademacher DJ. Retrieval of contextual memories increases activity-regulated cytoskeleton-associated protein in the amygdala and hippocampus. Brain structure & function. 2013;218:1177–1196. doi: 10.1007/s00429-012-0453-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Knapp S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nat Rev Drug Discov. 2014;13:337–356. doi: 10.1038/nrd4286. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltynie T, Cheeran B, Williams-Gray CH, Edwards MJ, Schneider SA, Weinberger D, Rothwell JC, Barker RA, Bhatia KP. BDNF val66met influences time to onset of levodopa induced dyskinesia in Parkinson’s disease. Journal of neurology, neurosurgery, and psychiatry. 2009;80:141–144. doi: 10.1136/jnnp.2008.154294. [DOI] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradinaru V, Mogri M, Thompson KR, Henderson JM, Deisseroth K. Optical deconstruction of parkinsonian neural circuitry. Science. 2009;324:354–359. doi: 10.1126/science.1167093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandas F, Galiano ML, Tabernero C. Risk factors for levodopa-induced dyskinesias in Parkinson’s disease. J Neurol. 1999;246:1127–1133. doi: 10.1007/s004150050530. [DOI] [PubMed] [Google Scholar]
- Halder R, Hennion M, Vidal RO, Shomroni O, Rahman RU, Rajput A, Centeno TP, van Bebber F, Capece V, Vizcaino JC, Schuetz AL, Burkhardt S, Benito E, Sala MN, Javan SB, Haass C, Schmid B, Fischer A, Bonn S. DNA methylation changes in plasticity genes accompany the formation and maintenance of memory. Nat Neurosci. 2016;19:102–110. doi: 10.1038/nn.4194. [DOI] [PubMed] [Google Scholar]
- Heiman M, Heilbut A, Francardo V, Kulicke R, Fenster RJ, Kolaczyk ED, Mesirov JP, Surmeier DJ, Cenci MA, Greengard P. Molecular adaptations of striatal spiny projection neurons during levodopa-induced dyskinesia. Proc Natl Acad Sci U S A. 2014;111:4578–4583. doi: 10.1073/pnas.1401819111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Johnston TH, Huot P, Damude S, Fox SH, Jones SW, Rusche JR, Brotchie JM. RGFP109, a histone deacetylase inhibitor attenuates L-DOPA-induced dyskinesia in the MPTP-lesioned marmoset: a proof-of-concept study. Parkinsonism Relat Disord. 2013;19:260–264. doi: 10.1016/j.parkreldis.2012.07.001. [DOI] [PubMed] [Google Scholar]
- Korb E, Herre M, Zucker-Scharff I, Darnell RB, Allis CD. BET protein Brd4 activates transcription in neurons and BET inhibitor Jq1 blocks memory in mice. Nat Neurosci. 2015;18:1464–1473. doi: 10.1038/nn.4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Massart R, Barnea R, Dikshtein Y, Suderman M, Meir O, Hallett M, Kennedy P, Nestler EJ, Szyf M, Yadid G. Role of DNA methylation in the nucleus accumbens in incubation of cocaine craving. J Neurosci. 2015;35:8042–8058. doi: 10.1523/JNEUROSCI.3053-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadjar A, Gerfen CR, Bezard E. Priming for l-dopa-induced dyskinesia in Parkinson’s disease: a feature inherent to the treatment or the disease? Prog Neurobiol. 2009;87:1–9. doi: 10.1016/j.pneurobio.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ, Barrot M, Self DW. DeltaFosB: a sustained molecular switch for addiction. Proc Natl Acad Sci U S A. 2001;98:11042–11046. doi: 10.1073/pnas.191352698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas AP, Lubin FD, Hallett PJ, Vattem P, Ravenscroft P, Bezard E, Zhou S, Fox SH, Brotchie JM, Sweatt JD, Standaert DG. Striatal histone modifications in models of levodopa-induced dyskinesia. J Neurochem. 2008;106:486–494. doi: 10.1111/j.1471-4159.2008.05417.x. [DOI] [PubMed] [Google Scholar]
- Picconi B, Centonze D, Hakansson K, Bernardi G, Greengard P, Fisone G, Cenci MA, Calabresi P. Loss of bidirectional striatal synaptic plasticity in L-DOPA-induced dyskinesia. Nat Neurosci. 2003;6:501–506. doi: 10.1038/nn1040. [DOI] [PubMed] [Google Scholar]
- Pisani A, Centonze D, Bernardi G, Calabresi P. Striatal synaptic plasticity: implications for motor learning and Parkinson’s disease. Mov Disord. 2005;20:395–402. doi: 10.1002/mds.20394. [DOI] [PubMed] [Google Scholar]
- Poewe W. Treatments for Parkinson disease--past achievements and current clinical needs. Neurology. 2009;72:S65–73. doi: 10.1212/WNL.0b013e31819908ce. [DOI] [PubMed] [Google Scholar]
- Rascol O, Brooks DJ, Korczyn AD, De Deyn PP, Clarke CE, Lang AE. A five-year study of the incidence of dyskinesia in patients with early Parkinson’s disease who were treated with ropinirole or levodopa. N Engl J Med. 2000;342:1484–1491. doi: 10.1056/NEJM200005183422004. [DOI] [PubMed] [Google Scholar]
- Santini E, Valjent E, Usiello A, Carta M, Borgkvist A, Girault JA, Herve D, Greengard P, Fisone G. Critical involvement of cAMP/DARPP-32 and extracellular signal-regulated protein kinase signaling in L-DOPA-induced dyskinesia. J Neurosci. 2007;27:6995–7005. doi: 10.1523/JNEUROSCI.0852-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini E, Alcacer C, Cacciatore S, Heiman M, Herve D, Greengard P, Girault JA, Valjent E, Fisone G. L-DOPA activates ERK signaling and phosphorylates histone H3 in the striatonigral medium spiny neurons of hemiparkinsonian mice. J Neurochem. 2009;108:621–633. doi: 10.1111/j.1471-4159.2008.05831.x. [DOI] [PubMed] [Google Scholar]
- Sartor GC, Powell SK, Brothers SP, Wahlestedt C. Epigenetic Readers of Lysine Acetylation Regulate Cocaine-Induced Plasticity. J Neurosci. 2015;35:15062–15072. doi: 10.1523/JNEUROSCI.0826-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell. 2014;54:728–736. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenningsson P, Nishi A, Fisone G, Girault JA, Nairn AC, Greengard P. DARPP-32: an integrator of neurotransmission. Annual review of pharmacology and toxicology. 2004;44:269–296. doi: 10.1146/annurev.pharmtox.44.101802.121415. [DOI] [PubMed] [Google Scholar]
- West AE, Griffith EC, Greenberg ME. Regulation of transcription factors by neuronal activity. Nat Rev Neurosci. 2002;3:921–931. doi: 10.1038/nrn987. [DOI] [PubMed] [Google Scholar]
- Wu SY, Lee AY, Lai HT, Zhang H, Chiang CM. Phospho switch triggers Brd4 chromatin binding and activator recruitment for gene-specific targeting. Mol Cell. 2013;49:843–857. doi: 10.1016/j.molcel.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La Russa M, Tsai JC, Weissman JS, Dueber JE, Qi LS, Lim WA. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 2015;160:339–350. doi: 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]