Abstract
Noncoding sequences in plant viral genomes are well-known to control viral replication and gene expression in cis. However, plant viral and viroid noncoding RNA (ncRNA) sequences can also regulate gene expression acting in trans, often acting like “sponges” that bind and sequester host cellular machinery to favor viral infection. Noncoding sequences of small subgenomic RNAs (sgRNAs) of barley yellow dwarf virus (BYDV) and red clover necrotic mosaic virus (RCNMV) contain a cap-independent translation element that binds translation initiation factor eIF4G. We provide new evidence that an sgRNA of BYDV can globally attenuate host translation, probably by “sponging” eIF4G. Subgenomic ncRNA of RCNMV is generated via 5′ to 3′ degradation by a host exonuclease. The similar noncoding subgenomic flavivirus RNA (sfRNA), inhibits the innate immune response, enhancing viral pathogenesis. Cauliflower mosaic virus transcribes massive amounts of a 600 nt ncRNA, which is processed into small RNAs that overwhelm the host’s RNA interference (RNAi) system. Viroids use the host RNAi machinery to generate viroid-derived ncRNAs that inhibit expression of host defense genes by mimicking a microRNA. More examples of plant viral and viroid ncRNAs are likely to be discovered, revealing fascinating new weaponry in the host-virus arms race.
Plant viruses have small and compact genomes. Thus, noncoding regions are limited to a very small portion of a typical plant virus genome. In fact, sequence space is at such a premium that many plant viral genomes encode overlapping genes (e.g., Figure 1). Because so little sequence is noncoding, few noncoding RNAs (ncRNAs) are known in plant viruses. Viroids also generate ncRNAs, which are discussed here. Noncoding satellite RNAs are discussed elsewhere in this Molecular Plant-Microbe Interactions Focus issue (Palukaitis, 2016) and by Shimura and Masuta (2016). In contrast to plant viral RNAs, ncRNAs that regulate host and viral gene expression are abundant in herpes viruses of animals. Their large (>100 kbp) DNA genomes encode numerous microRNAs and other ncRNAs that manipulate expression of host and viral genes (Guo and Steitz, 2014). For reasons that are beyond the scope of this article, no such large viruses exist in plants (Dolja and Koonin, 2011). The major known roles of noncoding regions in plant viral RNA are to control RNA synthesis, encapsidation or translation in cis, as part of the viral genomic RNA or viral mRNAs (Newburn and White, 2015). Some examples of plant viral RNAs that act in trans, via their noncoding regions are known in the luteoviruses, Tombusviridae, and pararetroviruses, and are the subject of this review, as are viroids which are entirely noncoding. We also provide new data supporting the role of plant viral ncRNAs as inhibitors of host translation. It is likely that many more plant viral ncRNAs remain to be discovered.
Fig. 1.
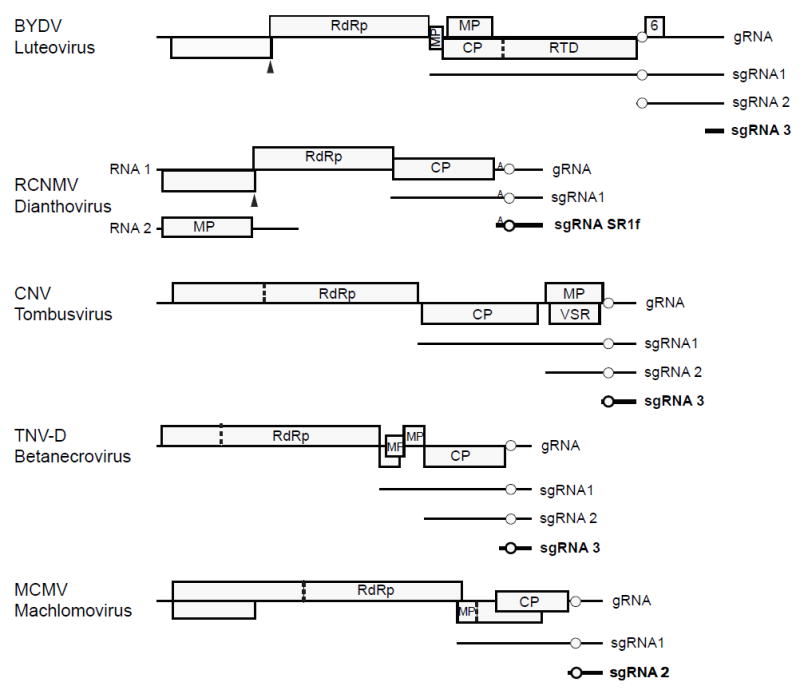
Genome organizations of RNA viruses known to produce noncoding subgenomic RNAs. Noncoding subgenomic RNAs are indicated in bold. Luteoviruses SbDV and BLRV lack ORF 6 and sgRNA2, while luteovirus RSDaV has two additional small ORFs downstream of ORF 6. 5′ ends of Cucumber necrosis virus (Johnston and Rochon, 1995) and Tobacco necrosis virus-D (Jiwan et al. 2011) ncsgRNAs have not been mapped precisely. Circle indicates position of 3′ CITE. Small A on RCNMV RNAs indicates the A-rich sequence needed for maximal CITE activity. Solid triangle: ribosomal frameshift site; dashed line: leaky stop codon. Abbreviations: RdRp, RNA-dependent RNA polymerase; MP, movement protein; CP, coat protein; RTD, coat protein readthrough domain; VSR, viral suppressor of RNAi.
Barley yellow dwarf virus and other luteoviruses
Viruses in genus Luteovirus of the Luteoviridae family, and viruses in the related Tombusviridae family produce 3′ coterminal subgenomic RNAs (sgRNAs) (Miller and Koev, 2000; Jiwan and White, 2011; Domier, 2012; Rochon et al. 2012). Most of these sgRNAs serve as messenger RNAs to allow translation of 5′-distal genes in the genomic RNA. However, others correspond only to the 3′ untranslated region (UTR) and are thus noncoding sgRNAs (ncsgRNAs). Also, noncoding regions of some of these sgRNAs that do contain coding regions, i.e., open reading frames (ORFs) may regulate host and/or viral RNA gene expression in trans, in the manner of a ncRNA.
In the infected cell, luteoviruses generate two or three sgRNAs, depending on the virus (Fig. 1). All share the 3′ terminus of genomic RNA (Kelly et al. 1994; Yamagishi et al. 2003). Subgenomic RNA 1 (sgRNA1) of barley yellow dwarf virus (BYDV), and other luteoviruses consists of the 3′ half of the viral genome and serves as mRNA for four ORFs, translated by noncanonical means, that code for coat protein (CP) and other proteins involved in virus movement in the plant or its aphid vector (Dinesh-Kumar and Miller, 1993; Brown et al. 1996; Smirnova et al. 2015). Thus sgRNA1 is a supercoding mRNA. On the other hand, 800 nt sgRNA2 of BYDV encodes only the small ORF 6, the product of which varies in size from 4.3 to 7.2 kDa, depending on the virus isolate (Chaloub et al. 1994). This protein (P6) has not been detected in infected cells (Shen et al. 2006). A construct designed to express P6 was shown to suppress RNA silencing (Liu et al. 2012), but the authors did not detect the P6 protein, so they did not rule out the possibility that the RNA encoding P6, which includes the 5′ end of sgRNA2, is the actual silencing suppressor, rather than the predicted protein. sgRNA3 encodes no ORFs and consists of the 3′-terminal 330 nts of the BYDV genome (Kelly et al. 1994). sgRNA2 and sgRNA3 are present in many tens-fold molar excess over sgRNA1 and genomic RNA (Kelly et al. 1994; Koev and Miller, 2000).
RNAs of luteoviruses and the tombusvirids lack a 5′ cap and a poly(A) tail. Instead, for protein synthesis, they rely on a cap-independent translation element (CITE) located near the 5′ end of the 3′ UTR of the genomic RNA (Simon and Miller, 2013). The 3′ CITE of BYDV, called the BYDV-like translation element (BTE), is located between ORFs 5 and 6, about 800 nt from the 3′ end of the genome (Fig. 1). The BTE powerfully stimulates translation of the viral genomic RNA and subgenomic RNA1 (Wang et al. 1997; Rakotondrafara et al. 2006; Fan et al. 2012) by binding with high affinity to eIF4G, which is the scaffolding subunit of the key translation initiation heterodimer, eIF4F (Treder et al. 2008; Kraft et al. 2013). The BTE is in the 5′ UTR of sgRNA2 (Fig. 1). sgRNA2 regulates translation of viral genomic RNA and subgenomic RNA1 in trans, via its BTE (Wang et al. 1999; Shen et al. 2006). Because of the BTE at its 5′ end, which binds eIF4G, sgRNA2 strongly trans-inhibits translation of genomic RNA, but only slightly inhibits translation of sgRNA1 (Wang et al. 1999). In infected cells this interaction may serve as a switch to favor translation of late genes (virus movement and packaging) from sgRNA1 over translation of early (RNA synthesis) genes from the genomic RNA (Shen and Miller, 2004; Shen et al. 2006). This differential effect on translation of BYDV genomic RNA and sgRNA1 by sgRNA2 is conferred by their different 5′ UTRs. Genomic RNA has a highly structured 5′ UTR (Guo et al. 2001) whereas sgRNA1 has a relatively unstructured and thus less eIF4F-dependent 5′ UTR (Shen et al. 2006). To summarize, the noncoding portion of sgRNA2 regulates translation of other viral genes, and it acts like a regulatory ncRNA (even though it contains a small ORF).
Owing to its high abundance and high affinity for eIF4G, we expect sgRNA2 to attenuate translation of host mRNAs as well as the other viral RNAs. Indeed, we showed that sgRNA2 inhibits translation (in trans) of a capped, polyadenylated luciferase mRNA, lacking any viral sequence (Shen and Miller, 2004). To determine if translation inhibition occurs in infected cells, this reporter mRNA was electroporated into oat protoplasts 24 h after infection with wild type BYDV-PAV (PAV6) or mutant BYDV-PAV with a point mutation preventing synthesis of sgRNA2 (PAV6ΔSG2) (Koev and Miller, 2000), and luciferase activity was measured after 4 h as in Shen and Miller (2004; 2007). Both sets of cells accumulated similar amounts of viral genomic RNA and sgRNA1 (Fig. 2A, right panel). Importantly, luciferase expression averaged one-half as much in the PAV6-infected cells compared with the PAV6ΔSG2-infected cells (Fig. 2A, left panel). Thus, sgRNA2 accumulation inhibits translation of nonviral mRNA, leading us to predict that host mRNAs would be similarly inhibited.
Fig. 2.
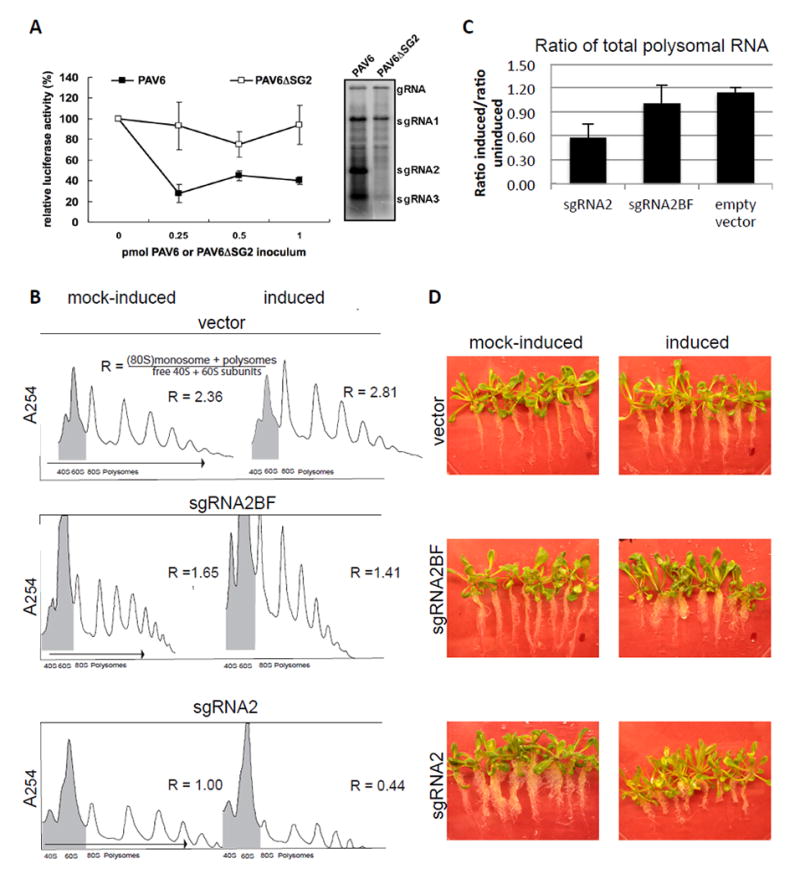
Effect of expression of BYDV sgRNA2 on translation and plant phenotype. A. Effect of sgRNA2 on translation of capped, polyadenylated, nonviral luciferase mRNA in infected oat protoplasts. Oat protoplasts infected with the indicated transcript were electroporated with firefly luciferase mRNA, then assayed for luciferase activity after 4 h. B. Polysome profiles of plants expressing sgRNA2. The polysome profiles are representative of profiles collected in duplicate (pER8 vector and sgRNA2BF) or quadruplicate (sgRNA2). To calculate the ratio (R) of translating to nontranslating RNA, the combined areas of the monosome (80S) and polysome peaks (unshaded areas) were divided by the areas of the 40S plus 60S peaks (shaded). (R indicated beside polysome profiles.) C. The changes R (panel B) due to expression of the indicated RNA are plotted showing averages (with standard error) from four separate experiments such as the one shown in panel B. D. Transgenic Arabidopsis seedlings after twelve days of growth on media containing estradiol (induced) or DMSO buffer only (mock-induced).
To detect global reduction of host translation, we transgenically expressed sgRNA2 via an estradiol-inducible promoter in Arabidopsis. Arabidopsis Col-0 plants were transformed with either empty vector, vector expressing full-length sgRNA2, or full-length sgRNA2 containing a four base duplication in the natural BamHI site that completely inactivates the BTE (sgRNA2BF), driven by an estradiol-inducible promoter. Binary vectors pERSG2 and pERSG2BF were constructed by inserting PCR-amplified BYDV sgRNA2 and sgRNA2BF into Xho I /Spe I-cut pER8 (Zuo et al. 2000), respectively. Transformation of Agrobacterium tumefaciens strain GV3101∷pMP90 was done as in (Shen and Forde, 1989) by using a MicroPulser (Bio-Rad). Transformation of Arabidopsis thaliana Col-0 ecotype was carried out by floral dip as in (Clough and Bent, 1998). T3 or T4 seeds were used for experiments. Polysomes were obtained from seedlings treated with dimethyl sulfoxide (DMSO) alone (mock-induced) or with β-17-estradiol (10mM) in DMSO (induced) by grinding 0.7 to 2 grams of plant material under liquid nitrogen. Extraction buffer (200 mM Tris-HCl pH 8.5, 200 mM KCl, 30 mM MgCl2, 10 mM EGTA, 200 mM sucrose, 10 mM beta mercaptoethanol, 2.5 mM DTT, 0.5 mg/mL heparin, 5 μg/mL proteinase K, 100 μg/ml chloramphenicol, 50 μg/ml cyclohexamide) was added to the pulverized tissue at a ratio of 0.5 mL to 1 gram of plant material (Hollingsworth et al. 1998). Upon thawing, the homogenates were centrifuged at 16,000 g for 10 min, supernatants were layered on a sucrose cushion (1.75 M sucrose, 40 mM Tris-HCl pH 9, 30 mM MgCl2, 200 mM KCl, 5 mM EGTA, 10 mM β-mercaptoethanol) and centrifuged in a SW50.1 rotor at 234,000 g for 18 h. Pelleted polysomes were washed and resuspended in 50 μl resuspension buffer (200 mM Tris-HCl pH 8.5, 60 mM KCl, 30 mM MgCl2, 100 μg/mL chloramphenicol, 50 μg/ml cyclohexamide, 10 mM β-mercaptoethanol) and then layered on a 20 to 60% sucrose gradient (40 mM Tris-HCl pH 8.5, 30 mM KCl, 10 mM MgCl2, 10 mM β-mercaptoethanol) through which they were centrifuged in a SW41 rotor at 200,000 g for 2.5 h. Gradients were fractionated with a Brandel pump and an Isco Apparatus (UA-6) absorbance monitor with a 254 nm UV filter.
High levels of sgRNA2 were detected after induction with estradiol, and total translating RNA (monosomes and polysomes relative to 40S and 60S ribosomal subunits) was reduced by about 50%, indicating a reduction in translation of host mRNAs (Fig. 2B, C). Plants expressing empty vector, and negative control plants expressing sgRNA2BF with a nonfunctional BTE, showed little change in polysomal mRNAs (Fig. 2B, C). Such a global reduction in translation caused by sgRNA2 would be expected to have an adverse effect on plant health. Indeed, twelve days after induction of sgRNA2, seedlings showed a phenotype of decreased root length, and ultimately appeared stunted (Fig. 2D). The plants expressing sgRNA2BF showed only slight, if any, decrease in root length, and those expressing empty vector RNA were unchanged in phenotype (Fig. 2D).
The biological role of inhibition of host translation by sgRNA2 is uncertain. It may inhibit translation of host defense genes. sgRNA2 and sgRNA3 are not essential for RNA replication in protoplasts: mutations in the promoters that prevent synthesis of these sgRNAs did not greatly reduce viral RNA accumulation (Koev & Miller, 2000; Shen et al. 2004). Surprisingly, virus containing these mutations was able to infect oat plants and accumulate to levels similar to those of wild type virus (Miller et al. 2015). However, after passaging the sgRNA double knockout mutants, a new sgRNA that is slightly larger than sgRNA3 appeared in some infected plants. Thus, natural selection seems to favor the presence of a small ncsgRNA similar to sgRNA3, but BYDV can replicate in the host (at least in highly susceptible oats, c.v. Clintland 64) under the controlled conditions of our laboratory in the absence of both sgRNA2 and sgRNA3. We speculate that the wild type virus would be more successful in direct competition with the mutant virus, or in different hosts and/or field conditions sgRNA2 and sgRNA3 may provide a distinct advantage. In support of their importance, these sgRNAs were found in all 22 field isolates tested by Kelly et al. (1994).
Another luteovirus, soybean dwarf virus (SbDV) generates a highly abundant 320 nt subgenomic ncRNA, similar to sgRNA3 of BYDV (Yamagishi et al. 2003). This virus appears not to produce a homolog of BYDV sgRNA2, although a faint band of about this size is visible on northern blot hybridizations of some isolates (Yamagishi et al. 2003). Also, for the closely related bean leafroll virus (BLRV), only sgRNA3-sized (319 nt) noncoding sgRNA was detected, although this virus contained a potential promoter at the expected site for sgRNA2 synthesis (Domier et al. 2002). Interestingly, neither SbDV nor BLRV encode a homolog of ORF 6 of BYDV (Domier et al. 2002). Yet these viruses also harbor a BTE at the 5′ end of a long (700 nt) 3′ UTR. In contrast, like BYDV, Rose spring dwarf-associated luteovirus (RSDaV) generates an ~850 nt sgRNA2 which encodes an ORF 6. Unlike other luteoviruses, RSDaV sgRNA2 also contains two small ORFs downstream of ORF 6 (Salem et al., 2008). Whether RSDaV generates sgRNA3 is unknown. Thus the regions of the luteovirus genome downstream of ORF 5 remain somewhat of a mystery. Depending on the virus it can encode between zero and three ORFs, and it may or may not generate the ~850 nt sgRNA2. In all luteoviruses, the region downstream of ORF 5 contains the BTE and a 25 nt stem-loop downstream required for -1 frameshifting (Barry and Miller, 2002) at the 5’ end, and a structure required for RNA replication at the 3’ end (Koev et al. 2002). The intervening 300 – 400 bases perform no known function in trans or in cis.
Red clover necrotic mosaic virus and other Tombusviridae
One of the few other plant viruses for which trans-acting ncsgRNAs have been identified is red clover necrotic mosaic virus (RCNMV, genus Dianthovirus, family Tombusviridae). Although in a different family, it is closely related to the luteoviruses (Miller et al. 2002). Unlike the luteoviruses and other tombusvirids, RCNMV has a bipartite genome consisting of genomic RNAs 1 and 2 (Okuno and Hiruki, 2013). Like other tombusvirids and luteoviruses, both of these positive sense RNAs have neither a 5′ cap (Mizumoto et al. 2003) nor a poly(A) tail (Lommel et al. 1988; Xiong et al. 1989). RNA1 encodes a 27-kDa protein, p27, and an 88-kDa protein, p88, which contains an RNA-dependent RNA polymerase motif (Fig. 1) (Lommel et al. 1988; Xiong et al. 1989). As in the luteovirus genome, the RdRp is translated via -1 ribosomal frameshifting directed by an RNA structure that requires base-pairing of a stem-loop in the 3′ UTR with a bulged stem-loop structure adjacent to the frameshift site (Tajima et al. 2011). A 3′-coterminal subgenomic RNA, CPsgRNA, is generated from RNA1, and serves as mRNA for the 37 kDa coat protein (Zavriev et al. 1996). RNA2 encodes a 35 kDa movement protein (Fig. 1) (Xiong et al. 1993).
RCNMV RNA1 contains a BTE in its 3′ UTR to facilitate cap-independent translation (Mizumoto et al. 2003). This BTE, called 3′TE-DR1, includes the 17 nt sequence conserved among all BTEs and folds into a similar secondary structure. It differs from the luteovirus BTE in three key ways: (i) it has five rather than three stem-loops radiating from the central hub, (ii) an adjacent upstream A-rich sequence (ARS), enhances the activity of 3′TE-DR1 by recruiting poly(A)-binding protein (PABP) (Iwakawa et al. 2012), and (iii) no base pairing to the 5′ UTR is required (Mizumoto et al. 2003; Iwakawa et al. 2012). The 3′ end of the 3′ UTR of RNA1 contains other stem loop (SL) structures: SLDE, SLF and intervening sequence SeqB, which are essential for negative strand RNA synthesis. 3′TE-DR1 is dispensable for RCNMV RNA1 negative strand synthesis (Iwakawa et al. 2007).
There are two examples of trans-acting viral RNAs that control RCNMV gene expression. The first is a 34-nt RNA2 trans-activator (TA) that is essential for the transcription of CPsgRNA from RNA1 (Sit et al. 1998), RNA2 replication (Tatsuta et al. 2005; Basnayake et al. 2006), and virion assembly (Basnayake et al. 2006). This sequence in RNA2 forms a stem-loop and the 8 base loop base pairs to a region in RNA1 two bases upstream of the start site of CPsgRNA (Sit et al. 1998). It is proposed that this base pairing sometimes blocks the replicase as it synthesizes (-) strand of RNA1, creating a truncated negative strand that serves as template for synthesis of (+) sense CPsgRNA (Sit et al. 1998). The TA of RNA2 is also essential for virion assembly as it acts as origin of assembly sequence and its interaction with RNA1 facilitates coencapsidation of RNA1 and RNA2 in the virion (Basnayake et al. 2009). Although this is a trans-acting RNA, it is not a noncoding RNA because the TA resides in the MP coding region.
RNA1 of RCNMV generates a 431 nt ncsgRNA, called SR1f, which comprises the 3′ UTR (Iwakawa et al. 2008). SR1f is resembles BYDV sgRNA2 as follows: (i) it is highly abundant, (ii) it contains the BTE and the downstream frameshift element near its 5′ end and the replication origin at its 3′ end, (iii) it inhibits translation of viral genomic RNA in trans, in vitro and in vivo, (iv) it inhibits translation of capped, polyadenylated, nonviral RNA in vitro and in vivo, (v) it is not required for the virus to infect protoplasts or plants, although genomic RNA levels are lower when unable to produce SR1F (Iwakawa et al. 2008), and (vi) the BTE in SR1f binds the eIF4G subunit of eIF4F with high affinity (Kraft et al. 2013). SR1f differs from BYDV sgRNA2 in that it (i) is packaged in virions, (ii) is half as long, (iii) encodes no ORF, (iv) contains the ARS upstream of the BTE, and (v) is fully occupied with sequence required for translation or RNA synthesis, i.e., it lacks the tracts of “mystery” sequence of unknown function in the luteovirus 3′ UTR.
A remarkable feature of SR1f is the mechanism by which it arises. SR1f is generated in the absence of RNA replication by a host exonuclease that degrades RNA1 in the 5′ to 3′ direction until the nuclease is blocked 431 nt from the 3′ end by a 58 nt structure called Seq1f58 (Fig. 3) (Iwakawa et al. 2008). This mechanism of SR1f generation may be identical to that of the ncsgRNA of flaviviruses (sfRNA), which is generated by exonuclease XRN1 (Pijlman et al. 2008). For both RCNMV and the flaviviruses, the RNA structure (xrRNA) that blocks the exonuclease is highly specialized. Mutations that disrupted secondary structure, removed the blockage, and thus the sfRNA/SR1f RNA production, but compensating mutations predicted to restore the xrRNA structure, did not restore the sfRNA or SRf1 RNA accumulation (Iwakawa et al. 2008; Chapman et al. 2014b). Thus, specific structures that are not easily predicted are required to block the exonuclease. X-ray crystallography revealed that the xrRNA of Murray valley encephalitis virus (MVEV) RNA forms a looped pseudoknot that binds tightly to XRN1 and is pulled tightly like a noose as the nuclease attempts to proceed in the 3′ direction (Chapman et al. 2014b) Somewhat different structures achieve the same results in other flaviviruses (Chapman et al. 2014a; Clarke et al. 2015). In fact, xrRNA binds XRN1 so tightly and is so abundant that it sequesters the nuclease, preventing it from performing its normal duties in host mRNA turnover (Moon et al. 2012).
Fig. 3.
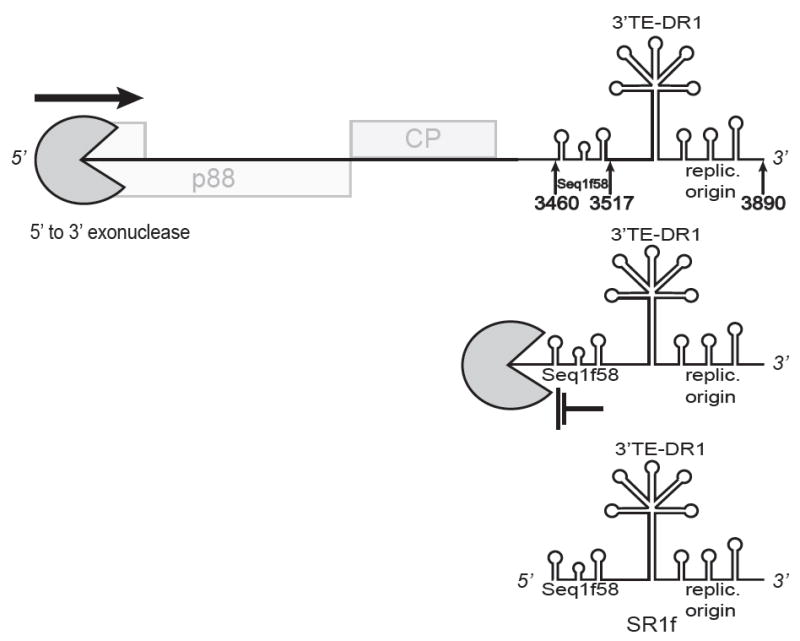
Generation of the small noncoding RNA, SR1f, spanning the 3′ UTR of RCNMV RNA1 (not to scale). Host exonuclease degrades RNA1 from the 5′ end until it is blocked by the Seq1f58 structure, leaving the stable product, SR1f, comprising the 3′ end of the genome: bases 3460-3890 (Iwakawa et al. 2008). Stem-loops shown in SR1f RNA, indicate (i) Seq1f58 which is necessary and sufficient to block the exonuclease, (ii) the downstream stem-loop required for frameshifting (fs), (iii) the 3′ BTE (3′ TE-DR1), and (iv) the 3′-proximal stem-loops required for initiation of replication (replic. origin). Not shown: it is possible that CPsgRNA generated from RCNMV RNA1 may also be a substrate for the exonuclease to generate SR1f RNA.
sfRNA has been shown to act as a sponge to sequester many host and viral proteins (Roby et al. 2014) including those needed to mount an interferon response (Schuessler et al. 2012; Bidet et al. 2014; Manokaran et al. 2015). WNV sfRNA also inhibits the RNA interference response (Schnettler et al. 2012). sfRNA is not required for virus replication in cells or in mice but it greatly increases pathogenesis (Manokaran et al. 2015). While plants don’t have an interferon response, it is an intriguing possibility that RCNMV SR1f RNA could affect the RNAi-based antiviral defense system, given the importance of RNAi in plant innate immunity.
RCNMV RNA lacks a 5′ cap, so it may be particularly vulnerable to exonucleolytic degradation. Sequestration of the exonuclease (most likely XRN4 in plants) by SR1f may thus protect full-length viral RNAs from degradation. Finally, sfRNA may function like sgRNA2 of BYDV (Shen and Miller, 2004; Shen et al. 2006) and possibly SR1f RNA of RCNMV (Iwakawa et al. 2012), by regulating viral translation in trans, as Fan et al. (2011) provided evidence that sfRNA modulates both translation and negative strand synthesis of Japanese encephalitis virus in trans.
In addition to the SR1f RNA of RCNMV, ncsgRNAs corresponding to the 3′ UTR have been identified in the following tombusvirids (Fig. 1): the machlomovirus, maize chlorotic mottle virus (MCMV)(Scheets, 2000), the tombusvirus, cucumber necrosis virus (Johnston and Rochon, 1995), and the betanecrovirus, tobacco necrosis virus-D (Jiwan et al. 2011). Similar to RCNMV RNA1, and unlike luteoviruses, these viruses have shorter (around 300-400 nt) 3′ UTRs, which have the CITE near the 5′ end, and the replication origin at the extreme 3′ end (Fig. 1). Whether these ncsgRNAs are required for virus infection, and their role in the virus life cycle is unknown.
It is possible that these ncsgRNAs, as well as sgRNA3 of BYDV are generated by the same host exonuclease mechanism that produces SR1f RNA of RCNMV. The larger sgRNAs of BYDV, RCNMV and other tombusvirids require RNA synthesis to accumulate and have sequences at their 5′ termini that resemble the 5′ end of the genome, presumably origins of (+) strand synthesis on the negative strand (Sit et al. 1998; Koev et al. 1999; Jiwan and White, 2011; Jiwan et al. 2011; Newburn and White, 2015). In contrast, the small ncsgRNAs often lack any recognizable promoter-like sequences near their 5′ ends (Johnston and Rochon, 1995; Koev and Miller, 2000; Scheets, 2000), supporting a different mechanism of production, such as via host exonuclease. In addition to genomic RNA, sgRNA1 and especially the abundant sgRNA2 of BYDV could also serve as substrates from which sgRNA3 is generated. This would explain why a point mutation, which completely prevents synthesis of sgRNA2, also reduces accumulation of sgRNA3 (Fig. 2A and (Shen et al. 2006). It is quite possible that other tombusvirids produce such ncsgRNAs but that they have been obscured by degradation products in low-resolution northern blots, or were simply ignored because they are too small to serve as subgenomic mRNAs and too large to be small RNAs involved in host RNA-mediated defense.
Cauliflower mosaic virus RNA jams the RNAi machinery
A DNA plant virus produces ncRNAs that appear to inhibit the RNA interference system, as mentioned above for the flaviviruses and proposed as a possibility for RCNMV SR1f RNA. Cauliflower mosaic virus (CaMV) is a pararetrovirus with a nicked, double-stranded circular DNA genome (Hohn and Rothnie, 2013). In infected cells, massive amounts of 20-25 nt viral small RNAs (vsRNAs) mapping to both strands of the highly structured 600 nt leader sequence of the viral 35S RNA accumulate (Blevins et al. 2011). This leader is transcribed as a separate 600 nt 8S RNA whose function has been a mystery since its discovery in 1982 (Guilley et al. 1982). The 8S RNA is likely generated by initiation of transcription at the 35S promoter and termination by polymerase run-off at a nick in the negative strand DNA located 600 nt downstream (Blevins et al. 2011).
CaMV levels do not increase in plants with the RNAi defense system eliminated: rdr1/2/6 triple knockouts and dcl1/2/3/4 quadruple knockouts had no effect on CaMV levels (Blevins et al.l., 2011). Thus the RNA-based defense system is ineffective against CaMV. The 8S transcript is highly structured and may resemble viroid RNA (Hemmings-Mieszczak et al. 1997), which is recognized and replicated by host DNA-dependent RNA polymerase II (pol II) (Rackwitz et al. 1981). Thus, Blevins et al. (2011) speculate that the negative strand of the 8S RNA is also generated by pol II. They propose that the massive amounts of vsRNAs generated from the 8S dsRNA serve as decoys to overwhelm the RNAi silencing complex (RISC) and prevent it from generating enough vsRNAs that target the rest of the genome. Indeed, immunoprecipitation of AGO1, the major component of the RISC, revealed that almost all of the coimmunoprecipitating RNA associated with AGO1 mapped to the 8S RNA derived from the 600 nt leader sequence, and almost none to the rest of the genome (Blevins et al. 2011). Thus, the antiviral defense system is targeted almost entirely to the abundant, noncoding 8S transcript. Owing to its high degree of secondary structure, few vsRNAs gain access to their complementary targets in 8S RNA (or the same sequences in the leader of the 35S RNA), rendering the virus unscathed by the RNAi machinery (Hohn, 2015). This leaves the genomic 35S RNA and the other viral mRNA (19S RNA) free to express viral genes, and allows the 35S RNA to serve as template for viral genome replication.
The role of the 8S RNA in jamming host RNAi-mediated defense was further supported by transferring the 8S DNA sequence into a completely different kind of virus, cabbage leaf curl virus (CaLCuV), a geminivirus. CaLCuV expressing this leader sequence replicated to higher levels than virus expressing only vector RNA, and large amounts of vsRNAs corresponding to the 8S RNA sequence in the modified CaLCuV accumulated (Blevins et al. 2011). To summarize, the authors propose a model in which the nicked, circular form of CaMV DNA is transcribed to produce the 8S RNA (noncoding leader sequence of 35S RNA), which is then copied by pol II into dsRNA that is processed by the dicer-like proteins into massive amounts of vsRNAs which overload the RISC machinery (AGO1 protein), preventing the RNAi response from inhibiting virus infection (Fig. 4). For CaMV genome replication, host DNA repair enzymes remove the nicks in virion DNA, preventing termination of transcription that generates the 8S RNA, while allowing transcription of the normal 35S and 19S viral RNAs needed for virus gene expression and replication (Fig. 4). Given the high structure and abundance of the ncsgRNAs of tombusvirid and luteovirus 3′ UTRs (above), we speculate that they too have the potential to overwhelm the RNAi-based defense system in a similar fashion.
Fig. 4.
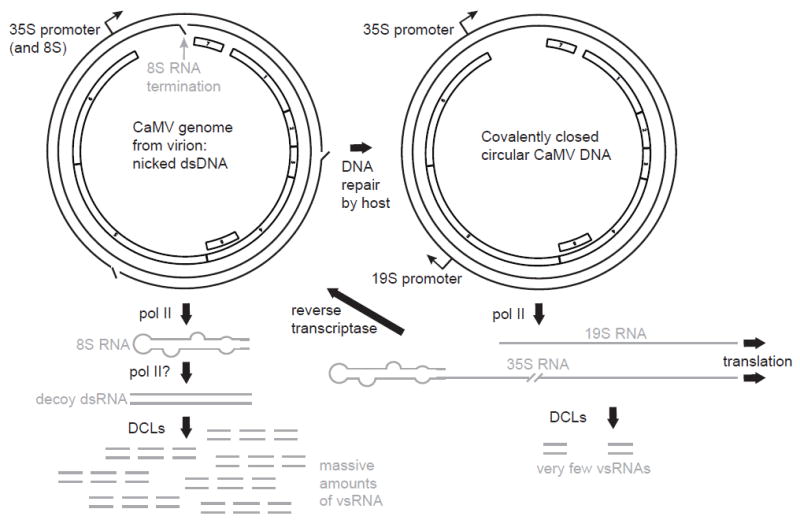
Transcription of massive levels of ncRNA from the 5′ leader of CaMV 35S RNA (modified from Blevins et al. 2011). Circular, double-stranded DNA genome is shown with ORFs inside. Left: transcription of the nicked, double stranded circular genome of CaMV from the 35S promoter results in early termination at the first nick, giving large quantities of 8S RNA comprising the highly structured untranslated leader sequence. This RNA may be copied into dsRNA via pol II owing to its resemblence to viroid RNA. Any of the DCL proteins then cleave this RNA into 20 – 25 nt vsRNAs that flood the RISC, preventing the defense system from making effective amounts of siRNA to traget the larger viral RNAs. At right, host enzymes repair the nicks, creating the covalently closed dsDNA template from which full-length 35S and 19S RNAs are transcribed. Both serve as mRNAs, and 35S RNA also serves as the genomic replication intermediate which is reverse transcribed to make more nicked, ds genomic DNA.
Viroid-derived small RNAs can target specific host mRNAs to reduce their expression and enhance pathogenesis
Viroids consist of 250 to 400 nt noncoding, circular RNAs with a high degree of self-complementarity causing them to form rod-shaped, partially double-stranded structures. As with viral infections, small (21-24 nt) viroid RNAs (vd-sRNAs) accumulate in plants infected with viroids, owing to processing by Dicer-like (DCL) and AGO proteins in the RNAi-based antiviral defense system (Navarro et al. 2012; Minoia et al. 2015; Tsushima et al. 2015). Emerging evidence indicates that some vd-sRNAs (Adkar-Purushothama et al. 2015; Flores et al. 2015), as well as a satellite RNA of cucumber mosaic virus (Shimura et al. 2011; Smith et al. 2011) can function like host microRNAs (miRNA) to specifically target and degrade host mRNAs.
Strains of Peach latent mosaic viroid (PLMVd) that cause a “peach calico” (PC) or albino symptom contain a 12-nt hairpin insertion. In infected tissue that has the albino phenotype, a 21 nt vd-sRNA, which includes this 12 nt insertion, accumulates. This vd-sRNA can base pair to mRNA encoding the chloroplast heat-shock protein 90 (cHSP90). This results in cleavage of the mRNA as predicted for miRNA-mediated cleavage (Navarro et al. 2012). The vd-sRNA accumulates and cHSP90 mRNA is cleaved only in the albino tissue. This degradation of cHSP90 mRNA may favor viroid accumulation, as somewhat higher levels of PLMVd RNA accumulate in the albino tissue relative to the green tissue in which cHSP90 mRNA is not cleaved (Flores et al. 2015). Thus, the PC strains of PLMVd may be turning the RNAi-mediated defense system of the host to their own advantage.
A more clear cut example of this exploitation of the RNAi system by a viroid was discovered recently for Potato spindle tuber viroid (PSTVd) (Adkar-Purushothama et al. 2015). As few as two base differences in the pathogenicity-determining domain of PSTVd RNA can greatly affect symptom severity and replication efficiency (Tsushima et al. 2015). Adkar-Purushothama et al. (2015) showed that this region is processed by the host DCL proteins and RISC to produce a vd-sRNA that modulates host gene expression to the advantage of the viroid. One of the vd-sRNAs in tomato plants infected with a severe strain of PSTVd (PSTVd-I) has partial complementarity to mRNA encoding the callose synthase 11-like protein (CalS11-like), whereas the homologous vd-sRNA from a mild strain of PSTVd (PSTVd-M) has much weaker complementarity to CalS11-like mRNA. Expression of CalS11-like mRNA was reduced more in PSTVd-I-infected than in PSTVd-M-infected plants. Expression of the severe 21 nt vd-sRNA sequence in the context of a microRNA (miRNA), in the absence of PSTVd infection, reduced expression of a GFP reporter gene containing the predicted target sequence in its 3′ UTR. These data, plus results of additional experiments, support the hypothesis that the vd-sRNA generated by the host antiviral RNAi machinery, functionally mimics a miRNA, which targets host callose synthesis mRNAs, thereby reducing mRNA levels and presumably callose levels. This, in turn, enhances viroid accumulation and movement in the plant because callose synthesis at the plasmodesmata is a host defense mechanism known to reduce virus movement from cell to cell through the plasmodesmata (Li et al. 2012). Indeed, it had been shown previously that the sequence of the pathogenicity domains that generate these key vd-sRNAs controls efficiency of viroid cell-to-cell movement (Zhong et al. 2008). Thus in both PLMVd and PSTVd infections, ncRNA generated from the viroid genome simply incorporates itself into the RNAi system of the host plant to negatively regulate expression of a presumed host defense gene in order to facilitate more efficient infection.
It remains a mystery why, to our knowledge and that of Flores et al. (2015), only viroids and satellite RNAs are known to produce small RNAs that knock down expression of host genes, while plant viral genomes are not known to (naturally) produce small RNAs (vsRNAs) that directly target host mRNAs, reducing their expression. This is despite observations that (i) vsRNAs accumulate to substantial levels in most plant viral infections, (ii) some of these vsRNAs are complementary to host mRNAs (Qi et al. 2009), and (iii) artificial insertion of host sequences into viral genomes allows many plant viruses to serve as efficient tools for virus-induced gene silencing (VIGS) of host genes (Mysore and Senthil-Kumar, 2015).
Conclusion: Viral RNAs are often sponges of proteins
All viral RNAs, coding and noncoding, compete with host RNAs and with each other for interactions with host and viral proteins, complexes and organelles in a way that results in productive infection. Thus viral RNAs can be considered sponges of host proteins and larger complexes (Charley and Wilusz, 2014). In most cases, such as positive strand RNA virus genomic RNAs and subgenomic mRNAs, the RNAs efficiently bind translation factors, effectively “sponging” them away from host mRNAs, and they bind other host proteins and membranes (Pathak et al. 2011; Nagy et al. 2012), sponging them away from their normal cellular processes (Clarke et al. 2015). This can hinder the ability of the host to mount a defense response (Bidet et al. 2014; Charley and Wilusz, 2014). Only recently have plant virologists become aware that ncRNAs of plant viruses, often unnoticed despite their high abundance, may play such a role in plant virus infection. Here we have provided examples of such RNAs that can reduce the amount of translation machinery, RNA interference machinery, or defense gene mRNA available to the cell, thus favoring infection.
A common feature of ncRNAs is that they are not essential for virus infection. Knockout mutations that prevent expression of viral ncRNAs do not prevent virus replication (Fig. 2A) (Iwakawa et al. 2008; Clarke et al. 2015; Miller et al. 2015). Similarly, viroids with sequence variations that prevent vd-sRNAs from mimicking defense-gene targeted miRNAs, are still able to replicate in plants (Adkar-Purushothama et al. 2015; Flores et al. 2015). However viruses and viroids that generate functional ncRNAs are known (or likely) to have a competitive advantage for optimal accumulation by attenuating the defense response of the host. This lack of essentiality and lack of coding regions may explain why so few ncRNAs have been identified or discussed much in publications that reveal them. For example, an sfRNA of a flavivirus was first reported in 1997 (Urosevic et al. 1997), but its significance was not realized until 2008 (Pijlman et al. 2008; Roby et al. 2014). For these reasons, we may have missed some other published ncRNAs of plant viruses (for that we apologize). We predict that many more trans-acting ncRNAs exist for many plant viruses, and still await discovery.
Acknowledgments
The authors thank K.A. White for imparting some of his vast knowledge of viral RNAs, and S. Whitham for valuable discussions on RNAi and VIGS. This work was funded by NIH grant no. R01GM067104 to WAM . This journal paper of the Iowa Agriculture and Home Economics Experiment Station, Ames, IA, Project No. 3708 was supported in part by Hatch Act and State of Iowa funds.
Literature Cited
- Adkar-Purushothama CR, Brosseau C, Giguère T, Sano T, Moffett P, Perreault JP. Small RNA Derived from the Virulence Modulating Region of the Potato spindle tuber viroid Silences callose synthase Genes of Tomato Plants. Plant Cell. 2015;27:2178–2194. doi: 10.1105/tpc.15.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry JK, Miller WA. A -1 ribosomal frameshift element that requires base pairing across four kilobases suggests a mechanism of regulating ribosome and replicase traffic on a viral RNA. Proc Natl Acad Sci USA. 2002;99:11133–11138. doi: 10.1073/pnas.162223099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basnayake VR, Sit TL, Lommel SA. The genomic RNA packaging scheme of Red clover necrotic mosaic virus. Virology. 2006;345:532–539. doi: 10.1016/j.virol.2005.10.017jrn. [DOI] [PubMed] [Google Scholar]
- Basnayake VR, Sit TL, Lommel SA. The Red clover necrotic mosaic virus origin of assembly is delimited to the RNA-2 trans-activator. Virology. 2009;384:169–178. doi: 10.1016/j.virol.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Bidet K, Dadlani D, Garcia-Blanco MA. G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. PLoS Pathog. 2014;10:e1004242. doi: 10.1371/journal.ppat.1004242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins T, Rajeswaran R, Aregger M, Borah BK, Schepetilnikov M, Baerlocher L, Farinelli L, Meins F, Jr, Hohn T, Pooggin MM. Massive production of small RNAs from a non-coding region of Cauliflower mosaic virus in plant defense and viral counter-defense. Nucleic Acids Res. 2011;39:5003–5014. doi: 10.1093/nar/gkr119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CM, Dinesh-Kumar SP, Miller WA. Local and distant sequences are required for efficient readthrough of the barley yellow dwarf virus PAV coat protein gene stop codon. J Virol. 1996;70:5884–5892. doi: 10.1128/jvi.70.9.5884-5892.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub BA, Kelly L, Robaglia C, Lapierre HD. Sequence variability in the genome-3′-terminal region of BYDV for 10 geographically distinct PAV-like isolates of barley yellow dwarf virus: analysis of the ORF6 variation. Arch Virol. 1994;139:403–416. doi: 10.1007/BF01310801. [DOI] [PubMed] [Google Scholar]
- Chapman EG, Costantino DA, Rabe JL, Moon SL, Wilusz J, Nix JC, Kieft JS. The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production. Science. 2014b;344:307–310. doi: 10.1126/science.1250897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman EG, Moon SL, Wilusz J, Kieft JS. RNA structures that resist degradation by Xrn1 produce a pathogenic Dengue virus RNA. eLife. 2014a;3:e01892. doi: 10.7554/eLife.01892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charley PA, Wilusz J. Sponging of cellular proteins by viral RNAs. Curr Opin Virol. 2014;9:14–18. doi: 10.1016/j.coviro.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke BD, Roby JA, Slonchak A, Khromykh AA. Functional non-coding RNAs derived from the flavivirus 3′ untranslated region. Virus Res. 2015;206:53–61. doi: 10.1016/j.virusres.2015.01.026. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Dinesh-Kumar SP, Miller WA. Control of start codon choice on a plant viral RNA encoding overlapping genes. Plant Cell. 1993;5:679–692. doi: 10.1105/tpc.5.6.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolja VV, Koonin EV. Common origins and host-dependent diversity of plant and animal viromes. Curr Opin Virol. 2011;1:322–331. doi: 10.1016/j.coviro.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domier LL. Family Luteoviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy: Ninth Report of the International Committee on the Taxonomy of Viruses. Elsevier Academic Press; Amsterdam: 2012. pp. 1045–1053. [Google Scholar]
- Domier LL, McCoppin NK, Larsen RC, D’Arcy CJ. Nucleotide sequence shows that Bean leafroll virus has a Luteovirus-like genome organization. J Gen Virol. 2002;83:1791–1798. doi: 10.1099/0022-1317-83-7-1791. [DOI] [PubMed] [Google Scholar]
- Fan Q, Treder K, Miller WA. Untranslated regions of diverse plant viral RNAs vary greatly in translation enhancement efficiency. BMC Biotechnol. 2012;12:22. doi: 10.1186/1472-6750-12-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y-H, Nadar M, Chen C-C, Weng C-C, Lin Y-T, Chang R-Y. Small noncoding RNA modulates Japanese encephalitis virus replication and translation in trans. Virol J. 2011;8:492. doi: 10.1186/1743-422X-8-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores R, Minoia S, Carbonell A, Gisel A, Delgado S, López-Carrasco A, Navarro B, Di Serio F. Viroids, the simplest RNA replicons: How they manipulate their hosts for being propagated and how their hosts react for containing the infection. Virus Res. 2015;209:136–145. doi: 10.1016/j.virusres.2015.02.027. [DOI] [PubMed] [Google Scholar]
- Guilley H, Dudley RK, Jonard G, Balàzs E, Richards KE. Transcription of Cauliflower mosaic virus DNA: detection of promoter sequences, and characterization of transcripts. Cell. 1982;30:763–773. doi: 10.1016/0092-8674(82)90281-1. [DOI] [PubMed] [Google Scholar]
- Guo L, Allen EM, Miller WA. Base-pairing between untranslated regions facilitates translation of uncapped, nonpolyadenylated viral RNA. Mol Cell. 2001;7:1103–1109. doi: 10.1016/S1097-2765(01)00252. [DOI] [PubMed] [Google Scholar]
- Guo YE, Steitz JA. Virus meets host microRNA: the destroyer, the booster, the hijacker. Mol Cell Biol. 2014;34:3780–3787. doi: 10.1128/MCB.00871-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmings-Mieszczak M, Steger G, Hohn T. Alternative structures of the cauliflower mosaic virus 35 S RNA leader: implications for viral expression and replication. J Mol Biol. 1997;267:1075–1088. doi: 10.1006/jmbi.1997.0929. [DOI] [PubMed] [Google Scholar]
- Hohn T. RNA based viral silencing suppression in plant pararetroviruses. Front Plant Sci. 2015;6:398. doi: 10.3389/fpls.2015.00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn T, Rothnie H. Plant pararetroviruses: replication and expression. Curr Opin Virol. 2013;3:621–628. doi: 10.1016/j.coviro.2013.08.013. [DOI] [PubMed] [Google Scholar]
- Hollingsworth MJ, Kim J-K, Stollar NE. Heelprinting analysis of in vivo ribosome pause sites. In: Martin R, editor. Protein Synthesis: Methods and Protocols. Humana Press; Totowa, NJ: 1998. pp. 153–165. [DOI] [PubMed] [Google Scholar]
- Iwakawa HO, Kaido M, Mise K, Okuno T. cis-Acting core RNA elements required for negative-strand RNA synthesis and cap-independent translation are separated in the 3′-untranslated region of Red clover necrotic mosaic virus RNA1. Virology. 2007;369:168–181. doi: 10.1016/j.virol.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Iwakawa HO, Mizumoto H, Nagano H, Imoto Y, Takigawa K, Sarawaneeyaruk S, Kaido M, Mise K, Okuno T. A viral noncoding RNA generated by cis-element-mediated protection against 5′->3′ RNA decay represses both cap-independent and cap-dependent translation. J Virol. 2008;82:10162–10174. doi: 10.1128/JVI.01027-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa HO, Tajima Y, Taniguchi T, Kaido M, Mise K, Tomari Y, Taniguchi H, Okuno T. Poly(A)-binding protein facilitates translation of an uncapped/nonpolyadenylated viral RNA by binding to the 3′ untranslated region. J Virol. 2012;86:7836–7849. doi: 10.1128/JVI.00538-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiwan SD, White KA. Subgenomic mRNA transcription in Tombusviridae. RNA Biol. 2011;8:287–294. doi: 10.4161/rna.8.2.15195. [DOI] [PubMed] [Google Scholar]
- Jiwan SD, Wu B, White KA. Subgenomic mRNA transcription in tobacco necrosis virus. Virology. 2011;418:1–11. doi: 10.1016/j.virol.2011.07.005. [DOI] [PubMed] [Google Scholar]
- Johnston JC, Rochon DM. Deletion analysis of the promoter for the cucumber necrosis virus 0.9-kb subgenomic RNA. Virology. 1995;214:100–109. doi: 10.1006/viro.1995.9950. [DOI] [PubMed] [Google Scholar]
- Kelly L, Gerlach WL, Waterhouse PM. Characterisation of the subgenomic RNAs of an Australian isolate of barley yellow dwarf luteovirus. Virology. 1994;202:565–573. doi: 10.1006/viro.1994.1378. [DOI] [PubMed] [Google Scholar]
- Koev G, Liu S, Beckett R, Miller WA. The 3′-terminal structure required for replication of barley yellow dwarf virus RNA contains an embedded 3′ end. Virology. 2002;292:114–126. doi: 10.1006/viro.2001.1268. [DOI] [PubMed] [Google Scholar]
- Koev G, Miller WA. A positive-strand RNA virus with three very different subgenomic RNA promoters. J Virol. 2000;74:5988–5996. doi: 10.1128/JVI.74.13.5988-5996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koev G, Mohan BR, Miller WA. Primary and secondary structural elements required for synthesis of barley yellow dwarf virus subgenomic RNA1. J Virol. 1999;73:2876–2885. doi: 10.1128/jvi.73.4.2876-2885.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft JJ, Treder K, Peterson MS, Miller WA. Cation-dependent folding of 3′ cap-independent translation elements facilitates interaction of a 17-nucleotide conserved sequence with eIF4G. Nucleic Acids Res. 2013;41:3398–3413. doi: 10.1093/nar/gkt026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhao Y, Liu C, Yao G, Wu S, Hou C, Zhang M, Wang D. Callose deposition at plasmodesmata is a critical factor in restricting the cell-to-cell movement of Soybean mosaic virus. Plant Cell Rep. 2012;31:905–916. doi: 10.1007/s00299-011-1211-y. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhai H, Zhao K, Wu B, Wang X. Two suppressors of RNA silencing encoded by cereal-infecting members of the family Luteoviridae. J Gen Virol. 2012;93:1825–1830. doi: 10.1099/vir.0.042135-0. [DOI] [PubMed] [Google Scholar]
- Manokaran G, Finol E, Wang C, Gunaratne J, Bahl J, Ong EZ, Tan HC, Sessions OM, Ward AM, Gubler DJ, Harris E, Garcia-Blanco MA, Ooi EE. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science. 2015;350:217–221. doi: 10.1126/science.aab3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WA, Jackson J, Feng Y. Cis- and trans-regulation of luteovirus gene expression by the 3′ end of the viral genome. Virus Res. 2015;206:37–45. doi: 10.1016/j.virusres.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WA, Koev G. Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology. 2000;273:1–8. doi: 10.1006/viro.2000.0421. [DOI] [PubMed] [Google Scholar]
- Miller WA, Liu S, Beckett R. Barley yellow dwarf virus: Luteoviridae or Tombusviridae? Mol Plant Pathol. 2002;3:177–183. doi: 10.1046/j.1364-3703.2002.00112.x. [DOI] [PubMed] [Google Scholar]
- Minoia S, Navarro B, Delgado S, Di Serio F, Flores R. Viroid RNA turnover: characterization of the subgenomic RNAs of potato spindle tuber viroid accumulating in infected tissues provides insights into decay pathways operating in vivo. Nucleic Acids Res. 2015;43:2313–2325. doi: 10.1093/nar/gkv034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizumoto H, Tatsuta M, Kaido M, Mise K, Okuno T. Cap-independent translational enhancement by the 3′ untranslated region of red clover necrotic mosaic virus RNA1. J Virol. 2003;77:12113–12121. doi: 10.1128/JVI.77.22.12113-12121.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon SL, Anderson JR, Kumagai Y, Wilusz CJ, Akira S, Khromykh AA, Wilusz J. A noncoding RNA produced by arthropod-borne flaviviruses inhibits the cellular exoribonuclease XRN1 and alters host mRNA stability. RNA. 2012;18:2029–2040. doi: 10.1261/rna.034330.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysore KS, Senthil-Kumar M, editors. Plant Gene Silencing: Methods & Protocols. Humana Press; New York: 2015. [DOI] [Google Scholar]
- Nagy PD, Barajas D, Pogany J. Host factors with regulatory roles in tombusvirus replication. Curr Opin Virol. 2012;2:691–698. doi: 10.1016/j.coviro.2012.10.004. [DOI] [PubMed] [Google Scholar]
- Navarro B, Gisel A, Rodio ME, Delgado S, Flores R, Di Serio F. Small RNAs containing the pathogenic determinant of a chloroplast-replicating viroid guide the degradation of a host mRNA as predicted by RNA silencing. Plant J. 2012;70:991–1003. doi: 10.1111/j.1365-313X.2012.04940.x. [DOI] [PubMed] [Google Scholar]
- Newburn LR, White KA. Cis-acting RNA elements in positive-strand RNA plant virus genomes. Virology. 2015;479-480:434–443. doi: 10.1016/j.virol.2015.02.032. [DOI] [PubMed] [Google Scholar]
- Okuno T, Hiruki C. Molecular biology and epidemiology of dianthoviruses. Adv Virus Res. 2013;87:37–74. doi: 10.1016/B978-0-12-407698-3.00002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palukaitis P. Satellite RNAs and satellite viruses. Mol Plant-Microbe Interact. 29 doi: 10.1094/MPMI-10-15-0232-FI. n press. [DOI] [PubMed] [Google Scholar]
- Pathak KB, Pogany J, Nagy PD. Non-template functions of the viral RNA in plant RNA virus replication. Curr Opin Virol. 2011;1:332–338. doi: 10.1016/j.coviro.2011.09.011. [DOI] [PubMed] [Google Scholar]
- Pijlman GP, Funk A, Kondratieva N, Leung J, Torres S, van der Aa L, Liu WJ, Palmenberg AC, Shi PY, Hall RA, Khromykh AA. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe. 2008;4:579–591. doi: 10.1016/j.chom.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Qi X, Bao FS, Xie Z. Small RNA deep sequencing reveals role for Arabidopsis thaliana RNA-dependent RNA polymerases in viral siRNA biogenesis. PLoS One. 2009;4:e4971. doi: 10.1371/journal.pone.0004971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rackwitz HR, Rohde W, Sänger HL. DNA-dependent RNA polymerase II of plant origin transcribes viroid RNA into full-length copies. Nature. 1981;291:297–301. doi: 10.1038/291297a0. [DOI] [PubMed] [Google Scholar]
- Rakotondrafara AM, Polacek C, Harris E, Miller WA. Oscillating kissing stem-loop interactions mediate 5′ scanning-dependent translation by a viral 3′-cap-independent translation element. RNA. 2006;12:1893–1906. doi: 10.1261/rna.115606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roby JA, Pijlman GP, Wilusz J, Khromykh AA. Noncoding subgenomic flavivirus RNA: multiple functions in West Nile virus pathogenesis and modulation of host responses. Viruses. 2014;6:404–427. doi: 10.3390/v6020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochon D, Lommel SA, Martelli GP, Rubino L, Russo M. Tombusviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy: Ninth report of the International Committee on Taxonomy of Viruses. Elsevier; San Diego: 2012. pp. 1111–1138. [Google Scholar]
- Salem NM, Miller WA, Rowhani A, Golino DA, Moyne A-L, Falk BW. Rose spring dwarf-associated virus has RNA structural and gene-expression features like those of Barley yellow dwarf virus. Virology. 2008;375:354–360. doi: 10.1016/j.virol.2008.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheets K. Maize chlorotic mottle machlomovirus expresses its coat protein from a 1.47-kb subgenomic RNA and makes a 0.34-kb subgenomic RNA. Virology. 2000;267:90–101. doi: 10.1006/viro.1999.0107. [DOI] [PubMed] [Google Scholar]
- Schnettler E, Sterken MG, Leung JY, Metz SW, Geertsema C, Goldbach RW, Vlak JM, Kohl A, Khromykh AA, Pijlman GP. Noncoding flavivirus RNA displays RNA interference suppressor activity in insect and Mammalian cells. J Virol. 2012;86:13486–13500. doi: 10.1128/JVI.01104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuessler A, Funk A, Lazear HM, Cooper DA, Torres S, Daffis S, Jha BK, Kumagai Y, Takeuchi O, Hertzog P, Silverman R, Akira S, Barton DJ, Diamond MS, Khromykh AA. West Nile virus noncoding subgenomic RNA contributes to viral evasion of the type I interferon-mediated antiviral response. J Virol. 2012;86:5708–5718. doi: 10.1128/JVI.00207-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen R, Miller WA. Subgenomic RNA as a riboregulator: negative regulation of RNA replication by Barley yellow dwarf virus subgenomic RNA 2. Virology. 2004;327:196–205. doi: 10.1016/j.virol.2004.06.025. [DOI] [PubMed] [Google Scholar]
- Shen R, Rakotondrafara AM, Miller WA. trans regulation of cap-independent translation by a viral subgenomic RNA. J Virol. 2006;80:10045–10054. doi: 10.1128/JVI.00991-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen WJ, Forde BG. Efficient transformation of Agrobacterium spp. by high voltage electroporation. Nucleic Acids Res. 1989;17:8385. doi: 10.1093/nar/17.20.8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura H, Pantaleo V, Ishihara T, Myojo N, Inaba J, Sueda K, Burgyán J, Masuta C. A viral satellite RNA induces yellow symptoms on tobacco by targeting a gene involved in chlorophyll biosynthesis using the RNA silencing machinery. PLoS Pathog. 2011;7:e1002021. doi: 10.1371/journal.ppat.1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon AE, Miller WA. 3′ cap-independent translation enhancers of plant viruses. Annu Rev Microbiol. 2013;67:21–42. doi: 10.1146/annurev-micro-092412-155609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sit TL, Vaewhongs AA, Lommel SA. RNA-mediated trans-activation of transcription from a viral RNA. Science. 1998;281:829–832. doi: 10.1126/science.281.5378.829. [DOI] [PubMed] [Google Scholar]
- Smirnova E, Firth AE, Miller WA, Scheidecker D, Brault V, Reinbold C, Rakotondrafara AM, Chung BY, Ziegler-Graff V. Discovery of a Small Non-AUG-Initiated ORF in Poleroviruses and Luteoviruses That Is Required for Long-Distance Movement. PLoS Pathog. 2015;11:e1004868. doi: 10.1371/journal.ppat.1004868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith NA, Eamens AL, Wang MB. Viral small interfering RNAs target host genes to mediate disease symptoms in plants. PLoS Pathog. 2011;7:e1002022. doi: 10.1371/journal.ppat.1002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima Y, Iwakawa HO, Kaido M, Mise K, Okuno T. A long-distance RNA-RNA interaction plays an important role in programmed -1 ribosomal frameshifting in the translation of p88 replicase protein of Red clover necrotic mosaic virus. Virology. 2011;417:169–178. doi: 10.1016/j.virol.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuta M, Mizumoto H, Kaido M, Mise K, Okuno T. The red clover necrotic mosaic virus RNA2 trans-activator is also a cis-acting RNA2 replication element. J Virol. 2005;79:978–986. doi: 10.1128/JVI.79.2.978-986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treder K, Kneller EL, Allen EM, Wang Z, Browning KS, Miller WA. The 3′ cap-independent translation element of Barley yellow dwarf virus binds eIF4F via the eIF4G subunit to initiate translation. RNA. 2008;14:134–147. doi: 10.1261/rna.777308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsushima D, Adkar-Purushothama C, Taneda A, Sano T. Changes in relative expression levels of viroid-specific small RNAs and microRNAs in tomato plants infected with severe and mild symptom-inducing isolates of Potato spindle tuber viroid. J Gen Plant Pathol. 2015;81:49–62. doi: 10.1007/s10327-014-0566-7. [DOI] [Google Scholar]
- Urosevic N, van Maanen M, Mansfield JP, Mackenzie JS, Shellam GR. Molecular characterization of virus-specific RNA produced in the brains of flavivirus-susceptible and -resistant mice after challenge with Murray Valley encephalitis virus. J Gen Virol. 1997;78:23–29. doi: 10.1099/0022-1317-78-1-23. [DOI] [PubMed] [Google Scholar]
- Wang S, Browning KS, Miller WA. A viral sequence in the 3′-untranslated region mimics a 5′ cap in facilitating translation of uncapped mRNA. EMBO J. 1997;16:4107–4116. doi: 10.1093/emboj/16.13.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Guo L, Allen E, Miller WA. A potential mechanism for selective control of cap-independent translation by a viral RNA sequence in cis and in trans. RNA. 1999;5:728–738. doi: 10.1017/S1355838299981979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z, Kim KH, Giesman-Cookmeyer D, Lommel SA. The roles of the red clover necrotic mosaic virus capsid and cell-to-cell movement proteins in systemic infection. Virology. 1993;192:27–32. doi: 10.1006/viro.1993.1004. [DOI] [PubMed] [Google Scholar]
- Yamagishi N, Terauchi H, Kanematsu S, Hidaka S. Characterization of the small subgenomic RNA of Soybean dwarf virus. Arch Virol. 2003;148:1827–1834. doi: 10.1007/s00705-003-0137-2. [DOI] [PubMed] [Google Scholar]
- Zavriev SK, Hickey CM, Lommel SA. Mapping of the red clover necrotic mosaic virus subgenomic RNA. Virology. 1996;216:407–410. doi: 10.1006/viro.1996.0076. [DOI] [PubMed] [Google Scholar]
- Zhong X, Archual AJ, Amin AA, Ding B. A genomic map of viroid RNA motifs critical for replication and systemic trafficking. Plant Cell. 2008;20:35–47. doi: 10.1105/tpc.107.056606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH. Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 2000;24:265–273. doi: 10.1046/j.1365-313x.2000.00868.x. [DOI] [PubMed] [Google Scholar]


