Abstract
Increasing hepatic stellate cell (HSC) death is a very attractive approach for limiting liver fibrosis. Tyrosine kinase inhibitors have been shown to have anti-fibrotic properties, but the mechanisms are poorly understood. Here, we identified the mechanism of action of the second-generation tyrosine kinase inhibitor nilotinib in inducing HSC death. Human HSC line (LX-2) and rat HSCs were treated with nilotinib and its predecessor, imatinib, in the absence or presence of various blockers, known to interfere with death signaling pathways. Nilotinib, but not imatinib, induced progressive cell death of activated, but not quiescent, HSCs in a dose-dependent manner. Activated HSCs died through apoptosis, as denoted by increased DNA fragmentation and caspase activation, and through autophagy, as indicated by the accumulation of autophagic markers, light chain (LC)3A-II and LC3B-II. Although inhibition of caspases with Z-VAD-FMK suppressed nilotinib-induced HSCs apoptosis, there was no increase in HSCs survival, because autophagy was exacerbated. However, blocking the mitochondrial permeability transition pore (mPTP) opening with cyclosporin A completely abolished both apoptosis and autophagy due to nilotinib. Moreover, nilotinib treatment decreased the protein expression of histone deacetylases 1, 2 and 4. Interestingly, pretreament with C646, a selective p300/CBP histone acetyl transferase inhibitor, resulted in diverting nilotinib-induced apoptosis and autophagy towards necrosis. In conclusion, the identification of mPTP as a target of nilotinib in activated HSCs suggests a coordination with histone deacetylases inhibition to induce apoptosis and autophagy. Thus, our study provides novel insights into the anti-fibrotic effects of nilotinib.
Keywords: Imatinib, Nilotinib, Hepatic fibrosis, Autophagy, Apoptosis, Histone Deacetylases
Introduction
Hepatic fibrosis is a wound-healing response characterized by the accumulation of extracellular matrix (ECM) proteins as result of chronic liver injury. Activation of hepatic stellate cells (HSCs), the main source of ECM proteins, represents a crucial event in the pathogenic sequence of fibrosis and thus, provides an important framework to define potential strategies for anti-fibrotic therapy [1]. Beside the inhibition of fibrogenic cytokines, the induction of HSC death is now considered an effective strategy for the resolution of hepatic fibrosis [2, 3]. The ideal cell death strategy should not alert the hepatic inflammatory response and should selectively target activated HSCs while maintaining quiescent HSCs. In contrast to the unorganized way of death “necrosis” that triggers the inflammatory response, apoptosis and autophagy proceed with the participation of organized and controlled cellular processes, which achieve a cleaner cellular execution [4].
Death by apoptosis commonly signals through the activation of initiator and executioner caspases, resulting in the formation of apoptotic bodies that are removed by phagocytes [5]. The autophagic pathway involves the formation of a membrane around a targeted region of the cell, resulting in the autophagosome formation, which fuses with a lysosome to form an autophagolysosome where the contents are degraded via acidic lysosomal hydrolases [6, 7]. Autophagy has been reported to be involved in the degradation of long-lived proteins and organelles in order to promote survival and activation of HSCs [8]. Most recently, autophagy has been implicated in the release of lipid droplets from HSCs that further promoted fibrogenesis of activated HSCs [9]. Paradoxically, Rickmann et al. have reported that tocotrienols counteracted pancreatic fibrogenesis through the induction of apoptosis and autophagy in activated pancreatic stellate cells [10]. Thus, the contribution of autophagy to cell survival or death depends on the type of stimulus and the cell context.
Nilotinib is a second-generation tyrosine kinase inhibitor that was originally developed for treatment of chronic myeloid leukemia resistant to treatment with its predecessor, imatinib [11]. There has been a recent interest regarding the potential efficacy of nilotinib in the treatment of the most common and progressive forms of fibrosis. These include treatment of pulmonary fibrosis [12], renal fibrosis [13], dermal fibrosis [14] and hepatic fibrosis [15–17]. However, the molecular mechanisms involved in nilotinib anti-fibrotic effects have not been completely characterized.
In the present study, we provide novel insights into the anti-fibrotic effects of nilotinib in vitro using human HSC line (LX-2) and rat HSCs. Herein, we report that nilotinib, but not imatinib, induces apoptosis and autophagic cell death of activated HSCs through the intrinsic mitochondrial pathway. Notably, when nilotinib-induced apoptosis was blocked by caspase inhibition, cells failed to survive not due to a switch towards necrosis, but because autophagy was exacerbated. Thus, it is presumable that nilotinib is triggering autophagy as an alternative process of cell death rather than a survival mechanism. Besides, we reveal novel targets for nilotinib, including histone deacetylases (HDACs) 1, 2 and 4 that we believe to be involved in nilotinib-induced cytotoxicity of HSCs.
Materials and methods
Cell culture and treatments
LX-2 cells, immortalized human HSCs, were obtained from Dr. Scott Friedman (Mount Sinai School of Medicine) and were cultured in Dulbecco’s modified eagle medium (DMEM, high glucose) plus 5% (v/v) FBS and 1% (v/v) penicillin-streptomycin [18]. Primary rat HSCs were routinely prepared by collagenase/pronase digestion of liver using a perfusion system and were isolated using the Optiprep gradient as previously described [19, 20]. Primary rat HSCs were cultured in M-199 medium [10% v/v FBS + 1% v/v penicillin-streptomycin + 2mM L-glutamine + amphotericin B (2.5μg/ml) + gentamicin (20 μg/ml)]. Our studies were conducted according to institutional ethical guidelines of Yale University. Imatinib and nilotinib were freshly prepared as 20 mM stock solution in DMSO and filter sterilized using a syringe filter (0.2 μm). After 24 hr of fasting at 0.2% (v/v) FBS, LX-2 cells were treated with imatinib or nilotinib (10 and 20 μM) with 0.2% (v/v) FBS for 24 hr, in the absence or presence of various agents dissolved in DMSO and applied 1 hr before, such as Z-VAD-FMK (50 μM), cyclosporin A (20 μM), 3-methyladenine (10 mM), AEBSF (100 μM), UCF-101 (2 μM) and C646 (1 μM). In clinical practice, steady-state peak plasma concentration of nilotinib was shown to be 4.27 μM after oral administration of 400 mg nilotinib twice daily [21]. We observed that 5 μM of nilotinib produced the same effects as 20 μM after longer time (48 hr rather than 24 hr). Thus, the used concentrations of nilotinib are in the same in vivo range.
Chemotaxis assay
The migration of LX-2 cells was studied using a 24-well plate equipped with transwell inserts (8-μm-pore polycarbonate-free filters, purchased from Costar®, Corning, NY, USA) as previously described [22] with a slight modification. Briefly, 250 μl of the cell suspension was added to the upper chamber of the transwell (2 × 104 cells/well). To the lower chamber, 500 μl of DMEM (containing 0% v/v FBS) was added per well. After 2 hr, LX-2 cells were pretreated with imatinib or nilotinib (10 and 20 μM) 1 hr before the addition of PDGF (10 ng/ml). After 24 hr, the cells were fixed and the lower surface of the membrane was stained using hematoxylin-eosin, and the upper surface of the membrane was gently scraped. Thereafter, 6 random fields per transwell were photographed using the high-power field (200x), and counted for the migrated cells.
Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, California, USA) according to the manufacturer’s instructions. Briefly, LX-2 cells were lysed with the reagent, chloroform was added, and the cellular RNA was precipitated by isopropyl alcohol. After washing with 75% ethanol, the RNA pellet was dissolved in RNAse free water. Total RNA was reverse transcribed to cDNA using the transcriptor first strand cDNA (Roche Applied Science, Mannheim, Germany). RNA (1 μg) in 13 μl of RNAse free water was added to 1 μl of oligo(dT)18 primer. After incubating at 65 °C for 10 min in the thermal cycler, 4 μl of 5× reverse transcriptase buffer, 1 μl of deoxynucleotide mix, 0.5 μl of protector RNAse inhibitor and 0.5 μl of transcriptor reverse transcriptase were added (total volume of 20 μl). The reaction was performed for 60 minutes at 50°C (cDNA synthesis) and 5 minutes at 85°C (enzyme denaturation). The previously prepared cDNA samples were prediluted first 1:10 by PCR-grade water. For every cDNA amplification, 4.9 μl of the diluted sample was loaded as duplicate in the 384-multiwell plate, and 5 μl of LightCycler® 480 probes master 2× (Roche Applied Science, Mannheim, Germany) in addition to 0.1 μl of human TGF-β1 or COL1A1 TaqMan primer-probe mixture (Applied Biosystems, Foster City, California, USA). The housekeeping gene GAPDH was used as a reference gene for normalisation and PCR-grade water was used as a negative control. Comparative quantitative real time PCR was performed with a LightCycler® 480 Real-Time PCR System (Roche Applied Science, Mannheim, Germany) using the “Mono Color Hydrolysis Probe” format, and the ΔΔCt method was used to determine the Relative expression.
Western Blotting
At the end, the cells were washed twice with ice-cold PBS and lysed in 100 μl of 1X RIPA buffer (Cell Signaling, Danvers, MA, USA) containing MiniEDTA Protease inhibitor (Roche Applied Science Mannheim, Germany) for 20 min on ice. The cell lysates were centrifuged at 12,000g for 10 min at 4 °C. Protein concentrations were measured with the Bradford assay. After denaturation, equal amounts of protein (20 μg) were subjected to gel electrophoresis (150 V, 400 mA) using NuPAGE® 4%–12% Bis-Tris mini gel (Invitrogen, Grand Island, NY, USA) and transferred to PVDF membranes (Millipore, MA, USA) at 4° C during applying a transfer voltage of (120 V, 400mA) for 90 min. After transfer, the PVDF membranes were blocked with TBST buffer containing 5% (w/v) BSA at room temperature for 1 hour. Without washing, one of the primary antibodies (specified in the supplementary table 1) was added in the TBST buffer containing containing 5% (w/v) BSA overnight (16 hr) at 4 °C. Thereafter, the blot was washed 3 times in TBST buffer to get rid of the extra primary antibody and was incubated with secondary antibody (HRP-conjugated) diluted with TBST buffer containing 5% (w/v) BSA for 1 hr. The blot was washed 3 times in TBST buffer to get rid of the extra secondary antibody, and an enhanced chemiluminscent kit substrate (Thermo Scientific, Waltham, MA, USA) was added for developing. Then, the blot was developed against a sensitive film in the dark room for certain time, and the film was visualized by Kodak Image Reader. Quantitation of protein expression was determined by densitometry of digitalized images by using Totallab software v6 (Newcastle, Tyne, England).
Flow cytometric analysis of apoptosis
LX-2 cells were treated with imatinib or nilotinib (10 and 20 μM) for 24 hr, in the absence or presence of the previously mentioned agents, but in the medium containing 1% rather than 0.2% (v/v) FBS. HSCs were labeled with annexin V–fluorescein isothiocyanate (AV-FITC, BD Pharmingen, San Joes, CA) and propidium iodide (PI) (Sigma) for flow cytometric analysis (Becton Dickinson, USA) using Cell Quest software as previously described [16].
Detection of Autophagic Vacuoles
After growing LX-2 cells to 70% confluence on glass coverslips in 12 well-plates, the medium was changed from 5% DMEM to 1% DMEM. After 24 hr of fasting, LX-2 cells were treated with 20 μM imatinib or nilotinib for further 24 hr. Thereafter, LX-2 cells were incubated with 0.05 mM monodansyl cadaverine in PBS at 37°C for 10 minutes. The cells were next washed 4 times with PBS and observed under a fluorescence microscope at excitation wavelength of 380 nm and emission wavelength of 525 nm as previously described [23].
Immunocytochemistry
LX-2 cells were cultured on glass coverslips in 24 well-plates and grown to approximately 60% confluence for three days to promote the adherence of cells. After 24 hr of fasting at 0.2% (v/v) FBS, LX-2 cells were treated with imatinib or nilotinib (10 and 20 μM) with 0.2% (v/v) FBS for 3 hr. Then, the cells were then washed twice with serum free media (DMEM) and then fixed in 4% paraformaldehyde in PBS for 30 minutes. After fixation, the cells were washed twice with PBS and then permeabilised with PBS containing 0.1% (v/v) triton X-100 for 30 minutes. They were next washed twice with PBS and incubated with blocking solution (5% w/v BSA in PBS) for 1 hr. Primary antibody (NF-κB, 1:200 in blocking solution) was added to the cells and incubated at 4°C overnight. In the next day, the cells were washed three times with DPBST and incubated with the secondary antibody (1:2000) at room temperature for 1 hr. Then, the cells were washed three times with DPBST, and DAB peroxidase substrate kit (Vector Laboratories, Burlingame CA, USA) was then added for 5 minutes according the manufacturer instructions. Then, the slides were washed one time with water, stained with haematoxylin and Permount® was used as the mounting media. After 15 min, the edges were coated with nail polish to prevent dryness of the mounting medium. Thereafter, 8 random fields per slide were photographed (100x), and counted for the dark brown stained nuclei.
Statistical analysis
Data were expressed as mean ± standard error (SE) of mean. Statistical evaluations of the results were carried out by means of one way analysis of variance, followed by Tukey-Kramer multiple comparison test. The level of significance was set at P<0.05. Statistical tests were performed with GraphPad Instat V 3.05 (GraphPad Software Inc, San Diego, CA, USA). Column bar graphs were performed with GraphPad Prism V 5.00 (GraphPad Software Inc, San Diego, CA, USA).
Results
Nilotinib inhibits chemotaxis, upregulation of COL1A1 and TGF-β1 mRNA, activation of HSCs and exhibits selective cytotoxicity towards activated HSCs, but not quiescent cells
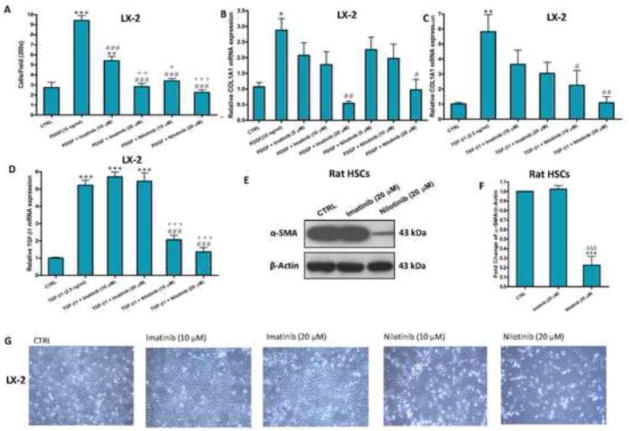
To examine whether nilotinib and its predecessor, imatinib, could inhibit the chemotaxis of LX-2 cells, PDGF-BB-induced migration of LX-2 cells using the transwell inserts was studied. Nilotinib (20 μM) showed the most prominent inhibitory effect against PDGF-BB-induced chemotaxis of LX-2 cells (Figure 1A). We next investigated the effects of nilotinib and imatinib on type I collagen synthesis in LX-2 cells in response to PDGF-BB and TGF-β1 stimulations. Quantitative RT-PCR demonstrated that both imatinib and nilotinib equally reduced PDGF-BB-induced COL1A1 mRNA expression in a dose-dependent manner (Figure 1B). On the other hand, TGF-β1-induced elevation in COL1A1 mRNA expression was significantly reduced only by treatment with nilotinib (20 μM) and nilotinib (10 μM) at P<0.01 and P<0.05, respectively, compared with the TGF-β1-treated LX-2 cells (Figure 1C). TGF-β1-induced elevation in TGF-β1 mRNA expression was reduced significantly (P<0.001) by nilotinib (10 and 20 μM), but not imatinib (10 and 20 μM), compared with the TGF-β1-treated LX-2 cells (Figure 1D). Nilotinib (20 μM), but not imatinib, completely suppressed the protein expression of α-SMA, a marker of HSCs activation, due to spontaneous activation of rat HSCs in culture media (day 11) (Figure 1E and F). Besides, nilotinib, but not imatinib, produced progressive cell death in LX-2 cells over a period of 24 hr in a dose-dependent fashion, as observed by phase-contrast microscopy (Figure 1G). To be considered a beneficial therapy for liver fibrosis, nilotinib and, to a lesser extent, imatinib demonstrated selective cytotoxicity toward transformed rat HSCs (i.e. activation HSCs to MFBs) at various timing (day 5, 8 and 11 after isolation, Supplemenatry Figure 1), but failed to induce any apoptosis for quiescent cells (day 1–3), which are not involved in fibrogenesis.
Figure 1. Nilotinib inhibits chemotaxis, upregulation of COL1A1 and TGF-β1 mRNA, and activation of HSCs, and induces progressive cell death of activated HSCs.
Effects of imatinib and nilotinib treatments for 24hr on chemotaxis (A), upregulation of COL1A1(B–C) and TGF-β1 (D) mRNA, activation (E–F) and viability (G) of HSCs. Data are means ± SE of at least 4 independent experiments. Statistically significant differences are indicated as: *P<0.05, **P<0.01 and ***P<0.001 versus CTRL; #P<0.05, ##P<0.01 and ###P<0.001 versus PDGF (10 ng/ml) or TGF-β1 (2.5 ng/ml); +P<0.05, ++P<0.01 and +++P<0.001 versus imatinib (10 μM); ΔΔΔP<0.001 versus imatinib (20 μM).
Nilotinib-induced cytotoxicity of HSCs is mainly apoptosis associated with cleavage of caspases
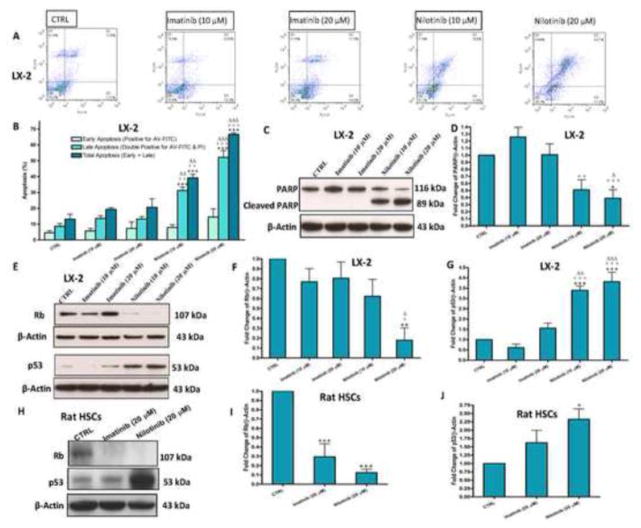
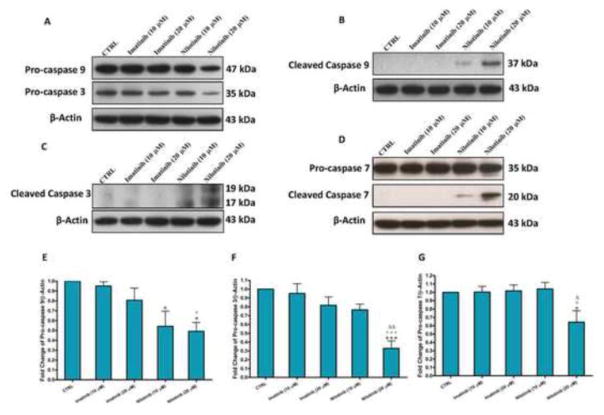
Flow cytometric analysis using AV-FITC and PI stainings (Figure 2A and B) showed that nilotinib when used at 10 and 20 μM for 24 hr resulted in approximately 40% and 65% total apoptosis, respectively. Besides, nilotinib induced cleavage of poly (ADP-ribose) polymerase (PARP) in LX-2 cells in a dose dependent manner (Figure 2C). Nilotinib also increased the protein expression of p53 in both LX-2 and transformed rat HSCs (day 11), which correlated with the decreased expression of the hypophosphorylated retinoblastoma (Rb) protein (Figure 2D–J). To examine whether nilotinib-induced apoptosis was related to caspases, we evaluated the effect of nilotinib on the expression of pro-caspases in LX-2 cells. Compared with the control untreated and imatinib treated cells, nilotinib (20 μM) treatment resulted in the reduction of the expression of pro-caspase 9, pro-caspase 3 and pro-caspase 7 in LX-2 cells, indicating a cleavage of these substrates (Figure 3). To confirm that the apoptotic effects produced by nilotinib on LX-2 cells were mediated by caspases cleavage, we verified that the cleaved products of caspase 9, caspase 3 and caspase 7 were present in the cells treated only with nilotinib (20 μM) and, to a lesser extent, nilotinib (10 μM).
Figure 2. Nilotinib, but not imatinib, induces apoptosis and DNA damage of activated HSCs in a dose-dependent manner.
Effects of imatinib and nilotinib treatments for 24hr on early and late (lower and upper right quadrant, respectively) apoptosis percentage (A–B) as assessed by flow cytometric analysis using AV-FITC (FL1-H) and PI (FL2-H) stainings, and the protein expression of PARP/cleaved PARP (C–D), Rb, p53 and their fold changes (E–J) in HSCs as assessed by the western blot for the total lysates. Data are means ± SE of at least 4 independent experiments. Statistically significant differences are indicated as: *P<0.05, **P<0.01 and ***P<0.001 versus the corresponding CTRL group; +P<0.05, ++P<0.01 and +++P<0.001 versus the corresponding imatinib (10 μM) group; ΔP<0.05, ΔΔP<0.01 and ΔΔΔP<0.001 versus the corresponding imatinib (20 μM) group.
Figure 3. Nilotinib-induced apoptosis of HSCs is associated with activation of caspases.
Effects of imatinib and nilotinib treatments for 24hr on the protein expression of pro-caspase 9/pro-caspase 3 (A), cleaved caspase 9 (B), cleaved caspase 3 (C), pro-caspase 7/cleaved caspase 7 (D) and the fold changes of procaspases (E–G) as assessed for the total lysates of LX-2 cells. Data are means ± SE of at least 4 independent experiments. Statistically significant differences are indicated as: *P<0.05 and ***P<0.001 versus CTRL; +P<0.05 and +++P<0.001 versus imatinib (10 μM); ΔΔP<0.01 versus imatinib (20 μM); ΔP<0.05 and ΔΔP<0.01 versus imatinib (20 μM).
Nilotinib-induced cytotoxicity of HSCs is completely inhibited by pretreatment with cyclosporin A, but not Z-VAD-FMK
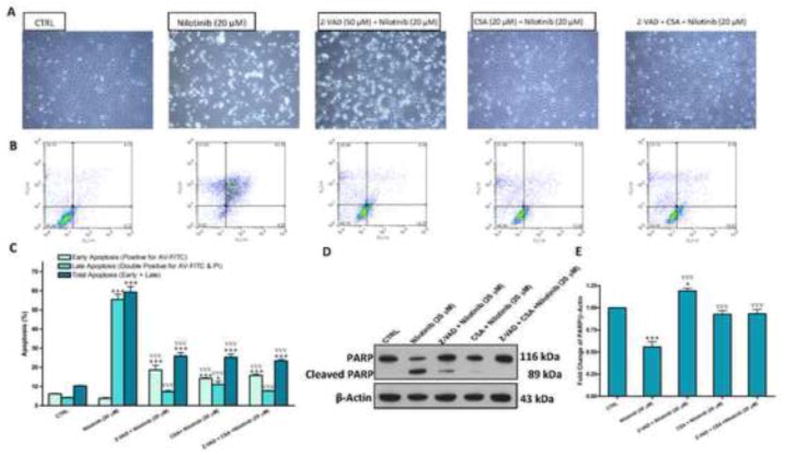
Next, we sought to investigate whether the blockade of this apoptosis with the pancaspase inhibitor Z-VAD-FMK (50 μM) would prevent nilotinib-induced cytotoxicity. As assessed by flow cytometric analysis, Z-VAD-FMK seemed to prevent nilotinib-induced apoptosis. However, by observing LX-2 Cells under the phase-contrast microscopy, Z-VAD-FMK hardly hampered nilotinib-induced cytotoxicity of LX-2 (Figure 4A, B and C), indicating that there are caspase independent pathways involved in nilotinib-induced cytotoxicity of LX-2. A key event of the apoptotic cell death mediated by the intrinsic mitochondrial pathway is the mitochondrial permeability transition pore (mPTP) opening. To investigate whether the cytotoxic effect of nilotinib is mediated by mPTP opening, we examined the effect of cyclosporin A, a mPTP blocker, on nilotinib-induced cytotoxicity. Cyclosporin A completely hampered nilotinib-induced cytotoxicity, as confirmed by the flow cytometric analysis and the normal appearance of LX-2 cells. We also examined pretreatment with both Z-VAD-FMK and cyclosporin A on nilotinib-induced, and we found that Z-VAD-FMK did not add any additive effect to cyclosporin A. By Western blotting, we observed that nilotinib-induced DNA damage, indicated by PARP cleavage, is significantly inhibited by Z-VAD-FMK, cyclosporin A and their combination (Figure 4D and E).
Figure 4. Blockade of the mPTP opening, but not inhibition of caspases, prevents HSCs death induced by nilotinib.
Effects of Z-VAD-FMK (Z-VAD, 50 μM) and cyclosporin A (CSA,20 μM) on nilotinib (20 μM)-induced apoptosis as evaluated under the phase contrast microscope (A) and by flow cytometric analysis using AV-FITC (FL1-H) and PI (FL2-H) staining (B–C), and the protein expression of PARP/cleaved PARP (D–E) as assessed by Western blotting for the total lysates of LX-2 cells. Data are means ± SE of at least 4 independent experiments. Statistically significant differences are indicated as: *P<0.05 and ***P<0.001 versus CTRL; ∇∇∇P<0.001 versus nilotinib (20 μM).
Nilotinib induced-cytotoxicity of HSCs involves autophagic cell death in addition to apoptosis
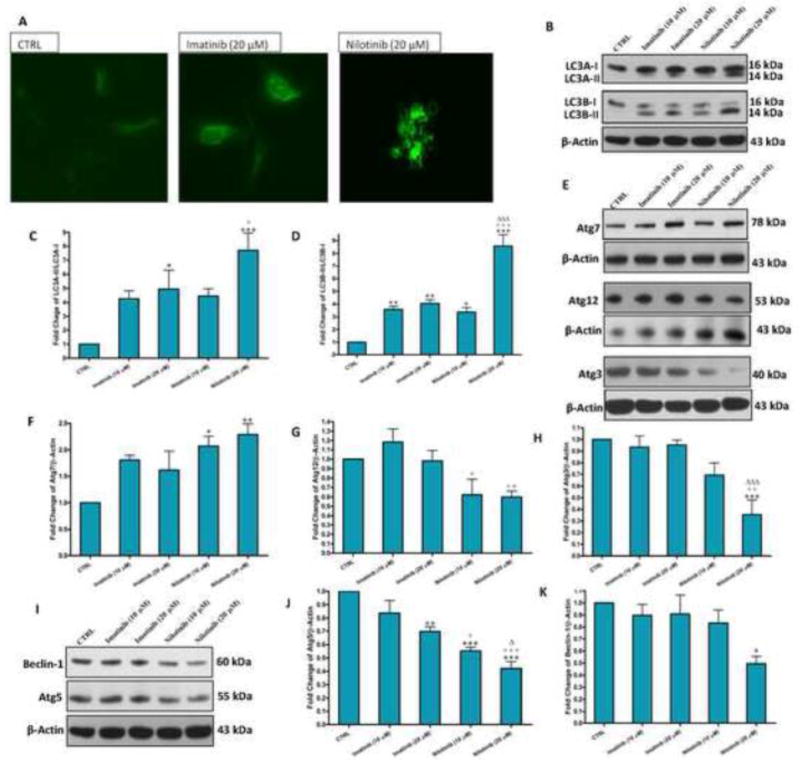
Autophagy has also been implicated in a type of programmed cell death type II called “autophagic cell death” that is different from apoptosis, which is programmed cell death type I [24]. We next investigated whether, beside apoptosis, nilotinib induced-cytotoxicity involves autophagic cell death. By using the autofluorescent monodansyl cadaverine dye, we observed that nilotinib and to less extent imatinib induced the formation of large autophagic vacuoles in LX-2 cells (Figure 5A). Moreover, nilotinib, and to a lesser extent, imatinib resulted in a pronounced accumulation of autophagic markers, light chain 3 (LC3)A-II and LC3B-II, as determined by Western blotting (Figure 5B–D). To further confirm our findings, we analyzed the protein expression of Atg7, which is involved in the conversion both LC3A-I and LC3B-I to LC3A-II and LC3B-II, respectively. As determined by Western blotting, nilotinib (10 and 20 μM) resulted in more than 2 folds increase in Atg7 (Figure 5E and F). The autophagic pathway is known to be initiated by an increase in the protein expression of beclin-1 that is correlated with an increase in the protein expression autophagy-related genes (Atgs) 3, 5 and 12, resulting in conversion of LC3-I into LC3-II. Unexpectedly, nilotinib (20 μM) resulted in almost 0.5 folds decrease, rather than increase, of the protein expression of beclin-1, Atgs 3, 5 and 12 (Figure 5E–K).
Figure 5. Nilotinib induced-cytotoxicity of HSCs involves autophagic cell death along with apoptosis.
Effects of imatinib and nilotinib treatments for 24hr on the monodansyl cadaverine-labeled autophagolysosome vacuoles by fluorescence microscopy (200×, A), and the protein expression of LC3A-I/II and LC3B-I/II (B–D), Atgs 7, 3, 12 and 5 (E–J), and beclin-1 (I, K) as assessed by Western blotting for the total lysates of LX-2 cells. Data are means ± SE of at least 4 independent experiments. Statistically significant differences are indicated as: *P<0.05, **P<0.01 and ***P<0.001 versus CTRL; +P<0.05, ++P<0.01 and +++P<0.001 versus imatinib (10 μM); ΔP<0.05 and ΔΔP<0.01 versus imatinib (20 μM).
Nilotinib-induced autophagy of HSCs is potentiated by Z-VAD-FMK, but inhibited by cyclosporin A
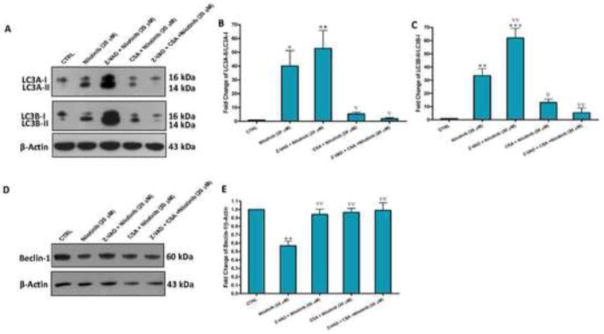
We next hypothesized that the mPTP opening is the upstream trigger of the autophagic cell death induced by nilotinib. To prove this hypothesis, we tested again pretreatment with Z-VAD-FMK, cyclosporin A and their combination on nilotinib-induced changes of autophagic markers, LC3A-II/I and LC3B-II/I, as well as beclin-1. Interestingly, we observed a severe increase in both LC3A-II/I and LC3B-II/I protein expressions upon pretreatment with Z-VAD-FMK (Figure 6A, B and C), which correlated with an increase in beclin-1 protein expression. On the other hand, cyclosporin A clearly suppressed the increase in both LC3A-II/I and LC3B-II/I protein expressions due to either nilotinib or nilotinib+Z-VAD-FMK and restored the decrease in beclin-1 protein expression, suggesting that the mPTP opening is involved in nilotinib-induced autophagic cell death (Figure 6D and E).
Figure 6. Nilotinib-induced autophagy of HSCs is exacerbated by caspases inhibition, but inhibited by the mPTP opening blockade.
Effects of Z-VAD-FMK (Z-VAD, 50 μM) and cyclosporin A (CSA, 20 μM) on nilotinib (20 μM)-induced changes of the protein expression of LC3A-I/II and LC3B-I/II (A–C), and beclin-1 (D–E) as assessed by Western blotting for the total lysates of LX-2 cells. Data are means ± SE of at least 4 independent experiments. Statistically significant differences are indicated as: *P<0.05, **P<0.01 and ***P<0.001 versus CTRL; ∇P<0.05 and ∇∇P<0.01 versus nilotinib (20 μM).
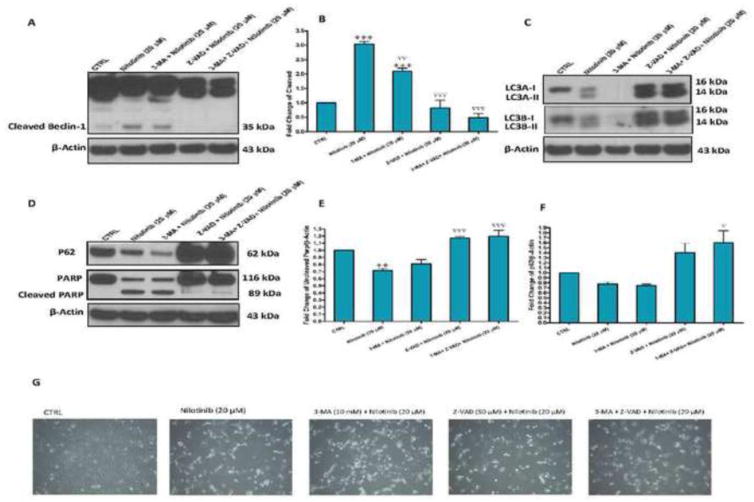
Nilotinib-induced cleavage of caspases is implicated in the cleavage of beclin-1
Next, we hypothesized that nilotinib-induced cleavage of caspases might be involved in the cleavage of beclin-1. To prove this hypothesis, we tested again pretreatment with Z-VAD-FMK, the autophagy inhibitor 3-methyladenine and their combination on nilotinib-induced changes of cleaved beclin-1. Interestingly, we observed a cleavage of beclin-1 in LX-2 cells treated with nilotinib, which was inhibited only by Z-VAD-FMK, indicating that beclin-1 is a substrate for cleaved caspases induced by nilotinib (Figure 7A and B). On the other hand, 3-methyladenine clearly suppressed the increase in both forms of LC3-I and II, but did not affect cleaved beclin-1 and Parp, and did not increase the viability (Figure 7C, D, E and G). Notably, autophagic flux was increased by nilotinib, as indicated by the decreased protein expression of p62 (Figure 7D and F).
Figure 7. Nilotinib-induced cleavage of caspases is implicated in the cleavage of beclin-1.
Effects of Z-VAD-FMK (Z-VAD, 50 μM) and 3-methyladenine (3-MA, 10 mM) on nilotinib (20 μM)-induced changes of the protein expression of cleaved beclin-1 (A–B), LC3A-I/II and LC3B-I/II (C), PARP/cleaved PARP and p62 (D–F) as assessed by Western blotting for the total lysates of LX-2 cells, as well as the viability(G). Data are means ± SE of at least 4 independent experiments. Statistically significant differences are indicated as: **P<0.01 and ***P<0.001 versus CTRL; ∇P<0.05, ∇∇P<0.01 and ∇∇∇P<0.001 versus nilotinib (20 μM).
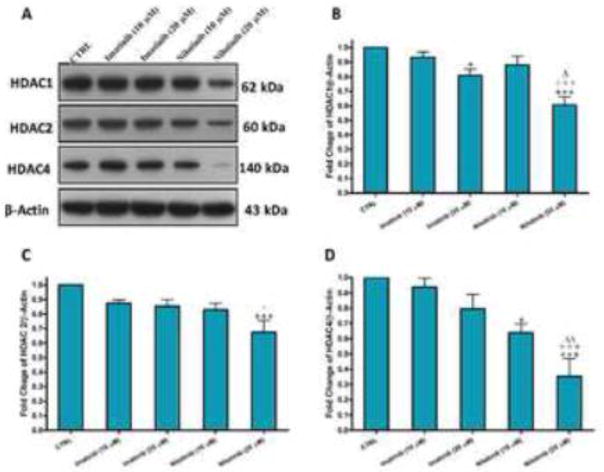
Nilotinib decreases the protein expression of histone deacetylases (HDACs) 1, 2 and 4 in HSCs
Inhibition of HDACs has been shown to mediate cell death via different mechanisms, including apoptosis by the intrinsic and extrinsic pathways, and autophagic cell death, as well as other types of cell death. To investigate whether nilotinib induced apoptosis might be related the inhibition of HDACs as the upstream signaling pathway, we examined effect of nilotinib on the protein expression of HDACs 1, 2 and 4 in LX-2 cells. Interestingly, nilotinib treatment decreased the protein expression of HDAC1, HDAC2 and more pronouncedly HDAC4 in a dose dependent manner (Figure 8A–D). Imatinib (20 μM) also resulted in a slight decrease in HDACs 1, 2 and 4 protein expressions, when compared with untreated control. We also investigated the protein expression of HDACs 3, 5 and 7 in LX-2 cell, but they could not be detected.
Figure 8. Nilotinib decreases the protein expression of HDACs 1, 2 and 4 in HSCs.
Effects of imatinib and nilotinib treatments for 24hr on the protein expression of HDACs 1, 2 and 4 as assessed by Western blotting for the total lysates of LX-2 cells. Data are means ± SE of at least 4 independent experiments. Statistically significant differences are indicated as: *P<0.05 and ***P<0.001 versus CTRL; +P<0.05 and +++P<0.001 versus imatinib (10 μM); ΔP<0.05 and ΔΔP<0.01 versus imatinib (20 μM).
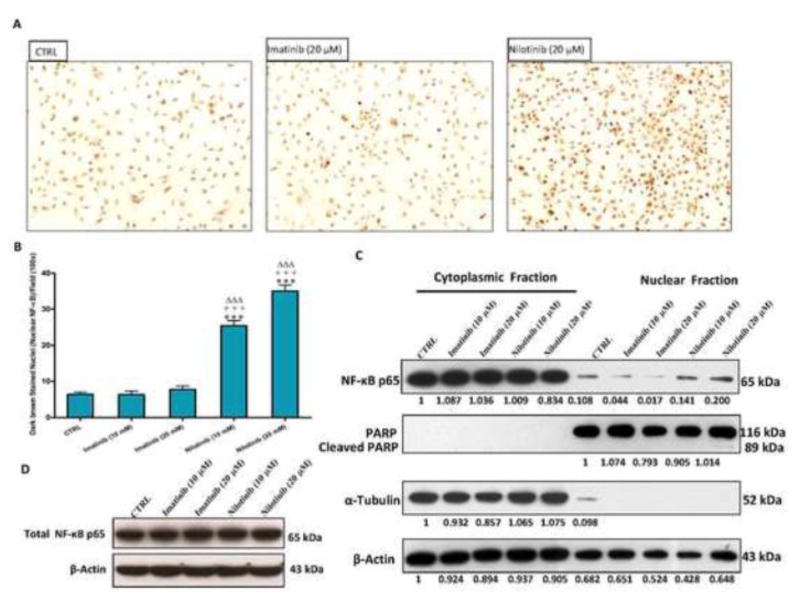
Nilotinib triggers NF-κB p65 nuclear translocation in HSCs
To examine whether nilotinib-induced apoptosis might be related to the inhibition of the nuclear translocation of the anti-apoptotic transcription factor nuclear factor-kappaB (NF-κB) p65, we evaluated the expression of total, cytoplasmic and nuclear NF-κB p65 in LX-2 cells. Nilotinib (10 and 20 μM) activated, rather than inhibited, NF-κB p65 nuclear translocation in LX-2 cells after 3 hr, as observed by immunocytochemistry (Figure 9A and B) and Western blotting for different cell fractions (Figure 9C and D). By counting the dark brown stained nuclei per field (100x), nilotinib at 10 and 20 μM resulted in a 4 and 5.5 fold increase in NF-κB p65 nuclear translocation, respectively. Unlike nilotinib, imatinib did not produce any significant increase on the nuclear translocation of NF-κB p65 in LX-2 cells with the same concentrations.
Figure 9. Nilotinib, but not imatinib, triggers the nuclear translocation of NF-κB p65 in HSCs.
Effects of imatinib and nilotinib treatments for 3 hr on NF-κB p65 nuclear translocation in LX-2 as evaluated by immunocytochemistry and counting the dark brown nuclei per field (100×, A–B), and by Western blot for different cell fractions (C–D). Data are means ± SE of at least 4 independent experiments. Statistically significant differences are indicated as: ***P<0.001 versus CTRL; +++P<0.001 versus imatinib (10 μM); ΔΔΔP<0.001 versus imatinib (20 μM).
Nilotinib induced-cytotoxicity of HSCs does not involve serine proteases, but histone acetylase transferases
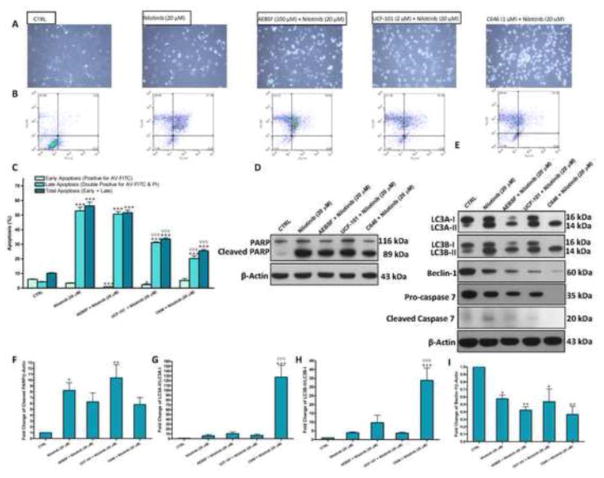
In addition to caspase-dependent apoptosis and autophagic cell death, we next hypothesized that serine proteases, including high temperature requirement protein A2 (HTRA2), might be involved in nilotinib cytotoxicity. To test this hypothesis, we tried the use of AEBSF (100 μM), a broad spectrum serine protease inhibitor, and UCF-101 (2 μM), a selective HTRA2 inhibitor, on nilotinib-induced cell death. Pretreatment with AEBSF failed to block nilotinib-induced cell death, but UCF-101 showed a slight protection (Figure 10A). To investigate whether the imbalance produced in histone acetylation due to nilotinib inhibition of HDACs might be involved in cell death, we tested pretreatment with C646 (1 μM), a selective p300/CBP histone acetyl transferase inhibitor. We observed that C646 slightly inhibited nilotinib-induced cell death, but its use was associated with morpholigcal changes in LX-2 cells (become rounded rather star shaped). As assessed by flow cytometric analysis (Figure 10B and C), it seemed that both UCF-101 and C646 significantly resulted in reduction of nilotinib-induced total apoptosis by approximately 41% and 55%, respectively. However, these results should be cautiously interpreted, because we observed that pretreatment with either UCF-101 or C646 resulted in 60% necrosis versus 30% necrosis for untreated and nilotinib (20 μM) treated cell populations. Both AEBSF (100 μM) and C646 (1 μM), but not UCF-101 (2 μM), slightly decreased nilotinib-induced PARP cleavage by 24% and 29%, respectively (Figure 10D and F). We observed that pretreatment with C646 produced a severe increase in LC3A-II/I and LC3B-II/I ratios, which was significant (P<0.001) not only from control, but also from nilotinib (20 μM)-treated cells (Figure 10E, G and H). Thus, the severe increase due to pretreatment with C646 in LC3A-II/I and LC3B-II/I ratios is due comparing the LC3A-II and LC3B-II to the background. These results suggest that the upstream signaling of LC3A-I and LC3B-I is not working, as confirmed by decreased expression of beclin-1 (Figure 10I). Pretreatment with AEBSF, but not UCF-101, decreased the expression of both LC3A-I+LC3A-II and LC3B-I+ LC3B-II, but the ratios of both LC3A-II/LC3A-I and LC3B-II/LC3B-I remained slightly higher than nilotinib (20 μM). Notably, we observed that pretreatment with C646 repressed constitutive basal caspases and there was no caspase cleavage, as denoted by pro-caspase 7 and cleaved caspase 7, respectively.
Figure 10. Nilotinib-induced apoptosis and autophagy are diverted towards necrosis by inhibition of histone acetyl transferases.
Effects of pretreatment with AEBSF (100 μM), UCF-101 (2 μM) and C646 (1 μM) on nilotinib (20 μM)-induced cytotoxicity as evaluated under the phase contrast microscope (A) and by flow cytometric analysis using AV-FITC (FL1-H) and PI (FL2-H) staining (B–C), and the protein expression of PARP/cleaved PARP, LC3A-I/II and LC3B-I/II (A–C), beclin-1 (D–E) and pro-caspase 7/cleaved caspase 7 (D–I) as assessed by Western blotting for the total lysates of LX-2 cells. Data are means ± SE of at least 4 independent experiments. Statistically significant differences are indicated as: *P<0.05, **P<0.01 and ***P<0.001 versus CTRL; ∇∇∇P<0.001 versus nilotinib (20 μM).
Discussion
A large amount of evidence supports the importance of HSCs apoptosis during the regression of liver fibrosis [25, 26]. Thus, selective removal of activated HSCs through induction of programmed death is a therapeutic strategy in hepatic fibrosis. With respect to the inhibitory effects of imatinib and nilotinib on chemotaxis, activation and collagen synthesis pathways in HSCs, we observed that the application of nilotinib, but not imatinib, induces apoptosis of HSCs. As an attempt to elucidate the molecular mechanism involved in nilotinib-induced apoptosis, we observed that application of nilotinib resulted in the cleavage of caspases and PARP, as well as extensive DNA damage associated with elevation of p53 protein expression [27]. The decrease in Rb protein expression due to nilotinib suggests deregulation of the cell cycle and subsequently, apoptosis of HSCs [28, 29]. This was further supported by the stimulation of NF-κB nuclear translocation in LX-2 cells by nilotinib, because activation of the transcription factor NF-κB results in up-regulation of not only anti-apoptotic genes, but also pro-apoptotic genes [30, 31].
Interestingly, pretreatment with cyclosporin A, but not Z-VAD-FMK, blocked nilotinib-induced cytotoxicity. Most recently, Liu et al. have shown that application of nilotinib with the same doses induced upregulation of TRAIL receptors and ligands in HSCs as confirmed by flow cytometric analysis and Western blot, respectively [32]. However, there was no impressive difference in TRAIL ligand expression levels between nilotinib (5, 10 and 20 μM) treated cells and the untreated control by observing their data. According to our data, the mPTP opening plays a crucial role in the cytotoxic effect of nilotinib as confirmed by the use of cyclosporin A, whereas the extrinsic pathway mediated by the death ligand TRAIL has a minor role in this apoptosis.
A novel aspect revealed by this study is that nilotinib triggers HSCs death not only by the classical apoptosis, but also through the autophagic cell death pathway, as denoted by accumulation autophagic vacuoles and LC3A-II and LC3B-II, as well as the increase in autophagic flux, as indicated by p62 decrease. Paradoxically, beclin-1, Atgs 3, 5 and 12 protein expressions were decreased, rather than increased. It was recently suggested that beclin-1 is cleaved by caspases during apoptosis in different cell lines [33, 34]. This finding is in agreement with our results, because pretreatment with Z-VAD-FMK reversed the decline in the uncleaved beclin-1 and prevented its cleavage due to nilotinib and resulted in both LC3A-II/I and LC3B-II/I accrual. In addition to blocking nilotinib-induced apoptosis, cyclosporin A efficiently blocked nilotinib-induced autophagic cell death, as well as the potent induction observed by combining nilotinib with Z-VAD-FMK. However, the autophagy inhibitor 3-methyladenine succeeded in preveting nilotinib-induced increase LC3A-II/I and LC3B-II/I, but failed to inhibit this increase when Z-VAD-FMK was included along with nilotinib.
One of the key events in the mitochondrial death pathway is the mitochondrial outer membrane permeabilization, which unleashes death-promoting factors that reside in the mitochondrial intermembrane space, including cytochrome c, apoptosis-inducing factor (AIF), endonuclease G (endo G) and HTRA2 [35, 36]. AIF and endo G function in a caspase-independent manner by relocating to the nucleus and mediating large-scale DNA fragmentation [37]. Our results suggest that AIF and endo G have a minor role in DNA fragmentation induced by nilotinib, because we could see only slight cleavage of PARP after pretreatment with Z-VAD-FMK. On the other hand, HTRA2 facilitates caspase activation by sequestering and/or degrading several members of XIAP, the inhibitor protein of apoptosis [37], which can block both the initiation phase, by inhibiting caspase-9, and the execution phase, by blocking caspase-3 and caspase-7, of the cascade [38]. Besides, HTRA2 also contributes to caspase-independent apoptosis by cleaving a wide array of cellular substrates, the most important are cytoskeletal proteins [39]. Accordingly, we investigated the effect of inhibition of HTRA2 on nilotinib induced cytotoxicity by the use of the general serine protease inhibitor AEBSF and the selective HTRA2 inhibitor UCF-101. Our results support that HTRA2 effect in HSCs cells is not involved in autophagy, which contradicts a recently published report [40].
We observed also for the first time that application of nilotinib to LX-2 cells resulted in the inhibition of the protein expression of HDAC 1, HDAC 2 and more pronouncedly HDAC 4 in a dose dependent manner. Accordingly, nilotinib induced cytotoxicity for HSCs might be related to the inhibition of HDACs. It has been reported that HDAC1, but not HDAC 2, is required for TGF- β1-induced epithelial-mesenchymal transition, and its high expression level in majority of hepatocellular carcinoma samples suggests that HDAC1 can be related with the invasiveness of hepatocellular carcinoma [41]. Most recently, it has been reported that the expression level of HDAC4 profoundly increased during HSCs transdifferentiation and acted as a repressor for the MMPs 9 and 13 [42]. Otherwise, Liu et al. have found that nilotinib inhibited not only Smad2 signaling induced by TGF-β1, but also phosphorylation of ERK and Akt stimulated by TGF-β1 in vivo during CCl4 and BDL-induced liver fibrosis. Akt is involved in TGF-β1-induced down-regulation of MMP 13 expression [43]. Moreover, HDAC inhibitors have been shown to repress TGF-β1-induced ERK and PI3K/Akt in fibroblasts without altering the Smad pathway [44]. Taken these findings together, nilotinib-induced inhibition of HDACs, and in particular HDAC4, might be the upstream for the up-regulation of MMPs and subsequently, resolution of liver fibrosis via degradation of ECM proteins.
An unresolved question is how HDAC inhibitors induce cell death. Recent studies suggest that acetylation of nonhistone proteins may play an important role in the biological effects of this class of compounds, and may explain lack of correlation between histone acetylation and induction of cell death by HDAC inhibitors in some circumstances [45]. Pretreatment with the selective p300/CBP histone acetyl transferase inhibitor C646 resulted in decreasing nilotinib-induced apoptosis and diversion of apoptosis to necrosis (as summarized in Supplementary Figure 2 and 3). Besides, C646 resulted in a slight decrease in PARP cleavage and a complete suppression of both LC3A-I and LC3B-I, as well as beclin-1. Besides, C646 repressed constitutive basal caspases and there was no caspase cleavage. The last observation suggests that the upstream signalling pathways of nilotinib-induced autophagy and apoptosis are inhibited by C646, which resulted in caspases-independent postlytic DNA damage rather than the caspases-dependent prelytic DNA damage. When a cell is undergoing death and the mitochondria is not ensuring its energy needs, caspase inhibition can switch apoptosis to necrosis because of insufficient ATP to support the active process of apoptosis [46], and this was the case with C646 pretreatment to block-nilotinib induced cell death. However, this was not the case when we blocked caspase activation in nilotinib-treated LX-2 cells because Z-VAD-FMK has no effect on mitochondria, and there was no significant increase in necrotic cells.
Conclusions
Our results indicate that the inhibition of HDACs by nilotinib is involved in the apoptosis and autophagic cell death. Overall, this study suggests that nilotinib may be valuable in the treatment of hepatic fibrosis in humans.
Supplementary Material
Highlights.
We recently reported the antifibrotic activity of nilotinib in vivo.
Here, we observed in vitro that nilotinib induces death of activated hepatic stellate cells.
We identified the mechanism of nilotinib in inducing HSC death, which involves apoptosis and autophagy.
Both effects were mediated through mitochondrial permeability transition pore opening.
The upstream signaling trigger for both effects was the inhibition of histone deacetylases 1, 2 and 4.
Acknowledgments
This work was supported by a grant (JS-2586 for M.E. Shaker) from the Egyptian Cultural Affairs and Missions Sector (Cairo, Egypt) in cooperation with the Egyptian Cultural and Educational Bureau (Washington DC, USA) in the Joint Supervision Program, and a grant from the Science and Technology Development Fund (STDF no. 1739 to G.E. Shiha), as well as a VA Merit award and NIH grant (R01DK076674-01A2 for W.Z. Mehal).
Footnotes
Conflict of interest
The authors have no conflict of interest to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Friedman SL. Hepatic fibrosis -- overview. Toxicology. 2008;254:120–129. doi: 10.1016/j.tox.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 2.Mehal W, Imaeda A. Cell death and fibrogenesis. Semin Liver Dis. 2010;30:226–231. doi: 10.1055/s-0030-1255352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kisseleva T, Brenner DA. Anti-fibrogenic strategies and the regression of fibrosis. Best Pract Res Clin Gastroenterol. 2011;25:305–317. doi: 10.1016/j.bpg.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edinger AL, Thompson CB. Death by design: apoptosis, necrosis and autophagy. Curr Opin Cell Biol. 2004;16:663–669. doi: 10.1016/j.ceb.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 5.Taylor RC, Cullen SP, Martin SJ. Apoptosis: controlled demolition at the cellular level. Nat Rev Mol Cell Bio. 2008;9:231–241. doi: 10.1038/nrm2312. [DOI] [PubMed] [Google Scholar]
- 6.Fengsrud M, Erichsen ES, Berg TO, Raiborg C, Seglen PO. Ultrastructural characterization of the delimiting membranes of isolated autophagosomes and amphisomes by freeze-fracture electron microscopy. Eur J Cell Biol. 2000;79:871–882. doi: 10.1078/0171-9335-00125. [DOI] [PubMed] [Google Scholar]
- 7.Rubinsztein DC, DiFiglia M, Heintz N, Nixon RA, Qin ZH, Ravikumar B, Stefanis L, Tolkovsky A. Autophagy and its possible roles in nervous system diseases, damage and repair. Autophagy. 2005;1:11–22. doi: 10.4161/auto.1.1.1513. [DOI] [PubMed] [Google Scholar]
- 8.Thoen LF, Guimaraes EL, Dolle L, Mannaerts I, Najimi M, Sokal E, van Grunsven LA. A role for autophagy during hepatic stellate cell activation. J Hepatol. 2011;55:1353–1360. doi: 10.1016/j.jhep.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 9.Hernandez-Gea V, Ghiassi-Nejad Z, Rozenfeld R, Gordon R, Fiel MI, Yue Z, Czaja MJ, Friedman SL. Autophagy Releases Lipid That Promotes Fibrogenesis by Activated Hepatic Stellate Cells in Mice and in Human Tissues. Gastroenterology. 2012 doi: 10.1053/j.gastro.2011.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rickmann M, Vaquero EC, Malagelada JR, Molero X. Tocotrienols induce apoptosis and autophagy in rat pancreatic stellate cells through the mitochondrial death pathway. Gastroenterology. 2007;132:2518–2532. doi: 10.1053/j.gastro.2007.03.107. [DOI] [PubMed] [Google Scholar]
- 11.Deremer DL, Ustun C, Natarajan K. Nilotinib: a second-generation tyrosine kinase inhibitor for the treatment of chronic myelogenous leukemia. Clin Ther. 2008;30:1956–1975. doi: 10.1016/j.clinthera.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 12.Rhee CK, Lee SH, Yoon HK, Kim SC, Lee SY, Kwon SS, Kim YK, Kim KH, Kim TJ, Kim JW. Effect of nilotinib on bleomycin-induced acute lung injury and pulmonary fibrosis in mice. Respiration. 2011;82:273–287. doi: 10.1159/000327719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iyoda M, Shibata T, Hirai Y, Kuno Y, Akizawa T. Nilotinib attenuates renal injury and prolongs survival in chronic kidney disease. J Am Soc Nephrol. 2011;22:1486–1496. doi: 10.1681/ASN.2010111158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akhmetshina A, Dees C, Pileckyte M, Maurer B, Axmann R, Jungel A, Zwerina J, Gay S, Schett G, Distler O, Distler JH. Dual inhibition of c-abl and PDGF receptor signaling by dasatinib and nilotinib for the treatment of dermal fibrosis. FASEB J. 2008;22:2214–2222. doi: 10.1096/fj.07-105627. [DOI] [PubMed] [Google Scholar]
- 15.Shaker ME, Salem HA, Shiha GE, Ibrahim TM. Nilotinib counteracts thioacetamide-induced hepatic oxidative stress and attenuates liver fibrosis progression. Fundam Clin Pharmacol. 2011;25:248–257. doi: 10.1111/j.1472-8206.2010.00824.x. [DOI] [PubMed] [Google Scholar]
- 16.Shaker ME, Shiha GE, Ibrahim TM. Comparison of early treatment with low doses of nilotinib, imatinib and a clinically relevant dose of silymarin in thioacetamide-induced liver fibrosis. Eur J Pharmacol. 2011;670:593–600. doi: 10.1016/j.ejphar.2011.08.041. [DOI] [PubMed] [Google Scholar]
- 17.Shaker ME, Zalata KR, Mehal WZ, Shiha GE, Ibrahim TM. Comparison of imatinib, nilotinib and silymarin in the treatment of carbon tetrachloride-induced hepatic oxidative stress, injury and fibrosis. Toxicol Appl Pharmacol. 2011;252:165–175. doi: 10.1016/j.taap.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu L, Hui AY, Albanis E, Arthur MJ, O’Byrne SM, Blaner WS, Mukherjee P, Friedman SL, Eng FJ. Human hepatic stellate cell lines, LX-1 and LX-2: new tools for analysis of hepatic fibrosis. Gut. 2005;54:142–151. doi: 10.1136/gut.2004.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiskirchen R, Gressner AM. Isolation and culture of hepatic stellate cells. Methods Mol Med. 2005;117:99–113. doi: 10.1385/1-59259-940-0:099. [DOI] [PubMed] [Google Scholar]
- 20.Ma J, Li F, Liu L, Cui D, Wu X, Jiang X, Jiang H. Raf kinase inhibitor protein inhibits cell proliferation but promotes cell migration in rat hepatic stellate cells. Liver Int. 2009;29:567–574. doi: 10.1111/j.1478-3231.2009.01981.x. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka C, Yin OQ, Sethuraman V, Smith T, Wang X, Grouss K, Kantarjian H, Giles F, Ottmann OG, Galitz L, Schran H. Clinical pharmacokinetics of the BCR-ABL tyrosine kinase inhibitor nilotinib. Clin Pharmacol Ther. 2010;87:197–203. doi: 10.1038/clpt.2009.208. [DOI] [PubMed] [Google Scholar]
- 22.Hashmi AZ, Hakim W, Kruglov EA, Watanabe A, Watkins W, Dranoff JA, Mehal WZ. Adenosine inhibits cytosolic calcium signals and chemotaxis in hepatic stellate cells. Am J Physiol Gastrointest Liver Physiol. 2007;292:G395–401. doi: 10.1152/ajpgi.00208.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biederbick A, Kern HF, Elsasser HP. Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles. Eur J Cell Biol. 1995;66:3–14. [PubMed] [Google Scholar]
- 24.Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, Blagosklonny MV, El-Deiry WS, Golstein P, Green DR, Hengartner M, Knight RA, Kumar S, Lipton SA, Malorni W, Nunez G, Peter ME, Tschopp J, Yuan J, Piacentini M, Zhivotovsky B, Melino G. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009;16:3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iredale JP. Hepatic stellate cell behavior during resolution of liver injury. Semin Liver Dis. 2001;21:427–436. doi: 10.1055/s-2001-17557. [DOI] [PubMed] [Google Scholar]
- 26.Issa R, Williams E, Trim N, Kendall T, Arthur MJ, Reichen J, Benyon RC, Iredale JP. Apoptosis of hepatic stellate cells: involvement in resolution of biliary fibrosis and regulation by soluble growth factors. Gut. 2001;48:548–557. doi: 10.1136/gut.48.4.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valenzuela MT, Guerrero R, Nunez MI, Ruiz De Almodovar JM, Sarker M, de Murcia G, Oliver FJ. PARP-1 modifies the effectiveness of p53-mediated DNA damage response. Oncogene. 2002;21:1108–1116. doi: 10.1038/sj.onc.1205169. [DOI] [PubMed] [Google Scholar]
- 28.Fattman CL, Delach SM, Dou QP, Johnson DE. Sequential two-step cleavage of the retinoblastoma protein by caspase-3/-7 during etoposide-induced apoptosis. Oncogene. 2001;20:2918–2926. doi: 10.1038/sj.onc.1204414. [DOI] [PubMed] [Google Scholar]
- 29.Katsuda K, Kataoka M, Uno F, Murakami T, Kondo T, Roth JA, Tanaka N, Fujiwara T. Activation of caspase-3 and cleavage of Rb are associated with p16-mediated apoptosis in human non-small cell lung cancer cells. Oncogene. 2002;21:2108–2113. doi: 10.1038/sj.onc.1205272. [DOI] [PubMed] [Google Scholar]
- 30.Ravi R, Bedi GC, Engstrom LW, Zeng Q, Mookerjee B, Gelinas C, Fuchs EJ, Bedi A. Regulation of death receptor expression and TRAIL/Apo2L-induced apoptosis by NF-kappaB. Nat Cell Biol. 2001;3:409–416. doi: 10.1038/35070096. [DOI] [PubMed] [Google Scholar]
- 31.Jin F, Liu X, Zhou Z, Yue P, Lotan R, Khuri FR, Chung LW, Sun SY. Activation of nuclear factor-kappaB contributes to induction of death receptors and apoptosis by the synthetic retinoid CD437 in DU145 human prostate cancer cells. Cancer Res. 2005;65:6354–6363. doi: 10.1158/0008-5472.CAN-04-4061. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Wang Z, Kwong SQ, Lui EL, Friedman SL, Li FR, Lam RW, Zhang GC, Zhang H, Ye T. Inhibition of PDGF, TGF-beta, and Abl signaling and reduction of liver fibrosis by the small molecule Bcr-Abl tyrosine kinase antagonist Nilotinib. J Hepatol. 2011;55:612–625. doi: 10.1016/j.jhep.2010.11.035. [DOI] [PubMed] [Google Scholar]
- 33.Cho DH, Jo YK, Hwang JJ, Lee YM, Roh SA, Kim JC. Caspase-mediated cleavage of ATG6/Beclin-1 links apoptosis to autophagy in HeLa cells. Cancer Lett. 2009;274:95–100. doi: 10.1016/j.canlet.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 34.Wirawan E, Vande Walle L, Kersse K, Cornelis S, Claerhout S, Vanoverberghe I, Roelandt R, De Rycke R, Verspurten J, Declercq W, Agostinis P, Vanden Berghe T, Lippens S, Vandenabeele P. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis. 2010;1:e18. doi: 10.1038/cddis.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daugas E, Nochy D, Ravagnan L, Loeffler M, Susin SA, Zamzami N, Kroemer G. Apoptosis-inducing factor (AIF): a ubiquitous mitochondrial oxidoreductase involved in apoptosis. FEBS Lett. 2000;476:118–123. doi: 10.1016/s0014-5793(00)01731-2. [DOI] [PubMed] [Google Scholar]
- 36.van Loo G, Schotte P, van Gurp M, Demol H, Hoorelbeke B, Gevaert K, Rodriguez I, Ruiz-Carrillo A, Vandekerckhove J, Declercq W, Beyaert R, Vandenabeele P. Endonuclease G: a mitochondrial protein released in apoptosis and involved in caspase-independent DNA degradation. Cell Death Differ. 2001;8:1136–1142. doi: 10.1038/sj.cdd.4400944. [DOI] [PubMed] [Google Scholar]
- 37.Galluzzi L, Vitale I, Abrams JM, Alnemri ES, Baehrecke EH, Blagosklonny MV, Dawson TM, Dawson VL, El-Deiry WS, Fulda S, Gottlieb E, Green DR, Hengartner MO, Kepp O, Knight RA, Kumar S, Lipton SA, Lu X, Madeo F, Malorni W, Mehlen P, Nunez G, Peter ME, Piacentini M, Rubinsztein DC, Shi Y, Simon HU, Vandenabeele P, White E, Yuan J, Zhivotovsky B, Melino G, Kroemer G. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2011 doi: 10.1038/cdd.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riedl SJ, Shi Y. Molecular mechanisms of caspase regulation during apoptosis. Nat Rev Mol Cell Biol. 2004;5:897–907. doi: 10.1038/nrm1496. [DOI] [PubMed] [Google Scholar]
- 39.Vande Walle L, Van Damme P, Lamkanfi M, Saelens X, Vandekerckhove J, Gevaert K, Vandenabeele P. Proteome-wide Identification of HtrA2/Omi Substrates. J Proteome Res. 2007;6:1006–1015. doi: 10.1021/pr060510d. [DOI] [PubMed] [Google Scholar]
- 40.Li B, Hu Q, Wang H, Man N, Ren H, Wen L, Nukina N, Fei E, Wang G. Omi/HtrA2 is a positive regulator of autophagy that facilitates the degradation of mutant proteins involved in neurodegenerative diseases. Cell Death Differ. 2010;17:1773–1784. doi: 10.1038/cdd.2010.55. [DOI] [PubMed] [Google Scholar]
- 41.Lei W, Zhang K, Pan X, Hu Y, Wang D, Yuan X, Shu G, Song J. Histone deacetylase 1 is required for transforming growth factor-beta1-induced epithelial-mesenchymal transition. Int J Biochem Cell Biol. 2010;42:1489–1497. doi: 10.1016/j.biocel.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 42.Qin L, Han YP. Epigenetic repression of matrix metalloproteinases in myofibroblastic hepatic stellate cells through histone deacetylases 4: implication in tissue fibrosis. Am J Pathol. 2010;177:1915–1928. doi: 10.2353/ajpath.2010.100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lechuga CG, Hernandez-Nazara ZH, Dominguez Rosales JA, Morris ER, Rincon AR, Rivas-Estilla AM, Esteban-Gamboa A, Rojkind M. TGF-beta1 modulates matrix metalloproteinase-13 expression in hepatic stellate cells by complex mechanisms involving p38MAPK, PI3-kinase, AKT, and p70S6k. Am J Physiol Gastrointest Liver Physiol. 2004;287:G974–987. doi: 10.1152/ajpgi.00264.2003. [DOI] [PubMed] [Google Scholar]
- 44.Barter MJ, Pybus L, Litherland GJ, Rowan AD, Clark IM, Edwards DR, Cawston TE, Young DA. HDAC-mediated control of ERK- and PI3K-dependent TGF-beta-induced extracellular matrix-regulating genes. Matrix Biol. 2010;29:602–612. doi: 10.1016/j.matbio.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 45.Rosato RR, Grant S. Histone deacetylase inhibitors: insights into mechanisms of lethality. Expert Opin Ther Targets. 2005;9:809–824. doi: 10.1517/14728222.9.4.809. [DOI] [PubMed] [Google Scholar]
- 46.Leist M, Jaattela M. Four deaths and a funeral: from caspases to alternative mechanisms. Nat Rev Mol Cell Biol. 2001;2:589–598. doi: 10.1038/35085008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.