Interactions between bacteria and roots are critical to the terrestrial ecosystem. The zone of soil immediately surrounding roots is known as the rhizosphere and the surface of the root the rhizoplane (1, 2). This region is of paramount importance to the growth and productivity of plants because it is the main area where they interact with an enormously complex microbial community, the microbiome. Soil may contain up to 104 bacterial species and 109 bacteria per g as well as a diverse archaeal and eukaryotic population (3, 4). Although the diversity of bacteria in the rhizosphere is less than in soil, it is much more active because it is under strong plant selection and has access to root exudation. Plants may release up to 20% of their photosynthate via their roots, which is critical in shaping and possibly farming the root microbiome. This microbiome consists of symbionts, commensals, and pathogens that will interact with roots, the soil, and one another to form a root holobiont that determines much of biological and agricultural productivity. Despite the immense importance of the root environment, it is often overlooked because it is challenging to image and study roots that are underground and are difficult to observe without disturbance. For example, compare the ease of sampling shoots relative to roots. One of the principal ways of overcoming this is to image roots with X-ray computed tomography scanning, a powerful way of building 3D images of root growth in a spatial and temporal fashion (5). However, this approach is not suited to examine the critical interaction of microbes with roots. To overcome this, plants can be grown between thinly separated glass sheets, known as rhizotrons, on agar plates or similar artificial media and imaged by a wide range of physical and microscopic techniques. This tends to be done at one time point and does not allow dynamic mapping of the spatial and temporal interaction of microbes with roots. A further recent advance is the use of a microfluidics-based RootChip to monitor root development (6). However, a breakthrough in the live imaging of roots and bacteria is described in the paper by Massalha et al. (7) using a technique called TRIS (tracking root interactions system). For TRIS analysis, roots are grown into a narrow root chamber (160 μM high) contained in a microfluidics device. Bacteria can then be introduced into this chamber and imaged with a wide variety of techniques, including dark-field and confocal microscopy. The microfluidics chamber was made by using soft lithography to etch a single sheet of polydimethylsiloxane with nine separate chambers, each of which contains a single Arabidopsis root, with separate input and output channels through which bacteria can be introduced. Bright-field microscopy can be used to image unlabeled bacteria but the technique is most powerful when fluorescently labeled bacteria are added and imaged by confocal microscopy. Using Bacillus subtilis as their test bacterium, there was a remarkable accumulation of bacteria observable in just 20 min, behind the root tip in the root elongation zone (Fig. 1).
Fig. 1.
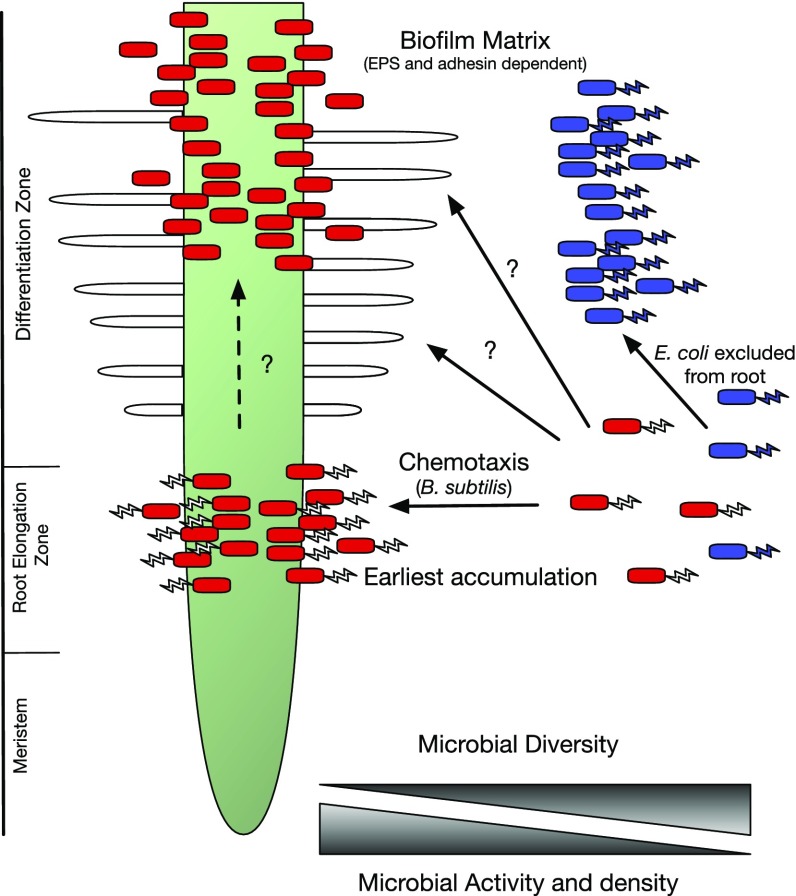
A cartoon of a root is shown where two bacteria, B. subtilis (in red) and E. coli (in blue), are competing to attach to A. thaliana. B. subtilis has been shown by TRIS to accumulate rapidly at the root elongation zone with later accumulation occurring higher up the root. Arrows and question marks highlight what might happen in steps subsequent to accumulation at the root elongation zone because the order of events is not clear, particularly in soil where the root will be occupied by existing bacteria in biofilms. Bacteria in biofilms are shown without flagella to indicate that motility is usually suppressed at the sessile stage of biofilm formation.
This could be further imaged every 30 min up to 12 h, with later colonization visible further up the root. The root elongation zone was elegantly and precisely mapped as the region of initial accumulation using GFP-marked lines of Arabidopsis (COBL9, CORTEX, WER, SCR, WOL, and PET111) and mKate-labeled bacteria. From these results the root elongation zone seems to be a very special hotspot in the initial interaction of roots and microbes. Some of this may result from this region’s being a major site of exudation, and such rapid accumulation of bacteria at the root elongation zone must require chemotaxis of motile bacteria to plant metabolites (8). Given the broad range of metabolites secreted by Arabidopsis thaliana roots it is not possible to say whether this is a reaction to a specific metabolite or to multiple metabolites. However, preliminary metabolomics conducted on root secretions in this study revealed the presence of several amino acids that are known chemoattractants. Following on from this, an exciting use of TRIS will be to screen mutant bacteria to determine whether specific chemotactic signals are required and whether there are specific recognition and signaling mechanisms involved. It is known that early attachment of B. subtilis requires the chemoreceptors McpB and McpC and the orphan receptor TlpC (9); therefore, it will be of interest to test these mutants in the TRIS system.
Bacterial chemotaxis is almost certainly involved in bacterial migration to roots, followed by attachment and biofilm formation (10–12). Many processes in initial attachment and biofilm formation are conserved among bacteria. B. subtilis requires exopolysaccharide (EPS) and proteins such as TasA to form a biofilm matrix on roots, and it has also been shown that induction of a biofilm is stimulated by the plant polysaccharides pectin, xylan, and arabinogalactan, with the latter important in A. thaliana (10). Furthermore, the regulatory proteins SinA and SpoA and up to five Kin sensor kinases seem to mediate the stimulation of biofilm formation by plant polysaccharides. Likewise, in pseudomonads, biofilms also require EPS but in addition involves the 6,310-aa LapF protein to adhere to plant roots (13). In the well-studied case of Rhizobium, attachment to root hairs at mildly acid pH (6.5) is initially mediated by end-on attachment of polar-located glucomannan to pea lectin (14). Rhizobium adhering proteins then become essential, possibly by binding to EPS to initiate a biofilm complex (15). In addition, cellulose microfibrils are made that reinforce the biofilm matrix. In rhizobia, binding to root hairs can lead to root-hair curling, entrapment, and infection-thread development through which bacteria traverse the developing nodule before being engulfed by plant cells in the nodule cortex (16). The details of rhizobial attachment have been intensively studied because N2 fixation by rhizobia contributes a substantial portion of the biosphere’s available nitrogen. A critical point concerning attachment in different bacteria is that it occurs at later stages in root colonization than accumulation at the root elongation zone. For example, much attention has been focused on attachment in rhizobia, which occurs on differentiated root-hair cells, in contrast to the root elongation zone, which contains undifferentiated cells. This implies there are likely to be multiple stages in attraction to roots, initial colonization, and biofilm formation, as well as attachment to root hairs. The work of Massalha et al. (7) suggests we may have underestimated the spatial and temporal separation of steps in root colonization. Indeed, the first step in colonization may be to the newly divided cells of the root elongation zone that are not already colonized by bacteria that have formed a highly resistant biofilm.
A further, exciting development in the work of Massalha et al. (7) was the introduction of B. subtilis (labeled with mKate) and Escherichia coli (labeled with GFP) and the demonstration that B. subtilis actively excluded E. coli from colonization of the root. It is known that B. subtilis can secrete the antiadhesion compound surfactin that inhibits biofilm formation by pathogens such as E. coli CFT073 (17), and Massalha et al. (7) speculate that it might induce the exclusion zone around roots. This opens up a large area of biology with the
A breakthrough in the live imaging of roots and bacteria is described in the paper by Massalha et al. using a technique called TRIS (tracking root interactions system).
ability to look at a myriad of combinations of bacteria that are differentially marked to look for antagonistic or symbiotic interactions. This could even be extended beyond two or three bacteria to larger synthetic communities, where keystone species are marked with fluorescent labels and the effects on root colonization by multiple bacteria investigated. Furthermore, it would be of interest to see whether TRIS could be used to study potential three-way interactions between bacteria, mycorrhizae, and roots.
Finally, Massalha et al. (7) developed a two-root chamber where roots from two plants are separated by a semipermeable membrane (pore size 50 μΜ) that permits solute and bacteria to move but prevents roots on different plants from contacting one another physically. This allows bacterial preference for specific plant root genotypes to be assayed in one chamber. It was observed that the hairless caprice/triptychon (cpc/trt) and the excessive root hair werewolf/myb23 (wer/myb23) mutants had peak attraction of bacteria occurring about 1 h earlier than wild-type plants. There are obviously a large range of Arabidopsis mutants that could be tested in this way, including those involved in immune signaling (18) and root secretion such as ABC exporters (19). The alternative approach will be to use bacteria labeled with inducible fluorescent, luminescent (e.g., lux) (20–22), or FRET biosensors (23) to monitor root secretion and the bacterial response to roots.
TRIS has manifold uses in plant and microbial science to examine interactions of both partners. It is particularly valuable because it can give a spatial and temporal map to the earliest events in root attachment and colonization. Whereas it has a vast range of applications to classical reductionist biology, where the interactions of a single microbe and plant are considered, it also has powerful application to the new wave of integrative community biology. This has been led by high-throughput sequencing that has shown how plants can shape their rhizosphere and root-associated communities (4, 24, 25) and how specific or groups of bacteria can suppress pathogens (26). However, much of this work to date has been descriptive, where community composition is counted. Techniques such as TRIS, where single or multiple bacteria can be assayed with a range of plants, offers an exciting way forward to investigate more complex dynamic community-level interactions. It will be particularly informative to move to other plant species, particularly those that can be infected by mycorrhizae (most plants) and/or nodulate (legumes) via the common SYM signaling pathway, which has also recently been shown to alter the microbial community (27). With a light such as TRIS the future of root–microbe interactions is bright.
Footnotes
The author declares no conflict of interest.
See companion article on page 4549.
References
- 1.Turner TR, James EK, Poole PS. The plant microbiome. Genome Biol. 2013;14:209. doi: 10.1186/gb-2013-14-6-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Philippot L, Raaijmakers JM, Lemanceau P, van der Putten WH. Going back to the roots: The microbial ecology of the rhizosphere. Nat Rev Microbiol. 2013;11:789–799. doi: 10.1038/nrmicro3109. [DOI] [PubMed] [Google Scholar]
- 3.Weinert N, et al. PhyloChip hybridization uncovered an enormous bacterial diversity in the rhizosphere of different potato cultivars: Many common and few cultivar-dependent taxa. FEMS Microbiol Ecol. 2011;75:497–506. doi: 10.1111/j.1574-6941.2010.01025.x. [DOI] [PubMed] [Google Scholar]
- 4.Turner TR, et al. Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants. ISME J. 2013;7:2248–2258. doi: 10.1038/ismej.2013.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zappala S, et al. Effects of X-ray dose on rhizosphere studies using X-ray computed tomography. PLoS One. 2013;8:e67250. doi: 10.1371/journal.pone.0067250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grossmann G, et al. The RootChip: An integrated microfluidic chip for plant science. Plant Cell. 2011;23:4234–4240. doi: 10.1105/tpc.111.092577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Massalha H, et al. Live imaging of root–bacteria interactions in a microfluidics setup. Proc Natl Acad Sci USA. 2017;114:4549–4554. doi: 10.1073/pnas.1618584114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strehmel N, Böttcher C, Schmidt S, Scheel D. Profiling of secondary metabolites in root exudates of Arabidopsis thaliana. Phytochemistry. 2014;108:35–46. doi: 10.1016/j.phytochem.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Allard-Massicotte R, et al. Bacillus subtilis early colonization of Arabidopsis thaliana roots involves multiple chemotaxis receptors. MBio. 2016;7:e01664-16. doi: 10.1128/mBio.01664-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beauregard PB, Chai Y, Vlamakis H, Losick R, Kolter R. Bacillus subtilis biofilm induction by plant polysaccharides. Proc Natl Acad Sci USA. 2013;110:E1621–E1630. doi: 10.1073/pnas.1218984110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Downie JA. The roles of extracellular proteins, polysaccharides and signals in the interactions of rhizobia with legume roots. FEMS Microbiol Rev. 2010;34:150–170. doi: 10.1111/j.1574-6976.2009.00205.x. [DOI] [PubMed] [Google Scholar]
- 12.Branda SS, Vik S, Friedman L, Kolter R. Biofilms: The matrix revisited. Trends Microbiol. 2005;13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 13.Martínez-Gil M, Yousef-Coronado F, Espinosa-Urgel M. LapF, the second largest Pseudomonas putida protein, contributes to plant root colonization and determines biofilm architecture. Mol Microbiol. 2010;77:549–561. doi: 10.1111/j.1365-2958.2010.07249.x. [DOI] [PubMed] [Google Scholar]
- 14.Williams A, et al. Glucomannan-mediated attachment of Rhizobium leguminosarum to pea root hairs is required for competitive nodule infection. J Bacteriol. 2008;190:4706–4715. doi: 10.1128/JB.01694-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdian PL, Caramelo JJ, Ausmees N, Zorreguieta A. RapA2 is a calcium-binding lectin composed of two highly conserved cadherin-like domains that specifically recognize Rhizobium leguminosarum acidic exopolysaccharides. J Biol Chem. 2013;288:2893–2904. doi: 10.1074/jbc.M112.411769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oldroyd GE, Murray JD, Poole PS, Downie JA. The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet. 2011;45:119–144. doi: 10.1146/annurev-genet-110410-132549. [DOI] [PubMed] [Google Scholar]
- 17.Rivardo F, Turner RJ, Allegrone G, Ceri H, Martinotti MG. Anti-adhesion activity of two biosurfactants produced by Bacillus spp. prevents biofilm formation of human bacterial pathogens. Appl Microbiol Biotechnol. 2009;83:541–553. doi: 10.1007/s00253-009-1987-7. [DOI] [PubMed] [Google Scholar]
- 18.Lebeis SL, et al. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science. 2015;349:860–864. doi: 10.1126/science.aaa8764. [DOI] [PubMed] [Google Scholar]
- 19.Badri DV, et al. An ABC transporter mutation alters root exudation of phytochemicals that provoke an overhaul of natural soil microbiota. Plant Physiol. 2009;151:2006–2017. doi: 10.1104/pp.109.147462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramachandran VK, East AK, Karunakaran R, Downie JA, Poole PS. Adaptation of Rhizobium leguminosarum to pea, alfalfa and sugar beet rhizospheres investigated by comparative transcriptomics. Genome Biol. 2011;12:R106. doi: 10.1186/gb-2011-12-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tett AJ, Karunakaran R, Poole PS. Characterisation of SalRAB a salicylic acid inducible positively regulated efflux system of Rhizobium leguminosarum bv viciae 3841. PLoS One. 2014;9:e103647. doi: 10.1371/journal.pone.0103647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frederix M, et al. Mutation of praR in Rhizobium leguminosarum enhances root biofilms, improving nodulation competitiveness by increased expression of attachment proteins. Mol Microbiol. 2014;93:464–478. doi: 10.1111/mmi.12670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bourdès A, Rudder S, East AK, Poole PS. Mining the Sinorhizobium meliloti transportome to develop FRET biosensors for sugars, dicarboxylates and cyclic polyols. PLoS One. 2012;7:e43578. doi: 10.1371/journal.pone.0043578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lundberg DS, et al. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488:86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bulgarelli D, et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature. 2012;488:91–95. doi: 10.1038/nature11336. [DOI] [PubMed] [Google Scholar]
- 26.Mendes R, et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–1100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- 27.Zgadzaj R, et al. Root nodule symbiosis in Lotus japonicus drives the establishment of distinctive rhizosphere, root, and nodule bacterial communities. Proc Natl Acad Sci USA. 2016;113:E7996–E8005. doi: 10.1073/pnas.1616564113. [DOI] [PMC free article] [PubMed] [Google Scholar]