Significance
Marine cone snails provide one of the highest toxin diversities among venomous animals, the conotoxins, which often target ion channels. In addition, cone snail venoms contain short conopeptides, the molecular targets of which are mostly unknown. We identified a new conopeptide from the venom of the cone snail Conus textile that targets the proton-gated ion channel ASIC3, which detects painful acidosis. This peptide belongs to the subgroup of cono-RFamides and potentiates ASIC3 currents, thereby increasing the excitability of nociceptors. Injection of the cono-RFamide in mice muscle enhanced acid-induced muscle pain. Our results thus identify ASIC3 as the first molecular target of cono-RFamides. These findings underline the role of ASIC3 in pain perception and the importance of nontoxic venom components.
Keywords: acid-sensing ion channel, ASIC, cone snail, venom, pain
Abstract
Acid-sensing ion channels (ASICs) are proton-gated Na+ channels that are expressed throughout the nervous system. ASICs have been implicated in several neuronal disorders, like ischemic stroke, neuronal inflammation, and pathological pain. Several toxins from venomous animals have been identified that target ASICs with high specificity and potency. These toxins are extremely useful in providing protein pharmacophores and to characterize function and structure of ASICs. Marine cone snails contain a high diversity of toxins in their venom such as conotoxins, which are short polypeptides stabilized by disulfide bonds, and conopeptides, which have no or only one disulfide bond. Whereas conotoxins selectively target specific neuronal proteins, mainly ion channels, the targets of conopeptides are less well known. Here, we perform an in vitro screen of venoms from 18 cone snail species to identify toxins targeting ASICs. We identified a small conopeptide of only four amino acids from the venom of Conus textile that strongly potentiated currents of ASIC3, which has a specific role in the pain pathway. This peptide, RPRFamide, belongs to the subgroup of cono-RFamides. Electrophysiological characterization of isolated dorsal root ganglion (DRG) neurons revealed that RPRFamide increases their excitability. Moreover, injection of the peptide into the gastrocnemius muscle strongly enhanced acid-induced muscle pain in mice that was abolished by genetic inactivation of ASIC3. In summary, we identified a conopeptide that targets the nociceptor-specific ion channel ASIC3.
Acid-sensing ion channels (ASICs) are ligand-gated Na+ channels that are expressed throughout the CNS and the peripheral nervous system (PNS), where they contribute to synaptic transmission (1–3) and detection of painful acidosis (4). ASICs play an important role in numerous physiological and pathological conditions such as synaptic plasticity and learning (5), fear conditioning (6), and neurodegeneration associated with ischemic stroke (7). Four genes code for at least six ASIC subunits (1a, 1b, 2a, 2b, 3, and 4) that assemble into homo- or heterotrimeric channels (8, 9). An increase of the extracellular proton concentration activates ASICs to cause rapidly desensitizing inward currents, predominantly conducted by Na+ (10). Their activation leads to a short-lasting membrane depolarization, modulating neuronal activity (11–13). Homomeric ASICs with a proton sensitivity in the physiological range are formed by ASIC1a, ASIC1b and ASIC3 (3). Whereas ASIC1a is expressed in the CNS and the PNS (10), ASIC1b and ASIC3 are mainly expressed in the PNS (14, 15). Several studies provided evidence that ASIC3 is directly involved in pain perception (4, 16, 17).
ASICs are related to peptide-gated ion channels of the Hydra nervous system, which are directly gated by RFamide neuropeptides (18, 19). RFamide neuropeptides do not activate ASICs but modulate the proton-activated current by directly binding to the channel (20), suggesting that ASICs evolved from ion channels gated by small neuropeptides (18).
In the last 15 y, four animal toxins have been identified that specifically target ASICs: the tarantula toxin PcTx1, which potently inhibits ASIC1a (21); the sea anemone peptide APETx2, which inhibits ASIC3 (22); the heteromeric coral snake toxin MitTx, which slowly activates all ASIC subtypes (23); and the black and green mamba toxins mambalgins, which inhibit ASIC1a and 1b (24). These toxins have been extremely useful in unraveling the physiological functions and the structure of ASICs (25, 26).
Marine cone snails are predatory snails living predominantly in tropical waters. Each of the more than 800 species produces its individual and peptide-rich venom, which is used for predation and/or defense (27). The sting of a cone snail is extremely painful and can even be fatal to humans (27, 28). Cone snail venoms consist of two major components: (i) the disulfide-rich conotoxins, which are further classified depending on their cysteine patterns and their target; and (ii) the conopeptides, which have no or only one disulfide bond and are so far less well characterized (27). Conotoxins are comparatively small peptides of 10–40 aa that target diverse receptors mainly of the nervous system with high potency and specificity, such as voltage- and ligand-gated ion channels, G-protein-coupled receptors, and neurotransmitter transporters (27), making cone snail venoms an outstanding source of small, highly specific and highly potent peptides.
In the present study, we screened cone snail venoms for components targeting ASICs. We identified a small conopeptide from Conus textile that belongs to the subgroup of cono-RFamides and specifically enhanced ASIC3 currents, increasing the excitability of sensory neurons. Injection of the cono-RFamide into mice muscle strongly enhanced acid-induced muscle pain. In summary, our study identifies a molecular target of a cono-RFamide. This peptide may be useful to further explore the role of ASIC3 in pain perception and underlines the importance of nontoxic components of cone snail venoms.
Results
Identification and Purification of a Cono-RFamide Targeting ASIC3.
To identify conotoxins or conopeptides targeting ASICs, we screened crude venoms from 18 different cone snail species (Materials and Methods) for their ability to modulate the current of three different ASICs of the peripheral nervous system, ASIC1a, ASIC1b, and ASIC3, expressed in Xenopus oocytes. The venom of C. textile, a predatory snail from the Indo-Pacific that is toxic also to mammals, potentiated the current of ASIC3 but not of ASIC1a or ASIC1b.
We fractionated the C. textile venom using reverse-phase HPLC (RP-HPLC) (Fig. 1A) and tested each fraction for its effect on ASIC3 currents. Three consecutive fractions containing venom components potentiating ASIC3 currents with decreasing potency (Fig. 1 B and C) were further purified, several small peptides were identified, and their amino acid sequence was elucidated: fraction 10, which had the highest potency (Fig. 1C), mainly contained RPRFamide (RPRFa); fraction 11 contained VGRPRFa; and fraction 12 AIVGRPRFa. These three short peptides all share a common C-terminal sequence of four amino acids and an amidated C terminus (Fig. 1D). Their sequence is identical to the C-terminal sequence of cono-neuropeptide Y from Conus betulinus (29) (Fig. 1D). The short length, the C-terminal Arg–Phe–NH2 (RFa) motif, and the lack of cysteines clearly distinguish these peptides from conotoxins and categorize them as cono-RFamides (CNFs).
Fig. 1.
Fractionation of crude C. textile venom. (A) HPLC chromatogram. Active peaks are indicated with arrows. (B) Representative current trace illustrating the potentiation of ASIC3 current by fraction 10, which was applied in the conditioning period at pH 7.4 (red bar). ASIC3 was activated by pH 6.3 (black bars). (C) Quantification of sustained currents, 10 s after activation, induced by fractions 10, 11, and 12. Currents were normalized to the respective peak currents (n = 6 per fraction). (D) Sequence alignment of the RFamide neuropeptides isolated from fractions 10–12 and of NPY1 from C. betulinus. Systematic names of CNFs isolated in this study are CNF-Tx1.1 (RPRFa), CNF-Tx1.2 (VGRPRFa), and CNF-Tx1.3 (AIVGRPRFa).
RPRFa and VGRPRFa Slow ASIC3 Desensitization.
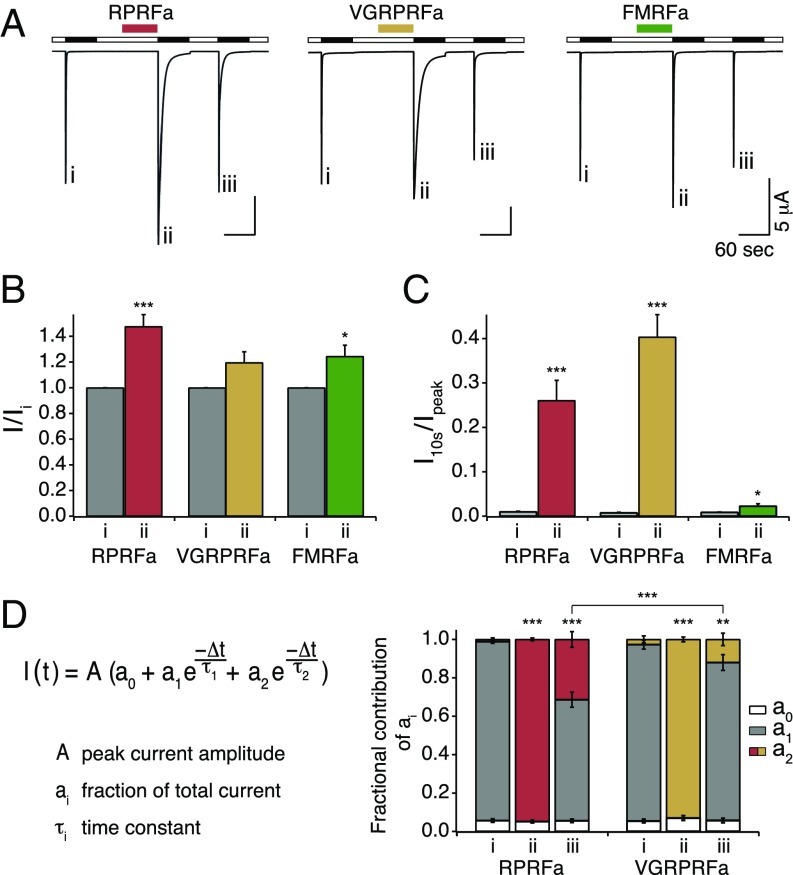
RFamide peptides form a large and evolutionary old group of neuropeptides (30). The prototypical RFamide, FMRFa (31), also potentiates ASIC3 currents (20). Therefore, we analyzed the modulation of ASIC3 by the CNFs RPRFa and VGRPRFa from C. textile in more detail and compared it to the modulation by FMRFa (Fig. 2). For this analysis, we used chemically synthesized peptides. ASIC3 was repeatedly activated for 60 s by an acidic solution (pH 6.3), generating rapidly desensitizing currents (Fig. 2A). Like for FMRFa, preapplication of RPRFa or VGRPRFa (50 μM) did not activate ASIC3 at pH 7.4 but potentiated the proton-evoked ASIC3 current (Fig. 2 A and B). RPRFa increased the peak current amplitude 1.48-fold (n = 10; P < 0.001, Student’s t test), whereas VGRPRFa and FMRFa increased the peak amplitude 1.2- and 1.24-fold, respectively.
Fig. 2.
Potentiation of ASIC3 currents by RPRFa, VGRPRFa, and FMRFa. (A) Representative current traces illustrating potentiation of ASIC3 current by RPRFa, VGRPRFa, and FMRFa. RFa peptides (50 μM) were preapplied in the conditioning period at pH 7.4 (colored bars). ASIC3 was activated by pH 6.3 (black bars). Roman numerals mark specific activations. (B) Quantification of peak currents. Peak current amplitudes of the second activation with RFa preapplication were normalized to the first activation without RFa preapplication. (C) Quantification of current amplitudes 10 s after activation, normalized to the peak current amplitude (n ≥ 9). *P < 0.05; ***P < 0.001 (Student’s t test). (D, Left) Double-exponential function describing desensitization of ASIC3 currents; ai indicates the relative contribution of the various current components (∑ai = 1). (D, Right) Quantification of ai (n ≥ 9). **P < 0.01; ***P < 0.001 (one-way repeated-measures ANOVA).
More strikingly, RPRFa and VGRPRFa slowed the desensitization kinetics of ASIC3 much stronger than FMRFa (Fig. 2C), leading to a strong increase of the current amplitude at the end of a 10-s proton pulse: RPRFa and VGRPRFa raised the 10-s-to-peak ratio (I10s/Ipeak) from 1% in their absence to 26 and 40% in their presence, respectively (n = 10; P < 0.001, t test vs. control). In contrast, FMRFa increased I10s/Ipeak to only 2% (n = 9; P = 0.02, t test). For a more precise and quantitative analysis of the desensitization kinetics, we fitted current traces with a double-exponential function I(t) (Fig. 2D), assuming two channel populations contributing to the channel kinetics. Under control conditions, without peptide, the slow time constant did not contribute to the fit (a2 = 0) (Fig. 2D), whereas at saturating peptide concentrations, the fast time constant did not contribute (a1 = 0), in both cases resulting in single exponential functions well describing the current decay phase (Fig. 2D). Under control conditions, the desensitization time constant was τ1 = 0.41 ± 0.02 s (n = 28), consistent with the literature (32). When RFamides (50 μM) were applied during the conditioning period, the desensitization time constant τ increased strongly for RPRFa (τ2 = 5.99 ± 0.23 s; n = 9) and for VGRPRFa (τ2 = 8.58 ± 0.49 s; n = 10) but less for FMRFa (τ2 = 1.54 ± 0.17 s; n = 9). Thus, VGRPRFa most efficiently slowed desensitization of ASIC3. Interestingly, τ2 did not change significantly using various RFamide concentrations; only the ratio of a2 to a1 increased with higher peptide concentration until a concentration of 50 μM was sufficient to saturate peptide binding and thus almost abolished a1 (Figs. 2D and 4C). Thus, it appears that the population of peptide-bound channels was homogenous and that the two populations of ASIC3, with RFamide bound and with no RFamide bound, could be separated based on the exponential fit of desensitization kinetics. At intermediate peptide concentrations, both time constants were required for an adequate fit, suggesting a mixed population of channels with RFamide bound and with no RFamide bound; the relative contribution of both channel populations to the total current is described by the coefficients a1 and a2. For RPRFa and for VGRPRFa, 50 μM was sufficient to saturate peptide binding as after preapplication all channels could be fit solely with the second exponential term (Fig. 2D); 110 s after washout of the RFamides, both exponential terms were needed for a proper fit (Fig. 2 A and D), indicating that RPRFa was still bound to 31 ± 4% of all channels and VGRPRFa to 12 ± 3%. These differences were highly significant (n ≥ 9; P < 0.001, ANOVA) and suggest a slower dissociation rate (higher affinity) for RPRFa than for VGRPRFa. Based on these results, we decided to focus on the modulation of ASIC3 by RPRFa. Like the crude venom, synthesized RPRFa (50 μM) did not modulate homomeric ASIC1a or ASIC1b, respectively (Fig. S1A).
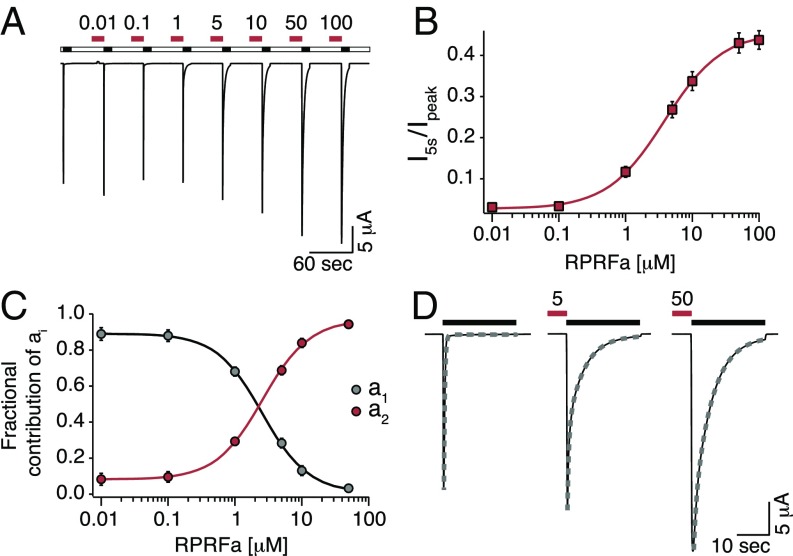
Fig. 4.
RPRFa potentiates ASIC3 currents with a high potency. (A) Representative current trace illustrating the concentration-dependent potentiation of ASIC3 currents by RPRFa. ASIC3 was repeatedly activated by pH 6.3 (black bars), and RPRFa was preapplied for 30 s in concentrations as indicated (in μM; red bars). (B) Quantification of the potentiation as measured by the currents 5 s after activation, normalized to the corresponding peak currents. Line represents a fit to the Hill equation. (C) Quantification of the potentiation as measured by a biexponential fit to the desensitization phase. As a1, representing channels without RFa, decreased, a2, representing channels with RFa bound, increased. Lines represent fits to the Hill equation (n = 11). (D) Individual ASIC3 responses from A are shown on an expanded scale to illustrate the exponential fits used in C (dotted lines).
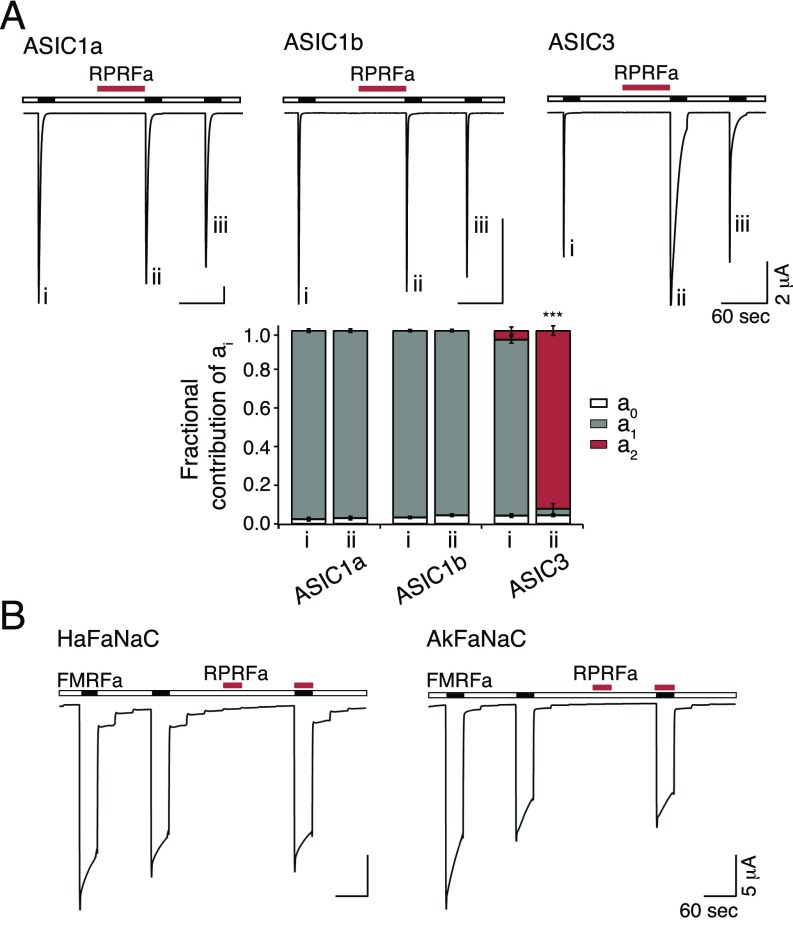
Fig. S1.
RPRFa specifically potentiates ASIC3 currents. (A, Top) Representative current traces of homomeric ASIC1a, ASIC1b, and ASIC3, activated with pH 6.5, pH 5.8, and pH 6.3, respectively. Roman numerals mark specific activations. RPRFa (50 μM; red bars) was preapplied at pH 7.4 for 60 s. (A, Bottom) Quantification of ai (n = 10). ***P < 0.001 (Student’s t test). (B) FaNaCs are not affected by RPRFa. Representative current traces illustrating activation of HaFaNaC and AkFaNaC by FMRFa (10 μM; black bars). RPRFa (10 μM; red bars) neither activated FaNaC nor inhibited activation by FMRFa.
RPRFa Slows Desensitization of ASIC3-Containing Heteromers.
To test whether RPRFa modulated heteromeric ASICs containing ASIC3, we coexpressed ASIC3 and one other ASIC subunit (either ASIC1a, ASIC1b, ASIC2a, or ASIC2b) in oocytes and repeatedly activated ASICs by an acidic solution with a pH that corresponds roughly to the EC50 expected for the respective heteromer (pH 6.3, 6.0, 5.7, or 6.5, respectively; Fig. 3A) (33, 34). RPRFa significantly increased peak current amplitudes of ASIC1a/3 1.25-fold (n = 10; P < 0.001, t test) and of ASIC2b/3 1.4-fold (n = 17; P < 0.05; Fig. 3B). Like for homomeric ASIC3, preapplication of RPRFa (50 μM) slowed desensitization of heteromeric ASICs, such that the 10-s-to-peak ratio (I10s/Ipeak) was strongly raised from 0.3 to 6% for ASIC1a/3 (n = 10; P = 0.001, t test), from 1 to 15% for ASIC1b/3 (n = 18; P < 0.001), from 9 to 32% for ASIC2a/3 (n = 12; P < 0.001), and from 1 to 18% for ASIC2b/3 (n = 17; P < 0.001; Fig. 3C), respectively. Although we cannot exclude a contribution by homomeric ASIC3 to these ASICs (9), these results suggest that all four heteromers were modulated by RPRFa.
Fig. 3.
Potentiation of ASIC3-containing heteromers by RPRFa. (A) Representative current traces of heteromeric ASIC1a/3, ASIC1b/3, ASIC2a/3, and ASIC2b/3, activated with pH 6.3, 6.0, 5.7, and 6.5, respectively. Roman numerals mark specific activations. RPRFa (50 μM; red bars) was preapplied at pH 7.4 for 60 s. (B) Quantification of peak currents. Peak current amplitudes of the second activation with RFa preapplication were normalized to the first activation without RFa preapplication. (C) Quantification of current amplitudes 10 s after activation, normalized to the peak current amplitude (n = 10–18). *P < 0.05; ***P < 0.001 (Student’s t test).
RPRFa Does Not Affect FMRFa-Gated Na+ Channels.
The FMRFa-gated Na+ channel (FaNaC) shares sequence homology to ASICs, is directly activated by FMRFa, and is expressed in the nervous system of various snails (35). Because C. textile is a molluscivorous snail, we considered that FaNaCs might be the biological target of RPRFa and investigated the peptide’s effect on FaNaC of two different snails. We applied RPRFa to the FaNaC of the common garden snail Helix aspersa (35) and of the marine snail Aplysia kurodai (36). Strikingly, although both peptides are RFamides and have equal length, RPRFa (10 μM) did not activate the two FaNaCs. Moreover, coapplication of RPRFa (10 μM) together with the specific ligand FMRFa (10 μM) did not reduce the FMRFa-evoked currents (n = 5; Fig. S1B), suggesting that RPRFa does not act as a competitive antagonist of FMRFa. We conclude that RPRFa does not bind to FaNaC and that FaNaCs are probably not the biological target of RPRFa.
RPRFa Binds with High Potency to ASIC3 in the Closed State.
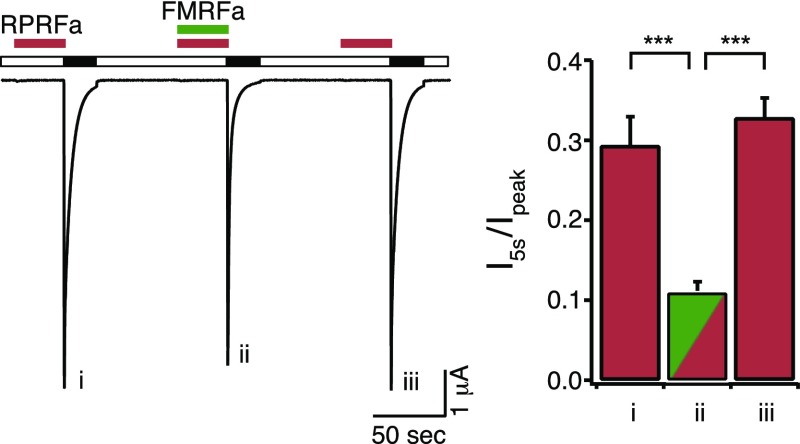
We determined the concentration needed for the half-maximal effect (EC50) by analyzing (Fig. 4 A and D) I5s/Ipeak (Fig. 4B) and the values of a1 and a2 of the double-exponential fit (Fig. 4C) at different concentrations of RPRFa. Either result was fit to a Hill equation, leading to similar results: EC50 was 4.23 ± 0.44 μM for I5s/Ipeak and 2.90 ± 0.35 μM for a2 (n = 11). Hill coefficients were h = 1.07 ± 0.05 and h = 1.29 ± 0.05, respectively. These data reveal an apparent affinity of RPRFa to ASIC3 of 3–4 μM, ∼10-fold lower than the EC50 of FMRFa on ASICs (12, 20), identifying RPRFa as the most potent RFamide modulating ASICs known so far. To test whether RPRFa binds to the same site on ASIC3 as FMRFa, we preapplied 30 μM RPRFa either alone or together with a high concentration (500 μM) of FMRFa. I5s/Ipeak was significantly smaller when FMRFa was coapplied (I5s/Ipeak = 0.11 ± 0.01 for RPRFa plus FMRFa and I5s/Ipeak = 0.29 ± 0.04 for RPRFa alone before and I5s/Ipeak = 0.33 ± 0.02 for RPRFa alone after the coapplication of RPRFa plus FMRFa; n = 8; P < 0.001, ANOVA; Fig. S2), suggesting competition of FMRFa with RPRFa for a common binding site.
Fig. S2.
FMRFa competes with RPRFa. (Left) Representative current traces of homomeric ASIC3, activated with pH 6.3. RPRFa (30 μM; red bars) was preapplied at pH 7.4 for 30 s, either alone or together with 500 μM FMRFa. (Right) Quantification of currents 5 s after activation, normalized to the respective peak currents (n = 8). ***P < 0.001 (one-way repeated-measures ANOVA).
To determine to which state of ASIC3 RPRFa binds, we applied RPRFa (10 μM) and acidic solution (pH 6.3) at various times (Fig. 5). When RPRFa was applied during the conditioning period at pH 7.4, it did not open the channel itself. Strikingly, however, even when acid was applied 30 s after washout of RPRFa, currents were still modulated (Fig. 5A, activation ii). RPRFa was still bound to 20% of channels, as judged by an a2 value of 0.20 ± 0.03 [n = 10; activation ii vs. i, P < 0.001 (ANOVA); Fig. 5C], indicating that RPRFa binds to and slowly dissociates from channels in the closed state. Coapplication of peptide and acidic solution (Fig. 5A, activation iv) led to a relatively small population of RPRFa-bound channels [a2 = 0.12 ± 0.02; n = 10; activation iv vs. ii, P = 0.229 (ANOVA); Fig. 5 A–C], suggesting slow peptide binding to the open state. Likewise, when comparing preapplication (Fig. 5A, activation iii) and continuous pre- and coapplication (Fig. 5A, activation v) of RPRFa, the results were not significantly different: under both conditions, the peptide was bound to about 80% of channels [a2 = 0.81 ± 0.05 for activation iii, a2 = 0.82 ± 0.04 for activation v; n = 10; activation iii vs. v, P = 1 (ANOVA); Fig. 5 A–C]. The fact that the population of peptide bound channels did not increase with the additional coapplication confirms that RPRFa does only slowly bind to and unbind from channels in the open state. Thus, efficient modulation of ASIC3 requires RPRFa application and binding in the closed state.
Fig. 5.
RPRFa strongly potentiates ASIC3 when applied in the closed state. (A) Representative current trace of ASIC3 repeatedly activated with pH 6.3 (black bars). Roman numerals mark specific activations. RPRFa (10 μM) was applied at different times, as indicated by red bars. (B) Quantification of currents 10 s after activation, normalized to the respective peak currents. (C) Quantification of ai (n =10); significance vs. the first control activation i is indicated. *P < 0.05; ***P < 0.001 (one-way repeated-measures ANOVA). (D) Kinetic scheme illustrating basic states during gating of ASIC3.
These results are in agreement with a simplified kinetic model previously proposed for the interaction of ASICs and FMRFa (34) (Fig. 5D). In this model, activation of ASIC3 can be described by three sequential states: closed (C), opened (O), and desensitized (D). Opening of the channel depends on H+ binding, whereas desensitization depends only indirectly on H+. For simplicity, other possible states are ignored in this model. RPRFa binds to all three states with varying dissociation constants. Desensitization of peptide-bound channels would be slower than desensitization of channels that have no peptide bound, and gating of ASICs without bound peptide is described by variables a1 and τ1 and the upper linear reaction scheme in Fig. 5D, whereas gating of ASICs with RPRFa bound is described by variables a2 and τ2 and the lower linear reaction scheme.
RPRFa Slightly Changes the Apparent H+ Affinity of ASIC3.
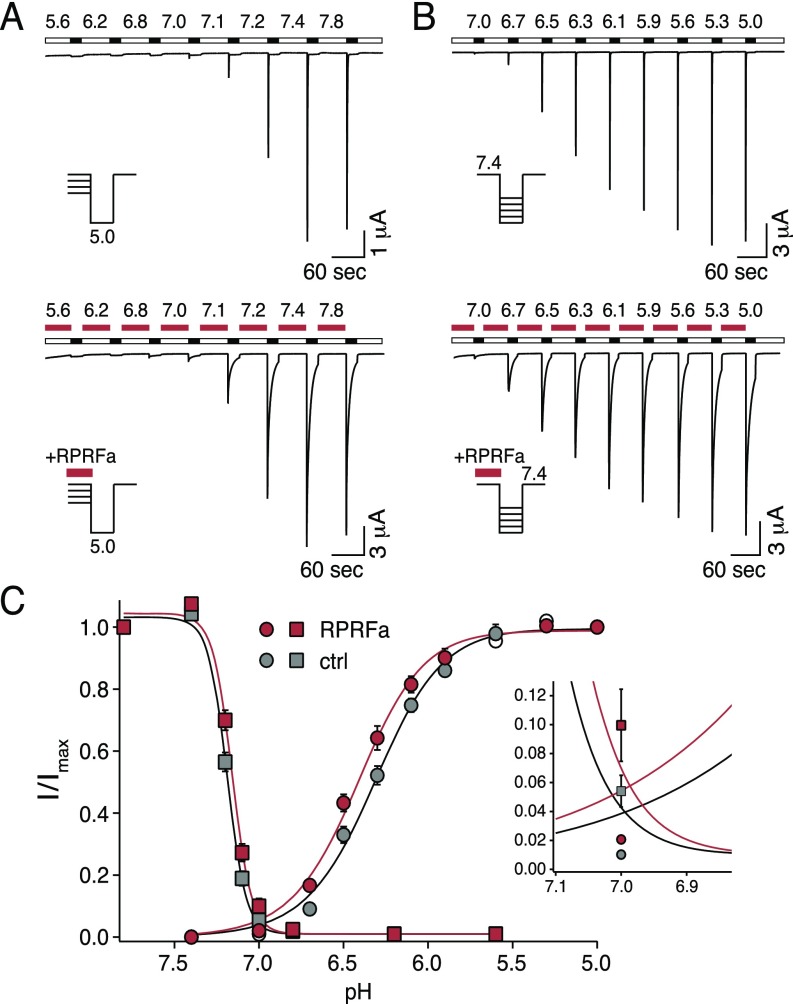
In addition to slowing desensitization, FMRFa also decreases proton sensitivity of some ASICs (34, 37). Therefore, we tested whether RPRFa changed proton sensitivity of ASIC3. RPRFa (10 μM) indeed slightly shifted the pH needed for half-maximal steady-state desensitization toward higher proton concentrations (lower pH) (from pH 7.19 ± 0.01 to pH 7.16 ± 0.01; n = 10; P < 0.05, t test; Fig. S3), but it shifted the pH needed for half-maximal activation slightly toward lower proton concentrations (higher pH) (from pH 6.32 ± 0.02 to pH 6.41 ± 0.02; n = 10; P < 0.05, t test; Fig. S3). Hill coefficients h were not significantly changed. These results are expected if RPRFa decreased proton sensitivity and at the same time slowed desensitization (3).
Fig. S3.
RPRFa slightly shifts curves of H+ activation and steady-state desensitization. (A) Representative current traces of ASIC3, illustrating current amplitudes at different conditioning pH without (Top) and with RPRFa (10 μM; red bars) (Bottom). Activation was with pH 5.0. (B) Representative current traces of ASIC3, illustrating activation by different pH without (Top) and with RPRFa (10 μM; red bars) (Bottom). Conditioning pH was 7.4. (C) Quantification of peak current amplitudes at different pH values, normalized to the amplitude at conditioning pH 7.8 (steady-state desensitization) or pH 5.0 (activation), respectively (n = 10). The “window current” of ASIC3 is shown on an expanded scale. RPRFa would slightly increase window currents.
RPRFa Affects Excitability of mDRG Neurons.
To address whether RPRFa had similar effects on native ASIC3 in nociceptors as in Xenopus oocytes, we tested the effect of the peptide on ASICs of isolated mouse dorsal root ganglia (DRGs) neurons. DRG neurons with rapidly desensitizing (τ < 500 ms) transient inward currents to stimulations with pH 6.3 were chosen and RPRFa was applied in concentrations of 50 or 10 μM, respectively (n = 5; Fig. S4). Preapplication of RPRFa both potentiated the peak amplitude and slowed down desensitization of ASIC3-like currents (SI Results), suggesting that RPRFa also modulates native ASIC3-containing channels in sensory neurons.
Fig. S4.
RPRFa potentiates ASIC3-like currents in DRG neurons. (A) Representative current traces illustrating potentiation of ASIC3-like currents (τ < 500 ms) in DRG neurons by pre- and coapplication of 50 or 10 μM RPRFa (red bars). Activation was with pH 6.3 (black bars). (B) Quantification of currents 10 s after activation, normalized to the respective peak currents. Note that the ASIC3-like currents are probably heterogenous in DRG neurons, because they may comprise homomeric and different heteromeric ASIC3-containing channels (n = 5). *P < 0.05 (Student’s t test).
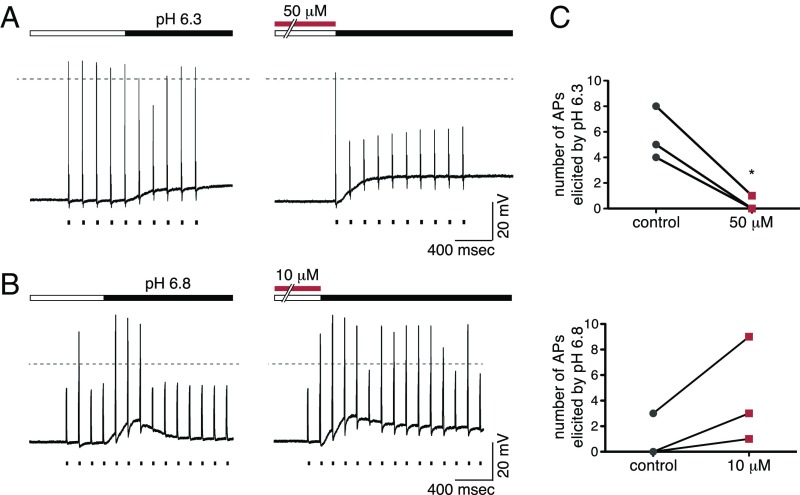
The effects of RPRFa on ASIC3—increase of the current amplitude and slowing of the desensitization—suggest that RPRFa might alter the excitability of ASIC3-expressing neurons to acidic stimuli. To test this possibility directly, we used the current-clamp technique and applied RPRFa together with acidic stimuli. First, we stimulated DRG neurons with ASIC3-like currents using suprathreshold electrical pulses (10–15 pulses of 10-ms duration every 200 ms), eliciting single action potentials (APs) (Fig. 6A). Additional application of an acidic solution (pH 6.3) led to a short and slight decrease of AP amplitudes, suggesting a weak depolarization block by pH 6.3. However, neurons quickly recovered (n = 3; Fig. 6A, Left). The membrane potential depolarized from −63.3 ± 0.9 to −55.1 ± 0.7 mV when the cells were perfused with pH 6.3 bath solution (n = 3; P = 0.001, t test). When we preapplied RPRFa in a high concentration (50 μM), application of pH 6.3 strongly reduced APs for 2 s (P < 0.05; Fig. 6C), suggesting a strong depolarization block. In agreement, membrane potential was significantly more strongly depolarized (from −64.2 ± 0.4 to −51.1 ± 0.5 mV; n = 3; P < 0.01, t test). This finding shows that preapplication of RPRFa more strongly depolarized the membrane potential than an acidic solution alone, decreasing excitability under these conditions.
Fig. 6.
RPRFa increases excitability of ASIC3-expressing sensory neurons. (A) DRG neurons with ASIC3-like currents were recorded in current-clamp mode and repeatedly stimulated by superthreshold electrical stimuli (n = 3). Application of pH 6.3 slightly decreased excitability (Left) and additional preapplication of 50 μM RPRFa strongly decreased excitability (Right). (B) DRG neurons with ASIC3-like currents were repeatedly stimulated by subthreshold electrical stimuli (n = 3); pH 6.8 transiently increased excitability (Left) and additional preapplication of 10 μM RPRFa more persistently increased excitability (Right). (C, Top) Number of APs elicited by pH 6.3 in three DRG neurons with an ASIC3-like current, in the absence and the presence of 50 μM RPRFa. (C, Bottom) Number of APs elicited by pH 6.8 in three DRG neurons with an ASIC3-like current, in the absence and the presence of 10 μM RPRFa. Note that the number of current pulses varied from neuron to neuron. *P < 0.05 (Student’s t test).
To better mimic a physiological situation, we used subthreshold electrical stimuli, a less acidic solution (pH 6.8) and lower concentrations of RPRFa (10 μM) (Fig. 6B). Without preapplication of RPRFa, acidification led to a short burst of APs (Fig. 6B, Left). With preapplication of 10 μM RPRFa, however, more electric stimuli elicited APs (Fig. 6B, Right), indicating an increased excitability of the neuron (Fig. 6C). Under these conditions, membrane depolarization induced by pH 6.8 did not change significantly upon preapplication of RPRFa (without RPRFa, membrane potential depolarized from −62.9 ± 1.4 to −50.1 ± 2.1 mV, with RPRFa from −62.5 ± 1.1 to −49.5 ± 3.5 mV; n = 3; P = 0.12, t test). These results indicate that RPRFa increases neuronal excitability under more physiological conditions, potentially enhancing or eliciting pain.
RPRFa Increases Acid-Induced Transient Muscle Pain and Chronic Hyperalgesia Mediated via ASIC3.
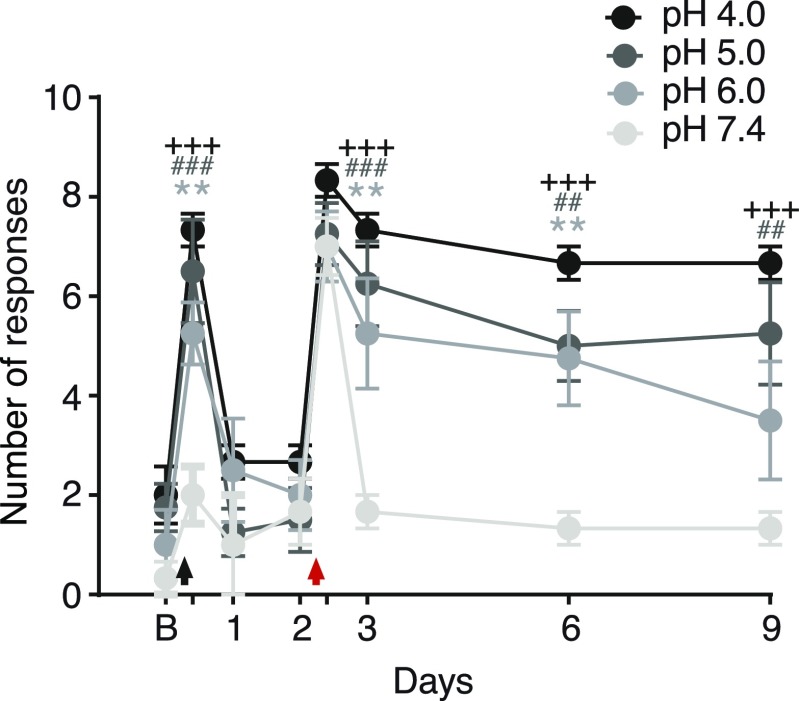
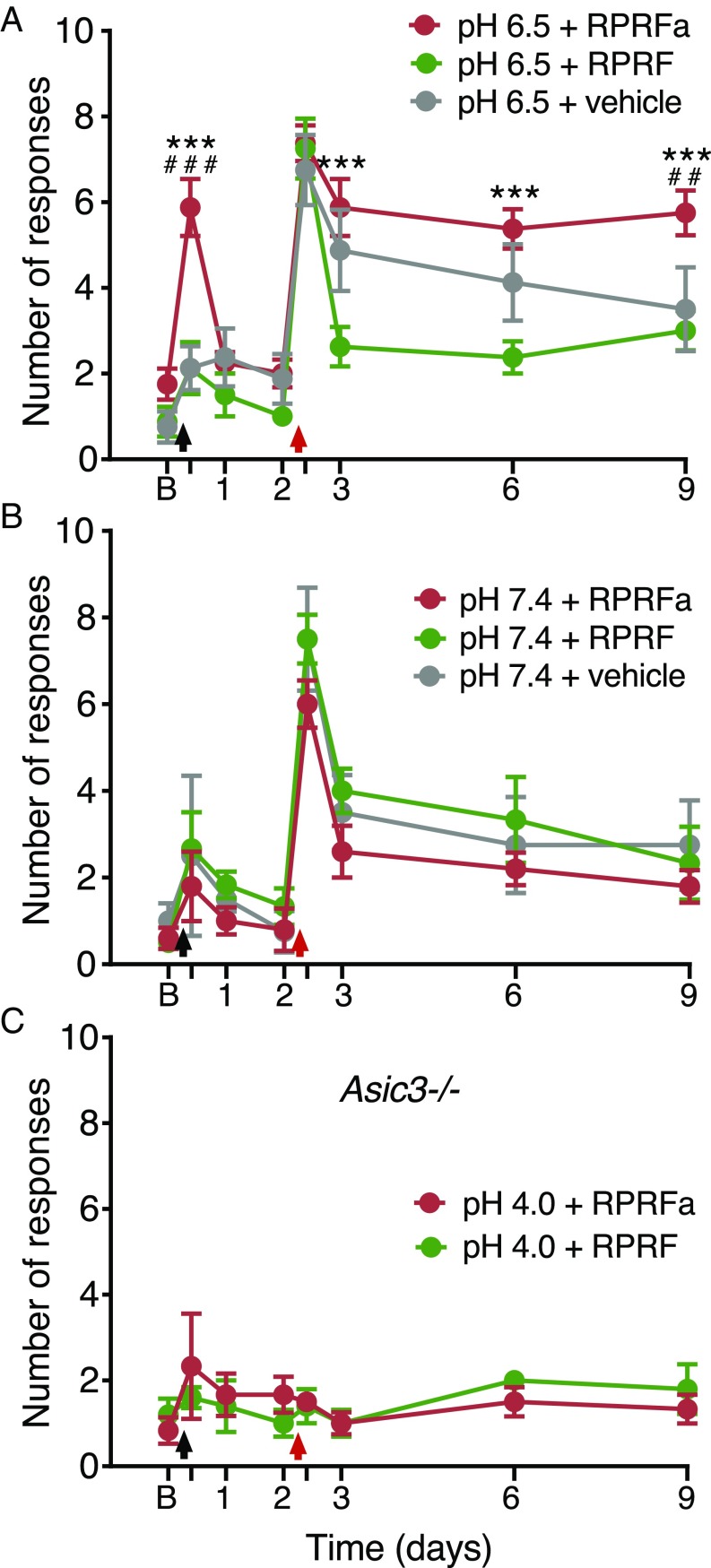
ASIC3 is expressed in muscle nociceptors (38) and has an established role in the development of widespread hyperalgesia in a rodent model of noninflammatory muscle pain (16). In this model, repeated injections of acidic solution to one side of the gastrocnemius muscle cause bilateral, long-lasting mechanical hyperalgesia in the hind paw (39) and ASIC3 triggers this acid-induced chronic muscle pain (16). We therefore tested whether RPRFa might enhance muscle pain upon acid injection by potentiating the response of ASIC3. In rats, injection of the gastrocnemius muscle with pH 4.0 saline reduces pH in the tissue on average to pH 6.5 (39), a pH that is sufficient to activate ASIC3 and induce strong mechanical hyperalgesia (16, 40). To have a stimulus that is closer to the threshold of ASIC3 activation and that does not itself induce strong hyperalgesia (Fig. S5), we injected the gastrocnemius muscle of mice on day 0 with pH 6.5 saline either without peptide, with RPRFa (500 pmol) or with RPRF (500 pmol), a peptide that lacks C-terminal amidation and does not potentiate ASIC3 currents (Fig. S6). As expected from the relatively low proton concentration in the saline, saline alone did not induce transient hyperalgesia in these mice. Strikingly, however, coinjection with RPRFa evoked a robust mechanical hyperalgesia 4 h after the first injection, an effect that was not seen when we coinjected RPRF [RPRFa vs. RPRF or vehicle, P < 0.001 (ANOVA); n = 8 for each treatment group; Fig. 7A]. Thus, RPRFa strongly potentiated the response to a near-threshold pH stimulus. This first injection can prime the muscle nociceptors so that the second injection of acidic saline (pH 4.0) on day 2 induces long-lasting hyperalgesia. Mechanical hyperalgesia was therefore again tested by determining the withdrawal response to a mechanical stimulus 4 h after the second injection and on days 3, 6, and 9 (n = 8 for each treatment group; Fig. 7A). As expected from the injection of pH 4 saline into muscle, 4 h after the second acidic injection, the response to a mechanical stimulus was similarly strong among the three groups. However, mechanical hyperalgesia lasted longer in mice that had been injected with RPRFa on day 0. On day 9, mice injected with RPRFa showed an enhanced response to mechanical stimulation compared with the two control groups (P < 0.001, RPRFa vs. RPRF; P < 0.01, RPRFa vs. vehicle; ANOVA), in which responses to mechanical stimuli were near baseline. On day 3 and day 6, there were also small differences in the mechanical hyperalgesia between the two control groups, which we attribute to some variability in the animals’ responses to the first stimulus near the threshold of ASIC3 activation (Fig. S5).
Fig. S5.
Acidic saline with a pH from 4.0 to 6.0, but not neutral saline, induces mechanical hyperalgesia in mouse muscle. On day 0, the gastrocnemius muscle of mice was injected with saline with a pH of 7.4 (n = 3), 6.0 (n = 4), 5.0 (n = 4), or 4.0 (n = 3). On day 2, the muscle was uniformly injected with a pH 4.0 saline. Transient and long-lasting mechanical hyperalgesia was observed for each condition except when the muscle was injected with pH 7.4 on day 0; for the pH 6.0 group, the hyperalgesia did not sustain beyond day 6. The black arrow indicates the first intramuscular injection of saline (pH 7.4, 6.0, 5.0, or 4.0, respectively); the red arrow indicates the second intramuscular injection of acidic saline (pH 4.0). B, baseline on day 0. +++P < 0.001, pH 4.0 vs. 7.4; ###P < 0.001; ##P < 0.01, pH 5.0 vs. 7.4; **P < 0.01, pH 6.0 vs. 7.4 (two-way repeated-measures ANOVA).
Fig. S6.
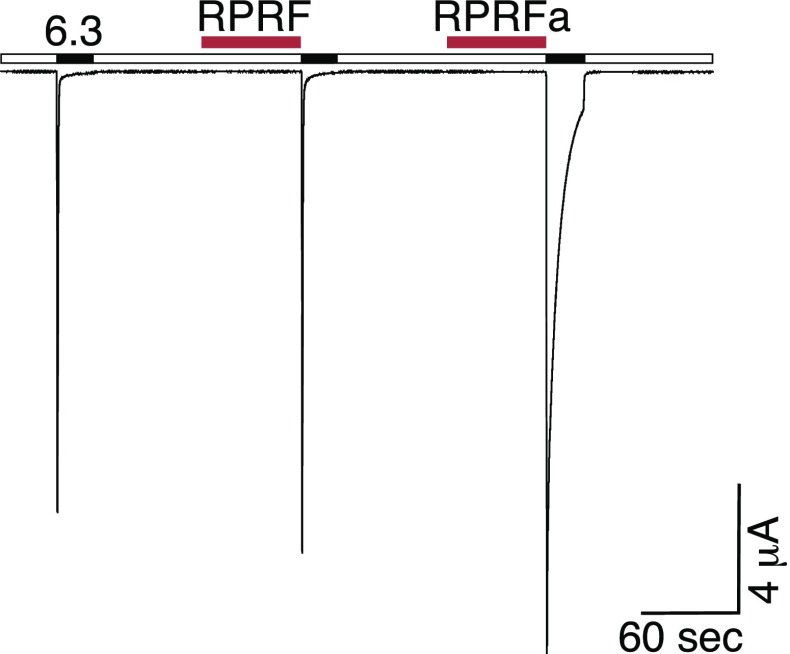
Amidation of RPRFa is essential for high-affinity potentiation of ASIC3 currents. Representative current trace illustrating the lack of potentiation of ASIC3 current by 50 μM RPRF, lacking the C-terminal amidation. In contrast, RPRFa (50 μM) potentiated ASIC3 currents. Peptides were applied in the conditioning period at pH 7.4 for 1 min (red bars). ASIC3 was activated by pH 6.3 (black bars).
Fig. 7.
RPRFa potentiates acid-induced transient muscle pain and chronic hyperalgesia mediated by ASIC3. (A) Enhancing effect of RPRFa on acid-induced mechanical hyperalgesia (n = 8 for each group). Transient and long-lasting hyperalgesia was only observed after the first injection of pH 6.5 saline with RPRFa (day 0) and after the second injection of acidic saline (pH 4.0) (day 2), respectively (n = 8 for each group). ***P < 0.001, RPRFa vs. RPRF; ###, P < 0.001, RPRFa vs. vehicle; ##, P < 0.01, RPRFa vs. vehicle (two-way repeated-measures ANOVA). (B) Injection of RPRFa (n = 5), RPRF (n = 6), or vehicle (n = 4) with neutral pH 7.4 saline showed no significant potentiation of transient or long-lasing hyperalgesia. (C) Asic3−/− mice did not show an increased response to mechanical stimuli after coinjection of RPRFa and pH 4.0 acidic saline (n = 6). Black arrows indicate the first intramuscular injection of saline (pH 6.5, 7.4, or 4.0, respectively) with or without RPRFa/RPRF; red arrows indicate the second intramuscular injection of acidic saline (pH 4.0). B, baseline on day 0.
We asked whether the induction by RPRFa of the transient hyperalgesia after the first injection of pH 6.5 saline and the long-lasting hyperalgesia after the second injection was dependent on acid and repeated the same experiment but with pH 7.4 saline in the first injection. Under these conditions, there were no significant differences among the experimental groups (n = 4–6 for each treatment group; Fig. 7B), confirming that the induction by RPRFa of enhanced mechanical hyperalgesia was acid-dependent.
It has been shown that ASIC3 is necessary for the development of mechanical hyperalgesia in this animal model (16, 40). To specifically test the role of ASIC3 in the potentiation of hyperalgesia by RPRFa, we repeated these experiments with Asic3−/− mice using pH 4.0 for the first injection. As described previously, mice lacking ASIC3 showed no increased response to mechanical stimuli after injection of acidic saline alone (16, 40). Similarly and in strong contrast to wild-type mice, these mice did not show enhanced response to mechanical stimuli after coinjection of acidic solution and RPRFa (n = 5–6 for each treatment group; Fig. 7C). These results indicate that the induction by RPRFa of enhanced mechanical hyperalgesia depends on ASIC3.
In experiments described above, we measured behavioral responses to fixed mechanical stimuli. To determine whether RPRFa also decreased the threshold to mechanical stimuli, we repeated the abovementioned experiment using pH 6.5 for the first injection and pH 4.0 for the second injection and determined the threshold to mechanical stimuli with an electrical von Frey Anesthesiometer 4 h after each injection and on days 0, 1, 2, 3, 6, and 9 (Fig. 8). This new experiment showed that RPRFa strongly reduced thresholds to mechanical stimuli transiently 4 h after the first injection and also lastingly for 7 d after the second injection. In contrast, in the two control groups, only the second injection (pH 4.0) transiently reduced mechanical thresholds. In both control groups, there was no long-lasting reduction of mechanical thresholds as in the RPRFa group (Fig. 8).
Fig. 8.
RPRFa potentiates acid-induced transient and long-lasting pain responses in wild-type mice. Enhancing effect of RPRFa on acid-induced pain responses. A first injection of pH 6.5 saline with RPRFa (day 0) strongly but transiently decreased mechanical withdrawal thresholds, whereas RPRF and vehicle had no effect on thresholds. A second injection of pH 4.0 saline (day 2) strongly reduced thresholds in all three groups. However, only after initial injection of RPRFa, the second injection of pH 4.0 produced a long-lasting (7 d) decrease of thresholds. ***P < 0.001, RPRFa vs. vehicle (two-way repeated-measures ANOVA). No significant difference was observed between RPRF and vehicle (n = 6 for RPRFa and RPRF, n = 4 for vehicle). The black arrow indicates the first intramuscular injection of pH 6.5 saline with or without RPRFa/RPRF; the red arrow indicates the second intramuscular injection of acidic saline (pH 4.0). B, baseline on day 0.
SI Results
RPRFa Slows ASIC3-Like Current Desensitization in Mouse DRG Neurons.
Out of 17 isolated mouse DRG neurons that were stimulated with pH 6.3, 10 responded with a rapidly desensitizing transient inward current (τ < 500 ms) that is typical for ASIC3, 3 responded with a transient inward current that desensitized more slowly (τ > 500 ms), and 4 had no transient currents but small sustained inward currents. Neurons with ASIC3-like currents to stimulations with pH 6.3 were chosen and RPRFa was applied in concentrations of 50 or 10 μM, respectively (n = 5; Fig. S3). With 50 μM RPRFa, the peak amplitude increased more than twofold from 287 ± 59 pA to 653 ± 90 pA (n = 7; P < 0.05, Student’s t test), the 10-s-current/peak ratio increased from 0.28 ± 0.05 to 0.63 ± 0.08 (n = 5) and the desensitization time constant τ strongly increased from 0.25 ± 0.06 to 6.03 ± 1.08 s. With 10 μM RPRFa, the peak amplitude did not change significantly (996 ± 375 pA for control vs. 1,006 ± 410 pA for RPRFa; n = 5; P = 0.92, t test), but the 10-s current/peak ratio increased from 0.16 ± 0.08 to 0.27 ± 0.08 (n = 5) and the desensitization time constant τ increased from 0.24 ± 0.04 to 3.80 ± 0.88 s (data were fit with a single exponential function). These results reproduced the modulation of ASIC3 by RPRFa in Xenopus oocytes.
We also applied RPRFa to those neurons that did not respond with a rapidly desensitizing current that is typical for ASIC3. Of the three neurons with transient ASIC currents that desensitized with τ > 500 ms, two were not influenced by 50 μM RPRFa (τ was 1.61 ± 0.56 s before RPRFa and 1.68 ± 0.31 s after RPRFa), and for the third, desensitization was strongly slowed by RPRFa (τ was 1.32 s before RPRFa and 10.77 s after RPRFa), suggesting that this neuron expressed ASIC3-containing channels. The 10-s-to-peak ratio (I10s/Ipeak) was not significantly increased. Of the four neurons without transient currents, three were not influenced by 50 μM RPRFa (the amplitude of the sustained currents was 146 ± 53 pA before RPRFa and 144 ± 13 pA after RPRFa; P = 0.98), whereas for the fourth current, amplitude was strongly increased by preapplication of RPRFa (19 pA before RPRFa and 324 pA after RPRFa). It is unclear whether this neuron might have expressed an ASIC3-containing channel that was silent under control conditions.
Discussion
In the present study, we identified three short RFamides in the venom of C. textile that potentiate ASIC3 currents, leading to enhanced excitability of ASIC3-expressing sensory neurons and increasing muscle pain. A peptide of just four amino acids, RPRFa, exhibited the highest potency. RFamides had previously been isolated from the venom gland of three other Conus species: cono-RFamide (CNF)-Vc1 from Conus victoriae (41), CNF-Sr1, and -Sr2 from Conus spurius (42, 43), and cono-neuropeptide Y (cono-NPY) from C. betulinus (29), suggesting that RFamides are widespread in Conus venoms. In molluscs, there are five genes coding for RFamides (44): the FMRFa-related gene, the LFRFa, the luqin, the cholecystokinin/sulfakinin, and the neuropeptide F (NPF) gene. The latter gene is related to the vertebrate NPY superfamily, because in many molluscs the C-terminal tyrosine-amide, characteristic for NPY, is replaced by a phenylalanine-amide. Because the sequence of the three C. textile CNFs that we isolated is identical to the C-terminal sequence of cono-NPY (NPF) from C. betulinus (Fig. 1D), it is rather likely that they are a product of the C. textile NPF gene, most likely by proteolytic cleavage from NPF. In contrast to cono-NPY1, CNF-Vc1 and CNF-Sr1, which consist of 18 and 12 aa, exhibit no sequence similarity to CNFs from C. textile apart from the C-terminal RFa motif. Thus, although our study suggests that RFamides might be a common component of Conus venoms, their sequence is variable.
CNFs belong to the group of conopeptides that are distinguished from conotoxins by a lack of disulfide bonds. Whereas conotoxins mainly target ion channels with high specificity and potency, the targets of conopeptides are often unknown. Intrathecal injection of CNF-Vc1 and CNF-Sr1 in mice either leads to hyperactivity (41, 42) or complete incapacitation (41), indicating that they are bioactive. It had been speculated that ASICs or FaNaCs might be their molecular targets. In the present work, we could demonstrate that ASIC3 is indeed a target for CNFs but that the molluscan FaNaCs are not a common target of CNFs.
However, is ASIC3 also the biological target of CNFs from C. textile? Because we used extracts of dissected venom glands for screening assays, we cannot exclude that CNFs are not part of the venom of C. textile, but act, for example, as endogenous neurotransmitters in the venom duct. CNF-Vc1, however, is present in milked venom (41) and, therefore, it is conceivable that CNFs are also part of the ejected venom of C. textile. Increasing evidence suggests that hormones/neuropeptides are biologically active components of the Conus venom, contributing to envenomation (45, 46). For example, NPY as a component of a venom may target neuroendocrine processes facilitating capture of a prey. As we focused on fractions that were active on ASIC3, we cannot confirm the presence of cono-NPY1 in the venom of C. textile. Moreover, cone snails can rapidly switch between a defensive and a predatory venom composition depending on the stimulus they receive (47). It is therefore intriguing to speculate that C. textile uses differentially processed forms of NPY for hunting and for defense by inducing pain or discomfort in predators.
Pain is a powerful strategy for defense and ASICs are targets of toxins inducing pain. The tarantula Psalmopoeus cambridgei, for example, uses toxins in its venom to induce pain by activating TRPV1 (48) and ASIC1a (21). Similarly, the Texas coral snake uses the toxin MitTx to target ASICs and induce pain (23). ASIC3, but not ASIC1a, has a crucial role in the induction of noninflammatory pain in rodent models (16, 40, 49, 50). We found that RPRFa increases DRG neuron excitability as well as muscle pain in mice by targeting ASIC3. Strikingly, RPRFa coinjected with pH 6.5 saline into the muscle of mice was able to induce strong transient hyperalgesia as well as to prime nociceptors to induce long-lasting hyperalgesia. Because pH 6.5 will only slightly reduce the average pH in muscle (39), this finding underscores the potential of RPRFa to induce pain under physiological conditions. This result, together with the high potency of RPRFa for ASIC3 compared with vertebrate RFamide neuropeptides, indeed suggests that RPRFa has been tailored by C. textile to enhance pain or discomfort in predators by targeting ASIC3. RPRFa might also be a useful tool to further unravel the role of ASIC3 in pain.
In summary, in the present study, we isolated short CNFs from the venom of C. textile that potentiate currents of the vertebrate pain receptor ASIC3, identifying a molecular target of CNFs. This CNF enhances excitability of ASIC3-expressing sensory neurons and increases muscle pain in mice. These results underscore the importance of ASIC3 for the development of muscle pain and reveal the potential significance of nontoxic components of cone snail venoms.
Materials and Methods
Preparation of Venoms and Peptides.
Whole venom ducts were prepared from the following cone snail species, which had been collected in the coral reefs of the Philippines: Conus bandanus, Conus capitaneus, Conus carinatus, Conus circumcisus, Conus flavidus, Conus generalis, Conus imperialis, Conus litteratus, Conus miles, Conus mustelinus, Conus omaria, Conus planorbis, Conus quercinus, Conus striatus, Conus tessulatus, C. textile, Conus virgo, and Conus vulpinus. Immediately after preparation, samples were stored in 10% acetic acid and were later homogenized using ultrasound. After centrifugation, the supernatant was lyophilized and stored at −20 °C until use. For the in vitro screening, lyophilized crude venom was dissolved in 1 mL standard bath solution (see below) containing 0.1% BSA to avoid unspecific peptide binding to the tubing. pH was adjusted to pH 7.4. The peptides RPRFa, VGRPRFa, and FMRFa were chemically synthesized with a purity of >95% (GeneCust).
Isolation, Purification, and Sequencing of Cono-RFamides.
Crude C. textile venom was extracted three times with 30% acetonitrile/water acidified with 0.1% TFA and centrifuged. Soluble material was lyophilized and stored at −20 °C before use. For fractionation, the crude venom extract was dissolved in 4.5 mL of 0.055% TFA and centrifuged (11,000 × g for 5 min). The supernatant was filtered (pore size, 45 μm) and fractionated on a semipreparative RP-HPLC column (5-μm C18; Nucleosil), elution was at 2.5 mL/min with a linear gradient of 0–80% solvent B (80% acetonitrile, 0.05% TFA) over 30 min. Fractions containing individual peaks were collected and each fraction was tested for its effect on ASIC3 currents. The active fractions 10 (12.50–13.50 min), 11 (13.50–14.30 min), and 12 (14.30–15.80 min) were first analyzed by MALDI-TOF-MS. Dominant [M + H]+ mass peaks m/z 574.4 (fraction 10), m/z 730.4 (fraction 11), and m/z 914.6 (fraction 12) were detected. These fractions were then further characterized by liquid chromatography–MS (LC/MS) analysis.
Fractions were desalted on a C18 column (ZipTip; Millipore) and eluted peptides analyzed on a linear-trap quadrupole Orbitrap XL mass spectrometer (Thermo Fisher Scientific) coupled to Nano-HPLC NanoLC 2D (Eksigent) with a nanoflow electrospray ionization source. Identified peptides were searched against the UniProt protein sequence database (www.uniprot.org/), using the Mascot server (v. 2.51; Matrix Science).
The resulting sequenced peptides were chemically synthesized and used for coelution using an analytical HPLC System with a linear gradient over 40 min (5-μm C18, 4.6 × 150 mm Reprosil at 1 mL/min with 0.055% TFA and solvent B). Moreover, the LC/MS analysis with the synthetic peptides showed complete identity in elution time and corresponding mass spectra.
Electrophysiology with Xenopus Oocytes.
Animal care and experiments were conducted under protocols approved by the State Office for Nature, Environment and Consumer Protection (LANUV) of North Rhine-Westphalia (NRW), Germany, and were performed in accordance with LANUV NRW guidelines. cRNA synthesis and preparation of Xenopus laevis oocytes were performed as described previously (19). Homomeric ASICs were expressed in Xenopus oocytes by injection of 0.03–0.05 ng of cRNA per oocyte for rat ASIC1a, 1–2 ng for rat ASIC1b, and 4–7 ng for rat ASIC3. For expression of heteromeric ASICs, we coinjected equal amounts of cRNAs coding for two different subunits, except for ASIC1a/3, for which we coinjected a 1:5 ratio (ASIC1a:ASIC3) to account for the better expression of ASIC1a. Total amounts of cRNA per oocyte were 2.4 ng for rat ASIC1a/ASIC3, 5 ng for rat ASIC1b/ASIC3 and ASIC2b/ASIC3, and 10 ng for rat ASIC2a/ASIC3. After 24–48 h, whole-cell ASIC currents were recorded at room temperature with a Turbo TEC-03X amplifier (npi Electronic) at a holding potential of −70 mV. Peak amplitudes varied from 2.13 to 23.15 μA for ASIC1a, from 1.38 to 8.38 μA for ASIC1b, from 1.46 to 33.34 μA for ASIC3, from 4.38 to 21.16 μA for ASIC1a/3, from 0.41 to 11.20 μA for ASIC1b/3, from 0.37 to 15.37 μA for ASIC2a/3, and from 0.23 to 7.31 μA for ASIC2b/3. Data acquisition was managed by the software Cellworks (npi Electronic). Microelectrodes had a resistance of <2 MΩ. Data were filtered at 20 Hz and the sampling rate was 100 Hz. For fast solution exchange, a pump-driven programmable pipetting workstation (Screening Tool; npi Electronic) controlled by the software Robosoft was used (51). Standard bath solution contained (in mM) 140 NaCl, 1.8 CaCl2, 1.0 MgCl2, and 10 Hepes; pH was adjusted by titration with NaOH or HCl, as appropriate. For acidic test solutions with pH <6.8, Hepes was replaced by Mes. Other substances were added to the bath solution, as indicated. Two-electrode voltage clamp data were analyzed using the software Cellworks Reader (npi Electronic) and Igor Pro (WaveMetrics).
Electrophysiology with mDRG Neurons.
DRG neurons from 6- to 10-wk-old mice were mechanically dissociated in DMEM containing 0.5 mg/mL pronase E (Serva) and 1 mg/mL collagenase A and were seeded on poly-l-lysine–coated coverslips and maintained for 7 d in DMEM supplemented with 2 g/L NaHCO3, 5% FBS, 5% horse serum, 50 ng/mL NGF, and 1% penicillin/streptomycin. For patch clamp measurements, coverslips were mounted in a perfused bath on the stage of an inverted microscope (IX71; Olympus). The bath solution contained (in mM) 128 NaCl, 5.4 KCl, 10 Hepes, 5.5 glucose, 1 MgCl2, and 2 CaCl2 (pH 7.4). For bath solution with pH <6.0, Hepes was replaced by Mes. Patch-clamp experiments were performed in the whole-cell configuration at room temperature. Patch pipettes had an input resistance of 4–6 MΩ, when filled with an intracellular like solution containing (in mM) 10 NaCl, 121 KCl, 10 Hepes, 5 EGTA, 2 MgCl2 (pH 7.2). Currents and voltage were recorded using a patch-clamp amplifier (Axopatch 200B), the Axon-CNS (Digidata 1440A), and Clampex software (Molecular Devices). Data were filtered at 1 kHz with low-pass filter and stored continuously on a computer hard disk and were analyzed using PCLAMP software. For voltage-clamp, membrane voltage was clamped to −70 mV, and data were sampled at a rate of 4 kHz. For current-clamp, membrane current was clamped to 0 pA, and data were sampled at a rate of 20 kHz. Current pulses were given for 2 s with 5 Hz and a pulse width of 10 ms.
Pain Behaviors.
Adult (8- to 12- week-old) C57BL/6J male mice were used. All procedures were approved by the Institutional Animal Care and Use Committee of Academia Sinica and followed the Guide for the Care and Use of Laboratory Animals (52). Asic3−/− mice were generated and genotyped as previous described (53). Mechanical hyperalgesia of hind paws was measured by using von Frey test. A 0.2-mN von Frey filament was applied to the plantar surface of both hind paws. A positive response was defined as foot lifting or licking. The filament was applied 5 times at 30 s intervals for each paw. The mechanical threshold was measured by using an electrical von Frey Anesthesiometer (IITC Life Science). A semiflexible tip was chosen and applied five times at 30-s intervals for each paw to get an average mechanical threshold. The experimenter was blinded to the mouse genotypes or drug injections.
To test the effect of RPRFa on enhancement of acid-induced hyperalgesia, 20 μL of acidic saline (pH 6.5) with or without 500 pmol of peptide was injected into the left gastrocnemius muscle on day 0. A control study with RPRF (500 pmol) was also conducted. Mice received a second injection of 20 μL of acidic saline (pH 4.0) (without peptide) on day 2. von Frey assay was performed at baseline and 4, 24, and 48 h after the first acid injection and 4 h, 24 h, 4 d, and 7 d after the second acid injection. In a second set of experiments, we aimed to test the peptide-only effect without acidic saline. Neutral pH 7.4 saline with or without 500 pmol of RPRFa (or RPRF) was injected on day 0, followed by pH 4.0 acidic saline (without peptide) injection on day 2. von Frey tests were conducted at the same time points as during the first set of experiments. Finally, the same experiment was conducted with Asic3−/− mice to test the role of ASIC3 in the potentiating effect of RPRFa. Acidic saline (pH 4.0) with RPRFa or RPRF was injected on day 0, followed by acidic saline (pH 4.0) on day 2. Pain behavioral tests were conducted at the same time points as before.
Data Analysis.
Peak currents from each measurement were normalized to the peak amplitude of the first activation. Currents remaining 10 s after activation were determined if not specified otherwise and normalized to the peak current of the same activation. This ratio is reported as the 10s-current-to-peak ratio (I10s/Ipeak) and describes current desensitization.
ASIC3 currents were fit with a single or a double-exponential function I(t):
A is the absolute peak current amplitude, ai (i = 0, 1, 2) indicates the relative contribution of the various current components (), and τi (i = 1, 2) is the desensitization time constant. Almost all data could be well fit using this function.
Concentration–response curves for peptides or protons were fit with a Hill function:
where a represents the baseline current and Imax the maximal normalized current, xhalf is the concentration to elicit a half-maximal effect, and h represents the Hill coefficient.
Statistical analysis was done with Student’s t tests if comparing two results or else with one-way repeated-measures ANOVA (referred to as “ANOVA”), followed by Bonferroni post hoc tests if not indicated otherwise. In behavioral studies, two-way repeated-measures ANOVA was used to compare responses in mice with different treatment at different time points. The following P values were considered significant: *P < 0.05; **P < 0.01; and ***P < 0.001. Data from oocytes represent the mean values of n individual measurements on different oocytes from at least two different animals. All data are reported as means ± SEM.
Acknowledgments
We thank E. Bressan for advice on isolation and maintenance of dorsal root ganglion (DRG) neurons, S. Lenz for isolation of mouse DRG neurons, H.-J. Förster for help with artwork, E. Lingueglia for the gift of cDNA for H. aspersa FaNaC, and Y. Furukawa for the gift of cDNA for A. kurodai FaNaC. C.-H.L. and C.-C.C. are supported by intramural grants from Academia Sinica. C.-H.H. is supported by Kaohsiung Medical University Hospital.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1616232114/-/DCSupplemental.
References
- 1.Du J, et al. Protons are a neurotransmitter that regulates synaptic plasticity in the lateral amygdala. Proc Natl Acad Sci USA. 2014;111:8961–8966. doi: 10.1073/pnas.1407018111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kreple CJ, et al. Acid-sensing ion channels contribute to synaptic transmission and inhibit cocaine-evoked plasticity. Nat Neurosci. 2014;17:1083–1091. doi: 10.1038/nn.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gründer S, Pusch M. Biophysical properties of acid-sensing ion channels (ASICs) Neuropharmacology. 2015;94:9–18. doi: 10.1016/j.neuropharm.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 4.Deval E, Lingueglia E. Acid-sensing ion channels and nociception in the peripheral and central nervous systems. Neuropharmacology. 2015;94:49–57. doi: 10.1016/j.neuropharm.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 5.Wemmie JA, et al. The acid-activated ion channel ASIC contributes to synaptic plasticity, learning, and memory. Neuron. 2002;34:463–477. doi: 10.1016/s0896-6273(02)00661-x. [DOI] [PubMed] [Google Scholar]
- 6.Wemmie JA, et al. Acid-sensing ion channel 1 is localized in brain regions with high synaptic density and contributes to fear conditioning. J Neurosci. 2003;23:5496–5502. doi: 10.1523/JNEUROSCI.23-13-05496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiong ZG, et al. Neuroprotection in ischemia: Blocking calcium-permeable acid-sensing ion channels. Cell. 2004;118:687–698. doi: 10.1016/j.cell.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 8.Jasti J, Furukawa H, Gonzales EB, Gouaux E. Structure of acid-sensing ion channel 1 at 1.9 A resolution and low pH. Nature. 2007;449:316–323. doi: 10.1038/nature06163. [DOI] [PubMed] [Google Scholar]
- 9.Bartoi T, Augustinowski K, Polleichtner G, Gründer S, Ulbrich MH. Acid-sensing ion channel (ASIC) 1a/2a heteromers have a flexible 2:1/1:2 stoichiometry. Proc Natl Acad Sci USA. 2014;111:8281–8286. doi: 10.1073/pnas.1324060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waldmann R, Champigny G, Bassilana F, Heurteaux C, Lazdunski M. A proton-gated cation channel involved in acid-sensing. Nature. 1997;386:173–177. doi: 10.1038/386173a0. [DOI] [PubMed] [Google Scholar]
- 11.Baron A, Waldmann R, Lazdunski M. ASIC-like, proton-activated currents in rat hippocampal neurons. J Physiol. 2002;539:485–494. doi: 10.1113/jphysiol.2001.014837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deval E, et al. Effects of neuropeptide SF and related peptides on acid sensing ion channel 3 and sensory neuron excitability. Neuropharmacology. 2003;44:662–671. doi: 10.1016/s0028-3908(03)00047-9. [DOI] [PubMed] [Google Scholar]
- 13.Vukicevic M, Kellenberger S. Modulatory effects of acid-sensing ion channels on action potential generation in hippocampal neurons. Am J Physiol Cell Physiol. 2004;287:C682–C690. doi: 10.1152/ajpcell.00127.2004. [DOI] [PubMed] [Google Scholar]
- 14.Waldmann R, et al. Molecular cloning of a non-inactivating proton-gated Na+ channel specific for sensory neurons. J Biol Chem. 1997;272:20975–20978. doi: 10.1074/jbc.272.34.20975. [DOI] [PubMed] [Google Scholar]
- 15.Chen CC, England S, Akopian AN, Wood JN. A sensory neuron-specific, proton-gated ion channel. Proc Natl Acad Sci USA. 1998;95:10240–10245. doi: 10.1073/pnas.95.17.10240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sluka KA, et al. Chronic hyperalgesia induced by repeated acid injections in muscle is abolished by the loss of ASIC3, but not ASIC1. Pain. 2003;106:229–239. doi: 10.1016/S0304-3959(03)00269-0. [DOI] [PubMed] [Google Scholar]
- 17.Deval E, et al. ASIC3, a sensor of acidic and primary inflammatory pain. EMBO J. 2008;27:3047–3055. doi: 10.1038/emboj.2008.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Golubovic A, et al. A peptide-gated ion channel from the freshwater polyp Hydra. J Biol Chem. 2007;282:35098–35103. doi: 10.1074/jbc.M706849200. [DOI] [PubMed] [Google Scholar]
- 19.Assmann M, Kuhn A, Dürrnagel S, Holstein TW, Gründer S. The comprehensive analysis of DEG/ENaC subunits in Hydra reveals a large variety of peptide-gated channels, potentially involved in neuromuscular transmission. BMC Biol. 2014;12:84. doi: 10.1186/s12915-014-0084-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Askwith CC, et al. Neuropeptide FF and FMRFamide potentiate acid-evoked currents from sensory neurons and proton-gated DEG/ENaC channels. Neuron. 2000;26:133–141. doi: 10.1016/s0896-6273(00)81144-7. [DOI] [PubMed] [Google Scholar]
- 21.Escoubas P, et al. Isolation of a tarantula toxin specific for a class of proton-gated Na+ channels. J Biol Chem. 2000;275:25116–25121. doi: 10.1074/jbc.M003643200. [DOI] [PubMed] [Google Scholar]
- 22.Diochot S, et al. A new sea anemone peptide, APETx2, inhibits ASIC3, a major acid-sensitive channel in sensory neurons. EMBO J. 2004;23:1516–1525. doi: 10.1038/sj.emboj.7600177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bohlen CJ, et al. A heteromeric Texas coral snake toxin targets acid-sensing ion channels to produce pain. Nature. 2011;479:410–414. doi: 10.1038/nature10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diochot S, et al. Black mamba venom peptides target acid-sensing ion channels to abolish pain. Nature. 2012;490:552–555. doi: 10.1038/nature11494. [DOI] [PubMed] [Google Scholar]
- 25.Baconguis I, Gouaux E. Structural plasticity and dynamic selectivity of acid-sensing ion channel-spider toxin complexes. Nature. 2012;489:400–405. doi: 10.1038/nature11375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baconguis I, Bohlen CJ, Goehring A, Julius D, Gouaux E. X-ray structure of acid-sensing ion channel 1-snake toxin complex reveals open state of a Na(+)-selective channel. Cell. 2014;156:717–729. doi: 10.1016/j.cell.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terlau H, Olivera BM. Conus venoms: A rich source of novel ion channel-targeted peptides. Physiol Rev. 2004;84:41–68. doi: 10.1152/physrev.00020.2003. [DOI] [PubMed] [Google Scholar]
- 28.Gray WR, Olivera BM, Cruz LJ. Peptide toxins from venomous Conus snails. Annu Rev Biochem. 1988;57:665–700. doi: 10.1146/annurev.bi.57.070188.003313. [DOI] [PubMed] [Google Scholar]
- 29.Wu X, Shao X, Guo ZY, Chi CW. Identification of neuropeptide Y-like conopeptides from the venom of Conus betulinus. Acta Biochim Biophys Sin (Shanghai) 2010;42:502–505. doi: 10.1093/abbs/gmq042. [DOI] [PubMed] [Google Scholar]
- 30.Jékely G. Global view of the evolution and diversity of metazoan neuropeptide signaling. Proc Natl Acad Sci USA. 2013;110:8702–8707. doi: 10.1073/pnas.1221833110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price DA, Greenberg MJ. Structure of a molluscan cardioexcitatory neuropeptide. Science. 1977;197:670–671. doi: 10.1126/science.877582. [DOI] [PubMed] [Google Scholar]
- 32.Sutherland SP, Benson CJ, Adelman JP, McCleskey EW. Acid-sensing ion channel 3 matches the acid-gated current in cardiac ischemia-sensing neurons. Proc Natl Acad Sci USA. 2001;98:711–716. doi: 10.1073/pnas.011404498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hesselager M, Timmermann DB, Ahring PK. pH dependency and desensitization kinetics of heterologously expressed combinations of acid-sensing ion channel subunits. J Biol Chem. 2004;279:11006–11015. doi: 10.1074/jbc.M313507200. [DOI] [PubMed] [Google Scholar]
- 34.Chen X, Paukert M, Kadurin I, Pusch M, Gründer S. Strong modulation by RFamide neuropeptides of the ASIC1b/3 heteromer in competition with extracellular calcium. Neuropharmacology. 2006;50:964–974. doi: 10.1016/j.neuropharm.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 35.Lingueglia E, Champigny G, Lazdunski M, Barbry P. Cloning of the amiloride-sensitive FMRFamide peptide-gated sodium channel. Nature. 1995;378:730–733. doi: 10.1038/378730a0. [DOI] [PubMed] [Google Scholar]
- 36.Furukawa Y, Miyawaki Y, Abe G. Molecular cloning and functional characterization of the Aplysia FMRFamide-gated Na+ channel. Pflugers Arch. 2006;451:646–656. doi: 10.1007/s00424-005-1498-z. [DOI] [PubMed] [Google Scholar]
- 37.Sherwood TW, Askwith CC. Endogenous arginine-phenylalanine-amide-related peptides alter steady-state desensitization of ASIC1a. J Biol Chem. 2008;283:1818–1830. doi: 10.1074/jbc.M705118200. [DOI] [PubMed] [Google Scholar]
- 38.Molliver DC, et al. ASIC3, an acid-sensing ion channel, is expressed in metaboreceptive sensory neurons. Mol Pain. 2005;1:35. doi: 10.1186/1744-8069-1-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sluka KA, Kalra A, Moore SA. Unilateral intramuscular injections of acidic saline produce a bilateral, long-lasting hyperalgesia. Muscle Nerve. 2001;24:37–46. doi: 10.1002/1097-4598(200101)24:1<37::aid-mus4>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 40.Chen WN, et al. Roles of ASIC3, TRPV1, and NaV1.8 in the transition from acute to chronic pain in a mouse model of fibromyalgia. Mol Pain. 2014;10:40. doi: 10.1186/1744-8069-10-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson SD, et al. Discovery by proteogenomics and characterization of an RF-amide neuropeptide from cone snail venom. J Proteomics. 2015;114:38–47. doi: 10.1016/j.jprot.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maillo M, et al. Conorfamide, a Conus venom peptide belonging to the RFamide family of neuropeptides. Toxicon. 2002;40:401–407. doi: 10.1016/s0041-0101(01)00176-3. [DOI] [PubMed] [Google Scholar]
- 43.Aguilar MB, et al. Conorfamide-Sr2, a gamma-carboxyglutamate-containing FMRFamide-related peptide from the venom of Conus spurius with activity in mice and mollusks. Peptides. 2008;29:186–195. doi: 10.1016/j.peptides.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zatylny-Gaudin C, Favrel P. Diversity of the RFamide peptide family in mollusks. Front Endocrinol (Lausanne) 2014;5:178. doi: 10.3389/fendo.2014.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Safavi-Hemami H, et al. Specialized insulin is used for chemical warfare by fish-hunting cone snails. Proc Natl Acad Sci USA. 2015;112:1743–1748. doi: 10.1073/pnas.1423857112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson SD, et al. Hormone-like peptides in the venoms of marine cone snails. Gen Comp Endocrinol. 2015;244:11–18. doi: 10.1016/j.ygcen.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dutertre S, et al. Evolution of separate predation- and defence-evoked venoms in carnivorous cone snails. Nat Commun. 2014;5:3521. doi: 10.1038/ncomms4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siemens J, et al. Spider toxins activate the capsaicin receptor to produce inflammatory pain. Nature. 2006;444:208–212. doi: 10.1038/nature05285. [DOI] [PubMed] [Google Scholar]
- 49.Sluka KA, Gregory NS. The dichotomized role for acid sensing ion channels in musculoskeletal pain and inflammation. Neuropharmacology. 2015;94:58–63. doi: 10.1016/j.neuropharm.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin SH, Sun WH, Chen CC. Genetic exploration of the role of acid-sensing ion channels. Neuropharmacology. 2015;94:99–118. doi: 10.1016/j.neuropharm.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 51.Baburin I, Beyl S, Hering S. Automated fast perfusion of Xenopus oocytes for drug screening. Pflugers Arch. 2006;453:117–123. doi: 10.1007/s00424-006-0125-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.National Research Council . Guide for the Care and Use of Laboratory Animals. 8th Ed National Academies Press; Washington, DC: 2011. [Google Scholar]
- 53.Chen CC, et al. A role for ASIC3 in the modulation of high-intensity pain stimuli. Proc Natl Acad Sci USA. 2002;99:8992–8997. doi: 10.1073/pnas.122245999. [DOI] [PMC free article] [PubMed] [Google Scholar]