Significance
The ability to manipulate signaling proteins with high spatiotemporal and genetic precision holds great promise for understanding the role of specific molecules in physiology and disease. In this study, we have provided mechanistic characterization of an optogenetic method for the control of G protein-coupled metabotropic glutamate receptors (mGluRs) using tethered photoswitches. This method provides substantial advantages, including improved labeling efficiency, photoswitch efficacy, and specificity. We harness this information to improve photoswitch efficacy, expand the approach throughout the mGluR family, thereby vastly expanding the toolbox, and design orthogonal photoswitches that permit dual optical control of multiple receptors. The engineering principles and approaches defined in this study should facilitate the extension of orthogonal photopharmacology to other signaling proteins.
Keywords: metabotropic glutamate receptor, optogenetics, photopharmacology, G protein-coupled receptor, photoswitchable ligand
Abstract
G protein-coupled receptor (GPCR) signaling occurs in complex spatiotemporal patterns that are difficult to probe using standard pharmacological and genetic approaches. A powerful approach for dissecting GPCRs is to use light-controlled pharmacological agents that are tethered covalently and specifically to genetically engineered receptors. However, deficits in our understanding of the mechanism of such photoswitches have limited application of this approach and its extension to other GPCRs. In this study, we have harnessed the power of bioorthogonal tethering to SNAP and CLIP protein tags to create a family of light-gated metabotropic glutamate receptors (mGluRs). We define the mechanistic determinants of photoswitch efficacy, including labeling efficiency, dependence on photoswitch structure, length dependence of the linker between the protein tag and the glutamate ligand, effective local concentration of the glutamate moiety, and affinity of the receptor for the ligand. We improve the scheme for photoswitch synthesis as well as photoswitch efficiency, and generate seven light-gated group II/III mGluRs, including variants of mGluR2, 3, 6, 7, and 8. Members of this family of light-controlled receptors can be used singly or in specifically labeled, independently light-controlled pairs for multiplexed control of receptor populations.
G protein-coupled receptors (GPCRs) provide a link between extracellular stimuli and intracellular signaling pathways in many complex biological systems, including the central and peripheral nervous systems (1). Due to the inherent dynamic spatial and temporal complexity of GPCR signaling, as well as the vast diversity of subtypes, it remains difficult to probe the role and mechanism of specific receptors in physiological processes ranging from molecular and cellular events to neural circuits and behavior. Conventional probes such as pharmacological agonists, antagonists, or allosteric modulators are both difficult to apply and remove rapidly and to target spatially to either specific cell types or subcellular processes. Furthermore, designing fully subtype-selective ligands remains a major challenge, despite the increasing abundance of X-ray crystal structures (2). To overcome these limitations, a number of approaches have recently been developed based on either chemical engineering or optical control, which allow receptor activation to be tightly controlled in space and/or time and, when combined with genetic encoding, allow for targeting of subsets of cellular populations.
The most widely used manipulators of G-protein signaling are the DREADDs. These engineered receptors are selectively activated by an othogonal ligand, allowing G-protein activation to be initiated in genetically defined cell types (3) but with slow on and off kinetics and limited spatial precision. Efforts to overcome these limitations led to the use of rhodopsin (4), cone opsin (5), and melanopsin (6, 7). Although these tools permit improved spatial and temporal precision compared with DREADDs, it remains unclear whether they can be used to fully recapitulate the complex signaling properties of specific receptors, despite clever chimeric approaches to impart regions of different GPCRs to the light-sensitive opsin core (8–11). Efforts to develop optical control of GPCRs also led to the development of fully native GPCRs from the class C metabotropic glutamate receptor (mGluR) family, which are controlled by a photoswitchable tethered ligand (PTL) and enable both biophysical analysis of molecular mechanism and cellular analysis of neuronal signaling (12, 13). We recently complemented cysteine-attaching PTLs with photoswitchable orthogonal remotely tethered ligands (PORTLs) that attach via a linker to a SNAP domain that is fused to the ligand-binding domain of the receptor, thereby gaining robust and high-selectivity targeting of the photoswitchable ligand for the receptor of interest (12, 14).
The PORTL system offers the opportunity for parallel targeting of distinct photoswitched ligands to distinct receptors for multiplexed optical control, an opportunity that we seize upon here. The PORTL system differs in design due to the remote attachment of the photoswitch to a fused protein domain rather than the direct attachment of a small PTL directly to the ligand-binding domain. We describe experiments that define the local concentration of the ligand in the photoactivated state, the photoactivation and deactivation kinetics, and the system’s efficacy limits. We harness this mechanistic knowledge to improve photoswitch efficacy, design orthogonal CLIP-targeting PORTLs, and extend photocontrol to five of the six members of the group II and III mGluR subfamilies. Significantly, we show that spectral variants of SNAP-targeting and CLIP-targeting PORTLs may be multiplexed to achieve independent optical control of two distinct receptor populations, advancing the power of optogenetic analysis of GPCR signaling from molecular mechanism to neural circuit and behavior. The engineering principles and approaches defined in this study should facilitate the extension of orthogonal photopharmacology to other signaling proteins.
Results
SNAP-Tethered (PORTL) and Cysteine-Conjugated (PTL) Glutamate Photoswitches for mGluR2.
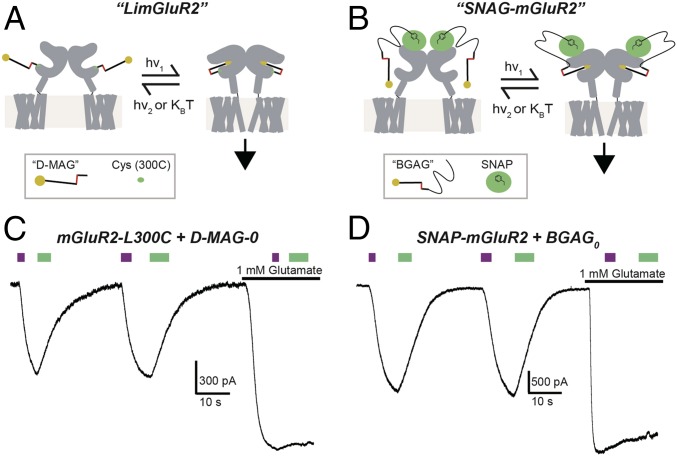
We previously developed two alternative mechanisms for optical control of mGluR2 using azobenzene-glutamate–based compounds. “LimGluR2” relies on the attachment of a maleimide-containing PTL to a cysteine that is introduced to the surface of the ligand-binding domain of the receptor (mGluR2-L300C) (12), whereas “SNAG-mGluR2” attaches an O6-benzylguanine (BG)–containing PORTL to an N-terminal SNAP-tagged receptor (SNAP-mGluR2) (12, 14). LimGluR2 conjugated to its PTL d-MAG-0 (Fig. 1A and SI Appendix, Fig. S1) shows photoagonism in the cis state (under 380-nm illumination), which activates coexpressed GIRK channels in HEK 293T cells. The photocurrent amplitude is 57.8 ± 6.9% (n = 8 cells) of those induced by saturating glutamate (Fig. 1B). Following photoactivation, LimGluR2 remains on in the dark until it is turned off by visible-light illumination (≥480 nm) (Fig. 1B). In contrast, SNAP-mGluR2 is conjugated to a long photoswitch that has a SNAP-reactive BG at one end, a linker, and an azobenzene-glutamate (AG) at the other end, and so is called “BGAG” (Fig. 1C and SI Appendix, Fig. S1). Once conjugated to BGAG, SNAP-mGluR2 shows photoagonism in the cis state (under 380-nm illumination). The photocurrent amplitude is 65.6 ± 5.0% (n = 12 cells) relative to the current evoked by saturating glutamate (Fig. 1D). A red-shifted variant of BGAG, BGAG460, yields maximal activation under 460-nm light and undergoes spontaneous deactivation in the dark (SI Appendix, Fig. S1). Importantly, SNAG-mGluR2 is efficiently labeled by BGAG at <1% of the concentration required for d-MAG-0 labeling of LimGluR2 (∼1 μM versus ∼200 μM, respectively) due to its bioorthogonality and the lack of BG hydrolysis compared with the rapidly hydrolyzed maleimide (14).
Fig. 1.
Optical control of mGluR2 through complementary photoswitch conjugation strategies: LimGluR2 and SNAG-mGluR2. (A and B) Schematics showing optical control of mGluR2 through either conjugation of a cysteine-reactive maleimide-containing photoswitch (“d-MAG-0”) directly to the ligand-binding domain (LimGluR2; A) or conjugation of an O6-benzylguanine–containing photoswitch (BGAG) to an N-terminal SNAP tag (SNAG-mGluR2; B). Both photoswitches use an azobenzene core tethered to a glutamate moiety via a 4′d linkage. (C and D) Representative traces showing photoactivation (380 nm, violet bars) and deactivation (500 nm, green bars) of mGluR2 in HEK 293T cells coexpressing GIRK channels by either LimGluR2 (C) or SNAG-mGluR2 (D). Both approaches produce 40 to 70% efficacy relative to saturating 1 mM glutamate.
Analysis of Photoswitch Mechanism: Linker Length, Efficacy, and Intrasubunit Photocontrol.
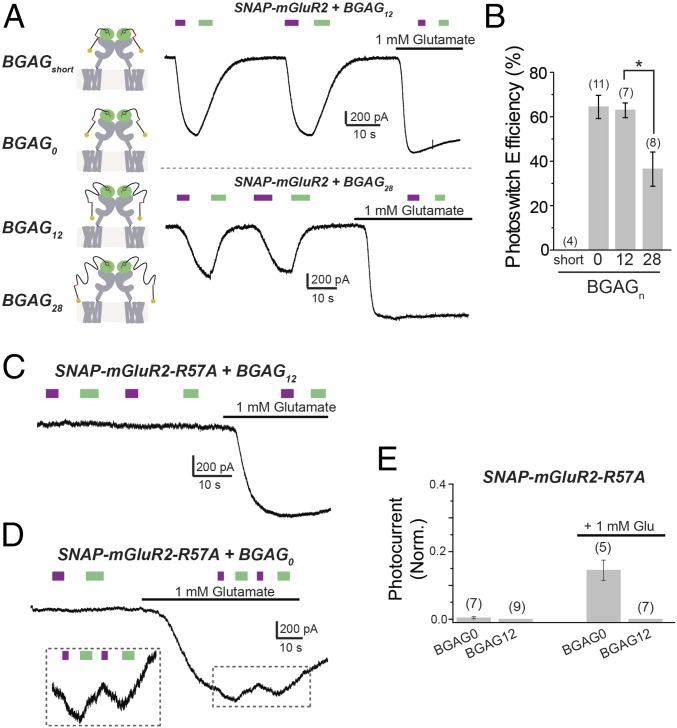
We sought to gain a deeper understanding of the photoswitch mechanism to optimize and tune the design and adapt photoswitching to other receptors. To improve the synthetic route and facilitate synthesis of novel photoswitches, we searched for an alternative to Cu(I)-catalyzed alkyne-azide “click chemistry,” which requires overstoichiometric amounts of toxic copper to drive the reaction to completion. We switched to strain-promoted alkyne-azide click chemistry together with peptide couplings to make the synthesis more environmentally friendly and easier to reproduce (SI Appendix, Schemes S1–S5). The family of BGAG PORTLs that we produced enabled us to compare the mechanisms of photoactivation for a PTL such as d-MAG-0, whose conjugation site is through a very short linker directly to a cysteine introduced into the lower lobe of the ligand-binding domain, near the orthosteric binding site in LimGluR2 versus a BGAG, which attaches via a long polyethylene glycol (PEG) linker to a separate N-terminal fused SNAP domain in SNAG-mGluR2. Interestingly, LimGluR2 is highly sensitive to d-MAG-0 length: The addition of a single glycine between the azobenzene and glutamate, to produce d-MAG-1, converts photoagonism to photoantagonism (12). In contrast, BGAGs with linkers consisting of different numbers of PEG repeats—BGAG0, BGAG4, BGAG8, and BGAG12—showed similar photoagonism in SNAG-mGluR2, suggesting that, unlike MAGs, whose photoisomerization points the glutamate into the binding site, photoisomerization of BGAG removes an obstruction from the glutamate that allows it to bind in the manner of a photochromic ligand on a string (12, 14). To explore this photoswitch mechanism model, we synthesized a version of BGAG with an even shorter BG-to-azobenzene linker than BGAG0 (“BGAGshort”) (SI Appendix, Fig. S1 and Scheme S1), as well as one with a longer linker containing 28 PEG repeats (BGAG28) (SI Appendix, Fig. S1 and Scheme S2). It is important to note that due to the flexibility of the many single bonds within the PEG linker, we can model BGAG as a worm-like chain (15), where there is a sublinear increase in end-to-end length of the compound as PEG repeats are added. Following conjugation to SNAP-mGluR2, BGAGshort produced no detectable photoactivation, whereas BGAG28 showed clear cis agonism but had significantly reduced efficacy compared with BGAG12 or BGAG0 (Fig. 2 A and B). We also synthesized a shortened version of BGAG460 that lacked PEG repeats (“BGAGshort,460”) (SI Appendix, Fig. S2 and Scheme S3) and observed a complete lack of photoactivation, compared with ∼50% photoswitch efficiency for BGAG12,460 (SI Appendix, Fig. S3). It is worth noting that GIRK activation provides a readout that agrees closely with mGluR activation as determined from both downstream second-messenger assays and a direct measure of the receptor’s activation-associated conformational changes (16).
Fig. 2.
Mechanism of SNAG-mGluR2 photoswitching: BGAG length dependence, glutamate affinity, and subunit specificity. (A) Characterization of BGAG length dependence. BGAG variants of differential PEG linker lengths (depicted in cartoons; Left) were tested on SNAP-mGluR2. (A, Right) Representative traces showing photoactivation of SNAG-mGluR2 by BGAG12 (Top) or BGAG28 (Bottom). (B) Summary of photoswitch efficiency relative to saturating 1 mM glutamate for BGAG variants on SNAP-mGluR2. * indicates statistical significance (unpaired t test between BGAG12 and BGAG28, P = 0.004). (C and D) Reduction of glutamate affinity with the mutation R57A abolishes photoswitching of SNAG-mGluR2 in the absence of glutamate for both BGAG12 (C) and BGAG0 (D). (D, Inset) Photoswitching in the presence of 1 mM glutamate for BGAG0, indicating cooperativity between glutamate binding and photoactivation. (E) Summary of R57A experiments. Photocurrent amplitude is normalized to the amplitude of the response to 1 mM glutamate. The numbers of HEK 293T cells tested are shown in parentheses. Error bars show standard errors.
Together, the above observations show that the efficacy of BGAG depends on linker length. The broad range of acceptable lengths suggests that BGAG potency depends on a low effective concentration of the azobenzene-glutamate on its long tether, rather than a precise angle-dependent high effective concentration (>10 mM) docking that operates in MAG PTLs (17–19). To test this model, we introduced the mGluR2 ligand binding site mutation R57A to reduce glutamate affinity by ∼30-fold (20). We showed earlier that in LimGluR2, photoactivation by MAG is not affected by the R57A mutation, even though sensitivity to synaptically released glutamate is expected to be drastically reduced (21). We show here that the same logic applies to mGluR3, where the homologous mutation, R64A, was used to generate a low-affinity version of LimGluR3 (“LA-LimGluR3”) that also maintained efficient photoactivation (SI Appendix, Fig. S4 A and B).
In striking contrast to the lack of effect of binding-site mutations on photoactivation by MAGs, BGAG0 and BGAG12 produced no photoactivation of the low-affinity SNAP-mGluR2-R57A (Fig. 2C). We considered that the loss of photoactivation may reflect the low stability, and hence short lifetime, of the BGAG-bound state, which would be expected to result in very little synchronous liganding across the receptor population in the cell, and rare simultaneous activation of the two subunits in an mGluR2 dimer that is needed for full activation (21). To test this hypothesis, we applied a subsaturating (∼EC50 to 70) 1 mM dose of glutamate to SNAP-mGluR2-R57A and asked whether photoswitching would have an effect on top of this partially liganded and, hence, incompletely activated background. Consistent with this expectation, we found that BGAG0 photoisomerization produced a cis agonism on top of the submaximal glutamate-activated background in SNAP-mGluR2-R57A (Fig. 2D). However, BGAG12 had no effect (Fig. 2 C and E), suggesting that the azobenzene-glutamate of BGAG0 is in a higher local concentration near the glutamate binding site than it is in BGAG12, consistent with the expectation that the longer linker would allow for the glutamate to wander more widely.
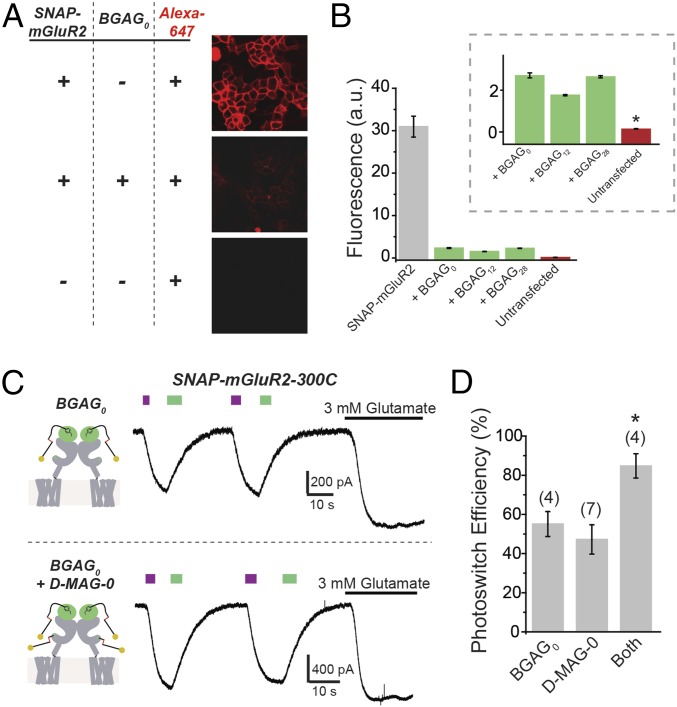
The implication of the above observations—that the glutamate in BGAG0 is present in a higher effective concentration but that the activation of wild-type affinity SNAP-mGluR2 is the same for BGAG0 and BGAG12—is that normally high affinity of the binding site for glutamate assures saturated (and thus equal) binding in both cases. In such a case, one would expect to be able to account quantitatively for the efficacy of photoactivation compared with saturating free glutamate. To assess this, we first sought to estimate the labeling efficiency of BGAGs. We took advantage of the ability of SNAP domains to be efficiently labeled with BG-conjugated fluorophores and developed a labeling assay where SNAP-mGluR2–expressing cells are exposed to a BGAG, washed, and then exposed to the BG-Alexa 647 dye to determine what percentage of the SNAP domains on the membrane have not been conjugated to BGAG and so remain free for dye labeling. Compared with control SNAP-mGluR2–expressing cells that were only exposed to dye, cells pretreated with either BGAG0, BGAG12, or BGAG28 (10 μM for 45 min) before dye labeling showed a reduction in fluorophore labeling by ∼90% (Fig. 3 A and B). Untransfected cells that were not pretreated with BGAG had an even lower level of nonspecific dye labeling, indicating that the residual ∼10% of BG-Alexa 647 dye labeling in SNAP-mGluR2–expressing cells represented availability of the SNAP-mGluR2 due to incomplete labeling by BGAG (Fig. 3B, Inset). Importantly, the BGAG variants had similar labeling efficiencies (Fig. 3B), indicating that differences in photoswitch efficiency (Fig. 2) reflect differences in occupancy of the binding site and not in labeling efficiency. We asked whether we could account for the photoswitch efficiency observed with SNAG-mGluR2 conjugated to either BGAG0 or BGAG12. The observed ∼90% BGAG labeling efficiency would mean that ∼80% of SNAP-mGluR2 dimers would have two labeled subunits and ∼18% would have one labeled subunit. Considering that at 380 nm ∼90% of azobenzenes isomerize to the cis state (17) and that binding of glutamate to one of the subunits in the dimer activates mGluR2 by ∼20% relative to the full activation that is induced by binding to both subunits (21), we estimate an overall photoswitch efficiency of ∼70%. This is close to the observed values of ∼65% (Figs. 1 and 2). Following this, we reasoned that we should be able to overcome the ∼70% efficacy limit imposed by the combination of incomplete labeling and incomplete photoisomerization if only we could double the number of photoswitchable ligands per subunit. To achieve this, we combined BGAG and MAG labeling using SNAP-mGluR2-300C. When labeled only with BGAG0 or only with d-MAG-0, we obtained an efficacy of ∼50 to 60%; however, when both photoswitches were added, we obtained near-complete photoactivation (85 to 95%) relative to saturating glutamate (Fig. 3 C and D), supporting the interpretation.
Fig. 3.
Mechanism of SNAG-mGluR2 photoswitching: labeling and BGAG efficacy. (A) Analysis of BGAG labeling efficiency. HEK 293T cells were treated with BG-Alexa 647 without (Top) or with (Middle) preincubation with BGAG0. BGAG0 labeling drastically reduced BG-Alexa 647 labeling because they attach via the same reactive cysteine of SNAP. In the absence of SNAP-mGluR2, no fluorescence was observed (Bottom). (B) Summary of fluorescence intensity for SNAP-mGluR2 labeled with BG-Alexa 647 without (gray) or with (green) prelabeling with BGAG variants or in untransfected cells (red). (B, Inset) Labeling of SNAP-mGluR2 with BG-Alexa 647 was above the background levels observed in the untransfected control. * indicates statistical significance (unpaired t test between BGAG0 and untransfected, P = 0.0001). a.u., arbitrary units. (C) Photoactivation of SNAP-mGluR2-300C with either BGAG0 (Top) or BGAG0 and d-MAG-0 (Bottom) shows an additive effect of combining photoswitches with colabeling, producing near-complete photoactivation. (D) Summary of photoswitch efficiency for photoswitch combination. * indicates statistical significance (unpaired t test between both and d-MAG-0, P = 0.0027; unpaired t test between both and BGAG, P = 0.017). The numbers of cells tested are shown in parentheses. Error bars show standard errors.
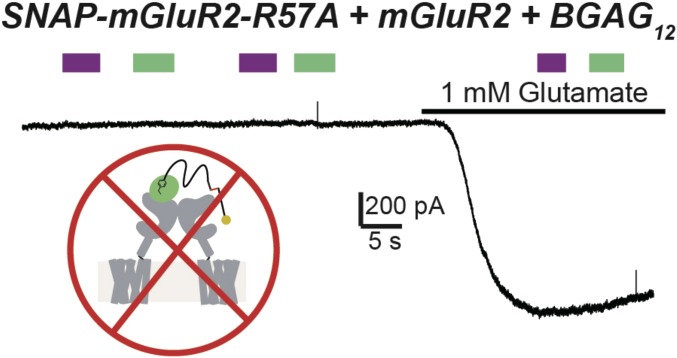
A key aspect of photocontrol of multisubunit receptors is the ability to selectively control the ligand occupancy of a particular (photoswitch-labeled) subunit. In the case of PTL compounds, such as d-MAG-0, this specificity is assured by their small size (∼1 nm). However, in the case of a PORTL, especially one with a long PEG linker, it is important to determine whether the photocontrol is limited to the conjugated subunit or, alternatively, is able to reach the adjacent subunit within a dimer. We tested this by coexpressing mGluR2wt with a low-affinity subunit that contained the SNAP attachment site (SNAP-mGluR2-R57A). Following exposure to BGAG12, we observed no photoagonism in the absence or presence of glutamate (Fig. 4). This observation indicates that a linker of 12 PEG repeats is insufficient to reach from SNAP in one subunit to the glutamate-binding pocket in the partner subunit of the dimer.
Fig. 4.
Photoagonism of SNAP-mGluR2 by BGAG proceeds through the conjugated subunit. Representative trace from HEK 293T cells showing that coassembly of BGAG-labeled but photoactivation-deficient mutant subunits (SNAP-mGluR2-R57A + BGAG12) with wild-type subunits does not allow for photoactivation. This result argues that BGAG activates the same subunit to which it is labeled and is unable to bind the other ligand-binding domain within a dimer.
Together, these results indicate that the PORTL approach is photoselective for the conjugated subunit and demonstrate that we have an adequate understanding of the mechanism of photoactivation and a means for boosting photoactivation to near completion.
Extension of PORTLs in the Group II and III mGluR Families.
To expand the light-gated mGluR toolbox, we next sought to adapt the PORTL approach to the other group II member, mGluR3, and to the members of group III. We used N-terminally SNAP-tagged versions of each of these mGluRs (22) and tested a series of BGAG ligands. In SNAP-mGluR3, BGAG0 produced a moderate (∼20%) photoagonism, whereas longer and shorter variants showed weaker or no photoswitching (SI Appendix, Fig. S4 B and C). The lower potency of BGAG photoactivation of SNAP-mGluR3 compared with SNAP-mGluR2 suggests differences in structure and pharmacology, despite the ∼70% sequence identity.
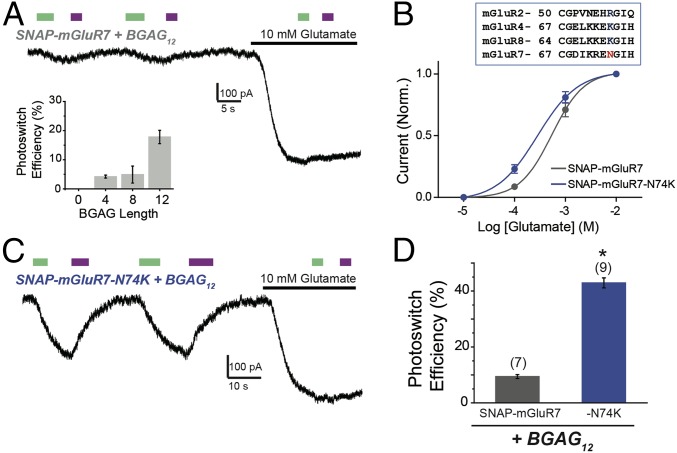
We next turned to group III mGluRs, which are challenging to study because of a limited repertoire of specific pharmacological agents. We first sought to extend optical control to mGluR7, which is especially hard to study because of its extremely low apparent glutamate affinity compared with all other mGluRs (∼1 mM versus ∼1 to 10 µM, respectively). SNAP-mGluR7 showed weak photoagonism that was highly BGAG length-dependent, with peak photoswitch efficiency exerted by BGAG12 (∼15 to 20%) (Fig. 5A, Inset). Strikingly, activation was evoked by illumination with 500-nm light and turned off by illumination at 380 nm (Fig. 5A). This trans photoagonism was seen for each of the BGAGs that produced a photoresponse, suggesting that it is inherent to the way that the azobenzene-glutamate moiety interacts with the ligand-binding domain.
Fig. 5.
Molecular engineering of SNAG-mGluR7: trans agonism with BGAG12. (A) Photoactivation of SNAP-mGluR7 by BGAGs in HEK 293T cells. Unlike SNAG-mGluR2, visible light (500 nm) activates mGluR7, and near UV (380 nm) deactivates mGluR7. (A, Inset) Summary of BGAG length dependence of photoactivation. (B, Top) Alignment between mGluRs highlighting the conserved charged residue that is present in all group II/III mGluRs except for mGluR7 (N74). (B, Bottom) Glutamate dose–response curves showing a small increase in apparent affinity for SNAP-mGluR7-N74K compared with wild type. (C and D) SNAP-N74K shows enhanced photoactivation by BGAG12. * indicates statistical significance (unpaired t test, P = 0.00002). The numbers of cells tested are shown in parentheses. Error bars show standard errors.
To improve the efficacy of photoswitching beyond 20% in mGluR7, we reasoned that it would help if we could compensate for the relatively low local concentration of BGAG at the ligand-binding pocket, as seen above in SNAG-mGluR2 (Fig. 3), which is exacerbated by the low intrinsic glutamate affinity of mGluR7. We introduced a mutation at N74 in the glutamate binding site of mGluR7 to change its identity to that of the other high glutamate affinity group III mGluRs (Fig. 5B, Inset). The N74K mutation (23) (SI Appendix, Fig. S5A) increased the apparent glutamate affinity of mGluR7 (Fig. 5B) and increased photoswitch efficiency to ∼40% with BGAG12 (Fig. 5 C and D) and to ∼20% with BGAG28. SNAP-mGluR7-N74K retained normal surface expression (SI Appendix, Fig. S5C), and its photoactivation could be fully blocked by the competitive antagonist LY341495 (SI Appendix, Fig. S5D).
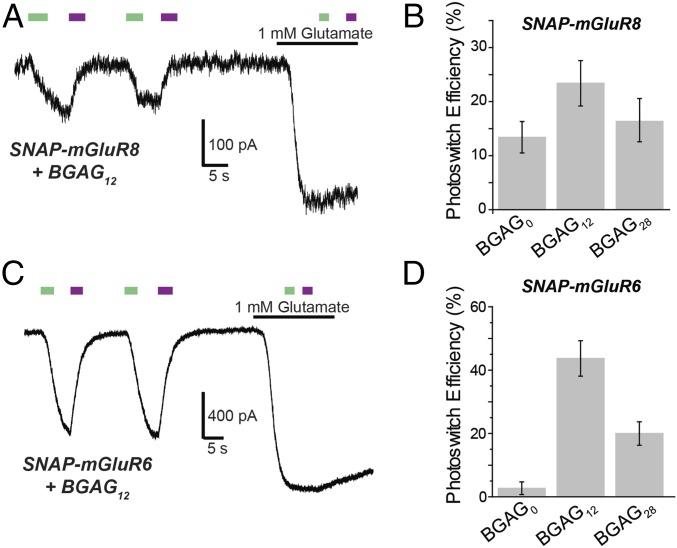
We next turned to mGluR8, which is ubiquitously expressed throughout the brain and spinal cord and is prominent as a drug target for pain but is difficult to study because of a limited repertoire of specific drugs (24). As with SNAP-mGluR7, we observed trans photoagonism of SNAP-mGluR8 that was most potent with BGAG12 (Fig. 6 A and B). SNAP-mGluR8 had only a moderate (∼20%) photoactivation relative to activation by saturating glutamate.
Fig. 6.
Extension of optical control throughout the group III subfamily: mGluR8 and mGluR6. (A) Representative trace showing trans agonism of SNAP-mGluR8 by BGAG12. (B) BGAG12 shows improved efficiency compared with BGAG0. (C) Representative trace showing robust trans agonism of SNAP-mGluR6 by BGAG12. (D) BGAG12 shows maximal photoswitch efficiency compared with BGAG0 and BGAG28. All recordings were performed in HEK 293T cells. Error bars show standard errors.
The other remaining group III mGluR that is widely expressed throughout the brain is mGluR4. SNAP-mGluR4 showed no photoactivation with either BGAG0 or BGAG12 (n = 4 cells for each variant). In contrast, mGluR6, which has an essential role in the visual cascade of ON-bipolar cells of the retina (25), was robustly photoactivated (∼50%). As with mGluR7 and 8, photoactivation was in trans and was maximal by BGAG12 (Fig. 6 C and D). Thus, three of the four group III mGluRs could be photoactivated by SNAP-BGAG, and in each case by the trans state of the photoswitch, as opposed to the cis-state activation seen in group II members mGluR2 and 3.
Multiplexed Orthogonal Optical Control.
Having established a tool chest of photoswitchable mGluRs, we next sought to develop another PORTL that would label orthogonally to SNAP to allow for two receptors to be controlled independently. We turned to the related protein tag system of CLIP. CLIP is an engineered variant of SNAP, which can be efficiently conjugated to O2-benzylcytosine–containing compounds rather than O6-benzylguanine–containing compounds (26). We designed and synthesized a family of PORTLs that would target CLIP-mGluRs, termed BCAGs, where the benzylguanine moiety is replaced by benzylcytosine (BC) (SI Appendix, Fig. S6 and Scheme S4).
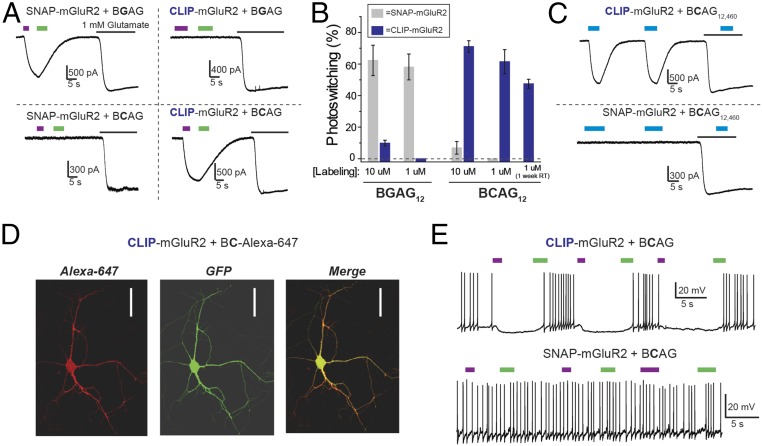
We tested BCAG12 on CLIP-mGluR2 and found robust cis photoactivation (Fig. 7A). Importantly, there was no cross-conjugation between the PORTLs targeted separately to the two protein tags at optimal PORTL concentration. Labeling with BCAG12 at 1 µM did not produce any photosensitization of SNAP-mGluR2 and, similarly, labeling with BGAG12 at 1 µM did not produce any photosensitization of CLIP-mGluR2 (Fig. 7 A and B). It is worth noting that at 10 µM some weak cross-labeling was observed between subtypes (Fig. 7B), emphasizing the importance of performing labeling controls for any new context in which these orthogonal domains are used. We also found that, like the BGAG family (14), BCAG12 is stable in aqueous buffer at physiological pH. When we diluted BCAG12 to 1 µM in labeling solution and left it at room temperature for 1 wk before applying to cells, we maintained large photoactivation of CLIP-mGluR2 (Fig. 7B).
Fig. 7.
Further engineering of SNAG-mGluR2: orthogonal targeting of CLIP tags with BCAG. (A) Representative traces showing photocontrol of SNAP- or CLIP-mGluR2 with either BGAG12 (BGAG) or BCAG12 (BCAG) show specific photoswitch conjugation. (B) Summary of BGAG and BCAG photoswitching. For all conditions, 45-min labeling periods and HEK 293T cells were used. “1 week RT” indicates that BCAG was maintained in aqueous buffer at room temperature for 1 wk before labeling, demonstrating the stability of the photoswitch. Error bars show standard errors. (C) Representative traces showing that BCAG12,460 labeling (at 1 μM) is also specific for CLIP- over SNAP-mGluR2 in HEK 293T cells. (D) Images showing expression and labeling (with BG-conjugated Alexa 647 fluorophore; red) of CLIP-mGluR2 in dissociated hippocampal neurons. GFP coexpression is shown in green. (Scale bars: 50 μM.) (E) CLIP-mGluR2 photoactivation by BCAG12 hyperpolarizes and reduces action-potential firing (Top), and BCAG labeling remains specific for CLIP over SNAP in dissociated hippocampal neurons (Bottom).
We further expanded the photoswitchable mGluR2 family by synthesizing BCAG12,460 (SI Appendix, Fig. S6 and Scheme S4), which produced robust photoactivation of CLIP-mGluR2 but not of SNAP-mGluR2 (Fig. 7C). In contrast to the intermediate linker length BCAG, BCAG0,460 showed no photoactivation of CLIP-mGluR2. We confirmed that BCAG labeling works efficiently in neuronal cultures, a key system for understanding neural signaling. CLIP-mGluR2 expressed efficiently on the surface of hippocampal neurons, where it could be visualized through conjugation of the BC-Alexa 647 fluorophore (Fig. 7D). Following conjugation with BCAG12, CLIP-mGluR2 photoactivation induced neuronal hyperpolarization that led to a reversible and repeatable decrease in action-potential firing, consistent with activation of native GIRK channels (Fig. 7E, Top). Importantly, in neurons expressing SNAP-mGluR2, BCAG12 did not produce any photosensitization (Fig. 7E, Bottom), demonstrating that photoswitch labeling retains specificity in neurons.
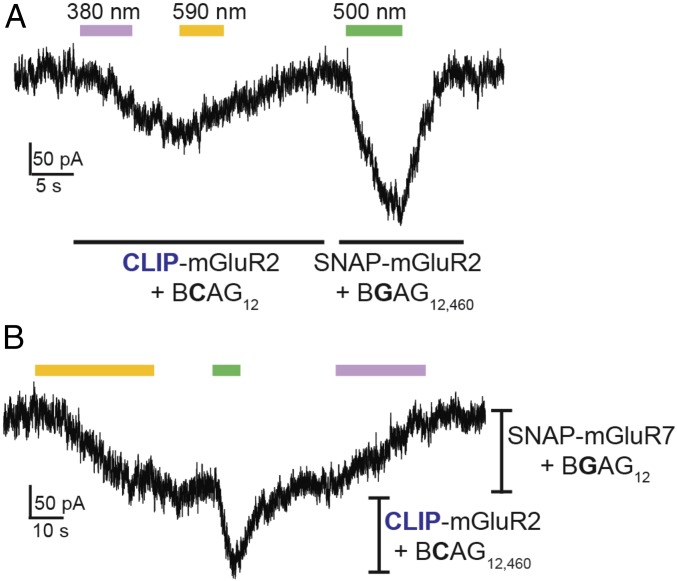
After establishing that BCAG variants allow efficient photoactivation of CLIP-mGluR2 with very similar properties to SNAG-mGluR2, we wondered whether we could combine these tools in the same preparation to independently drive two different receptor populations with light. To do this, we needed not only orthogonal strategies for photoswitch attachment but spectral differences between the photoswitches that would enable them to be separately controlled. We therefore coexpressed SNAP-mGluR2 and CLIP-mGluR2 and labeled cells with both BCAG12 and BGAG12,460 (1 µM). Photoactivation of SNAP-mGluR2 (500-nm ON and dark OFF to photoswitch BGAG12,460) was then done separately or in combination with photoactivation of CLIP-mGluR2 (380-nm ON and 590-nm OFF to photoswitch BCAG12) (Fig. 8A).
Fig. 8.
Dual photoactivation of SNAP- and CLIP-mGluRs. (A) Dual optical control of CLIP- and SNAP-mGluR2 in the same cell using BCAG12 (activated with ∼0.01 mW/mm2 380-nm light and deactivated with ∼1 mW/mm2 590-nm light) and BGAG12,460 (activated with ∼1 mW/mm2 500-nm light), respectively. (B) Sequential coactivation of SNAG-mGluR7 (with BGAG12) and CLIP-mGluR2 (with BCAG12,460); 590-nm illumination (∼1 mW/mm2) fully activates SNAG-mGluR7 followed by 500-nm illumination (∼1 mW/mm2), which activates CLIP-mGluR2 without altering SNAG-mGluR7 because it is already fully activated (i.e., in trans). Dim 380-nm illumination (∼0.01 mW/mm2) is able to turn off SNAG-mGluR7 without activation of CLIP-mGluR2. All experiments were performed in HEK 293T cells.
In an alternative logic for multiplexing, we took advantage of the difference between cis photoagonism of group II mGluRs and trans photoagonism of group III mGluRs. By combining CLIP-mGluR2 conjugated to BCAG460 with SNAP-mGluR7, we could sequentially activate SNAG-mGluR7 with 590-nm illumination followed by a flash of 500-nm illumination to activate SNAG-mGluR2 (Fig. 8B). Because 590-nm illumination fully activates SNAG-mGluR7, 500-nm illumination only produces activation of mGluR2 without altering mGluR7. Photoactivation was then terminated for SNAG-mGluR7 by illumination with dim 380-nm light (∼0.1 mW/mm2), without altering SNAG-mGluR2. This demonstrates that depending on the combination of photoswitches and the directionality of photoagonism, two different receptor populations can be photoactivated sequentially with or without simultaneous activation.
Discussion
Optical and genetic control of ligand binding and activation of GPCRs is a powerful approach that has shown great promise for enhancing our understanding of the molecular mechanisms and physiological roles of this important family of signaling proteins and for probing the function of neural circuits. In this study, we have expanded the toolset of light-controlled metabotropic glutamate receptors, obtained insight into the mechanisms of photoswitchable activation to enable rational tuning, and deployed two bioorthogonal attachment systems to permit independent optical control or different receptors. Because these tools are based on the covalent attachment of one of a diverse array of rapidly photoswitchable ligands to genetically encoded receptor variants, the receptor target and location of action are defined genetically.
PORTL vs. PTL Approaches: Mechanisms and Advantages.
In this study, we have provided insight into the photoswitch mechanism of PORTL photoagonism by synthesizing and testing a battery of compounds on a range of mGluR variants. Our observations allow for a number of distinctions between PORTL and PTL optical control to emerge. In the PORTL approach, because of the long linker between the relatively remote protein tag attachment moiety and the ligand moiety—in our case the glutamate of BGAG—the ligand is present in a relatively low local concentration (<100 μM) around the binding site. This is in contrast to small PTLs that are directly attached to the ligand-binding domain, which we find here to have a much higher effective local concentration (>1 mM), consistent with an earlier estimate of ∼10 mM in an ionotropic glutamate receptor (17). The minimal length dependence observed in the 0 to 12 PEG repeat range is consistent with previous work on tethered ligands that modeled PEG linkers as random-coil polymers, which maintain conformational mobility when the ligand is bound (27). Outside of the optimal linker length range, PORTL linkers that are too short do not support photoactivation, likely because the ligand cannot reach its binding site and, among ones that do reach, the longest lose efficacy, as predicted for lower local concentration of the ligand around the binding site. Across the mGluR members the optimal length PORTL had 12 PEG repeats (BGAG12), and this proved too short to reach from an attachment site in one subunit to the ligand-binding domain of its dimeric partner, providing for the same subunit-specific photocontrol that characterizes the much smaller PTLs. Thus, as previously proposed (14), unlike the photoswitching mechanism of PTLs, in which azobenzene isomerization to cis points the ligand directly into the binding pocket and trans isomerization removes the ligand, in PORTLs the ligand is always within equal reach of the binding site but its ability to bind and agonize is reversibly blocked by the azobenzene, as is the case in free photochromic ligands. Consistent with this interpretation, the cysteine conjugation position of LimGluRs is quite deep within the cleft of the ligand-binding domain, which may constrain the ability of the longer trans form, but not the shorter cis form, to reach the binding site.
The high local concentration of the PTL provides a tight control of ligand occupancy, an advantage for biophysical studies of receptor activation mechanisms, as shown recently in both ionotropic and metabotropic receptors (21, 28). On the other hand, the similarity in binding of the PORTL to that of free ligand may provide a better model for receptor activation in vivo. A major additional advantage of the PORTLs is that because they virtually do not react with water and are bioorthogonal, there are no competing reactions with the one that attaches them covalently to their protein tag. As a result, they can be used at far lower concentrations, well below their ability to bind significantly in the free form, and are stable for days in solution and so should be effective in reaching their protein target in vivo. Another advantage is that the protein tag can also be labeled selectively with a fluorophore, providing a view of the site of protein expression and subcellular localization and a gauge of the efficiency of PORTL conjugation. We show that PORTLs can now be simultaneously applied and separately targeted to more than one protein target using the SNAP and CLIP tags. Other systems, such as the HaloTag (29), afford additional possibilities (30). Labeling specificity when both SNAP and CLIP are used together is maintained even when concentrations are high enough to ensure a labeling efficiency of ∼90%.
We asked whether tethered photoswitches could achieve the efficacy of the native ligand. Although selective labeling in cells is very high (∼90%) it is not stoichiometric, and, moreover, maximal photoisomerization to either the trans or cis state is high but incomplete (85 to 90%), resulting in submaximal photoactivation. We show that this limit can be overcome by doubling the number of photoswitches per ligand-binding domain to raise photoswitch efficiency to near 100%.
A Group II/III mGluR Optogenetic Toolset.
We have generated a family of PORTL-operated mGluRs and expanded the family of PTL-operated ones to cover the group II and III subfamilies (Table 1). The group II mGluR subfamily consists of mGluR2 and mGluR3, which show ∼70% sequence identity and both couple to the Gi/o G-protein family (31). It has been difficult to distinguish their roles in neurological functions due to limited pharmacological tools, especially among orthosteric compounds. Further complicating the analysis of mGluR2 and mGluR3, it has recently been shown that these subunits can coassemble to produce heterodimeric receptors with unique basal activity and cooperativity (21, 22). This finding emphasizes the need for ligands that can unambiguously distinguish between subtypes. In contrast to LimGluR3, which shows very robust photoactivation with d-MAG-0 (∼70%), we show in this study that SNAP-mGluR3 is weakly agonized by BGAG (∼20%). This is consistent with pharmacological studies that have shown that many 4-functionalized glutamate compounds have a higher affinity for mGluR2 than mGluR3 (32). In addition, SNAP-mGluR3 has a more pronounced dependence on BGAG length than SNAP-mGluR2. Presumably, in the context of LimGluR3, the high local concentration of the glutamate moiety of d-MAG-0 allows for very efficient photoactivation despite the presumed weak activity of the parent 4′-substituted glutamate. Furthermore, the spontaneous dynamics of mGluR3 (16), which results in enhanced single-subunit activation in mGluR3-containing receptors (21), may contribute to the enhanced efficacy of LimGluR3 relative to LimGluR2.
Table 1.
Summary of all photoswitchable mGluR variants using either PTL or PORTL switches
| Receptor | Construct | Photoswitch (n) | Efficiency, % | Wavelength, nm | Note | 2-Photon | Source |
| mGluR2 | SNAP-mGluR2 | BGAGn (0 to 12) | ∼60 | ∼380: ON | SNAG-mGluR2 | No | (14) |
| ∼500: OFF | |||||||
| BGAGn,460 (0 to 12) | ∼50 | ∼460: ON | SNAG460-mGluR2 | Yes | (14) | ||
| Dark: OFF | |||||||
| CLIP-mGluR2 | BCAG12 | ∼60 | ∼380: ON | Orthogonal to SNAP | No | This paper | |
| ∼500: OFF | |||||||
| BCAG12,460 | ∼460: ON | Orthogonal to SNAP | Yes | This paper | |||
| Dark: OFF | |||||||
| mGluR2-L300C | d-MAG-0 | ∼50 | ∼380: ON | LimGluR2 | No | (12) | |
| ∼500: OFF | |||||||
| mGluR2-L300C-R57A | d-MAG-0 | ∼40 | ∼380: ON | LA-LimGluR2; 50-fold reduced Glu affinity | No | (20) | |
| ∼500: OFF | |||||||
| mGluR2-S302C | d-MAG-1 | ∼40 | ∼380: antagonist | LimGluR2 block | No | (12) | |
| ∼500: OFF | |||||||
| mGluR3 | SNAP-mGluR3 | BGAG0 | ∼20 | ∼380: ON | SNAG-mGluR3 | No | This paper |
| ∼500: OFF | |||||||
| mGluR3-Q306C | d-MAG-0 | ∼75 | ∼380: ON | LimGluR3 | No | (12) | |
| ∼500: OFF | |||||||
| d-MAG-0460 | ∼30 | ∼460: ON | Yes | (39) | |||
| Dark: OFF | |||||||
| mGluR3-Q306C-R64A | d-MAG-0 | ∼75 | ∼380: ON | LA-LimGluR3; >100-fold reduced Glu affinity | No | This paper | |
| ∼500: OFF | |||||||
| mGluR6 | SNAP-mGluR6 | BGAG12 | ∼50 | ∼500: ON | SNAG-mGluR6 | No | This paper |
| ∼380: OFF | |||||||
| mGluR7 | SNAP-mGluR7-N74K | BGAG12 | ∼40 | ∼500: ON | SNAG-mGluR7; subtle increase in apparent Glu affinity vs. WT (∼2-fold) | No | This paper |
| ∼380: OFF | |||||||
| mGluR8 | SNAP-mGluR8 | BGAG12 | ∼25 | ∼500: ON | SNAG-mGluR8 | No | This paper |
| ∼380: OFF |
The group III mGluR subfamily contains mGluR4, mGluR6, mGluR7, and mGluR8, which are thought to play distinct roles in a variety of neural functions, with especially prominent synaptic modulatory roles in sensory processing, Parkinson’s disease, and pain pathophysiology (24, 31). For this reason, significant effort has been put into the development of specific drugs for these receptors. Unfortunately, a relatively limited repertoire of specific compounds has been developed for group III mGluRs, making it difficult to proceed clinically as well as to precisely study these receptors in their native contexts. mGluR7 is particularly interesting, due to its uniquely low apparent glutamate affinity (∼1 mM versus ∼1 to 10 μM for other mGluRs), which has classically been interpreted as consistent with its synaptic localization compared with the perisynaptic localization of other mGluR subtypes (33). This extreme low glutamate affinity has made it difficult to study mGluR7. Existing synthetic orthosteric agonists remain low-affinity ligands for mGluR7 and are not able to distinguish between group III subtypes. Furthermore, because of the high concentrations required to activate mGluR7, pharmacological experiments are especially sensitive to off-target effects and are limited in the temporal parameters of drug application and removal.
BGAG showed trans photoagonism of mGluR6, 7, and 8 but no effect on mGluR4. The conserved trans agonism for all subtypes and all BGAG lengths indicates that group III mGluRs have a fundamental preference for trans over cis azobenzene-glutamate. This is consistent with recent pharmacological studies showing differences between group II and III mGluRs in their sensitivity to 4-functionalized glutamate analogs (32). We interpret this result as an indication that the precise shape of the linker-azobenzene moiety determines the ability to produce either trans or cis agonism in group II versus group III mGluRs. The steeper length dependence of BGAG photoagonism of group III mGluRs contrasts with what was observed with group II mGluRs. An attractive interpretation is that due to structural differences, either in the binding pocket or the N terminus of the ligand-binding domain, a longer BGAG is required for sufficient reach to the binding site. As BGAG length extends beyond 12 PEG repeats, presumably the local concentration drops to a point that photoswitch efficiency is impaired, as observed with SNAP-mGluR2 and BGAG28. The photoagonism of mGluR6 was most notable because it was the most efficient BGAG/group III mGluR combination and approached the efficacy of SNAP-mGluR2. Optical control of mGluR6 opens the door to manipulation of one of the central players in the visual transduction cascade of ON bipolar cells of the retina.
When two cis agonists were used, this allowed for sequential independent activation, whereas combination of a cis agonist with a trans agonist allowed for sequential, combined activation. This approach opens the door for precise probing of the contribution of individual receptors in intact circuits, including at synapses where multiple receptors are expressed together or apart pre- and postsynaptically or in nearby glia.
Together, this toolset should provide a means of probing a wide range of biological phenomena that are mediated by G-protein signaling, generally, and mGluR signaling, specifically. The relative ease of application should also facilitate the use of these tools. SNAP-mGluR variants can be expressed robustly using standard gene delivery techniques, and photoswitch application is straightforward in culture and should be efficient in vivo using targeted injection, as previously shown for MAG-based compounds (19). Controls that omit receptor expression or photoswitch application serve as helpful confirmation of the specificity of the measured optical effect. This approach adds significantly to other methods for manipulating GPCRs, as summarized in SI Appendix, Table S1.
Whereas optogenetic control with exogenous signaling proteins has yielded major scientific advances, optical manipulation of native receptors holds great promise for understanding their specific biological roles. For example, nearly all of the mGluR subtypes are coexpressed at hippocampal synapses, including in presynaptic, postsynaptic, and glial compartments (33). The limited set of subtype-specific pharmacological agents and inability to selectively aim them at a specific compartment represent a great barrier to understanding the individual role of receptor and compartment in long-term plasticity. These challenges can be overcome by the ability to target expression of photoswitchable mGluRs to a specific cell and point light at a specific synaptic compartment, thereby controlling ligand occupancy with millisecond precision. At the circuit and behavioral level, photoswitchable mGluRs should enable identification of the brain regions and cell types involved in the mechanisms of mGluR2/3-targeting ligands for the treatment of psychiatric disorders (34).
Ultimately, the overall design principles established in this study will facilitate the transfer of the PORTL approach to many other signaling systems. In the future it will be key to continue to spectrally tune switches to improve the ability to orthogonally manipulate multiple receptors. Major advances in photoswitch red shifting have recently been published, which should lead to the expansion of the BGAG repertoire (35–37). Furthermore, it will be important to develop a richer pharmacological repertoire of PORTLs including antagonists, allosteric modulators, and biased ligands and to use either genetic manipulation (38) or chemical targeting (39) to adapt this approach to control native receptors.
Materials and Methods
Chemical Synthesis.
Chemical synthesis was performed as previously described (14). See SI Appendix, Materials and Methods for more details.
Molecular Cloning and Gene Expression.
Experiments were performed using rat (mGluR2, mGluR3, mGluR6, mGluR7) or human (mGluR4, mGluR8) isoforms of mGluRs. HEK 293T cell transfection of mGluR constructs and GIRK reporters was performed as previously described using Lipofectamine 2000 (12).
Whole-Cell Patch Clamp.
HEK 293T cell electrophysiology and photoswitch labeling were performed as previously described (14). Briefly, recordings were performed in high-potassium solution containing 120 mM KCl, 25 mM NaCl, 1 mM MgCl2, 2 mM CaCl2, and 10 mM Hepes (pH 7.4) using glass pipettes (3 to 6 MΩ) filled with solution containing 140 mM KCl, 10 mM Hepes, 5 mM EGTA, and 3 mM MgCl2 (pH 7.4). Cells were patch-clamped to −60 using an Axopatch 200B (Molecular Devices) amplifier in whole-cell mode. Drugs were applied using a gravity-driven perfusion system, and illumination was controlled using a DG-4 system (Sutter) in combination with excitation filters.
Dissociated hippocampal neuron experiments were performed as previously described (14).
Data were analyzed with Clampfit (Molecular Devices) and Prism (GraphPad).
Fluorescence Imaging.
Cells were imaged using a scanning confocal Zeiss LSM 780 microscope. All dyes were purchased from New England Biolabs and applied at 1 to 2 μM for 45 min in standard extracellular buffers. Fluorescence intensities were calculated from small clusters of cells using Zen software (Zeiss).
Approval.
The experiments, use of human cell lines, and animal use were approved by the University of California Berkeley Biosafety Office and Animal Care and Use Committee.
Supplementary Material
Acknowledgments
We thank P. Donthamsetti and M. Berry for helpful discussions, and Zhu Fu for technical assistance. We kindly thank Prof. K. Johnsson for providing BG- and BC-containing compounds. The work was supported by National Institutes of Health Nanomedicine Center for the Optical Control of Biological Function (2PN2EY018241) and Instrumentation Award S10 RR028971, and National Science Foundation Major Research Instrumentation Award 1041078 and EAGER Award IOS-1451027 (to E.Y.I.). We are grateful to the Center for Integrated Protein Science Munich (J.B., P.L., D.K., and D.T.) and SFB 1032 (P.L. and D.T.). J.B. acknowledges support from the “EPFL Fellows” Fellowship Programme cofunded by Marie Skłodowska-Curie, Horizon 2020 Grant Agreement 665667. D.T. was supported by an Advanced Grant from the European Research Council (268795).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1619652114/-/DCSupplemental.
References
- 1.Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 2.Katritch V, Cherezov V, Stevens RC. Structure-function of the G protein-coupled receptor superfamily. Annu Rev Pharmacol Toxicol. 2013;53:531–556. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roth BL. DREADDs for neuroscientists. Neuron. 2016;89:683–694. doi: 10.1016/j.neuron.2016.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X, et al. Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc Natl Acad Sci USA. 2005;102:17816–17821. doi: 10.1073/pnas.0509030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masseck OA, et al. Vertebrate cone opsins enable sustained and highly sensitive rapid control of Gi/o signaling in anxiety circuitry. Neuron. 2014;81:1263–1273. doi: 10.1016/j.neuron.2014.01.041. [DOI] [PubMed] [Google Scholar]
- 6.Qiu X, et al. Induction of photosensitivity by heterologous expression of melanopsin. Nature. 2005;433:745–749. doi: 10.1038/nature03345. [DOI] [PubMed] [Google Scholar]
- 7.Melyan Z, Tarttelin EE, Bellingham J, Lucas RJ, Hankins MW. Addition of human melanopsin renders mammalian cells photoresponsive. Nature. 2005;433:741–745. doi: 10.1038/nature03344. [DOI] [PubMed] [Google Scholar]
- 8.Kim JM, et al. Light-driven activation of beta 2-adrenergic receptor signaling by a chimeric rhodopsin containing the beta 2-adrenergic receptor cytoplasmic loops. Biochemistry. 2005;44:2284–2292. doi: 10.1021/bi048328i. [DOI] [PubMed] [Google Scholar]
- 9.Spoida K, Masseck OA, Deneris ES, Herlitze S. Gq/5-HT2c receptor signals activate a local GABAergic inhibitory feedback circuit to modulate serotonergic firing and anxiety in mice. Proc Natl Acad Sci USA. 2014;111:6479–6484. doi: 10.1073/pnas.1321576111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Airan RD, Thompson KR, Fenno LE, Bernstein H, Deisseroth K. Temporally precise in vivo control of intracellular signalling. Nature. 2009;458:1025–1029. doi: 10.1038/nature07926. [DOI] [PubMed] [Google Scholar]
- 11.Oh E, Maejima T, Liu C, Deneris E, Herlitze S. Substitution of 5-HT1A receptor signaling by a light-activated G protein-coupled receptor. J Biol Chem. 2010;285:30825–30836. doi: 10.1074/jbc.M110.147298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levitz J, et al. Optical control of metabotropic glutamate receptors. Nat Neurosci. 2013;16:507–516. doi: 10.1038/nn.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reiner A, Levitz J, Isacoff EY. Controlling ionotropic and metabotropic glutamate receptors with light: Principles and potential. Curr Opin Pharmacol. 2015;20:135–143. doi: 10.1016/j.coph.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Broichhagen J, et al. Orthogonal optical control of a G protein-coupled receptor with a SNAP-tethered photochromic ligand. ACS Cent Sci. 2015;1:383–393. doi: 10.1021/acscentsci.5b00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.French AC, Thompson AL, Davis BG. High-purity discrete PEG-oligomer crystals allow structural insight. Angew Chem Int Ed Engl. 2009;48:1248–1252. doi: 10.1002/anie.200804623. [DOI] [PubMed] [Google Scholar]
- 16.Vafabakhsh R, Levitz J, Isacoff EY. Conformational dynamics of a class C G-protein-coupled receptor. Nature. 2015;524:497–501. doi: 10.1038/nature14679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorostiza P, et al. Mechanisms of photoswitch conjugation and light activation of an ionotropic glutamate receptor. Proc Natl Acad Sci USA. 2007;104:10865–10870. doi: 10.1073/pnas.0701274104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Numano R, et al. Nanosculpting reversed wavelength sensitivity into a photoswitchable iGluR. Proc Natl Acad Sci USA. 2009;106:6814–6819. doi: 10.1073/pnas.0811899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levitz J, Popescu AT, Reiner A, Isacoff EY. A toolkit for orthogonal and in vivo optical manipulation of ionotropic glutamate receptors. Front Mol Neurosci. 2016;9:2. doi: 10.3389/fnmol.2016.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malherbe P, et al. Identification of essential residues involved in the glutamate binding pocket of the group II metabotropic glutamate receptor. Mol Pharmacol. 2001;60:944–954. doi: 10.1124/mol.60.5.944. [DOI] [PubMed] [Google Scholar]
- 21.Levitz J, et al. Mechanism of assembly and cooperativity of homomeric and heteromeric metabotropic glutamate receptors. Neuron. 2016;92:143–159. doi: 10.1016/j.neuron.2016.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doumazane E, et al. A new approach to analyze cell surface protein complexes reveals specific heterodimeric metabotropic glutamate receptors. FASEB J. 2011;25:66–77. doi: 10.1096/fj.10-163147. [DOI] [PubMed] [Google Scholar]
- 23.Rosemond E, et al. Molecular basis for the differential agonist affinities of group III metabotropic glutamate receptors. Mol Pharmacol. 2004;66:834–842. doi: 10.1124/mol.104.002956. [DOI] [PubMed] [Google Scholar]
- 24.Acher F, Goudet C. Therapeutic potential of group III metabotropic glutamate receptor ligands in pain. Curr Opin Pharmacol. 2015;20:64–72. doi: 10.1016/j.coph.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 25.Nakajima Y, et al. Molecular characterization of a novel retinal metabotropic glutamate receptor mGluR6 with a high agonist selectivity for L-2-amino-4-phosphonobutyrate. J Biol Chem. 1993;268:11868–11873. [PubMed] [Google Scholar]
- 26.Gautier A, et al. An engineered protein tag for multiprotein labeling in living cells. Chem Biol. 2008;15:128–136. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 27.Krishnamurthy VM, Semetey V, Bracher PJ, Shen N, Whitesides GM. Dependence of effective molarity on linker length for an intramolecular protein-ligand system. J Am Chem Soc. 2007;129:1312–1320. doi: 10.1021/ja066780e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reiner A, Isacoff EY. Tethered ligands reveal glutamate receptor desensitization depends on subunit occupancy. Nat Chem Biol. 2014;10:273–280. doi: 10.1038/nchembio.1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Los GV, et al. HaloTag: A novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 30.Xue L, Karpenko IA, Hiblot J, Johnsson K. Imaging and manipulating proteins in live cells through covalent labeling. Nat Chem Biol. 2015;11:917–923. doi: 10.1038/nchembio.1959. [DOI] [PubMed] [Google Scholar]
- 31.Niswender CM, Conn PJ. Metabotropic glutamate receptors: Physiology, pharmacology, and disease. Annu Rev Pharmacol Toxicol. 2010;50:295–322. doi: 10.1146/annurev.pharmtox.011008.145533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huynh TH, et al. New 4-functionalized glutamate analogues are selective agonists at metabotropic glutamate receptor subtype 2 or selective agonists at metabotropic glutamate receptor group III. J Med Chem. 2016;59:914–924. doi: 10.1021/acs.jmedchem.5b01333. [DOI] [PubMed] [Google Scholar]
- 33.Shigemoto R, et al. Differential presynaptic localization of metabotropic glutamate receptor subtypes in the rat hippocampus. J Neurosci. 1997;17:7503–7522. doi: 10.1523/JNEUROSCI.17-19-07503.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker AG, Conn PJ. Group I and group II metabotropic glutamate receptor allosteric modulators as novel potential antipsychotics. Curr Opin Pharmacol. 2015;20:40–45. doi: 10.1016/j.coph.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Konrad DB, Frank JA, Trauner D. Synthesis of redshifted azobenzene photoswitches by late-stage functionalization. Chemistry. 2016;22:4364–4368. doi: 10.1002/chem.201505061. [DOI] [PubMed] [Google Scholar]
- 36.Lerch MM, Hansen MJ, Velema WA, Szymanski W, Feringa BL. Orthogonal photoswitching in a multifunctional molecular system. Nat Commun. 2016;7:12054. doi: 10.1038/ncomms12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong M, Babalhavaeji A, Samanta S, Beharry AA, Woolley GA. Red-shifting azobenzene photoswitches for in vivo use. Acc Chem Res. 2015;48:2662–2670. doi: 10.1021/acs.accounts.5b00270. [DOI] [PubMed] [Google Scholar]
- 38.Lin WC, et al. A comprehensive optogenetic pharmacology toolkit for in vivo control of GABA(A) receptors and synaptic inhibition. Neuron. 2015;88:879–891. doi: 10.1016/j.neuron.2015.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Izquierdo-Serra M, et al. Optical control of endogenous receptors and cellular excitability using targeted covalent photoswitches. Nat Commun. 2016;7:12221. doi: 10.1038/ncomms12221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.