Significance
A century ago, it was noted that damage to the lateral hypothalamic (LH) brain area caused people to be motionless most of the time. Therefore, it is thought that this brain area emits essential signals for promoting physical activity. However, it remained a mystery what these signals are. Using newly developed genetic and deep-brain recording methods, we found that, unexpectedly, a critical signal for movement comes from a subset of LH cells that are molecularly distinct from neurons previously implicated in locomotion control. These movement-promoting cells were switched on by the local peptide orexin, a key signal of stress and hunger. These findings shed new light on deep-brain signals that maintain healthy levels of physical activity.
Keywords: hypothalamus, GAD65, orexin, hypocretin, locomotion, stress
Abstract
Damage to the lateral hypothalamus (LH) causes profound physical inactivity in mammals. Several molecularly distinct types of LH neurons have been identified, including orexin cells and glutamic acid decarboxylase 65 (GAD65) cells, but their interplay in orchestrating physical activity is not fully understood. Here, using optogenetic circuit analysis and cell type-specific deep-brain recordings in behaving mice, we show that orexin cell activation rapidly recruits GAD65LH neurons. We demonstrate that internally initiated GAD65LH cell bursts precede and accompany spontaneous running bouts, that selective chemogenetic silencing of natural GAD65LH cell activity depresses voluntary locomotion, and that GAD65LH cell overactivation leads to hyperlocomotion. These results thus identify a molecularly distinct, orexin-activated LH submodule that governs physical activity in mice.
The lateral hypothalamus (LH) is thought to provide an essential drive for diverse vital behaviors, including locomotion. Peri-LH lesions in mammals disrupt context-appropriate physical activity, and are associated with the human disorder encephalitis lethargica, which has been noted to make humans “sit motionless…all day” (1–6). The LH is not a homogeneous entity, but contains many molecularly distinct classes of neurons that are thought to have different physiological roles (7). However, the interrelations of distinct LH cell classes in computing and driving context-appropriate physical activity are not fully understood.
For example, LH neurons expressing orexins/hypocretins (8, 9) become activated in diverse stressful contexts, including acute auditory stimulation, hypoglycemia, hypercapnia, and physical capture (3, 9–13). OrexinLH cell activity may thus represent an important input variable for computing context-appropriate locomotor outputs (3, 14), and orexin peptides have been proposed to increase arousal and locomotion by actions on extrahypothalamic projections to areas such as the locus coeruleus (15, 16). However, hypoactivity phenotypes caused by orexinLH cell deletion are milder than the hypoactivity phenotypes caused by broader LH lesions (1–3, 5, 12, 13, 17–19), whereas melanin-concentrating hormone LH (MCHLH) cell deletion causes hyperactivity, implying that MCHLH cells suppress locomotion (20). These findings suggest that additional drivers of physical activity may exist in the LH.
It was recently found that LH neurons expressing glutamic acid decarboxylase 65 (GAD65) are distinct from orexin and MCH neurons (21). We hypothesized that these cells may be a source of natural LH signals underlying normal levels of physical activity. Here, we investigate this hypothesis by using a combination of cell type-specific manipulation and recording techniques. We find that GAD65LH cells operate as a stress- and orexin-activated LH module whose physiological activity is essential for normal locomotion, and whose hyperactivity causes hyperlocomotion.
Results
Orexin and Stress Rapidly Recruit GAD65LH Neurons.
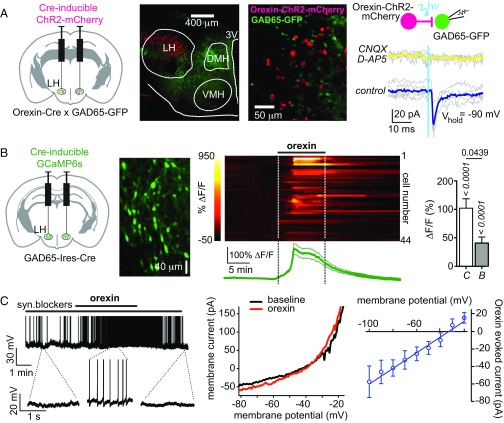
To examine whether orexin cells communicate with GAD65LH cells, we targeted the light-activated excitatory actuator channelrhodpsin-2 (ChR2) to orexin cells, enabling selective optical activation of orexin-ChR2 cells (22). We then examined resulting optically induced postsynaptic responses in GAD65LH neurons (Fig. 1A; group data and/or statistics for findings illustrated as figures are described in the figure legends). We found that GAD65LH cells received time-locked excitatory glutamatergic inputs when orexinLH cells were stimulated (Fig. 1A). To determine if orexin peptide signaling drives the GAD65LH network, we also investigated the effect of applying exogenous orexin peptide. We used confocal network imaging of a Cre-dependent calcium indicator, GCaMP6s (23), expressed in LH slices from GAD65-Ires-Cre mice (Fig. 1B). Orexin-A peptide robustly excited the GAD65LH network, and this excitation persisted (but was slightly reduced) in the presence of a mix of synaptic blockers (Fig. 1B). In parallel, electrophysiological recordings showed that orexin excites individual GAD65LH cells by activating a current with a reversal potential consistent with a nonselective cationic conductance (Fig. 1C). Overall, >98% of GAD65LH cells (64 of 65 cells) were activated by orexin peptide in vitro.
Fig. 1.
Evidence for orexinLH → GAD65LH excitatory circuit. (A) Optogenetic interrogation of orexinLH → GAD65LH signaling. (Left) Targeting strategy. (Center, Left and Right) Localization of orexin-ChR2-mCherry and GAD65-GFP expression (representative example of five brains). (Right) Orexin cell photostimulation (vertical blue line) evokes a CNQX/D-AP5–sensitive (i.e., glutamate receptor-mediated) inward current in a GAD65LH cell. Gray lines are individual trials, and colored lines are trial averages [representative example of five cells; ≈80% cells (n = 5/6 cells) were connected]. (B) Effects of orexin peptide on GAD65LH network activity. Scheme for targeting GCaMP6s (Left), example of GCaMP6s expression in GAD65LH cells (representative example of five brains) (Center Left), GCaMP6s response of GAD65LH cells to 300 nM orexin-A with corresponding mean ± SEM plot (n = 44) (Center Right), and data summary without (C) and with (B) synaptic blockers (n = 44 and n = 13 cells, respectively, mean ± SEM responses of cells during 2–20 min after orexin-A infusion) (Right). P values (italics) are from sign tests of whether the response within each group is different from zero, and P value (regular font) is from a two-tailed Mann–Whitney test comparing response amplitude between groups. (C) Effects of orexin peptide on GAD65LH cell electrical activity. (Left) Membrane potential effect of 300 nM orexin-A on a GAD65LH cell (representative example of six cells). syn., synaptic. (Center) Example of whole-cell current–voltage relation showing effect of orexin-A (representative example of five cells). (Right) Net orexin-activated current (mean and SEM of n = 5 cells).
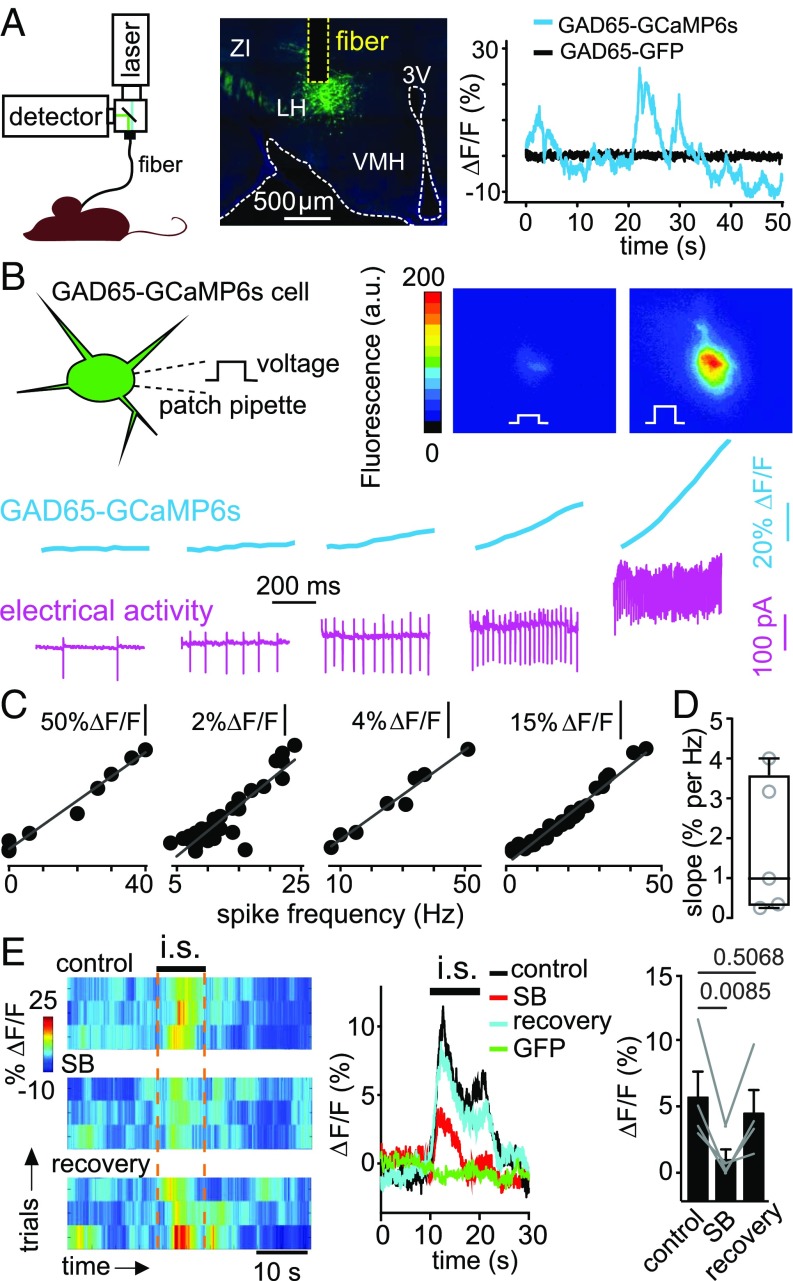
If orexin cells also activate GAD65LH cells in vivo, then stress stimuli that elicit orexinLH cell activity should also activate GAD65LH cells in vivo. Acute immobilization stress rapidly and robustly activates orexinLH cells in awake mice (13). We therefore used this paradigm, combined with deep-brain fiber photometry recording of GAD65LH cell activity and pharmacological antagonism of orexin receptors, to determine if stress and orexin drive GAD65LH cell activity in vivo. We expressed Cre-dependent GCaMP6s, or control protein EGFP, in the LH of GAD65-Ires-Cre mice, and recorded fluorescence intensity using a fiberoptic probe implanted in the LH (Fig. 2A). We observed bursts of activity in GAD65LH-GCaMP6s cells, but not in GAD65LH-EGFP cells, of freely moving mice (Fig. 2A). These bursts were similar (≈5–30% ΔF/F) to the bursts recorded using photometry in other deep networks, and are thought to represent network firing bursts (24–26). Using simultaneous electrical recordings and GCaMP6s imaging from the same GAD65LH-GCaMP6s cells in vitro, we confirmed that GCaMP6s linearly reports spike firing rate (Fig. 2 B–D), with a mean slope (≈1% ΔF/F per hertz; Fig. 2D) similar to the mean slope observed in other LH neurons (12, 13). Upon acute immobilization, we observed rapid activation of GAD65LH-GCaMP6s cells, but not control GAD65LH-GFP cells (Fig. 2E; peak amplitude of stress-induced GAD65LH-GCaMP6s activity transient was 12.3 ± 2.7%; P = 0.0174 by one-sample t test: t, df = 4.782, 3; n = 4 mice). The competitive orexin receptor antagonist SB-334867 (Materials and Methods) reversibly reduced this stress-induced activation of GAD65LH cells (Fig. 2E).
Fig. 2.
Stress-induced, orexin-dependent control of GAD65LH cells in vivo. (A, Left) Schematic of fiber photometry recording. (A, Center) GCaMP6s expression in GAD65LH cells, and localization of recording site and optical fiber (representative example of five brains). (A, Right) Example fluorescence trace during free cage exploration for mice expressing GCaMP6s (blue) or EGFP (black) in GAD65LH neurons (typical examples of n = 5 and n = 5 mice, respectively). (B) Simultaneous measurement of membrane electrical activity and calcium level in GAD65LH-GCaMP6s cells. (Top) Schematic of recording and examples of two video frames from a cell injected with depolarizing current steps. (Bottom) Example of simultaneous cell-attached and fluorescence recording from a GAD65LH cell (typical example of n = 5 cells). (C) Examples of quantification of data in B, showing the linear relationship between calcium level and spike frequency in four different cells. (D) Group plot of slopes (taken from linear regression fits) of relations between spike frequency and calcium level (n = 5 cells). (E, Left) GAD65LH-GCaMP6s signals aligned to immobilization stress (i.s.). Heat maps of individual trials from one mouse, in the presence and absence of orexin receptor antagonist SB-334867 (SB; 30 mg/kg i.p.; Materials and Methods), representative example of n = 4 mice. (E, Center) Averaged data for bouts such as those bouts shown on the left. Each trace is the average response of n = 4 mice (each n is a mean of four trials such as those trials shown on the left). The GFP trace shows responses of GAD65-GFP mice (negative controls). (E, Right) Mean calcium signals during i.s. (10–20 s) per mouse (each point represents one mouse, n = 4 mice). One-way repeated measures ANOVA: F(2,6) = 11.61; P = 0.0087, P values on the plot are from Tukey’s posttests.
Collectively, these data identify the orexin system as a physiologically relevant upstream activator of GAD65LH neurons.
GAD65LH Cell Activity Drives (and Correlates with) Voluntary Locomotion.
We next tested whether the GAD65LH module could act as an LH effector of physical activity. To probe causal links between GAD65LH cell activity and locomotion, we explored locomotor effects of selective silencing or activation of GAD65LH cell activity in freely moving mice.
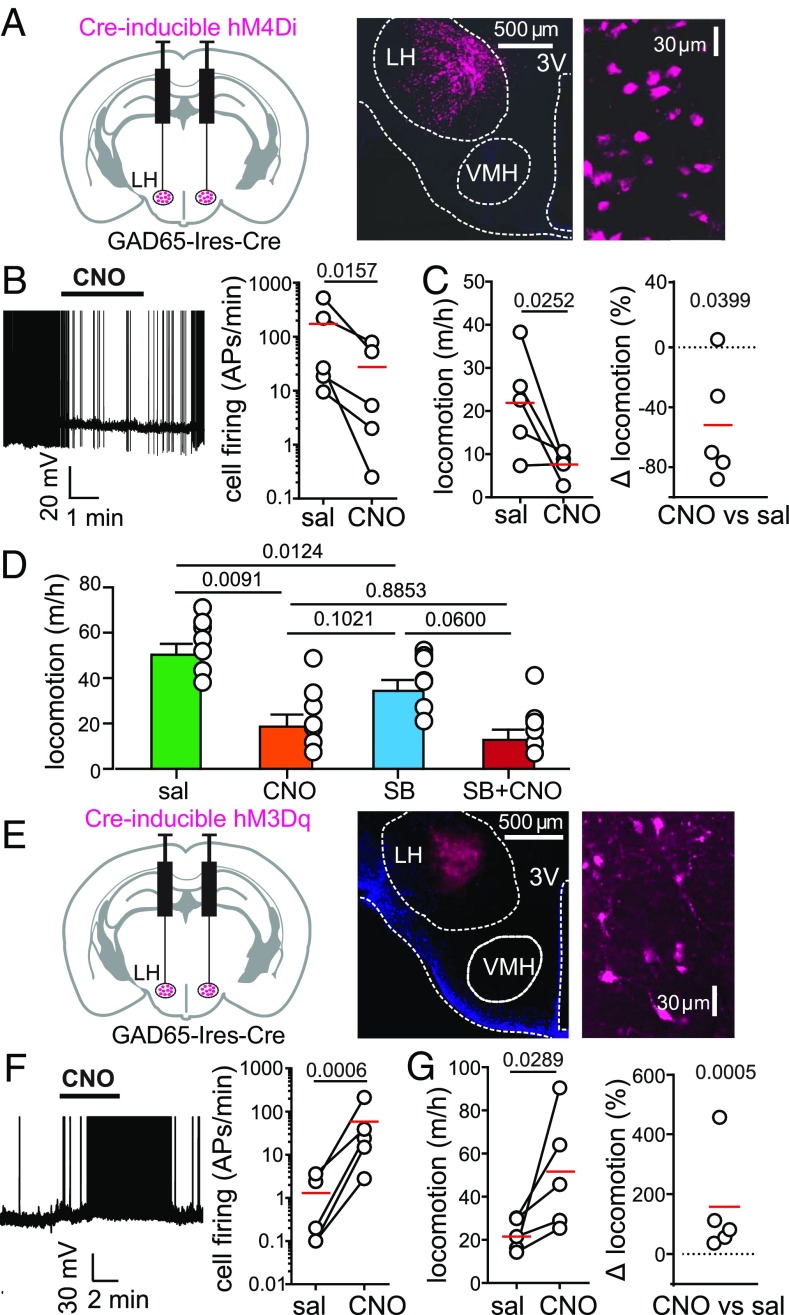
First, we targeted the neuroinhibitory clozapine-N-oxide (CNO) receptor hM4Di (27) to the LH of GAD65-Ires-Cre transgenic mice (Fig. 3A). This procedure enabled GAD65LH cell inhibition by CNO (Fig. 3B). Inhibition of GAD65LH-hM4Di cells in freely behaving mice substantially reduced their voluntary locomotion (by an average of ≈50%, as shown in Fig. 3 C and D; no such effect was seen in control GAD65LH-ChR2 mice, in which the CNO-induced change in locomotion was 4.6 ± 8.2%, n = 5, P = 0.6016 by one-sample t test: t, df = 0.0566, 4). Interestingly, orexin receptor blockade with SB-334867 reduced voluntary locomotion under control conditions, but not when GAD65LH-hM4Di cells were inactivated with CNO (Fig. 3D and Discussion).
Fig. 3.
Effect of natural and evoked GAD65LH cell activity on locomotion. (A, Left) Targeting scheme for hM4Di-mCherry. (A, Center) Localization of hM4Di-mCherry. (A, Right) GAD65LH-hM4Di-mCherry cells at high zoom (representative example of five brains). (B, Left) Effect of CNO on GAD65LH-hM4Di cell firing. Spikes are truncated at 0 mV (representative example of n = 5 cells). (B, Right) Group data, raw values, and means (red); the on-plot P value is from a one-tailed, ratio-paired t test (t, df = 3.251, 4) (n = 5 cells). APs, action potentials. (C) Effect of CNO on locomotion of GAD65LH-hM4Di mice. Raw data and means are shown in red (Left), and the same data are shown as % change (Right). On-plot P values are from a one-tailed paired t test (t, df = 2.767, 4; Left) and from a two-tailed one-sample t test (t, df = 3.001, 4; Right) (n = 5 mice). Corresponding negative control data are described in Results. (D) Effects of SB-334867 (30 mg/kg i.p.; Materials and Methods) on locomotion of GAD65LH-hM4Di mice in the absence and presence of CNO. Repeated measures one-way ANOVA with Geisser–Greenhouse correction [F(1.977,11.86) = 16.15; P = 0.0004, on-plot values are P values from Tukey’s multiple comparison tests]. (E, Left) Targeting scheme for hM3Dq-mCherry. (E, Center) Localization of hM3Dq-mCherry. (E, Right) GAD65LH-hM3Dq-mCherry cells at high zoom (representative example of five brains). (F, Left) Effect of CNO on GAD65LH-hM3Dq cell firing. Spikes are truncated at 0 mV (representative example of n = 5 cells). (F, Right) Group data, raw values, and means (red); on-plot P value is from a one-tailed ratio-paired t test (t, df = 8.043, 4; n = 5 cells). (G) Effect of CNO on locomotion of GAD65LH-hM3Dq mice. Raw data and means are shown in red (Left), and the same data are shown as % change (Right). On-plot P values are from a one-tailed paired t test (t, df = 2.638, 4; Left) and from a two-tailed one-sample t test (t, df = 10.31, 4; Right) (n = 5 mice). Corresponding negative control data are described in Results.
Next, we targeted the neuroexcitatory CNO receptor hM3Dq (27) to the LH of GAD65-Ires-Cre transgenic mice (Fig. 3 E and F). Stimulation of GAD65LH-hM3Dq cells with CNO in freely-behaving mice substantially increased their voluntary locomotion (by ≈150%; Fig. 3G; no such effect was found in control WT mice injected with the same viral vector, in which the CNO-induced change in locomotion was 49 ± 48.7%, n = 5, P = 0.3684 by one-sample t test; t, df = 1.013, 4). Collectively, these data demonstrate that the activity of GAD65LH cells is necessary for maintaining a normal level of locomotion (Fig. 3 C and D), and sufficient for stimulating locomotion (Fig. 3G).
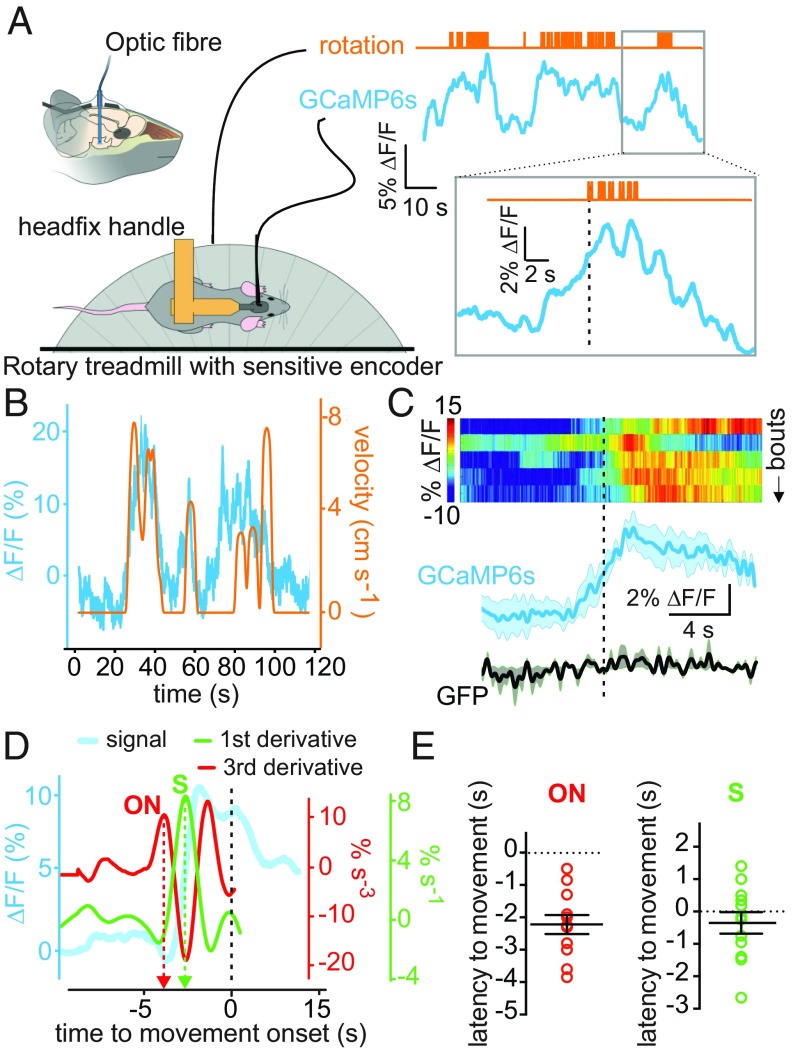
Finally, we reasoned that if GAD65LH cells naturally promote locomotion, then their activity should be associated with spontaneous/voluntary locomotion. To test this hypothesis, we measured GAD65LH-GCaMP6s network activity using fiber photometry, and quantified locomotion using a sensitive rotary treadmill, which could resolve distances traveled as low as 1.8 mm (Fig. 4A and Materials and Methods). We found that spontaneously initiated locomotion bouts (defined as locomotion with a speed of >0.17 cm⋅s−1; Materials and Methods) were associated with increased natural GAD65LH cell activity (Fig. 4 B and C). Locomotion-associated GAD65LH cell activity appeared to increase before movement was detected, and persisted throughout the locomotion bouts (Fig. 4 A–C). To estimate the temporal relationship between the natural GAD65LH cell signals and locomotion initiation, we defined signal onset time using a third derivative method (28), and also defined the steepest point of GAD65LH signal rise as the peak in the first derivative (Fig. 4D). The steepest rise of the perilocomotion GAD65LH signal occurred approximately at movement onset, whereas the onset of GAD65LH signal rise occurred ≈2 s before movement onset (Fig. 4E and Discussion).
Fig. 4.
Natural GAD65LH cell dynamics underlying voluntary movement. (A) Scheme of experimental setup, with example raw data illustrating temporal alignment of locomotion initiation (upward deflections on the orange trace) and GAD65LH signal (typical example of n = 4 mice). (B) Example of rotary encoder pulses converted to locomotion velocity (orange), illustrating coquantification of natural GAD65LH cell dynamics (blue) and locomotion (typical example of n = 4 mice). (C) Peri-event plots aligned to onset of locomotion bouts [dashed black line indicates the threshold locomotion criterion (0.17 cm·s−1) for bout onset]. (Top) Heat maps of individual trials from one mouse. (Bottom) Group data for bout averages in GAD65LH-GCaMP6s and GAD65LH-GFP (mean ± SEM). Statistics for data binned in 2-s bins: one-way repeated measures ANOVAs for control GFP group [n = 3 mice; F(9,18) = 0.8308, P = 0.5974] and for GCaMP6s group [n = 4 mice; F(9,27) = 4.630, P = 0.0009]. (D) Illustration of the third and first derivative methods used to measure the timing of GAD65LH-GCaMP6s signal onset (ON) and steepest rise (S) relative to locomotion onset (dashed black line, defined as in B) (typical example of n = 4 mice). (E) Quantification of latencies of GAD65LH-GCaMP6s signal onsets and steepest rises to locomotion initiation. Signal onset occurred at −2.22 ± 0.29 s (P < 0.0001 by two-tailed one-sample t test; t, df = 7.572, 11). The steepest rise occurred at −0.35 ± 0.22 s (P = 0.3139 by two-tailed one-sample t test; t, df = 1.055, 11).
Collectively, these data provide correlative (Fig. 4) and causal (Fig. 3) evidence that GAD65LH cell activity drives locomotion in mice.
Discussion
Despite the long-recognized importance of the LH for maintaining normal physical activity in mammals, including humans (1–6, 29, 30), the functional relations of molecularly distinct LH subnetworks remained poorly understood. Our study identifies several properties of the GAD65LH subnetwork that reveal a previously overlooked interplay between molecularly distinct LH signals relevant for locomotion control. First, GAD65LH cells are activated by orexin neurons, and are rapidly recruited by stress in vivo in an orexin receptor-dependent manner. Second, natural GAD65LH cell activity is necessary for maintaining normal levels of locomotion, whereas experimentally induced GAD65LH cell activity is sufficient to promote locomotion. Third, internally initiated GAD65LH network bursts are associated with initiation and maintenance of voluntary movement. When GAD65LH cells were inactivated, reducing orexin receptor activity no longer affected locomotion (Fig. 3D), consistent with the idea that GAD65LH cells act as mediators of locomotion driven by natural orexin signaling. These multiple lines of evidence demonstrate a critical role for GAD65LH cells in driving normal locomotion, and in acting as effectors of the orexin system.
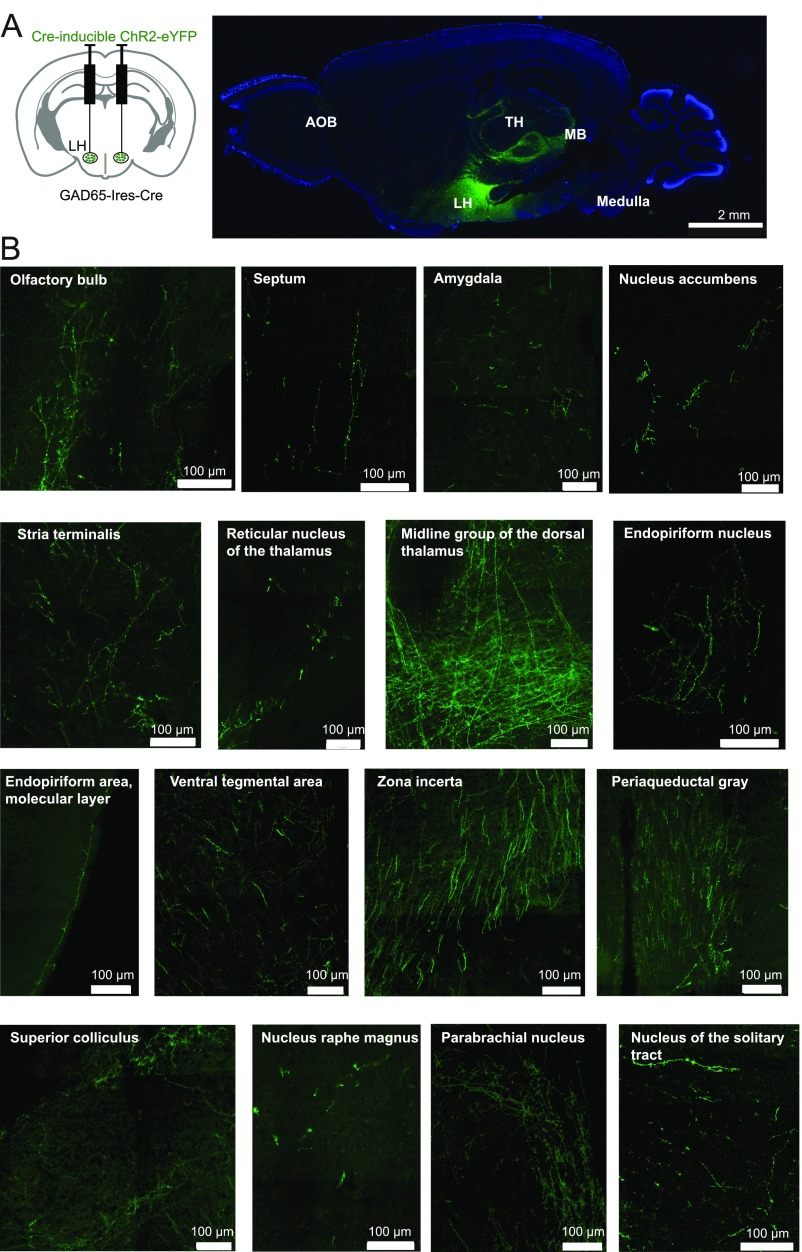
In known brain controllers of voluntary action, such as the basal ganglia, neural signals implicated in movement control appear seconds before action initiation (e.g., figure 3 E and F of ref. 24). These delays are comparable to the delays we observed between the onset of the GAD65LH network bursts and movement initiation (≈2 s; Fig. 4E). These long delays presumably imply multiple downstream processing stages between GAD65LH cell signals and motor commands that drive muscle activity underlying locomotion. When we attempted to determine downstream targets of GAD65LH neurons by tracing their axon distribution in brains of GAD65LH-ChR2-YFP mice, we found projections to multiple neural systems, extending from the LH to the thalamus, ventral tegmental area, cortical sensory areas, brainstem, and many hypothalamic nuclei (Fig. S1). A comprehensive analysis of these downstream effectors of GAD65LH cells will be included in future studies, because it is a large task requiring the relative locomotor impacts of GAD65LH cell targets to be deciphered. A specific question for such future work would be whether the locomotion drivers described here, and the classical basal ganglia pathways, operate in series or in parallel to promote physical activity.
Fig. S1.
Axonal projections of GAD65LH cells. (A, Left) Scheme for targeting ChR2-YFP to GAD65LH cells. (A, Right) DAPI-stained (blue) 30-μm sagittal section of the whole brain showing GAD65LH projections (green) to brain areas like the accessory olfactory bulb (AOB), LH, thalamus (TH), midbrain (MB), and medulla (representative example of three brains). (B) High magnification of brain areas where dense GAD65-ChR2-YFP axons are found (representative example of three brains).
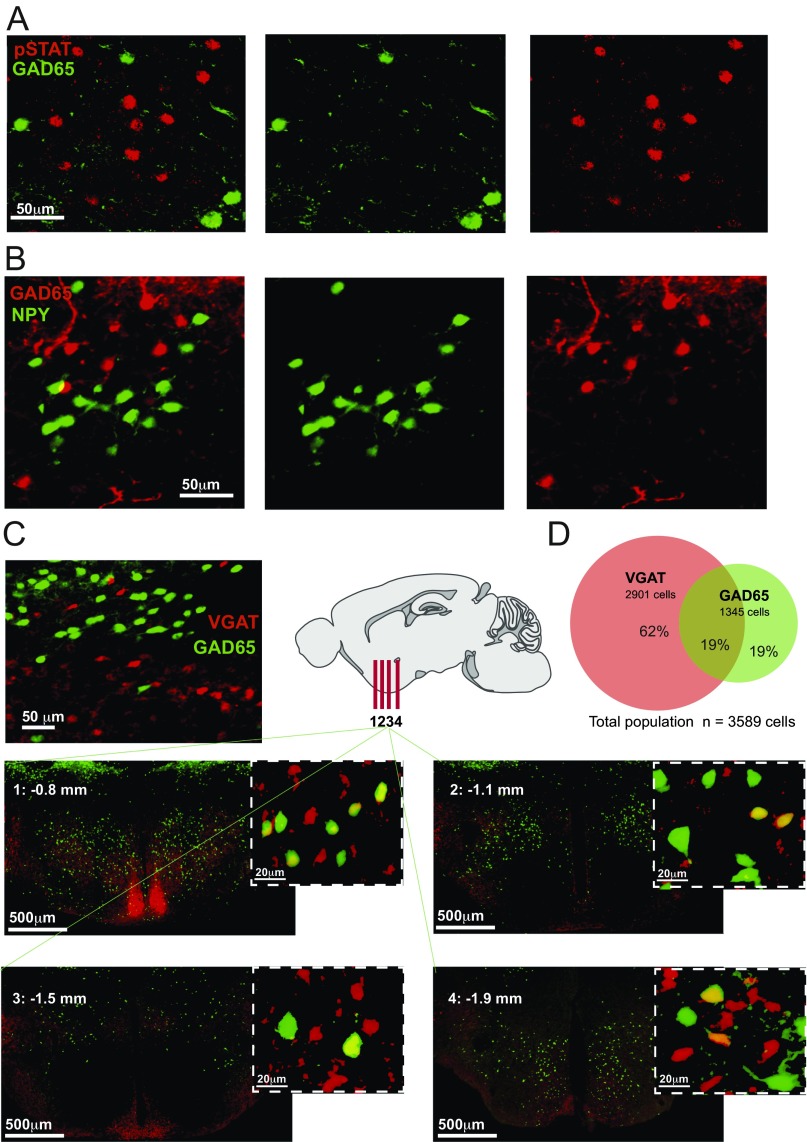
Related questions for future studies are what neurotransmitters GAD65LH cells release at their many projection targets, and what regulatory receptors they express. GAD65LH cells do not express orexin or MCH neurotransmitters (21); note that this finding does not contradict previous reports of GAD67 expression in MCH cells, because GAD65 and GAD67 are not always coexpressed in the hypothalamus (e.g., refs. 31, 32). We also found that the GAD65LH cells are distinct from the previously studied LH neurons expressing neuropeptide Y (NPY) or leptin receptor (33, 34) (Fig. S2 A and B). It remains to be determined whether GAD65LH cells release excitatory and/or inhibitory transmitters, because vesicular GABA transporter (VGAT) is not always expressed in GAD65 neurons, and some hypothalamic GAD65 neurons contain glutamatergic markers (31, 32, 35). Indeed, we found that only 50% of the GAD65LH cells contain VGAT, and that most (≈80%) VGATLH cells are distinct from GAD65LH cells (Fig. S2 C and D). Thus, the previous studies of VGATLH cells (36–39) likely targeted larger and more heterogeneous cell populations than our study, which was selective for GAD65LH cells. This fact may explain why the VGATLH cell stimulation drove different behaviors [increased eating and insignificant effect on locomotion (36, 39)] than the GAD65LH cell stimulation (increased locomotion and no significant effect on eating; Fig. 3 and Fig. S3). The latter findings presumably imply that GAD65LH-stimulated locomotion is unlikely to be specifically motivated by hunger or directed toward a food goal. We note that in some contexts, it may be evolutionarily advantageous to develop a locomotion drive not directed toward a specific goal (e.g., food), but toward a more general goal (e.g., change of place, physical escape). It is thus tempting to speculate that the orexin-activated GAD65LH cells may provide such a general locomotion drive that serves, for example, to move the animal away from stresses that activate orexin neurons (3, 10, 11, 13). Such an escape-like behavioral drive would also be consistent with the observed GAD65LH cell dynamics during capture, when the mouse is immobilized but the drive is high.
Fig. S2.
Neurochemical characteristics of GAD65LH cells. (A) Colocalization of GAD65LH cells and pSTAT3 (leptin receptor) LH cells. GAD65LH cells were labeled with GFP (GAD65-GFP transgenic mouse), and pSTAT3 neurons were labeled with Alexa Fluor 555 antibodies (Materials and Methods). A total of 11.45% of pSTAT3LH neurons contained GAD65 (analysis of 1,544 pSTAT neurons from three GAD65-GFP/pSTAT3-Alexa555 brains), and 6.89% of GAD65LH cells contained pSTAT3 (analysis of 819 GAD65 neurons from three GAD65-GFP/pSTAT3-Alexa555 brains). The % colocalization values are averages per hemisphere. (B) Colocalization of GAD65LH cells and NPYLH cells. NPY cells were labeled with GFP (NPY-hrGFP transgenic mouse), and GAD65-Ires-Cre cells were labeled with ChR2-mCherry. A total of 2.06% of NPYLH neurons contained GAD65 (analysis of 705 NPYLH neurons from three NPY-hrGFP/GAD65-mCherry brains), and 0.78% of GAD65LH cells contained NPY (analysis of 1,645 GAD65LH neurons from three NPY-hrGFP/GAD65-mCherry brains). The % colocalization values are averages per hemisphere. (C) Colocalization of GAD65LH cells and VGATLH cells. GAD65LH cells were labeled with GFP (GAD65-GFP transgenic mouse), and VGAT-Ires-Cre cells were labeled with tdTomato (CAG-tdTomato;VGAT-Ires-Cre transgenic mouse) or ChR2-mCherry (Materials and Methods). (Left) Example of LH colocalization. (Right and Bottom) More examples in coronal slices from different anteroposterior LH locations (bregma coordinates indicated on the slides). (D) Quantification of data in A (combined cell counts from three brains).
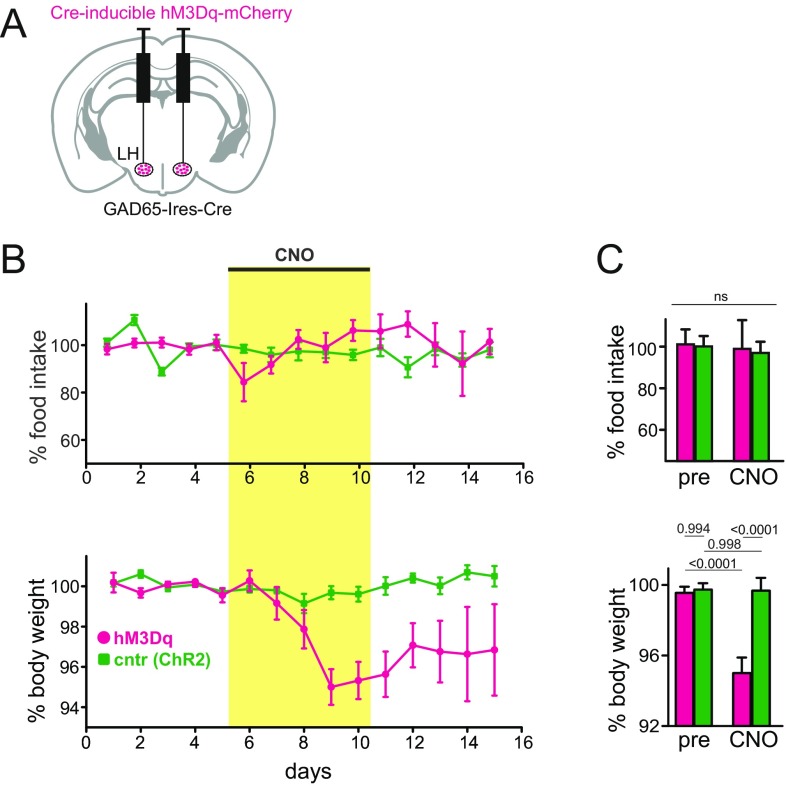
Fig. S3.
Effects of GAD65LH cell stimulation on eating and body weight. (A) Scheme for targeting the excitatory CNO receptor hM3Dq (or control protein ChR2-YFP) to GAD65LH cells. Targeting was confirmed by histology, as shown in Fig. 3E. (B) Effect of CNO on food intake and body weight in GAD65-Ires-Cre mice expressing ChR2 or hM3Dq in GAD65LH cells (five mice per group). Values are mean ± SEM. CNO was injected daily (0.5 mg/kg i.p.) during the indicated days. (C) Data from B, analyzed with two-way repeated measures ANOVA. pre, day 5; CNO, day 9. (Top) Interaction [F(1,8) = 0.01474, P = 0.9064]. ns, not significant. (Bottom) Interaction [F(1,8) = 13.47, P = 0.0063; numbers above bars are P values from Tukey’s multiple comparison test].
In summary, we found that physiological activity of GAD65LH cells is vital for normal locomotion and identified orexin as a physiological upstream regulator of GAD65LH activity. These findings establish the GAD65LH cells as key players in intra-LH information processing. Together with recent findings on roles of defined LH neurons in eating and arousal (7, 36, 38), our results substantiate specific LH circuits as orchestrators of key voluntary actions.
Materials and Methods
Gene Transfer.
All procedures followed United Kingdom Home Office regulations and were approved by the Animal Welfare and Ethical Review Panel of the Francis Crick Institute. Adult male and female mice (at least 8 wk old) were used for in vitro experiments. Male mice were used for behavioral experiments, and kept on a standard 12-h/12-h light/dark cycle and on standard mouse chow and water ad libitum. Mice were anesthetized with isoflurane and injected with meloxicam (2 mg/kg of body weight, s.c.) for analgesia. After placement into a stereotaxic frame (David Kopf Instruments), a craniotomy was performed and a borosilicate glass pipette was used to inject viral vectors bilaterally into the LH. Two injections (each 75 nL, delivered at 75 nL⋅min−1) were made into the LH in each hemisphere (bregma: −1.30 mm, midline: ±1 mm, from brain surface: 5.20 mm and 5.25 mm). Mice were allowed to recover for at least 1 wk after surgery while single-housed. Cre-dependent designer receptors exclusively activated by designer drugs (DREADD) constructs (40, 41), rAAV8/hSvn-DIO-hm3D(Gq)-mCherry [2.2 × 1012 genome copies (gc)/mL; UNC Vector Core] or rAAV8/hSyn-DIO-hM4(Gi)-mCherry (5.3 × 1012 gc/mL; UNC Vector Core), were used to measure the effect of cell type-specific manipulation on locomotion. For ChR-assisted circuit mapping, “FLEX switch” ChR2 constructs (42) were used. These constructs were either AAV1.EF1.flox.hChR2(H134R)-mCherry.WPRE.hGH (8.78 × 1012 gc/mL) or AAV1.EF1.DIO.hChR2(H134R)-YFP.WPRE.hGH (6.2 × 1012 gc/mL). For calcium imaging, we used a GCaMP6s construct (23), rAAV9.CAG.Flex.GCaMP6s.WPRE.SV40 (2.74 × 1013 gc/mL). Previously characterized and validated transgenic mouse lines (or their crosses) were used where indicated: GAD65-Ires-Cre mice (43) bred in homozygous (hom)-WT pairs with C57BL/6 mice, orexin-Cre mice (22) bred in heterozygous (het)-WT pairs with C57BL/6 mice, GAD65-GFP mice (21) bred in het-WT pairs with C57BL/6 mice, NPY-hrGFP mice (44) bred in het-WT pairs with C57BL/6 mice, VGAT-Ires-Cre mice (45) bred in hom-WT breeding pairs with C57BL/6 mice, and CAG-tdTomato mice (46) bred in hom-hom pairs.
Cell Type-Specific Deep-Brain Recordings in Awake Behaving Mice.
After injection of Cre-dependent GCaMP6s into GAD65-Ires-Cre mice, the fiberoptic implants were stereotaxically installed with the fiber tip above the LH (1.38 mm caudal from bregma, 0.95 mm lateral from midline, and 5 mm ventral from brain surface) and fixed to the skull as previously described (12, 13); this method is expected to capture signals only from within 500 μm of the fiber tip (12). Fiber tip locations were verified in each mouse by examining slices with a visible fiber tract. For locomotion experiments and immobilization, mice also had a custom-built, stainless-steel head-restraint implant glued to their skulls using tissue-friendly cyanoacrylate (Histoacryl; TissueSeal). Fiber photometry was performed as described by González et al. (13). Fluorescence signals were converted to ΔF/F (%) values as follows: ΔF/F = 100 * (Fr − F)/F, where Fr is the raw signal and F is the trial mean.
In Fig. 2E (immobilization stress experiments), mice were placed in a recording cage and left for 10 min; then, a trial consisting of 10 s of restraint stress (holding the mouse head stationary inside the cage by clamping the head-fixed restraint) was performed, based the method of González et al. (13). For each mouse, four trials per day were performed (with intertrial intervals >5 min). Trial blocks for drug condition (control, SB-334867, and recovery) were separated by 48 h. The competitive orexin receptor antagonist SB-334867 (or 0.9% NaCl and 10% DMSO as a control vehicle) was given i.p. 1 h before immobilization stress. This antagonist has a higher affinity for orexin type-1 receptor than for orexin type-2 receptor, but at higher concentrations, it antagonizes orexin binding to both receptors (47). We used a single high dose of SB-334867 (30 mg/kg), based on previous studies with this compound (48, 49). To control for circadian factors, all experiments were performed during the dark phase.
Quantifying Physical Activity.
Locomotor activity was assessed after a single i.p. dose of CNO or saline during the dark-phase in open home cages using either video-tracking (Anymaze or Ethovision) or a rotary treadmill. Mice of the same genotype were grouped according to vectors injected into their brains, and all groups had a similar composition based on sex (male), age, and body weight. For treadmill locomotion quantification, mice were head-restrained in a sound-attenuated chamber where they could run on a disk treadmill with a rotary encoder. The center of the animal’s head was such that 1° of rotation equated 1.8 mm of forward locomotion. The encoder generated a transistor-transistor logic (TTL) pulse every time the animal moved 1.8 mm. The pulses were collected by the same analogue-to-digital converter as the photodetector signals for accurate time-stamping of GAD65-GCaMP6s signals and locomotion. Locomotion bout onset was defined as the time of crossing a velocity threshold of 0.17 cm⋅s−1, and locomotion bouts of at least 5 s in duration from this onset are illustrated in the analyses shown in Fig. 4C.
Brain Slice Optogenetics, Electrophysiology, and Imaging.
Patch-clamp recordings combined with optogenetics were carried out as reported by Schöne et al. (50). Briefly, LH slices were prepared at least 2 months after virus injection. Coronal brain slices of 250-μm thickness containing the LH were cut while immersed in ice-cold slicing solution. Slices were incubated for 1 h in artificial cerebrospinal fluid (ACSF) at 35 °C, and then transferred to a submerged-type recording chamber. Neurons containing fluorescent markers were visualized with an Olympus BX61WI microscope with an oblique condenser and fluorescence filters. Excitation light for ChR2 was delivered from a LAMBDA DG-5 beam switcher (Sutter) with a xenon lamp and ET470/40-nm bandpass filter. A 40× 0.8-N.A. objective was used to deliver 1-ms flashes of blue light (∼10 mW/mm2) onto ChR2-containing axons around the recorded cell, and postsynaptic responses were recorded at a range of potentials (0 to −90 mV) in each cell. For studying effects of orexin on membrane current (Fig. 1C), voltage-clamp ramps from 0 to −120 mV were performed at 0.1 mV⋅ms−1, and orexin-evoked current was then derived in each cell by subtracting the baseline current from the orexin current at each potential and plotted at 10-mV increments for visual clarity (Fig. 1C, Right). Functional ChR2 expression was confirmed by recording light-activated action potentials in the target cells (n = 3 cells per group). For calcium imaging, brain slices were placed in a recording chamber of a BX61WI Olympus microscope controlled by Olympus Fluoview software (FV10-ASW version 4.0), perfused at 35 °C with ACSF. Confocal imaging was performed at 0.5 Hz with an Olympus 20× 0.50-N.A. objective, 488-nm argon laser excitation, and 500- to 545-nm emission collection. Motion and bleach corrections were applied if needed (StackReg plug-in, ImageJ; NIH). A region of interest (ROI) containing each GCaMP6s-positive neuron was selected via ROI manager in ImageJ. The mean fluorescence of each ROI was extracted for each frame. These raw fluorescence (Fr) values were normalized to produce ΔF/F values, where F is the mean baseline before orexin application and ΔF = (Fr − F). Simultaneous brain slice cell-attached recordings and calcium imaging (Fig. 2 B and C) were performed as reported by González et al. (13). Because the cell-attached experiments, which were necessary to preserve cytosolic integrity and thus in vivo-like GCaMP signals, do not measure transmembrane potential, it is possible that the GCaMP signal is increased by interspike depolarization as well as by the depolarization during the spikes.
Chemicals and Solutions.
For brain slice recordings, ACSF and ice-cold slicing solution were gassed with 95% O2 and 5% CO2, and contained the following: 125 mM NaCl ACSF, 2.5 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 1.2 mM NaH2PO4, 21 mM NaHCO3, 2 mM d-(+)-glucose, 0.1 mM Na+-pyruvate, and 0.4 mM ascorbic acid. The slicing solution contained 2.5 mM KCl, 1.3 mM NaH2PO·H20, 26.0 mM NaHCO3, 213.3 mM sucrose, 10.0 mM d-(+)-glucose, 2.0 mM MgCl2, and 2.0 mM CaCl2. For standard whole-cell recordings, pipettes were filled with intracellular solution containing the following: 120 mM K-gluconate, 10 mM KCl, 10 mM Hepes, 0.1 mM EGTA, 4 mM K2ATP, 2 mM Na2ATP, 0.3 mM Na2GTP, and 2 mM MgCl2 (pH 7.3) with KOH. Concentrations used were as follows: 300 nM orexin-A; 5 μM CNO; synaptic blocker mix: 50 μM D-AP5, 10 μM picrotoxin, 10 μM CNQX, and 10 μM CGP 35348; and 3 μM gabazine. For in vivo chemogenetic manipulations, CNO was injected i.p. at 5 mg/kg (experiments involving hM4Di) or at 0.5 mg/kg (experiments involving hM3Dq). All chemicals were from Sigma or Tocris Bioscience.
Histochemistry.
For the immunolabeling of pSTAT3 in Fig. S2, 30-μm cryosections of leptin-injected GAD65-GFP mice were pretreated with H2O2, NaOH, glycine, and SDS. Staining for pSTAT3 was done with rabbit anti-pSTAT3 lgG (no. 9131, 1:500; Cell Signaling Technology) sera overnight as the primary antibody and Alexa 555-conjugated donkey to rabbit IgG (A-31572, 1:500; Life Technologies) as the secondary antibody (51). In chemogenetic experiments, actuator targeting to LH (Fig. S4) was verified in fixed sections prepared as reported by González et al. (12).
Fig. S4.
Examples of coronal LH sections from four GAD65-Ires-Cre mice injected with Cre-dependent DREADD-mCherry, illustrating the typical extent of LH labeling in our study.
Statistical Analysis.
Statistical tests and descriptive statistics were performed as specified in the figure legends. In each experimental dataset at the cellular level, each n was a different cell (no repeated trials from the same cell were used as n values) and cells from at least three mice were analyzed. Before performing parametric tests, data were assessed for normality with a D’Agostino–Pearson omnibus test or Kolmogorov–Smirnov test for small sample sizes, and variances were assessed for homogeneity with a Brown–Forsyth test. To compare interactions within normally distributed data with repeated measurements, repeated measures ANOVA was used, with multiple comparison tests where appropriate. Analysis was performed with GraphPad Prism, MATLAB (The MathWorks, Inc.), and ImageJ.
Acknowledgments
We thank Prof. Troy Margrie for advice on head-fixed experimental paradigms and Martyn Stopps for building the rotary encoder. This work was funded by The Francis Crick Institute, which receives its core funding from Cancer Research UK, the UK Medical Research Council, and the Wellcome Trust.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1619700114/-/DCSupplemental.
References
- 1.Saper CB, Chou TC, Scammell TE. The sleep switch: Hypothalamic control of sleep and wakefulness. Trends Neurosci. 2001;24:726–731. doi: 10.1016/s0166-2236(00)02002-6. [DOI] [PubMed] [Google Scholar]
- 2.Hara J, et al. Genetic ablation of orexin neurons in mice results in narcolepsy, hypophagia, and obesity. Neuron. 2001;30:345–354. doi: 10.1016/s0896-6273(01)00293-8. [DOI] [PubMed] [Google Scholar]
- 3.Yamanaka A, et al. Hypothalamic orexin neurons regulate arousal according to energy balance in mice. Neuron. 2003;38:701–713. doi: 10.1016/s0896-6273(03)00331-3. [DOI] [PubMed] [Google Scholar]
- 4.Sacks O. Awakenings. Duckworth & Co; London: 1973. [Google Scholar]
- 5.Levitt DR, Teitelbaum P. Somnolence, akinesia, and sensory activation of motivated behavior in the lateral hypothalamic syndrome. Proc Natl Acad Sci USA. 1975;72:2819–2823. doi: 10.1073/pnas.72.7.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von Economo C. Sleep as a problem of localization. J Nerv Ment Dis. 1930;71:249–259. [Google Scholar]
- 7.Stuber GD, Wise RA. Lateral hypothalamic circuits for feeding and reward. Nat Neurosci. 2016;19:198–205. doi: 10.1038/nn.4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Lecea L, et al. The hypocretins: Hypothalamus-specific peptides with neuroexcitatory activity. Proc Natl Acad Sci USA. 1998;95:322–327. doi: 10.1073/pnas.95.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sakurai T, et al. Orexins and orexin receptors: A family of hypothalamic neuropeptides and G protein-coupled receptors that regulate feeding behavior. Cell. 1998;92:573–585. doi: 10.1016/s0092-8674(00)80949-6. [DOI] [PubMed] [Google Scholar]
- 10.Mileykovskiy BY, Kiyashchenko LI, Siegel JM. Behavioral correlates of activity in identified hypocretin/orexin neurons. Neuron. 2005;46:787–798. doi: 10.1016/j.neuron.2005.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams RH, Jensen LT, Verkhratsky A, Fugger L, Burdakov D. Control of hypothalamic orexin neurons by acid and CO2. Proc Natl Acad Sci USA. 2007;104:10685–10690. doi: 10.1073/pnas.0702676104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.González JA, et al. Inhibitory interplay between orexin neurons and eating. Curr Biol. 2016;26:2486–2491. doi: 10.1016/j.cub.2016.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.González JA, Iordanidou P, Strom M, Adamantidis A, Burdakov D. Awake dynamics and brain-wide direct inputs of hypothalamic MCH and orexin networks. Nat Commun. 2016;7:11395. doi: 10.1038/ncomms11395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sutcliffe JG, de Lecea L. The hypocretins: Setting the arousal threshold. Nat Rev Neurosci. 2002;3:339–349. doi: 10.1038/nrn808. [DOI] [PubMed] [Google Scholar]
- 15.Peyron C, et al. Neurons containing hypocretin (orexin) project to multiple neuronal systems. J Neurosci. 1998;18:9996–10015. doi: 10.1523/JNEUROSCI.18-23-09996.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagan JJ, et al. Orexin A activates locus coeruleus cell firing and increases arousal in the rat. Proc Natl Acad Sci USA. 1999;96:10911–10916. doi: 10.1073/pnas.96.19.10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anand BK, Brobeck JR. Localization of a “feeding center” in the hypothalamus of the rat. Proc Soc Exp Biol Med. 1951;77:323–324. doi: 10.3181/00379727-77-18766. [DOI] [PubMed] [Google Scholar]
- 18.Sakurai T. The neural circuit of orexin (hypocretin): Maintaining sleep and wakefulness. Nat Rev Neurosci. 2007;8:171–181. doi: 10.1038/nrn2092. [DOI] [PubMed] [Google Scholar]
- 19.Gerashchenko D, et al. Hypocretin-2-saporin lesions of the lateral hypothalamus produce narcoleptic-like sleep behavior in the rat. J Neurosci. 2001;21:7273–7283. doi: 10.1523/JNEUROSCI.21-18-07273.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whiddon BB, Palmiter RD. Ablation of neurons expressing melanin-concentrating hormone (MCH) in adult mice improves glucose tolerance independent of MCH signaling. J Neurosci. 2013;33:2009–2016. doi: 10.1523/JNEUROSCI.3921-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karnani MM, Szabó G, Erdélyi F, Burdakov D. Lateral hypothalamic GAD65 neurons are spontaneously firing and distinct from orexin- and melanin-concentrating hormone neurons. J Physiol. 2013;591:933–953. doi: 10.1113/jphysiol.2012.243493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schöne C, et al. Optogenetic probing of fast glutamatergic transmission from hypocretin/orexin to histamine neurons in situ. J Neurosci. 2012;32:12437–12443. doi: 10.1523/JNEUROSCI.0706-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen TW, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui G, et al. Concurrent activation of striatal direct and indirect pathways during action initiation. Nature. 2013;494:238–242. doi: 10.1038/nature11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gunaydin LA, et al. Natural neural projection dynamics underlying social behavior. Cell. 2014;157:1535–1551. doi: 10.1016/j.cell.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y, Lin YC, Kuo TW, Knight ZA. Sensory detection of food rapidly modulates arcuate feeding circuits. Cell. 2015;160:829–841. doi: 10.1016/j.cell.2015.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sternson SM, Roth BL. Chemogenetic tools to interrogate brain functions. Annu Rev Neurosci. 2014;37:387–407. doi: 10.1146/annurev-neuro-071013-014048. [DOI] [PubMed] [Google Scholar]
- 28.Henze DA, Buzsáki G. Action potential threshold of hippocampal pyramidal cells in vivo is increased by recent spiking activity. Neuroscience. 2001;105:121–130. doi: 10.1016/s0306-4522(01)00167-1. [DOI] [PubMed] [Google Scholar]
- 29.Elmquist JK, Elias CF, Saper CB. From lesions to leptin: Hypothalamic control of food intake and body weight. Neuron. 1999;22:221–232. doi: 10.1016/s0896-6273(00)81084-3. [DOI] [PubMed] [Google Scholar]
- 30.Saper CB, Scammell TE, Lu J. Hypothalamic regulation of sleep and circadian rhythms. Nature. 2005;437:1257–1263. doi: 10.1038/nature04284. [DOI] [PubMed] [Google Scholar]
- 31.Romanov RA, et al. Molecular interrogation of hypothalamic organization reveals distinct dopamine neuronal subtypes. Nat Neurosci. 2017;20:176–188. doi: 10.1038/nn.4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeong JH, Woo YJ, Chua S, Jr, Jo YH. Single-cell gene expression analysis of cholinergic neurons in the arcuate nucleus of the hypothalamus. PLoS One. 2016;11:e0162839. doi: 10.1371/journal.pone.0162839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leinninger GM, et al. Leptin acts via leptin receptor-expressing lateral hypothalamic neurons to modulate the mesolimbic dopamine system and suppress feeding. Cell Metab. 2009;10:89–98. doi: 10.1016/j.cmet.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marston OJ, Hurst P, Evans ML, Burdakov DI, Heisler LK. Neuropeptide Y cells represent a distinct glucose-sensing population in the lateral hypothalamus. Endocrinology. 2011;152:4046–4052. doi: 10.1210/en.2011-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jarvie BC, Hentges ST. Expression of GABAergic and glutamatergic phenotypic markers in hypothalamic proopiomelanocortin neurons. J Comp Neurol. 2012;520:3863–3876. doi: 10.1002/cne.23127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jennings JH, et al. Visualizing hypothalamic network dynamics for appetitive and consummatory behaviors. Cell. 2015;160:516–527. doi: 10.1016/j.cell.2014.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nieh EH, et al. Decoding neural circuits that control compulsive sucrose seeking. Cell. 2015;160:528–541. doi: 10.1016/j.cell.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herrera CG, et al. Hypothalamic feedforward inhibition of thalamocortical network controls arousal and consciousness. Nat Neurosci. 2016;19:290–298. doi: 10.1038/nn.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Navarro M, et al. Lateral hypothalamus GABAergic neurons modulate consummatory behaviors regardless of the caloric content or biological relevance of the consumed stimuli. Neuropsychopharmacology. 2016;41:1505–1512. doi: 10.1038/npp.2015.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Armbruster BN, Li X, Pausch MH, Herlitze S, Roth BL. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc Natl Acad Sci USA. 2007;104:5163–5168. doi: 10.1073/pnas.0700293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krashes MJ, et al. Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. J Clin Invest. 2011;121:1424–1428. doi: 10.1172/JCI46229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Atasoy D, Aponte Y, Su HH, Sternson SM. A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J Neurosci. 2008;28:7025–7030. doi: 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taniguchi H, et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van den Pol AN, et al. Neuromedin B and gastrin-releasing peptide excite arcuate nucleus neuropeptide Y neurons in a novel transgenic mouse expressing strong Renilla green fluorescent protein in NPY neurons. J Neurosci. 2009;29:4622–4639. doi: 10.1523/JNEUROSCI.3249-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vong L, et al. Leptin action on GABAergic neurons prevents obesity and reduces inhibitory tone to POMC neurons. Neuron. 2011;71:142–154. doi: 10.1016/j.neuron.2011.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smart D, et al. SB-334867-A: The first selective orexin-1 receptor antagonist. Br J Pharmacol. 2001;132:1179–1182. doi: 10.1038/sj.bjp.0703953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adamantidis AR, Zhang F, Aravanis AM, Deisseroth K, de Lecea L. Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature. 2007;450:420–424. doi: 10.1038/nature06310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karnani MM, et al. Activation of central orexin/hypocretin neurons by dietary amino acids. Neuron. 2011;72:616–629. doi: 10.1016/j.neuron.2011.08.027. [DOI] [PubMed] [Google Scholar]
- 50.Schöne C, Apergis-Schoute J, Sakurai T, Adamantidis A, Burdakov D. Coreleased orexin and glutamate evoke nonredundant spike outputs and computations in histamine neurons. Cell Reports. 2014;7:697–704. doi: 10.1016/j.celrep.2014.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu L, et al. Leptin signaling modulates the activity of urocortin 1 neurons in the mouse nonpreganglionic Edinger-Westphal nucleus. Endocrinology. 2011;152:979–988. doi: 10.1210/en.2010-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]