Significance
Without a vaccine or cost-effective antivirals against hepatitis C virus (HCV) there is a need to understand better the molecular mechanisms underlying the establishment of productive HCV infection and chronic liver disease. Recently, the Crohn's disease and tuberculosis risk factor immunity-related GTPase M (IRGM) was found to promote HCV replication, but the mechanisms involved are unknown. Here we provide mechanistic insight into how IRGM stimulates the two membrane-remodeling pathways, Golgi fragmentation and autophagy, to facilitate HCV replication. Our findings offer insights into the replication strategies used by HCV that should be useful for antiviral approaches. Also, these findings might aid understanding how IRGM functions in infection and inflammation in the setting of other diseases, e.g., in Crohn’s disease.
Keywords: HCV, Golgi fragmentation, autophagy, IRGM, membranous web
Abstract
Positive-stranded RNA viruses, such as hepatitis C virus (HCV), assemble their viral replication complexes by remodeling host intracellular membranes to a membranous web. The precise composition of these replication complexes and the detailed mechanisms by which they are formed are incompletely understood. Here we show that the human immunity-related GTPase M (IRGM), known to contribute to autophagy, plays a previously unrecognized role in this process. We show that IRGM is localized at the Golgi apparatus and regulates the fragmentation of Golgi membranes in response to HCV infection, leading to colocalization of Golgi vesicles with replicating HCV. Our results show that IRGM controls phosphorylation of GBF1, a guanine nucleotide exchange factor for Arf-GTPases, which normally operates in Golgi membrane dynamics and vesicle coating in resting cells. We also find that HCV triggers IRGM-mediated phosphorylation of the early autophagy initiator ULK1, thereby providing mechanistic insight into the role of IRGM in HCV-mediated autophagy. Collectively, our results identify IRGM as a key Golgi-situated regulator that links intracellular membrane remodeling by autophagy and Golgi fragmentation with viral replication.
Hepatitis C virus (HCV) is a positive-sense RNA virus in the family Flaviviridae that is a major cause of chronic liver disease. All positive-strand RNA viruses studied until now, including HCV, replicate their genomes in association with cellular membrane rearrangements. In this process, viruses remodel intracellular membranes [e.g., of mitochondria, endoplasmic reticulum (ER), and plasma membrane] to generate membrane structures such as single- or double-membrane vesicles that contribute to viral replication complexes (VRCs). HCV replication takes place at a unique subcellular compartment, the membranous web (MW), which has been proposed to be derived from the ER (1, 2). The HCV MW has a complex morphology consisting of clusters of single-, double-, and multimembrane vesicles and probably includes autophagosomes and lipid droplets (1, 3, 4). Recent findings reveal that the MW is produced by distinct HCV nonstructural (NS) proteins acting through sequential interaction with several host factors, such as the virus-targeted phosphatidylinositol-4 kinase III α (PI4KIIIα) (2, 3), but the full spectrum of host components and precise membrane composition that supports HCV replication are not fully defined.
Autophagy is an evolutionarily conserved cellular mechanism that involves intracellular membrane trafficking and degradation to maintain cell homeostasis. Viruses, including HCV, have been reported to exploit autophagy for replication purposes (4–6), but the mechanism by which this exploitation occurs is largely unknown. De novo synthesis of autophagosomes is a complex process that involves the formation of a phagophore membrane and its elongation. Initiation of autophagy is regulated by the mammalian target of rapamycin complex 1 (mTORC1), which negatively controls the kinase by phosphorylation at S757 (7). In further processing of the membrane, the two ubiquitin-like conjugation systems involving coupling of ATG12 with ATG5 and LC3 (microtubule-associated protein 1A/1B-light chain 3) to phosphatidylethanolamine are instrumental in the elongation of the membrane and closure of the autophagosome. It is believed that HCV may use autophagy to generate cytoplasmic membrane structures required for genome replication. The mechanisms by which HCV induces autophagy are currently debated (for a review, see ref. 8).
The immunity-related GTPases (IRGs) belong to a large superfamily of IFN-inducible GTPases. Previous work has shown that rodent IRGs play a particularly important role in innate immune resistance to intracellular bacteria and protozoa (e.g., Salmonella typhimurium and Mycobacterium tuberculosis) (9, 10). Mouse immunity-related GTPase M (Irgm) has been shown to bind to various intracellular membrane compartments and upon infection to relocalize rapidly to the phagocytic cup after bacterial uptake (11, 12). In contrast, the functions of human IRGM are largely undefined. Nevertheless, Singh et al. (13, 14) showed that human IRGM stimulates autophagy and proposed that IRGM localized to mitochondria, aiding mitochondrial fission, an event that was suggested to be required for IRGM-mediated autophagic restriction of mycobacteria. In relation to viruses, Grégoire et al. (15) generated two-hybrid–based protein interaction maps and found that human IRGM was highly targeted by RNA viral proteins (from HCV, measles virus, and HIV-1) and interacted with autophagy components, thereby suggesting that IRGM is manipulated for promotion of virus replication. Recently, IRGM was found to regulate ULK1 and to promote the formation of autophagy initiation complexes (16). Hence, IRGM contributes to autophagy, but the mechanisms by which IRGM participates during viral infections are unknown.
Positive-strand RNA viruses, such as HCV, assemble their viral replication machinery by remodeling host intracellular membranes to provide the structural foundation of their replication complexes. These membrane-based replication sites may be formed by harnessing the secretory pathway to recruit lipid and protein components to optimize viral replication (17). Enteroviruses, which belong to the family Picornaviridae, rely on both protein and lipid components of the host secretory pathway to provide the structural foundation of their replication complexes (17). Recently, the guanine nucleotide exchange factor GBF1 and its effector ADP ribosylation factor 1 (Arf1), known to regulate Golgi membrane trafficking and organelle structure in the secretory pathway, were identified as host factors required for efficient HCV replication (18–21). This requirement suggests that Golgi components have a role in HCV replication.
In this study we show that IRGM controls HCV-triggered membrane remodeling via the kinase ULK1 to trigger autophagy and also that IRGM regulates the vesicular transport proteins GBF1 and Arf1, thereby leading to Golgi fragmentation. Our results suggest that HCV exploits IRGM to induce two distinct strategies of cellular membrane remodeling that promote its replication. Overall, our data extend the current knowledge of the functions of the disease risk factor IRGM and highlight its role in HCV infection.
Results
The ULK1/2 Complex and IRGM Are Required for HCV-Induced Autophagy to Promote HCV Replication.
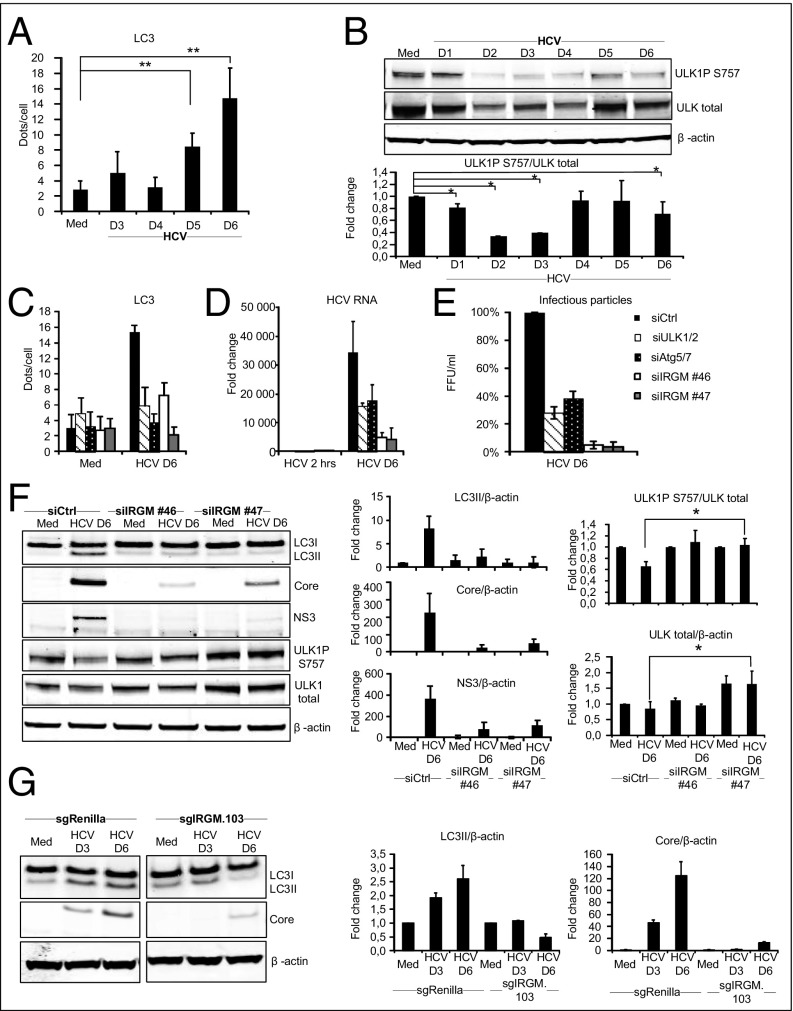
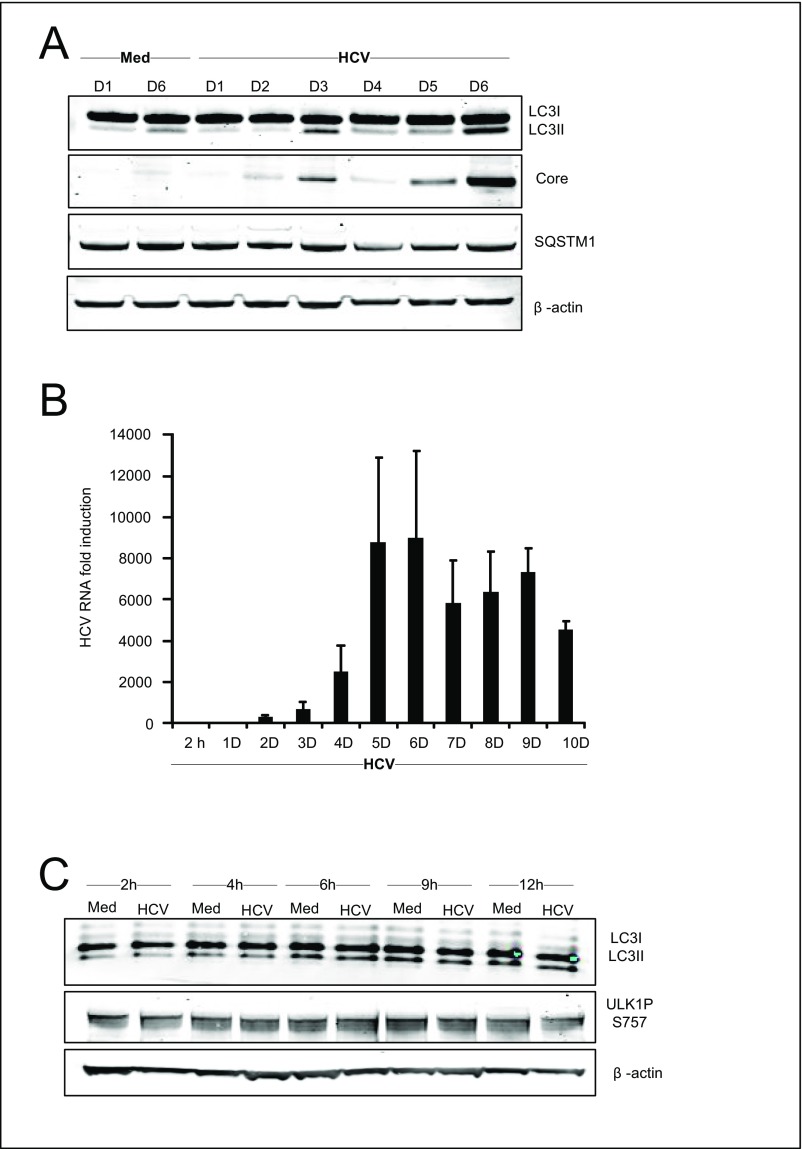
Several studies have shown that autophagosomes are formed during HCV infection (22), but the temporal regulation is debated, and the host components involved have not been determined. To establish the effect of HCV on autophagy, we initially infected Huh7.25CD81 cells with HCV and examined autophagy markers at different time points post infection (p.i.). Determination of endogenous LC3-II puncta formation by immunofluorescence and confocal microscopy showed that HCV infection increases LC3-II dot formation compared with uninfected cells, starting from day 3 p.i. with increased LC3-II dot formation at day 5 p.i. and day 6 p.i. (Fig. 1A). Immunoblot analysis of the conversion of endogenous LC3-I to LC3-II confirmed these results (Fig. S1A). Efficient expression of the HCV core protein and HCV RNA correlated well with the strong increase in LC3-II formation at day 6 p.i. (Fig. S1 A and B). We noted that the kinetics of autophagy triggered by HCV differed from that in a previous report by Dreux et al. (23) in which autophagy was detected a few hours p.i. (23), but we did not detect significant changes in LC3 lipidation at early time points (within 2–12 h p.i.) (Fig. S1C). The difference between the two studies may reflect the higher multiplicity of infection (MOI) used by Dreux et al. (MOI 6 compared with 0.3) (23), which would result in a higher degree of infected cells at early time points p.i. and higher viral gene expression that could cause autophagy induction at early time points. Taken together, our results confirm that HCV infection triggers the formation of autophagosomes along with an increase in HCV RNA and HCV proteins.
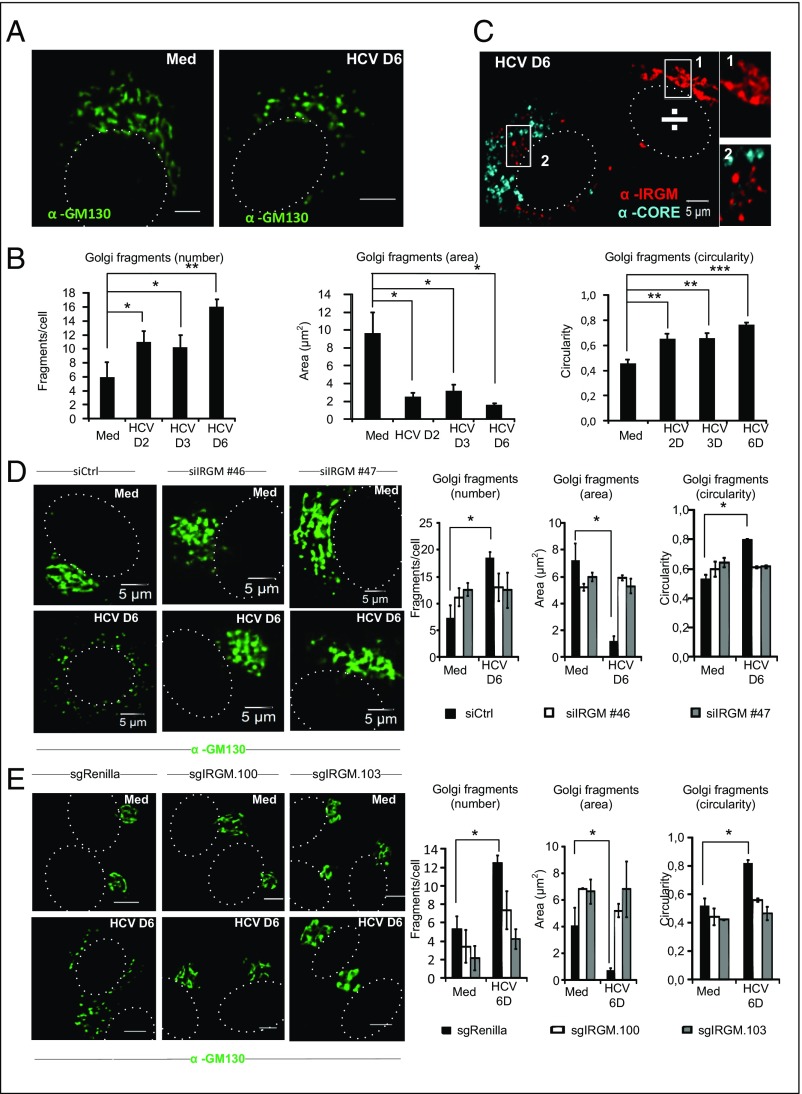
Fig. 1.
The ULK1/2 complex and IRGM are required for autophagy upon HCV infection and promote HCV replication. (A) Endogenous LC3 puncta were calculated in cells immunostained for LC3. (B) ULK1 phosphorylated and total protein levels were examined in HCV-infected cells by immunoblotting. A blot from one representative experiment is shown. ULK1PS757 levels were normalized to total ULK1 levels and visualized as means ± SD of three independent experiments. (C–E) LC3 dot formation, calculated as LC3 puncta per cell (C), HCV RNA levels determined by qRT-PCR (D), and production of infectious HCV particles calculated as focus-forming units per milliliter (E) were analyzed in cells transfected with siRNAs targeting ATG5, ATG7, ULK1, ULK2, or control siRNA before HCV infection for 6 d. (F) ULK1 phosphorylated and total protein levels, LC3II levels, and HCV protein levels were examined in cells treated with two different siRNAs against IRGM or control siRNA. Fold change values were calculated relative to siControl in noninfected cells. ULK1PS757 levels were normalized to total ULK1 levels, and HCV protein levels were normalized to β-actin. (G) LC3II and HCV protein levels were examined in the CRISPR/Cas9 IRGM knocked-down clone Huh7.25CD81 sgIRGM.103 and in control sgRenilla cells. Protein levels were normalized to β-actin. Data are shown as the mean ± SD of three independent experiments; *P < 0.05, **P < 0.005, paired t test. Med, uninfected hepatocytes.
Fig. S1.
Kinetics of HCV-induced autophagy and HCV replication. Huh7.25CD81 cells were infected with HCV (MOI = 0.3) for the indicated number of days. (A) Lysates were prepared, and proteins were separated by LDS-PAGE before immunoblotting as indicated. The decreased levels of LC3 at day 3 in this figure coincide with the passage of the cells at this time point. (B) Total RNA was isolated, and HCV RNA levels were determined by qRT-PCR. Data are expressed as the fold induction relative to corresponding RNA levels in cells infected with HCV for 2 h. (C) Huh7.25CD81 cells were infected with HCV (MOI 0.3) for the indicated days. Lysates were prepared, and proteins were separated by LDS-PAGE before immunoblotting as indicated. Data represent means ± SD from three independent experiments. Med, uninfected hepatocytes.
The ULK complex is a critical upstream regulator of autophagy induction (24), and its activity is controlled by phosphorylation at multiple sites (25). Dephosphorylation of ULK1S757 is associated with increased ULK activity and ULK-mediated autophagosome formation (25). However, whether ULK contributes to virus-mediated autophagy is largely unexplored. We found that HCV infection induces a time-dependent dephosphorylation of ULK1S757, starting between day 1 and day 2 p.i. (Fig. 1B). Notably, the level of total ULK1 was decreased upon HCV infection (Fig. 1B), possibly because the stability of ULK1 is decreased upon autophagy induction (24). Another study showed findings similar to ours, i.e., that total ULK1 levels were reduced upon infection with herpes simplex virus 1 (26).
ULK1/2 is known to direct the initiation of autophagy at the phagophore, whereas the autophagy proteins ATG5 and ATG7 act downstream in the process of elongation and closure of the autophagosome (27). We next examined whether these autophagy regulators control HCV-induced autophagy. Efficient siRNA-mediated knockdown of ULK1/2 and ATG5/7 (Fig. S2) resulted in marked reductions of LC3 dot numbers (Fig. 1C), which suggests that HCV-triggered autophagy is dependent on ULK1/2 and ATG5/7.
Fig. S2.
Effect of siRNA-mediated knockdown of autophagy-related proteins. Huh7.25CD81 cells were transfected with different siRNAs (ULK1/2, ATG5/7, and AllStar as control) for 6 d. Lysates were prepared, and proteins were separated by LDS-PAGE before immunoblotting with antibodies as indicated.
Autophagy has been suggested to represent a viral advantage (5, 28, 29). Accordingly, depletion of ULK1/2 and ATG5/ATG7 significantly reduced HCV replication and the formation of HCV infectious particles (Fig. 1 D and E). Collectively, our results show that HCV-mediated autophagy is dependent on the ULK1/2 complex and that ATG5/7 and promotes HCV replication.
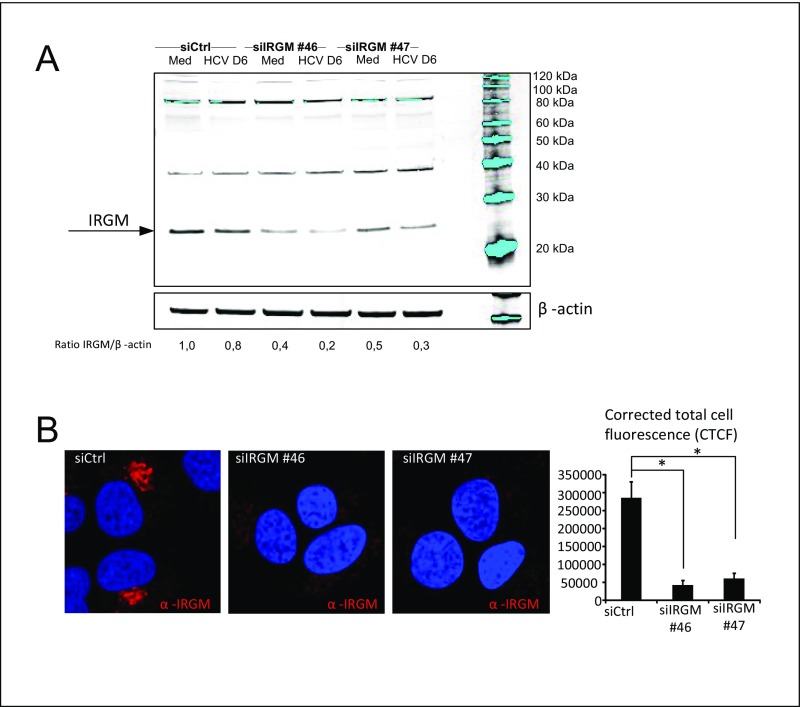
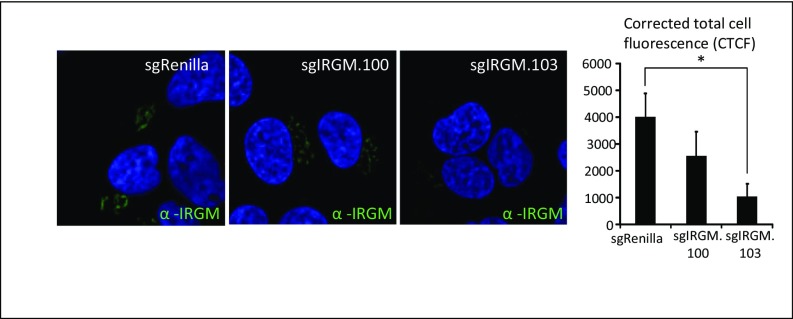
IRGM was identified in a two-hybrid interaction assay as an interactor with specific proteins of three distinct RNA viruses, including HCV, and with autophagy components (15). To characterize the role of IRGM in HCV-mediated autophagy and HCV infection, we first studied whether IRGM mediates HCV-stimulated autophagy by assessing the formation of endogenous LC3-II. Immunoblotting and immunofluorescence staining confirmed that knockdown of IRGM by two different siRNAs caused reduced expression of the expected IRGM ∼20 kDa band and reduced fluorescence signal upon IRGM staining (Fig. S3). Importantly, specificity of the specificity of the IRGM siRNAs (hereafter, siIRGMs) was verified, because they silenced only the target gene and did not affect the intensity of nonspecific bands (Fig. S3A). This IRGM knockdown caused a dramatic reduction of endogenous LC3 lipidation as detected by LC3 dot formation and LC3-II immunoblot conversion (Fig. 1 C and F). To confirm this observation, we targeted the IRGM locus using CRISPR/Cas9 in Huh7.25 CD81 cells to generate IRGM-deficient cells. Cells with reduced expression of IRGM (based on immunofluorescence staining) (Fig. S4) were selected, and the formation of LC3-II in response to HCV was examined. We found that the HCV-stimulated LC3-II formation in CRISPR/Cas9-single-guide IRGM (sgIRGM)–targeted cells was strongly reduced compared with the Huh7.25CD81 single-guide Renilla (sgRenilla) used as control cells (Fig. 1G), hence corroborating our results obtained with two different siRNAs targeting IRGM. Reduction of IRGM by siRNA or by CRISPR/Cas9-mediated targeting resulted in a strong reduction in the levels of HCV proteins and HCV RNA and also in the secretion of extracellular HCV infectious particles. Taken together, these results suggest that IRGM is required for autophagy during HCV infection and that it supports continuous HCV replication.
Fig. S3.
Control of IRGM knockdown in Huh7.25CD81 cells. Huh7.25CD81 cells were treated with two different siRNAs against IRGM or Control siRNA. (A) Total protein levels were examined by immunoblotting with an IRGM antibody. The migration of the molecular weight markers is presented to point out that the anti-IRGM antibodies specifically recognize IRGM at its expected position. The specificity of the siIRGMs was verified, because they silenced only the target gene and did not affect the intensity of nonspecific bands. (B) Intracellular staining of endogenous IRGM. Cells were fixed with 4% PFA, blocked, and immunostained with an antibody against IRGM. Cells were examined by confocal microscopy (magnification: 63×), and the corrected total cell fluorescence (CTCF) was calculated using ImageJ software. *P < 0.05. Med, uninfected hepatocytes.
Fig. S4.
IRGM fluorescence in CRISPR/Cas9-based IRGM-knockout and control Huh7.25CD81 cells. The IRGM levels in Huh7.25CD81 sgRenilla, sgIRGM.100, and sgIRGM.103 were examined by immunofluorescence staining. Cells were fixed with 4% PFA, blocked, and immunostained with an antibody against IRGM. Cells were examined by confocal microscopy (magnification: 63×), and the corrected total cell fluorescence (CTCF) was calculated using ImageJ software. *P < 0.05.
The role of IRGM in autophagy is largely unknown. A recent study showed that IRGM interacts with key autophagy regulators including ULK1 (16), but the mechanisms by which IRGM contributes during viral infections are not known. To examine if IRGM triggered ULK1 activation, we assessed the effect of siIRGM on the phosphorylation of ULK1S757. In siIRGM-treated cells, HCV infection did not induce dephosphorylation of ULK1S757 (Fig. 1F). Notably, we also repeatedly observed that ULK1 total levels were increased in cells depleted of IRGM. As discussed above, we propose that these findings may indicate that autophagy is reduced in cells deprived of IRGM. Another study reported that ULK1 levels were reduced upon starvation, presumably as a mechanism to restrain prolonged autophagy (30). Collectively, our results suggest that IRGM regulates ULK activation and ULK-mediated autophagy upon HCV infection.
The Immunity-Related GTPase IRGM Localizes to the Golgi Apparatus and Triggers Golgi Fragmentation upon HCV Infection.
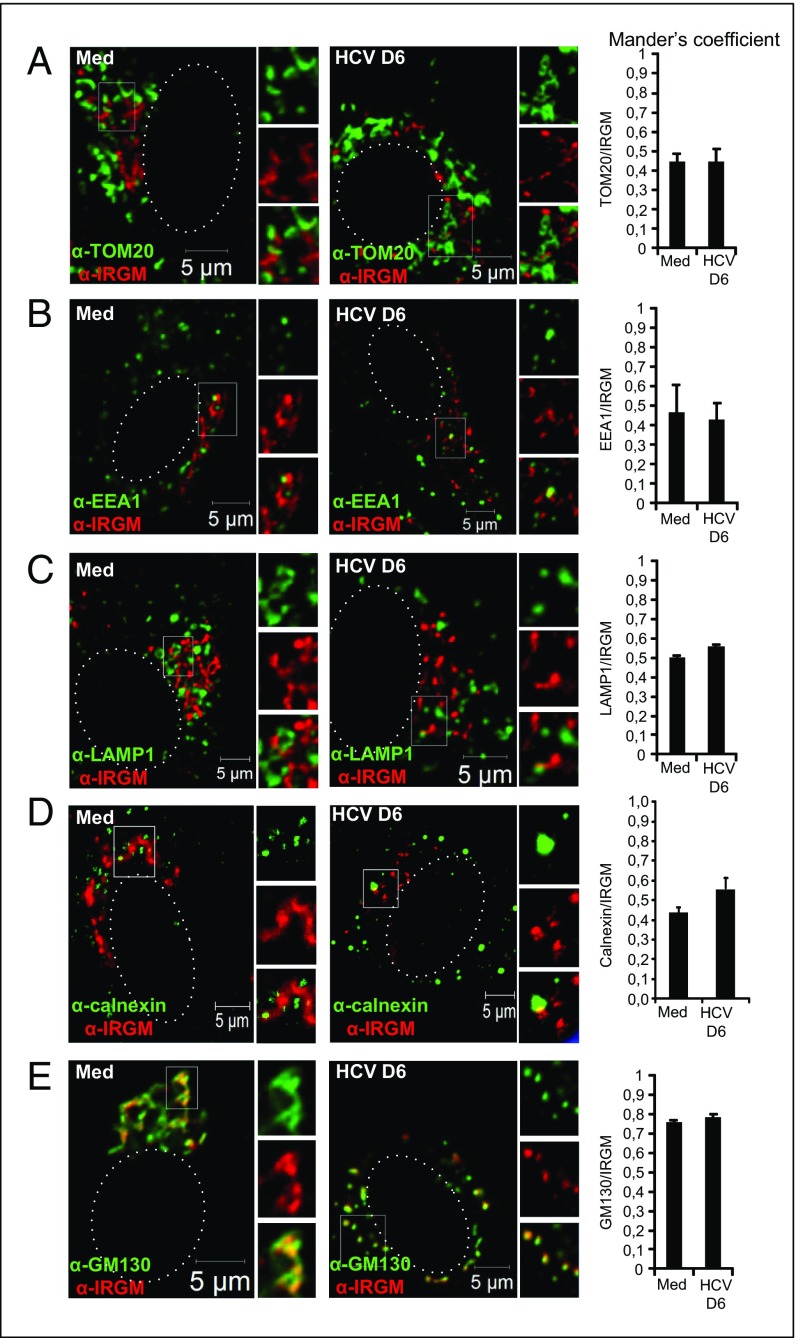
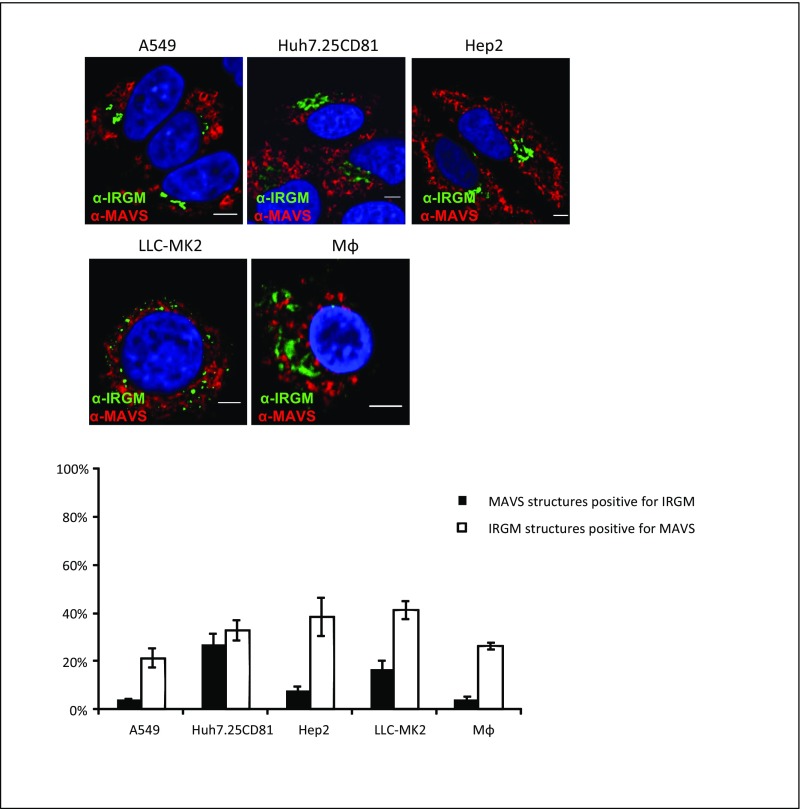
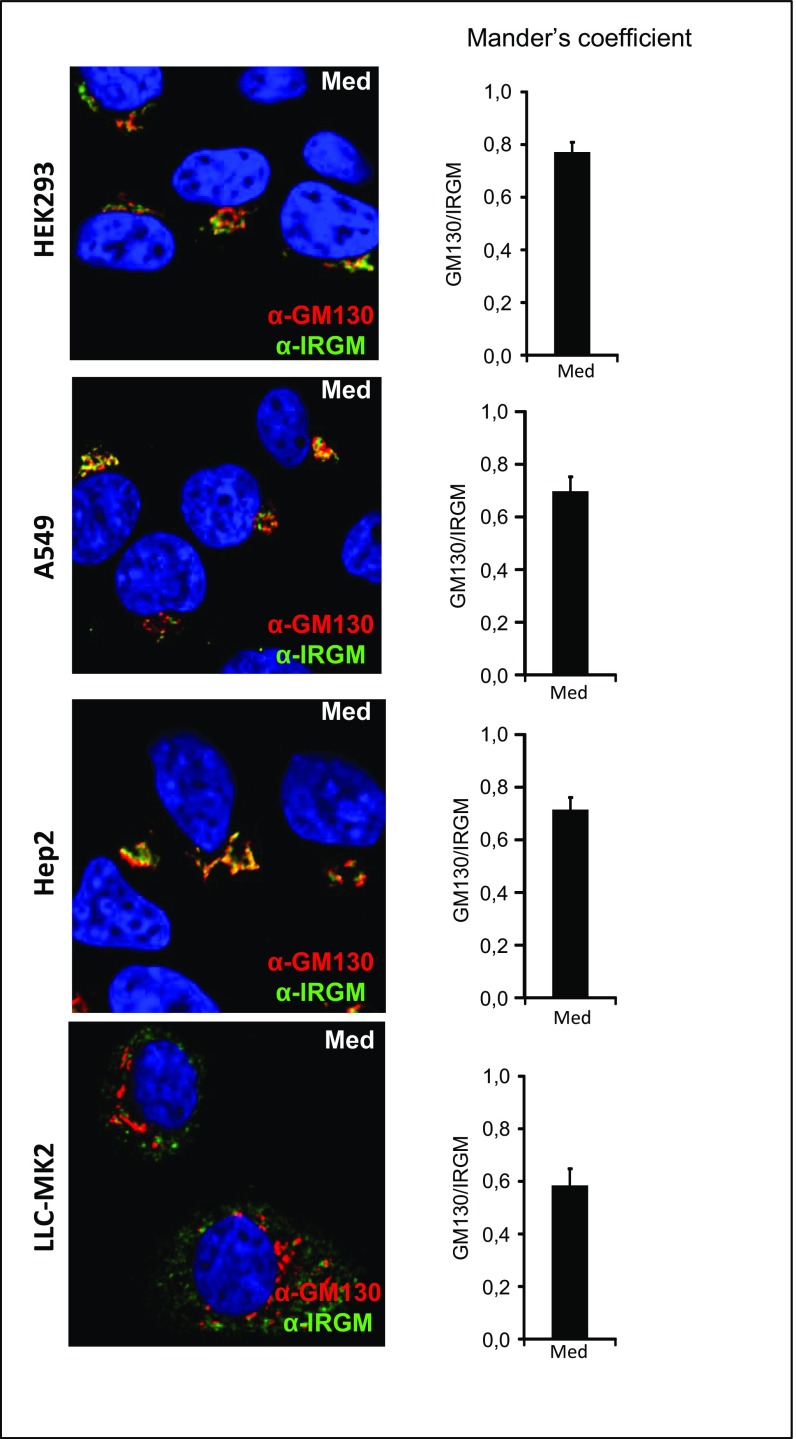
Although the subcellular localization of mouse IRGs in various inflammatory settings has been reported (13, 31, 32), the localization of endogenous human IRGM has not been fully explored. To assess the subcellular localization of IRGM in human hepatocytes, we performed confocal microscopy using an IRGM antibody together with antibodies for various intracellular markers. Using the mitochondrial markers TOM20 and MAVS, we did not find a significant degree of overlap between IRGM and mitochondria staining in uninfected or in HCV-infected cells, as assessed by Mander’s coefficient analysis (Fig. 2A and Fig. S5). Because this finding was in contrast with a report showing significant expression of IRGM at the mitochondria (14), we examined additional cell lines and similarly found only moderate overlap between IRGM and mitochondrial staining (Fig. S5). The mouse IRGM homolog Irgm1 is recruited to phagosomes (11, 12), and autophagic components may connect to lysosomal compartments during bacterial infection (33) and in vesicular trafficking (31). We found that antibodies to markers for early endosomes (EEA1) and late endosomes (LAMP1) did not appear to colocalize significantly with IRGM (Fig. 2 B and C). Using the ER marker calnexin, we observed sparse overlap with IRGM-positive staining (Fig. 2D). In uninfected cells, we observed that IRGM signals produced continuous staining in a twisted ribbon-like network (Fig. 2, Left) resembling the Golgi apparatus. Staining with GM130, a Golgi marker, revealed a striking colocalization of GM130 with IRGM in both resting and HCV-infected cells (Fig. 2E). Mander’s coefficient analysis revealed that the fluorescent signals for IRGM and GM130 overlapped to similarly high extents in uninfected and in HCV-infected cells. We confirmed the localization of IRGM to the Golgi membrane in four additional cell lines (Fig. S6). Collectively, these results suggest that human IRGM is localized mainly to the Golgi membrane in several human cell lines. Interestingly, we also observed that the staining pattern of GM130 was changed markedly upon HCV infection, and discrete tubulovesicular structures were dispersed into discrete vesicles (Fig. 2E). Previously, an increased level of Golgi fragmentation in HCV-infected cells was observed (34), but the cellular factors implicated in this process have not been addressed.
Fig. 2.
IRGM localizes to the Golgi apparatus in uninfected (Med) and in HCV-infected hepatocytes. (A–E) Intracellular staining of IRGM in combination with the following subcellular markers: TOM20 (A), EEA1 (B), LAMP1 (C), Calnexin (D), and GM130 (E). The dotted lines in the left subpanels mark the nucleus. White boxes in the left subpanels mark the zoomed area shown in the right subpanels. The cells were examined by confocal microscopy followed by Mander’s coefficient analysis. Data represent the means ± SD of ∼200 cells for each subcellular marker from two independent experiments.
Fig. S5.
IRGM localization to mitochondria in different cell types. Human macrophages were prepared by differentiation with M-CSF. Intracellular staining of IRGM together with MAVS, a subcellular marker for mitochondria in different cell lines. Cells were fixed with 4% PFA, blocked, and immunostained with antibodies against IRGM and MAVS. The cells were examined by confocal microscopy followed by Mander’s coefficient analysis. Data are shown as mean ± SD from two independent experiments. (Scale bars: 5 µm.)
Fig. S6.
IRGM localization to the Golgi apparatus in different cell types. Intracellular staining of IRGM together with the Golgi marker GM130 in different cell lines. Cells were fixed with 4% PFA, blocked, and immunostained with antibodies against IRGM and GM130. The cells were examined by confocal microscopy (magnification: 63×) followed by Mander’s coefficient analysis. Data are shown as means ± SD from two independent experiments. Med, uninfected hepatocytes.
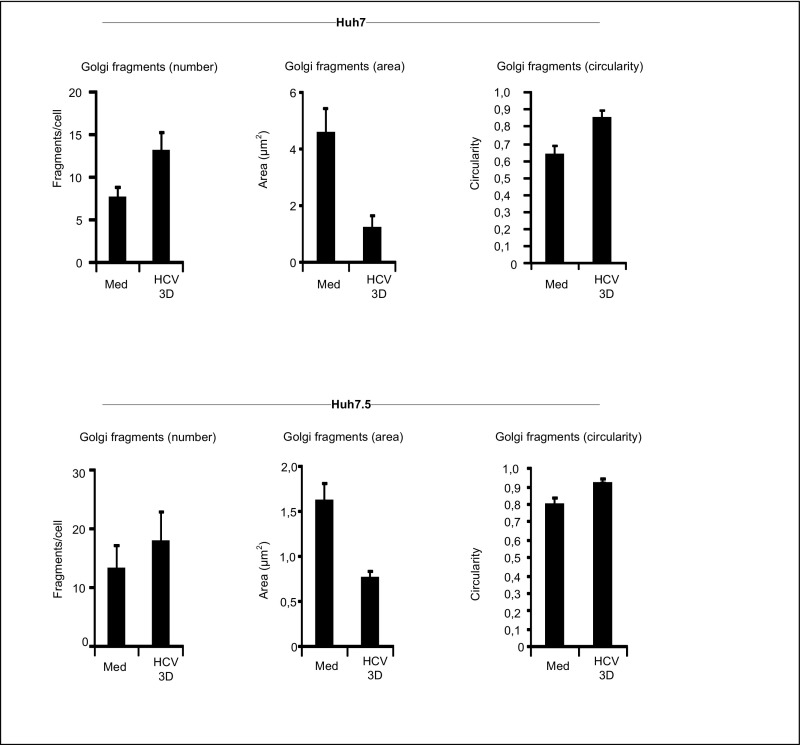
We next performed a detailed analysis and quantification of the dispersed Golgi phenotype observed by confocal imaging. This assessment revealed a marked increase in Golgi fragments in HCV-infected cells from day 2 p.i. (Fig. 3 A and B). Also, the area of the Golgi fragments was reduced, and the circularity of Golgi fragments increased upon HCV infection (Fig. 3 A and B). A very similar Golgi fragmentation pattern was observed in HCV-infected Huh7 and Huh7.5 cells (Fig. S7), thus excluding the possibility that Golgi fragmentation is a phenotype specific for the Huh7.25CD81 cell line. Notably, upon costaining of HCV-infected cells with IRGM and HCV core protein, we found that tubulovesicular structures were present in uninfected (HCV core-negative) cells, whereas clustered vesicles were observed in HCV-infected (core-positive) cells (Fig. 3C). This finding strongly suggests that the change in Golgi morphology between the uninfected and HCV-infected cells is caused by the viral infection. Because we found that IRGM localized to the Golgi apparatus, we next examined if IRGM contributed to Golgi fragmentation. Cells were treated with siIRGM or control siRNA before HCV infection and further analysis. Importantly, depletion of IRGM resulted in reduced numbers of Golgi fragments per cell, corresponding to the increased Golgi fragment area in HCV-infected cells relative to that in cells treated with control siRNA (Fig. 3D). To strengthen our results, we next assessed Golgi fragmentation in two of the CRISPR/Cas9-based IRGM-knockout clones. Indeed, we found that HCV-stimulated Golgi fragmentation was strongly reduced in the IRGM-deleted cells compared with the sgRenilla control cells (Fig. 1E). These results confirmed our data obtained with two different siIRGMs. Collectively, our results show that IRGM is localized to the Golgi apparatus and is required for HCV-triggered fragmentation of the Golgi membrane.
Fig. 3.
HCV stimulates IRGM-dependent fragmentation of Golgi membranes. (A) Representative images of uninfected or HCV-infected (6 d) cells immunostained with an antibody against GM130. (B) Quantification of Golgi fragments. (C) Representative images of cells immunostained with antibodies against IRGM and HCV Core. The ÷ symbol indicates a noninfected cell; “1” and “2” indicate the locations of the enlarged boxes. (D, Left) Representative images of cells treated with two different siRNAs against IRGM or with control siRNA and immunostained with an antibody against GM130. (Right) Quantification of Golgi fragments. (E, Left) Representative images of Huh7.25CD81 sgIRGM.100, sgIRGM.103, and control sgRenilla cells immunostained with antibodies against GM130 and quantification of Golgi fragments. The dotted lines in A and C–E mark the nucleus. (Right) Quantification of Golgi fragments. Data are shown as means ± SD for 20 cells in each condition calculated from two independent experiments; *P < 0.05, **P < 0.005, ***P < 0.0005, paired t test. (Scale bars: 5 μm.) Med, uninfected hepatocytes.
Fig. S7.
HCV-induced Golgi fragmentation in Huh7 and Huh7.5 cells. Huh7 and Huh7.5 cells were infected with HCV (MOI 0.3) for 3 d. Cells were fixed with 4% PFA, blocked, and immunostained with antibodies against GM130 and NS5A. The NS5A staining was used to include only infected cells in the calculations for HCV at 3 d. The number of Golgi fragments (defined as the number of GM130-positive structures per cell), the mean area of Golgi fragments, and the circularity of the Golgi fragments were calculated in 20 cells for each data point using ImageJ software. Data represent means ± SD from three independent experiments. Med, uninfected hepatocytes.
GBF1 and Arf1 Regulate HCV-Triggered Golgi Fragmentation.
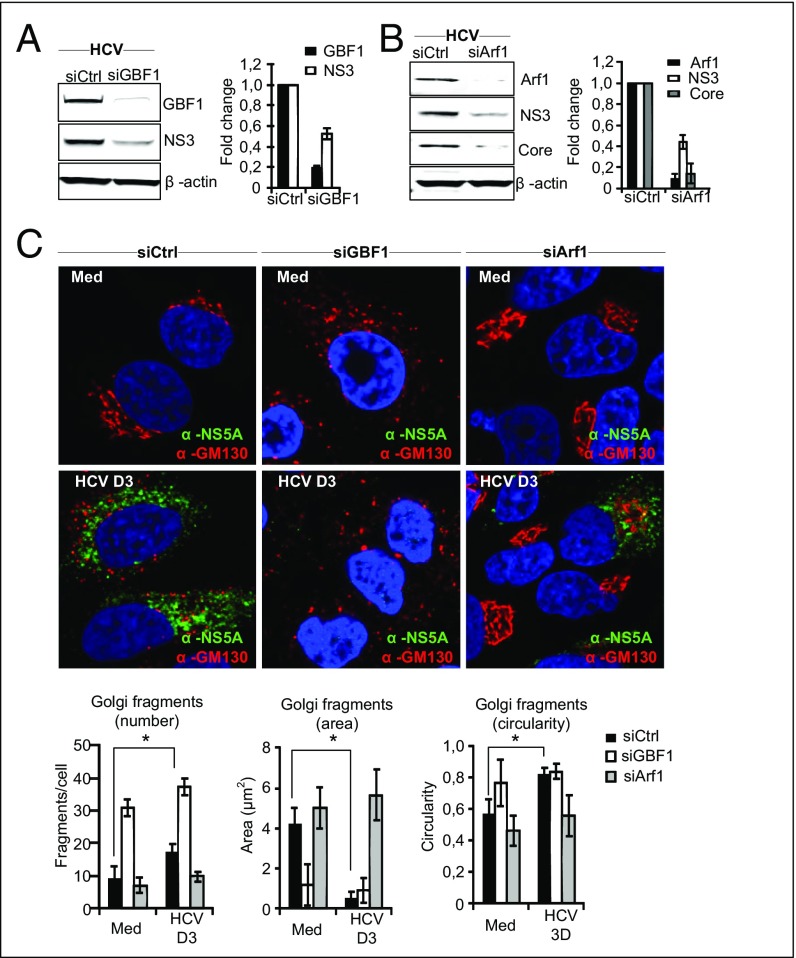
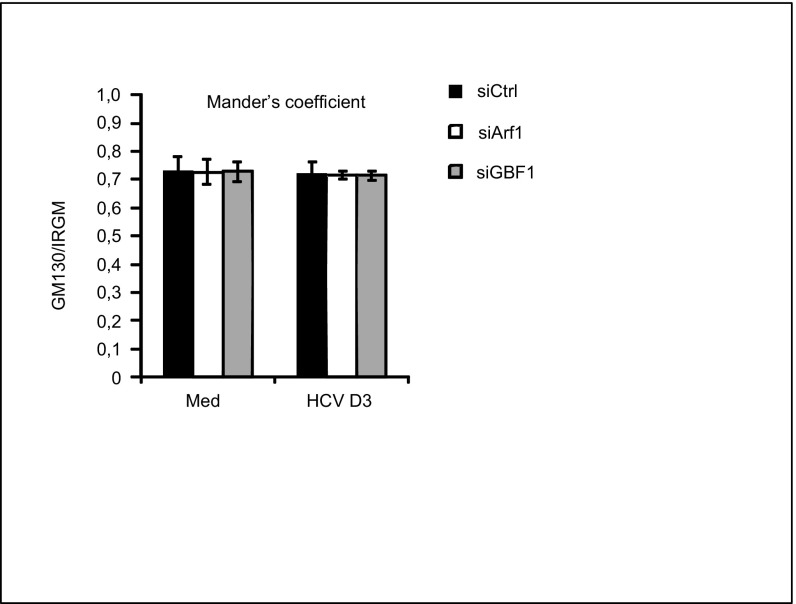
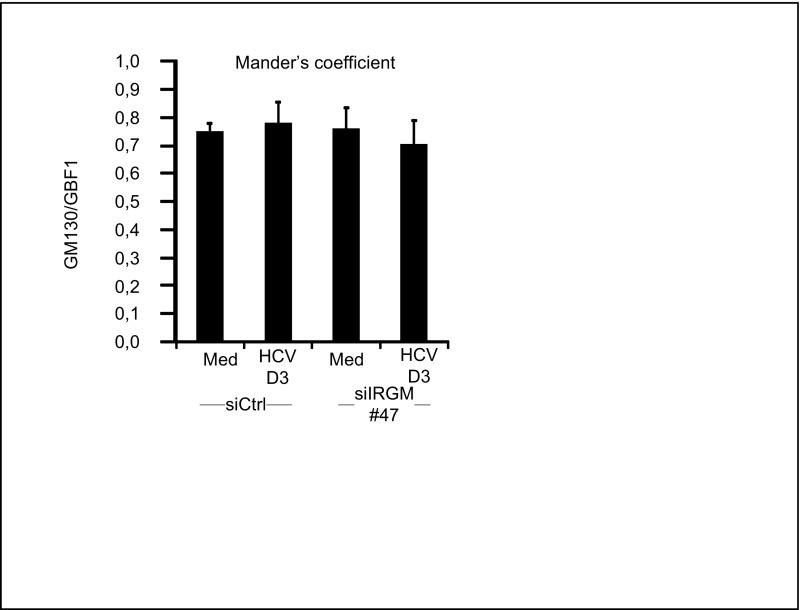
GBF1 and Arf1, proteins that conserve Golgi maintenance and function, are known to be crucial for the replication of certain RNA viruses, such as enteroviruses (17), possibly by recruitment of PI4KIIIβ to viral replication sites. Indeed, recent data have revealed that the Golgi membrane and Golgi proteins may have a previously unappreciated role in the replication of Enterovirus and Flavivirus (35, 36). Moreover, GBF1 and Arf1 have been suggested to be host factors for HCV (18–20). To determine the role of GBF1 and Arf1 in relation to Golgi fragmentation, hepatocytes were depleted of GBF1 and Arf1 by siRNA transfections before HCV infection. Reduction of GBF1 or Arf1 decreased HCV protein levels (Fig. 4 A and B), as is consistent with these proteins having a role as HCV host factors. Next, we examined the impact of depleting GBF1 or Arf1 on HCV-triggered Golgi fragmentation. Cells transfected with GBF1 or Arf1 siRNAs were costained with antibodies against GM130 and NS5A. Staining of the HCV nonstructural 5A (NS5A) protein was used as a marker to identify HCV-infected cells. Confocal analysis and quantification of the Golgi phenotype revealed that reduction of GBF1 led to a marked increase in circular and small Golgi fragments, even in noninfected cells (Fig. 4C), as is consistent with GBF1 having a role in the maintenance of the Golgi structure (37). HCV failed to induce significant changes in Golgi dispersion in siArf1-treated cells compared with siControl-treated cells (Fig. 4C). Thus Arf1 is required for HCV-induced Golgi fragmentation. The localization of IRGM to the Golgi was unaffected by siArf1 and siGBF1 treatments (Fig. S8), precluding an effect of siGBF1 and siArf1 on binding of IRGM to the Golgi. Taken together these results show that GBF1 and Arf1 are essential host factors for HCV and that their function as such may be related to Golgi fragmentation.
Fig. 4.
GBF1 and Arf1 control Golgi fragmentation induced by HCV infection. (A and B) Cells were transfected with siRNAs against GBF1, Arf1, or control siRNA. GBF1 and Arf1 knockdown were controlled by immunoblotting. HCV protein levels after GBF1 or Arf1 knockdown were analyzed by immunoblotting against NS3 or Core proteins. Data from one representative experiment are shown. Fold change values have been calculated relative to siControl (noninfected) cells and are normalized to β-actin. (C) Representative images of cells depleted of GBF1 or Arf1 and immunostained with antibodies against GM130 and NS5A or DAPI (blue) to mark the nucleus (magnification: 63×). The characteristics of Golgi fragments were calculated from two independent experiments as described in Materials and Methods. Data are shown as means ± SD; *P < 0.05, paired t test. Med, uninfected hepatocytes.
Fig. S8.
Golgi localization of IRGM in Huh7.25CD81 cells depleted of GBF1 or Arf1. Huh7.25CD81 cells were transfected with siRNAs against Arf1 and GBF1 for 6 d. Cells were fixed with 4% PFA, blocked, and immunostained with antibodies against GM130 and IRGM. The cells were examined by confocal microscopy followed by Mander’s coefficient analysis. Data are shown as mean ± SD from three independent experiments. Med, uninfected hepatocytes.
IRGM Directs HCV-Stimulated Phosphorylation of AMPK and GBF1.
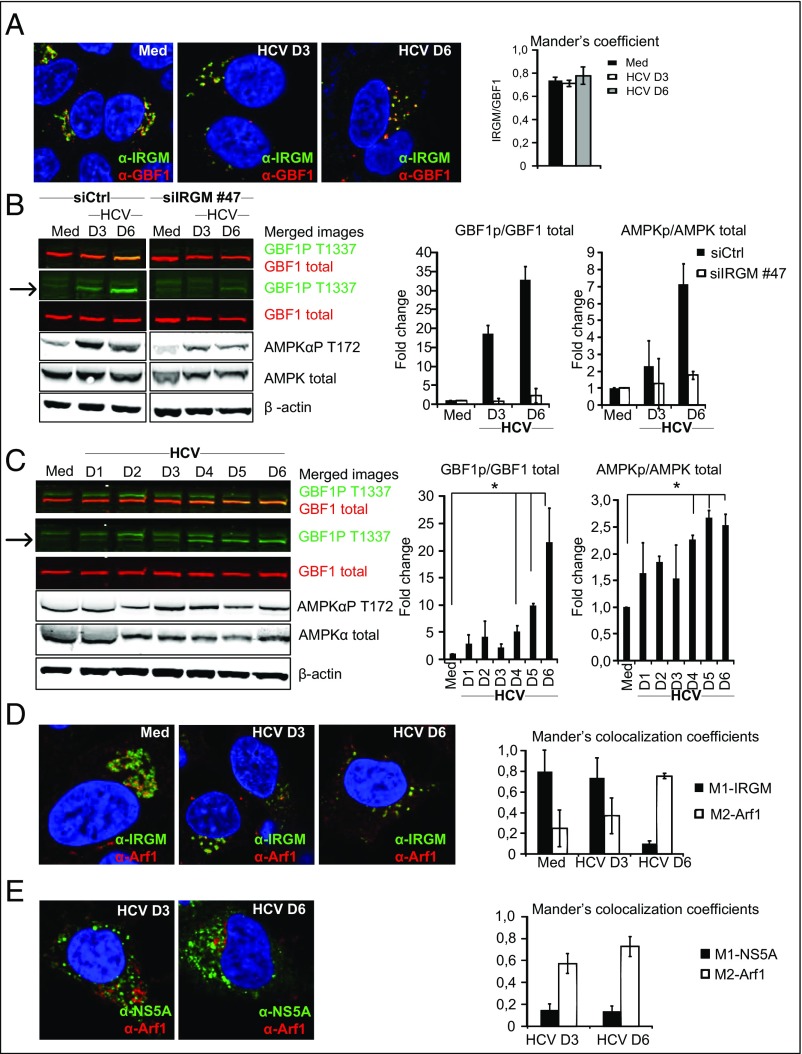
GBF1 localizes to the Golgi complex (38, 39). To explore the relationship between IRGM and GBF1, we initially examined the colocalization of these proteins. We found that IRGM and GBF1 colocalized to a high extent in both uninfected (intact Golgi structure) and HCV-infected (fragmented Golgi) cells, as assessed by Mander’s coefficient analysis (Fig. 5A). Next, we examined if IRGM affected GBF1. It recently was found that in normal cells undergoing mitosis Golgi fragmentation is induced by AMPKα-dependent phosphorylation of GBF1 at T1337 (40, 41). Another study showed that IRGM stimulates AMPKα by stabilization of the kinase in its T172 phosphorylated form (16). Hence, we examined the effect of siRNA-mediated depletion of IRGM on HCV-triggered phosphorylation of GBF1T1337 and AMPKαT172, phosphorylation events that mediate the activation of AMPK and GBF1. We found that siRNA-mediated depletion of IRGM efficiently reduced HCV-triggered phosphorylation of GBF1T1337 and AMPKT172 (Fig. 5B), but depletion of IRGM did not influence the expression levels of total GBF1 or AMPKα. We ensured that the siRNA-mediated reduction of IRGM did not affect the localization of GBF1 to the Golgi apparatus (Fig. S9) in noninfected or in HCV-infected cells. Therefore, we suggest that the impact of IRGM on GBF1 is mediated by AMPKα activation rather than by affecting the Golgi residence of GBF1. Next, we examined kinetics of phosphorylation and found that HCV induced phosphorylation of GBF1T1337 starting between 3 and 4 d p.i. (Fig. 5C). We observed a strong increase in GBF1 phosphorylation, and this increase correlated well with the kinetics of AMPKα phosphorylation, which was stimulated from day 3 p.i. (Fig. 5C). Throughout the time course, the total amount of GBF1 and AMPKα remained unchanged (Fig. 5C). Also, the presence of phosphorylated GBF1 at later times in HCV infection correlated well with the increased amount of Golgi fragments at these times (Fig. 3B), supporting the role of phosphorylated GBF1 in Golgi dispersion by HCV. This finding demonstrates that HCV stimulates the phosphorylation of GBF1. Overall, our results suggest that HCV uses IRGM-AMPKα to induce the phosphorylation of GBF1 at T1337, presumably as a signaling axis of HCV-triggered Golgi fragmentation. Notably, our imaging data showed that in HCV-infected cells GBF1 remains localized to fragmented Golgi structures, whereas our immunoblot analysis shows that GBF1 is phosphorylated at T1337 during HCV infection (Fig. 5 A and C). This result corroborates a previous study reporting that GBF1 colocalizes with Golgi markers both in control cells and in cells treated with inducers of AMPK activation to cause Golgi fragmentation (42).
Fig. 5.
IRGM mediates GBF1 phosphorylation and associates with Arf1 at HCV replication sites. (A) Representative images of cells immunostained with antibodies against IRGM and GBF1 (magnification: 63×). DAPI (blue) staining marks the nucleus. The MOC represents observations from three independent experiments. (B) The effect of siIRGM on GBF1 and AMPKα phosphorylation was examined by immunoblotting. A merged image in the first row reveals overlap of signals (seen as orange) between phosphorylated (green) and total GBF1 (red) protein levels. The band corresponding to phospho-GBF1 (GBF1PT1337) is indicated by an arrow. GBF1PT1337 or AMPKαPT172 protein levels were normalized to total GBF1 or AMPKα protein levels. (C) The effect of HCV infection on GBF1 and AMPKα phosphorylation were examined by immunoblotting as in B. (D and E) Representative images of cells immunostained with antibodies against IRGM and Arf1 (D) or NS5A and Arf1 (E). (Magnification: 63×.) DAPI staining (blue) marks the nucleus. The MCC quantification method was used because of the high level of IRGM or NS5A protein compared with the Arf1 protein level. Data shown are the mean ± SD of three independent experiments; *P < 0.05, paired t test. Med, uninfected hepatocytes.
Fig. S9.
Golgi localization of GBF1 in Huh7.25CD81 cells depleted of IRGM. The effect of siGBF1 or siArf1 on the intracellular localization of IRGM to Golgi was examined. Huh7.25CD81 cells were transfected with siRNAs against IRGM or control siRNA and were infected with HCV (MOI 0.3) for the indicated number of days. Cells were fixed with 4% PFA, blocked, and immunostained with antibodies against GM130 and IRGM and were examined by confocal microscopy followed by Mander’s coefficient analysis. Data are shown as mean ± SD from three independent experiments. Med, uninfected hepatocytes.
HCV Redirects Arf1 to IRGM-Containing Vesicles and HCV Replication Sites.
Having established that IRGM is necessary for phosphorylation of GBF1, which is an activator of the small GTPase Arf1, we next examined whether Arf1 was present together with IRGM-containing membranes. Arf1 is a small GTPase that exists in a GDP-bound cytosolic form and in a GTP-bound form that is mainly associated with the Golgi. Cells were infected with HCV and stained with antibodies against IRGM or Arf1 at 3 or 6 d p.i. In uninfected cells, both IRGM and Arf1 showed a ribbon-like Golgi pattern (Fig. 5D). In HCV-infected cells, IRGM and Arf1 displayed a dispersed, fragmented pattern. To assess the IRGM–Arf1 interplay in more detail, we quantified the fraction of IRGM that colocalizes with Arf1 and vice versa. To do so, we used Mander's colocalization coefficients (MCC) M1 and M2. MCC strictly measures the cooccurrence of proteins independently of signal proportionality and was useful because the IRGM signal was more intense than that of Arf1. We found that 20% of the total fraction of Arf1 colocalizes with IRGM (M2-Arf1 in Fig. 5D, Right). Interestingly, the fraction of Arf1 that colocalized with IRGM was markedly greater in HCV-infected cells than in uninfected cells, showing that around 80% of Arf1 was localized with IRGM at 6 d p.i. (Fig. 5D, Right). Hence, our results suggest that HCV infection induces an increased association of Arf1 with fragmented Golgi membranes and IRGM.
Arf1 is a host factor for HCV [Fig. 4B and other studies (18, 20)], has been suggested to generate a PI4P-eniched environment supportive of HCV replication by recruiting effectors such as PI4KIIIβ to Golgi membranes (18, 43, 44), and is associated with Golgi fragments (45). Based on these data and because our results suggest that Golgi fragments might contribute to HCV replication, we next examined if Arf1 is present at replication sites in HCV-infected cells. Accordingly, we examined the colocalization of Arf1 with the HCV NS5A protein (an essential component of the HCV replication complex) in cells infected with HCV for 3 or 6 d. Because of the high signal level of NS5A signal intensity compared with that of Arf1, the colocalization was quantified using MCCs M1, which measures the fraction of protein A in compartments containing protein B, and M2, which measures the fraction of protein B in compartments containing protein A. We found that up to 80% of Arf1 colocalized with NS5A, with a slight increase with time p.i. (Fig. 5E). This result could suggest that Arf1 is targeted to HCV replication sites. Therefore we speculate that HCV stimulates IRGM-dependent Golgi fragmentation and thereby facilitates the association of Arf1-containing vesicles with HCV replication sites.
Golgi-Derived Vesicles Associate with ER Membranes at HCV Replication Sites.
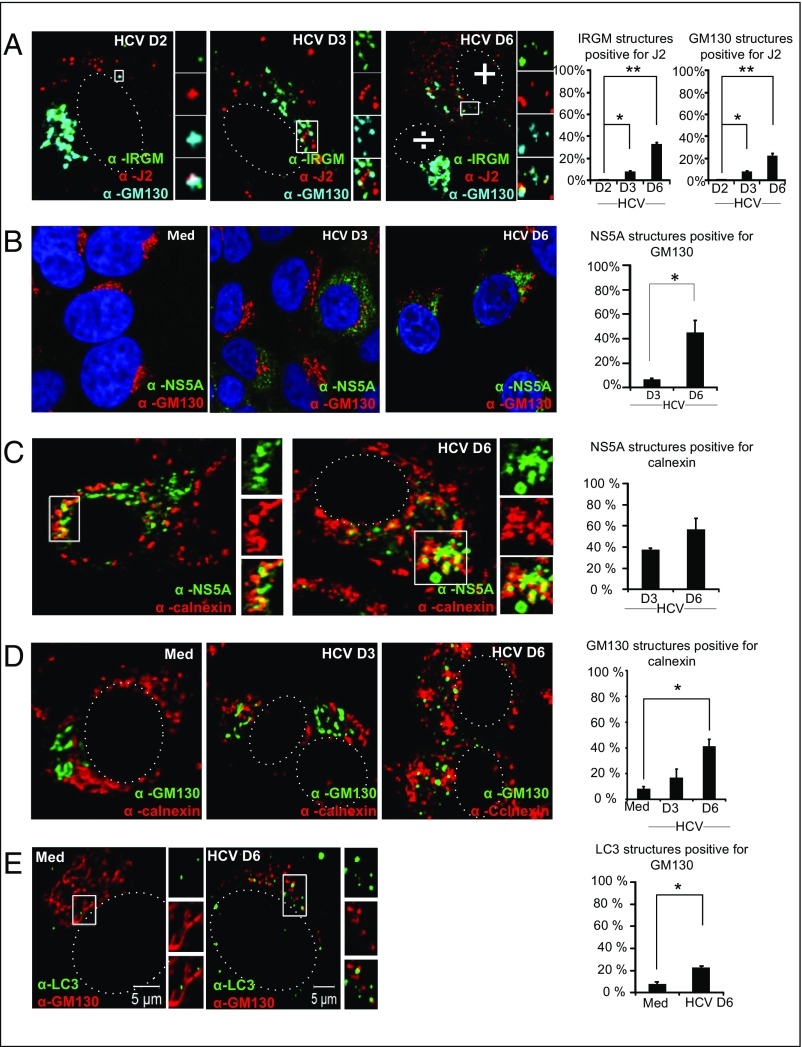
To probe if fragmented Golgi membranes are found in connection with viral replication sites, we examined the localization of IRGM and Golgi fragments in relation to newly generated HCV RNA. Cells were costained with J2 (a monoclonal antibody that binds to dsRNA) and with IRGM and GM130 antibodies. The confocal imaging showed an increase in the number of Golgi vesicles and IRGM-positive structures that were colocalized with dsRNA-positive structures at 6 d p.i. (Fig. 6A), thus indicating the relocation of IRGM and Golgi vesicles within close proximity with HCV RNA. Although the precise localization of the HCV replication complex at the different membranous structures that constitute the MW has not yet been explicitly determined, these clustered vesicles morphologically resembled structures that have previously been identified as membrane-bound VRCs for HCV (1). To confirm and extend our data, we examined the localization of Golgi fragments with NS5A, which is used as a marker for viral replication. The increase we observed in the NS5A structures positive for the Golgi marker GM130 (Fig. 6B) strongly suggests that HCV replication is associated with IRGM and Golgi fragments.
Fig. 6.
Golgi and IRGM relocalize to viral replication sites and to the ER in response to HCV infection. (A) Representative images of cells immunostained with antibodies against IRGM, J2 (double-stranded RNA), and GM130. White boxes mark the zoomed area. The ÷ symbol indicates a noninfected cell, and the + symbol indicates an infected cell. (B–E) HCV NS5A and GM130 (B), HCV NS5A and Calnexin (C), GM130 and Calnexin (D), and LC3 and GM130 (E). All colocalization quantifications were done using the JaCoP (66) plugin in ImageJ. (Magnification: 63×.) Data represent means ± SD; 20 cells for each data point were calculated in two independent experiments; *P < 0.05, **P < 0.005, paired t test. Med, uninfected hepatocytes.
Previous results have suggested that HCV replication at the MW occurs in association with the ER membrane (1, 2). To address this association in our cellular model of HCV infection, we examined the localization of calnexin, an ER marker, with NS5A, a marker for HCV replication sites. We observed an increased association between NS5A and the ER marker, with ∼60% of NS5A structures staining positive for calnexin at day 6 p.i. (Fig. 6C). Because our results indicated that Golgi membranes could contribute to the HCV replication complex, we next examined if HCV affected the colocalization of Golgi and ER membranes. HCV infection led to a marked increase in Golgi structures positive for ER staining (Fig. 6D), suggesting that HCV infection leads to the relocation of Golgi fragments to membranes derived from the ER.
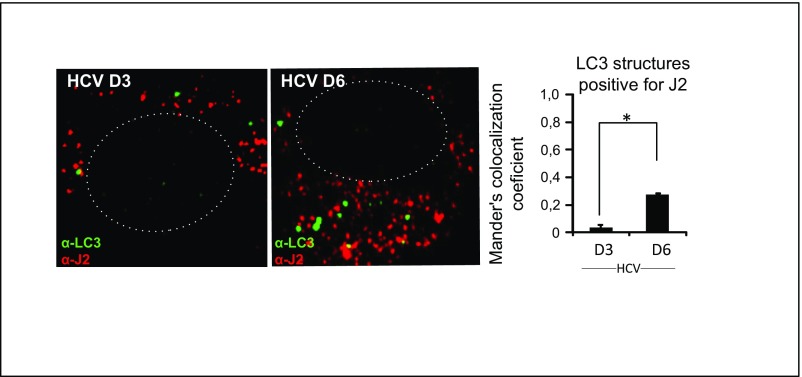
We found that HCV induces both Golgi fragmentation and autophagy. It has been proposed that autophagosomes may be generated from Golgi membranes (46). Therefore we explored if autophagosomes were present with Golgi-derived vesicles. Indeed, HCV infection resulted in a significant increase in LC3 dots that overlapped with GM130 staining in cytoplasmic vesicles that were juxtaposed to the nucleus (Fig. 6E). Moreover, we found that a significant number of HCV-induced LC3 dots at 6 d p.i. were positive for the dsRNA marker J2 (Fig. S10). These results indicate that viral replication intermediates can be located transiently at the autophagosomal membranes and could suggest that autophagosomal membranes are components of the MW, corroborating a previous report (4).
Fig. S10.
Colocalization of LC3 and the dsRNA marker J2 in Huh7.25CD81 cells. Huh7.25CD81 cells were infected with HCV (MOI 0.3) for the indicated number of days. Cells were fixed with 4% PFA and were blocked and immunostained with antibodies against LC3 and J2, a marker for dsRNA. Representative images are shown. The dotted lines mark the nucleus. The cells were examined by confocal microscopy followed by Mander’s coefficient analysis. Data are shown as the mean ± SD from two independent experiments. *P < 0.05. (Magnification: 63×.)
Discussion
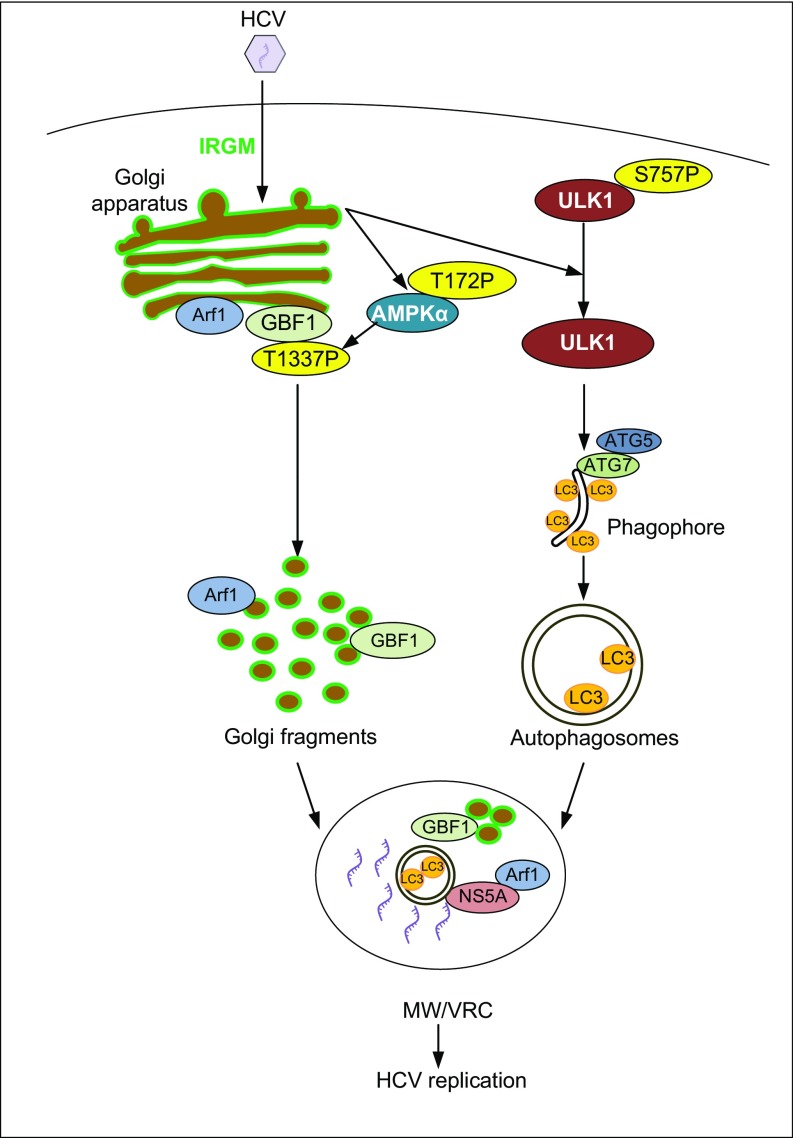
Human IRGM regulates autophagy (16) and has been identified as a risk factor for Crohn’s disease and tuberculosis risk. Recently, IRGM was reported to interact with proteins from RNA viruses, including HCV (15), thereby promoting HCV replication. However, the mechanism by which IRGM supports HCV replication is unknown. Here we provide insights into the mechanism of HCV MW formation and show that HCV exploits IRGM for autophagy and viral replication (summarized in Fig. 7). Importantly, we show that IRGM is required for the fragmentation of Golgi through the regulation of AMPKα and GBF1, a guanine nucleotide exchange factor (GEF) for Arf-GTPases that normally operates in Golgi membrane dynamics in resting cells. Moreover, IRGM regulates HCV-triggered activation of ULK1, one of the most upstream regulators of autophagosome formation.
Fig. 7.
Proposed model for the role of IRGM in Golgi fragmentation, autophagy, and viral replication. Based on our results, we propose that HCV exploits Golgi-situated IRGM (in green) to regulate two important pathways required for HCV replication. IRGM regulates the phosphorylation of ULK1 to enhance the formation of autophagosomes. Also, HCV induces phosphorylation of AMPKα and GBF1 through IRGM to trigger Golgi fragmentation. We propose that Golgi fragments and autophagosomes contribute to the formation of MW and VRCs. Arf1 associates with IRGM-containing vesicles that associate with NS5A at the HCV replication sites, possibly to recruit effector proteins required for efficient HCV replication.
We found that human IRGM was localized almost exclusively to Golgi membranes, both in resting and HCV-infected hepatocytes and in other human cells examined. Previously, it was suggested that human IRGM localizes to the mitochondria and directs autophagy by promoting depolarization and fission of mitochondria (14). In contrast, mouse Irgm1 is found in the Golgi apparatus and on the endolysosomal system and stains only weakly positive for mitochondrial markers (31, 32). The reason for the discrepancy in human IRGM localization between our study and the studies by Singh et al. (13, 14) is unknown but could relate to the different experimental approaches and cells studied. Singh and coworkers mainly studied overexpressed GFP-tagged IRGM and endogenous IRGM in macrophages, which could differ from endogenous IRGM in hepatocytes. Of note, it has been reported that the Golgi localization of mouse Irgm1 was lost upon N- or C-terminal tagging of IRGM with EGFP (31), and this tagging likewise might impact the localization of human IRGM.
We observed a marked fragmentation of the Golgi apparatus during HCV infection. Flaviviruses, such as HCV, dengue virus, and West Nile virus, induce massive cellular membrane rearrangements during infection. Previous studies have considered ER-derived membranes as being the main membrane source supporting HCV and dengue virus replication. Extending these studies, our results suggest that IRGM-containing Golgi membranes contribute to the generation of the MW formed upon HCV infection. This suggestion is based on an increased colocalization of Golgi fragments and IRGM with NS5A and dsRNA, markers for HCV replication sites. In line with this suggestion, we found enhanced colocalization of Golgi fragments with ER membranes in response to HCV infection. We propose that this increased association between the ER and Golgi membrane compartments reflects the contribution of both types of membranes to the formation of the HCV MW/VRC. In accordance with this notion a recent study showed that the Golgi-associated protein ER13 is a dengue virus host factor that relocates to ER membranes during viral infection (36). These results demonstrate that Golgi-derived membranes/proteins are important for replication of Flaviviruses other than HCV and highlight the complexity of the cellular membrane structures supporting viral replication. Similarly, other studies of membrane rearrangements during infection with Kunjin and West Nile virus suggest that Golgi proteins are indeed recruited to replication sites (36, 47, 48). Moreover, based on electron microscopy studies, it was suggested that Golgi membranes are the initial sites of poliovirus replication complexes (35). In this context our results highlight a previously unexplored role for IRGM as a Golgi protein that contributes to HCV replication.
Although it is known that other RNA viruses (e.g., Picornaviruses such as rhinovirus) make use of similar rearrangements of Golgi membranes to support the formation of VRCs (49, 50), the mechanisms by which Golgi fragmentation occur are not well understood for any virus. Addressing this issue, we found that HCV infection triggers the phosphorylation of AMPKα and GBF1 at T1337 in an IRGM-dependent manner. Although it was previously reported that Golgi dispersal is induced by AMPKα-dependent phosphorylation of GBF1 at T1337 during mitosis (40, 41), this mechanism has not previously been known to be used by viruses. During Enterovirus infections GBF1 is known to be recruited to VRCs by the poliovirus 3A protein (51), but it is unclear how GBF1 functions in viral replication. GBF1 is normally required for the assembly of the Coat protein I (COPI) vesicle at the ER–Golgi complex interface, and GBF1 activates Arf proteins. Arf1 is required for coatomer binding and bud formation on Golgi membranes to generate vesicles (52). We observed that GBF1 was phosphorylated during HCV infections up to 6 d p.i., and we suggest that this phosphorylation may lead to prolonged Arf1 activation. Indeed, this notion is supported by our findings that the association of Arf1 with IRGM-containing vesicles was enhanced at day 6 of HCV infection (Fig. 6A) and is correlated with increased Golgi fragmentation. Others have shown that Arf1 membrane targeting depends on active GBF1 to induce the Arf1–GTP binding (39). Our results also show that most of the Arf1 protein expressed was associated with VRCs in HCV-infected cells. We propose that the fragmentation of Golgi into vesicles and the localization of Arf1 to vesicular membranes might promote the recruitment of Arf1 to VRCs or supply suitable membrane components to support replication. It will be interesting in future studies to explore if IRGM contributes to the replication of other viruses using Golgi membranes during steps of their replication cycle. Of note, it was proposed recently that the GBF1 interactome is altered in virally infected cells compared with uninfected cells (53), and identifying the interaction partners may elucidate these GBF1-dependent mechanisms in the future.
We speculate that IRGM acts in concert with other membrane-remodeling proteins to achieve Golgi membrane fragmentation. The rationale for this suggestion is that IRGM belongs to the IFN-inducible GTPase superfamily that is part of the dynamin-like family of proteins. This family consists of membrane-remodeling GTPases and is known to direct the budding and fusion of transport vesicles (54). IRGM is one of the smaller IRG proteins that, because of low intrinsic rates of GTP hydrolysis, have been suggested to make use of other GEFs to drive membrane remodeling (54). Furthermore, it has been proposed that the IRG subclass to which IRGM belongs comprises intrinsic regulators of membrane-shaping factors, controlling respective effectors (e.g., SNARE adaptors and fission and autophagy proteins) (54). In our study, we found that the IRGM colocalized with the GEF GBF1, which is known to function in Golgi fragmentation (17), at the Golgi membrane and mediated GBF1 phosphorylation. Based on this finding, we propose that IRGM and GBF1 may cooperate and stimulate Golgi fragmentation together with the GBF1 effector Arf1.
In addition to its being required for bud formation on Golgi membranes to generate vesicles (52) another function of Arf1 that has been proposed to support viral replication is the recruitment of PI4KIIIβ to Golgi membranes to promote the production of PI4P (55). It was reported that poliovirus (another positive-stranded RNA virus) assembles its VRCs on the Golgi network through the interaction of the viral replication protein 3A with Arf1 and GBF1. This interaction preferentially led to recruitment of PI4KIIIβ rather than vesicle coat proteins to membranes forming the VRC (17). Hence, host proteins that normally are present at these membrane organelles in uninfected cells are replaced by the effector PI4KIIIβ in poliovirus-infected cells, leading to membrane reorganization and to the enrichment of PI4P at these sites. Likewise, PI4P is important for HCV replication (2), and, although the suggestion is still controversial, both PI4KIIIα (2, 56, 57) and PI4KIIIβ (17, 58) may be involved in generating PI4P to support HCV replication.
Our finding that IRGM orchestrates the formation of HCV replication membranes provides unique insights into the replication mechanism of an important human pathogen. It is possible that viral targeting of IRGM for the benefit of replication is a common theme for viruses that modify the Golgi membranes, where IRGM is situated. Influenza virus replicates in the nucleus and does not depend on Golgi-membrane fragmentation for the formation of VRCs. Interestingly, Grégoire et al. (15) found that influenza virus, although modulating the autophagic pathway, did not require IRGM for its replication. We propose that this finding could be attributed to the Golgi-independent replication strategy of influenza virus. Overall, it is possible that IRGM may be important for additional viral or bacterial pathogens that depend on the fragmentation of the Golgi apparatus and/or on the acquisition of lipids from host cell membranes, such as Chlamydia trachomatis, Toxoplasma gondii, picornavirus, and rhinovirus (59–61).
Materials and Methods
Reagents.
The HCV-NS5A antibody was provided by the T.W. Laboratory. Other antibodies used are given in Table S1.
Table S1.
Overview of antibodies used in this study
| Antibodies | Supplier | Catalog no. |
| AMPKα | Cell Signaling Technology | #5832 |
| AMPKα-P | Cell Signaling Technology | #2531 |
| Arf1 | SantaCruz | sc-53168 |
| ATG5 | Cell Signaling Technology | #2630 |
| ATG7 | Cell Signaling Technology | #2631 |
| Calnexin | Abcam | ab31290 |
| EEA1 | BD Bioscience | 610456 |
| GBF1 | BD Bioscience | 612116 |
| GBF1-P | Immuno-Biological Laboratories | 28065 |
| GM130 | BD Bioscience | 610822 |
| GM130-A647 | BD Bioscience | 558712 |
| HCV-Core | SantaCruz | sc-57800 |
| HCV-NS3 | Abcam | ab13830 |
| HCV-NS5A | BioFront | HCV-2F6 |
| IRGM | Abcam | Ab69494 |
| J2 | Scicons | J2-1406 |
| LAMP1 | BD Bioscience | 555798 |
| LC3 | Cell Signaling Technology | #3868 |
| SQSTM1 | BD Bioscience | 610832 |
| TOM20 | Santa Cruz | sc-17764 |
| ULK1 | Cell Signaling Technology | #8054 |
| ULK1S757 | Cell Signaling Technology | #6888 |
| β-actin | Sigma | A1978 |
Cell Culture, Transfection, and Virus.
Huh7.25CD81 (62, 63) cells were maintained in DMEM supplemented with (vol/vol) 10% FBS, 1% nonessential amino acids, 20 µg/mL gentamicin, and 0.4 mg/mL geneticin (G418) and were cultured at 37 °C in 5% CO2. Huh7 cells were cultured in the same medium without G418. JFH1 HCV stocks were prepared and titrated as previously described (64). HCV infections were performed with an MOI of 0.3 in complete medium for 2 h before the removal of virus and incubation for the indicated time points.
RNAi.
The following siRNA oligonucleotides were used: ATG7 (Dharmacon; L-020112-00-0005), ULK1 (Dharmacon; J-005049-06-0020), ULK2 (Dharmacon; LQ-005396-00-0005), ATG5 (Cell Signaling Technology; 6345), IRGM (Ambion; s51146 and s51147), Arf1 (Dharmacon; J-011580-06-0005), GBF1 (Dharmacon; J-019783-06-0005), and AllStar negative control siRNA (Qiagen). siRNA duplexes were reverse transfected into cells using Lipofectamine RNAiMAX (Invitrogen; 17-0618-0) according to the manufacturer`s instructions. Huh7.25CD81 cells were trypsinized 72 h post transfection, and a second (repeated) reverse transfection was performed before analysis of HCV-mediated effects at 6 d p.i.
Immunoblotting.
Following treatment, cells were washed once in PBS and lysed in CHAPS lysis buffer containing 50 mM Tris (pH 7.5), 140 mM NaCl, 5% glycerol, 1% CHAPS, 2 mM EDTA, 40 mM glycerophosphate, 100 mM NaF, 200 µM Na3VO4, 10 µg/mL leupeptin, 1 µM pepstatin, and 1 mM PMSF. Cell lysates were clarified by centrifugation, separated by lithium dodecyl sulfate (LDS)-PAGE, and electrophoretically transferred to nitrocellulose membranes using the iBlot transfer system (Invitrogen, Life Technologies). Membranes were blocked in LI-COR Odyssey blocking buffer at a 1:1 ratio with Tris-buffered saline for 1 h at room temperature before incubation with the indicated antibodies overnight at 4 °C. After three washings with Tris-buffered saline containing 0.25% Tween 20, immunoreactive proteins were detected with LI-COR IRDye secondary antibodies and were visualized on the LI-COR Odyssey system.
Immunofluorescence.
Cells were fixed in PBS containing 4% paraformaldehyde (PFA) for 15 min on ice and were permeabilized with PBS containing 10% FBS and 0.3% Triton X-100. Nonspecific antibody sites were blocked with PBS containing 2.5% BSA and 10% FBS for 30 min at room temperature. For staining, cells were incubated with antibodies diluted in blocking solution. Primary antibodies were added overnight, and secondary antibodies were added for 30 min. Nuclei were visualized by incubation with DAPI or DRAQ5 (1,5-bis{[2-(di-methylamino) ethyl]amino}-4, 8-dihydroxyanthracene-9,10-dione) for 5 min at room temperature. For staining of IRGM proteins, cells were permeabilized with PBS containing 0.01% digitonin for 30 min at room temperature.
Autophagy Assay.
Upon induction of autophagy the cytosolic LC3I form is conjugated to phosphatidylethanolamine to generate LC3II, which binds tightly to autophagosomal membranes. The LC3II form can be monitored as dots by immunofluorescence or by immune blotting. Formation of autophagic vesicles was assessed by endogenous LC3 aggregation. The number of LC3 puncta in LC3-stained cells was determined from 300 cells per condition.
Confocal Fluorescence Microscopy.
Confocal fluorescence microscopy studies were performed with a Zeiss Axiovert 100-M inverted microscope equipped with an LSM 510 laser-scanning unit and a 1.4 NA, 63× Plan-Apochromat oil-immersion objective. Cells were seeded in μ-Slide eight-well, ibiTreat tissue culture-treated plates (Inter Instrument AS). To minimize photobleaching, laser power typically was 20% under maximum, and the pinhole was set to 0.8–1.5. Multitracking was used for dual- or triple-color imaging. Quantitative confocal image analyses were performed using Image J software. The fluorescence intensity of Golgi objects (based on GM130 staining) with or without HCV infection was determined by background subtraction and using a fixed threshold. Then the particle area was quantified using the Analyze Particle plugin in ImageJ (https://imagej.nih.gov/ij).To apply circularity measurements to the Golgi object, a freehand selection option in ImageJ software was used to outline the Golgi based on GM130 staining. Otsu’s thresholding algorithm was applied to convert images to binary images and to create ROIs based on pixel intensity. Circularity index values were assigned to Golgi outlines by the ImageJ circularity plugin in which circularity = 4 π (area/perimeter2). A circularity value of 1 corresponds to a perfect circle (65). Colocalization analysis was performed using the colocalization plugin integrated in the Zen Image software or the JaCoP plugin in ImageJ (66). Image acquisition parameters remained constant during imaging, and threshold values were kept the same from image to image during image analysis. Mander’s overlap coefficient (MOC) and MCC were used. MOC is primarily sensitive to cooccurrence, i.e., the fraction of pixels with positive values for both channels, regardless of signal levels. MCCs measure the fraction of one protein that colocalizes with a second protein: M1 measures the fraction of protein A in compartments containing protein B, and M2 measures the fraction of protein B in compartments containing protein A (67).
CRISPR/Cas9 IRGM-Knockout Cell Lines.
LentiCRISPRv2 plasmid (a gift from Feng Zhang, Broad Institute of MIT and Harvard, Cambridge, MA) (68) (Addgene no. 52961, a kind gift from the Feng Zhang laboratory) was ligated with the following single-guide RNA (sgRNA) oligos targeting the IRGM locus: 5′-CACCGCCAGTTAACATCACTATGGC-3′ and 5′-AAACGCCATAGTGATGTTAACTGGC-3′ for cell_line_P_100 and 5′- CACCGTTCCATCAGGTAGTTCTCCA-3′ and 5′-AAACTGGAGAACTACCTGATGGAAC-3′ for cell_line_P_103. LentiCRISPR_v2 expressing sgRNA 5′-GGTATAATACACCGCGCTAC-3′ targeting sgRenilla (a gift from Giulio Superti-Furga, Center for Molecular Medicine, Vienna was used as control. Second-generation packaging plasmids pMD2.G and psPAX2 were used for producing lentivirus by cotransfecting HEK293T cells with the packaging and lentiCRISPRv2 plasmids and washing the cells after 16 h. Supernatants containing the lentivirus were collected after 48 h, and the Huh7.25CD81 cells were transduced along with protamine sulfate (final concentration, 8 µg/mL). The Huh7.25CD81 clones were selected with puromycin (10 μg/mL) and geneticin/G418 (0.4 mg/mL).
Statistical Analyses.
For quantification of immunofluorescence microscopy images, at least 20 cells were counted for each condition in each experiment. Unless otherwise stated, at least three independent experiments were performed for all figures. Data are shown as mean ± SD. P values were calculated by using paired Student t test, and a P value <0.05 was considered statistically significant.
Acknowledgments
We thank Drs. Marlène Dreux (International Center for Infectiology Research–INSERM U1111-CNRS UMR5308) and Frank van den Kuppeveld (Utrecht University) for critical reading of the manuscript and valuable discussion, and Kristin Rian for excellent technical assistance. Confocal imaging was performed at the Cellular and Molecular Imaging Core Facility at NTNU. This work was supported by the Liaison Committee between the Central Norway Regional Health Authority and NTNU, the Cancer Fund at St. Olav’s Hospital, the Norwegian Cancer Society, the Research Council of Norway (to M.W.A. and CEMIR through Centres of Excellence Funding Scheme Project 223255/F50), an Onsager fellowship from NTNU (to R.K.K.), and Institut Pasteur, Paris.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1616683114/-/DCSupplemental.
References
- 1.Romero-Brey I, et al. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog. 2012;8:e1003056. doi: 10.1371/journal.ppat.1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reiss S, et al. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe. 2011;9:32–45. doi: 10.1016/j.chom.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagy PD, Pogany J. The dependence of viral RNA replication on co-opted host factors. Nat Rev Microbiol. 2011;10:137–149. doi: 10.1038/nrmicro2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferraris P, Blanchard E, Roingeard P. Ultrastructural and biochemical analyses of hepatitis C virus-associated host cell membranes. J Gen Virol. 2010;91:2230–2237. doi: 10.1099/vir.0.022186-0. [DOI] [PubMed] [Google Scholar]
- 5.Ke PY, Chen SS. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J Clin Invest. 2011;121:37–56. doi: 10.1172/JCI41474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiramel AI, Brady NR, Bartenschlager R. Divergent roles of autophagy in virus infection. Cells. 2013;2:83–104. doi: 10.3390/cells2010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamb CA, Yoshimori T, Tooze SA. The autophagosome: Origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14:759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- 8.Ke PY, Chen SS. Autophagy in hepatitis C virus-host interactions: Potential roles and therapeutic targets for liver-associated diseases. World J Gastroenterol. 2014;20:5773–5793. doi: 10.3748/wjg.v20.i19.5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henry SC, et al. Impaired macrophage function underscores susceptibility to Salmonella in mice lacking Irgm1 (LRG-47) J Immunol. 2007;179(10):6963–6972. doi: 10.4049/jimmunol.179.10.6963. [DOI] [PubMed] [Google Scholar]
- 10.MacMicking JD, Taylor GA, McKinney JD. Immune control of tuberculosis by IFN-gamma-inducible LRG-47. Science. 2003;302(5645):654–659. doi: 10.1126/science.1088063. [DOI] [PubMed] [Google Scholar]
- 11.Martens S, et al. Mechanisms regulating the positioning of mouse p47 resistance GTPases LRG-47 and IIGP1 on cellular membranes: Retargeting to plasma membrane induced by phagocytosis. J Immunol. 2004;173(4):2594–2606. doi: 10.4049/jimmunol.173.4.2594. [DOI] [PubMed] [Google Scholar]
- 12.Tiwari S, Choi HP, Matsuzawa T, Pypaert M, MacMicking JD. Targeting of the GTPase Irgm1 to the phagosomal membrane via PtdIns(3,4)P(2) and PtdIns(3,4,5)P(3) promotes immunity to mycobacteria. Nat Immunol. 2009;10(8):907–917. doi: 10.1038/ni.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313(5792):1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 14.Singh SB, et al. Human IRGM regulates autophagy and cell-autonomous immunity functions through mitochondria. Nat Cell Biol. 2010;12(12):1154–1165. doi: 10.1038/ncb2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grégoire IP, et al. IRGM is a common target of RNA viruses that subvert the autophagy network. PLoS Pathog. 2011;7:e1002422. doi: 10.1371/journal.ppat.1002422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chauhan S, Mandell MA, Deretic V. IRGM governs the core autophagy machinery to conduct antimicrobial defense. Mol Cell. 2015;58:507–521. doi: 10.1016/j.molcel.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu NY, et al. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, et al. ARF1 and GBF1 generate a PI4P-enriched environment supportive of hepatitis C virus replication. PLoS One. 2012;7:e32135. doi: 10.1371/journal.pone.0032135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goueslain L, et al. Identification of GBF1 as a cellular factor required for hepatitis C virus RNA replication. J Virol. 2010;84:773–787. doi: 10.1128/JVI.01190-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matto M, et al. Role for ADP ribosylation factor 1 in the regulation of hepatitis C virus replication. J Virol. 2011;85:946–956. doi: 10.1128/JVI.00753-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farhat R, et al. Identification of class II ADP-ribosylation factors as cellular factors required for hepatitis C virus replication. Cell Microbiol. 2016;18(8):1121–1133. doi: 10.1111/cmi.12572. [DOI] [PubMed] [Google Scholar]
- 22.Hillaire MLB, Décembre E, Dreux M. Autophagy: A home remodeler for hepatitis C virus. In: Jackson WT, Swanson MS, editors. Autophagy, Infection, and the Immune Response. John Wiley & Sons, Inc; Hoboken, NJ: 2014. [Google Scholar]
- 23.Dreux M, Gastaminza P, Wieland SF, Chisari FV. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci USA. 2009;106:14046–14051. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong PM, Puente C, Ganley IG, Jiang X. The ULK1 complex: Sensing nutrient signals for autophagy activation. Autophagy. 2013;9:124–137. doi: 10.4161/auto.23323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russell RC, Yuan HX, Guan KL. Autophagy regulation by nutrient signaling. Cell Res. 2014;24:42–57. doi: 10.1038/cr.2013.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konno H, Konno K, Barber GN. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 2013;155:688–698. doi: 10.1016/j.cell.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 28.Ait-Goughoulte M, et al. Hepatitis C virus genotype 1a growth and induction of autophagy. J Virol. 2008;82:2241–2249. doi: 10.1128/JVI.02093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sir D, et al. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology. 2008;48:1054–1061. doi: 10.1002/hep.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allavena G, et al. Suppressed translation and ULK1 degradation as potential mechanisms of autophagy limitation under prolonged starvation. Autophagy. 2016;12:2085–2097. doi: 10.1080/15548627.2016.1226733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao YO, Konen-Waisman S, Taylor GA, Martens S, Howard JC. Localisation and mislocalisation of the interferon-inducible immunity-related GTPase, Irgm1 (LRG-47) in mouse cells. PLoS One. 2010;5(1):e8648. doi: 10.1371/journal.pone.0008648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Springer HM, Schramm M, Taylor GA, Howard JC. Irgm1 (LRG-47), a regulator of cell-autonomous immunity, does not localize to mycobacterial or listerial phagosomes in IFN-gamma-induced mouse cells. J Immunol. 2013;191(4):1765–1774. doi: 10.4049/jimmunol.1300641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujita N, et al. Recruitment of the autophagic machinery to endosomes during infection is mediated by ubiquitin. J Cell Biol. 2013;203:115–128. doi: 10.1083/jcb.201304188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bishé B, Syed GH, Field SJ, Siddiqui A. Role of phosphatidylinositol 4-phosphate (PI4P) and its binding protein GOLPH3 in hepatitis C virus secretion. J Biol Chem. 2012;287:27637–27647. doi: 10.1074/jbc.M112.346569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Belov GA, et al. Complex dynamic development of poliovirus membranous replication complexes. J Virol. 2012;86:302–312. doi: 10.1128/JVI.05937-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ward AM, et al. The Golgi associated ERI3 is a Flavivirus host factor. Sci Rep. 2016;6:34379. doi: 10.1038/srep34379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manolea F, Claude A, Chun J, Rosas J, Melançon P. Distinct functions for Arf guanine nucleotide exchange factors at the Golgi complex: GBF1 and BIGs are required for assembly and maintenance of the Golgi stack and trans-Golgi network, respectively. Mol Biol Cell. 2008;19:523–535. doi: 10.1091/mbc.E07-04-0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao X, Lasell TK, Melançon P. Localization of large ADP-ribosylation factor-guanine nucleotide exchange factors to different Golgi compartments: Evidence for distinct functions in protein traffic. Mol Biol Cell. 2002;13:119–133. doi: 10.1091/mbc.01-08-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawamoto K, et al. GBF1, a guanine nucleotide exchange factor for ADP-ribosylation factors, is localized to the cis-Golgi and involved in membrane association of the COPI coat. Traffic. 2002;3:483–495. doi: 10.1034/j.1600-0854.2002.30705.x. [DOI] [PubMed] [Google Scholar]
- 40.Mao L, et al. AMPK phosphorylates GBF1 for mitotic Golgi disassembly. J Cell Sci. 2013;126:1498–1505. doi: 10.1242/jcs.121954. [DOI] [PubMed] [Google Scholar]
- 41.Morohashi Y, Balklava Z, Ball M, Hughes H, Lowe M. Phosphorylation and membrane dissociation of the ARF exchange factor GBF1 in mitosis. Biochem J. 2010;427:401–412. doi: 10.1042/BJ20091681. [DOI] [PubMed] [Google Scholar]
- 42.Miyamoto T, et al. AMP-activated protein kinase phosphorylates Golgi-specific brefeldin A resistance factor 1 at Thr1337 to induce disassembly of Golgi apparatus. J Biol Chem. 2008;283:4430–4438. doi: 10.1074/jbc.M708296200. [DOI] [PubMed] [Google Scholar]
- 43.Donaldson JG, Honda A, Weigert R. Multiple activities for Arf1 at the Golgi complex. Biochim Biophys Acta. 2005;1744:364–373. doi: 10.1016/j.bbamcr.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 44.D’Souza-Schorey C, Chavrier P. ARF proteins: Roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 45.Xiang Y, Seemann J, Bisel B, Punthambaker S, Wang Y. Active ADP-ribosylation factor-1 (ARF1) is required for mitotic Golgi fragmentation. J Biol Chem. 2007;282:21829–21837. doi: 10.1074/jbc.M611716200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geng J, Klionsky DJ. The Golgi as a potential membrane source for autophagy. Autophagy. 2010;6:950–951. doi: 10.4161/auto.6.7.13009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mackenzie JM, Jones MK, Westaway EG. Markers for trans-Golgi membranes and the intermediate compartment localize to induced membranes with distinct replication functions in flavivirus-infected cells. J Virol. 1999;73:9555–9567. doi: 10.1128/jvi.73.11.9555-9567.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Westaway EG, Mackenzie JM, Kenney MT, Jones MK, Khromykh AA. Ultrastructure of Kunjin virus-infected cells: Colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J Virol. 1997;71:6650–6661. doi: 10.1128/jvi.71.9.6650-6661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou Z, Mogensen MM, Powell PP, Curry S, Wileman T. Foot-and-mouth disease virus 3C protease induces fragmentation of the Golgi compartment and blocks intra-Golgi transport. J Virol. 2013;87:11721–11729. doi: 10.1128/JVI.01355-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mousnier A, et al. Human rhinovirus 16 causes Golgi apparatus fragmentation without blocking protein secretion. J Virol. 2014;88:11671–11685. doi: 10.1128/JVI.01170-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Belov GA, et al. Hijacking components of the cellular secretory pathway for replication of poliovirus RNA. J Virol. 2007;81:558–567. doi: 10.1128/JVI.01820-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moss J, Vaughan M. Structure and function of ARF proteins: Activators of cholera toxin and critical components of intracellular vesicular transport processes. J Biol Chem. 1995;270:12327–12330. doi: 10.1074/jbc.270.21.12327. [DOI] [PubMed] [Google Scholar]
- 53.Belov GA, Sztul E. Rewiring of cellular membrane homeostasis by picornaviruses. J Virol. 2014;88:9478–9489. doi: 10.1128/JVI.00922-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim BH, Shenoy AR, Kumar P, Bradfield CJ, MacMicking JD. IFN-inducible GTPases in host cell defense. Cell Host Microbe. 2012;12:432–444. doi: 10.1016/j.chom.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Godi A, et al. ARF mediates recruitment of PtdIns-4-OH kinase-beta and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat Cell Biol. 1999;1:280–287. doi: 10.1038/12993. [DOI] [PubMed] [Google Scholar]
- 56.Berger KL, Kelly SM, Jordan TX, Tartell MA, Randall G. Hepatitis C virus stimulates the phosphatidylinositol 4-kinase III alpha-dependent phosphatidylinositol 4-phosphate production that is essential for its replication. J Virol. 2011;85:8870–8883. doi: 10.1128/JVI.00059-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lim YS, Hwang SB. Hepatitis C virus NS5A protein interacts with phosphatidylinositol 4-kinase type IIIalpha and regulates viral propagation. J Biol Chem. 2011;286:11290–11298. doi: 10.1074/jbc.M110.194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Borawski J, et al. Class III phosphatidylinositol 4-kinase alpha and beta are novel host factor regulators of hepatitis C virus replication. J Virol. 2009;83:10058–10074. doi: 10.1128/JVI.02418-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heuer D, et al. Chlamydia causes fragmentation of the Golgi compartment to ensure reproduction. Nature. 2009;457:731–735. doi: 10.1038/nature07578. [DOI] [PubMed] [Google Scholar]
- 60.Roulin PS, et al. Rhinovirus uses a phosphatidylinositol 4-phosphate/cholesterol counter-current for the formation of replication compartments at the ER-Golgi interface. Cell Host Microbe. 2014;16:677–690. doi: 10.1016/j.chom.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 61.Romano JD, Sonda S, Bergbower E, Smith ME, Coppens I. Toxoplasma gondii salvages sphingolipids from the host Golgi through the rerouting of selected Rab vesicles to the parasitophorous vacuole. Mol Biol Cell. 2013;24:1974–1995. doi: 10.1091/mbc.E12-11-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arnaud N, et al. Hepatitis C virus reveals a novel early control in acute immune response. PLoS Pathog. 2011;7:e1002289. doi: 10.1371/journal.ppat.1002289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Akazawa D, et al. CD81 expression is important for the permissiveness of Huh7 cell clones for heterogeneous hepatitis C virus infection. J Virol. 2007;81:5036–5045. doi: 10.1128/JVI.01573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arnaud N, et al. Hepatitis C virus controls interferon production through PKR activation. PLoS One. 2010;5:e10575. doi: 10.1371/journal.pone.0010575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miller PM, et al. Golgi-derived CLASP-dependent microtubules control Golgi organization and polarized trafficking in motile cells. Nat Cell Biol. 2009;11:1069–1080. doi: 10.1038/ncb1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bolte S, Cordelières FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 67.Dunn KW, Kamocka MM, McDonald JH. A practical guide to evaluating colocalization in biological microscopy. Am J Physiol Cell Physiol. 2011;300:C723–C742. doi: 10.1152/ajpcell.00462.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11(8):783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]