The intricacies of protein-mediated signaling continue to be revealed in exquisite detail. It therefore seems fitting to return to the fundamental and almost magical feature of protein molecules that makes it all possible: allostery, or action at a distance. This long-held concept arose and was refined during the explosive expansion of the structural biology of proteins. The resulting models of allostery through discrete two-state structural transitions have held sway for over half a century. However, they are incomplete. Even for the paradigm allosteric protein hemoglobin, alternate functionally relevant structures that are dynamically averaged (1) force a broader ensemble description of the thermodynamics of allostery (2). Furthermore, the view seen through the lens of structure is a very enthalpic one that focuses analysis on the details of specific interactions within and between proteins and their ligands. However, it is free energy that guides the allosteric response. Entropy, which is what makes the energy free, has rarely been examined experimentally in the same microscopic detail. Although it was known decades ago that protein molecules should (3, 4) and do (5–7) dynamically fluctuate, it remains uncertain to what degree conformational entropy (Sconf) actually does contribute to the free energy of ligand binding and allostery. This uncertainty is because Sconf has largely resisted experimental measurement. Obviously, motion between various states that a protein visits reports, albeit indirectly, on its Sconf. Developments in solution NMR spectroscopy have recently converged to allow detailed analysis of protein internal motion on many time scales (8), and thus open the door to the elusive thermodynamic variable. In an elegant and unusually comprehensive study reported in PNAS, Capdevila et al. (9) illuminate the dynamical aspects of ligand binding and the contributions of ∆Sconf to allostery in a transcription factor that is negatively regulated by zinc. Their results are not comprehensible within the confines of interconversions between static structures and require a context of an ensemble of states. How they achieved this insight requires some explanation.
With the development of advanced multidimensional heteronuclear NMR spectroscopy has come the capability to measure NMR relaxation phenomena in proteins in a site-resolved way (8). NMR relaxation is fundamentally linked to motion, and a wide range of times scales is accessible. Important here are the fast picosecond-nanosecond and slower microsecond-millisecond motions that are sampled at the backbone amide N-H and the methyl groups of side chains. Relatively simple experiments using cross-correlated relaxation in methyl groups lead to a particularly direct analytical path for fast motion (10). This advance has greatly accelerated the pace of detailed dynamical studies of protein motion and is used extensively by Capdevila et al. (9). Relaxation due to fast internal protein motions is usually interpreted using the popular Lipari–Szabo “model-free” formalism (11) that captures the disorder of some “interaction vector” in terms of the squared generalized order parameter (S2). The S2 is related to the populations of the various states that are visited by the NMR probe during equilibrium fluctuations of the protein (11). Hence, in principle, changes in motion are related to changes in Sconf. Indeed, as long ago as the mid-1990s, it was pointed out that if one assumed a specific energy potential describing motion of an NMR-accessible vector (e.g., the N-H bond, the symmetry axis of a methyl group), then one could solve the parametric relationship between S2 and the associated entropy (12–14). These model-dependent interpretations gave comfort to the idea that “more motion means more entropy” and vice versa. Early studies of calmodulin binding of various regulatory domains found a significant redistribution and reduction of side motion (15) that correlated with the total binding entropy (∆Stot) (16). This finding suggested that ∆Sconf was a major component of ∆Stot. Unfortunately, several problems complicate making this view quantitative through a local model-dependent interpretation (17, 18). A breakthrough was to establish an empirical relationship between changes of motion and changes in Sconf that avoided the main problems with a model-based interpretation (18, 19). The resulting NMR-based dynamical proxy or “entropy meter” approach provides measures of ∆Sconf for the entire protein molecule. Capdevila et al. (9) use the entropy meter to probe the thermodynamic origins of the allosteric response in Zn-induced allosteric inhibition of DNA binding by the homodimeric repressor CzrA.
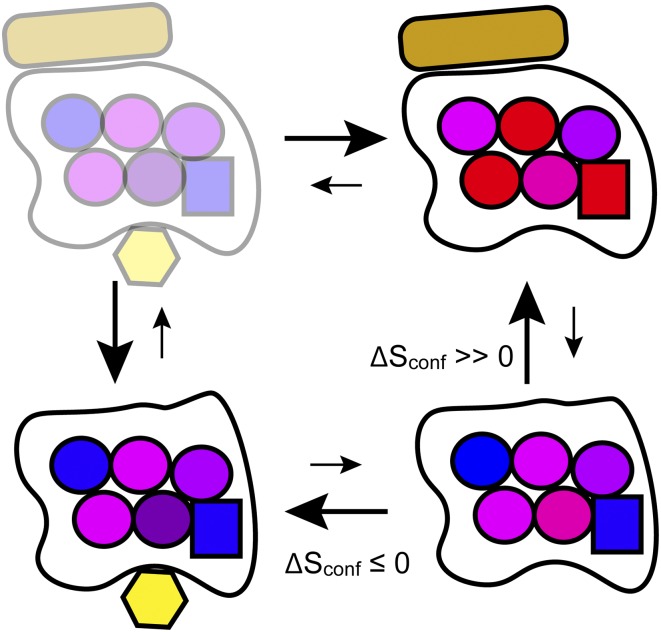
The role of Sconf in CzrA function was examined by comparing fast backbone and methyl side-chain motion in three functional states: CzrA2, CzrA2:DNA, and CzrA2:Zn2. The backbone of the protein was found to be largely unresponsive to the binding of operator DNA, whereas the methyl side-chain motion was broadly influenced and, on average, increased upon forming the complex. The decoupling of backbone and side-chain motion is not unprecedented and warns strongly that both need to be examined (17). The net increase in side-chain dynamics on forming the CzrA:DNA complex was found to correspond to nearly two-thirds of the large and favorable ∆Stot. This determination set the stage for understanding how negative allosteric regulation by zinc is achieved. Binding of zinc to the CzrA causes only localized diminishment of methyl side-chain motion, corresponding to a small unfavorable change in ∆Sconf. Importantly, binding of zinc appears to abolish the ability of the protein to unlock the large favorable change of Sconf necessary to bind the operator DNA with high affinity. This observation broadly explains the thermodynamic origins of the negative allostery (Fig. 1).
Fig. 1.
Schematic illustration of the basic features of an allosteric mechanism regulated by Sconf. Sconf directs binding by a protein (black envelope) of a ligand (rectangles), which is suppressed by the binding of a negative heterotropic allosteric regulator (hexagons). Detailed dynamical studies by Capdevila et al. (9) support this model for the binding of DNA to the CzrA transcription factor and its allosteric regulation by zinc. In the zinc-bound states, side-chain (circles) motion is, on average, suppressed (more blue), whereas in the DNA-bound states, it is, on average, elevated (more red). A number of residues in CzrA show extraordinary influence on the overall the entropic response (squares). The slightly transparent ternary complex, which is strongly disfavored, was not directly examined in this way. For graphical simplicity, the CzrA2 and Crz2:Zn2 species are represented as the monomer and 1:1 complexes, respectively.
To reveal how the allosteric signal is coordinated by the protein, Capdevila et al. (9) use the temperature dependence of methyl side-chain dynamics to highlight residues with unusual motion. Using an analytical strategy introduced by Massi and Palmer (20), they find a remarkable degree of motional coupling in the core of CzrA. They then take advantage of a pioneering effort by Tzeng and Kalodimos (21), who showed that seemingly innocuous point mutations in the catabolite activator protein often profoundly influenced its dynamics and the thermodynamics of DNA binding. Introduction of cavity-creating mutations in CzrA illuminated a strong correlation between binding affinity and dynamic (entropic) contributions. Through these two approaches, a large network of residues was found to provide a reservoir of Sconf that is controlled through a smaller number of critical side chains. The influence of Zn binding on these “hot spot” residues is the key to locking away the contribution of ∆Sconf to the binding of DNA. Importantly, this work also clearly
In an elegant and unusually comprehensive study reported in PNAS, Capdevila et al. illuminate the dynamical aspects of ligand binding and the contributions of ∆Sconf to allostery in a transcription factor that is negatively regulated by zinc.
reaffirms that the seductive concept of restricted pathways of dynamic connectivity need not be operative for allostery involving Sconf (17).
Capdevila et al. (9) also address the mechanism of molecular recognition and binding of DNA to CzrA. Structural transitions are very often critical to the recognition and binding of a ligand by a protein. Kinetic aspects of molecular recognition give rise to “conformational selection” and “induced fit” ligand-binding mechanisms (22), which can lead to fundamental insights of both biological and medical importance (23). Dispersion relaxation NMR techniques (8) were used to show that, in the absence of ligands, CzrA fluctuates on the millisecond time scale to a minor conformation that is likely structurally compatible with DNA binding. Zn binding quenches this motion, suggesting that conformational selection is the operative mechanism for recognition and binding of operator DNA and is intimately connected to CzrA allostery.
In closing, it is interesting to note that only a few dozen examples of comprehensive NMR relaxation studies of the dynamical effects of ligand binding and allostery have been reported in the literature. The contribution by Capdevila et al. (9) exemplifies the richness of the dynamical-entropy axis in protein function that has been exposed by this small number of examples. It seems likely that there are many more arenas in which internal motion, and the Sconf that it reflects, will prove to be critical elements of the physical basis of protein function. Indeed, as molecular dynamics reaches reliable accuracy (24) and crystallography more fully adopts the notion of conformational heterogeneity (25), the combination of NMR spectroscopy, crystallography, and simulation will dance across the landscape of relationships between protein internal motion, structure, and function.
Acknowledgments
Work in this area by A.J.W. is supported by NIH Grants GM102447 and GM100910 and by the Mathers Foundation.
Footnotes
The author declares no conflict of interest.
See companion article on page 4424.
References
- 1.Lukin JA, et al. Quaternary structure of hemoglobin in solution. Proc Natl Acad Sci USA. 2003;100:517–520. doi: 10.1073/pnas.232715799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Motlagh HN, Wrabl JO, Li J, Hilser VJ. The ensemble nature of allostery. Nature. 2014;508:331–339. doi: 10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooper A. Thermodynamic fluctuations in protein molecules. Proc Natl Acad Sci USA. 1976;73:2740–2741. doi: 10.1073/pnas.73.8.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper A, Dryden DTF. Allostery without conformational change. A plausible model. Eur Biophys J. 1984;11:103–109. doi: 10.1007/BF00276625. [DOI] [PubMed] [Google Scholar]
- 5.Lakowicz JR, Weber G. Quenching of protein fluorescence by oxygen. Detection of structural fluctuations in proteins on the nanosecond time scale. Biochemistry. 1973;12:4171–4179. doi: 10.1021/bi00745a021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Englander SW, Kallenbach NR. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q Rev Biophys. 1983;16:521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- 7.Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 8.Mittermaier AK, Kay LE. Observing biological dynamics at atomic resolution using NMR. Trends Biochem Sci. 2009;34:601–611. doi: 10.1016/j.tibs.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 9.Capdevila DA, Braymer JJ, Edmonds KA, Wu H, Giedroc DP. Entropy redistribution controls allostery in a metalloregulatory protein. Proc Natl Acad Sci USA. 2017;114:4424–4429. doi: 10.1073/pnas.1620665114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tugarinov V, Sprangers R, Kay LE. Probing side-chain dynamics in the proteasome by relaxation violated coherence transfer NMR spectroscopy. J Am Chem Soc. 2007;129:1743–1750. doi: 10.1021/ja067827z. [DOI] [PubMed] [Google Scholar]
- 11.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J Am Chem Soc. 1982;104:4546–4559. [Google Scholar]
- 12.Akke M, Brüschweiler R, Palmer AG. NMR order parameters and free-energy: An analytical approach and its application to cooperative Ca2+ binding by calbindin-D(9k) J Am Chem Soc. 1993;115:9832–9833. [Google Scholar]
- 13.Li Z, Raychaudhuri S, Wand AJ. Insights into the local residual entropy of proteins provided by NMR relaxation. Protein Sci. 1996;5:2647–2650. doi: 10.1002/pro.5560051228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang D, Kay LE. Contributions to conformational entropy arising from bond vector fluctuations measured from NMR-derived order parameters: Application to protein folding. J Mol Biol. 1996;263:369–382. doi: 10.1006/jmbi.1996.0581. [DOI] [PubMed] [Google Scholar]
- 15.Lee AL, Kinnear SA, Wand AJ. Redistribution and loss of side chain entropy upon formation of a calmodulin-peptide complex. Nat Struct Biol. 2000;7:72–77. doi: 10.1038/71280. [DOI] [PubMed] [Google Scholar]
- 16.Frederick KK, Marlow MS, Valentine KG, Wand AJ. Conformational entropy in molecular recognition by proteins. Nature. 2007;448:325–329. doi: 10.1038/nature05959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Igumenova TI, Frederick KK, Wand AJ. Characterization of the fast dynamics of protein amino acid side chains using NMR relaxation in solution. Chem Rev. 2006;106:1672–1699. doi: 10.1021/cr040422h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kasinath V, Sharp KA, Wand AJ. Microscopic insights into the NMR relaxation-based protein conformational entropy meter. J Am Chem Soc. 2013;135:15092–15100. doi: 10.1021/ja405200u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marlow MS, Dogan J, Frederick KK, Valentine KG, Wand AJ. The role of conformational entropy in molecular recognition by calmodulin. Nat Chem Biol. 2010;6:352–358. doi: 10.1038/nchembio.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Massi F, Palmer AG., 3rd Temperature dependence of NMR order parameters and protein dynamics. J Am Chem Soc. 2003;125:11158–11159. doi: 10.1021/ja035605k. [DOI] [PubMed] [Google Scholar]
- 21.Tzeng SR, Kalodimos CG. Protein activity regulation by conformational entropy. Nature. 2012;488:236–240. doi: 10.1038/nature11271. [DOI] [PubMed] [Google Scholar]
- 22.Hammes GG, Chang YC, Oas TG. Conformational selection or induced fit: A flux description of reaction mechanism. Proc Natl Acad Sci USA. 2009;106:13737–13741. doi: 10.1073/pnas.0907195106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson C, et al. Kinase dynamics. Using ancient protein kinases to unravel a modern cancer drug’s mechanism. Science. 2015;347:882–886. doi: 10.1126/science.aaa1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Brien ES, Wand AJ, Sharp KA. On the ability of molecular dynamics force fields to recapitulate NMR derived protein side chain order parameters. Protein Sci. 2016;25:1156–1160. doi: 10.1002/pro.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van den Bedem H, Fraser JS. Integrative, dynamic structural biology at atomic resolution--it’s about time. Nat Methods. 2015;12:307–318. doi: 10.1038/nmeth.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]