Significance
Algae account for a large proportion of global primary productivity, and the carbon that they fix supports many ecosystems and associated services used by humans. Green algae overcome various physical limitations on the rate of CO2 supply for photosynthesis by the action of carbon-concentrating mechanisms (CCM) that deliver CO2 to the carboxylating enzyme, Rubisco. However, marine systems are dominated by algae that contain chloroplasts of red algal origin, and we know relatively little about their biology at the molecular level. Here we characterize a carbonic anhydrase as part of a simple CCM in the oleaginous photosynthetic stramenopile, Nannochloropsis oceanica. This work expands our understanding of the diversity in CCMs, and may lead to biotechnological improvements in this potential biofuel-producing alga.
Keywords: photosynthesis, carbon-concentrating mechanism, carbonic anhydrase, heterokont, algae
Abstract
Aquatic photosynthetic organisms cope with low environmental CO2 concentrations through the action of carbon-concentrating mechanisms (CCMs). Known eukaryotic CCMs consist of inorganic carbon transporters and carbonic anhydrases (and other supporting components) that culminate in elevated [CO2] inside a chloroplastic Rubisco-containing structure called a pyrenoid. We set out to determine the molecular mechanisms underlying the CCM in the emerging model photosynthetic stramenopile, Nannochloropsis oceanica, a unicellular picoplanktonic alga that lacks a pyrenoid. We characterized CARBONIC ANHYDRASE 1 (CAH1) as an essential component of the CCM in N. oceanica CCMP1779. We generated insertions in this gene by directed homologous recombination and found that the cah1 mutant has severe defects in growth and photosynthesis at ambient CO2. We identified CAH1 as an α-type carbonic anhydrase, providing a biochemical role in CCM function. CAH1 was found to localize to the lumen of the epiplastid endoplasmic reticulum, with its expression regulated by the external inorganic carbon concentration at both the transcript and protein levels. Taken together, these findings show that CAH1 is an indispensable component of what may be a simple but effective and dynamic CCM in N. oceanica.
Rubisco is the principal carboxylation enzyme in photosynthetic carbon fixation. In aquatic photosynthetic organisms, the supply of Rubisco’s substrate, CO2, can be restricted by the slow diffusion of CO2 in water and the hydration/dehydration reaction that interconverts CO2 with other forms of dissolved inorganic carbon (DIC) that are unavailable to Rubisco, such as bicarbonate (HCO3−) (1). The limitations of physical chemistry are compounded by the biochemical properties of Rubisco, which has a moderately slow turnover rate and exhibits a counterproductive oxygenase activity, leading to photorespiration (2, 3). As a result, many aquatic photosynthetic organisms operate a carbon-concentrating mechanism (CCM) to elevate the concentration of CO2 near Rubisco, thereby enhancing the rate of carboxylation and suppressing photorespiration (4, 5). In the model green alga, Chlamydomonas reinhardtii, the CCM includes active DIC transporters that accumulate bicarbonate within the cell (HCO3− being charged and relatively cell-impermeant compared with CO2) and a suite of carbonic anhydrases that catalyze the otherwise sluggish equilibration of CO2 and HCO3− (6, 7).
In cells of Chlamydomonas (and numerous other algae) grown under CO2-limiting conditions, the majority of Rubisco is localized to a central structure within the chloroplast known as a pyrenoid (8, 9), which is traversed by thylakoid minitubules (10). It is thought that bicarbonate is ultimately transported to the lumen of these transpyrenoidal thylakoids, where the acidic pH and activity of the carbonic anhydrase CAH3 leads to the rapid formation of CO2 (11, 12).
Although our understanding of CCM function has been greatly advanced by research on C. reinhardtii and the carboxysome-based system of cyanobacteria (5), extensive experimental studies have been limited to these select taxa. The timing and origins of CCM evolution are topics of active research and debate, and it has been proposed that fluctuating atmospheric CO2 and O2 concentrations over geologic time resulted in multiple instances of convergent evolution and possibly serial gains and losses of CCMs (13, 14). This dynamic evolutionary history implies that diverse CCM configurations could exist, and thus studying the CCMs of other algae could illustrate how different organisms have evolved to cope with this common challenge (8).
Algae harboring secondary plastids of red algal origin constitute most of the diversity in marine algae (15), and collectively they provide most of the total primary production of the oceans (16). Along with their ecological importance, the different membrane structure and biochemistry of secondary red plastid-bearing species suggests the strong possibility of novel spatial configurations of CCM components, making these taxa attractive for CCM research.
Species of the genus Nannochloropsis constitute an emerging model for fundamental research on photosynthesis and algal biology. Like diatoms, for which the CCM is beginning to be elucidated at the molecular level (17–19), Nannochloropsis spp. belong to the stramenopile (heterokont) clade and have a plastid of red algal origin that is separated from the cytoplasm by a total of four membranes, the outermost being contiguous with the endoplasmic reticulum (ER) and outer nuclear envelope (20–22). However, the cells of these species are remarkably small in both morphology and genome size (∼29 Mb; ∼10,000 estimated genes) (23, 24). Importantly, they appear to lack pyrenoids or other CO2-concentrating structures, which are central features of known CCMs (9). Physiological studies have demonstrated that cells of Nannochloropsis gaditana primarily take up HCO3− and, strikingly, release CO2 into the surrounding media in excess of the chemical equilibrium (25, 26). Energization of this phenomenon was dependent on mitochondrial respiration rather than on the chloroplastic light reactions that are thought to drive the more intensively studied CCMs (27). This indicates that Nannochloropsis spp. may operate what has been termed a “pump-leak” type of CCM, whereby bicarbonate transporter and carbonic anhydrase activity deliver CO2 in excess of what photosynthesis can use, resulting in a sizeable leakage back to the surroundings.
Recent technical advances have made molecular genetic studies of Nannochloropsis feasible. The genomes of several Nannochloropsis species have been sequenced, making genomic comparisons possible (23, 24, 28), and transformation and homologous recombination have been demonstrated in Nannochloropsis oceanica CCMP1779 (29) and N. gaditana (30). Nannochloropsis spp. also have attracted attention for potential applications as a biofuel feedstock, given their remarkable ability to accumulate lipids and potential for producing bioengineered high-value compounds (23, 30, 31). Thus, the Nannochloropsis system is poised to provide a rich subject of study for a breadth of fundamental and applied research, including CCM biology.
In this work, we aimed to expand on our understanding of the unusual CCM in Nannochloropsis spp. by beginning to identify the underlying molecular players. We generated mutants for the CARBONIC ANHYDRASE 1 (CAH1) gene (gene ID: 6698-mRNA) by homologous recombination, and characterized it at as an essential component of the CCM in N. oceanica CCMP1779. The results presented here provide a beginning framework for understanding the CCM not only in N. oceanica and closely related species, but also in other algae that lack a pyrenoid.
Results
The cah1 Mutant Displays Severe DIC-Dependent Defects in Growth and Photosynthesis.
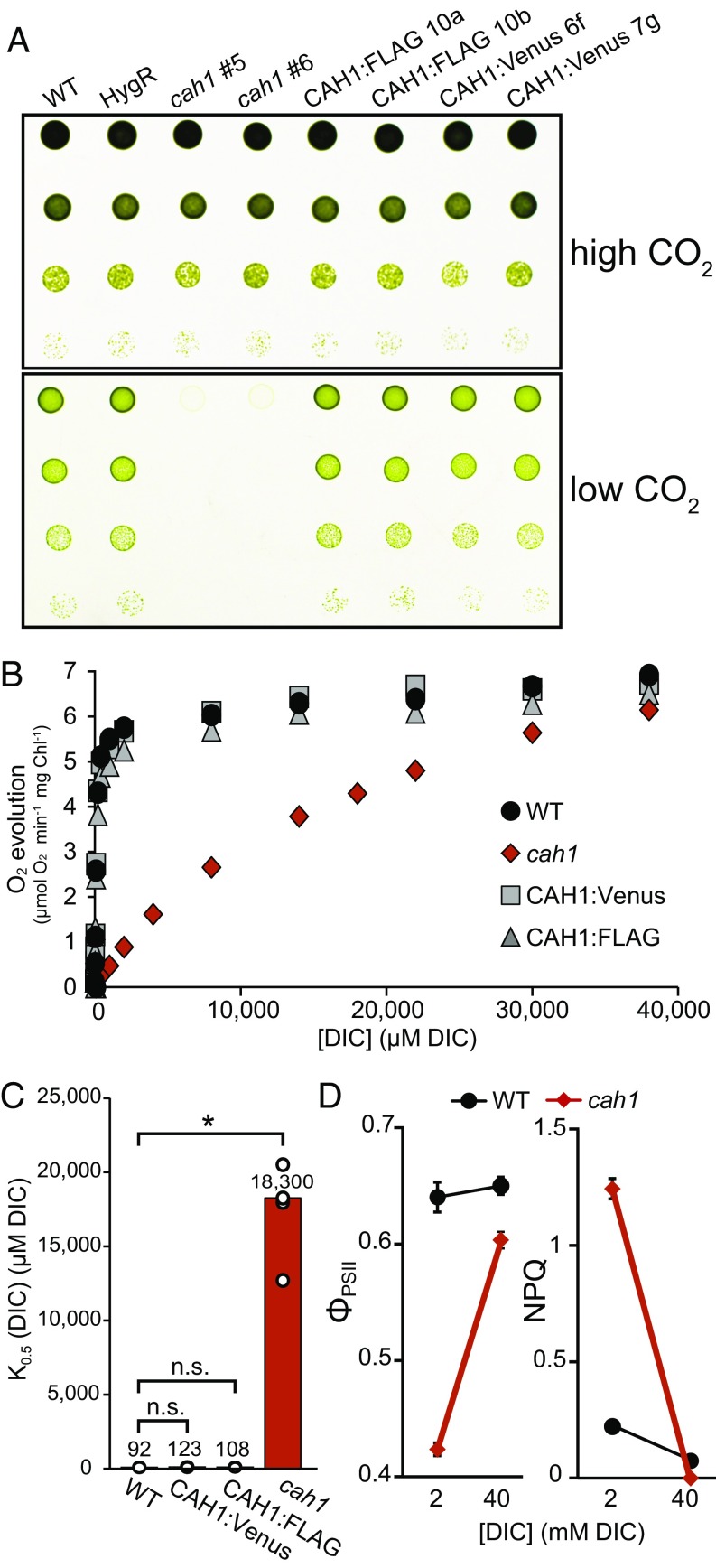
Based on the genome analysis of N. oceanica and identification of potential CCM genes by Vieler et al. (23), we generated loss-of-function mutants for CAH1 by replacing the first exon and part of the first intron with a 2.1-kb hygromycin resistance cassette via homologous recombination (SI Appendix, Fig. S1A). The cah1 mutants were positively identified by PCR screening for a size increase of an amplicon generated by primers flanking the insertion site (SI Appendix, Fig. S1B). These mutants were unable to grow at ambient CO2, a defect that was fully rescued when the cells were grown in 3% CO2 (Fig. 1A; additional lines shown in SI Appendix, Fig. S1C). To confirm that CAH1 was the causal gene and test the functionality of tagged constructs, we transformed the cah1 mutant with the CAH1 coding sequence fused with a FLAG epitope or Venus fluorescent protein tag at the C terminus, driven by the native promoter (SI Appendix, Fig. S1A). Both of the tagged constructs successfully rescued the growth phenotype, and the genotypes of these lines were again confirmed by PCR (Fig. 1A; additional lines shown in SI Appendix, Fig. S1D).
Fig. 1.
CAH1 is required for normal growth and photosynthesis at low CO2. (A) Spot growth phenotype of WT, cah1 mutant, and tagged complementation lines. High CO2, 3% CO2; low CO2, 0.04% CO2; HygR, control line expressing the hygromycin resistance cassette; CAH1:FLAG or CAH1:Venus, cah1 (#5) complemented with CAH1 CDS with a C-terminal tag. Two independent lines are shown for cah1 and complemented lines. (B) Whole-cell photosynthetic DIC affinity assay. Cells were acclimated to low CO2 for 24 h, after which O2 evolution was measured in response to increasing concentrations of DIC. Chl, chlorophyll. (C) Calculated [DIC] for half maximal rate = K0.5 (DIC). Bar height and numbers above bars indicate the mean. Individual data points are plotted as white circles. *P < Bonferroni-corrected α (0.05/3 = 0.0167), Welch’s t test (n = 4). n.s., not significant. (D) Quantum yield of ΦPSII and photoprotection (NPQ) of WT and cah1 cells at two [DIC]. Data markers indicate the mean. Error bars indicate SD (n = 9). Two-factor ANOVA yielded significant genotype × [DIC] interaction terms (P < 0.001).
To test whether carbon assimilation was specifically impaired in the cah1 mutant, we measured photosynthesis (oxygen evolution) in response to varying concentrations of DIC. Although the maximum rate of photosynthesis was similar in all lines, the cah1 mutant exhibited a dramatic reduction in DIC affinity, as seen in the shape of the response curve as well as in the estimated DIC concentration required for half-saturation, K0.5[DIC] (Fig. 1 B and C). The wild-type (WT) DIC affinity was recapitulated in the complemented lines (Fig. 1B).
To further assess photosynthetic performance with a different method, we measured chlorophyll fluorescence using pulse-amplitude modulated (PAM) fluorometry (32) to calculate the quantum yield of photosystem II (ΦPSII) and nonphotochemical quenching (NPQ) of WT and cah1 cells. The test was carried out in medium containing either 2 mM or 40 mM DIC (as NaHCO3) to approximate that found at ambient or 3% CO2, respectively. In the low-DIC test condition, cah1 cells showed a severe reduction in ΦPSII and a large increase in NPQ compared with WT, and this effect was largely abolished when the cells were assayed at the high-DIC condition (P < 0.001 for the genotype × [DIC] interaction term in a two-factor ANOVA for both parameters) (Fig. 1D).
CAH1 Is an α-Type Carbonic Anhydrase.
Carbonic anhydrases are nearly ubiquitous metalloenzymes that function in a diverse set of organisms and physiological processes. Several families of carbonic anhydrases are thought to have evolved independently, including the α, β, γ, δ, and ζ types, as well as a recently described θ type (18). A whole-genome analysis reported by Vieler et al. (23) identified one α-type carbonic anhydrase (CAH1) and one β-type carbonic anhydrase in N. oceanica CCMP1779. Building a working model for CCM function in this organism would be aided by a complete list of possible carbonic anhydrases, so we independently searched for orthologs belonging to each of the families using blastp and tblastn. We identified two possible γ-type enzymes (SI Appendix, Table S1), corroborating recent findings reported by Wei et al. (33); however, when aligned to known γ-type carbonic anhydrases, not all of the conserved residues typically necessary for activity were present (SI Appendix, Fig. S2).
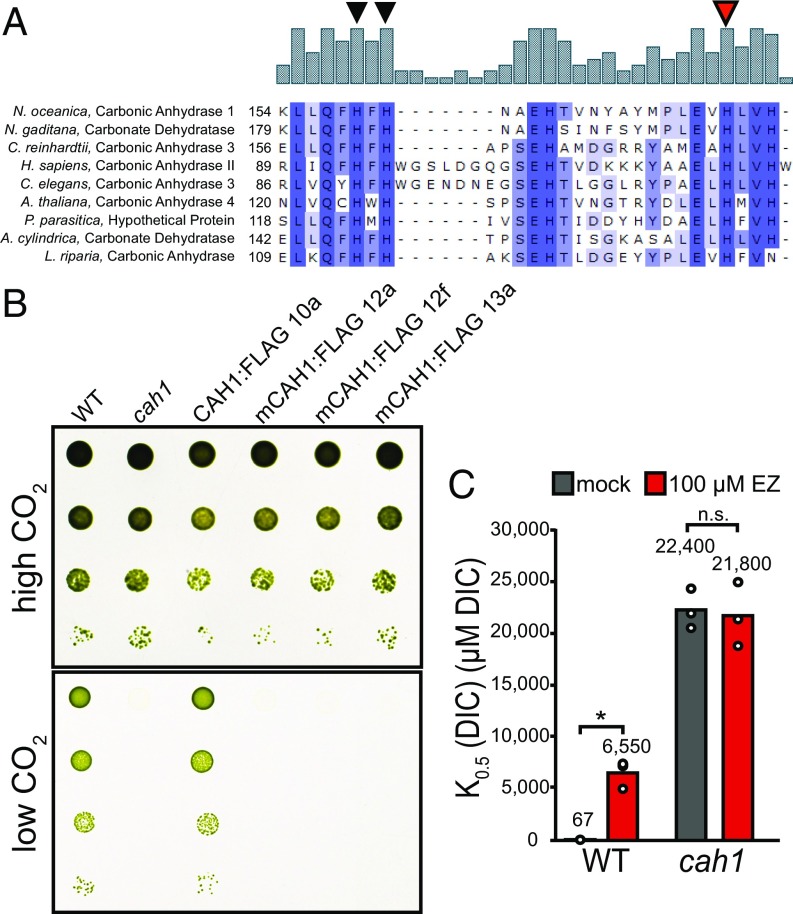
We performed a multiple sequence alignment with CAH1 and known α-type carbonic anhydrases from disparate parts of the tree of life and found areas of highly conserved sequence (SI Appendix, Fig. S3). Notably, the trio of conserved histidine residues responsible for binding of the catalytic zinc ion was present in CAH1 (Fig. 2A). Given that mutation of these critical histidine residues is known to lead to compromised carbonic anhydrase activity (34), to test whether or not CAH1 acts through this canonical carbonic anhydrase activity, we altered histidine residue 177 to an alanine and attempted to complement the cah1 mutant. These H177A “mCAH1” lines, which were confirmed for protein expression by immunoblot analysis (SI Appendix, Fig. S4), failed to grow at ambient CO2, but maintained normal growth at 3% CO2 (Fig. 2B).
Fig. 2.
CAH1 is an α-type carbonic anhydrase. (A) Multiple sequence alignment of α-type carbonic anhydrases. Catalytic zinc-binding histidine residues are denoted by triangles; the red triangle indicates the target of the His to Ala site-directed mutagenesis to produce the mCAH1 lines. (B) Spot growth assay of H177A mCAH1 lines. The assay was performed as in Fig. 1A. High CO2, 3%; low CO2, 0.04%. (C) The carbonic anhydrase inhibitor ethoxyzolamide reduces DIC affinity in WT cells, but not in cah1 cells. WT and cah1 liquid cultures were incubated with 100 μM ethoxyzolamide before assessment of photosynthetic DIC affinity by oxygen evolution. *P < 0.05/2, Welch’s t test. n.s., not significant.
Ethoxyzolamide is a cell-permeant sulfonamide inhibitor of carbonic anhydrase activity (8, 26). If CAH1 enhances carbon assimilation through its carbonic anhydrase activity, we predicted that treatment of cells with ethoxyzolamide would cause defects in photosynthetic carbon assimilation similar to those seen in the cah1 mutant. In accordance with this prediction, incubation with 100 µM ethoxyzolamide reduced photosynthetic DIC affinity in WT, whereas the cah1 mutant was not further impaired by treatment with ethoxyzolamide (Fig. 2C).
CAH1 Protein Is Localized to the Epiplastid ER Lumen.
In both C. reinhardtii (green alga) and Phaeodactylum tricornutum (pennate diatom), a carbonic anhydrase located within transpyrenoidal thylakoids is important for CCM function, and the acidic microenvironment of this compartment is thought to aid the formation of CO2 from HCO3− in the immediate vicinity of Rubisco (11, 19). We did not observe a pyrenoid in N. oceanica CCMP1779 (SI Appendix, Fig. S5), and the absence of a pyrenoid was previously reported for N. gaditana (9), which leaves open the possibility of a different spatial configuration of CCM components.
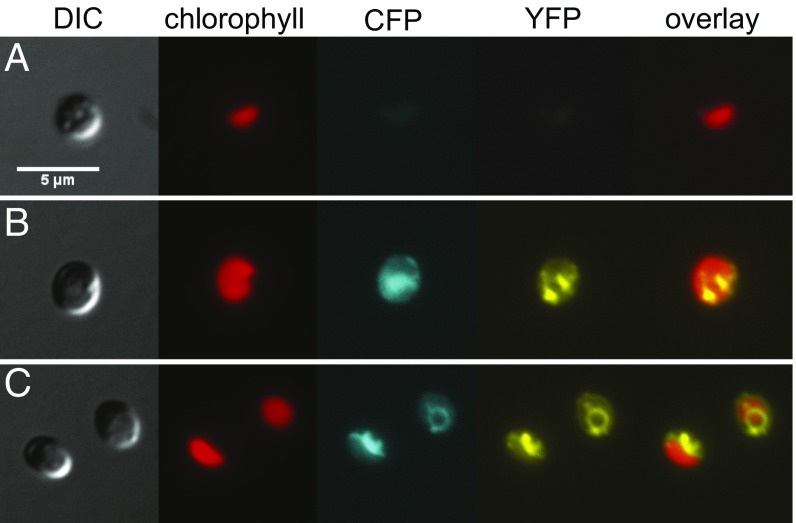
A signal peptide in CAH1 was predicted by both TargetP 1.1 (SP score, 0.941; cutoff, 0.430; RC, 1, indicating strong confidence; SP length, 23 amino acids) (35) and HECTAR, a program designed for organisms with secondary plastids enclosed by additional membranes (signal peptide score, 0.8311; chloroplast, 0.0167; mitochondrion, not available; other, not available) (36). To experimentally determine the subcellular localization of CAH1 protein, we used the Venus fusion complementation lines (denoted as CAH1:Venus).
To refine the visualization of a given cell’s morphology, we transformed the CAH1:Venus strain with a cyan-fluorescent protein (CFP) marker (37) that was directed either to the ER lumen with a known ER targeting signal (20) or to the cytosol (no targeting peptide). Through fluorescence microscopy, it was apparent that although the cytoplasmic CFP marker filled much of the cell (excluding the chloroplast), the CAH1:Venus signal was restricted to a reticulate distribution that matched the ER-targeted CFP (Fig. 3 and SI Appendix, Fig. S6). Because endomembrane localization of an essential CCM carbonic anhydrase is unprecedented, we tested the effect of ectopically targeting CAH1 to the chloroplast. Using the bipartite chloroplast targeting sequence (BTS) from a photosynthetic antenna protein (20), we observed chloroplast-localized signals in the BTS:CAH1:Venus lines, yet growth in ambient air was rescued only partially (SI Appendix, Fig. S7).
Fig. 3.
Subcellular localization of CAH1:Venus fusion protein by fluorescence microscopy. (A) WT cell showing baseline plastid autofluorescence in the CFP and YFP channels. (B) A CAH1:Venus fusion protein (YFP channel) was coexpressed with a cytoplasmic mCerulean marker (CFP channel). (C) Similar to B, but with an ER-luminal mCerulean marker. Cells were grown in liquid culture in 3% CO2 and then transferred to ambient air for 24 h before imaging. (Scale bar: 5 µm.)
The CCM and CAH1 Expression Are Inducible by Environmental [DIC].
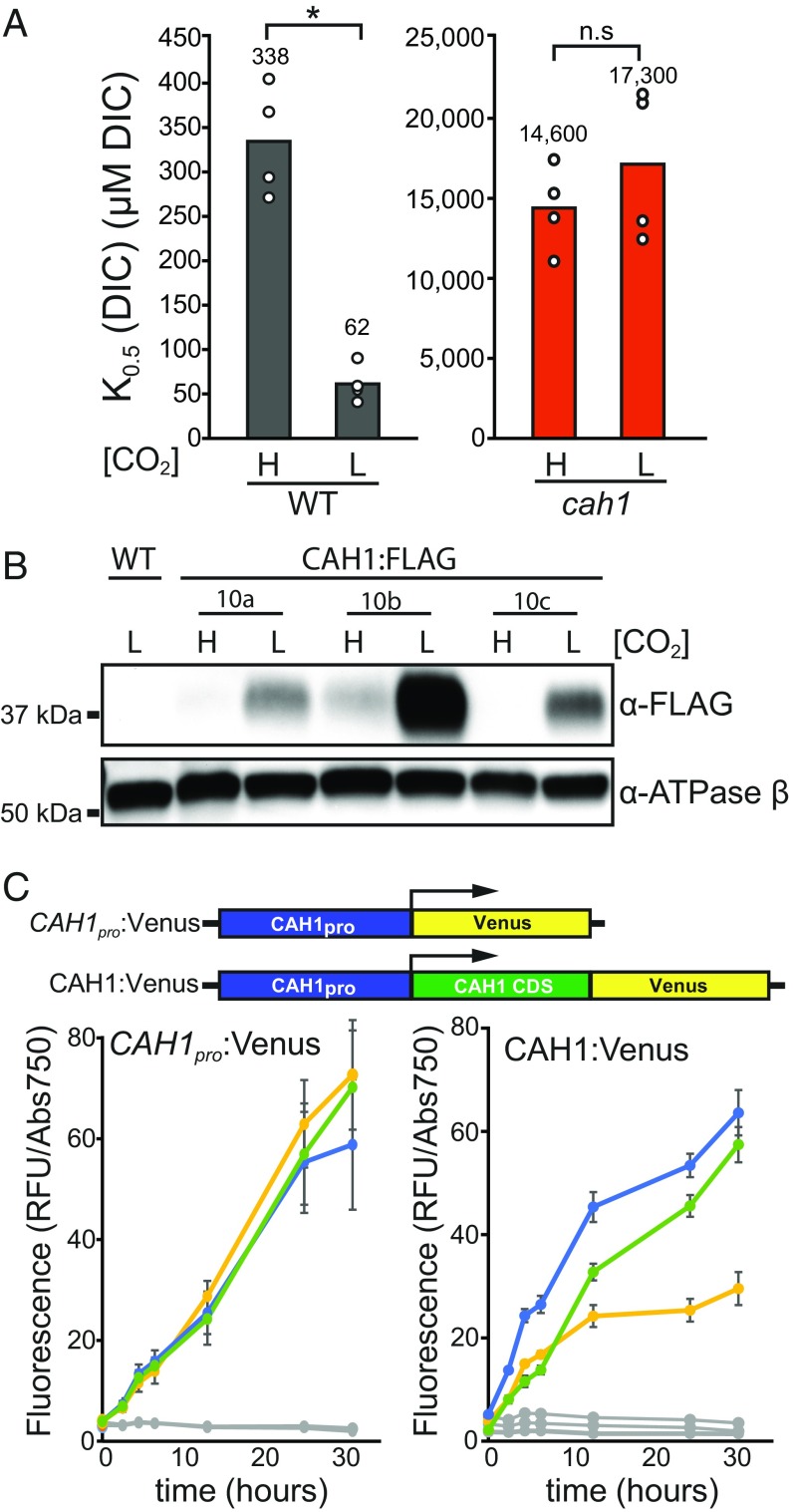
Modulation of CCM gene expression and function by environmental DIC concentration has been observed in several organisms (38, 39). To test the inducibility of the CCM in N. oceanica, we compared cells that had been acclimated to ambient (low CO2) conditions to those kept at 3% CO2. Photosynthetic DIC affinity was increased in low-CO2-acclimated WT cells, but not in cah1 cells (Fig. 4A). Immunoblot analysis of CAH1:FLAG complementation lines showed that this physiological acclimation of the CCM is associated with increased CAH1 protein accumulation after 24 h in low CO2 (Fig. 4B). To assess CAH1 induction over time as cells acclimated to a transfer from 3% CO2 to ambient air, we used easily quantified fluorescent reporter lines. To monitor protein accumulation, we used the aforementioned CAH1pro:CAH1:Venus complementation lines, and for a transcriptional reporter we used a similar construct without the CAH1 CDS (CAH1pro:Venus). In both reporter lines, fluorescence signal increased by ∼5-fold within 4.5 h and by ∼20-fold by 24 h after transfer from 3% CO2 to low CO2 (Fig. 4C), suggesting that induction of the CCM in N. oceanica is accompanied by transcriptional up-regulation of CAH1 as well as CAH1 protein accumulation.
Fig. 4.
Increased DIC affinity and CAH1 expression are associated with low CO2. (A) K0.5 (DIC) calculated from whole-cell O2 evolution assays of WT and cah1 cells after acclimation to low CO2 for 24 h. (B) CAH1 protein expression was assessed by immunoblot analysis of CAH:FLAG complementation lines (three independent lines shown). Protein samples were taken from cells placed at low CO2 for 24 h or kept at high CO2. (C) Fluorescent reporter lines for CAH1 transcription and protein level were grown at high CO2 (3%) and transferred to low CO2 (0.04%). The transcriptional reporter consisted of the native 1-kb promoter region driving Venus (CAH1pro:Venus). The CAH1 protein reporter was similar but included the CAH1 CDS in frame (CAH1:Venus). Normalized fluorescence, in relative fluorescence units (RFU), divided by absorbance at 750 nm, was quantified at the indicated time points for three independent lines (colored lines). Error bars show SD of n = 6 wells per line. Replicate plates were kept at 3% CO2 as controls (gray lines).
Discussion
A pump-leak type of CCM comprising bicarbonate transport and internal carbonic anhydrase activity has been proposed for Nannochloropsis based on measurements of CO2 and O2 fluxes (25–27). The severe DIC-dependent phenotype of the cah1 mutant and apparent regulation of CAH1 expression by environmental DIC strongly suggest that CAH1 provides this critical CCM-related carbonic anhydrase activity. The low photosynthetic DIC affinity of the cah1 mutant (K0.5[DIC] of ∼18,000 μM DIC) compared with that of the WT (∼90 μM DIC) (Fig. 1) is remarkable when considered alongside the values reported for CCM mutants of C. reinhardtii. A double mutant lacking two bicarbonate transporters, HLA3 (plasma membrane) and LCIA (chloroplast envelope), exhibited a K0.5[DIC] of ∼900 μM DIC at pH 9.0, compared with ∼250 μM DIC for WT in this condition (40). Even the cia5 mutant with a compromised “master regulator” of the CCM has a relatively milder defect, K0.5[DIC] ∼480 μM DIC, compared with ∼50 μM DIC for WT (41). One possible explanation is that N. oceanica relies entirely on bicarbonate as an inorganic carbon source (25, 26), yet achieves only a relatively small DIC fold accumulation compared with the environment (26). In this case, disruption of CAH1 may be compensated for only by extremely high concentrations of external bicarbonate that enters the cell and slowly converts to CO2 in the absence of catalysis.
In contrast to the chloroplast-localized carbonic anhydrases that are critical for CCM function in C. reinhardtii (11, 12) and P. tricornutum (18), CAH1 localizes to the lumen of the epiplastid ER (Fig. 3 and SI Appendix, Fig. S6), which forms the outermost membrane surrounding the chloroplast and is contiguous with the outer nuclear envelope and the ER (20, 21, 42). Directing CAH1 to the chloroplast with a different targeting signal rescued the mutant only weakly (SI Appendix, Fig. S7), indicating that CAH1 normally functions outside of the plastid. External carbonic anhydrase activity was not detected in N. gaditana (25), suggesting that the site of CO2 release is within the endomembrane network adjacent to the plastid, and not from a secreted protein. This position implies the existence of HCO3− transporters or channels in both the plasma membrane and the epiplastid ER to deliver HCO3− to CAH1. In this model, CAH1 in the epiplastid ER luminal space catalyzes the formation of CO2, which then either traverses the remaining membranes by diffusion to reach Rubisco or leaks back out of the cell (Fig. 5). In diatoms, carbonic anhydrases in the spaces between the multiple membranes surrounding the plastid are hypothesized to recover leaking CO2 from the chloroplast (43), suggesting an alternative model in which CAH1 traps leaking CO2 formed by some other carbonic anhydrase in the chloroplast. However, the substantial CO2 leakiness observed in other Nannochloropsis species by membrane-inlet mass spectrometry (25, 26) and stable isotope discrimination (44) does not support the existence of an effective CO2 retention mechanism, and this scenario would necessitate additional HCO3− transporters and a chloroplast-localized carbonic anhydrase.
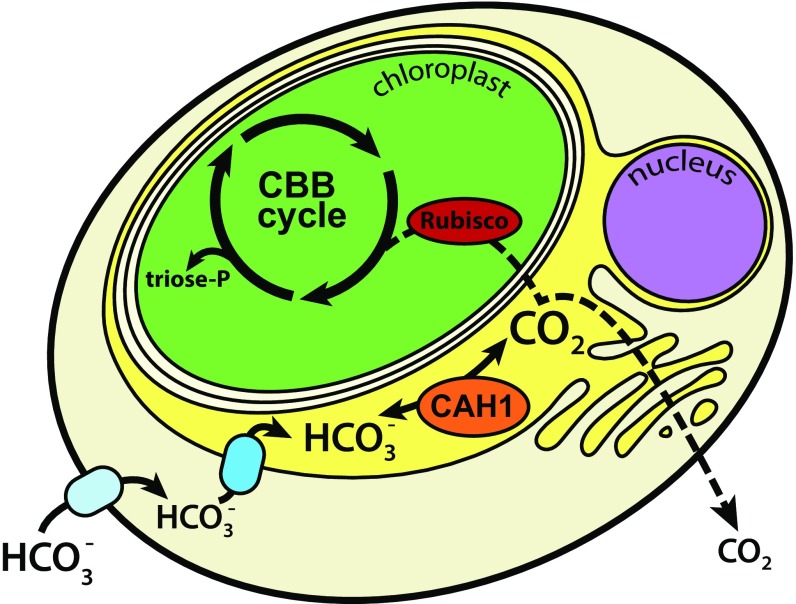
Fig. 5.
A proposed model for the CCM of N. oceanica. The plastid is separated from the cytoplasm by a total of four membranes, the outermost of which is contiguous with the ER and outer nuclear envelope (called the epiplastid ER). Transporters pump bicarbonate into the cytoplasm and then into the lumen of the epiplastid ER, where the carbonic anhydrase CAH1 also accumulates. CAH1 catalyzes the formation of CO2, which diffuses (dotted line) either into the chloroplast stroma to be fixed by Rubisco in the Calvin–Benson–Bassham cycle, or back out into the environment. Although leaky, this CCM is necessary for growth and photosynthesis at ambient (400 ppm CO2) conditions, likely by enhancing the carboxylation rate and suppressing photorespiration. CBB, Calvin–Benson–Bassham cycle.
Along with CAH1 (α type), there are at least two γ-type and two β-type carbonic anhydrases in N. oceanica CCMP1779 (SI Appendix, Table S1) (33). Knockdown lines of the β-type carbonic anhydrase (11263-mRNA in CCMP1779) showed no defect in growth at ambient CO2, whereas experiments with RNAi lines exhibiting reduced transcript levels of a γ-type carbonic anhydrase (g2209 in the N. oceanica strain IMET1) indicate a possible role in repressing the CCM under low pH (33). The strong phenotype of the cah1 mutant (Fig. 1) indicates that the other carbonic anhydrases in N. oceanica are nonredundant with CAH1, and the lack of effect of the carbonic anhydrase inhibitor ethoxyzolamide on the cah1 mutant (Fig. 2) suggests that there is little to no remaining carbonic anhydrase activity contributing to DIC affinity independently of CAH1. In addition, transcriptomic data from synchronous cultures of N. oceanica CCMP1779 show CAH1 to be relatively highly expressed with peak mRNA levels near the beginning of the light phase, in accordance with a role in carbon assimilation, whereas the other carbonic anhydrases show lower mRNA expression with peaks in the dark phase (45). Compared with C. reinhardtii and P. tricornutum, both of which have at least 12 carbonic anhydrases (7), N. oceanica apparently has a smaller gene repertoire of these enzymes. In C. reinhardtii, LCIB/C protein associates with the pyrenoid periphery and is thought to possibly recapture CO2 (46, 47). Curiously, the LCIB/C homolog in Phaeodactylum has recently been characterized as a novel type of carbonic anhydrase localized not to the pyrenoid periphery, but rather within transpyrenoidal thylakoids, functioning analogously to CAH3 in C. reinhardtii (18). In N. oceanica, no clear ortholog was found (SI Appendix, Table S1), which is consistent with the lack of a pyrenoid in this species.
What emerges from these observations is a possible model that describes a simple (and perhaps as a consequence, leaky) CCM in Nannochloropsis spp. that is nonetheless responsive to environmental DIC concentration and required for normal growth and physiology under ambient CO2 conditions. In N. oceanica, CAH1 plays a central role in this CCM, likely by facilitating the release of CO2 adjacent to the plastid to enhance carboxylation by Rubisco and suppress photorespiration (Fig. 5). This model likely applies to other species within this genus, and may provide a general framework for understanding CCMs in other algae that lack a pyrenoid. Identifying the other CCM components (e.g, HCO3− transporters), characterizing the other carbonic anhydrases, and determining the regulatory cue and signaling pathway responsible for activating/repressing the transcription of CAH1 (and presumably other CCM genes) will refine this model and further increase our understanding of the diversity of CCM form and function.
Nannochloropsis spp. have attracted considerable attention for biofuel and high-value compound production because of their rapid growth, genetic tractability, and ability to accumulate large amounts of lipids (23, 24, 48). Genetic manipulation and improvement of the CCM may enhance carbon uptake and growth in production settings, because the leaky CCM can be viewed as inefficient. However, loss-of-function mutants, such as cah1, may be useful as well, because such strains would constitute a safeguarding genetic barrier to accidental release of these algae into the environment (49), given their ability to thrive under exogenously supplied CO2 but not in the DIC concentrations found in natural settings.
Materials and Methods
Strains and Culture Conditions.
The sequenced strain of N. oceanica CCMP1779 was used. For all experiments, media consisted of artificial seawater with nutrient enrichment based on full-strength f medium (50) buffered by 10 mM Tris⋅HCl, pH 8.1. Growth chambers were set to continuous 28 °C and 100 μmol photons m−2 s−1. High CO2 conditions were maintained at 3% CO2 by a gas mixer and compressed CO2; low CO2 was ambient air (0.04% CO2). Because the cah1 mutant requires elevated CO2 conditions for growth, cultures were kept at 3% CO2 and grown to midlog phase (1 × 106 – 2 × 107 cells mL−1) for most experiments, and then either kept at 3% CO2 (control) or transferred to ambient air for 24 h before the assay, unless stated otherwise.
Cloning of the Homologous Recombination, Complementation, and Cell Marker Constructs.
Mutations in the gene NannoCCMP1779_6698-mRNA-1, which we named CARBONIC ANHYDRASE 1 (CAH1), were generated through insertion of a hygromycin resistance cassette (23). Insertion of this cassette was directed by homologous recombination (29) using 1-kb flanking homology regions to replace 250 bp beginning at the ATG start codon. All DNA sequences from CCMP1779 were obtained from the version 1 genome assembly (23).
For the complementation constructs, the native 1-kb CAH1 promoter region, the CAH1 coding sequence, and a C-terminal tag (either FLAG or Venus) was assembled via the Gibson method (51). To generate the His to Ala mutation of the active site (H177 in N. oceanica CAH1, corresponding to H119 in Homo sapiens CA-II), mutagenic primers were used in conjunction with Gibson assembly. The cah1 mutant line #5 was used as the recipient strain for all complementation experiments. Transformation was performed as described previously (29).
DIC Affinity Curves and Half-Saturation Determination.
DIC-dependent O2 evolution was measured as described previously (26, 52). O2 evolution assays were carried out in f medium (pH 8.1) without added DIC, and allowed to reach the DIC compensation point before beginning the experiment. Chlorophyll was quantified (53) and used to normalize the O2 evolution rates. The resulting normalized DIC-dependent photosynthesis curves were processed with a Python script to fit a Michaelis–Menten equation to the data and solve for Vmax and K0.5(DIC). For photosynthesis inhibition experiments, cells were incubated for 1 h in standard medium with a final concentration of 100 μM ethoxyzolamide or an equivalent volume of DMSO.
Chlorophyll Fluorescence by PAM Fluorometry.
Cells were grown in liquid culture at 3% CO2 and then acclimated for 24 h in ambient air before harvesting by centrifugation. Cells were resuspended in fresh medium with a final concentration of either 2 mM or 40 mM DIC added as NaHCO3, capped, and placed in the dark for 30 min before the beginning of the experiment. Chlorophyll fluorescence was analyzed by PAM fluorometry with red actinic light of 100 μmol photons m−2 s−1. A two-factor ANOVA analysis was performed using JASP version 0.7.5.5 to determine a P value for the interaction term genotype × [DIC].
Fluorescent Reporter Induction.
Cultures were grown in duplicate 24-well tissue culture plates to midlog phase at 3% CO2. Bulk fluorescence (Venus excitation, 515 nm; emission, 535 nm) was measured with a plate reader and normalized to absorbance at 750 nm. The experiment was initiated when one plate was transferred to ambient air, and readings were taken at the indicated times.
Supplementary Material
Acknowledgments
We thank Christoph Benning for the sequenced strain of N. oceanica CCMP1779 and the hygromycin resistance cassette, Kent McDonald and Reena Zalpuri for technical assistance with TEM sample prep and imaging, Steve Ruzin and Denise Schichnes for technical assistance with fluorescence microscopy, Victor Chubukov for assistance with Python scripting, and Matthew Brooks and Patrick Shih for identification and cloning of expression vector promoters and terminators. C.W.G. was supported by the National Science Foundation (Graduate Student Research Fellowship Grant DGE 1106400). K.K.N. is an investigator of the Howard Hughes Medical Institute and is supported by the Gordon and Betty Moore Foundation (Grant GBMF3070).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700139114/-/DCSupplemental.
References
- 1.Riebesell U, Wolf-Gladrow D, Smetacek V. Carbon dioxide limitation of marine phytoplankton growth rates. Nature. 1993;361:249–251. [Google Scholar]
- 2.Ogren WL. Photorespiration: Pathways, regulation, and modification. Annu Rev Plant Physiol. 1984;35:415–442. [Google Scholar]
- 3.Bauwe H, Hagemann M, Fernie AR. Photorespiration: Players, partners and origin. Trends Plant Sci. 2010;15:330–336. doi: 10.1016/j.tplants.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Reinfelder JR. Carbon-concentrating mechanisms in eukaryotic marine phytoplankton. Annu Rev Mar Sci. 2011;3:291–315. doi: 10.1146/annurev-marine-120709-142720. [DOI] [PubMed] [Google Scholar]
- 5.Price GD, Badger MR, Woodger FJ, Long BM. Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): Functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J Exp Bot. 2008;59:1441–1461. doi: 10.1093/jxb/erm112. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Stessman DJ, Spalding MH. The CO2-concentrating mechanism and photosynthetic carbon assimilation in limiting CO2: How Chlamydomonas works against the gradient. Plant J. 2015;82:429–448. doi: 10.1111/tpj.12829. [DOI] [PubMed] [Google Scholar]
- 7.Moroney JV, et al. The carbonic anhydrase isoforms of Chlamydomonas reinhardtii: Intracellular location, expression, and physiological roles. Photosynth Res. 2011;109:133–149. doi: 10.1007/s11120-011-9635-3. [DOI] [PubMed] [Google Scholar]
- 8.Badger MR, et al. The diversity and coevolution of Rubisco, plastids, pyrenoids, and chloroplast-based CO2-concentrating mechanisms in algae. Can J Bot. 1998;76:1052–1071. [Google Scholar]
- 9.Mackinder LCM, et al. A repeat protein links Rubisco to form the eukaryotic carbon-concentrating organelle. Proc Natl Acad Sci USA. 2016;113:5958–5963. doi: 10.1073/pnas.1522866113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engel BD, et al. Native architecture of the Chlamydomonas chloroplast revealed by in situ cryo-electron tomography. eLife. 2015;4:1–29. doi: 10.7554/eLife.04889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karlsson J, et al. A novel alpha-type carbonic anhydrase associated with the thylakoid membrane in Chlamydomonas reinhardtii is required for growth at ambient CO2. EMBO J. 1998;17:1208–1216. doi: 10.1093/emboj/17.5.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sinetova MA, Kupriyanova EV, Markelova AG, Allakhverdiev SI, Pronina NA. Identification and functional role of the carbonic anhydrase Cah3 in thylakoid membranes of pyrenoid of Chlamydomonas reinhardtii. Biochim Biophys Acta. 2012;1817:1248–1255. doi: 10.1016/j.bbabio.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 13.Giordano M, Beardall J, Raven JA. CO2-concentrating mechanisms in algae: Mechanisms, environmental modulation, and evolution. Annu Rev Plant Biol. 2005;56:99–131. doi: 10.1146/annurev.arplant.56.032604.144052. [DOI] [PubMed] [Google Scholar]
- 14.Raven JA, Cockell CS, De La Rocha CL. The evolution of inorganic carbon-concentrating mechanisms in photosynthesis. Philos Trans R Soc Lond B Biol Sci. 2008;363:2641–2650. doi: 10.1098/rstb.2008.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falkowski PG, et al. The evolution of modern eukaryotic phytoplankton. Science. 2004;305:354–360. doi: 10.1126/science.1095964. [DOI] [PubMed] [Google Scholar]
- 16.Rousseaux CS, Gregg WW. Interannual variation in phytoplankton primary production at a global scale. Remote Sens. 2013;6:1–19. [Google Scholar]
- 17.Hopkinson BM, Dupont CL, Matsuda Y. The physiology and genetics of CO2-concentrating mechanisms in model diatoms. Curr Opin Plant Biol. 2016;31:51–57. doi: 10.1016/j.pbi.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 18.Kikutani S, et al. Thylakoid luminal θ-carbonic anhydrase critical for growth and photosynthesis in the marine diatom Phaeodactylum tricornutum. Proc Natl Acad Sci USA. 2016;113:9828–9833. doi: 10.1073/pnas.1603112113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tachibana M, et al. Localization of putative carbonic anhydrases in two marine diatoms, Phaeodactylum tricornutum and Thalassiosira pseudonana. Photosynth Res. 2011;109:205–221. doi: 10.1007/s11120-011-9634-4. [DOI] [PubMed] [Google Scholar]
- 20.Moog D, Stork S, Reislöhner S, Grosche C, Maier U-G. In vivo localization studies in the stramenopile alga Nannochloropsis oceanica. Protist. 2015;166:161–171. doi: 10.1016/j.protis.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 21.Murakami R, Hashimoto H. Unusual nuclear division in Nannochloropsis oculata (Eustigmatophyceae, Heterokonta), which may ensure faithful transmission of secondary plastids. Protist. 2009;160:41–49. doi: 10.1016/j.protis.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Cavalier-Smith T. Genomic reduction and evolution of novel genetic membranes and protein-targeting machinery in eukaryote-eukaryote chimaeras (meta-algae) Philos Trans R Soc Lond B Biol Sci. 2003;358:109–133, discussion 133–134. doi: 10.1098/rstb.2002.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vieler A, et al. Genome, functional gene annotation, and nuclear transformation of the heterokont oleaginous alga Nannochloropsis oceanica CCMP1779. PLoS Genet. 2012;8:e1003064. doi: 10.1371/journal.pgen.1003064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radakovits R, et al. Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropis gaditana. Nat Commun. 2012;3:686. doi: 10.1038/ncomms1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huertas IE, Espie GS, Colman B, Lubian LM. Light-dependent bicarbonate uptake and CO2 efflux in the marine microalga Nannochloropsis gaditana. Planta. 2000;211:43–49. doi: 10.1007/s004250000254. [DOI] [PubMed] [Google Scholar]
- 26.Sukenik A, et al. Uptake, efflux, and photosynthetic utilization of inorganic carbon by the marine eustigmatophyte Nannochloropsis sp. J Phycol. 1997;33:969–974. [Google Scholar]
- 27.Huertas IE, Colman B, Espie GS. Mitochondrial-driven bicarbonate transport supports photosynthesis in a marine microalga. Plant Physiol. 2002;130:284–291. doi: 10.1104/pp.004598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang D, et al. Nannochloropsis genomes reveal evolution of microalgal oleaginous traits. PLoS Genet. 2014;10:e1004094. doi: 10.1371/journal.pgen.1004094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kilian O, Benemann CSE, Niyogi KK, Vick B. High-efficiency homologous recombination in the oil-producing alga Nannochloropsis sp. Proc Natl Acad Sci USA. 2011;108:21265–21269. doi: 10.1073/pnas.1105861108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dolch LJ, et al. A palmitic acid elongase affects eicosapentaenoic acid and plastidal monogalactosyldiacylglcerol levels in Nannochloropsis. Plant Physiol. 2016;173:742–759. doi: 10.1104/pp.16.01420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vieler A, Brubaker SB, Vick B, Benning C. A lipid droplet protein of Nannochloropsis with functions partially analogous to plant oleosins. Plant Physiol. 2012;158:1562–1569. doi: 10.1104/pp.111.193029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baker NR. Chlorophyll fluorescence: A probe of photosynthesis in vivo. Annu Rev Plant Biol. 2008;59:89–113. doi: 10.1146/annurev.arplant.59.032607.092759. [DOI] [PubMed] [Google Scholar]
- 33.Wei L, et al. RNAi-based targeted gene knockdown in the model oleaginous microalgae Nannochloropsis oceanica. Plant J. 2017;89:1236–1250. doi: 10.1111/tpj.13411. [DOI] [PubMed] [Google Scholar]
- 34.Kiefer LL, Fierke CA. Functional characterization of human carbonic anhydrase II variants with altered zinc-binding sites. Biochemistry. 1994;33:15233–15240. doi: 10.1021/bi00255a003. [DOI] [PubMed] [Google Scholar]
- 35.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 36.Gschloessl B, Guermeur Y, Cock JM. HECTAR: A method to predict subcellular targeting in heterokonts. BMC Bioinformatics. 2008;9:393. doi: 10.1186/1471-2105-9-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Markwardt ML, et al. An improved cerulean fluorescent protein with enhanced brightness and reduced reversible photoswitching. PLoS One. 2011;6:e17896. doi: 10.1371/journal.pone.0017896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsuda Y, Nakajima K, Tachibana M. Recent progresses on the genetic basis of the regulation of CO2 acquisition systems in response to CO2 concentration. Photosynth Res. 2011;109:191–203. doi: 10.1007/s11120-011-9623-7. [DOI] [PubMed] [Google Scholar]
- 39.Winck FV, et al. Genome-wide identification of regulatory elements and reconstruction of gene regulatory networks of the green alga Chlamydomonas reinhardtii under carbon deprivation. PLoS One. 2013;8:e79909. doi: 10.1371/journal.pone.0079909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamano T, Sato E, Iguchi H, Fukuda Y, Fukuzawa H. Characterization of cooperative bicarbonate uptake into chloroplast stroma in the green alga Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 2015;112:7315–7320. doi: 10.1073/pnas.1501659112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L, Yamano T, Kajikawa M, Hirono M, Fukuzawa H. Isolation and characterization of novel high-CO2-requiring mutants of Chlamydomonas reinhardtii. Photosynth Res. 2014;121:175–184. doi: 10.1007/s11120-014-9983-x. [DOI] [PubMed] [Google Scholar]
- 42.Hempel F, Bullmann L, Lau J, Zauner S, Maier UG. ERAD-derived preprotein transport across the second outermost plastid membrane of diatoms. Mol Biol Evol. 2009;26:1781–1790. doi: 10.1093/molbev/msp079. [DOI] [PubMed] [Google Scholar]
- 43.Samukawa M, Shen C, Hopkinson BM, Matsuda Y. Localization of putative carbonic anhydrases in the marine diatom, Thalassiosira pseudonana. Photosynth Res. 2014;121:235–249. doi: 10.1007/s11120-014-9967-x. [DOI] [PubMed] [Google Scholar]
- 44.Hanson DT, et al. On-line stable isotope gas exchange reveals an inducible but leaky carbon concentrating mechanism in Nannochloropsis salina. Photosynth Res. 2014;121:311–322. doi: 10.1007/s11120-014-0001-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poliner E, et al. Transcriptional coordination of physiological responses in Nannochloropsis oceanica CCMP1779 under light/dark cycles. Plant J. 2015;83:1097–1113. doi: 10.1111/tpj.12944. [DOI] [PubMed] [Google Scholar]
- 46.Yamano T, et al. Light- and low-CO2-dependent LCIB-LCIC complex localization in the chloroplast supports the carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Cell Physiol. 2010;51:1453–1468. doi: 10.1093/pcp/pcq105. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y, Spalding MH. LCIB in the Chlamydomonas CO2-concentrating mechanism. Photosynth Res. 2014;121:185–192. doi: 10.1007/s11120-013-9956-5. [DOI] [PubMed] [Google Scholar]
- 48.Simionato D, et al. The response of Nannochloropsis gaditana to nitrogen starvation includes de novo biosynthesis of triacylglycerols, a decrease of chloroplast galactolipids, and reorganization of the photosynthetic apparatus. Eukaryot Cell. 2013;12:665–676. doi: 10.1128/EC.00363-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clark RL, Cameron JC, Root TW, Pfleger BF. Insights into the industrial growth of cyanobacteria from a model of the carbon-concentrating mechanism. Am Inst Chem Eng J. 2014;60:1269–1277. [Google Scholar]
- 50.Guillard RRL, Ryther JH. Studies of marine planktonic diatoms, I: Cyclotella nana Hustedt, and Detonula confervacea (cleve) Gran. Can J Microbiol. 1962;8:229–239. doi: 10.1139/m62-029. [DOI] [PubMed] [Google Scholar]
- 51.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 52.Yamano T, Miura K, Fukuzawa H. Expression analysis of genes associated with the induction of the carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Physiol. 2008;147:340–354. doi: 10.1104/pp.107.114652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ritchie RJ. Consistent sets of spectrophotometric chlorophyll equations for acetone, methanol, and ethanol solvents. Photosynth Res. 2006;89:27–41. doi: 10.1007/s11120-006-9065-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.