Significance
Hybrids are of high value in agriculture. Hybrid vigor applies only to the F1 generation. Pure breeding lines, which maintain the hybrid phenotype, hybrid mimics, together with the small phenotype line have proven to be powerful in identifying genes and pathways critical for hybrid vigor. PHYTOCHROME-INTERACTING FACTOR (PIF4), regulating auxin biosynthesis and action, features in the development of the hybrid vigor phenotype. The homozygous hybrid mimic lines all have the same particular 12 chromosomal segments, 4 from the C24 parent and 8 from Ler. The small phenotype line has each of these segments derived from the genome of the alternative parent, so one or more loci needed for the large hybrid mimic phenotype are likely to be on each segment.
Keywords: heterosis, transcription factor, biomass, transregulation, germination
Abstract
F1 hybrids in Arabidopsis and crop species are uniform and high yielding. The F2 generation loses much of the yield advantage and the plants have heterogeneous phenotypes. We generated pure breeding hybrid mimic lines by recurrent selection and also selected a pure breeding small phenotype line. The hybrid mimics are almost completely homozygous with chromosome segments from each parent. Four particular chromosomal segments from C24 and 8 from Ler were present in all of the hybrid mimic lines, whereas in the F6 small phenotype line, the 12 segments were each derived from the alternative parent. Loci critical for promoting hybrid vigor may be contained in each of these 12 conserved segments. We have identified genes with similar altered expression in hybrid mimics and F1 plants but not in the small phenotype line. These genes may be critical for the generation of hybrid vigor. Analysis of transcriptomes indicated that increased expression of the transcription factor PHYTOCHROME-INTERACTING FACTOR (PIF4) may contribute to hybrid vigor by targeting the auxin biosynthesis gene YUCCA8 and the auxin signaling gene IAA29. A number of auxin responsive genes promoting leaf growth were up-regulated in the F1 hybrids and hybrid mimics, suggesting that increased auxin biosynthesis and signaling contribute to the hybrid phenotype. The hybrid mimic seeds had earlier germination as did the seeds of the F1 hybrids, indicating cosegregation of the genes for rosette size and the germination trait. Early germination may be an indicator of vigorous hybrids.
Two features common to hybrids in many crops are the increased yield and phenotypic uniformity of the F1 hybrid generation and the reduced yield and phenotypic heterogeneity of the F2 generation (1). These characteristics also apply to the F1 and F2 populations in Arabidopsis hybrids (2). The processes of capture of light and assimilation of CO2 into photosynthate are the same in the hybrid and parents and as the hybrids have larger leaves than their parents, they produce more photosynthate (3).
In a previous paper (2), transcriptome analyses showed that in the hybrids, most genes are expressed at the same levels as in the parents. Compared with the average level of gene expression in the parents, ∼2,000 genes (6% of the genome) have altered expression in the hybrids and are likely to be involved in the generation of the hybrid vigor phenotype. Many of the differentially expressed genes (DEGs) encode proteins in key metabolic pathways such as the plant hormone systems auxin and salicylic acid (SA) and the basal defense response, suggesting that these pathways contribute to the development of hybrid vigor (4). Altered hormone abscisic acid (ABA) and defense response have also been reported in rice hybrids (5). Despite the contrasting phenotypes of the F1 and F2 populations, we were able to develop “pure breeding” F5/F6 lines with phenotypes comparable to the F1 hybrids (hybrid mimics) (2). The hybrid mimic lines retained the production properties of the F1 hybrids in biomass and had phenotypic uniformity in the F5/F6 and subsequent generations. Transcriptome analysis of the hybrid mimics showed that many differentially expressed genes in the F1 hybrids maintain their expression levels in the hybrid mimics. In contrast to the heterozygous F1 hybrids, the genomes of the hybrid mimics are homozygous for chromosome segments contributed by both parents (2).
In the present paper, we characterized an additional F6 hybrid mimic line with F1 hybrid-like phenotype and three other F6 lines with smaller phenotypes. This allowed us to identify pathways and chromosome segments operating in the F1 hybrid and hybrid mimics but not in the small phenotype plants. Increased expression of a transcription factor gene, PHYTOCHROME-INTERACTING FACTOR 4 (PIF4), was associated with the large plant phenotype of the F1 hybrid and hybrid mimics through altered auxin biosynthesis and downstream signaling. In contrast, the F6 small line did not show increased PIF4 activity. The F1 hybrids and the three hybrid mimic lines had similar developmental characteristics in early as well as later stages of growth. The F1 hybrids and hybrid mimics had earlier seed germination than the parents; the small line germinated later than the parents. Rapid germination could be a precursor of vigor in the mature hybrid plant.
Results
Hybrid Mimic Lines Are Similar to the F1 Hybrids.
The F1 hybrid produced by crossing C24 and Landsberg erecta (Ler) accessions had substantial hybrid vigor in both biomass and seed yield phenotypes (6). The phenotype of the heterozygous F1 hybrid is highly uniform, but there were several phenotypes in the F2 population. We found that by using individual F2 plants in a stringent recurrent phenotypic selection program, plants with a phenotype similar to the selected F2 plant were produced in a few generations (2). We have generated six pure breeding F6 plant lines and assessed the growth patterns of these lines against those of the parents and F1 hybrids (Fig. S1). The reciprocal F1 hybrids showed growth heterosis in the first week, but they were also subject to maternal influence; by 15 d after sowing (DAS) the growth difference between the reciprocal hybrids was negligible (7) and they had similar phenotypes in rosette diameter, leaf morphology, and fresh weight (Fig. 1 A and B). The three F6 hybrid mimic lines: HM-W, HM-S, and HM-G had growth patterns resembling the F1 hybrids, and at 15 DAS were similar to the hybrids in plant size and aerial fresh weight (Fig. 1 A and B and Fig. S1A). The F1 hybrids and hybrid mimics flowered within the time frame of the two parents (17–38 DAS) (Dataset S1, Table S1) and were ∼60% larger than the larger parent C24 at 35 DAS (Fig. S1B). The small phenotype line, Sml-D was early flowering with fresh weight approximately half the average weight of the parents at 15 DAS (Fig. 1 A and B and Dataset S1, Table S1). By 35 DAS, the rosette diameter of Sml-D was about half the rosette diameter of the parents and was much smaller than the hybrids and hybrid mimics (Fig. S1B). Two other F6 medium lines: Med-E and Med-F had plant sizes similar to the parents until 23 DAS when Med-F and the Ler parent flowered. Med-E had a flowering time similar to the later flowering parent C24 (C24: 30 ± 5 DAS, Med-E: 33 ± 5 DAS) and was larger than both parents at 35 DAS (Fig. S1 and Dataset S1, Table S1).
Fig. S1.
Growth patterns of the parents C24 and Ler, F1 hybrids, three hybrid mimic lines (F7), and other F7 plant lines. (A) Growth course of the parents, hybrids, and F6 lines. More than 30 plants were measured for each line. Inset shows the rosette diameter of the parent, hybrids, and F6 lines at 7 DAS (cotyledon stage). (B) Box plot showing the range of plant size in each line at 35 DAS. Error bars = SEM. The black dotted line represents the rosette diameter of the F1 hybrid. n ≥ 30 plants for each line.
Fig. 1.
Hybrid mimic lines have phenotypes similar to the F1 hybrids. (A) Photographs of the two parents (C24 and Ler), the F1, and selected lines at 15 DAS. (Scale bar, 1 cm; applies to all images.) (B) Rosette diameter and aerial fresh weight of the two parents (C24 and Ler), the F1, and the selected lines at 15 DAS. * indicates significant differences from the mid-parent value (MPV) at P < 0.05. Error bars = SEM n ≥ 3. The black dotted line represents MPV. (C) Representation of the genotype of chromosome 1 of three sibling plants from each F6 plant line. Each line along the chromosomes represents a SNP. The red/blue/green bars represent the genotypes C24 homozygous, Ler homozygous, and C24/Ler heterozygous (Heter.). (D) Location of 12 chromosomal segments with the same parental genotype in three hybrid mimic lines (HM-W, HM-S, and HM-G), and not in the small line (Sml-D). The red/blue bars represent the genotypes C24 homozygous, Ler homozygous conserved in the hybrid mimic lines.
The similarity between the F1 hybrids and the three hybrid mimic lines was evident as early as seed germination. The C24/Ler F1 hybrids germinate earlier than the parents (7). In the present experiment, both reciprocal hybrids and the three hybrid mimic lines had germination times ∼10–12 h ahead of the parents [34 vs. 44–46 h after sowing (HAS)] (Fig. S2). At 36 h after sowing when parental seeds were 50% germinated, ∼90% of the seeds of the F1 hybrids and the three hybrid mimic lines had germinated. Plant lines Med-E and Med-F, with growth patterns similar to the parents until 23 DAS, had germination times close to those of the parents, although Med-E germinated slightly before either parent. The small plant line, Sml-D, germinated 21–23 h later than the parents, with 100% germination by 88 h after sowing (Table 1 and Fig. S2).
Fig. S2.
Germination time of the two parents C24 and Ler, F1 hybrids, three hybrid mimic lines (F7), and three smaller F7 lines. The values present the time [hours after sowing (HAS)] when 80% of seeds had germinated in each plant line. Three replicates were conducted. n ≥ 10 in each replicate. Error bars = SEM.
Table 1.
Proportion of germinated seeds from the two parents C24 and Ler, F1 hybrids, and F6 plant lines at 28, 36, 44, and 88 HAS
| Time after sowing | C24 (%) | Ler (%) | C24 × Ler (%) | Ler × C24 (%) | HM-W (%) | HM-S (%) | HM-G (%) | Med-E (%) | Med-F (%) | Sml-D (%) |
| 28 HAS | 2 | 2 | 30 | 35 | 2 | 42 | 17 | 2 | 0 | 0 |
| 36 HAS | 53 | 50 | 95 | 98 | 88 | 92 | 100 | 68 | 53 | 24 |
| 44 HAS | 80 | 71 | 95 | 100 | 93 | 97 | 100 | 88 | 75 | 34 |
| 88 HAS | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
Genomes of F6 Hybrid Mimic Lines.
The genomes of three siblings from each of the independent mimic lines were characterized by SNP analysis (Dataset S1, Tables S2 and S3). The mimic plants were almost completely homozygous with chromosomal segments from both parents, half from the C24 parent, and half from Ler (Fig. 1C and Fig. S3). In the mimic lines, residual heterozygosity ranged from 0.1% to 12.6% of the total SNPs in the genome; one HM-S plant was heterozygous for one arm of chromosome 1, accounting for 12.3% of the SNPs in the genome (Dataset S1, Table S3). Because this plant and its two siblings had the same phenotype for rosette diameter, the DNA segments carrying genes responsible for the large plant size of the HM-S line were unlikely to be present in this arm of chromosome 1 (Fig. 1B and Fig. S3). Approximately 60% of genes have the same genotypes between hybrid mimic lines HM-W and HM-S and between HM-S and HM-G, but not between HM-W and HM-G (47%); in lines Med-F and Sml-D, 70% of the genes had the same allelic makeup; the level of similarity in segmental genotype was reflected in similar growth patterns and germination times between these two lines (Figs. S1 and S2).
Fig. S3.
Genotypes of chromosomal segments of F6 lines. The genotypes of three siblings (sib.) of each F6 plant line were displayed along the five Arabidopsis chromosomes. Each line along the chromosomes represents a SNP. The red/blue/green bars represent the genotypes C24 homozygous, Ler homozygous, and C24/Ler heterozygous (Heter.). The black arrows show the chromosomal location of PIF4, FRI, YUC8, IAA29, and FLC.
The F6 plants selected on the basis of a phenotype resembling the F1 hybrids had similar phenotypes to the selected F2 plant and to each other (hybrid mimics). We identified 12 chromosomal segments (4 from C24 and 8 from Ler) present in all three hybrid mimic lines. The F6 line selected for a small phenotype (Sml-D) had the opposite parental source for each of the 12 segments (Fig. 1D, Fig. S3, and Dataset S1, Table S4). For example, one third of chromosome 1 was homozygous for Ler in the three hybrid mimic lines, but in the small line (Sml-D), the same region was derived from C24 (Fig. 1 C and D and Fig. S3). Approximately 6,400 genes are located in the 12 chromosome segments, and 21% of these genes had different expression levels in C24 and Ler (1,348 genes); in the hybrid mimic and small lines, the alternative genomic segments produce different expression levels. Of these 12 chromosomal segments in the hybrid mimic lines, 3 were present in the medium line Med-E and 5 were present in the Med-F line. The absence of some of the hybrid mimic conserved chromosomal segments could be associated with the smaller plant size of the Med-E and Med-F lines compared with the size of the hybrid mimics (Fig. S3 and Dataset S1, Table S4).
Transcriptome Analysis of Parents, Hybrid, and Hybrid Mimics.
The presence of the two genomes in the same nucleus of the hybrid makes interaction possible between genes and gene products from the two parental genomes. Transinteractions could result in the altered gene expression levels that generate hybrid vigor.
Transcriptomes of parents, hybrids, and hybrid mimics were analyzed to identify genes with the same altered patterns of expression in hybrids and hybrid mimics. Aerial tissue of 15-d-old-seedlings of the C24 and Ler parents, the reciprocal hybrids, and the six F6 plant lines was used for transcriptome analyses (Dataset S1, Table S2). A total of 6,169 genes were differentially expressed between the two parents, with approximately half expressed at a higher level in C24 than in Ler (Fig. 2A). A total of 2,059 genes were differentially expressed in the hybrids relative to the average expression level in the parents [mid-parent value (MPV)] (Fig. 2B). There were 157 genes with transcription factor activity in the nonadditively expressed genes in the hybrids (8) (Dataset S1, Table S5), suggesting that many of the differentially expressed genes in the F1 hybrids could have been subjected to transregulation by these transcription factors. Metabolic pathways that were enriched in the nonadditively expressed genes included “response to hormone stimulus” (75 genes), “photosynthesis” (18 genes), and “lipid transport” (14 genes) (Dataset S1, Table S5). Approximately half of the hormone-associated differentially expressed genes in the F1 hybrids were annotated as genes responding to auxin stimulus (34 genes). Of the 14 genes involved in lipid transport, 10 were down-regulated in the F1 hybrids relative to the parents (Dataset S1, Table S6), indicating that the hybrids and the parents differ in lipid metabolic activities (9). Down-regulated genes included 18 nuclear-encoded photosynthesis genes with products involved in photosystems I and II (Dataset S1, Table S6). The down-regulation of these genes did not alter photosynthesis efficiency, as the parents and hybrids have the same rate of photosynthesis per unit area (3). Previously we found that down-regulated defense response genes were significantly (P < 0.05) overrepresented in the F1 nonadditive genes (4), but in the present analysis, the down-regulated defense response genes were only marginally enriched.
Fig. 2.
Transcriptome analysis of parents, hybrid, and hybrid mimics. (A) Genes totaling 6,169 were differentially expressed between C24 and Ler parents. (B) The numbers of genes in the F1 hybrids and F6 lines differentially expressed compared with the average expression level of the parents (MPV). (C) Venn diagram of differentially expressed genes in the F1 hybrids and the F6 plant lines. (D) Venn diagram of up-regulated differentially expressed genes shared by the F1 hybrids and the three hybrid mimic lines. (E) Venn diagram of down-regulated differentially expressed genes shared by the F1 hybrids and the three hybrid mimic lines.
Between 3,170 and 5,318 genes were differentially expressed in the F6 plant lines relative to the MPV of the parents (Fig. 2B). There were more DEGs shared between the hybrids and the hybrid mimic lines than between the hybrids and the small plants (Fig. 2C). More DEGs were shared between the two hybrid mimic lines HM-W and HM-S than between HM-W and HM-G, or between HM-S and HM-G (Fig. 2 C–E); this finding is consistent with the similar growth patterns and cotyledon sizes at the early stage between HM-W and HM-S (Fig. S1A, Inset).
The 2,059 genes differentially expressed in the F1 hybrid (F1 DEGs) were scored for their expression levels in the hybrid mimics and the other F6 lines and shown in gene expression heat maps (Fig. 3A). The HM-W and HM-S hybrid mimic lines had the highest percentage of differentially expressed loci expressed similarly to the F1 DEGs (HM-W: 53%, HM-S: 65%); only 1–2% were changed in the opposite direction of expression. In the HM-G hybrid mimic line, 22% of the F1 DEGs retained their differential levels of expression, and 3% of the F1 DEGs had an opposite direction of changes to the DEGs in the HM-G line (Fig. 3). The lower number of F1 DEGs present in HM-G might indicate that these are the loci most critical to the development of the F1 hybrid phenotype.
Fig. 3.
Hybrid mimic lines show gene expression profiles similar to the hybrids. (A) Heat maps showing the expression levels of the 2,059 F1 differentially expressed genes (DEGs) relative to the MPV in the hybrids and F6 plant lines. The red/green colors indicate the up-/down-regulated fold change (FC) from the MPV. Each data point represents the mean of at least three biological replicates. (B) The proportion of genes in each expression category in the heat maps (A) in the various lines represented as a histogram.
In the other F6 lines, the majority of the F1 DEGs (71–80%) were expressed at parental levels. F1 DEGs were expressed in Med-E in a similar (9%) or opposite (11%) direction to the F1 hybrid (Fig. 3). The Med-E plants had a larger size in later stages of growth, but not at 15 DAS (Fig. S1); they showed more similarity of gene expression to the hybrid in the rosette leaves at 28 DAS (2). The Sml-D line had the fewest F1 DEGs (8%) expressed at levels similar to the hybrids, and 11% of the F1 DEGs had expression patterns opposite to that of the hybrids (Fig. 3).
In Arabidopsis, plant biomass and flowering time are correlated, with earlier flowering plants being smaller than those that flower later. Med-F and Sml-D with flowering times as early as 15–18 DAS had small rosette sizes at the mature plant stage (Fig. S1 and Dataset S1, Table S1). At 15 DAS, as in the HM-G line (22%), 21% of the F1 DEGs were expressed similarly in the F1 hybrids and the Med-F line (Fig. 3) with an expectation that Med-F should have a large phenotype; but early flowering may have prevented this line from reaching a large plant size (Dataset S1, Table S1).
F1 Hybrids and F6 Hybrid Mimic Lines Had Similar Changes in the Levels of Expression of Genes in Hormonal Pathways.
A total of 258 DEGs were shared between the F1 hybrids and the three hybrid mimic lines, both in the levels of expression and the direction of change (Fig. 2 D and E and Dataset S1, Table S7); of those in the small line, 195 genes (76%) were either differentially expressed in an opposite direction or expressed at MPV, suggesting that alternative alleles segregated with the two contrasting phenotypic selections. Of the 135 genes up-regulated in the F1 hybrids and all three hybrid mimics, the response-to-hormone-stimulus term was enriched (Dataset S1, Table S8); many genes were associated with indole-3-acetic acid (IAA) (auxin), ethylene (ET), brassinosteroid (BR), gibberellic acid (GA), and cytokinin (CK), emphasizing the role of hormones in the generation of the hybrid vigor phenotype (Dataset S1, Table S7). In the 123 down-regulated genes shared by the hybrids and the three hybrid mimics, there was an overrepresentation of the number of genes in the “carbohydrate biosynthetic process” term (six genes) (Dataset S1, Table S9). The altered gene activities in the F1 hybrids and in the three hybrid mimic lines were not present in the small or medium lines (Dataset S1, Table S10). Two genes involved in starch biosynthesis, ADP-GLUCOSE PYROPHOSPHORYLASE 3 (APL3) and BRANCHING ENZYME 1 (DBE1) were down-regulated in the hybrids and the three hybrid mimic lines, but were up-regulated or had no change in the medium or small phenotype lines (Dataset S1, Table S10).
Auxin Pathway Responses.
Auxin is critical for embryogenesis and leaf development (10) and it has been suggested that it contributes to hybrid vigor in both Arabidopsis and rice (4, 11). The YUCCA flavin monooxygenases are key enzymes catalyzing the rate-limiting step in auxin biosynthesis (12). Up-regulation of YUCCA genes results in elevated IAA levels (12). Of the 11 Arabidopsis YUCCA genes, YUC8 was up-regulated in the hybrids and in the three hybrid mimic lines, but was expressed at a level similar to the parents in the medium and small lines (Fig. 4A). YUC2 and YUC5 were also up-regulated in the hybrids and in two mimic lines: HM-W and HM-S (P < 0.05); both genes showed an up-regulation in the HM-G line (YUC2: P = 0.11, YUC5: P = 0.06) (Fig. S4A).
Fig. 4.
The F1 hybrids and F6 hybrid mimic lines had altered expression of auxin-related genes. (A) The relative expression of YUC8, IAA29, and PIF4. The transcriptome data for each gene are shown as normalized reads. The black dotted line represents MPV. (B) Heat maps showing the expression level of auxin-related differentially expressed genes in the F1 hybrids and F6 plant lines. Red/green colors indicate the up-/down-regulated fold change (FC) from the MPV. “>” or “<” indicates that expression shows an up or down trend but the change is not significant (P > 0.05 from the MPV). (C) PIF4 was up-regulated in the 3-, 5-, and 7-d-old seedlings of F1 hybrids and in the leaves of 28-d-old plants of the F1 hybrids compared with the MPV. ** indicates significant differences at P (Student’s t test) <0.01 from MPV. * indicates significant differences at P < 0.05 from MPV. Error bars = SEM. The percentages (in red) represent the increased levels of PIF4 expression in the hybrids relative to the MPV.
Fig. S4.
Expression patterns of auxin-associated genes in two parents C24 and Ler, F1 hybrids, and F6 plant lines. (A) YUC2 and YUC5 were up-regulated in the hybrids and in two mimic lines: HM-W and HM-S at 15 DAS. Black dotted line represents the MPV. (B) Expression patterns of auxin-associated genes in two parents C24 and Ler, F1 hybrids, and F6 plant lines. Different red/green colors indicate the fold change (up/down) from the MPV. Two genes [YUC8 (ATG28720) and IAA29 (AT4G32280)] highlighted in red were up-regulated in the hybrids and three hybrid mimic lines, but not in medium and small plant lines at 15 DAS. (C) BEE1 and BEE3 were up-regulated in the hybrids and in three mimic lines at 15 DAS. The transcriptome data for each gene are represented by normalized reads. Black dotted line represents MPV. The values for the parental lines are the mean of read counts from three biological replicates; the value for the F1 hybrids is the mean of six replicates. The values for the F6 lines are the mean of three siblings. Black dotted line represents the MPV. (D) IAA29 was up-regulated in the hybrids at 5 and 7 DAS relative to the MPV. The mean of read counts from biological replicates (n ≥ 2) of parental lines and F1 hybrids was normalized to the MPV. (E) Relative expression of YUC8 and IAA29 in the parents and hybrids at 28 DAS. The mean of read counts from biological replicates (n ≥ 2) of parental lines and F1 hybrids was normalized to the MPV. * indicates significant differences at P < 0.05 from MPV. Error bars = SEM.
Aux/IAA and the auxin response factors (ARFs) are two major protein families in the auxin signaling pathway (13). Aux/IAA proteins are short-lived transcription factors. At low auxin concentrations, Aux/IAA proteins form heterodimers with ARF proteins and repress the transcriptional activities of ARFs. Increasing auxin accelerates the degradation of the AUX/IAA proteins and the ARF proteins released from the heterodimers modulate down-stream target genes such as the auxin responsive genes (14). Of the 23 members of the ARF family, only one was differentially expressed in the F1 hybrids but it was not in the hybrid mimics (Fig. S4B). Among the 29 genes in the Aux/IAA family, only IAA29 showed up-regulated transcription in the hybrids and the hybrid mimics. It was expressed at low levels in the medium and small lines (Fig. 4A and Fig. S4B).
Two auxin-inducible genes BR ENHANCED EXPRESSION 1 and 3 (BEE1 and BEE3) (15) showed expression patterns consistent with increased expression of YUC8, producing increased levels of auxin (Fig. 4B and Fig. S4C). The up-regulation of BEE1 and BEE3 in the hybrids and hybrid mimics did not occur in the medium and small lines (Fig. 4B and Fig. S4C). A number of auxin-responsive genes associated with increasing cell expansion such as ARGOS-LIKE (AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE, ARGOS) (ARL), SMALL AUXIN UP-REGULATED 3 (SAUR3), SAUR15, SAUR50, SAUR78, EXPANSIN-LIKE A1 (EXLA1), EXLA3, and EXPANSIN A16 (EXPA16) were up-regulated in the F1 hybrids and the hybrid mimic lines, but not in the medium and/or small lines (Fig. 4B) (16–18). Reduced expression of ARL results in decreased size of cotyledons, whereas overexpression of ARL leads to an increase in cotyledon size (18). The cotyledon areas of SAUR50 and SAUR65 overexpression lines were significantly larger than wild-type cotyledons (16) and overexpression of SAUR76, SAUR77, or SAUR78 showed increased plant size (17). The phenotypes of the F6 lines were consistent with these results, the hybrid mimic lines having larger cotyledons than the medium and small lines (Fig. S1A, Inset). These results suggest a subset of auxin-responsive genes contributes to the large phenotypes of the hybrids and hybrid mimics.
PIF4 Regulation of Auxin May Contribute to Hybrid Leaf Biomass.
YUC8 and IAA29 were similar in their patterns of gene expression between different plant lines (Fig. 4A). Both genes were expressed at higher levels in hybrid mimic lines than in the hybrids. The promoters of YUC8 and IAA29 are bound and activated by the light perception transcription factor, phytochrome-interacting factor 4 (PIF4). Overexpression of PIF4 doubles the expression level of YUC8 and elevates endogenous-free IAA levels to 50% above wild type (19). The expression of IAA29 was increased in a 35S-PIF4 overexpression line, and decreased in the pif4 pif5 double mutant compared with the wild type (20). In our experiments, PIF4 was up-regulated in the F1 hybrids and the three hybrid mimics and down-regulated in the small line (Sml-D) (Fig. 4A). The alteration in transcript level of PIF4 was less than the change of the YUC8 and IAA29 transcript levels in the hybrids and the hybrid mimics, possibly due to PIF4 being a transcription factor where a minor change of expression can be amplified by signaling cascades resulting in changes in the activity of downstream targets. The activities of PIF4 target genes including 1-AMINO-CYCLOPROPANE-1-CARBOXYLATE SYNTHASE 8 (ACS8), ETHYLENE-INSENSITIVE 3 (EIN3), GIBBERELLIC ACID INSENSITIVE (GAI), ARABIDOPSIS THALIANA HOMBOX PROTEIN 2 (ATHB2) and CYTOKININ OXIDASE 5 (CKX5) (21) mostly corresponded to the transcript level of PIF4 in the different F6 lines (Fig. S5).
Fig. S5.
Altered expression of target genes of PIF4. Different red/green colors indicate the fold change (up/down) from the MPV (P < 0.05). “>” or “<” indicates that expression shows an up or down trend but the change is not significant (P > 0.05 from the MPV).
PIF4 May Contribute to F1 Heterosis at More than One Growth Stage.
The C24/Ler F1 hybrids showed growth vigor relative to parents as early as the first week after sowing (7). Heterosis was well established at 7 DAS in both reciprocal hybrids with cotyledon areas exceeding the MPV by 6.5% and 22.7% (7). At 3 DAS, when the hybrids first had growth vigor in cotyledon size, PIF4 had a 76% higher level of expression in the hybrids than the MPV and much higher than either parent (Fig. 4C). At 5 and 7 DAS, there was significant up-regulation of PIF4 in the hybrids over the MPV (5 DAS: 23%; 7 DAS: 13%), with PIF4 expressed in the C24 parent at levels close to that in the hybrids (Fig. 4C).
A higher level of the PIF4 target gene IAA29 relative to the MPV was found in the hybrids at 5 DAS and 7 DAS (Fig. S4D). Expression of IAA29 was not able to be detected at 3 DAS. The PIF4 target gene YUC8 showed no change of expression in the 5-d and 7-d-old-seedlings of F1 hybrids compared with the MPV. PIF4 is expressed mainly in the aerial tissues of young seedlings (22, 23), YUC8 is mainly expressed in the roots (24), so change in expression in the cotyledons may not have been detected.
Genes associated with “response to auxin stimulus” were overrepresented in the F1 nonadditive genes at 3 and 5 DAS (3 DAS: 47 genes; 5 DAS: 30 genes), but not at 7 DAS (Dataset S1, Tables S11 and S12). This finding is consistent with the cotyledons expanding rapidly before 7 DAS. Over 70% of these differentially expressed auxin-responsive genes were up-regulated in the F1 hybrids relative to the parents (3 DAS: 72%; 5 DAS: 76.6%) (Dataset S1, Table S13), suggesting elevated auxin signaling pathways in the F1 hybrids at early growth stages.
At 28 DAS, PIF4 was up-regulated ∼50% in the hybrids relative to either parent (Fig. 4C). Up-regulated expression of YUC8, IAA29, and downstream auxin-responsive genes such as the SAUR genes was detected in both reciprocal F1 hybrids (Fig. S4E and Dataset S1, Table S13). These results suggest that increased PIF4 expression through its control of auxin levels contributes to the F1 heterotic phenotype at a number of growth stages.
Intercrossing of Hybrid Mimics Increases Growth Vigor.
Although hundreds of differentially expressed genes were shared by the three hybrid mimic lines, large numbers were unique to each line (Fig. 2 C–E). During the recurrent selection process, different subsets of genes could have been included in the genomes of the independent hybrid mimic lines.
Up-regulated “auxin biosynthesis and signaling” is a metabolic pathway contributing to the large rosette size in all three hybrid mimic lines. This is not the only pathway contributing in this way, as the GO analysis (8) identified other biological processes and genes shared in the F1 hybrids and hybrid mimic lines. Photosynthesis was one of the metabolic pathways noted in the HM-W and HM-S lines, but not in the HM-G line. Genes associated with “defense response” and “flower development” were significantly altered in the HM-G line, but not in the other two hybrid mimic lines (Dataset S1, Tables S14–S16).
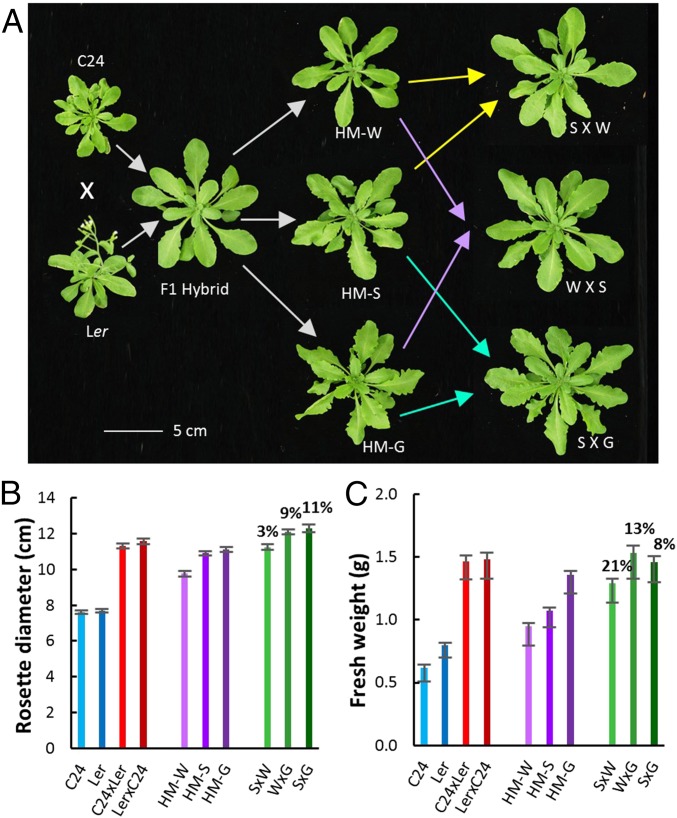
Because of these different subsets of DEGs, we examined whether crosses between the three F6 hybrid mimic lines could result in increased heterosis (HM-S × W, W × G, and S × G). In the progeny of the interhybrid mimic crosses rosette diameters exceeded the better parental hybrid mimic lines by 3–11%, but there was no major gain compared with the original F1 hybrids (Fig. 5 A and B). The aerial fresh weights of the intercross offspring were increased by 8–21% over its better hybrid mimic parent. Intercross offspring with HM-G as one parent had growth vigor in both rosette diameter and fresh weight equal to the F1 hybrids (Fig. 5).
Fig. 5.
Intercrossing of hybrid mimics generates small increases in growth vigor. The phenotypes (A), the rosette diameter measurements (B), and the fresh weight measurements (C) of the two parents C24 and Ler, F1 hybrids, three hybrid mimic lines, and the progeny of the intercrosses between the hybrid mimics at 30 DAS. n > 25. Percentage shows the increased vigor compared with the better parent hybrid mimic line.
Transregulation in the F6 Hybrid Mimic Lines.
Meiotic segregation of chromosomal segments delivers genes key to the generation of a particular phenotype, as suggested by the conservation of the 12 chromosomal segments in the hybrid mimics but not in the small line. Expression of genes in other parts of the genome could be under transregulation by loci in the 12 conserved segments. In the mimic lines, a gene expressed at a level different from its expression level in the parent of origin must be under transregulation. There were 2,518–4,066 genes expressed at a different level in the F6 lines compared with the relevant parent, accounting for 12–23% of the total genes defined by SNP analysis; equal numbers of genes were transregulated up or down (Fig. 6A and Dataset S1, Table S17). The locations of transregulated genes were distributed across the chromosomes in each line (Fig. S6). A total of 249 genes are classified in the response to auxin stimulus pathway, of which 113 were transregulated (Dataset S1, Table S18), suggesting a difference in auxin biosynthesis and downstream regulation of target genes in the hybrid mimic lines compared with the parental lines. These data are consistent with our finding of up-regulated auxin biosynthesis genes (YUCs) and signaling genes (IAA29) in the hybrid mimic lines (Fig. 4A).
Fig. 6.
Transregulation in the F6 hybrid mimic lines. (A) Genes (12–23%) with a defined genotype were under transregulation in the six F6 plant lines. Red/green represents up-/down-regulated genes compared with the expression of the parent of origin of the allele. (B and C) The expression of FRI (B) and FLC (C), in the two parents, F1 hybrids, three hybrid mimic lines, and the other F6 plant lines. The red/blue/green bars represent the genotypes C24 homozygous, Ler homozygous, and C24/Ler heterozygous (Heter.). The transcriptome data for each gene are shown as normalized reads from at least three biological replicates. Error bars = SEM. (D) Pictures of the plant lines showing plant size and flowering time. Photos were taken at 35 DAS.
Fig. S6.
The locations of transregulated genes on the five Arabidopsis chromosomes in each plant line. Red/green represents up-/down-regulated genes compared with the expression level of the parent of origin of the allele.
Our results suggest that selection of particular parental alleles with transregulation of transcriptional activity can contribute to the generation of hybrid mimics. An example of coselection of genes involved in transregulation is found in two genes regulating flowering time: FRIGIDA (FRI) and FLOWERING LOCUS C (FLC). FRI regulates the flowering repressor FLC (25). In the mimics FRI and FLC were selected, one from each of the parents. The active FRI allele from C24 (FRIC24) has been selected in the three hybrid mimics, but not in Sml-D and Med-F (Fig. 6B). The FLC allele from Ler (FLCLer) (26) is present in all of the hybrid mimic lines (Fig. 6C). The plant lines Med-F and Sml-D containing the nonfunctional friLer had low expression of FLC, and both lines showed an early flowering time similar to the Ler parent (Fig. 6 B–D).
Discussion
The F1 hybrids from crosses of the C24 and Ler ecotypes have a high level of hybrid vigor (heterosis) with uniform phenotypes in plant biomass (6). Recurrent selection of the phenotypes displayed by individual F2 plants produced F6 plants with phenotypes resembling the selected individual F2 plants. We produced three hybrid mimic lines, a contrasting small phenotype line, and two lines with intermediate rosette sizes. The hybrid mimic lines (F6) maintained hybrid vigor properties in the subsequent generation.
The three hybrid mimic lines shared 12 genomic segments, 4 from C24 and 8 from Ler. In the small line, these genomic segments were derived from the alternative parent. Each segment is likely to include one or more loci necessary for the generation of the hybrid phenotype. Each line differed in the end points of the segments, which must have been generated by recombination events occurring during the selection process.
A physiological feature common to the hybrids and hybrid mimics is that their seeds germinate earlier than the seeds of either parent or the F6 small and medium lines. The recurrent selection program based on the rosette diameter resulted in cosegregation of genes for the early germination trait. Loci concerned with early germination must also be present in one or more of the 12 conserved genomic segments identified in the hybrid mimics. The early germination character of the seeds of the F1 hybrids and the pure breeding hybrid mimics may be a useful diagnostic for the selection of high-performing hybrids.
Transregulation of genes as a consequence of the two parental genomes being in the same nucleus could produce the changes in gene expression, which promote hybrid vigor. In the hybrid mimics 12–23% of genes were expressed at different levels than they were in their parent of origin, suggesting the hybrid levels of expression were inherited by the hybrid mimics. Transregulation could also result from interaction with the epigenomes of the two parents (27). The underlying basis of hybrid vigor lies in the alteration of the levels of gene activity in the F1.
Methylation can be one of the epigenetic processes modifying expression levels of genes in the hybrid. In Columbia-0 (Col)/C24 hybrids, an active copy of the chromatin remodeler DECREASE IN DNA METHYLATION (DDM1), which affects DNA methylation in all sequence contexts, is required for a wild-type level of heterosis, whereas the alterations in gene expression produced by transchromosomal methylation via the RNA-directed DNA methylation (RdDM) pathway do not appear to contribute to hybrid vigor (28, 29).
Plant hormone networks play crucial roles in the growth of plants. Both hybrids and hybrid mimic lines showed changes in the levels of expression of genes in hormone pathways, and in particular, of genes in the auxin metabolic pathways. In Arabidopsis, auxin, in conjunction with other hormones, controls leaf size by regulating both cell proliferation and cell expansion (30). Increased auxin in early developmental stages of the hybrids could account for C24/Ler F1 hybrids and the hybrid mimic plants having large leaves due to both increased number and size of cells (2). The auxin biosynthesis gene YUC8 and the auxin-inducible gene IAA29, together with a number of auxin-responsive genes known to promote plant size, were up-regulated in both the hybrids and hybrid mimics, supporting the concept that an increased auxin signaling cascade is a contributor to the F1 hybrid phenotypes.
YUC8 and IAA29 are activated by the transcription factor PIF4 (19, 20). The expression of the auxin biosynthesis genes TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1 (TAA1) and YUC8 can be induced by transzeatin treatment in wild-type plants, but not in a pif4 mutant, indicating PIF4 is required for cytokinin-dependent auxin biosynthesis, which leads to cell division (22). PIF4 is a transacting regulator, which integrates light signals and plant hormone pathways, including auxin, ethylene, BR, GA, and CK to optimize plant growth and development (31). In our analyses, the activities of the hormone-associated PIF4 target genes (21) reflected the transcript level of PIF4 in the different F6 plant lines (Fig. S5); PIF4 was up-regulated in the plants with a F1 hybrid phenotype and down-regulated in the plants with a smaller plant phenotype. A change in plant hormone networks, particularly the auxin network, as a consequence of the altered expression of PIF4, may contribute to the increased biomass of the hybrids and hybrid mimic lines.
An altered circadian clock has been suggested to contribute to hybrid vigor in the allotetraploid between Arabidopsis thaliana and Arabidopsis arenosa and in the C24/Col F1 hybrid (32). The transcription of PIF4 is under the control of the circadian clock with high transcription levels at Zeitgeber time ZT 6 ± 3 (ZT = 0 refers to dawn) in long days (21). In our experiments, the altered expression of PIF4 was at ZT = 7 ± 0.5; the altered expression of the downstream target genes in the C24/Ler hybrids and hybrid mimics could reflect an altered circadian regulation by PIF4.
Unlike C24/Col hybrids which have been reported to accumulate starch to a higher level than the parents (32), six genes in the carbohydrate biosynthetic pathway were down-regulated in the C24/Ler hybrids and hybrid mimics. These include two genes involved in starch biosynthesis, ADP-GLUCOSE PYROPHOSPHORYLASE (APL3) and BRANCHING ENZYME 1 (DBE1), which were down-regulated in the hybrids and the three hybrid mimic lines, but were up-regulated or had no change in the medium or small phenotype lines. Cross et al. (33) found Arabidopsis accessions having a larger rosette size tend to accumulate lower amounts of starch; accessions with smaller rosettes are associated with a higher level of carbohydrate accumulation.
Eighteen nuclear-encoded photosynthesis genes, including six genes encoding light-harvesting proteins, had decreased expression in the C24/Ler F1 hybrid and in two hybrid mimic lines (HM-W and HM-S). This finding is unexpected because to ensure the energy supply for increased growth, more photosynthate is needed in the hybrids and hybrid mimics than in the parents. Parents and hybrids have the same photosynthesis capacity per unit leaf area (3), suggesting that the lower transcription level of the photosynthesis genes does not necessarily result in a decreased level of photosynthesis. A lower level of starch accumulation in the hybrids could indicate more sugar from photosynthesis is available for plant growth. When the production of sugar reaches a threshold, the transcriptional repression of photosynthesis pathway genes, including genes encoding light-harvesting proteins, occurs (34).
The use of hybrid mimics in a strategy to identify genes important for hybrid vigor has worked because the stringent recurrent selection process assembles all of the genes necessary for the large F1-like phenotype carried on a small number of homozygous segments distributed over the chromosomes of the genome. By using more hybrid mimic lines it should be possible to narrow down to key genes in the segments. This same strategy may be used to identify key genes and pathways in other plant species, including crops.
Materials and Methods
C24/Ler hybrids were produced by hand-pollination between Arabidopsis accessions C24 and Landsberg erecta (Ler). C24/Ler hybrid mimic lines, generated by the recurrent selection described previously (2), were renamed based on their phenotypes: L3-1-1-2 (HM-W), L4-2-1-2 (HM-S), and L2-1-1-1 (HM-G), L1-2-1-1 (Med-E), S2-1-1-1 (Med-F), and S1-1-1-1 (Sml-D). Plants were grown on Murashige and Skoog (MS) plates (3% sucrose and 0.8% wt/vol agar supplemented) under 16-h light (22 °C)/8-h dark (18 °C) conditions. The aerial tissues of 15 d MS plate-grown seedlings of two parents C24 and Ler, F1 hybrids, and F6 plant lines were collected at time ZT = 7 ± 0.5 (ZT = 0 refers to dawn) and were subjected to RNA extraction using the Plant DNeasy kit (Qiagen). mRNA-seq service was provided by the Australian Genome Research Facility (AGRF) on the Illumina platform. Raw and processed RNA-seq data are deposited in GEO (accession no. GSE94547). Sequenced reads alignment was performed using BioKanga 3.4.5 (https://sourceforge.net/projects/biokanga/). Mapped reads were normalization based on library size. The DEseq package was used to determine significant differences in gene expression between samples under the “RStudio” environment. Gene enrichment analysis was performed on the AgriGO platform (bioinfo.cau.edu.cn/agriGO/) (8). SNP analysis and parent-of-origin analysis were performed as in the previous paper (2) with slight change of parameters. Detailed information can be found in SI Materials and Methods.
SI Materials and Methods
Plant Materials and Growth Conditions.
Seeds of C24/Ler hybrids were produced by hand pollination between Arabidopsis accessions C24 and Landsberg erecta (Ler). Seeds of C24/Ler hybrid mimic lines (HM-W, HM-S, and HM-G) and three other F6 plant lines (Med-E, Med-F, and Sml-D) were produced from the recurrent selection protocol described previously (2). Surface sterilized seeds were sown on Murashige and Skoog (MS) medium supplemented with 3% (wt/vol) sucrose and 0.8% wt/vol agar. After stratification at 4 °C for 3 d, plants were grown under 16 h light (22 °C)/8 h dark (18 °C). Light intensity was controlled at 130–150 μmol photons m−2·s−1 under fluorescent light tubes (Sylvania Premium Extra FL36W/865 Super Daylight Deluxe). For later growth, the seedlings were transferred into soil at 15 d after sowing (DAS) and grown under the same light intensity and temperature. To minimize the effects of seed age difference on seed germination and plant growth, the parents C24 and Ler, and six F6 plant lines (HM-W, HM-S, HM-G, Med-E, Med-F, and Sml-D) (n > 3) were grown in one experiment and seeds were harvested at the same time. C24/Ler F1 hybrids were made in the same experiment using the C24 and Ler plants. All of the seeds (C24, Ler, F1 hybrids, and C24/Ler F7 seeds) used in the growth pattern and seed germination experiments were 3–4 mo old.
Transcriptome Sample Preparation and RNA Extraction.
The aerial tissues of 15 d MS plate-grown seedlings of the two parents C24 and Ler, F1 hybrids, and F6 plant lines were collected at time ZT = 7 ± 0.5 (ZT = 0 refers to dawn) and RNA extracted using the Plant DNeasy kit (Qiagen). Three biological replicates from each plant line were prepared. Each biological replicate for C24 and Ler, C24 × Ler, and Ler × C24, contained a pool of three seedlings. For each F6 plant line, three siblings were used as three biological replicates.
Rosette Diameter Measurement.
Rosette diameter data were obtained by image analysis using plant photos (ImageJ: National Institutes of Health). Hand measurements were applied when rosettes overlapped between adjacent plants.
Transcriptome Analysis.
For the transcriptomes of the parents C24 and Ler, F1 hybrids, and F6 plant lines at 15 DAS, the library preparation and mRNA sequencing were completed by the Australian Genome Research Facility (AGRF). A total of 41,600,826–53,397,900 100-bp sequenced reads were generated for different samples. The quality of raw data (.fastq files) was assessed by FastQC program (www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were mapped against the TAIR10 reference genome using BioKanga 3.4.5 (https://sourceforge.net/projects/biokanga/) under default settings. The percentage of aligned reads ranged from 85% to 89%. The DEseq package was used to determine significant differences in gene expression between samples. Normalization was performed on aligned reads across all of the samples based on the size of each library. To identify the differentially expressed genes between two samples, nonexpressed genes (genes with reads <100 in every sample) were filtered. A threshold of P ≤ 0.05 was applied for differentially expressed gene. Venn diagrams of shared differentially expressed genes between two or more samples were produced by Venny (bioinfogp.cnb.csic.es/tools/venny/). Gene expression heat maps were drawn in Microsoft Excel.
For the transcriptomes of parents C24 and Ler, and C24/Ler F1 hybrids at 3, 5, and 7 DAS, gene read counts of each sample were provided by the authors of ref. 7. For the 28 DAS transcriptomes data, mRNA-seq data of C24 (RepC and RepD), Ler (RepC and RepD), and Ler × C24 F1 Hybrid (RepA and RepB) were used (2). Read normalization and statistical tests of gene expression between samples were performed using the DEseq method with a “fit-only” parameter under the “RStudio” environment. Two reciprocal hybrids were treated as biological replicates of “hybrids” for datasets at 3, 5, and 7 DAS. For each dataset, genes that had reads of ≥100 in at least one sample were treated as expressed genes at this time point. A threshold of P ≤ 0.05 was applied for identification of the differentially expressed genes. Differentially expressed genes with fold change (FC) ≥ ±1.3 were used in the gene enrichment analysis (GO).
SNP Analysis.
SNP analysis was performed as in the previous paper (2) with a few modifications. These alterations are: SNP with a read coverage ≥4 were used for drawing genotype maps; Samtools v1.3 was used instead of Samtools v0.1.19; chromosome genotype maps were produced using “R” instead of Microsoft Excel. Conserved chromosome segments (Fig. 1D) were defined when a segment sourced from one particular parent was present in each of the three hybrid mimic lines, and in the F6 small phenotype plant line (Sml-D) the segment was derived from the alternative parent.
GO Analysis.
Gene enrichment analysis was performed on the AgriGO platform (bioinfo.cau.edu.cn/agriGO/) (8). Only the expressed genes (genes with reads ≥100 at least in one sample) were used as the background gene list, which was compared against the input lists. Gene enrichment was indicated when not adjusted P (Fisher’s test) <0.05 and the gene number ≥5 in one GO term unless specified.
Parent-of-Origin Analysis.
The allelic composition of each gene was identified by the SNPs (C24, Ler, or both) in each F6 plant. In most cases where three siblings have an identical genotype call for one gene, this genotype was determined as the genotype of this plant line for this gene. For parent-of-origin analysis in Fig. 6A, three siblings of each plant line were treated as biological replicates. For identifying the distribution of the transregulated genes on the chromosomes in each sibling of F6 plant lines (chromosome maps) (Fig. S6), at least three SNPs located in each gene were required to confirm the genotype of this gene. Gene expression levels were compared with the parent with the same allele. P values were generated by DEseq with default settings. Genes having ≥100 reads in at least one sample and with P value ≤0.05 were defined as transregulated genes in each sibling or in the F6 plant lines.
Supplementary Material
Acknowledgments
We thank Aihua Wang and Bjorg Sherman for technical assistance, Jose Barrero Sanchez for helpful discussion on the seed germination experiment, Ming-Bo Wang and Tina Liu for suggestions and advice, and Jean Finnegan for manuscript reviewing and correction.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE94547).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703179114/-/DCSupplemental.
References
- 1.Schnable PS, Springer NM. Progress toward understanding heterosis in crop plants. Annu Rev Plant Biol. 2013;64:71–88. doi: 10.1146/annurev-arplant-042110-103827. [DOI] [PubMed] [Google Scholar]
- 2.Wang L, et al. Hybrid mimics and hybrid vigor in Arabidopsis. Proc Natl Acad Sci USA. 2015;112:E4959–E4967. doi: 10.1073/pnas.1514190112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujimoto R, Taylor JM, Shirasawa S, Peacock WJ, Dennis ES. Heterosis of Arabidopsis hybrids between C24 and Col is associated with increased photosynthesis capacity. Proc Natl Acad Sci USA. 2012;109:7109–7114. doi: 10.1073/pnas.1204464109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Groszmann M, et al. Hormone-regulated defense and stress response networks contribute to heterosis in Arabidopsis F1 hybrids. Proc Natl Acad Sci USA. 2015;112:E6397–E6406. doi: 10.1073/pnas.1519926112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhai R, et al. Transcriptome analysis of rice root heterosis by RNA-Seq. BMC Genomics. 2013;14:19. doi: 10.1186/1471-2164-14-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Groszmann M, et al. Intraspecific Arabidopsis hybrids show different patterns of heterosis despite the close relatedness of the parental genomes. Plant Physiol. 2014;166:265–280. doi: 10.1104/pp.114.243998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu A, et al. Early changes of gene activity in developing seedlings of Arabidopsis hybrids relative to parents may contribute to hybrid vigour. Plant J. 2016;88:597–607. doi: 10.1111/tpj.13285. [DOI] [PubMed] [Google Scholar]
- 8.Du Z, Zhou X, Ling Y, Zhang Z, Su Z. agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38:W64–70. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyer RC, et al. Heterosis manifestation during early Arabidopsis seedling development is characterized by intermediate gene expression and enhanced metabolic activity in the hybrids. Plant J. 2012;71:669–683. doi: 10.1111/j.1365-313X.2012.05021.x. [DOI] [PubMed] [Google Scholar]
- 10.Cheng Y, Dai X, Zhao Y. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell. 2007;19:2430–2439. doi: 10.1105/tpc.107.053009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L, et al. Small RNAs as important regulators for the hybrid vigour of super-hybrid rice. J Exp Bot. 2014;65:5989–6002. doi: 10.1093/jxb/eru337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kasahara H. Current aspects of auxin biosynthesis in plants. Biosci Biotechnol Biochem. 2015;80:34–42. doi: 10.1080/09168451.2015.1086259. [DOI] [PubMed] [Google Scholar]
- 13.Liscum E, Reed JW. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol. 2002;49:387–400. [PubMed] [Google Scholar]
- 14.Wang R, Estelle M. Diversity and specificity: Auxin perception and signaling through the TIR1/AFB pathway. Curr Opin Plant Biol. 2014;21:51–58. doi: 10.1016/j.pbi.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedrichsen DM, et al. Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics. 2002;162:1445–1456. doi: 10.1093/genetics/162.3.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun N, et al. Arabidopsis SAURs are critical for differential light regulation of the development of various organs. Proc Natl Acad Sci USA. 2016;113:6071–6076. doi: 10.1073/pnas.1604782113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li ZG, et al. Three SAUR proteins SAUR76, SAUR77 and SAUR78 promote plant growth in Arabidopsis. Sci Rep. 2015;5:12477. doi: 10.1038/srep12477. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Hu Y, Poh HM, Chua NH. The Arabidopsis ARGOS-LIKE gene regulates cell expansion during organ growth. Plant J. 2006;47:1–9. doi: 10.1111/j.1365-313X.2006.02750.x. [DOI] [PubMed] [Google Scholar]
- 19.Sun J, Qi L, Li Y, Chu J, Li C. PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating arabidopsis hypocotyl growth. PLoS Genet. 2012;8:e1002594. doi: 10.1371/journal.pgen.1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun J, Qi L, Li Y, Zhai Q, Li C. PIF4 and PIF5 transcription factors link blue light and auxin to regulate the phototropic response in Arabidopsis. Plant Cell. 2013;25:2102–2114. doi: 10.1105/tpc.113.112417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nomoto Y, et al. A circadian clock- and PIF4-mediated double coincidence mechanism is implicated in the thermosensitive photoperiodic control of plant architectures in Arabidopsis thaliana. Plant Cell Physiol. 2012;53:1965–1973. doi: 10.1093/pcp/pcs141. [DOI] [PubMed] [Google Scholar]
- 22.Di DW, et al. Functional roles of Arabidopsis CKRC2/YUCCA8 gene and the involvement of PIF4 in the regulation of auxin biosynthesis by cytokinin. Sci Rep. 2016;6:36866. doi: 10.1038/srep36866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimizu H, Torii K, Araki T, Endo M. Importance of epidermal clocks for regulation of hypocotyl elongation through PIF4 and IAA29. Plant Signal Behav. 2016;11:e1143999. doi: 10.1080/15592324.2016.1143999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rawat R, et al. REVEILLE1, a Myb-like transcription factor, integrates the circadian clock and auxin pathways. Proc Natl Acad Sci USA. 2009;106:16883–16888. doi: 10.1073/pnas.0813035106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He Y, Amasino RM. Role of chromatin modification in flowering-time control. Trends Plant Sci. 2005;10:30–35. doi: 10.1016/j.tplants.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 26.Gazzani S, Gendall AR, Lister C, Dean C. Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol. 2003;132:1107–1114. doi: 10.1104/pp.103.021212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greaves IK, et al. Epigenetic changes in hybrids. Plant Physiol. 2015;168:1197–1205. doi: 10.1104/pp.15.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Q, et al. The chromatin remodeler DDM1 promotes hybrid vigor by regulating salicylic acid metabolism. Cell Discov. 2016;2:16027. doi: 10.1038/celldisc.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawanabe T, et al. Role of DNA methylation in hybrid vigor in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2016;113:E6704–E6711. doi: 10.1073/pnas.1613372113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perrot-Rechenmann C. Cellular responses to auxin: Division versus expansion. Cold Spring Harb Perspect Biol. 2010;2:a001446. doi: 10.1101/cshperspect.a001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi H, Oh E. PIF4 integrates multiple environmental and hormonal signals for plant growth regulation in Arabidopsis. Mol Cells. 2016;39:587–593. doi: 10.14348/molcells.2016.0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ni Z, et al. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature. 2009;457:327–331. doi: 10.1038/nature07523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cross JM, et al. Variation of enzyme activities and metabolite levels in 24 Arabidopsis accessions growing in carbon-limited conditions. Plant Physiol. 2006;142:1574–1588. doi: 10.1104/pp.106.086629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pego JV, Kortstee AJ, Huijser C, Smeekens SC. Photosynthesis, sugars and the regulation of gene expression. J Exp Bot. 2000;51:407–416. doi: 10.1093/jexbot/51.suppl_1.407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.