Significance
TRAF6 is essential for many biological processes, including the operation of the innate immune system and bone formation. The enzymatic activity of TRAF6, an E3 ubiquitin ligase, is thought to have a pivotal role in triggering these processes. In this paper, we reexamined this assumption by generating mice expressing a catalytically inactive mutant of TRAF6. The bone structure of the mice was normal and a major intracellular signaling pathway of the immune system could still be switched on. We found that other E3 ligases, Pellino1 and Pellino2, could generate the ubiquitin chains needed to switch on immune signaling in human cells expressing catalytically inactive TRAF6. Our findings identify essential roles of TRAF6 that are independent of its enzymatic activity.
Keywords: TRAF6, TAK1, Pellino, Ubiquitin, IL-1
Abstract
It is widely accepted that the essential role of TRAF6 in vivo is to generate the Lys63-linked ubiquitin (K63-Ub) chains needed to activate the “master” protein kinase TAK1. Here, we report that TRAF6 E3 ligase activity contributes to but is not essential for the IL-1–dependent formation of K63-Ub chains, TAK1 activation, or IL-8 production in human cells, because Pellino1 and Pellino2 generate the K63-Ub chains required for signaling in cells expressing E3 ligase-inactive TRAF6 mutants. The IL-1–induced formation of K63-Ub chains and ubiquitylation of IRAK1, IRAK4, and MyD88 was abolished in TRAF6/Pellino1/Pellino2 triple-knockout (KO) cells, but not in TRAF6 KO or Pellino1/2 double-KO cells. The reexpression of E3 ligase-inactive TRAF6 mutants partially restored IL-1 signaling in TRAF6 KO cells, but not in TRAF6/Pellino1/Pellino2 triple-KO cells. Pellino1-generated K63-Ub chains activated the TAK1 complex in vitro with similar efficiently to TRAF6-generated K63-Ub chains. The early phase of TLR signaling and the TLR-dependent secretion of IL-10 (controlled by IRAKs 1 and 2) was only reduced modestly in primary macrophages from knockin mice expressing the E3 ligase-inactive TRAF6[L74H] mutant, but the late-phase production of IL-6, IL-12, and TNFα (controlled only by the pseudokinase IRAK2) was abolished. RANKL-induced signaling in macrophages and the differentiation of bone marrow to osteoclasts was similar in TRAF6[L74H] and wild-type cells, explaining why the bone structure and teeth of the TRAF6[L74H] mice was normal, unlike TRAF6 KO mice. We identify two essential roles of TRAF6 that are independent of its E3 ligase activity.
TNF receptor-associated factor 6 (TRAF6) is essential for many biological processes (1). These include the myeloid differentiation primary response gene 88 (MyD88) signaling network of the innate immune system, RANK ligand (RANKL)-dependent signaling and osteoclast formation, lymph node organogenesis (2), and the development of hair follicles, sweat glands, and sebaceous glands (3). TRAF6 expression is also needed for CD40 signaling in B cells (4), the maturation and development of dendritic cells (5), and the regulation of T-cell function (6, 7).
In innate immunity, nearly all Toll-like receptors (TLRs), as well as the receptors of the interleukin 1 (IL-1) family of cytokines, initiate signaling by recruiting the adaptor protein MyD88. This is followed by the interaction of IL-1-receptor (IL-R)-associated kinase 4 (IRAK4) with MyD88 and then the interaction of other IRAK family members with IRAK4, to form an oligomeric complex, termed the Myddosome (8, 9). IRAK1 and IRAK2 can then interact with TRAF6 (10, 11) and induce TRAF6 dimerization (12), which triggers the activation of its E3 ligase activity (13).
TRAF6 catalyzes the formation of Lys63-linked ubiquitin (K63-Ub) chains in vitro in the presence of Ubc13-Uev1a (also called UBE2N-UBE2V1), an E2 conjugating enzyme that directs the formation of this type of ubiquitin linkage (14, 15). Although truncated forms of TRAF6 lacking the really interesting new gene (RING) domain were reported to restore IL-1-signaling to TRAF6 knockout (KO) mouse embryonic fibroblasts (MEFs) many years ago (16), other laboratories reported subsequently that the E3 ligase-inactive TRAF6[C70A] mutant could not (12, 17, 18). These reports led to widespread acceptance of the notion that the E3 ligase activity of TRAF6 is essential for IL-1 signaling. Moreover, TRAF6-generated K63-Ub oligomers were shown to activate the protein kinase TAK1 (also called MAP3K7) in vitro (14, 15). TAK1 has a critical role in the MyD88 signaling network, because IL-1 signaling is abolished in TAK1 KO MEFs (19) or in MEFs expressing catalytically inactive TAK1 (20). Cells express two TAK1 complexes, each comprising the TAK1 catalytic subunit and TAK1-binding protein 1 (TAB1) plus either TAB2 or the related TAB3 (21). The specific interaction of K63-Ub chains with the C-terminal Npl4 zinc finger (NZF) domains of TAB2 and TAB3 (22, 23) is thought to induce a conformational change that activates TAK1 (14, 15).
Two major roles of TAK1 are to activate the canonical IκB kinase (IKK) complex, and mitogen-activated protein (MAP) kinase kinases (MKKs) that switch on p38 MAP kinases and c-Jun N-terminal kinases 1 and 2 (JNK1 and JNK2). The IKKβ component of the IKK complex activates the transcription factors NF-κB and IFN-regulatory factor 5 (IRF5) (24, 25), which are essential for the transcription of genes encoding proinflammatory cytokines (26). IKKβ also activates the protein kinase Tpl2 (MAP3K8) by phosphorylating its p105/NFκB1 subunit (27, 28), enabling Tpl2 to activate MEK1 and MEK2 (MAP or ERK kinase), which in turn activate extracellular signal regulated kinase 1 (ERK1) and ERK2 (also called MAPK3 and MAPK1, respectively). ERK1 and ERK2, together with p38α MAP kinase, activate mitogen and stress-activated kinase 1 (MSK1) and MSK2, which phosphorylate the cAMP response element-binding protein (CREB) that controls the transcription of the antiinflammatory cytokine IL-10 (29). Thus, TAK1 regulates the production of anti-inflammatory as well as proinflammatory cytokines.
If TRAF6-generated K63-Ub chains are essential for MyD88 signaling, then the formation of K63-Ub chains should be reduced in TRAF6 KO cells, and signaling should be abolished when WT TRAF6 is replaced by an E3 ligase-inactive mutant. Here, we tested these predictions, which led to the interesting and unexpected findings reported in this paper.
Results
The IL-1–Dependent Formation of K63-Ub Chains Is Unimpaired in TRAF6 KO IL-1R* Cells.
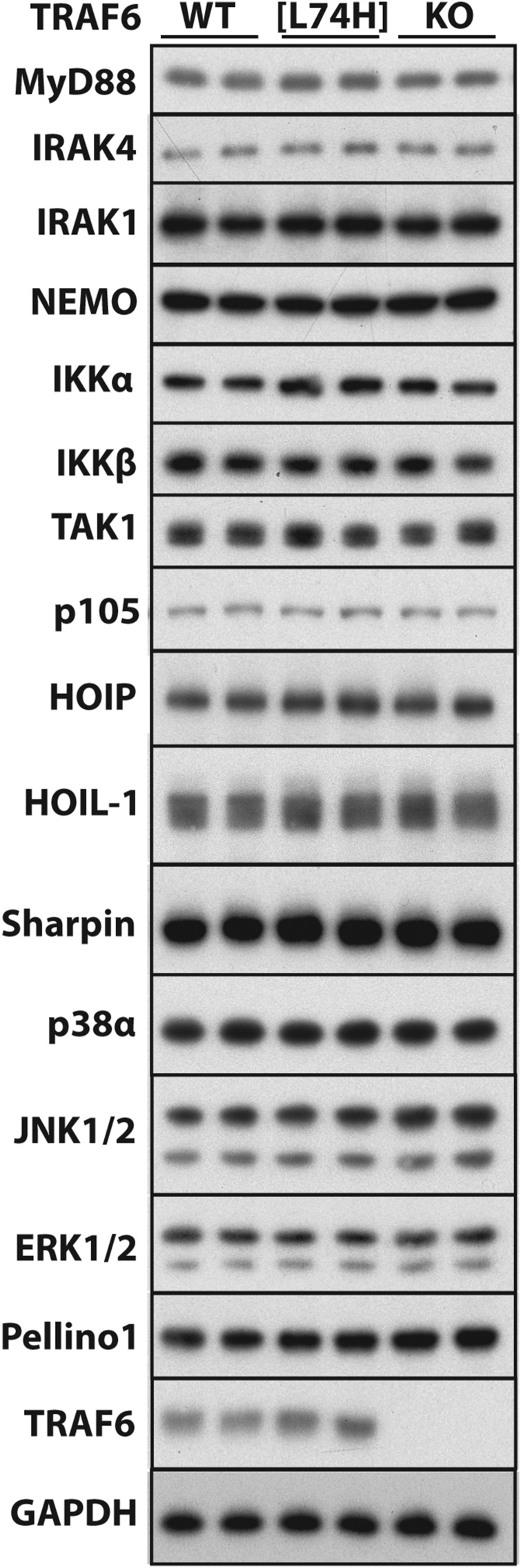
Our initial studies were performed in HEK293 cells stably expressing low levels of the IL-1R (IL-1R* cells) (Methods), which is a simple model system in which genes of interest can be disrupted easily by using CRISPR/Cas9 gene-editing technology. Similar to other mammalian cells, we found that IL-1β signaling in IL-1R* cells required the expression of TRAF6 and TAK1, as well the protein kinase activity of TAK1 (Fig. S1 A–C). The KO of TRAF6 did not affect the expression of any component of the MyD88 signaling pathway examined, apart from Pellino1, the expression of which was increased, as discussed later.
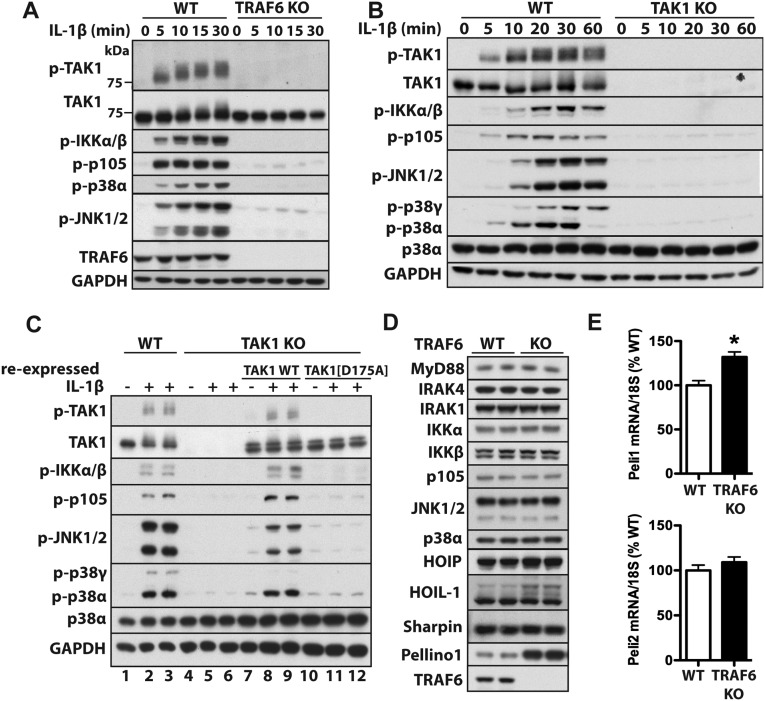
Fig. S1.
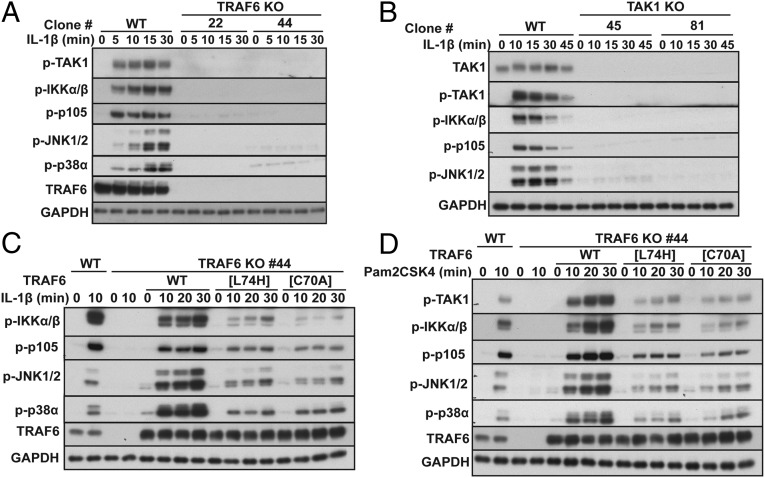
Characterization of TRAF6 and TAK1 KO cells. (A and B) WT IL-1R* and either TRAF6 KO (A) or TAK1 KO (B) IL-1R* cells were stimulated with IL-1β (5 ng/mL). Aliquots of the cell extracts (20 μg of protein) were subjected to SDS/PAGE, followed by immunoblotting with antibodies that recognize the active, phosphorylated (p) forms of TAK1, IKKα/β, JNK1/2, and p38α, as well as with antibodies that recognize p105 phosphorylated at Ser933, all forms of TAK1 and GAPDH as a loading control. (C) The reexpression of WT TAK1, but not the kinase-inactive TAK1[D175A] mutant, restores signaling to TAK1 KO IL-1R* cells. The cells were stimulated for 10 min with IL-1β. Cell extract protein (20 μg, lanes 1–6, or 40 μg, lanes 7–12) was subjected to SDS/PAGE, followed by immunoblotting with the antibodies in B. (D) The expression of many components of the MyD88 signaling network is similar in TRAF6 KO and WT IL-1R* cells, but Pellino1 expression is elevated in TRAF6 KO cells. (E) Total RNA was extracted from WT and TRAF6 KO IL-1R* cells not stimulated with IL-1β, and Pellino1 (A) and Pellino2 (B) mRNA were quantitated relative to 18S ribosomal mRNA and presented as a percentage of the Pellino1/2 mRNA present in WT IL-1R* cells. *P < 0.05. The results are presented as mean ± SEM for three independent experiments each carried out in quadruplicate (n = 12). Statistical analysis was performed using the unpaired Student t test.
To study the formation of IL-1β–dependent K63-Ub chains, we captured them from the cell extracts using Halo-NZF2 beads (30, 31) (Methods). Capture was quantitative because no further K63-Ub chains were pulled down when the supernatant obtained after the first Halo-NZF2 pull-down was subjected to a second treatment with these beads (Fig. 1A). We found that K63-Ub chains were present in cells not stimulated with IL-1β but increased after stimulation for 10 min with IL-1β. Importantly, the IL-1β–dependent formation of K63-Ub chains was similar in TRAF6 KO and TRAF6-expressing IL-1R* cells (Fig. 1A), implying that another IL-1–activated E3 ligase was generating K63-Ub chains in the TRAF6 KO cells.
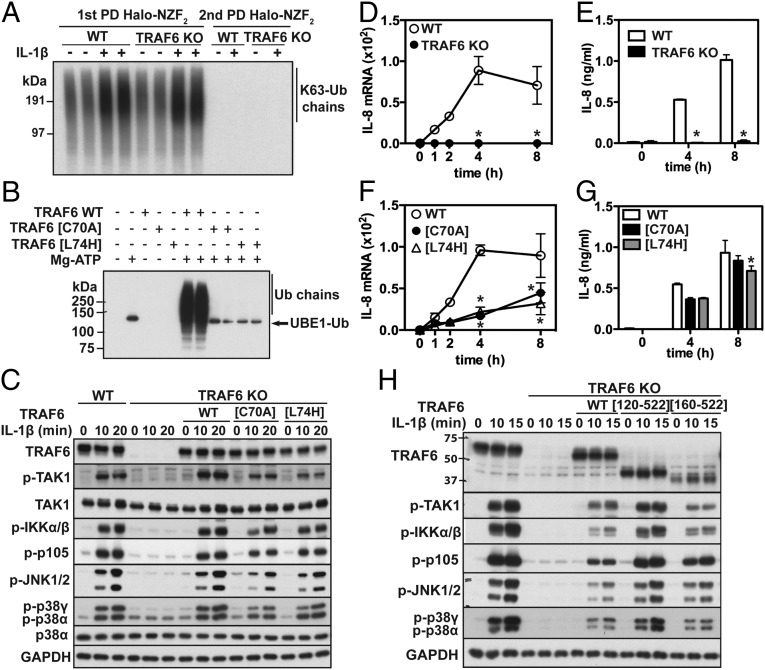
Fig. 1.
The E3 ligase activity of human TRAF6 is not required for IL-1β signaling in IL-1R* cells. (A) WT or TRAF6 KO IL-1R* cells were stimulated for 10 min with IL-1β, and K63-Ub chains were pulled down (PD) from the cell extracts on Halo-NZF2 beads. The supernatant was subjected to a second PD using fresh Halo-NZF2 beads. The K63-Ub chains were released with SDS and detected by immunoblotting with a specific antibody. (B) The activity of WT TRAF6 (5.0 nM) and the indicated TRAF6 mutants (50 nM) was assayed using FLAG-ubiquitin as in Methods. Reactions were terminated in SDS and Ub-chain formation detected by immunoblotting with anti-FLAG (which also detected Ub-loaded UBE1). (C) Untagged WT TRAF6, TRAF6[C70A], or TRAF6[L74H] were reexpressed in TRAF6 KO IL-1R* cells under the control of a doxycycline-inducible promoter. After stimulation with IL-1β, cell extracts were denatured in SDS and immunoblotted with antibodies recognizing all forms of TRAF6, TAK1, p38α MAPK, and GAPDH, and with antibodies recognizing the phosphorylated (p) forms of TAK1, IKKα/β, p105, JNK1/2, and p38α/γ. (D) WT and TRAF6 KO IL-1R* cells were stimulated with IL-1β, and then RNA was extracted and IL-8 mRNA measured by quantitative RT-PCR (q-RT-PCR). Results show fold increase in mRNA relative to the level in unstimulated cells and are presented as mean ± SEM (n = 3). (E) As in C, except that the secretion of IL-8 was determined by ELISA. Results are presented as mean ± SEM (n = 3). (F and G) As in D and E, except that WT TRAF6, TRAF6[C70A], or TRAF6[L74H] were reexpressed in TRAF6 KO IL-1R* cells. (H) As in C, except that TRAF6 KO IL-1R* cells were reconstituted with FLAG-tagged WT TRAF6, TRAF6[120-522], or TRAF6[160-522]. *P < 0.05 (Methods).
TRAF6 E3 Ligase Activity Is Not Essential for IL-1β Signaling in IL-1R* Cells.
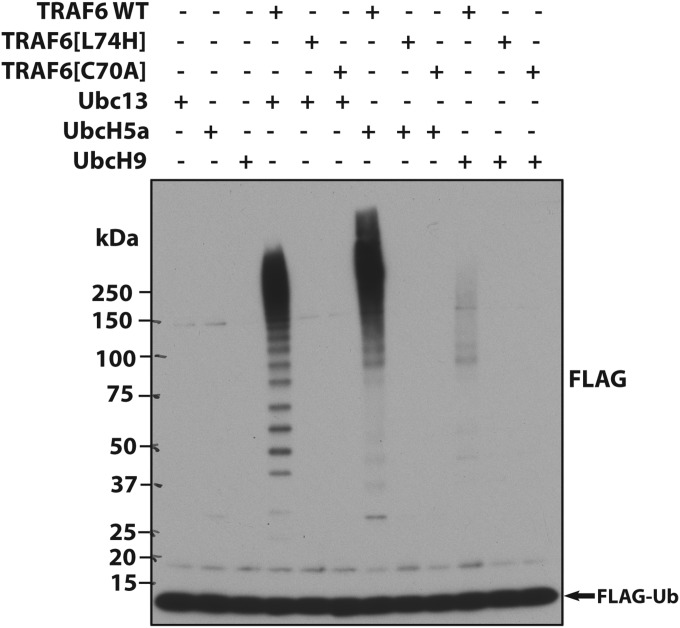
The results shown in Fig. 1 led us to reinvestigate whether TRAF6-generated K63-Ub chains were essential for IL-1 signaling. The TRAF6[L74H] mutation prevents interaction with E2 conjugating enzymes (12), whereas the TRAF6[C70A] mutation disrupts the RING domain structure. As expected, both mutants were devoid of E3 ligase activity in vitro, irrespective of whether the E2 conjugating enzyme was Ubc13-Uev1A (Fig. 1B), UbcH5a (also called UBE2D1), or UbcH9 (also called UBE2E3) (Fig. S2).
Fig. S2.
TRAF6[C70A] and TRAF6[L74H] lack E3 ligase activity in vitro. WT TRAF6 (5.0 nM) and the indicated TRAF6 mutants (50 nM) were incubated for 1 h with UBE1, the indicated E2-conjugating enzyme (Ubc13-Ubc1a, UbcH5a, or UbcH9), FLAG-ubiquitin, and Mg-ATP. The E3 ligase reaction was terminated in SDS, and an aliquot of the reaction was subjected to SDS/PAGE followed by immunoblotting with anti-FLAG.
Next, we engineered the TRAF6 KO IL-1R* cells to reexpress untagged E3 ligase-inactive TRAF6 mutants or WT TRAF6 under a doxycycline-inducible promoter at levels that were similar to the endogenous TRAF6 (Fig. 1C, Top). Interestingly, the reexpression of TRAF6[L74H] or TRAF6[C70A] partially restored and the reexpression of WT TRAF6 fully restored IL-1β signaling (Fig. 1C, panels 2–8 from Top) and IL-8 production (Fig. 1 D–G) to TRAF6 KO cells.
IL-1β signaling could also be restored to TRAF6 KO IL-1R* cells by the reexpression of TRAF6[120–522], which lacks the N-terminal 119 residues of TRAF6 containing the RING domain or by TRAF6[160–522], which lacks the RING domain plus the first zinc finger (Fig. 1H). This experiment confirmed that the E3 ligase activity of TRAF6 was not required for IL-1β signaling in IL-1R* cells.
The TRAF6 E3 Ligase Is Dispensable for IL-1 and TLR Signaling in HaCaT Cells.
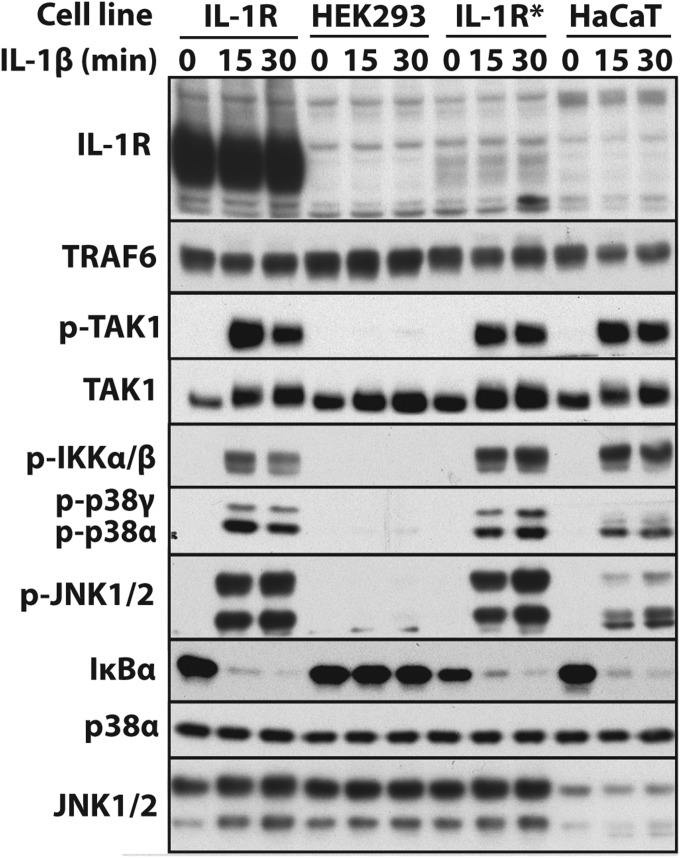
Because the results presented in Fig. 1 were unexpected, we repeated them in the human keratinocyte HaCaT cell line in which the IL-1R is not overexpressed and the level of the endogenous IL-1R is undetectable by immunoblotting (Fig. S3). Similar to IL-1R* cells, IL-1β–dependent signaling was abolished in TRAF6 KO (Fig. 2A) or TAK1 KO HaCaT cells (Fig. 2B), and IL-1-signaling could be partially restored to TRAF6 KO HaCaT cells by reexpressing E3 ligase-inactive TRAF6 and fully restored by reexpressing WT TRAF6 (Fig. 2C, top four panels).
Fig. S3.
Expression of the IL-1R in the human cell lines used in this study. The cells were stimulated with 5 ng/mL IL-1β for the times indicated, and aliquots of the cell extracts were analyzed by immunoblotting with the indicated antibodies.
Fig. 2.
The E3 ligase activity of TRAF6 is not required for IL-1β or Pam2CSK4 signaling in HaCaT cells. (A and B) IL-1β signaling is abolished in TRAF6 KO (A) and TAK1 KO (B) HaCaT cells. WT HaCaT cells and two independent clones of TRAF6 KO HaCaT cells (clones 22 and 44) or TAK1 KO HaCaT cells (clones 45 and 81) were stimulated with IL-1β followed by immunoblotting with the antibodies indicated. (C) WT TRAF6 or the E3 ligase-inactive TRAF6[L74H] or TRAF6[C70A] mutants were reexpressed in TRAF6 KO HaCaT cells (clone 44) under the control of a doxycycline-inducible promoter. Cells were stimulated with IL-1β and immunoblotted with the antibodies indicated. (D) As in C, except that the HaCaT cells were stimulated with the TLR2/6 agonist Pam2CSK4.
The expression of TRAF6 is also essential for MyD88 signaling induced by ligands that activate Toll-like receptors (TLRs). We found that HaCaT cells responded to Pam2CSK4, an activator of the TLR2/TLR6 heterodimer. Similar to IL-1β, Pam2CSK4-dependent signaling was abolished in TRAF6 KO HaCaT cells, partially restored by the reexpression of E3 ligase-inactive TRAF6 mutants, and fully restored by WT TRAF6 (Fig. 2D). Taken together, the results presented in Figs. 1 and 2 establish that TRAF6, but not its E3 ligase activity, is essential for MyD88 signaling in the two human cell lines studied.
Pellino1 and Pellino2 Generate the K63-Ub Chains Required for MyD88 Signaling in IL-1R* Cells Expressing E3 Ligase-Inactive TRAF6.
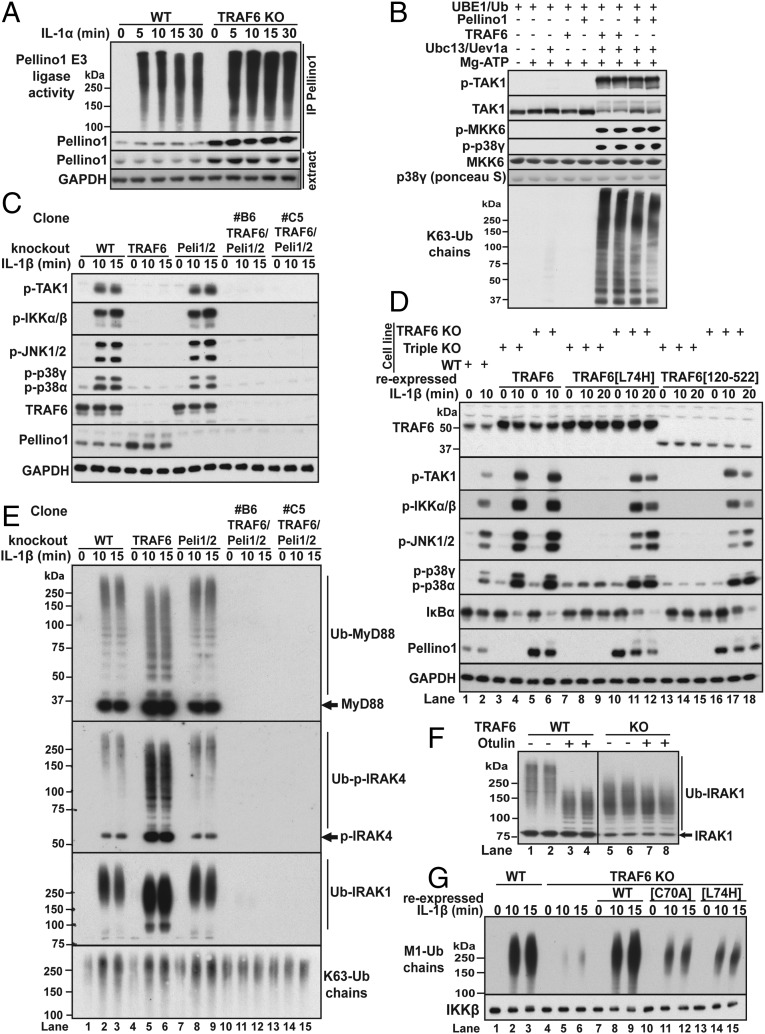
The results presented in the preceding sections suggested that another E3 ligase(s) was generating the K63-Ub chains required for MyD88 signaling in cells expressing E3 ligase-inactive mutants of TRAF6. Potential candidates to fulfill this role included members of the Pellino family of RING domain E3 ligases (32), which interact with IRAK1 (33, 34) and are converted from inactive to active E3 ligases by IRAK1-catalyzed phosphorylation in vitro (35, 36) and in cells (37). Like TRAF6, the Pellinos can also combine with Ubc13-Uev1a to produce K63-Ub chains in vitro (35, 36). Interestingly, the expression of Pellino1 (Fig. S1D) and hence IL-1β–dependent Pellino1 E3 ligase activity (Fig. 3A) was enhanced severalfold in TRAF6 KO cells (Fig. 3A). This appears to be an effect on Pellino1 stability because Pellino1 mRNA levels were only increased by 30% in TRAF6 KO cells, whereas Pellino2 mRNA levels were unaltered (Fig. S1E). We also found that Pellino1-generated K63-Ub chains were as effective as TRAF6-generated K63-Ub chains in triggering the activation of the TAB1–TAK1–TAB3 complex in vitro (Fig. 3B).
Fig. 3.
The E3 ligase activities of TRAF6 and Pellinos 1 and 2 function redundantly in the IL-1 signaling network. (A) Immortalized WT and TRAF6 KO MEFs were stimulated with IL-1α and Pellino1 immunoprecipitated from the cell extracts and assayed as described in Methods (Upper). The level of Pellino1 in the immunoprecipitates and cell extracts (second and third panels) and GAPDH in the cell extracts (Bottom) were detected by immunoblotting. (B) Activation of the TAB1–TAK1–TAB3 complex by TRAF6 or Pellino1-generated K63-Ub chains. The activation of TAK1 was studied in a coupled assay containing both its substrate MKK6 and p38γ MAP kinase, a substrate of MKK6 (Methods). (C) WT, TRAF6 KO, Pellino1/2 (Peli1/2) double KO, and two independent clones (B6, C5) of TRAF6 /Pellino1/2 triple-KO IL-1R* cells were stimulated with IL-1β, and the cell extracts immunoblotted with the antibodies indicated. (D) Flag-tagged WT TRAF6, TRAF6[L74H], or TRAF6[120-522] were reexpressed in TRAF6 KO or TRAF6/Peli1/2 triple-KO IL-1R* cells (clone B6). These cells (lanes 3–18) and untransfected WT IL-1R* cells (lanes 1 and 2) were stimulated with IL-1β, and the extracts immunoblotted with the antibodies indicated. (E) As in C, except that ubiquitylated forms of MyD88, p-IRAK4, IRAK1, and K63-Ub chains were captured on immobilized Halo-NZF2 beads and detected by immunoblotting with specific antibodies. (F) Ubiquitylated IRAK1 was captured from the extracts of IL-1–stimulated (10 min) WT and TRAF6 KO IL-1R* cells on Halo-TUBEs. Following incubation with λPPase with (+) or without (−) Otulin (Methods), IRAK1 was released with SDS and identified by immunoblotting. Due to the enhanced formation of Ub-IRAK1 in TRAF6 KO cells, the blots in lanes 1–4 were exposed for 15 s and those in lanes 5–8 for 5 s. (G) Halo-NEMO beads were used to capture M1-Ub chains from the extracts of WT IL-1R* cells or TRAF6 KO cells (Fig. 3G, lanes 1–6) or from TRAF6 KO cells reexpressing WT TRAF6 or the indicated E3 ligase-inactive TRAF6 mutants. The M1-Ub chains were identified by immunoblotting with a specific antibody. IKKβ, which binds to NEMO, was used as the loading control.
IL-1 stimulation induces the interaction of TRAF6 with Pro–Xaa–Glu motifs present in the C-terminal domain of IRAK1 (10), whereas the Forkhead-associated (FHA) domain present in Pellino isoforms interacts with phosphothreonine residues in IRAK1 (34). Consistent with the formation of a ternary complex between these proteins, Pellino1 and Pellino2 were both detected along with IRAK1 and TRAF6 when TRAF6 was immunoprecipitated from the extracts of IL-1R* cells, provided that the cells had been stimulated with IL-1β (Table S1). To investigate whether Pellino-generated K63-Ub chains trigger IL-1β signaling in cells that express E3 ligase-inactive mutants of TRAF6, we generated triple-KO IL-1R* cell lines lacking expression of TRAF6, Pellino1, and Pellino2. IL-1β signaling was similar in Pellino1/2 double-KO cells and WT IL-1R* cells, but abolished in either TRAF6 KO cells or TRAF6/Pellino1/2 triple-KO cells (Fig. 3C). IL-1β signaling could be partially restored by the reexpression of TRAF6[L74H] or TRAF6[120-522] in TRAF6 KO IL-1R* cells (Fig. 3D, lanes 10–12 and 16–18), but not by the reexpression of these mutants in the triple-KO cells (Fig. 3D, lanes 7–9 and 13–15). In contrast, the reexpression of WT TRAF6 fully restored IL-1β signaling to either TRAF6 KO or TRAF6/Pellino1/2 triple-KO IL-1R* cells (Fig. 3D, lanes 3–6). Taken together, these results indicate that the E3 ligase activities of TRAF6 and Pellino1/2 function redundantly to generate the K63-Ub chains required for IL-1 signaling.
Table S1.
Proteins captured by immunoprecipitation of TRAF6 from the extract of IL-1R* cells stimulated or not stimulated with IL-1β
| IL-1β (5 min) | ||
| + | − | |
| Protein | Unique peptides | Unique peptides |
| TRAF6 | 31 | 33 |
| Pellino1 | 3 | 0 |
| Pellino2 | 3 | 0 |
| IRAK1 | 24 | 0 |
| IRAK4 | 6 | 0 |
| MyD88 | 10 | 0 |
| IL-1R1 | 3 | 0 |
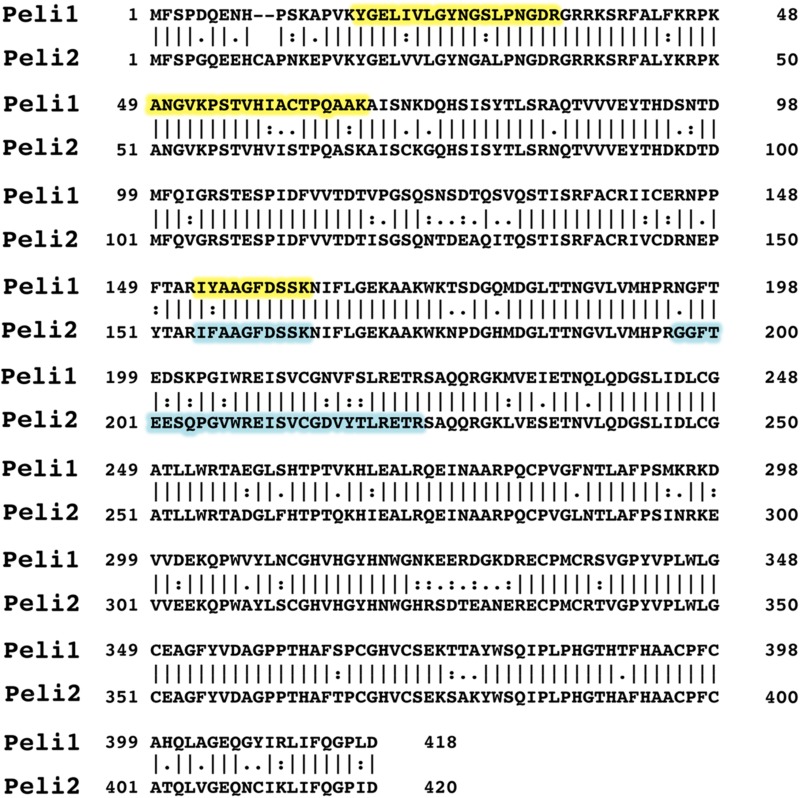
Proteins associated with TRAF6 immunoprecipitated from the extract of IL-1R* cells stimulated (+) or not stimulated (−) with IL-1β. The three unique peptides derived from Pellino1 and Pellino2 are highlighted in yellow and blue, respectively, in the sequence alignment of Pellino 1 and 2.
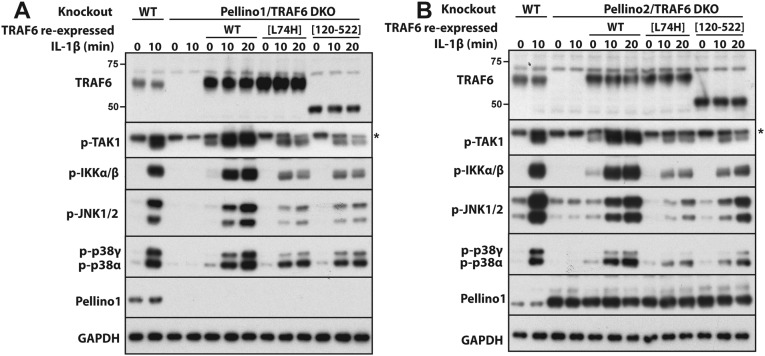
In contrast to TRAF6/Pellino1/2 triple-KO cells (Fig. 3D), the reexpression of E3 ligase-inactive TRAF6 mutants partially restored IL-1β signaling in either TRAF6/Pellino1 or TRAF6/Pellino2 double-KO cells (Fig. S4 A and B), indicating that Pellino1 or Pellino2 operate redundantly with one another to generate the K63-Ub chains needed to initiate IL-1β signaling in IL-1R* cells expressing E3 ligase-inactive mutants of TRAF6.
Fig. S4.
Pellino1 and Pellino2 function redundantly in the IL-1 pathway in IL-1R* cells. (A) WT IL-1R* cells (lanes 1 and 2) and Pellino1/TRAF6 double-KO cells (lanes 3 and 4) reexpressing Flag-tagged WT TRAF6, TRAF6[L74H], or TRAF6[120–522] (lanes 5–14) were stimulated with IL-1β, and the extracts were immunoblotted with antibodies that recognize the active, phosphorylated (p) forms of TAK1, IKKα/β, JNK1/2, p38α, and p38γ, as well as antibodies that recognize all forms of TRAF6. Pellino1, and GAPDH. (B) As in A, except that Pellino2/TRAF6 double-KO cells were used instead of Pellino1/TRAF6 double-KO cells. * denotes a nonspecific band detected by the antibody.
To study K63-Ub chain formation directly, we used Halo-NZF2 beads to capture them from the cell extracts (Fig. 1A). IL-1β stimulation induced the formation of K63-Ub chains in TRAF6 KO cells, Pellino1/2 double-KO cells, and WT cells, but not in TRAF6/Pellino1/2 triple-KO cells (Fig. 3E, Bottom, lanes 10–15). Some of the K63-Ub chains formed in response to IL-1 are attached covalently to the components of the Myddosome (30). Importantly, the IL-1β–dependent formation of Ub-IRAK1, Ub-IRAK4, or Ub-MyD88 was still robust in Pellino1/2 double-KO cells and TRAF6 KO IL-1R* cells, but abolished in TRAF6/Pellino1/2 triple-KO IL-1R* cells (Fig. 3E). Similar results were obtained with two different clones that were isolated independently. These findings confirm that TRAF6 and Pellino1 and 2 function redundantly in the IL-1β–dependent formation of K63-Ub chains, K63-Ub-IRAK1, K63-Ub-IRAK4, and K63-Ub-MyD88. MyD88 and IRAK4 form oligomeric complexes when they are recruited to the IL-1 receptor, explaining why the Halo-NZF2 beads capture the deubiquitylated as well as the ubiquitylated forms of these proteins from the cell extracts (Fig. 3E).
The ubiquitin chains attached to IRAK1, IRAK4, and MyD88 are “hybrid” molecules containing both Met-linked ubiquitin (M1-Ub) and K63-Ub linkages. The faster migration of the ubiquitylated-IRAK1 in TRAF6 KO cells compared with WT cells (Fig. 3E) is explained not by the generation of smaller K63-Ub chains, but by a drastic reduction in the number of M1-Ub linkages. Thus, incubation with Otulin, a deubiquitylase that only hydrolyses M1-Ub chains (38), increased the electrophoretic mobility of the Ub-IRAK1 formed in TRAF6-expressing cells (Fig. 3F, lanes 1–4), whereas the mobility of the Ub-IRAK1 produced in TRAF6 KO cells was only increased slightly by Otulin treatment (Fig. 3F, lanes 5–8). The greatly reduced formation of M1-Ub chains in TRAF6 KO cells was confirmed by immunoblotting with an M1-Ub chain-specific antibody (Fig. 3G). The IL-1–dependent formation of M1-Ub chains was restored by the reexpression of WT TRAF6 and partially restored by the reexpression of E3 ligase-inactive mutants of TRAF6 (Fig. 3G).
MyD88 Signaling in Primary Macrophages from TRAF6[L74H] Mice.
Few TRAF6 KO mice survive for more than 2 wk after birth (4), but the TRAF6[L74H] mice were still alive after 5 wk. However, at this age, they were culled because they had developed inflammation of the skin and other organs. A detailed analysis of the inflammatory phenotype will be presented in a separate publication.
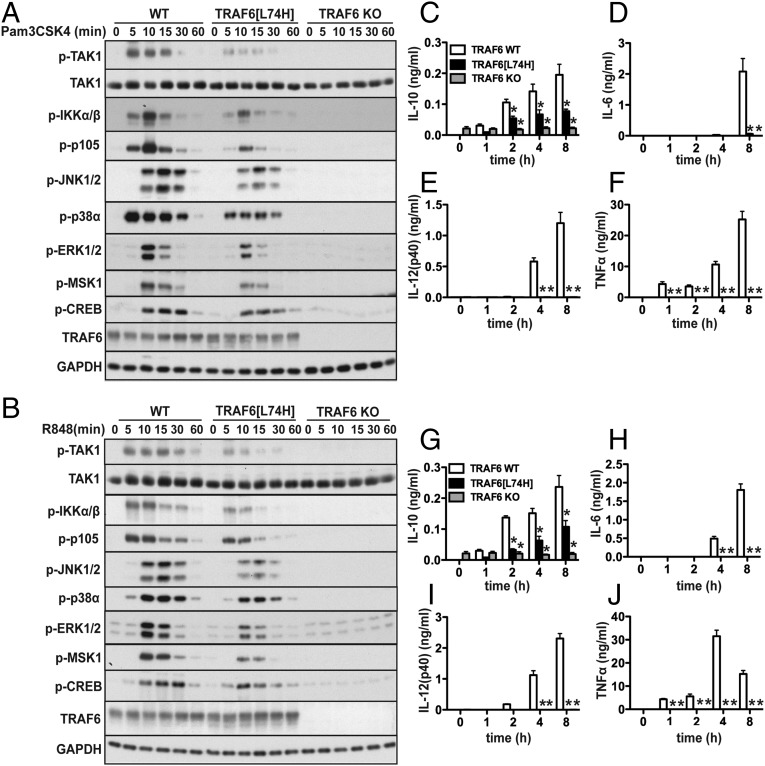
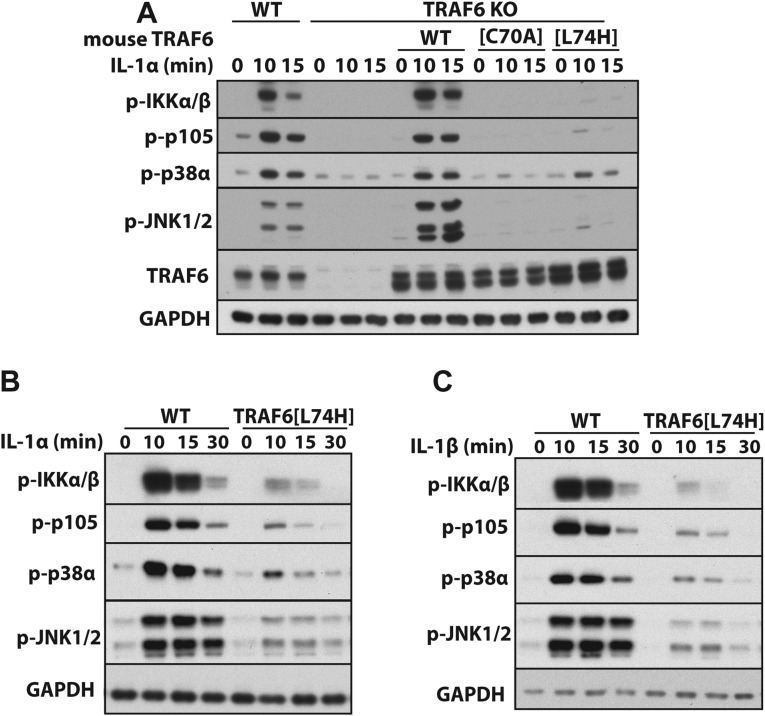
To investigate the role of the TRAF6 E3 ligase in primary macrophages, we generated a knockin mouse in which WT TRAF6 was replaced by the E3 ligase-inactive TRAF6[L74H] mutant (Fig. S5). The MyD88 signaling network in murine bone marrow-derived macrophages can be divided into an early phase up to 2 hr, which is characterized by robust but transient signaling and the production of antiinflammatory molecules, such as IL-10, and a late phase from 2 to 8 h, which is characterized by weaker signaling, but a huge acceleration in the production of proinflammatory cytokines (11). We found that early-phase signaling (Fig. 4 A and B) and IL-10 secretion (Fig. 4 C and G) induced by Pam3CSK4 (an activator of the TLR1/2 heterodimer) or R848 (a TLR7 agonist) was only reduced modestly in comparison with WT macrophages. The TLR-dependent transcription of il10 requires the MSK1/2-catalyzed phosphorylation of the transcription factor CREB (see Introduction), which was slightly reduced in TRAF6[L74H] macrophages (Fig. 4 A and B), consistent with the 50% reduction in IL-10 secretion observed in these cells (Fig. 4 C and G). In contrast, the late-phase production of TNFα, IL-6, and IL-12(p40) was reduced drastically in macrophages from TRAF6[L74H] mice (Fig. 4 D–F and H–J).
Fig. S5.
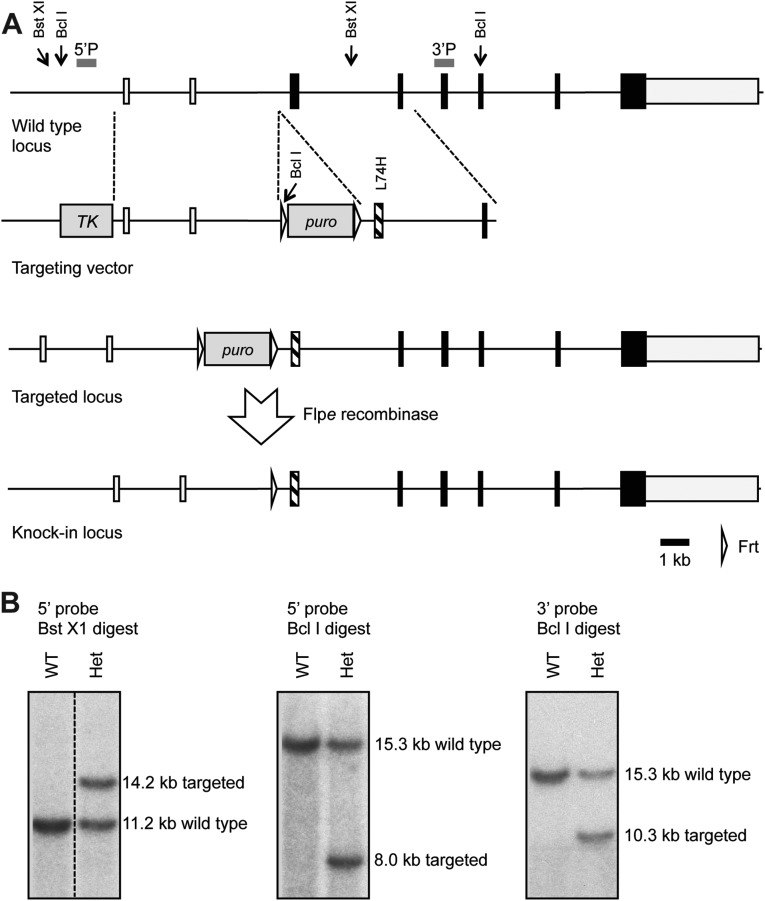
Generation of TRAF6[Leu74His] knockin mice. (A) The traf6 locus on chromosome 11 consists of eight exons with Leu74 encoded in exon 3. A targeting vector was constructed to introduce the desired Leu74His mutation. In addition to the regions of homology, an frt flanked neomycin resistance and f3 flanked puromycin resistance gene were included for positive selection and a thymidine kinase (TK) gene for negative selection. The positive selection markers were removed by crossing germline-transmitting chimeric mice to mice carrying a flp transgene. LoxP sites were also introduced around exon 6 to allow for Cre-mediated deletion of this exon. (B) Southern blots used to identify correctly targeted clones are shown. For a probe located 5′ to the targeting vector, genomic DNA was digested with BstXI or BclI, whereas for a probe 3′ to the targeting vector, a BclI digest was used. The positions of the BstXI and BclI sites relevant to the genomic Southern blots and the probes used are indicated in A. Restriction sites elsewhere in the traf6 locus have been omitted for clarity due to the size of this region. Het, heterozygous; puro, puromycin.
Fig. 4.
MyD88-dependent signaling in macrophages from TRAF6[L74H] mice. (A) Fetal liver macrophages from WT, TRAF6[L74H], and TRAF6 KO mice were stimulated with the TLR1/2 agonist, Pam3CSK4 (1 μg/mL). SDS-denatured cell extracts were separated by SDS/PAGE and immunoblotted with the indicated antibodies. (B) As in A, except that the cells were stimulated with the TLR7 agonist, R848 (1 μg/mL). (C–F) As in A, except that, after stimulation with Pam3CSK4, the concentrations of IL-10 (C), IL-6 (D), IL-12(p40) (E), and TNFα (F) in the culture medium were measured (mean ± SEM; n = 3). (G–J) As in C–F, except that the macrophages were stimulated with R848. *P < 0.05 (Methods).
Pam3CSK4 and R848 failed to elicit any signaling or cytokine secretion in fetal liver macrophages from TRAF6 KO mice, as expected (Fig. 4 A–J). A low basal level of IL-10 was detected consistently in the culture medium of TRAF6 KO macrophages incubated for 16 h before TLR stimulation. However, TLR ligation did not induce any increase in IL-10 secretion, as expected (Fig. 4 C and G).
Similar to IL-1R* cells (Fig. S1D), the expression of many components of the MyD88 signaling network was similar in the fetal liver macrophages of TRAF6[L74H], TRAF6 KO, and WT mice. However, the expression of Pellino1 was modestly enhanced in TRAF6 KO macrophages (Fig. S6).
Fig. S6.
Levels of expression of components of the MyD88 signaling network in fetal liver macrophages from WT, TRAF6[L74H], and TRAF6 KO mice. Macrophages were generated from the fetal livers of two mice of each genotype. The cells from each mouse were lysed and aliquots of the two extracts were immunoblotted with the antibodies indicated.
IL-1 Signaling in MEFs from TRAF6[L74H] Knockin Mice.
As reported previously by others in immortalized TRAF6 KO MEFs (12, 17, 18) or TRAF6 KO monocytes (39), we found that IL-1 signaling could be restored to immortalized TRAF6 KO MEFs by reexpressing the WT TRAF6 but not by the TRAF6[C70A] mutant, whereas only trace IL-1 signaling was restored by the reexpression of TRAF6[L74H] (Fig. S7A). In contrast, IL-1α (Fig. S7B) or IL-1β (Fig. S7C) signaling in primary MEFs from TRAF6[L74H] knockin mice was clearly detectable, although weaker than in WT MEFs. Why E3 ligase-inactive mutants of TRAF6 are unable to restore significant IL-1 signaling to immortalized TRAF6 KO MEFs, in contrast to human IL-1R* and HaCaT cells, and in contrast to the MyD88-dependent signaling that is clearly observed in primary macrophages and MEFs from TRAF6[L74H] mice, is unclear. It is possible that the TRAF6[C70A] mutant is unable to refold into a conformation capable of restoring IL-1 signaling in immortalized TRAF6 KO MEFs.
Fig. S7.
IL-1 signaling in MEFs from mice expressing WT and E3 ligase-inactive forms of TRAF6. (A) WT or the E3 ligase-inactive TRAF6[C70A] and TRAF6[L74H] mutants were reexpressed in immortalized MEFs from TRAF6 KO mice (Methods). The cells were stimulated with 5 ng/mL IL-1α for the times indicated, and the cell extracts were analyzed by immunoblotting with the antibodies indicated (Fig. S1). (B and C) Primary MEFs from WT and TRAF6[L74H] knockin mice were stimulated for the times indicated with IL-1α (B) or IL-1β (C) (5 ng/mL) followed by immunoblotting with the indicated antibodies.
Generation and Phenotypic Analysis of TRAF6[L74H] Knockin Mice.
TRAF6 is essential for many other cell functions, and the most striking phenotype of TRAF6 KO mice is independent of MyD88 signaling. TRAF6 KO mice are 20–30% smaller than WT mice, have deformed bones, and lack teeth (4). This phenotype is caused by osteopetrosis, due to the failure of RANKL to induce the production of osteoclasts (2, 4). RANKL, a TNF superfamily member, signals via its receptor RANK, which interacts directly with TRAF6 to initiate signaling (40, 41) by activating TAK1 (42, 43).
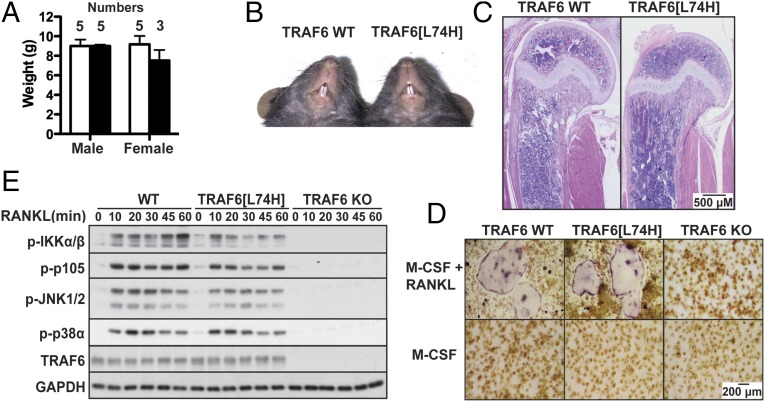
In contrast to TRAF6 KO mice, male or female TRAF6[L74H] mice were of similar weight to their WT littermates (Fig. 5A) and their teeth developed normally (Fig. 5B). The overall histological structure of their long bones was similar to WT mice, showing no evidence of osteopetrosis (Fig. 5C). Consistent with these observations, TRAF6[L74H] liver macrophages could be differentiated into osteoclasts in the presence of RANKL and macrophage colony-stimulating factor (M-CSF), but liver macrophages from TRAF6 KO mice could not (Fig. 5D). RANKL-dependent signaling was similar in macrophages from TRAF6[L74H] and WT mice, although the activation of IKKα/β and their substrate p105 was slightly reduced in TRAF6[L74H] macrophages (Fig. 5E). This was not due to a decrease in the expression of these proteins (Fig. S6). In contrast to TRAF6[L74H] mice, RANKL signaling was abolished in TRAF6 KO mice.
Fig. 5.
The phenotype and RANKL signaling in TRAF6[L74H] mice. (A) TRAF6[L74H] mice (black bars) are of similar size to their WT littermates (white bars). The indicated number of mice were weighed at 22 d of age, and weights are presented as mean ± SEM. An unpaired Student t test indicated that the differences were not significant. (B) The incisors of TRAF6[L74H] mice erupt similarly to WT littermate (33-d-old males). (C) H&E staining of bone (humerus) sections from WT and TRAF6[L74H] male mice aged 31 and 33 d, respectively (scale bar, 0.5 mm). Note normal microscopic architecture of the epiphysis, physeal growth plate cartilage, metaphysis, and diaphysis in the TRAF6[L74H] mouse. Similar results were obtained with bones from two further mice of each genotype. (D) Fetal liver macrophages from WT TRAF6 and TRAF6[L74H] mice, but not TRAF6 KO mice, differentiate into osteoclasts 6 d after stimulation with 100 ng/mL RANKL and M-CSF. Cells were fixed before staining for tartrate-resistant acid phosphatase and images were taken under bright-field microscopy at 4× magnification. (Scale bar, 0.2 mm.) (E) Fetal liver macrophages were stimulated with RANKL (100 ng/mL) and cell extract protein (20 μg) subjected to SDS/PAGE and immunoblotting with the indicated antibodies.
Other laboratories have reported that RANKL signaling cannot be restored to TRAF6 KO monocytes by the reexpression of TRAF6[C70A] (17, 39). This could be related to the problem discussed in the preceding section in refolding this E3 ligase-inactive mutant in MEFs to a conformation that can support signaling.
Discussion
The notion that TRAF6-generated K63-Ub oligomers initiate MyD88 signaling and other TRAF6-dependent processes has underpinned the field of innate immunity for many years. However, in the present study, we found that two different E3 ligase-inactive TRAF6 mutants (TRAF6[L74H] and TRAF6[C70A]) and even a TRAF6 mutant entirely lacking the RING domain, partially restored MyD88 signaling in both TRAF6 KO IL-1R* cells and HaCaT cells (Figs. 1 and 2). Further studies in IL-1R* cells revealed that the E3 ligase function of TRAF6 was not essential for IL-1 signaling because Pellino1 and Pellino2 were able to generate the K63-Ub chains required to activate TAK1 complexes in cells expressing E3 ligase-inactive mutants of TRAF6. These studies in model human cell lines led us to generate knockin mice expressing the E3 ligase-inactive TRAF6[L74H] mutant and to study MyD88 signaling in primary macrophages from these animals. These experiments confirmed that the TRAF6 E3 ligase contributes to, but is not required to trigger MyD88 signaling or IL-10 secretion by TLR ligands that signal via MyD88 (Fig. 4).
Although the early phase (0–2 h) of MyD88-dependent signaling and IL-10 secretion in TRAF6[L74H] macrophages was relatively intact, the production of proinflammatory cytokines (IL-6, IL-12, and TNFα), which takes place mainly during the late phase (2–8 h), was abolished (Fig. 4). During the early phase, IRAK1 and IRAK2 operate redundantly to induce signaling and IL-10 production, but during the late-phase IRAK1 expression is greatly decreased (11), and so IRAK2 becomes rate limiting. For this reason, the MyD88-dependent production of IL-6, IL-12, and TNFα is abolished in bone marrow-derived macrophages from IRAK2 KO mice (44) or in knockin mice expressing an IRAK2 mutant that cannot interact with TRAF6 (11). Like IRAK1, IRAK2 can induce the dimerization of TRAF6, which activates its E3 ligase activity (10, 12). However, unlike IRAK1, IRAK2 is a catalytically inactive “pseudokinase” (discussed in ref. 11) and unable to phosphorylate and activate Pellino isoforms. This could explain why the E3 ligase activity of TRAF6 is essential for late-phase signaling and proinflammatory cytokine production. In contrast, the IRAK1-catalyzed activation of Pellino isoforms may produce the K63-Ub chains required for early-phase signaling and IL-10 production in macrophages expressing the E3 ligase-inactive TRAF6[L74H] mutant. Therefore, whether the TRAF6 E3 ligase is essential is likely to depend on whether compensation by a Pellino isoform(s) or another K63-Ub-generating E3 ligase(s) is possible. This may in turn depend on the duration of the stimulus, as well as on the cell type and cell function.
We found that RANKL-induced osteoclast formation and RANKL signaling were similar in TRAF6[L74H] and WT macrophages (Fig. 5). These finding are consistent with a report that TRAF6 mutants lacking the RING domain restore RANKL-dependent differentiation to multinuclear osteoclasts when reexpressed in TRAF6 KO splenocytes (16). They are also consistent with the normal bone structure and teeth of the TRAF6[L74H] mice (Fig. 5). These findings demonstrate that the E3 ligase activity of TRAF6 is not required for RANKL signaling.
It will clearly be interesting to further exploit the TRAF6[L74H] mouse to investigate whether other essential roles of TRAF6 (see Introduction) require its E3 ligase activity and, if not, whether Pellino isoforms generate the K63-Ub chains required in these systems (Fig. 3). These studies will also require the TRAF6[L74H] mouse to be crossed to mice expressing E3 ligase-inactive mutants of Pellino1 and Pellino2, to generate triple-knockin mice deficient in all three E3 ligase activities.
An important outcome of this study is that it has identified two essential roles of TRAF6 in the MyD88 signaling network that are independent of its E3 ligase activity. First, although Pellino1 and Pellino2 generate the K63-Ub chains required for MyD88 signaling in cells expressing E3 ligase-inactive TRAF6 mutants, no signaling occurs in TRAF6 KO cells despite the unimpaired formation of K63-Ub chains. These findings imply that TRAF6, but not its E3 ligase activity, is required to couple K63-Ub chain formation to the activation of TAK1. Second, the IL-1–dependent formation of M1-Ub chains, catalyzed by the linear ubiquitin assembly complex (LUBAC), is greatly reduced in TRAF6 KO cells, but can be partially restored by reexpressing E3 ligase-inactive mutants of TRAF6. The M1-Ub chains are not required for the IL-1–dependent TAK1-catalyzed activation of MAP kinases, but for the TAK1-catalyzed activation of the canonical IKK complex (45). Because the intrinsic catalytic activity of LUBAC is not altered by IL-1 stimulation (30) and the expression of the components of LUBAC (HOIP, HOIL-1, and Sharpin; Fig. S1D) is unimpaired in TRAF6 KO cells, these observations suggest that TRAF6 could have a critical role in recruiting LUBAC to the Myddosome. Precisely how TRAF6 achieves these two essential “scaffolding” roles is an interesting problem for future research.
Methods
Phosphospecific Antibodies Used to Monitor Activation of the MyD88 Signaling Pathway.
All antibodies were from Cell Signaling Technologies, unless stated otherwise.
The activation of protein kinases in the pathway was monitored with phosphospecific antibodies that recognize phosphorylation sites in their activation loops, which are known to be required for activation. These were TAK1 phosphorylated at Thr187 (#4536), IKKα phosphorylated at Ser176 and Ser180 (#2697), IKKβ phosphorylated at Ser177 and Ser181 (#2697); p38α and p38γ MAP kinases phosphorylated at their Thr–Gly–Tyr motifs (#9211), ERK1 and ERK2 phosphorylated at their Thr–Glu–Tyr motifs (#9101), MKK6 phosphorylated at Ser207 (#12280), and JNK1 and JNK2 phosphorylated at their Thr–Pro–Tyr motifs (#4668). The activation of several protein kinases was also monitored by the phosphorylation of established substrates. The protein p105/NF-κB1 phosphorylated at Ser933 (#4806) is a physiological substrate of IKKβ (27, 28), whereas MSK1 is phosphorylated at Thr581 (#9595) by p38α MAP kinase, ERK1, and ERK2 in cells (46), and CREB is phosphorylated at Ser133 (#9198) by MSK1 in cells (29).
Generation of IL-1R* Cells.
HEK293 cells expressing FLAG-Cas9 under an inducible promoter were provided by Yosua Kristariyanto and John Rouse [MRC Protein Phosphorylation and Ubiquitylation Unit (MRC-PPU), Dundee, Scotland, UK] using the Flp-In T-Rex system (Invitrogen) (47). Retroviral particles containing the IL-1R with a neomycin-resistance gene (DU46481) were generated using a Murine Moloney Leukemia virus-based system prepared with the VSVG envelope protein, according to the manufacturer’s instructions (Clontech). The retroviral particles were incubated for 24 h with HEK293 FLAG-Cas9 cells with 2.0 μg/mL protamine sulfate (Sigma). Cells were selected by exposure to media containing 1.0 mg/mL G418 for 1–2 wk.
The IL-1 receptor is overexpressed in IL-1R* cells, although at far lower levels than in another widely used IL-1R–expressing cell line (Fig. S3) (48).
Generation of IL-1R* Cells Lacking Expression of TRAF6.
The guide RNAs (gRNAs) used to target the genes encoding TRAF6 are summarized in Table S2. The gRNA plasmids targeting TRAF6 were pooled and 10 µg was used to transfect IL-1R* cells for 8 h using the GeneJuice transfection reagent (Merck Millipore). Doxycycline was then added to the cells to a final concentration of 1.0 µg/mL, and a further 18 h later the cells were again transfected with the same amounts of gRNA plasmids. After 48 h, cells were single-cell plated into 96-well plates and left until colonies began to form (2–3 wk). The mutation efficiency was analyzed by immunoblotting of the cell extracts.
Table S2.
Sequences of the gRNAs used to knock out the indicated proteins by CRISPR/Cas9 gene-editing technology
| Gene | Exon | DNA sequence | DU number | Project number |
| TRAF6 | 2 | gTAAACTGTGAAAACAGCTG(TGG) | 48406 | CR45 |
| TRAF6 | 2 | gGCAGTCACTTTCAGACTGGC(TGG) | 48407 | CR45 |
| TRAF6 | 2 | gGCGCTACAGGAGCTGGCCA(TGG) | 48408 | CR45 |
| TRAF6 | 2 | gGAAGCAGTGCAAACGCCATG (sense) | 52382 | CR45 |
| TRAF6 | 2 | gGTCGTAATGCCATCAAGCAGA (antisense) | 52392 | CR45 |
| TAK1 | 1 | gCAAGGAGATCGAGGTGGAAGAGG (sense) | 52138 | CR146 |
| TAK1 | 1 | gAGGGGCTTCGATCATCTCACCGG (antisense) | 52141 | CR146 |
| PELI1 | 2 | gTATGGTGAACTCATTGTCTTAGGm (sense) | 52227 | CR180 |
| PELI1 | 2 | gTATTTTACTGGTGCTTTAGATGG (antisense) | 52239 | CR180 |
| PELI1 | 4 | gCAGCAACACCGATATGTTTCAGG (sense) | 52228 | CR180 |
| PELI1 | 4 | gATATTCAACCACCACAGTCTGGG (antisense) | 52240 | CR180 |
| PELI2 | 1 | gATAAGGAGCCAGTGAAATACGGG (sense) | 52229 | CR181 |
| PELI2 | 1 | gTGGGGGCGCAGTGTTCCTCCTGG (antisense) | 52241 | CR181 |
Generation of Other KO IL-1R* and HaCaT Cell Lines.
These cells were produced by CRISPR/Cas9 technology using an improved procedure. One pair of gRNAs was generated to target TRAF6, TAK1, IRAK1, Pellino1, or Pellino2. The antisense gRNA was introduced to the vector encoding the Cas9[D10A] mutant, which only cleaves one strand of the DNA molecule complementary to the gRNA. In contrast, the sense gRNA was inserted into a plasmid containing a puromycin resistance gene. Each gRNA plasmid (1.0 μg) was mixed with 1.0 mL of serum-free DMEM and 0.02 mL of polyethylenimine (1.0 mg/mL) (Fugene HD for HaCaT cells), and after incubation for 20 min at 20 °C, the solution was added to the cells dropwise for transfection. After 24 and 48 h, the medium was replaced with fresh medium containing 2.0 μg/mL puromycin. The cells were then single-cell plated into 96-well plates and left until colonies began to form (2–3 wk). The mutational efficiency was analyzed by immunoblotting of the cell extracts for the relevant proteins. Double-KO IL-1R* cells lacking expression of both TRAF6 and Pellino1 were generated by targeting TRAF6-null IL-1R* cells with gRNAs specific for Pellino1. Triple-KO IL-1R* cells lacking expression of TRAF6, Pellino1, and Pellino2 were generated by targeting the TRAF6/Pellino1 double-KO cells with gRNAs specific for Pellino2. The TRAF6/Pellino2 double-KO cells were generated by targeting the TRAF6 KO cells with gRNA specific for Pellino2. The Pellino1/Pellino2 double-KO cell lines were created in a similar fashion by targeting Pellino1 KO cells with gRNA specific to Pellino2. Due to the lack of an antibody that recognizes Pellino2, individual clones were screened for the absence of Pellino2 by PCR amplifying and sequencing a 342-bp region of genomic DNA containing the CRISPR target site (forward primer, ATTTGTTGCCGGCTCTGACT; reverse primer, AGGGACCCCAGGACTCAC), allowing the visualization of indels.
Reexpression of TRAF6 in TRAF6 KO IL-1R* Cells Using the Flp-In System.
TRAF6 KO IL-1R* cells were cotransfected using GeneJuice (Millipore) with 9 μg of POG44 recombinase (Invitrogen), and 1 μg of pcDNA5 FRT/TO vector containing WT human TRAF6 or TRAF6 mutants containing a puromycin-resistance gene (DU46785, DU46824, and DU46823). Forty-eight hours after transfection, cells were selected with 2.0 μg/mL puromycin. To induce TRAF6 expression at levels equivalent to the endogenous protein, reconstituted cells were incubated for 16 h with 0.03 ng/mL doxycycline for WT and TRAF6[L74H] and 0.3 ng/mL for TRAF6[C70A].
Reexpression of TRAF6 and TAK1 in IL-1R* Cells, HaCaTs, and MEFs.
Cells stably reexpressing TRAF6 or TAK1 were generated by retroviral transduction as described (45). Viruses encoding the gene of interest and the Tet-On protein were harvested 48 h after transfection, diluted fourfold with fresh medium, and incubated with the cells for 24 h in the presence of 2.0 μg/mL protamine sulfate (Sigma). Fresh medium containing 1 mg/mL G418 (Tet-On) and 1 μg/mL puromycin (gene of interest) was added to select the transduced cells. To induce gene expression, cells were cultured for 16 h with 0.1–1.0 μg/mL doxycycline. DNA encoding a TAK1 splice variant lacking DNA encoding amino acid residues 414–430 of the full-length protein were reexpressed in TAK1 KO IL-1R* cells (45).
Mouse TRAF6 and the mouse TRAF6[C70A] and TRAF6[L74H] (DU51028, DU51041, and DU51027) mutants were reexpressed in TRAF6 KO MEFs.
Human TRAF6, TRAF6[C70A], and TRAF6[L74H] (DU51583, DU51585, DU51584) were reexpressed in HaCaT cells.
Human FLAG-tagged TRAF6, TRAF6[L74H], TRAF6[120-522], and TRAF6[160-522] (DU32495, DU46743, DU51445, and DU51447) were reexpressed in IL-1R* cells. The vectors were expressed constitutively and did not require cotransfection of Tet-On or doxycycline induction.
Generation of TRAF6 Knockin Mice and Maintenance of Mouse Lines.
WT C57/BL6 mice were obtained from Charles River Labs, and knockin mice carrying the Leu74His mutation in TRAF6 were made by TaconicArtemis using conventional methods. Targeting vectors (Fig. S5) were constructed by recombinase-mediated cloning to generate the desired mutations. They were then used to target Art B6.3.5 (C57BL/6 NTac) ES cells. Positive colonies were identified by Southern blotting and the presence of the point mutation confirmed by PCR and sequencing of the appropriate genomic region. Correctly targeted ES cells were used to generate chimeric mice via blastocyst injection. To remove the neomycin and puromycin resistance selectable markers, mice were crossed to a constitutive flp transgenic line (on a C57BL/6 background). Once deletion had occurred, the TRAF6 allele was crossed away from the flp transgene. Routine genotyping was carried out by PCR of ear biopsies. The sequences of the primers used for TRAF6 were AATAGAAATCACCAGACTGGGC and CACACAACAGTCAATGTTTACTAGG, which resulted in a band of 283 bp for a WT allele and 357 bp for a knockin allele. Heterozygous TRAF6 KO mice were provided by Tak Mak. Mice were maintained on a C57BL/6 background and provided with free access to food and water. Animals were kept in ventilated cages under specific-pathogen–free conditions in accordance with UK and European Union regulations. Experiments were carried out subject to approval by the University of Dundee ethical review board under a UK Home Office project license.
Activation of the TAK1 Complex in Vitro.
The purified TAB1–TAK1–TAB3 complex (1.0 nM) was activated by incubation for 1 h at 30 °C with 0.2 µM UBE1, 1.0 µM Ubc13-Uev1a, 12.5 µM ubiquitin, and either 0.4 µM TRAF6 or 0.4 µM Pellino1 in 50 mM Tris/HCl, pH 7.5, 2.0 mM ATP, and 5 mM MgCl2 in a total reaction volume of 0.03 mL. MKK6 (0.1 μM) and p38γ MAP kinase (0.5 μM) were also included. Reactions were terminated by the addition of 5% (wt/vol) SDS and the activation of TAK1 measured by immunoblotting with the antibody recognizing TAK1 at Thr187 and by the phosphorylation of its substrate MAP kinase kinase 6 (MKK6) at Ser207. The activation of MKK6 was also assessed with an antibody recognizing the phosphorylated Thr–Gly–Tyr sequence in the activation loop of its substrate p38γ MAP kinase. The expression of TAK1 and MKK6 was checked by immunoblotting and the expression of p38γ MAP kinase by staining for protein with Ponceau S. The formation of K63-Ub chains was examined by immunoblotting with a specific antibody.
IL-1 Preparations.
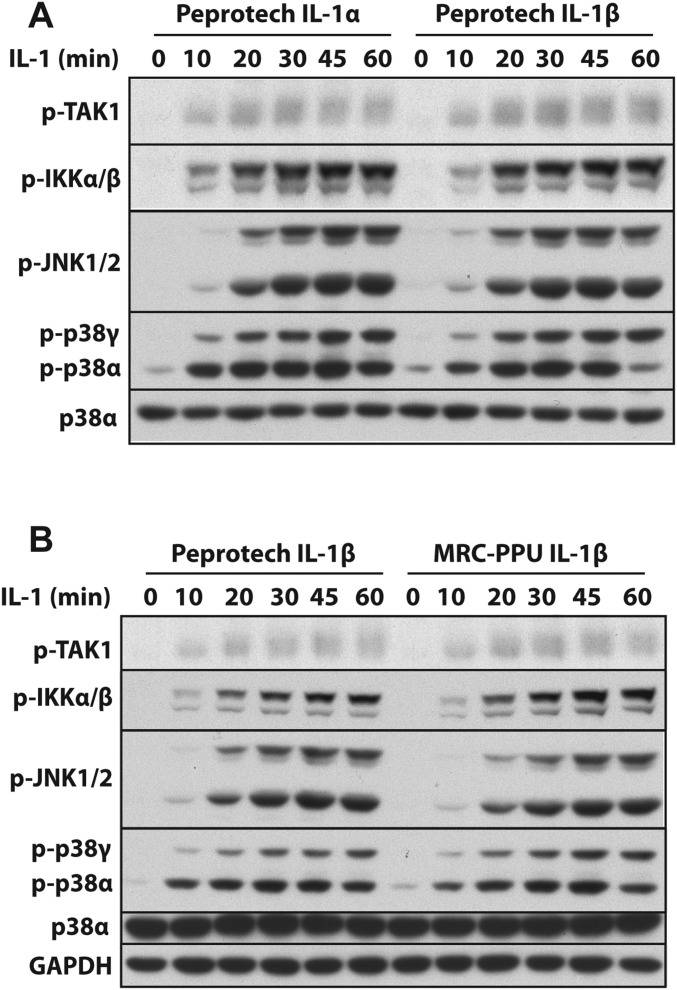
Human IL-1β (DU8685) (30) and mouse IL-1α (DU46302) were expressed in E. coli and purified by the PPT of the MRC-PPU. IL-1α was expressed as a GST-fusion protein separated by a PreScission protease cleavage site and purified on glutathione-Sepharose (GE Healthcare). GST-IL-1α was cleaved with PreScission protease to release IL-1α [115–270] and purified by gel filtration on Superdex G200. Endotoxin-free human IL-1α (200-01A), human IL-1β (200-01B) and murine IL-1β (211-11B) were all purchased from Peprotech. The commercial preparations of IL-1 were used to confirm that human IL-1α− and IL-β−dependent signaling were similar in human IL-1R* cells (Fig. S8A) and to establish that the commercial human IL-1β preparation induced signaling similarly to IL-1β produced in the MRC-PPU (Fig. S8B). The mouse IL-1α produced in the MRC-PPU and mouse IL-1β from Peprotech (211-11B) stimulated the MyD88 pathway similarly in primary MEFs (Fig. S7 B and C).
Fig. S8.
Comparison of IL-1α− and IL-β–induced activation of the MyD88 signaling network in IL-1R* cells. (A) IL-1R* cells were stimulated for the times indicated with 5 ng/mL IL-1α or IL-1β purchased from Peprotech followed by immunoblotting with the indicated antibodies. (B) As in A, except that IL-1R* cells were stimulated with Peprotech IL-1β or IL-1β generated in the MRC-PPU.
Reproducibility and Statistical Analysis.
All of the experiments reported in this paper were repeated at least three times with similar results. Statistical analyses were performed with GraphPad Prism Software, and quantitative data in graphs and bar charts are presented as the arithmetic mean ± SEM. Statistical significance of differences between experimental groups was assessed in all graphs and bar charts, using the two-way ANOVA with Bonferroni posttest, unless indicated otherwise. Differences in means were considered significant if P < 0.05 and indicated with an asterisk (*). Statistical differences that are not significant are not highlighted by asterisks in the figures.
SI Methods
DNA Constructs.
Recombinant DNA procedures, restriction digests, and ligations were performed using standard protocols. All PCRs were carried out using KOD Hot Start DNA polymerase (Novagen). DNA sequencing was performed by the DNA Sequencing Service, MRC Protein Phosphorylation and Ubiquitylation Unit (MRC-PPU) of Life Sciences, University of Dundee (https://www.dnaseq.co.uk/). All clones were human, unless stated otherwise. The pFastBac Dual DAC-TEV expression system was created by subcloning a BglII–BamH1 flanked PCR product encoding the full-length DAC tag (49) followed by a tobacco etch virus (TEV) protease cleavage site into the BamH1 site of pFastBac Dual (Life Technologies). In this vector, untagged proteins can be expressed from the p10 promoter in cassette 1 alongside N-terminally DAC-tagged proteins expressed from the polyhedron promoter in cassette 2. The ORF encoding TRAF6 (National Center for Biotechnology Information NM_145803.2) was cloned (50) and inserted into the BamHI NotI sites to create a DAC-TEV-TRAF6 expression cassette. Mutants were created using the QuikChange Site Directed Mutagenesis method (Agilent), but using KOD DNA Hotstart Polymerase (Merck Millipore). The MultiBac expression system (51) was used for expression of the multiprotein complex comprising TAB1, TAK1, and TAB3. The cDNAs coding for TAB1, TAK1, and TAB3 were reamplified by PCR from existing clones (accession numbers NM_006116.2, NM_003188.3, and NM_152787.3 GI:98991766, respectively), adding the appropriate restriction sites at the 5′ and 3′ ends of the ORFs. The 5′ PCR oligonucleotide for TAB3 included 6His and PreScission protease site sequences. TAB1 was cloned into pFBDM as a XhoI/KpnI insert. TAK1 was cloned into the resulting pFBDM TAB1 clone as a BamHI/NotI insert to create pFBDM TAB1/TAK1. 6His-PreScission-TAB3 was cloned into pFBDM as a BamHI/NotI insert to create pFBDM 6His-PreScission-TAB3. The triple-expression construct was created by cloning the PmeI/AvrII fragment from pFBDM 6His-PreScission-TAB3 into the SpeI/NruI sites of pFBDM TAB1/TAK1 (DU45364). Clones were sequence verified at every stage.
DNA clones encoding TRAF6 (DU46785), TRAF6[C70A] (DU46824), and TRAF6[L74H] (DU46823) were inserted in pcDNA5-FRT/TO vectors (47). DNA clones encoding Halo-NEMO (DU35939), Halo-NZF2[644-692] (DU23839) [expressing a tandem repeat of amino acid residues 644–692 of TAB2 (30)], and Halo-TUBEs (DU39178) were inserted into pFN18A vectors. DNA clones encoding TAK1 (DU51270), TAK1[D157A] (DU51293), TRAF6 (DU51583), TRAF6[C70A] (DU51585), TRAF6[L74H] (DU51584), mouse TRAF6 (DU51028), mouse TRAF6[C70A] (DU51027), and mouse TRAF6[L74H] (DU51041) were inserted into pRetroX-Tight-Puromycin vectors (45). The IL-1R1 (DU46481), FLAG-TRAF6 (DU32495), FLAG-TRAF6[L74H] (DU46743), FLAG-TRAF6[120–522] (DU51445), and FLAG-TRAF6[160–522] (DU51447) DNA clones were inserted into pBABE vectors (45). The DNA clones and proteins generated for the present study have been given the assigned [DU] numbers in the sections below and can be ordered from the reagents section of the MRC-PPU website (https://mrcppureagents.dundee.ac.uk/).
Proteins and Other Materials.
All proteins were expressed from human DNA, unless stated otherwise. TEV-protease, phage λ phosphatase (DU4170), PreScission Protease (DU34905), Ubc13 (UBE2N) (DU15705), Uev1a (UBE2V1) (DU20179), UbcH7 (UBE2L3) (DU12798), UBE1 (DU32888), ubiquitin (DU20027), FLAG-Ubiquitin (DU46789), Otulin (DU43487), p38γ MAP kinase (DU980), MKK6 (DU32175), and Pellino1 (DU8981) were expressed in Escherichia coli and purified by the Protein Production Team (PPT) of the MRC-PPU. IL-1α was expressed as a GST-fusion protein separated by a PreScission protease cleavage site and purified on glutathione-Sepharose (GE Healthcare). Murine RANKL (#315-11C) was purchased from Peprotech. TLR agonists Pam3CSK4 (tlrl-pms), Pam2CSK4 (tlrl-mp2s-1), and R848 (tlrl-r848) were purchased from Invivogen. Halo-NEMO beads, Halo-NFZ2 (previously called Halo-TAB2 (30), and Halo-TUBE beads were prepared as described (30).
Human TRAF6 (DU43249), or TRAF6 mutants in which Cys70 and Leu74 were changed to Ala (DU43527) and His (DU46742), respectively, were expressed in insect cells as Dac-tagged fusion proteins (49). The insect cells were sedimented by centrifugation, lysed in 50 mM Tris/HCl, pH 7.5, 0.2% (vol/vol) Triton X-100, 1 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP), 0.1 mM EDTA, 0.1 mM EGTA, 20 µg/mL leupeptin, and 1 mM Pefabloc, and insoluble material was removed by centrifugation. The supernatant made 250 mM in NaCl and incubated for 45 min at 21 °C with 2 mL of freshly prepared ampicillin-Sepharose. Contaminants were washed away by repeated (five times) resuspension of the ampicillin-Sepharose in 13 vol of 50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 0.03% (wt/vol) Brij35, and 1 mM tris(2-carboxyethyl)phosphine (Buffer C). To cleave TRAF6 from the Dac-tag, the ampicillin-Sepharose was resuspended in 0.5 vol of Buffer C and incubated for 3 h at 21 °C with 20 µg of TEV-protease with occasional agitation. TRAF6 was the major protein-staining band in the final preparation, the only significant contaminant being HSP70. Between 0.5 and 1.0 mg of TRAF6 was purified from each liter of cell culture medium.
Insect Sf21 cells were infected with baculovirus encoding the TAB1, TAK1, and TAB3 (DU45364) proteins, and the cells from 1 L of cell culture medium were pelleted and resuspended in lysis buffer [25 mM Tris/HCl, pH 7.5, 0.02 mM EDTA, 270 mM sucrose, 1% (vol/vol) Triton X-100, 0.03% (wt/vol) Brij-35, 1.0 mM 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride, protease inhibitor mixture (Roche; #11836170001; one tablet per 50 mL), and 0.5 mM tris(2-carboxyethyl)phosphine/HCl, pH 7.0]. After end-over-end rotation for 30 min at 4 °C, followed by centrifugation for 20 min at 15,000 × g at 4 °C, the supernatant was removed. NaCl was added to a final concentration of 0.3 M and the cell extract was then mixed with Talon cobalt-Sepharose resin (3-mL packed volume) (Clontech) followed by incubation for 60 min at 4 °C on a rotating wheel. The suspension was poured into a 30-mL column, and the resin was washed with 500 mL of 25 mM Tris/HCl, pH 7.5, 0.02 mM EDTA, 0.07% (vol/vol) 2-mercaptoethanol, 0.3 M NaCl, and 0.2 mM PMSF (Buffer A), containing 10 mM imidazole, and then with Buffer A. The TAB1–TAK1–TAB3 complex was eluted from the resin by cleaving the His6 tag attached to the N terminus of the TAB3 component with GST-tagged PreScission protease. The protease (0.1 mg in 10 mL of Buffer A) was incubated with the resuspended resin for 16 h at 4 °C and the supernatant retained. The resin was washed with a further 2 mL of Buffer A, and the two supernatants were combined and incubated with 0.3 mL of glutathione-Sepharose beads to remove the GST-Precission protease. The eluate was then concentrated to 0.5 mL using an Amicon Ultra tube (Millipore) with a 50-kDa cutoff limit to give a 0.05 mg/mL solution.
Antibodies.
A polyclonal antibody was raised against Pellino1 in rabbits (Biogenes) and is described in ref. 52. An antibody that recognizes M1-Ub chains specifically (53) was generously provided by Dr. Vishva Dixit (Genentech, South San Francisco, CA). A polyclonal antibody was raised in sheep against HOIP by the MRC PPU antibody production team (DU15972, S174D) and is described in ref. 30. An antibody that recognizes K63-Ub chains (#5621), antibodies recognizing all forms of TAK1 (#4505), IRAK1 (#4504), IRAK4 (#4363), MyD88 (#4823), IKKα (#2682), p38α MAP kinase (#9212), JNK1/2 (#9258), NEMO (#2685), ERK1/2 (#9102), IκBα (#4814) MKK6 (#8550), and GAPDH (#2118) were purchased from Cell Signaling Technology. Anti-ubiquitin was from Dako (#Z0458), anti-TRAF6 (#sc-7221) and anti-IL-1R (#sc-688) were from Santa Cruz Biotechnology, and anti-IKKβ (#05–535) was from Merck Millipore. Anti-p105 (#ab7971-1) was from Abcam. Anti-HOIL-1 (#HPA024185), anti-FLAG (#F3165), anti-FLAG coupled to agarose (A2220), and anti-HA coupled to agarose (A2095) were from Sigma-Aldrich, and anti-Sharpin (#14626-1-AP) was from Proteintech Group. An antibody recognizing IRAK4 phosphorylated at Thr345 and Ser346 (54) was a kind gift from Dr. Vikram Rao (Pfizer, Boston). Secondary antibodies coupled to HRP were from Bio-Rad.
Histological Evaluation of Mouse Tissues.
Mice were killed with CO2 asphyxiation and submitted to a full postmortem examination. Main organs from the thoracic and abdominal cavity, skin of the dorsum, skull (with brain), spine, forelimbs, and hindlimbs were collected in 10% neutral buffered formalin (NBF). Tissues containing bones were transferred in a decalcifying solution (10% EDTA in NBF) for 1 wk. Following adequate fixation (and decalcification where appropriate), tissues were trimmed and processed to paraffin blocks. Three-micrometer-thick sections were stained with H&E for microscopic evaluation. Slides were assessed by a veterinary pathologist (F.M.) blinded to the genotype of the mice. Histological findings were annotated. Representative microphotographs were acquired with an Olympus SC100 and imaging software (cellSens, version 1.15; Olympus).
Cell Culture, Cell Stimulation, and Cell Lysis.
Immortalized MEFs from TRAF6 KO and WT mice were generously provided by Tak Mak. HEK293 cells stably expressing IL-1R used as a control in Fig. S3 were provided by Xiaoxia Li and George Stark (Case Western Reserve University, Cleveland, OH). MEFs, HEK293, and HaCaT cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 2 mM glutamine, 10% (vol/vol) FBS, and the antibiotics streptomycin (0.1 mg/mL) and penicillin (100 U/mL). MEFs were stimulated with 5 ng/mL mouse IL-1α or mouse IL-1β and human IL-1R* cells with 5 ng/mL human IL-1α or IL-1β, and HaCaT cells with human IL-1β, washed in ice-cold PBS, and extracted in 50 mM Tris/HCl, pH 7.4, 1 mM EDTA, 1 mM EGTA, 50 mM NaF, 5 mM sodium pyrophosphate, 10 mM sodium 2-glycerol 1-phosphate, 1 mM sodium orthovanadate, 0.27 M sucrose, 1% (vol/vol) Triton X-100, 1 μg/mL aprotinin, 1 μg/mL leupeptin, and 1 mM phenylmethylsulphonyl fluoride. This buffer contained either 100 mM iodoacetamide to inactivate deubiquitylases (when studying ubiquitin chain formation) or 1.0 mM DTT and not iodoacetamide for all other studies. Cell extracts were clarified by centrifugation, and protein concentrations were determined by the Bradford assay.
Generation of Primary Macrophages, Osteoclasts, and MEFs.
Primary macrophages were generated by differentiating fetal livers from E14.5 embryos for 10 d in DMEM supplemented with 20% (vol/vol) L929 conditioned medium as a source of M-CSF, 2 mM glutamine, 10% (vol/vol) FCS, 1 mM sodium pyruvate, 0.2 mM 2-mercaptoethanol, penicillin, streptomycin, and nonessential amino acids at the concentrations recommended by the manufacturer (Life Technologies) (macrophage growth medium). Fetal liver-derived macrophages were differentiated for 10 d, harvested, and then replated at a density of 100,000 cells per cm2 in tissue culture-treated plastic containing 0.1 mL of fresh media before stimulation on day 11. To generate osteoclasts, fetal liver-derived macrophages were replated after 6 d into 24-well tissue culture-treated plastic plates, at a density of 200,000 cells per well in macrophage growth medium. The following day, the macrophage growth medium was supplemented with 50 ng/mL RANKL. Every 2 d, 50% of the media was replaced with fresh RANKL. Six days later, the cells were washed with PBS, fixed with 4% formaldehyde, and stained for TRAP according to the manufacturer’s instructions (Sigma; 387A-1KT). Primary MEFs were generated from E12.5 embryos. The embryos were washed in ice-cold PBS, minced, incubated with trypsin solution (Life Technologies) for 10 min at 37 °C, and plated into two 15-cm dishes. Primary MEFs were maintained in macrophage growth medium lacking L929-conditioned medium.
Capture of Ubiquitin Chains from Cell Extracts and Treatment with Phosphatase and Deubiquitylase.
Ubiquitin chains and ubiquitylated proteins were captured from cell extracts using Halo-NEMO beads to capture M1-Ub chains (30, 31), Halo-NZF2 beads to capture K63-Ub chains (23, 55), and Halo TUBEs (56) to capture all ubiquitin linkage types. Ten microliters of packed beads were used per milligram of cell extract protein, which was sufficient to capture all of the M1-Ub (Halo-NEMO beads), K63-Ub (Halo-NZF2 beads), or all Ub chains (Halo-TUBE beads) from the extracts. The beads were washed three times with 1 mL of 50 mM Tris/HCl, 0.5 M NaCl, and 1% (vol/vol) Triton X-100, and once with 1 mL of 50 mM Tris/HCl, pH 7.5, 50 mM NaCl, and 5 mM DTT. To dephosphorylate or deubiquitylate proteins captured by Halo-TUBE beads, they were resuspended in 30 μL of 50 mM Hepes, pH 7.5, 100 mM NaCl, 2 mM DTT, 1 mM MnCl2, and 0.01% (wt/vol) Brij-35 containing the protein phosphatase encoded by bacteriophage λgt10 phage (λPPase; 100 units per reaction) and the M1-Ub–specific deubiquitylase Otulin (5 μM). After 60 min at 37 °C, incubations were terminated, and proteins were released by denaturation in SDS followed by immunoblotting.
Immunoblotting.
Cell extracts (20 μg of protein), immunoprecipitated proteins, or proteins captured on immobilized Halo-tagged proteins were denatured in SDS, separated by SDS/PAGE, transferred to PVDF membranes, and immunoblotted with the antibodies indicated in the figure legends, using the ECL detection system (GE Healthcare).
Assay of E3 Ligase Activities in Vitro.
The endogenous Pellino1 was immunoprecipitated from 1.0 mg in IL-1R* cell extract protein by incubation for 16 h at 4 °C with 1 μg of anti-Pellino1 antibody bound to 7.5 μL of packed protein G-agarose beads. The beads were washed three times with 0.5 mL of 50 mM Tris/HCl, pH 7.5, 1% (vol/vol) Triton X-100, and 0.2 M NaCl, and once with 50 mM Tris/HCl, pH 7.5, and 5 mM MgCl2. The immunoprecipitates were resuspended in 20 μL of 50 mM Tris/HCl, pH 7.5, 5 mM MgCl2, 2 mM DTT, 0.1 μM UBE1, 0.4 μM Ubc13-Uev1a, 10 μM FLAG-ubiquitin (Pellino1), 5 mM MgCl2, and 2 mM ATP. After incubation for 1 h at 30 °C, reactions were stopped by denaturation in SDS and Ub-chain formation detected by immunoblotting with anti-FLAG (37).
Human TRAF6 (5 nM), TRAF6[L74H] (50 nM), or TRAF6[C70A] (50 nM) purified from insect Sf21 cells were incubated for 1 h at 30 °C with 20 μL of 50 mM Tris/HCl, pH 7.5, 5 mM MgCl2, 0.1 μM UBE1, 0.1 μM Ubc13-Uev1a or 0.1 μM UbcH5a or 0.1 μM UbcH9, 5 μM FLAG-ubiquitin, and 2 mM ATP. Reactions were stopped with SDS, subjected to SDS/PAGE, and immunoblotted with anti-FLAG.
Quantitative RT-PCR of IL-8, Pellino1, and Pellino2 mRNA.
RNA was extracted using RNA MicroElute kit from VWR (R6831-01) and reverse transcribed using the iScript cDNA synthesis kit (Bio-Rad; 170-8891). PCR was performed with SsoFast EvaGreen Supermix (Bio-Rad; 172-5204), and the results were normalized to 18S RNA as described (11). Primer sequences were as follows: IL-8, forward, 5′-ATAAAGACATACTCCAAACCTTTCCAC-3′, and IL-8 reverse, 5′-AAGCTTTACAATAATTTCTGTGTTGGC-3′; Pellino1, forward, 5′-CCTATGTCCCTCTGTGGCTTGG-3′, and Pellino1, reverse, 5′-GTGTGCGTACCATGAGGAAGTG-3′; Pellino2, forward, 5′-CGCGCGCGGATTTGACTCTT-3′, and Pellino2, reverse, 5′-CTGGGTGAAGCCCCCTCGTG-3′; 18S, forward, 5′-GTAACCCGTTGAACCCCATT-3′, and 18S, reverse, 5′-CCATCCAATCGGTAGTAGCG-3′.
Cytokine Secretion.
IL-1R* cells were seeded into 24-well plates, and primary macrophages were seeded into 48-well plates. After stimulation, the cell culture medium was collected. Human IL-8 secretion was measured using a kit from Peprotech (900-K18) and mouse IL-6, IL-10, IL-12(p40), and TNFα using the Bio-Plex Pro Assay system (Bio-Rad).
In-Solution Tryptic Digest and Mass-Spectrometric Analysis.
To identify proteins that interacted with TRAF6 in an IL-1–dependent manner, 15 mg of cell extract protein obtained from IL-1β–stimulated (5 min) or unstimulated TRAF6 KO IL-1R* cells, stably reexpressing Flag-TRAF6, were incubated for 3 h at 4 °C with 40 μL of immobilized anti-FLAG M2 affinity gel (Sigma). The resin was washed 5 times with buffer containing 50 mM Tris⋅HCl (pH 8.0), 0.5 M NaCl, and 0.5% Triton, and 10 times with 50 mM Tris⋅HCl (pH 8.0). Bound proteins were eluted from beads by incubation for 5 min at 60 °C with 1% Rapigest, 1 mM TCEP in 50 mM Tris⋅HCl (pH 8.0). Samples were then alkylated for 20 min at 20 °C in the dark with 10 mM iodoacetamide and then quenched with addition of DTT (10 mM). Samples were diluted with 50 mM Tris⋅HCl (pH 8.0) so that the Rapigest concentration was 0.1% (wt/vol). Trypsin (0.5 μg per sample) was then added, and the samples were incubated for 16 h at 30 °C. The digests were acidified by the addition of trifluoroacetic acid (TFA) to 2% (vol/vol) and incubated for 1 h at 37 °C to cleave the Rapigest. The samples were then centrifuged for 30 min at 21,000 × g and then desalted and cleaned up using microspin columns (The Nest Group #SEM SS18V) in 0.1% (vol/vol) TFA before being taken to dryness. The dried peptides were resuspended in 20 μL of 0.1% (vol/vol) TFA and separated on a Proxeon EASYn-LC system (Thermo Scientific) using a 15-cm-long C18 column. Mass spectra were acquired on an LTQ Orbitrap Velos mass spectrometer (Thermo Scientific) operating in data-dependent mode. After conversion to mzXML, the raw data were searched using X!Tandem with the K-score plug-in against version 3.87 of the International Protein Index human protein database using static carboxamido-methylation of cysteine residues and accounting for tryptic peptides with up to two missed cleavages. The Trans-Proteomic Pipeline was used to assign peptide and protein probabilities and to filter results at a 1% false-discovery rate. Peptide mass fingerprinting data analysis was performed using Scaffold (www.proteomesoftware.com/products/scaffold/).
Acknowledgments
We thank Tak Mak for immortalized TRAF6 KO MEFs and TRAF6 KO mice; Hao Wu for advice about TRAF6 mutagenesis; Rachel Toth for the vector expressing TAB1, TAK1, and TAB3 as a complex in insect cells; and Yogesh Kulathu for providing a protocol for the purification of this complex. We gratefully acknowledge the Proteomics and Mass Spectrometry Team based within the MRC-PPU and Lynn Stevenson and Lynn Oxford (Veterinary Diagnostic Services, University of Glasgow School of Veterinary Medicine) for their excellent technical assistance with tissue processing for histology. The research was supported by Wellcome Trust Grant WT100294 and by a core grant to the MRC-PPU from Boehringer Ingelheim, GlaxoSmithKline, and Merck-Serono. We also thank the UK Medical Research Council for PhD studentships (to S.S. and J.Z.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1702367114/-/DCSupplemental.
References
- 1.Wu H, Arron JR. TRAF6, a molecular bridge spanning adaptive immunity, innate immunity and osteoimmunology. BioEssays. 2003;25:1096–1105. doi: 10.1002/bies.10352. [DOI] [PubMed] [Google Scholar]
- 2.Naito A, et al. Severe osteopetrosis, defective interleukin-1 signalling and lymph node organogenesis in TRAF6-deficient mice. Genes Cells. 1999;4:353–362. doi: 10.1046/j.1365-2443.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- 3.Naito A, et al. TRAF6-deficient mice display hypohidrotic ectodermal dysplasia. Proc Natl Acad Sci USA. 2002;99:8766–8771. doi: 10.1073/pnas.132636999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lomaga MA, et al. TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev. 1999;13:1015–1024. doi: 10.1101/gad.13.8.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kobayashi T, et al. TRAF6 is a critical factor for dendritic cell maturation and development. Immunity. 2003;19:353–363. doi: 10.1016/s1074-7613(03)00230-9. [DOI] [PubMed] [Google Scholar]
- 6.King CG, et al. Cutting edge: Requirement for TRAF6 in the induction of T cell anergy. J Immunol. 2008;180:34–38. doi: 10.4049/jimmunol.180.1.34. [DOI] [PubMed] [Google Scholar]
- 7.King CG, et al. TRAF6 is a T cell-intrinsic negative regulator required for the maintenance of immune homeostasis. Nat Med. 2006;12:1088–1092. doi: 10.1038/nm1449. [DOI] [PubMed] [Google Scholar]
- 8.Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Motshwene PG, et al. An oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4. J Biol Chem. 2009;284:25404–25411. doi: 10.1074/jbc.M109.022392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ye H, et al. Distinct molecular mechanism for initiating TRAF6 signalling. Nature. 2002;418:443–447. doi: 10.1038/nature00888. [DOI] [PubMed] [Google Scholar]
- 11.Pauls E, et al. Two phases of inflammatory mediator production defined by the study of IRAK2 and IRAK1 knock-in mice. J Immunol. 2013;191:2717–2730. doi: 10.4049/jimmunol.1203268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin Q, et al. E2 interaction and dimerization in the crystal structure of TRAF6. Nat Struct Mol Biol. 2009;16:658–666. doi: 10.1038/nsmb.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng L, et al. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 14.Wang C, et al. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 15.Xia ZP, et al. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–119. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobayashi N, et al. Segregation of TRAF6-mediated signaling pathways clarifies its role in osteoclastogenesis. EMBO J. 2001;20:1271–1280. doi: 10.1093/emboj/20.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamothe B, et al. The RING domain and first zinc finger of TRAF6 coordinate signaling by interleukin-1, lipopolysaccharide, and RANKL. J Biol Chem. 2008;283:24871–24880. doi: 10.1074/jbc.M802749200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walsh MC, Kim GK, Maurizio PL, Molnar EE, Choi Y. TRAF6 autoubiquitination-independent activation of the NFkappaB and MAPK pathways in response to IL-1 and RANKL. PLoS One. 2008;3:e4064. doi: 10.1371/journal.pone.0004064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shim JH, et al. TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo. Genes Dev. 2005;19:2668–2681. doi: 10.1101/gad.1360605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sato S, et al. Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat Immunol. 2005;6:1087–1095. doi: 10.1038/ni1255. [DOI] [PubMed] [Google Scholar]
- 21.Cheung PC, Nebreda AR, Cohen P. TAB3, a new binding partner of the protein kinase TAK1. Biochem J. 2004;378:27–34. doi: 10.1042/BJ20031794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanayama A, et al. TAB2 and TAB3 activate the NF-kappaB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15:535–548. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 23.Kulathu Y, Akutsu M, Bremm A, Hofmann K, Komander D. Two-sided ubiquitin binding explains specificity of the TAB2 NZF domain. Nat Struct Mol Biol. 2009;16:1328–1330. doi: 10.1038/nsmb.1731. [DOI] [PubMed] [Google Scholar]
- 24.Lopez-Pelaez M, et al. Protein kinase IKKβ-catalyzed phosphorylation of IRF5 at Ser462 induces its dimerization and nuclear translocation in myeloid cells. Proc Natl Acad Sci USA. 2014;111:17432–17437. doi: 10.1073/pnas.1418399111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ben-Addi A, et al. IκB kinase-induced interaction of TPL-2 kinase with 14-3-3 is essential for Toll-like receptor activation of ERK-1 and -2 MAP kinases. Proc Natl Acad Sci USA. 2014;111:E2394–E2403. doi: 10.1073/pnas.1320440111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takaoka A, et al. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature. 2005;434:243–249. doi: 10.1038/nature03308. [DOI] [PubMed] [Google Scholar]
- 27.Waterfield MR, Zhang M, Norman LP, Sun SC. NF-kappaB1/p105 regulates lipopolysaccharide-stimulated MAP kinase signaling by governing the stability and function of the Tpl2 kinase. Mol Cell. 2003;11:685–694. doi: 10.1016/s1097-2765(03)00070-4. [DOI] [PubMed] [Google Scholar]
- 28.Beinke S, Robinson MJ, Hugunin M, Ley SC. Lipopolysaccharide activation of the TPL-2/MEK/extracellular signal-regulated kinase mitogen-activated protein kinase cascade is regulated by IkappaB kinase-induced proteolysis of NF-kappaB1 p105. Mol Cell Biol. 2004;24:9658–9667. doi: 10.1128/MCB.24.21.9658-9667.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ananieva O, et al. The kinases MSK1 and MSK2 act as negative regulators of Toll-like receptor signaling. Nat Immunol. 2008;9:1028–1036. doi: 10.1038/ni.1644. [DOI] [PubMed] [Google Scholar]
- 30.Emmerich CH, et al. Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains. Proc Natl Acad Sci USA. 2013;110:15247–15252. doi: 10.1073/pnas.1314715110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emmerich CH, Cohen P. Optimising methods for the preservation, capture and identification of ubiquitin chains and ubiquitylated proteins by immunoblotting. Biochem Biophys Res Commun. 2015;466:1–14. doi: 10.1016/j.bbrc.2015.08.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schauvliege R, Janssens S, Beyaert R. Pellino proteins are more than scaffold proteins in TLR/IL-1R signalling: A role as novel RING E3-ubiquitin-ligases. FEBS Lett. 2006;580:4697–4702. doi: 10.1016/j.febslet.2006.07.046. [DOI] [PubMed] [Google Scholar]
- 33.Grosshans J, Schnorrer F, Nüsslein-Volhard C. Oligomerisation of Tube and Pelle leads to nuclear localisation of dorsal. Mech Dev. 1999;81:127–138. doi: 10.1016/s0925-4773(98)00236-6. [DOI] [PubMed] [Google Scholar]
- 34.Huoh YS, Ferguson KM. The Pellino E3 ubiquitin ligases recognize specific phosphothreonine motifs and have distinct substrate specificities. Biochemistry. 2014;53:4946–4955. doi: 10.1021/bi5005156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ordureau A, et al. The IRAK-catalysed activation of the E3 ligase function of Pellino isoforms induces the Lys63-linked polyubiquitination of IRAK1. Biochem J. 2008;409:43–52. doi: 10.1042/BJ20071365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith H, et al. Identification of the phosphorylation sites on the E3 ubiquitin ligase Pellino that are critical for activation by IRAK1 and IRAK4. Proc Natl Acad Sci USA. 2009;106:4584–4590. doi: 10.1073/pnas.0900774106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goh ET, et al. Identification of the protein kinases that activate the E3 ubiquitin ligase Pellino 1 in the innate immune system. Biochem J. 2012;441:339–346. doi: 10.1042/BJ20111415. [DOI] [PubMed] [Google Scholar]
- 38.Keusekotten K, et al. OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin. Cell. 2013;153:1312–1326. doi: 10.1016/j.cell.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamothe B, et al. TRAF6 ubiquitin ligase is essential for RANKL signaling and osteoclast differentiation. Biochem Biophys Res Commun. 2007;359:1044–1049. doi: 10.1016/j.bbrc.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson DM, et al. A homologue of the TNF receptor and its ligand enhance T-cell growth and dendritic-cell function. Nature. 1997;390:175–179. doi: 10.1038/36593. [DOI] [PubMed] [Google Scholar]
- 41.Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. 2003;423:337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- 42.Mizukami J, et al. Receptor activator of NF-kappaB ligand (RANKL) activates TAK1 mitogen-activated protein kinase kinase kinase through a signaling complex containing RANK, TAB2, and TRAF6. Mol Cell Biol. 2002;22:992–1000. doi: 10.1128/MCB.22.4.992-1000.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang H, et al. Osteoclast differentiation requires TAK1 and MKK6 for NFATc1 induction and NF-kappaB transactivation by RANKL. Cell Death Differ. 2006;13:1879–1891. doi: 10.1038/sj.cdd.4401882. [DOI] [PubMed] [Google Scholar]
- 44.Kawagoe T, et al. Sequential control of Toll-like receptor-dependent responses by IRAK1 and IRAK2. Nat Immunol. 2008;9:684–691. doi: 10.1038/ni.1606. [DOI] [PubMed] [Google Scholar]
- 45.Zhang J, Clark K, Lawrence T, Peggie MW, Cohen P. An unexpected twist to the activation of IKKβ: TAK1 primes IKKβ for activation by autophosphorylation. Biochem J. 2014;461:531–537. doi: 10.1042/BJ20140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deak M, Clifton AD, Lucocq LM, Alessi DR. Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB. EMBO J. 1998;17:4426–4441. doi: 10.1093/emboj/17.15.4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Munoz IM, Szyniarowski P, Toth R, Rouse J, Lachaud C. Improved genome editing in human cell lines using the CRISPR method. PLoS One. 2014;9:e109752. doi: 10.1371/journal.pone.0109752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X, et al. Mutant cells that do not respond to interleukin-1 (IL-1) reveal a novel role for IL-1 receptor-associated kinase. Mol Cell Biol. 1999;19:4643–4652. doi: 10.1128/mcb.19.7.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee DW, et al. The Dac-tag, an affinity tag based on penicillin-binding protein 5. Anal Biochem. 2012;428:64–72. doi: 10.1016/j.ab.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 50.Windheim M, Peggie M, Cohen P. Two different classes of E2 ubiquitin-conjugating enzymes are required for the mono-ubiquitination of proteins and elongation by polyubiquitin chains with a specific topology. Biochem J. 2008;409:723–729. doi: 10.1042/BJ20071338. [DOI] [PubMed] [Google Scholar]
- 51.Fitzgerald DJ, et al. Protein complex expression by using multigene baculoviral vectors. Nat Methods. 2006;3:1021–1032. doi: 10.1038/nmeth983. [DOI] [PubMed] [Google Scholar]
- 52.Smith H, et al. The role of TBK1 and IKKε in the expression and activation of Pellino 1. Biochem J. 2011;434:537–548. doi: 10.1042/BJ20101421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matsumoto ML, et al. Engineering and structural characterization of a linear polyubiquitin-specific antibody. J Mol Biol. 2012;418:134–144. doi: 10.1016/j.jmb.2011.12.053. [DOI] [PubMed] [Google Scholar]
- 54.Cushing L, et al. Interleukin 1/Toll-like receptor-induced autophosphorylation activates interleukin 1 receptor-associated kinase 4 and controls cytokine induction in a cell type-specific manner. J Biol Chem. 2014;289:10865–10875. doi: 10.1074/jbc.M113.544809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sato Y, Yoshikawa A, Yamashita M, Yamagata A, Fukai S. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by NZF domains of TAB2 and TAB3. EMBO J. 2009;28:3903–3909. doi: 10.1038/emboj.2009.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hjerpe R, et al. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 2009;10:1250–1258. doi: 10.1038/embor.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]