Significance
Phosphate (Pi) deficiency constrains plant development and productivity in both natural and agricultural ecosystems. An interaction among Pi and Fe availability controls the developmental program that allows the Arabidopsis root system to more effectively explore the topsoil where Pi accumulates. Analysis of mutants unable to establish root architecture responses to low Pi allowed the identification of mutant alleles of STOP1 (a transcription factor) and ALMT1 (a malate transporter), two genes previously reported to play a role in the malate-mediated tolerance to toxic levels of aluminum. We show that these genes underlie a malate-exudation–dependent mechanism of Fe relocation in the root apical meristem that is essential for reprogramming root growth under low-Pi conditions.
Keywords: phosphate, root response, iron, cell differentiation, gene regulation
Abstract
Low phosphate (Pi) availability constrains plant development and seed production in both natural and agricultural ecosystems. When Pi is scarce, modifications of root system architecture (RSA) enhance the soil exploration ability of the plant and lead to an increase in Pi uptake. In Arabidopsis, an iron-dependent mechanism reprograms primary root growth in response to low Pi availability. This program is activated upon contact of the root tip with low-Pi media and induces premature cell differentiation and the arrest of mitotic activity in the root apical meristem, resulting in a short-root phenotype. However, the mechanisms that regulate the primary root response to Pi-limiting conditions remain largely unknown. Here we report on the isolation and characterization of two low-Pi insensitive mutants (lpi5 and lpi6), which have a long-root phenotype when grown in low-Pi media. Cellular, genomic, and transcriptomic analysis of low-Pi insensitive mutants revealed that the genes previously shown to underlie Arabidopsis Al tolerance via root malate exudation, known as SENSITIVE TO PROTON RHIZOTOXICITY (STOP1) and ALUMINUM ACTIVATED MALATE TRANSPORTER 1 (ALMT1), represent a critical checkpoint in the root developmental response to Pi starvation in Arabidopsis thaliana. Our results also show that exogenous malate can rescue the long-root phenotype of lpi5 and lpi6. Malate exudation is required for the accumulation of Fe in the apoplast of meristematic cells, triggering the differentiation of meristematic cells in response to Pi deprivation.
Phosphorus is an essential nutrient for plant development, a constituent of key molecules such as nucleic acids, ATP, and membrane phospholipids. Plants take up and metabolize phosphorus in the inorganic form of orthophosphate (Pi) (1). Pi is the least accessible macronutrient in many natural and agricultural ecosystems and its low availability in the soil often limits plant growth and productivity. Under phosphate-limiting conditions (−Pi), plants activate an array of genetic (2–6), biochemical (7, 8), and morphological modifications (9–11) that enhance their ability to cope with low Pi availability.
Arabidopsis responses to low Pi availability have been divided in systemic responses that depend upon the internal Pi status of the plant and local responses that depend upon the level of Pi available in the external medium (5, 12). A molecular dissection of local and systemic responses to Pi starvation using transcriptomic analysis has been reported (5). Systemic responses include the up-regulation of genes involved in the overall enhancement of Pi uptake and Pi internal use efficiency and are largely controlled by the master regulator PHR1 (a Myb transcription factor) (6, 13). Local responses include alterations of root traits such as the inhibition of primary root growth (14), an increase in lateral root density (10), and higher density and length of root hairs (9). These changes in root system architecture (RSA) have been proposed to enhance the soil exploration ability of the plant by increasing root surface area of exploration in the top layers of the soil where Pi tends to accumulate. Some evidence indicates that there is some degree of cross-talk between local and systemic responses to low Pi availability, as mutants altered in RSA responses to low Pi also have an altered expression of genes involved in systemic responses (15).
Growth of Arabidopsis seedlings under in vitro Pi-limiting conditions induces a determinate developmental program known as root apical meristem (RAM) exhaustion (11). RAM exhaustion consists of the loss of meristematic potential and the arrest of cell proliferation, leading to the inhibition of primary root growth. Meristematic potential is lost due to premature differentiation of the cells that constitute the stem cell niche (SCN), which includes the quiescent center (QC). Cell proliferation in the RAM is lost due to a gradual reduction in mitotic activity. Root-tip contact with low phosphate media (16) in the presence of iron (17) has been reported as essential for the inhibition of primary root growth in response to Pi-deficiency conditions in Arabidopsis thaliana. Arabidopsis mutants that fail to trigger root system morphological responses to low Pi have been reported previously (15, 16). Two of these mutants, low phosphate root 1 and 2 (lpr1 and lpr2) were found to be affected in genes encoding multicopper oxidases, suggesting that a metal with different levels of oxidation could be involved in the alteration of root system architecture in response to low Pi availability (16). A mutant that is hypersensitive to low Pi, phosphate deficiency response 2 (pdr2), and triggers the root system response to low Pi availability faster that the wild type (WT) was also reported (18). PDR2 encodes an endoplasmic reticulum (ER)-localized P5-type ATPase (19). PDR2 and LPR1 have been proposed to orchestrate RAM exhaustion in a genetically interacting route under low-Pi conditions (19). Whereas the precise function of PDR2 has not been determined, LPR1 is essential for primary root inhibition under low-Pi conditions and it has been shown to have ferroxidase activity (16, 19, 20). An LPR1-dependent accumulation of Fe+3 in the apoplast of cells in the elongation and meristematic regions of the primary root, that triggers the production of reactive oxygen species (ROS), is essential for meristem exhaustion in low-Pi media (20). ROS generation correlates with callose deposition in the RAM, which was proposed to activate meristem exhaustion by blocking the cell-to-cell movement of SHORT-ROOT (SHR), a transcription factor that is essential for stem cell maintenance in the RAM (20). However, the mechanism that regulates iron accumulation and relocation in the RAM remains largely unknown.
In this work, we characterized two low phosphate insensitive mutants (lpi5 and lpi6), which, in contrast to the short-root phenotype of WT Arabidopsis seedlings, show normal primary root elongation in low-Pi media. Mapping by sequencing revealed that the lpi5 corresponds to SENSITIVE TO PROTON RHIZOTOXICITY (STOP1) and lpi6 to ALUMINUM ACTIVATED MALATE TRANSPORTER 1 (ALMT1), the two genes previously reported to be responsible for activating malate efflux in the roots of Arabidopsis seedlings exposed to toxic concentrations of aluminum (21–23). Genetic, cellular, and transcriptomic analyses show that STOP1 and ALMT1 are required for a malate-dependent accumulation of iron in the root meristem, which leads to alterations in the redox balance that trigger primary root growth inhibition and RAM exhaustion in response to Pi-deficiency conditions in A. thaliana.
Results
Arabidopsis EMS Mutants lpi5 and lpi6 Show Indeterminate Primary Root Growth Under Phosphate-Deficiency Conditions.
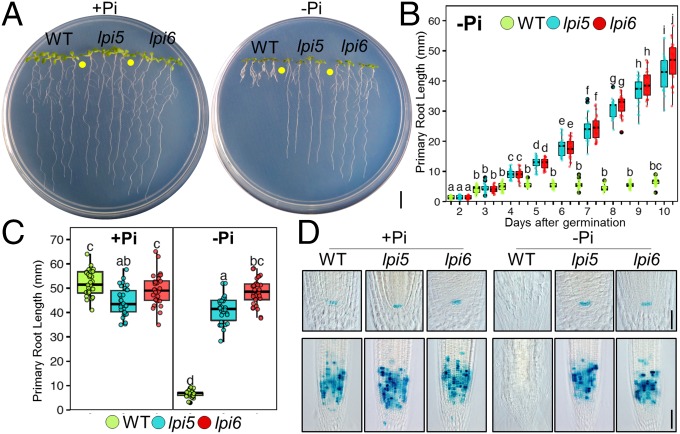
The A. thaliana Col-0 accession seedlings grown in media containing low-Pi concentrations (below 50 µM Pi) show a short-root phenotype defined by a determinate pattern of primary root growth and RAM differentiation. To identify mutants that are insensitive to the effect of low Pi on primary root growth, we screened an EMS-mutagenized Col-0 population for mutant lines presenting a long-root phenotype under low-Pi (−Pi, 5 µM Pi) conditions. We isolated ∼50 mutant lines with different alterations in the primary root growth in response to low Pi availability. Two lines that were insensitive to the effect of low Pi on primary root growth, which we named low phosphorus insensitive 5 and low phosphorus insensitive 6 (lpi5 and lpi6), were chosen for further characterization (Fig. 1 A–C). When grown under high Pi (+Pi; 1,000 µM Pi) conditions, both lpi5 and lpi6 seedlings presented a primary root length similar to the WT Arabidopsis Col-0 accession (Fig. 1 A and C). Under −Pi conditions at 10 d after germination (dag), WT plants had a visible reduction in primary root growth, whereas lpi5 and lpi6 seedlings had primary root elongation similar to that of WT seedlings grown in +Pi media, which is fourfold longer than that of Pi-deprived WT seedlings (Fig. 1 A and C). Although lpi5 seedlings had a long-root phenotype in −Pi media, they had a slightly shorter primary root than the WT and lpi6 in +Pi media and were also slightly shorter than lpi6 in −Pi medium (Fig. 1 A and C). Analysis of segregation frequencies under −Pi conditions showed that the long-root phenotype in lpi5 and lpi6 is the result of single recessive mutations (SI Appendix, Table S1).
Fig. 1.
Low-Pi insensitive mutants continue primary root growth and RAM maintenance under Pi-deficiency conditions. (A) Phenotypes of WT, lpi5, and lpi6 seedlings grown under high (1,000 μM Pi; +Pi)- and low-Pi (5 μM Pi; −Pi) conditions 10 dag. (Scale bar, 1 cm.) (B) Primary root growth kinetics of seedlings grown under −Pi conditions from 2 to 10 dag. Green, blue, and red dots depict WT, lpi5, and lpi6 individuals (n = 30 from three independent experiments), respectively. Statistical groups were determined using a Tukey honest significant difference (HSD) test (P value <0.05) and are indicated with letters. (C) Primary root length of seedlings grown under +Pi and −Pi conditions. Green, blue, and red dots depict WT, lpi5, and lpi6 individuals (n = 30 from three independent experiments), respectively. Statistical groups were determined using a Tukey HSD test (P value <0.05) and are indicated with letters. (D) GUS staining of proCycB1::GUS and proQC46::GUS expression in the RAM of WT, lpi5, and lpi6 seedlings 10 dag. (Scale bar, 100 µm.)
In Pi-deprived Arabidopsis seedlings, the short-root phenotype is accompanied by a reduction in cell proliferation and meristematic activity during the process of RAM exhaustion (11). To test for signs of RAM exhaustion in lpi5 and lpi6 seedlings grown in −Pi media, we examined the expression of the proCycB1::GUS cell cycle activity marker (24) and the proQC46::GUS QC identity marker (25). In +Pi media, WT, lpi5, and lpi6 seedlings showed clear cell proliferation activity as indicated by the proCycB1::GUS signal and an active QC as shown by the proQC46::GUS signal (Fig. 1D). In −Pi media at 10 dag, the cell cycle and QC marker genes were undetectable in WT seedlings, whereas GUS staining was clearly detectable for both the cell cycle (proCycB1::GUS) and QC (proQC46::GUS) markers in the primary root of lpi5 and lpi6 (Fig. 1D). These results show that, in contrast to the WT, low Pi does not trigger meristem exhaustion in the RAM of lpi5 and lpi6 seedlings.
lpi5 and lpi6 Have Mutations in the Transcription Factor STOP1 and the Malate Transporter ALMT1, Respectively.
We used a mapping-by-sequencing approach to identify the genes responsible for the lpi5 and lpi6 phenotypes (Materials and Methods). We identified 12 and 18 specific homozygous variants (missense, frameshift, and splice donor) potentially linked to lpi5 and lpi6 mutant phenotypes, respectively. Given the alterations of primary root of lpi5 and lpi6 seedlings in response to an environmental stress, we focused on the homozygous mutations that could potentially be linked to alterations in root morphology or root responses to abiotic stress. Among the potential candidates responsible for the root phenotype of lpi5 and lpi6 under low-Pi conditions, SENSITIVE TO PROTON RHIZOTOXICITY (STOP1) and ALUMINUM ACTIVATED MALATE TRANSPORTER 1 (ALMT1) were particularly interesting because both genes have been previously reported to participate in the tolerance of the Arabidopsis root to toxic concentrations of Al+3. STOP1 (At1g34370) encodes a zinc finger protein transcription factor that plays a critical role in Al+3 tolerance and acid soil tolerance in Arabidopsis (21). ALMT1 encodes a transmembrane protein that mediates malate efflux in the root in response to the presence of toxic Al+3 levels (22), and its expression has been shown to be activated by STOP1 in response to Al stress conditions (21).
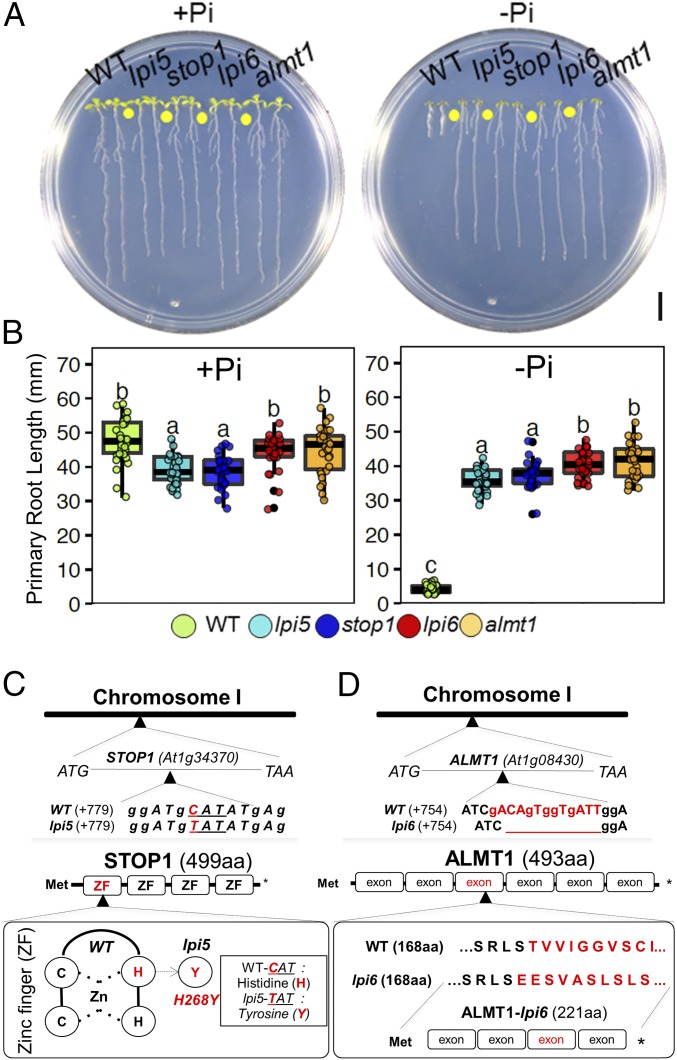
To test whether the long-root phenotype of lpi5 and lpi6 was indeed due to lesions in STOP1 and ALMT1, we tested the phenotype of T-DNA insertional mutants in STOP1 (SALK_114108) and ALMT1 (SALK_009629) in −Pi and +Pi media. In +Pi media, WT, lpi6, and almt1 seedlings had a similar primary root length, whereas lpi5 and stop1 had a slightly shorter root length than the WT (Fig. 2 A and B). In −Pi media, the T-DNA insertional mutants stop1 and almt1 had a long-root phenotype that contrasted with the short-root phenotype of the WT (Fig. 2 A and B). It has been reported that stop1 is sensitive to low pH (4.7) and levels of Al+3 (2 µM) that are not yet toxic for Arabidopsis WT seedlings and that almt1 is also sensitive to this Al+3 concentration (26). We also found that lpi5 was sensitive to low pH and Al (as is stop1) and that lpi6 was sensitive to Al but not to low pH (as observed for almt1) (SI Appendix, Fig. S1). Crosses between lpi5 and almt1 and between lpi6 and stop1 showed that lpi5 and lpi6 are mutant alleles of STOP1 and ALMT1, respectively (SI Appendix, Fig. S1).
Fig. 2.
Mapping by sequencing revealed lpi5 and lpi6 to be stop1 and almt1 mutants, respectively. (A) Phenotypes of Col-0, lpi5, stop1, almt1, and lpi6 seedlings grown under high-phosphate (+Pi) and low-phosphate (−Pi) conditions 10 dag. (Scale bar, 1 cm.) (B) Primary root length of WT, lpi5, stop1, almt1, and lpi6 seedlings grown under high-phosphate (+Pi) or low-phosphate (−Pi) conditions 10 dag. Dots depict WT, lpi5, stop1, almt1, and lpi6 individuals (n = 30 from three independent experiments). Statistical groups were determined using a Tukey HSD test (P value <0.05) and are indicated with letters. (C) C:T (CAT: TAT) substitution in the +84 base within the At1g34370 locus of the lpi5 genome. lpi5 mutation causes an amino acid substitution (H168Y) that replaces one of the two histidine residues of the four zinc fingers that constitute STOP1. (D) A 13-bp deletion in the +757 base pair position within the first exon of the At1g08430 locus of the lpi6 genome. The 13-bp deletion present in lpi6 causes a frameshift mutation that produces an aberrant protein with 200 amino acids less than the WT. Met, methionine.
Mapping by sequencing of lpi5 revealed a C:T (CAT:TAT) substitution in the +84 position of STOP1. In silico analysis predicted that the lpi5 mutation caused an amino acid substitution (H168Y) that replaces one of the two histidine residues that are crucial for the formation of the first of four zinc fingers of the DNA binding domain of STOP1 (Fig. 2C). The first zinc finger domain of STOP1 is critical to bind to the promoter of its target genes (27). Mapping by sequencing of the lpi6 mutant revealed a 13-base pair deletion in the first exon of ALMT1 starting at position +757 (Fig. 2D). In silico analysis predicted that this deletion causes a frameshift mutation that produces an aberrant protein that lacks 221 amino acids of the carboxyl-terminal moiety of ALMT1 (Fig. 2D). Our in silico sequence analysis further confirmed that lpi5 and lpi6 are EMS-induced mutant alleles of STOP1 and ALMT1, respectively.
ALMT1 Is Expressed in the RAM of Arabidopsis Under Pi-Deficiency Conditions Before Meristematic Exhaustion.
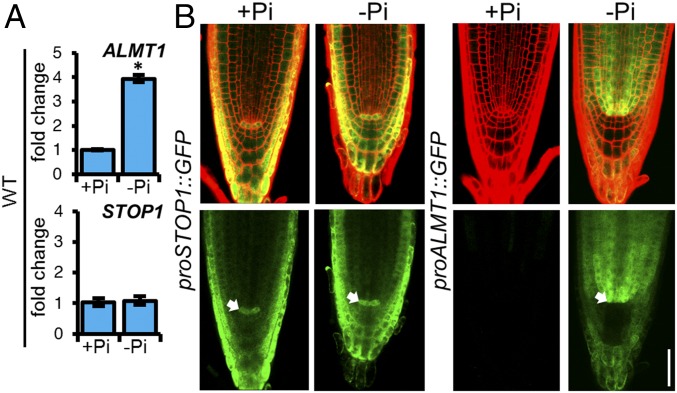
To determine whether the expression of STOP1 and ALMT1 is regulated by Pi availability in the root tip, we analyzed the expression of STOP1 and ALMT1 in the root apex of 5 dag WT and stop1 seedlings using qRT-PCR (Fig. 3A and SI Appendix, Fig. S3). In the WT, ALMT1 expression increased by approximately fourfold in response to −Pi conditions, whereas the level of expression of STOP1 was not significantly altered by Pi availability (Fig. 3A). We also found that in the stop1 background, the expression of ALMT1 was undetectable (SI Appendix, Fig. S2). Our results confirm a previous report (21) showing that STOP1 is essential for the expression of ALMT1, but also show that STOP1 is required for the induction of ALMT1 in response to low Pi.
Fig. 3.
STOP1 and ALMT1 are expressed in the RAM of Arabidopsis under Pi-deficiency conditions. (A) qRT-PCR analysis of STOP1 and ALMT1 expression in the root apex (2–3 mm) of WT (Col-0) plants. Bars represent the mean fold change ± SEM of two biological replicates with three technical replicates. WT +Pi samples were used as calibrator values. ACT2 and UBQ10 were used as internal controls. Asterisk indicates that the expression was significantly different between +Pi and −Pi conditions (Student t test; *P < 0.05). (B) Transgenic Col-0 plants harboring transcriptional gene fusions containing the STOP1 and ALMT1 promoter fused to a double GFP–GUS reporter gene, respectively (proSTOP1::GUS::GFP and proALMT1::GUS::GFP), were grown under +Pi and −Pi conditions and expression activity was observed at 5 dag using confocal microscopy. (Upper) Merged propidium iodine staining fluorescence (red) and GFP fluorescence (green). (Lower) GFP fluorescence. Arrow points to the stem cell niche region. (Scale bar, 100 µm.)
Because STOP1 and ALMT1 appear to be essential for RAM exhaustion under Pi-deficiency conditions, we examined the cell-specific expression pattern of these two genes in seedlings grown under +Pi and −Pi conditions 5 dag, a time point before full RAM exhaustion (Fig. 1C), but when primary root growth inhibition has already started (Fig. 1C). Confocal microscopy of proSTOP1::GUS::GFP seedlings grown in +Pi media revealed that STOP1 is expressed in the QC, columella, lateral root cap, and epidermis (Fig. 3B) and that its expression pattern is not altered in −Pi media (Fig. 3B). On the other hand, no detectable reporter activity of proALMT1::GUS::GFP was found in the RAM of seedlings grown under +Pi conditions, whereas in −Pi media expression of the reporter gene was clearly detectable in the proximal region of the SCN (QC, cortex, and stele initials), vascular bundle, pericycle, cortex, endodermis, columella, and lateral root cap (Fig. 3B).
Malate Treatment Rescues the Determinate Developmental Program in the Primary Root of stop1 and almt1 in Response to Low-Pi Conditions.
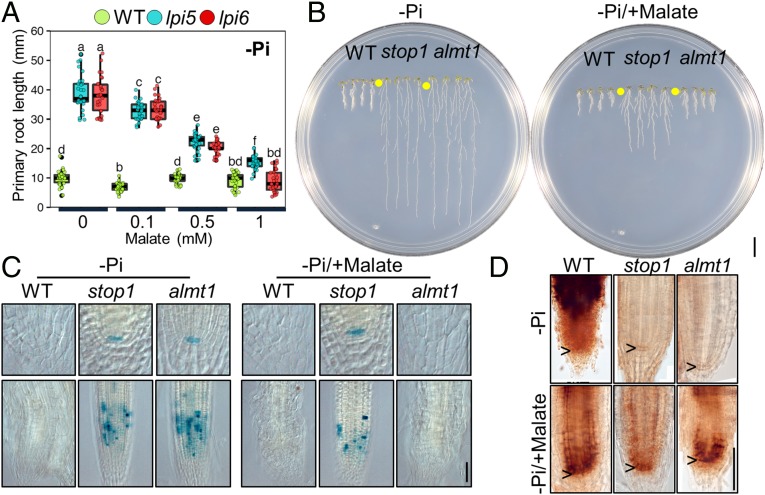
Because malate efflux has been shown to be affected in stop1 and almt1 mutants and both stop1 and almt1 present determinate primary root growth under Al+3 toxicity conditions (21, 22), we sought to determine whether malate exudation also played a role in the Arabidopsis primary root response to low Pi availability. To this end, we added increasing concentrations of malate to both +Pi and −Pi media and tested the effect of malate treatment on the primary root growth of WT, stop1, and almt1 seedlings (Fig. 4 and SI Appendix, Fig. S3). We observed that primary root growth was not altered by malate treatment under +Pi conditions in any of the tested lines (SI Appendix, Fig. S3). However, in −Pi media, treatment with malate restored the short-root phenotype in stop1 and almt1 seedlings in a concentration-dependent manner (Fig. 4 A and B). Although stop1 seedlings treated with 1 mM malate had significantly shorter roots than in media lacking malate, their primary roots were slightly, but statistically significantly, larger than those of the WT and almt1 seedlings grown in the same media (Fig. 4 A and B). Malate treatment of Pi-deprived WT seedlings showed a small effect at 0.1 mM; however, this effect was not observed at higher malate concentrations (Fig. 4 A and B) as WT seedlings remained short under all treatments (Fig. 4A).
Fig. 4.
Malate treatment rescues the mutant phenotype of stop1 and almt1 seedlings. (A) Primary root length of 10 dag WT, stop1, and almt1 seedlings grown under increasing concentration of malate supplemented into −Pi medium. Green, blue, and red dots depict WT, stop1, and almt1 individuals (n = 30 from three independent experiments), respectively. Statistical groups were determined using Tukey HSD test (P value <0.05) and are indicated with letters. (B) Phenotypes of 10 dag WT, stop1, and almt1 seedlings grown under low-phosphate medium (−Pi) and low-phosphate medium supplemented with 1 mM malate (−Pi/+malate). (Scale bar, 1 mm.) (C) proCycB1::GUS (Lower) and proQC46::GUS (Upper) expression. (D) Perls-DAB iron staining in the RAM of WT, stop1, and almt1 seedlings grown under −Pi and malate treatment (1 mM; −Pi/+malate) conditions 5 dag. Pictures show photographic reconstructions in which several images were spliced together to show the complete primary root meristem (Materials and Methods). (Scale bar, 50 µm.)
To determine whether malate treatment activates RAM exhaustion in stop1 and almt1 seedlings, we examined the expression of proCycB1::GUS and proQC46::GUS reporter genes in Pi-deprived/malate-treated stop1 and almt1 seedlings. Clear signs of cell differentiation in the RAM of Pi-deprived/malate-treated almt1 seedlings were observed and proCycB1::GUS and proQC46::GUS reporter activity was undetectable. In the case of Pi-deprived/malate-treated stop1 seedlings, although cell proliferation was reduced, it was not completely arrested and proQC46::GUS expression was still clearly detectable (Fig. 4C). No expression was found in the WT either in low-Pi media or low-Pi media supplemented with 1 mM malate (Fig. 4C).
Fe Accumulation Is Absent in the RAM of Pi-Deprived stop1 and almt1 Seedlings and Can Be Rescued by Malate Treatment.
Accumulation of Fe in the apoplast of cells in the RAM is required to activate the primary root response to low Pi availability (20). As carboxylate–iron complexes have been reported to participate in iron transport and acquisition in plants (28–30) and malate efflux is affected in stop1 and almt1 mutants (21, 22), we explored whether malate exudation plays a role in the Fe accumulation mechanism that is required to trigger primary root growth inhibition in response to low Pi availability. First, we tested whether malate is required for the accumulation of Fe in the apoplast of RAM cells using Perls-diaminobenzidine (DAB) histochemical Fe staining, which allows the detection of changes in labile Fe+3 (20), on the root tips of WT, stop1, and almt1 in low-Pi media with or without 1 mM malate (Fig. 4D and SI Appendix, Fig. S3). In Pi-deprived seedlings not exposed to malate, Fe staining was clearly observed in the roots of WT seedlings, whereas stop1 and almt1 seedlings showed a much lower Fe accumulation (Fig. 4D). In Pi-deprived WT seedlings treated with 1 mM malate, Fe staining was still clearly visible but in a more defined zone of the root apex, which included the QC. In contrast to stop1 and almt1 seedlings grown in −Pi media lacking malate, those treated with 1 mM of this organic acid showed a very similar pattern to that observed for the WT under the same conditions. Although malate-treated Pi-deprived stop1 seedlings show a clear Fe staining, accumulation of Fe+3 in the RAM was apparently lower around the QC than that observed for the WT and almt1 seedlings treated with malate (Fig. 4C). We did not observe significant differences in the patterns of Fe staining between the root tips of WT, stop1, and almt1 seedlings under +Pi conditions treated with 1 mM malate (SI Appendix, Fig. S3).
Citrate, as well as malate, is an organic acid that is released by plant roots in response to low Pi availability (31). Because organic acids are naturally occurring metal chelating agents, and if the malate chelating effect is responsible for the primary root growth inhibition in −Pi media, we wanted to test whether citrate treatment of Pi-deprived seedlings (1 mM citrate) could also phenocopy the short-root phenotype in Pi-deprived stop1 and almt1 seedlings. In −Pi media, we observed that citrate treatment slightly reduced primary root elongation of stop1 seedlings (10%) (SI Appendix, Fig. S4 A and B) and had no significant effect in the root growth of Pi-deprived almt1 seedlings (SI Appendix, Fig. S4 A and B). Interestingly, citrate treatment of Pi-deprived WT seedlings resulted in an average 2.5-fold increase in root length compared with that observed for WT seedlings grown in low-Pi media in the absence of citrate (SI Appendix, Fig. S4 A and B). These results suggest a specific role of malate in primary root growth inhibition by promoting the accumulation of Fe in the apoplast of root cells in the meristematic area.
As malate is capable of inducing root growth inhibition and meristem exhaustion in almt1 seedlings, linked to an effect on Fe accumulation in the RAM of almt1 seedlings, we hypothesized that malate has a chelating effect on Fe that contributes to its accumulation in the root tip. To test our hypothesis, we performed molecular dynamic calculations to simulate the effect of malate on the aggregation of metallic ions such as Fe+2, Fe+3, and Al+3. We built four different simulation sets using ascending malate:metal molecular ratios: 0:120–120:120 (SI Appendix, Fig. S5). We observed nonbonded interactions between malate and Fe+2 ions but the interactions did not induce Fe+2 aggregation in any of the simulated systems (SI Appendix, Fig. S5). In the case of the malate and Fe+3 system we observed nonbonded interactions and the formation of large malate–Fe+3 aggregates in all ratios tested with an increasing size of aggregates when an equimolar concentration of malate and Fe+3 was used (SI Appendix, Fig. S5). Metals did not aggregate when malate was not included in the simulation set (SI Appendix, Fig. S5). These results suggest that malate can form large aggregates with Fe+3 and Al+3 but not with Fe+2, which could be relevant for the activation of the Arabidopsis primary root response to low Pi.
Differential Expression Analysis Revealed a Preferential Loss of Local Transcriptional Responses to Pi Starvation in stop1 and almt1 Root Tips.
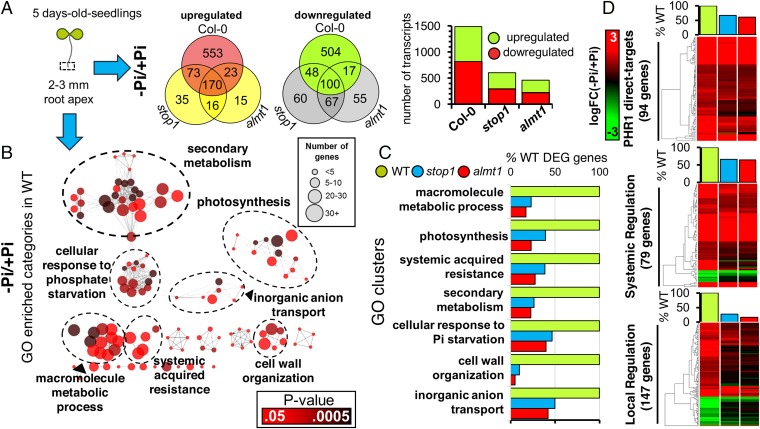
The root tip plays a fundamental role in the ability of the root system to sense and respond to Pi starvation (16). Therefore, to understand the role of STOP1 and ALMT1 in the local and systemic responses of the Arabidopsis root to low phosphate, we performed a whole transcriptome sequencing (RNA-seq) analysis of gene expression in root tips from WT, stop1, and almt1 seedlings grown under +Pi and −Pi conditions (Fig. 5). We performed pairwise comparisons of transcript abundances between −Pi and +Pi conditions to determine differentially expressed genes [−1.5 > logFC > 1.5; false discovery rate (FDR) < 0.05] in response to Pi deficiency in WT, stop1, and almt1 root tips (Fig. 5A). A total of 1,488 genes were found to be responsive to low phosphate in the WT (819 up; 669 down), whereas only 569 in stop1 (294 up; 275 down) and 463 in almt1 (224 up; 239 down) (Fig. 5A). To identify the biological processes whose expression is misregulated in the root apex of stop1 and almt1 in response to Pi availability, we performed a gene ontology (GO) enrichment analysis (Fig. 5). First, we performed a GO clusterization of all of the overrepresented categories that included genes that belong to the same biological process in the root tips of WT seedlings (Fig. 5B). We found seven clusters that were named after the most significantly overrepresented category of the cluster and included the cellular response to phosphate starvation (GO:0016036), secondary metabolism (GO:0019748), macromolecule metabolic process (GO:0044260), cell wall organization (GO:0071555), and systemic acquired resistance (GO:0009627). A full list of the GO categories that were enriched in the WT is included in Dataset S1. We then performed an analysis of the percentage of genes belonging to each cluster that were differentially expressed in stop1 and almt1 relative to the WT (Fig. 5C). Using such an approach, we determined that, overall, the percentage of genes that were activated in response to low phosphate in the root tip of the WT and misregulated in stop1 and almt1 ranged between 40% and 95% (Fig. 5C). The most affected biological process was “cell wall organization” as evidenced by the reduced number of transcripts from the cluster that were activated in response to low Pi in stop1 and almt1 (41 WT; 4 stop1; 2 almt1). Of the 46 differentially expressed genes included in the “response to phosphate starvation” that were regulated in WT root tips, 46% and 40% (21 stop1; 18 almt1) were differentially expressed in the root tips of stop1 and almt1, respectively.
Fig. 5.
Differential expression profiling of WT, stop1, and almt1 revealed a loss of transcriptional response to Pi starvation in the root apex of stop1 and almt1. (A) Venn diagram of differentially expressed genes (DEG) that are up- and down-regulated [−1.5 > logFC(−Pi/Pi) > 1.5; FDR < 0.05] in the root tip of WT, stop1, and almt1 seedlings in response to −Pi conditions. A bar graph illustrating the number of up-regulated and down-regulated transcripts in WT, stop1, and almt1 is presented. (B) Gene ontology (GO) enrichment analysis of overrepresented categories that are activated in the root apex of Pi-deprived WT seedlings. Each circle corresponds to a significantly enriched GO category (P value <0.05; hypergeometric test; Benjamini–Hochberg correction). Color code resembles P value and size resembles the number of genes that are associated with that respective GO category. GO categories that share genes are connected and clustered by the biological process that corresponds to the most significantly enriched category of the cluster. (C) Analysis of DEG by cluster in the root tip of stop1 and almt1. The number of genes that belong to each GO cluster, and are differentially expressed in stop1 and almt1, is presented as a percentage of the number of genes that are differentially expressed in WT (% DEG of WT). (D) Transcriptomic analysis of locally and systemically regulated genes in the root apex of WT, stop1, and almt1 revealed a key role of STOP1 and ALMT1 in the local response to Pi starvation in Arabidopsis. WT, stop1, and almt1 logFC values of genes that are differentially expressed in the root tips of WT seedlings [−1.5 > logFC(−Pi/Pi) > 1.5; FDR < 0.05] in response to −Pi conditions and were reported as PHR1-direct targets (6) and of those that were classified as part of the local or systemic transcriptional response as reported in ref. 5 is represented in a heatmap, respectively. Genes that are differentially expressed in WT root tips and their expression levels in WT, stop1, and almt1 seedlings grown under −Pi conditions are illustrated according to the key.
We found that the expression of SPX1 and SPX2, two key genes in the regulation of Pi-responsive genes that are systemically induced (32), were normally induced in the root tips of stop1 and almt1 mutants (Dataset S1). Therefore, we examined the transcription levels of genes that are known targets of the PHR1/SPX systemic regulatory node (6). Of the 94 PHR1 direct targets that we found induced in the root tips of WT plants, we found that 60% (63 stop1; 57 almt1) were also induced in stop1 and almt1 in response to low-Pi conditions (Fig. 5D). Because the activation of genes in the cell wall organization cluster is one of the most affected in stop1 and almt1 and its transcriptional regulation has been recently linked to the local response to low Pi availability (33), our results suggest that the local response to low Pi is largely lost in stop1 and almt1 and that the systemic response to low Pi is significantly less affected than the local response in these two mutants. To confirm that this was indeed the case, we performed a comparison of genes that had been previously defined to participate in local and systemic responses to low Pi (5) with those that were not activated in stop1 and almt1 (Fig. 5D). We found that among the 79 systemic and 147 local genes that were differentially expressed in the WT in response to Pi deficiency under our experimental conditions, over 60% of the systemically regulated genes remained responsive in stop1 (52 genes) and almt1 (51 genes), whereas less than 28% of locally regulated genes (40 stop1; 24 almt1) remained responsive to Pi starvation in the root tips of the mutants (Fig. 5D). These results confirm that STOP1 and ALMT1 have a key role in regulating the expression of genes in the local response to Pi deficiency. Nonetheless, STOP1 and ALMT1 also seem to have a significant effect on a group of genes that are systemically induced by low Pi availability.
Malate Treatment Rescued the Expression of Local-Response Genes Encoding Apoplast-Located Proteins.
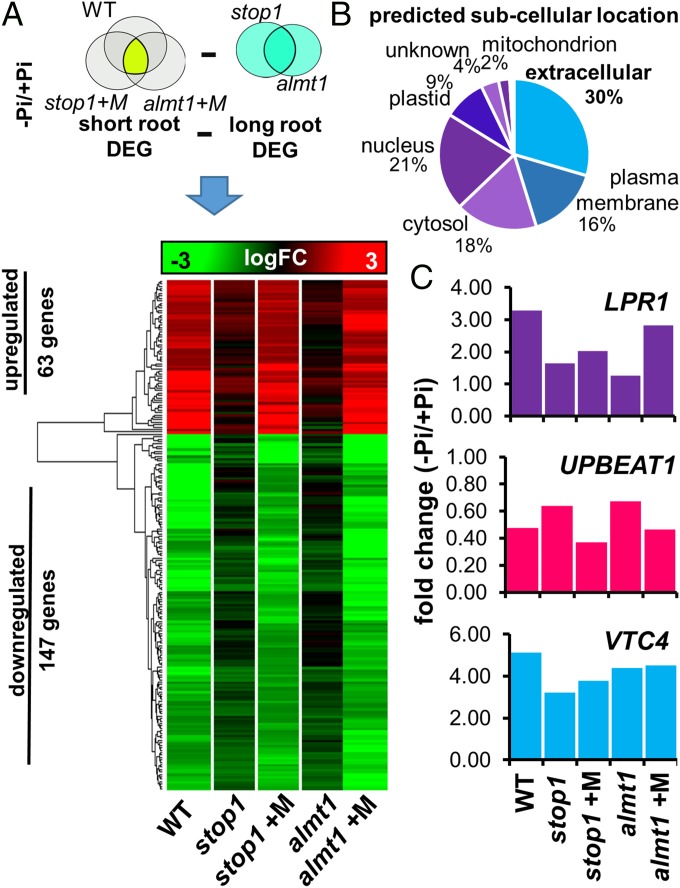
As we observed that malate treatment rescued iron accumulation in the RAM and the long-root phenotype of stop1 and almt1 mutants, we sought to identify the subset of genes that regulate primary root growth inhibition and whose expression under low-Pi conditions is reactivated by malate treatment in stop1 and almt1 seedlings. To this end, we isolated mRNA from the root tips of Pi-deprived stop1 and almt1 mutants that were treated with malate (1 mM) and carried out RNA-seq analysis (Fig. 6). Our rationale was that the common set of genes that are differentially expressed in the root tips of Pi-deprived seedlings that have a short-root phenotype (WT, stop1+M, and almt1+M) and that are not differentially expressed (induced or repressed) in the root tips of Pi-deprived stop1 and almt1 seedlings, which have a long-root phenotype in −Pi media, are linked to the malate-dependent mechanism that triggers primary root growth inhibition under Pi-deprivation conditions. A common set of 210 differentially expressed genes (63 up-regulated; 147 down-regulated) was found between the genotypes/treatment that induce a short-root phenotype under −Pi conditions and that are not differentially expressed in Pi-deprived seedlings with a long-root phenotype (Fig. 6A). Among the genes whose expression was rescued by malate treatment, we found several peroxidase family genes (PEROXIDASE2, PEROXIDASE37, AT3G01190, and PEROXIDASE4), which are closely related to the control of ROS homeostasis (34). A full list of the genes and description is included in Dataset S1. Interestingly, using SUBA, a subcellular prediction tool (35), we found that 30% of proteins encoded by genes whose responsiveness to low Pi is restored by malate treatment in stop1 and almt1 Pi-deprived seedlings are targeted to the apoplast or extracellular region (Fig. 6B), confirming a previous study in which a major role of the apoplast in the root response to Pi starvation was highlighted (33). Furthermore, an additional 16% of genes are targeted to the plasma membrane (Fig. 6B).
Fig. 6.
Malate treatment rescues the expression of transcripts whose products are targeted to the extracellular region. (A) Schematic of the Venn diagram analysis of common differentially expressed genes [DEG; −1.5 > logFC(−Pi/Pi) > 1.5; FDR < 0.05] in short-root phenotypes (WT∩stop1∩almt1) minus the DEG in long-root phenotypes (stop1∪almt1) under Pi-deficiency conditions, that was performed to determine the genes whose expression is linked to short-root phenotype and is rescued by malate treatment in stop1 and almt1 seedlings. Heatmap of the logFC values in WT, stop1/+M, and almt1/+M of the determined gene set is presented. (B) Predicted subcellular location of the DEG whose expression is restored by malate treatment. (C) LPR1, UPBEAT1, and VTC4 expression levels are presented in fold change (FC; FDR < 0.05) as revealed by our transcriptomic studies in the root apex of WT, stop1, and almt1 seedlings under −Pi conditions and −Pi and malate treatment (almt1+M, stop1+M) conditions.
LPR1 Acts Downstream of STOP1 and ALMT1.
In Arabidopsis, LPR1 has been reported to mediate the oxidation of Fe+2 to Fe+3 in the apoplast, which was correlated with ROS generation that triggers callose deposition in the root apex and disrupts SHR transport, which ultimately induces determinate primary root growth in response to Pi-deficiency conditions (20). As both Fe+3 accumulation and the regulation of peroxidases are lost and rescued by malate treatment in stop1 and almt1 mutants, using our RNA-seq data, we analyzed whether malate treatment had the same effect on the LPR1 expression in response to low-Pi conditions in the root apex of stop1 and almt1 (Fig. 6C). The expression of LPR1 was found to be enhanced in the root apex of WT seedlings (3.29-fold) exposed to low Pi, induction that was significantly lower in stop1 (1.64-fold) and almt1 (1.25-fold) Pi-deprived seedlings (Fig. 6C). Our data revealed that, indeed, malate treatment increased LPR1 transcript levels in Pi-deprived stop1 seedlings from 1.5- to 2.0-fold and in almt1 seedlings from 1.2- to 2.7-fold (Fig. 6C). The higher increase in LPR1 expression induced by malate treatment in almt1 than stop1 correlates with the capacity of the treatment to better restore the primary root response to low in almt1 than in stop1 (Fig. 6C). To test whether the effect of malate was dependent or independent of LPR1, we studied the effect of malate treatment on the primary root elongation of lpr1 seedlings grown in media lacking Pi. We observed that malate treatment did not rescue the lpr1 mutant phenotype (SI Appendix, Fig. S6), suggesting that the effect of malate to trigger the determinate root developmental program in response to Pi deficiency requires the presence of a functional LPR1 protein.
Because alterations in the ROS balance in the RAM of Arabidopsis are linked to the meristem exhaustion process observed in Pi-deprived seedlings (36) and LPR1 is essential for the low-Pi–dependent ROS signaling that takes place in the primary root of Arabidopsis, we decided to analyze the transcript levels of UPBEAT1 (UPB1) (Fig. 6C): the only transcription factor known to modulate ROS balance and control the transition from cell proliferation to cell differentiation in the RAM by modulating the transcription of peroxidase genes (37). We observed that UPB1 is down-regulated 0.48-fold in response to low Pi in the root apex of WT seedlings and that it is down-regulated to a lower degree in stop1 [0.64 fold change (FC)] and almt1 (0.67 FC). Malate treatment restored the down-regulation of UPB1 in Pi-deprived almt1 (0.46 FC) and stop1 (0.37 FC) seedlings to WT levels (Fig. 6C). Because LPR1 and UPB1 seem to be involved in modulating ROS balance in the RAM and their expression is misregulated in stop1 and almt1, we identified peroxidase genes that have been related to ROS homeostasis in the root (34) and analyzed their expression levels in WT, stop1, and almt1 seedlings. We found that 18 peroxidase genes (PRXS) were transcriptionally regulated by low Pi in the root apex of WT seedlings of which 13 PRXS were not responsive to low Pi in stop1 and almt1 (Dataset S1). Interestingly, the responsiveness of 11 peroxidase genes (PEROXIDASE52, AT4G08780, AT4G08780, PEROXIDASE2, AT5G06730, AT5G39580, PEROXIDASE37, AT5G15180, PEROXIDASE4, AT2G39040, and AT3G01190) was rescued by malate treatment in Pi-deprived stop1 and almt1 seedlings (SI Appendix, Fig. S7). Alterations of ROS balance have been related to callose deposition in the primary root of Arabidopsis (20). Given that the transcription of ROS-related genes (UPB1, LPR1, and PRXS) is disrupted in stop1 and almt1 under low-Pi conditions, we analyzed the expression values of genes coding for callose synthases (CALS). We found that the expression of 4 genes coding for CALS (CALLOSESYNTHASE7, CALLOSESYNTHASE9, GLUCANSYNTHASELIKE4, and GLUCANSYNTHASELIKE5) was induced (1.1 logFC; FDR < 0.05) in the root apex of WT seedlings and that it was induced to a lesser extent (logFC < 0.6) in stop1 and almt1 seedlings. It was observed that malate treatment also rescued the expression of these 4 CALS in Pi-deprived stop1 and almt1 (SI Appendix, Fig. S7). Our results suggested that malate efflux is required for the ROS signaling cascade that has been reported to be affected in low-Pi insensitive mutants (20, 36).
Given that Fe tends to its higher oxidation state (Fe+3) in natural environments and the iron uptake genes IRON REGULATED TRANSPORTER 1 (IRT1) and FERRIC REDUCTION OXIDASE 2 (FRO2) have been reported to be repressed in response to Pi-deficiency conditions (3, 5, 38) we asked whether an alternative Fe+3 reduction mechanism could provide Fe+2 to LPR1 to initiate the proposed ROS signaling cascade that is induced in the RAM in response to low Pi. An ascorbate-dependent Fe+2 reduction mechanism has been reported recently (29), in which VITAMINC4 (VTC4), a gene encoding a protein with dual myo-inositol-monophosphatase and ascorbate synthase activity (39), could play a central role. We found that VTC4 is induced in the root apex of WT (5 FC), almt1 (4.3 FC), and stop1 (3.22 FC) seedlings (Fig. 6C). Interestingly, VTC4 belongs to the set of genes that are direct targets of PHR1 (6), providing a potential link between local and systemic signaling in the primary root response to low Pi and the cross-talk between Fe and P in the Pi-deficiency response.
Discussion
LPR1 has been proposed to promote the accumulation of Fe in the apoplast of cells in the RAM, which in turn triggers an accumulation of callose that alters symplastic transport causing meristem differentiation (20). However, the precise mechanism by which Fe accumulates in the apoplast of RAM cells remained to be determined. Here we show that STOP1 and ALMT1 participate in the mechanism that triggers RAM exhaustion in response to low Pi availability by mediating the accumulation of Fe+3 in the apoplast of RAM cells. STOP1 and ALMT1 were originally described as genes responsible for the malate efflux that protects the Arabidopsis root from Al+3 toxicity (21, 22). AtSTOP1 is constitutively expressed in Arabidopsis, indicating that its involvement in the Al-dependent induction of gene expression must involve posttranslational processes of modification or the direct binding of Al+3. The finding that mutations in STOP1 and ALMT1 lead to long-root phenotypes in Pi-deprived seedlings suggests that malate excretion is also involved in the process of meristem exhaustion in response to low Pi availability. We found that STOP1 is expressed in the RAM in a Pi-independent fashion and that ALMT1 is expressed in the RAM but only in seedlings grown in media with low-Pi concentrations. Although the expression domains of STOP1 and ALMT1 do not completely overlap, we found that the expression directed by the ALMT1 promoter is completely dependent on an intact copy of STOP1 (SI Appendix, Fig. S2). The apparent inconsistency between the role of an activator with its target gene and the differences in patterns of expression of STOP1 and ALMT1 could be explained by the recent report that STOP1 mRNA is cell-to-cell mobile (40).
Pi and Fe availability have been shown to coordinately regulate RAM maintenance and primary root growth in vitro (11, 17, 20). Our results corroborate that, in Arabidopsis WT seedlings, Fe availability (SI Appendix, Fig. S8) in the medium is required for RAM exhaustion in media with a low Pi concentration and that Fe accumulation in the RAM is associated with the process of RAM exhaustion (Fig. 4). Apoplastic iron accumulation in the RAM was reported to be essential for primary root growth inhibition in response to −Pi conditions (20); however, the mechanism for iron accumulation in the root remained to be determined. We found that Fe failed to accumulate in the root apex of stop1 and almt1 seedlings grown in Pi-deficient media and that the treatment of stop1 and almt1 seedlings with malate restores both Fe accumulation in the RAM and the inhibition of primary root growth in Pi-deprived seedlings (Fig. 4). These data show that malate secretion is necessary and sufficient for iron accumulation in the RAM and to trigger cell differentiation in the RAM that is responsible for the meristem exhaustion process induced by Pi deficiency. We propose that such mechanism of iron accumulation happens in the apoplast as ALMT1 is reported to be a malate efflux protein (22) and thus, exogenous malate, which probably diffuses through the apoplast, can rescue iron accumulation in almt1 and stop1. Malate supplementation was found to fully rescue the short-root phenotype of almt1, whereas it only partially restored primary root growth inhibition in stop1, suggesting that, in addition to ALMT1, STOP1 regulates the expression of other genes whose activation by low Pi is required for full meristem exhaustion.
Molecular dynamic simulation of Fe+3 and Fe+2 interaction shows that malate can form large complexes with Fe+3 but not Fe+2 (SI Appendix, Fig. S5). These data suggest that malate promotes the accumulation of Fe+3 in the apoplast by forming these large molecular weight complexes, which by a still largely unknown mechanism that correlates with ROS generation (20), activate the processes required for meristem exhaustion. lpr1 seedlings also show a long-root phenotype in low-Pi media, suggesting that the ferroxidase activity of LPR1 is required to trigger cell differentiation during the primary root meristem exhaustion process triggered by Pi deprivation. We found that the long-root phenotype of lpr1 in low-Pi media cannot be rescued by malate treatment, suggesting that LPR1 acts downstream of STOP1 and ALMT1 and that most probably is required to activate the Fe-mediated mechanism involved in the process of RAM exhaustion observed in Pi-deprived seedlings. Therefore, cell-wall–localized LPR1 ferroxidase activity, which catalyzes Fe+2 to Fe+3 conversion (20), could act synergistically with malate efflux in the accumulation of Fe+3 in the apoplast of RAM cells. LPR1-dependent Fe+3 production in the apoplast could trigger ROS production by initiating a Fe redox cycle as previously proposed (41). Either ferric-chelate reductase oxidase activity, which reduces apoplast-diffusible Fe+3 chelates, or effluxed ascorbate (29), could reduce the Fe+3 produced by LPR1 to a redox-active Fe+2 to complete a cycle thereby triggering root cell differentiation. Supporting this notion, LPR1 overexpression causes ectopic Fe+3 and ROS generation in Pi-deprived seedlings (20). Our data suggest the existence of a STOP1, ALMT1, and LPR1 coordinated redox mechanism that involves Fe+3 deposition in the apoplast of RAM cells of seedlings exposed to low Pi. ALMT1 is transcriptionally up-regulated in a similar fashion in WT and lpr1 mutants (33), confirming that LPR1 acts downstream of the STOP1/ALMT1 low-Pi regulatory node.
The expression of up to 80% of the genes involved in the local response to low Pi and up to a 40% of the genes involved in the systemic response to low Pi was affected in stop1 and almt1 (Fig. 5D). The reduction in both local and systemic responses in stop1 and almt1, specifically in the PHR1-direct targets, points to a cross-talk between the signaling pathways that regulates the transcriptional activation or repression of the systemic and local responses to low Pi in the root apex. However, we cannot rule out the possibility that the internal concentration of Pi could be higher in the root tip of stop1 and almt1 than in the WT, which could lead to a down-regulation of the systemic response, as has been suggested for plants grown in low-Pi and low-Fe conditions (17). Further experiments regarding a possible STOP1 and ALMT1 interaction directly with Pi or PHR1- or SPX-domain proteins could shed light on the existence of a coordinated response to external and internal Pi levels in the root apex. The observation that PHR1 activates the expression of the ascorbate synthase VTC4 under Pi-deficiency conditions together with the recent report that ascorbate efflux contributes to Fe+3 reduction (29), support the notion that the redox cycle that generates ROS and triggers RAM exhaustion could be controlled by both local and systemic responses to Pi starvation.
Our transcriptomic analysis revealed that LPR1 is responsive to Pi deprivation in the root tips of WT plants (Fig. 6C) and that this response is significantly reduced in the root tips of stop1 and almt1 seedlings (Fig. 6C). These results suggest that a threshold level of LPR1 is required to activate meristem exhaustion in Pi-deprived seedlings. This notion is supported by the observation that the treatment with malate that reverts the long-root phenotype of Pi-deprived stop1 and almt1 seedlings (Fig. 4A) also leads to an increase in LPR1 transcript levels (Fig. 6C). Moreover, Arabidopsis accessions with higher LPR1 transcription levels have shorter primary roots under low-Pi conditions (16); and we observed that, in the case of malate-treated Pi-deprived stop1 and almt1 seedlings, a higher LPR1 expression in almt1 than in stop1 correlated with a shorter primary root in almt1 than in stop1 (Figs. 4B and 6C). A recent report on the interplay between the transcriptional activation of genes coding for extracellular enzymes and iron redistribution in the apoplast in response to Pi deprivation highlighted the role of the apoplast in the Pi-starvation response (33). Transcriptomic analysis of Pi-deprived stop1 and almt1 seedlings showed that malate treatment reactivates the low-Pi responsiveness of genes that encode extracellular proteins involved in cell-wall modification and ROS homeostasis, such as peroxidases (34). Our transcriptomic results confirm a previously reported role of apoplastic peroxidases in the Pi-starvation response (33), and highlight the role of malate secretion in the cell-wall remodeling processes potentially involved in the changes of Arabidopsis root system architecture induced by low Pi availability. In this context, the finding that UPBEAT1, a transcription factor that modulates the transition from cell proliferation to cell differentiation in the RAM by repressing peroxidase genes (37), is repressed in response to Pi-deficiency conditions and the fact its expression is altered in stop1 and almt1, support the notion that ROS generation plays an important role in the root response to Pi deprivation. Further experiments regarding the specific pattern of ROS signaling in the RAM under Pi-starvation conditions are required.
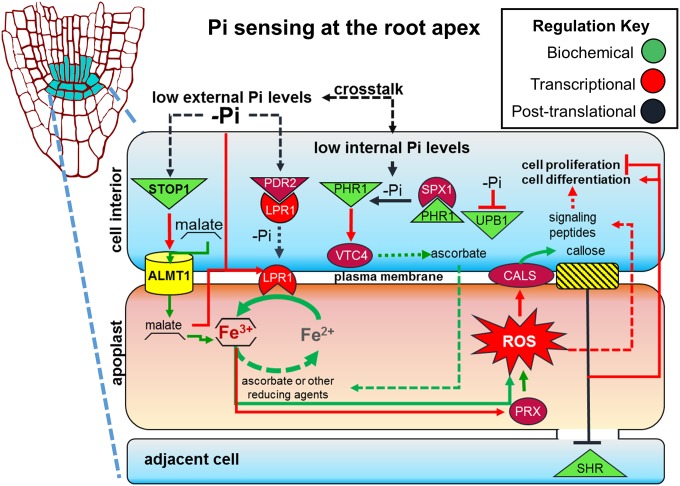
A model that summarizes what is known about the local response to Pi starvation and the proposed role of STOP1 and ALMT1 in the root response to Pi deprivation is presented in Fig. 7. Under Pi-deficiency conditions, expression of LPR1 is enhanced by a malate-dependent mechanism, and PDR2 activity is inhibited, facilitating LPR1 mobilization from the ER to the plasma membrane (20) where LPR1 ferroxidase activity catalyzes the conversion of Fe+2 into Fe+3 in the apoplast of RAM cells (Fig. 7). The mechanisms by which LPR1 is transported from the ER to the extracellular region and how PDR2 activity is regulated by Pi availability remain to be determined. STOP1, a constitutively expressed gene, up-regulates the expression of ALMT1 in seedlings exposed to low Pi, thereby activating the excretion of malate. LPR1 ferroxidase activity in the plasma membrane of cells in the meristem and elongation zones of the primary root, locally produces Fe+3, which forms large complexes with malate, leading to its accumulation. As mentioned above, we observed a cross-talk between local and systemic responses to Pi deficiency; therefore, we included the internal Pi-sensing module PHR1/SPX1 (6) in the model because it controls the majority of the systemic transcriptional responses to low Pi (5). PHR1-dependent induction of VTC4 could enhance ascorbate efflux, which represents a potential mechanism of Fe+3 reduction to produce the Fe+2 required to complete a redox cycle that generates ROS. Expression of the UPB1 repressor is reduced by a Fe/malate-dependent mechanism under low-Pi conditions, which enhances the transcription of peroxidase genes. Enhanced transcription of local response peroxidase genes likely triggers ROS generation in the root of Pi-deprived seedlings as previously reported (36). ROS generation triggers callose deposition as previously reported (20, 42). This notion is supported by the observation that the expression of several CALS genes is enhanced in the root apex of WT but not in stop1 and almt1 seedlings (SI Appendix, Fig. S7). Callose synthesis impairs symplastic transport by physically blocking plasmodesmatal pores, which reduces or inactivates the cell-to-cell movement of SHR (20). Because SHR cell-to-cell movement is required for stem cell niche maintenance, meristem exhaustion takes place in seedlings exposed to low Pi availability. However, because CLE-like peptide signaling is also required for stem cell niche maintenance in Arabidopsis (43), we cannot rule out that ROS or Fe+3 accumulation in the RAM could induce the transcription of CLE peptides that could also execute RAM exhaustion.
Fig. 7.
STOP1 and ALMT1 regulate the RAM response to low-Pi conditions in Arabidopsis. In response to limiting Pi levels in the medium, STOP1 induces ALMT1 (Fig. 3), which triggers malate efflux in the root apex. Low-Pi levels also inhibit PDR2 negative regulation over LPR1 and induce, in a largely unknown mechanism, LPR1 transport to the plasma membrane. Malate contributes to the aggregation of Fe+3 ions in the apoplast and enhances the expression of LPR1, callose synthase genes (CALS), and peroxidase genes (PRX) under Pi-deficiency conditions. LPR1 and PRX activity generates reactive oxygen species (ROS), which enhance callose deposition by CALS. Callose deposition closes the symplastic channels of communication, which ultimately impairs the transport of transcription factors, such as SHR, that are essential to maintain cell proliferation and organization in the RAM. Alternative factors induced by ROS, such as signaling peptides, could also induce cell differentiation in the RAM. A cross-talk between PHR1/SPX1-dependent internal Pi sensing and external Pi sensing could be possible because of the downgrade of systemic response in low-Pi insensitive mutants. Ascorbate efflux into the apoplast or another reducing agent, could reduce Fe+3 to Fe+2 and could restart the proposed redox cycle. Ascorbate efflux could be enhanced by VTC4 activity and whose gene is induced by PHR1 when internal Pi levels are limiting. Arrows represent relationships between the components. Dotted lines represent hypothetical relations, and the regulation key illustrates the type of evidence that has been provided for that relationship.
STOP1 controls ALMT1 transcription and its expression is not regulated by Pi availability, suggesting that STOP1 is most likely involved in sensing external Pi levels or an environmental cue that it is linked to low-Pi levels in the medium. Because STOP1 also controls low pH and Al+3 toxicity responses, it emerges as a possible master regulator/sensor that orchestrates the root responses to multiple environmental stresses.
The root tip is at the forefront of nutrient sensing and acquisition; therefore, it represents the first contact of the plant with the soil solution. Previous findings have shown that the root tip plays a critical role in Pi sensing (16) and that organic acids are secreted from the roots of Pi-deprived plants to solubilize Pi and facilitate its uptake (14). We describe an organic-acid–mediated mechanism that starts in the root tip and adjusts the root developmental response to low Pi. Organic acids have also been implicated in Fe uptake (30) and protection of the root from the entrance of toxic Al+3 ions (22, 23). Taken all together, the evidence suggests that organic-acid exudation is a critical checkpoint in plant nutrition as these molecules do not only protect the root from the entry of toxic ions and enhance nutrient uptake, but also contribute to the modifications of root development that increase the soil-exploration ability of the plant to enhance nutrient uptake. Biotechnological approaches, like the overexpression of ALMT1 or STOP1, could be coupled with the metabolic engineering of carbon metabolism in plants to optimize nutrient uptake in an agricultural environment.
Materials and Methods
Plant Material.
A. thaliana Col-0 accession (CS70000) was used in this work. stop1-SALK_114108 (N614108), almt1-SALK_009629 (N509629), and lpr1 (N516297) lines were provided by the Nottingham Arabidopsis Stock Center (NASC).
Growing Conditions.
Seeds were surface sterilized and sown in 1% agar, 0.1× Murashige and Skoog (MS) medium as described in ref. 10. A total of 1 mM KH2PO4 (high Pi; +Pi) or 5 µM KH2PO4 (low Pi; −Pi) Pi concentrations were used. A total of 1% (wt/vol) sucrose and 3.5 mM MES was added. Fe-free medium was prepared as described (15), and 100 µM ferrozine (Sigma, 82950) was added to reduce agar Fe availability. Malate and citrate (Sigma, M1000 and Sigma, C0759, respectively) were added to medium before sterilization. Seedlings were grown in a Percival chamber at 22 °C, under 16/8 h photoperiod with >200 µmol⋅m−2⋅s−1 luminous intensity.
Gene Mapping.
Plant mapping populations were built crossing lpi5 and lpi6 homozygous lines vs. Col-0 (CS70000), respectively, according to the Mutmap protocol (44). Heterozygous F1 plants were self-fertilized for seed propagation. The F2 segregating individuals were rescreened for WT (short root) and mutant (long root) phenotypes, respectively, for each of the two crosses. Then, DNA was extracted from 100 plants for each phenotype, pooled, and sequenced using Illumina Miseq technology (paired end, 250 base pair reads in length). Reads obtained were processed with fastQC and Trimmomatic (45) to improve their quality. Only paired-end reads were considered. Reads were mapped to the Col-0 reference genome (TAIR10) using BWA (46) and SAMtools (47). To identify and to evaluate specific variants related to the mutant phenotype (long root), a pipeline using GATK (https://software.broadinstitute.org/gatk/), VCFtools (48), SNPeff, and SNPshif (49) was implemented. IGV was used to visualize variants (50).
Histochemical Iron Staining.
Perls iron staining and DAB intensification were carried out as described in ref. 20 and analyzed using Nomarski optics on a Leica DMR microscope. To improve the resolution of the picture, optical sections of ∼40 µM were taken with a 100× objective and assembled into photographic reconstructions using Adobe Photoshop. No other manipulation of the images was used.
RNA-Seq High-Throughput Sequencing and Data Analysis.
High-throughput sequencing and data analysis were carried out as described (51).
qRT-PCR.
Total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Real-time PCR was performed with an Applied Biosystems 7500 real-time PCR system using SYBR Green detection chemistry (Applied Biosystems) and gene-specific primers. The relative expression levels were computed by the Ct method of relative quantification. Oligonucleotide primer sequences are available upon request.
A more detailed version of materials and methods is included in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank G. S. Gillmor for his advice on EMS seed mutagenesis and L. F. García-Ortega and O. Martínez for help in calculating the dispersion values for the normalization of transcriptomic data expression. J.M.-M. is indebted to Consejo Nacional de Ciencia y Tecnología (CONACyT) for a PhD fellowship. J.O.O.-R. is indebted to CONACyT for an MSc fellowship. This research was supported by CONACyT Ciencia Básica Grant 252039 (to L.H.-E.) and by Howard Hughes Medical Institute Grant 4367 (to L.H.-E.).
Footnotes
Conflict of interest statement: L.H.-E. and Leon V. Kochian were coauthors on a 2013 publication. This publication was a review article and did not involve any research collaboration.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE90061) and at National Center for Biotechnology Information Biosample database www.ncbi.nlm.nih.gov/biosample/ (accession nos. SAMN06013467; SAMN06013468; SAMN06013469; SAMN06013470).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1701952114/-/DCSupplemental.
References
- 1.Marschner H. Marschner’s Mineral Nutrition of Higher Plants. Elsevier Science; Amsterdam, The Netherlands: 2011. [Google Scholar]
- 2.Wu P, et al. Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol. 2003;132:1260–1271. doi: 10.1104/pp.103.021022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Misson J, et al. A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA. 2005;102:11934–11939. doi: 10.1073/pnas.0505266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morcuende R, et al. Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ. 2007;30:85–112. doi: 10.1111/j.1365-3040.2006.01608.x. [DOI] [PubMed] [Google Scholar]
- 5.Thibaud MC, et al. Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant J. 2010;64:775–789. doi: 10.1111/j.1365-313X.2010.04375.x. [DOI] [PubMed] [Google Scholar]
- 6.Bustos R, et al. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 2010;6:e1001102. doi: 10.1371/journal.pgen.1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffland E, Van Den Boogaard R, Nelemans J, Findenegg G. Biosynthesis and root exudation of citric and malic acids in phosphate-starved rape plants. New Phytol. 1992;122:675–680. [Google Scholar]
- 8.Cruz-Ramírez A, Oropeza-Aburto A, Razo-Hernández F, Ramírez-Chávez E, Herrera-Estrella L. Phospholipase DZ2 plays an important role in extraplastidic galactolipid biosynthesis and phosphate recycling in Arabidopsis roots. Proc Natl Acad Sci USA. 2006;103:6765–6770. doi: 10.1073/pnas.0600863103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bates TR, Lynch JP. Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorus availability. Plant Cell Environ. 1996;19:529–538. [Google Scholar]
- 10.López-Bucio J, et al. Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol. 2002;129:244–256. doi: 10.1104/pp.010934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sánchez-Calderón L, et al. Phosphate starvation induces a determinate developmental program in the roots of Arabidopsis thaliana. Plant Cell Physiol. 2005;46:174–184. doi: 10.1093/pcp/pci011. [DOI] [PubMed] [Google Scholar]
- 12.Péret B, Clément M, Nussaume L, Desnos T. Root developmental adaptation to phosphate starvation: better safe than sorry. Trends Plant Sci. 2011;16:442–450. doi: 10.1016/j.tplants.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 13.Rubio V, et al. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001;15:2122–2133. doi: 10.1101/gad.204401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raghothama KG. Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:665–693. doi: 10.1146/annurev.arplant.50.1.665. [DOI] [PubMed] [Google Scholar]
- 15.Sánchez-Calderón L, et al. Characterization of low phosphorus insensitive mutants reveals a crosstalk between low phosphorus-induced determinate root development and the activation of genes involved in the adaptation of Arabidopsis to phosphorus deficiency. Plant Physiol. 2006;140:879–889. doi: 10.1104/pp.105.073825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Svistoonoff S, et al. Root tip contact with low-phosphate media reprograms plant root architecture. Nat Genet. 2007;39:792–796. doi: 10.1038/ng2041. [DOI] [PubMed] [Google Scholar]
- 17.Ward JT, Lahner B, Yakubova E, Salt DE, Raghothama KG. The effect of iron on the primary root elongation of Arabidopsis during phosphate deficiency. Plant Physiol. 2008;147:1181–1191. doi: 10.1104/pp.108.118562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ticconi CA, Delatorre CA, Lahner B, Salt DE, Abel S. Arabidopsis pdr2 reveals a phosphate-sensitive checkpoint in root development. Plant J. 2004;37:801–814. doi: 10.1111/j.1365-313x.2004.02005.x. [DOI] [PubMed] [Google Scholar]
- 19.Ticconi CA, et al. ER-resident proteins PDR2 and LPR1 mediate the developmental response of root meristems to phosphate availability. Proc Natl Acad Sci USA. 2009;106:14174–14179. doi: 10.1073/pnas.0901778106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Müller J, et al. Iron-dependent callose deposition adjusts root meristem maintenance to phosphate availability. Dev Cell. 2015;33:216–230. doi: 10.1016/j.devcel.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 21.Iuchi S, et al. Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. Proc Natl Acad Sci USA. 2007;104:9900–9905. doi: 10.1073/pnas.0700117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoekenga OA, et al. AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc Natl Acad Sci USA. 2006;103:9738–9743. doi: 10.1073/pnas.0602868103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi Y, et al. Characterization of AtALMT1 expression in aluminum-inducible malate release and its role for rhizotoxic stress tolerance in Arabidopsis. Plant Physiol. 2007;145:843–852. doi: 10.1104/pp.107.102335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colón-Carmona A, You R, Haimovitch-Gal T, Doerner P. Technical advance: Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 1999;20:503–508. doi: 10.1046/j.1365-313x.1999.00620.x. [DOI] [PubMed] [Google Scholar]
- 25.Sabatini S, et al. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell. 1999;99:463–472. doi: 10.1016/s0092-8674(00)81535-4. [DOI] [PubMed] [Google Scholar]
- 26.Sawaki Y, et al. STOP1 regulates multiple genes that protect arabidopsis from proton and aluminum toxicities. Plant Physiol. 2009;150:281–294. doi: 10.1104/pp.108.134700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tokizawa M, et al. SENSITIVE TO PROTON RHIZOTOXICITY1, CALMODULIN BINDING TRANSCRIPTION ACTIVATOR2, and other transcription factors are involved in ALUMINUM-ACTIVATED MALATE TRANSPORTER1 expression. Plant Physiol. 2015;167:991–1003. doi: 10.1104/pp.114.256552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones DL. Organic acids in the rhizosphere: A critical review. Plant Soil. 1998;205:25–44. [Google Scholar]
- 29.Grillet L, et al. Ascorbate efflux as a new strategy for iron reduction and transport in plants. J Biol Chem. 2014;289:2515–2525. doi: 10.1074/jbc.M113.514828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rellán-Alvarez R, et al. Identification of a tri-iron(III), tri-citrate complex in the xylem sap of iron-deficient tomato resupplied with iron: New insights into plant iron long-distance transport. Plant Cell Physiol. 2010;51:91–102. doi: 10.1093/pcp/pcp170. [DOI] [PubMed] [Google Scholar]
- 31.Ziegler J, et al. Non-targeted profiling of semi-polar metabolites in Arabidopsis root exudates uncovers a role for coumarin secretion and lignification during the local response to phosphate limitation. J Exp Bot. 2016;67:1421–1432. doi: 10.1093/jxb/erv539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puga MI, et al. SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis. Proc Natl Acad Sci USA. 2014;111:14947–14952. doi: 10.1073/pnas.1404654111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoehenwarter W, et al. Comparative expression profiling reveals a role of the root apoplast in local phosphate response. BMC Plant Biol. 2016;16:106. doi: 10.1186/s12870-016-0790-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Francoz E, et al. Roles of cell wall peroxidases in plant development. Phytochemistry. 2015;112:15–21. doi: 10.1016/j.phytochem.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 35.Tanz SK, et al. SUBA3: A database for integrating experimentation and prediction to define the SUBcellular location of proteins in Arabidopsis. Nucleic Acids Res. 2013;41:D1185–D1191. doi: 10.1093/nar/gks1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chacón-López A, Ibarra-Laclette E, Sánchez-Calderón L, Gutiérrez-Alanis D, Herrera-Estrella L. Global expression pattern comparison between low phosphorus insensitive 4 and WT Arabidopsis reveals an important role of reactive oxygen species and jasmonic acid in the root tip response to phosphate starvation. Plant Signal Behav. 2011;6:382–392. doi: 10.4161/psb.6.3.14160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsukagoshi H, Busch W, Benfey PN. Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell. 2010;143:606–616. doi: 10.1016/j.cell.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 38.Li W, Lan P. Genome-wide analysis of overlapping genes regulated by iron deficiency and phosphate starvation reveals new interactions in Arabidopsis roots. BMC Res Notes. 2015;8:555. doi: 10.1186/s13104-015-1524-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torabinejad J, Donahue JL, Gunesekera BN, Allen-Daniels MJ, Gillaspy GE. VTC4 is a bifunctional enzyme that affects myoinositol and ascorbate biosynthesis in plants. Plant Physiol. 2009;150:951–961. doi: 10.1104/pp.108.135129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thieme CJ, et al. Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat Plants. 2015;1:15025. doi: 10.1038/nplants.2015.25. [DOI] [PubMed] [Google Scholar]
- 41.Kosman DJ. Redox cycling in iron uptake, efflux, and trafficking. J Biol Chem. 2010;285:26729–26735. doi: 10.1074/jbc.R110.113217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luna E, et al. Callose deposition: A multifaceted plant defense response. Mol Plant Microbe Interact. 2011;24:183–193. doi: 10.1094/MPMI-07-10-0149. [DOI] [PubMed] [Google Scholar]
- 43.Matsuzaki Y, Ogawa-Ohnishi M, Mori A, Matsubayashi Y. Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science. 2010;329:1065–1067. doi: 10.1126/science.1191132. [DOI] [PubMed] [Google Scholar]
- 44.Abe A, et al. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol. 2012;30:174–178. doi: 10.1038/nbt.2095. [DOI] [PubMed] [Google Scholar]
- 45.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Danecek P, et al. 1000 Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yong-Villalobos L, et al. Methylome analysis reveals an important role for epigenetic changes in the regulation of the Arabidopsis response to phosphate starvation. Proc Natl Acad Sci USA. 2015;112:E7293–E7302. doi: 10.1073/pnas.1522301112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.