Significance
Rapid access to defined, pure biomacromolecules is key to biochemical and biophysical investigations as evidenced by the impact automated solid-phase synthesis of oligonucleotides and oligopeptides had on basic research. Here, a strategy as well as a new instrument to routinely access oligosaccharides by automated synthesis is reported. The method produces glycans quickly and reliably to accelerate fundamental advances in glycobiology.
Keywords: oligosaccharide synthesis, automated synthesis, glycochemistry, glycobiology, Glyconeer
Abstract
Reliable and rapid access to defined biopolymers by automated DNA and peptide synthesis has fundamentally altered biological research and medical practice. Similarly, the procurement of defined glycans is key to establishing structure–activity relationships and thereby progress in the glycosciences. Here, we describe the rapid assembly of oligosaccharides using the commercially available Glyconeer 2.1 automated glycan synthesizer, monosaccharide building blocks, and a linker-functionalized polystyrene solid support. Purification and quality-control protocols for the oligosaccharide products have been standardized. Synthetic glycans prepared in this way are useful reagents as the basis for glycan arrays, diagnostics, and carbohydrate-based vaccines.
Progress in genomics and proteomics was enabled by automated synthesis (1, 2) and sequencing methods (3, 4) for oligopeptides and oligonucleotides. In contrast, the structure–function relationships of carbohydrates, the most abundant biopolymers on earth, are less well understood than those of nucleic acids and proteins. Because isolation of specific glycans using biochemical methods is difficult and cannot rely on amplification procedures, chemical and enzymatic syntheses, or combinations thereof, have served to procure these molecules (5). Efforts to accelerate traditional chemical syntheses, which is especially labor-intensive for glycans, has focused mainly on automated glycan assembly (AGA) on solid support and programmable one-pot synthesis methods (6, 7).
The advantages of solid-phase synthesis are well appreciated since the invention of peptide assembly by Merrifield (8), and oligonucleotide synthesis on polymer supports (9). Synthetic manipulations are executed by automated instrumentation and reactions can be driven to completion via the use of excess reagents that are simply removed by washing steps. Cleavage from the solid support and purification as postautomation steps are typically standardized. The keys to a successful automated synthesis are reliable building blocks and resins (based upon a comprehensive protecting group strategy and a resin linker, used as a “protecting group,” for anchoring the first monomer to the support), high-yielding couplings, and reliable instrumentation.
Since the introduction of AGA in 2001 (10), many aspects of the synthetic process have been systematically improved, ranging from strategic considerations relative to building blocks, linkers, purification methods, and quality control. Synthetic glycans of increasing length and complexity have been assembled by AGA (11–17). Quality control of the stereo- and regiochemical composition of synthetic products has been greatly accelerated by using ion mobility-mass spectrometry (IM-MS) (18).
The streamlined AGA process described herein relies upon the Glyconeer 2.1 automated oligosaccharide synthesizer and uses monosaccharide building blocks to rapidly assemble conjugation-ready oligosaccharides (Fig. 1A; the process is described in detail in the SI Appendix and the synthesizer is depicted in SI Appendix, Fig. S1).
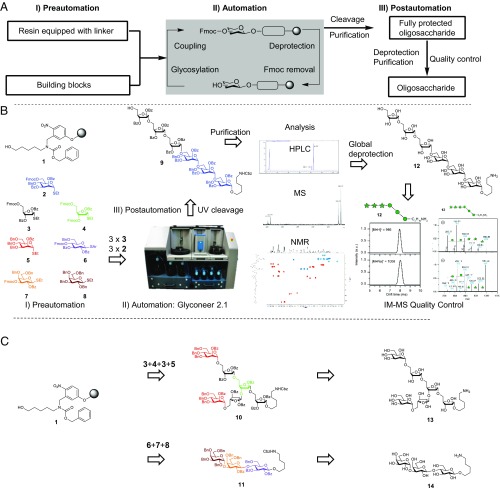
Fig. 1.
Automated glycan assembly illustrated for the syntheses of oligosaccharides 12–14. (A) Schematic overview of AGA. (B) Detailed description of the full workflow demonstrated for the synthesis of oligosaccharide 12. Preautomation phase: preparation of building blocks, resin with linker and the synthesizer. Automation phase: AGA using the Glyconeer 2.1 synthesizer. Postautomation phase: analysis and purification of fully protected oligosaccharide 9, procurement of the conjugation-ready oligosaccharide 12, and quality control using IM-MS. (C) Protected oligosaccharides 10 and 11 are synthesized by selecting from a set of building blocks. AGA protocols and postautomation steps are performed to yield the conjugation-ready oligosaccharides 13 and 14.
Results and Discussion
Design and Preparation of Automated Oligosaccharide Synthesis Process.
The standardized AGA process is composed of the preautomation, automation, and postautomation phases (Fig. 1A). The preautomation phase includes preparation of the Glyconeer 2.1 by loading the reaction vessel with a functionalized polystyrene resin and the loading of vials with the appropriate building blocks that are needed for the target oligosaccharide sequence. During the automation phase, the oligosaccharide is assembled as the Glyconeer 2.1 executes computer-controlled reaction cycles. The postautomation phase commences with cleavage of the resin-bound oligosaccharide to release protected products that are analyzed and purified by normal-phase HPLC. Finally, removal of the remaining protecting groups, a final purification step, and confirmation of the structure via NMR and IM-MS provide the target molecules as conjugation-ready oligosaccharides.
The Glyconeer 2.1 synthesizer (SI Appendix, Fig. S1) is designed with flexible hardware and software to accommodate the complexities of glycan assembly, where it is essential that both the stereo- and regiochemistry of the newly formed glycosidic bond must be controlled during each addition to the growing oligosaccharide. The reaction temperatures can be widely varied, sensitive and/or corrosive reagents can be used, and coupling cycles can be adjusted as needed. For fast and accurate reagent delivery, the pressure-driven system relies upon argon to push solvents and reagents through valves and tubes that are inert to volatile and corrosive reagents. Moisture is excluded from the reaction mixture by keeping the entire synthesizer under an argon atmosphere, thereby minimizing decomposition of activated building blocks via hydrolysis. Synthesis-ready building blocks are preweighed and either dissolved in the appropriate anhydrous solvent or kept as solids and placed in sealed vials on a 64-position carousel. “Approved” building blocks are those that can be synthesized in large quantities, are stable upon storage for extended periods of time, and upon activation result in very high coupling yields with excellent stereocontrol (SI Appendix, Fig. S2). To make the method more broadly accessible and to reduce the time for oligosaccharide preparation, a number of “approved building blocks” that can be used on the synthesizer are now commercially available from GlycoUniverse, the company that also sells the instrument. Solvents and reagents are placed in various sizes of glass bottles and kept under argon pressure. To decrease decomposition, the Glyconeer 2.1 also offers the relative flexibility to store bottles that contain sensitive reagent mixtures at 4 °C. A polystyrene-based resin functionalized with an appropriate linker is loaded into a triple-jacketed glass reaction vessel and can be mixed by a flow of argon from the bottom of the reactor. Building-block–specific cycles are then loaded by the operator in the Glyconeer 2.1 software environment to be executed during the synthesis to provide the desired structure (the detailed setup of the synthesizer is described in the SI Appendix).
The completion of one synthetic cycle, which extends the growing oligosaccharide chain by one unit, relies upon a successful glycosylation reaction followed by the removal of a specific temporary protecting group that reveals the nucleophile for glycosylation in the following cycle (for detailed module settings, see SI Appendix). Following glycosylation, the resin is washed free of excess reagents using different solvents. Thereafter, the removal of the desired temporary protecting group can be accomplished using a solution of 20% triethylamine (TEA) in N,N-dimethylformamide (DMF) for the removal of the 9-fluorenylmethoxycarbonyl (Fmoc) group (Fig. 2 B and D). The deprotection solution is subsequently drained from the reaction vessel through a UV-based flow cell for quantitation of the efficiency of glycosylation. In this manner, poor couplings can be detected so that the synthesis may be interrupted, thus avoiding the loss of valuable building blocks.
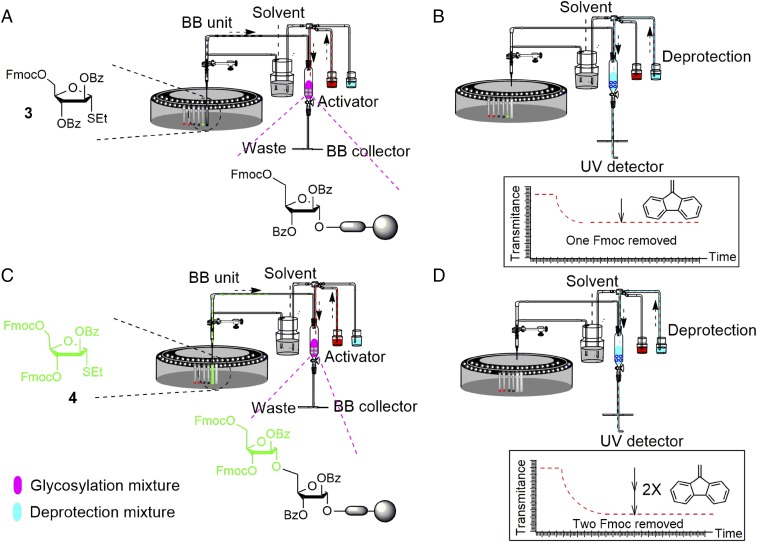
Fig. 2.
Delivery of reagents during glycosylation and removal of temporary protecting group during AGA of arabinomannan hexasaccharide 9. (A and C) Glycosylations: transparent tubes with colored bottoms represent building blocks (BB) stored in sealed vials before use, fully colored tubes represent dissolved building blocks, fully gray tubes represent empty vials after building-block delivery, red-colored bottles represents activator, and pink RV represents the activated glycosylation mixture. Below the reactor, the glycosylation product is shown. (B and D) Deprotection: light blue bottle represents the deprotection reagent; blue-colored RV represents the deprotection step. Dibenzofulvene formation as a result of Fmoc cleavage decreases UV transmittance as a quantitative measure of Fmoc release from the resin-bound glycan.
At the beginning of each glycosylation cycle, the reaction vessel (RV) is washed with anhydrous solvent and trimethylsilyl trifluoromethanesulfonate to quench any base left over from previous steps that would interfere with acid-mediated activation. After washing the resin, a fresh portion of anhydrous solvent is added to the reaction vessel while the RV cools to the desired temperature (Fig. 2 A and C). After purging the washing solvent from the RV, the building-block solution is transferred to the cooled RV and constant argon bubbling is maintained. Adjustment of the RV to the activation temperature is crucial for glycosylation; hence, the activator solution is delivered only after the RV temperature reaches the desired level (Fig. 2 A and C). Specific to each building block, the temperature is maintained throughout the glycosylation reaction, or may be gradually elevated to drive glycosylation to completion. After completing each glycosylation cycle, the reaction mixture is purged from the RV and transferred to the fraction collector, where it may be quenched using an appropriate base. This optional step allows the user to recover any hydrolyzed, excess building block for possible regeneration of the preactivated species. Analysis of the quenched building-block solution provides information regarding activation of a building block and therefore important feedback on unreactive building blocks and/or inefficient coupling conditions.
Automated Synthesis of Selected Oligosaccharide Motifs.
The streamlined AGA process is illustrated for the assembly of oligosaccharides representing motifs of the Mycobacterium tuberculosis cell surface (12 and 13) (19), and cancer cells (14) (20). The three oligosaccharides constitute various degrees of complexity and each requires several different building blocks. For example, linear hexamer 12 contains multiple 1,2-trans linkages, hexasaccharide 13 has a branched structure with 1,2-trans linkages, and trisaccharide 14 contains both 1,2-trans and 1,2-cis linkages. To synthesize hexamer 12, the Glyconeer 2.1 instrument was loaded with a polystyrene resin functionalized with a photolabile linker (1) (12, 21), and a set of approved monosaccharide building blocks 2–8 (Fig. 1B). This linear arabinomannan hexasaccharide was then assembled using thioglycoside building blocks 2 and 3. Activation of these thioglycosides was initiated by addition of a solution of N-iodosuccinimide and trifluoromethanesulfonic acid in a dichloromethane/dioxane mixture (vol/vol, 9:1). The reaction mixture was kept at an initial temperature of −40 °C for 5 min and then allowed to gradually warm to −10 °C over 25 min. Each glycosylation reaction was performed twice using five equivalents of a building block for each reaction. After removing the glycosylation mixture from the RV and washing the support, Fmoc removal was accomplished by treatment with TEA (20% in DMF). These glycosylation and deprotection cycles were repeated six times to assemble resin-bound hexasaccharide 9 (Fig. 1B and SI Appendix, Table S1).
Following automated assembly, the products were cleaved from the resin by UV irradiation in a continuous-flow reactor (12, 21). Fully protected hexasaccharide 9 (25.3 mg, 9.47 µmol, 38% overall yield based on resin loading, calc. 93% per coupling step: 12 on-resin steps) was obtained after preparative normal-phase HPLC (NP-HPLC) purification and was characterized by NMR and mass spectrometry (SI Appendix, Figs. S4, S16, and S17).
Branched hexasaccharide 13 and linear trisaccharide 14 (Fig. 1C) were assembled using building blocks 3–5 and 6–8, respectively (for detailed modules and sequence programs, see SI Appendix, Tables S2 and S3). Fully protected branched hexasaccharide 10 (15.1 mg, 5.88 µmol, 24% overall yield based on resin loading, 83% per step: 18 steps) and trisaccharide 11 (10.4 mg, 6.24 µmol, 25% overall yield based on resin loading, 80% per step: 6 steps) were obtained in pure form following preparative NP-HPLC to remove both deletion sequences and undesired stereoisomers (SI Appendix, Figs. S5, S6, and S18–S22).
All remaining protecting groups were removed from oligosaccharides 9–11 via methanolysis using sodium methoxide and subsequent hydrogenolysis using palladium on carbon and hydrogen gas to remove benzoyl ester and benzyl ether protecting groups, respectively (the full synthetic process is described in the SI Appendix, Fig. S3). Purification by semipreparative HPLC using a Hypercarb column furnished conjugation-ready linear hexasaccharide 12 (5.3 mg, 5.37 µmol, 56% over two steps), branched hexasaccharide 13 (3.1 mg, 3.24 µmol, 55% over two steps), and trisaccharide 14 (1.7 mg, 2.88 µmol, 46% over two steps). Compounds 12–14 were analyzed using Hypercarb HPLC (SI Appendix, Figs. S7–S9). NMR was used to confirm the identity the stereochemistry of the products (SI Appendix, Figs. S23–S28), whereas IM-MS–based quality control of oligosaccharides 12–14 revealed purities above 99% (SI Appendix, Figs. S10–S13).
Iterative Analysis and Feedback as Integral Part of Automated Oligosaccharide Synthesis.
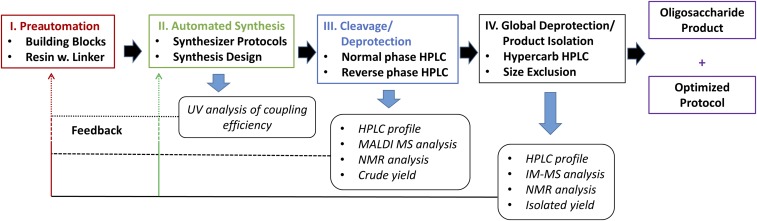
Direct feedback during organic synthesis in solution is straightforward. Spectral, chromatographic, and other analytical methods have been developed to provide information that is used to estimate the reaction progress and selectivity of each step. While developing streamlined AGA, a systematic approach to optimize building blocks and reaction conditions for reproducible glycan synthesis evolved (Fig. 3). This rational approach is based upon an ability to detect the steps that require optimization. This leads to appropriate modifications of building blocks, the automated cycle conditions, or both. The protocol and decision tree (Fig. 3) aims to provide maximum information for each step of the synthesis to establish a reproducible and optimized AGA protocol. During automated solid-phase synthesis, UV quantification after the Fmoc deprotection step provides crucial feedback regarding an estimation of glycosylation yields based on the release of temporary protecting groups. A sharp increase in UV transmission indicates failed glycosylation or deprotection events. The instrument stops the assembly process following UV feedback that falls below a threshold set by the operator to conserve valuable building blocks, as is common during peptide syntheses. To obtain feedback concerning stereo- or regioselectivity of coupling reactions, a more detailed analysis following completion of the automated assembly process is performed. Crude, semiprotected glycan products, which have been cleaved from the resin, are analyzed by HPLC, MS, and NMR. The HPLC analysis assesses the amount of product compared with shorter oligosaccharide by-products. The stereoselectivity of the reactions relies on an NMR analysis of the isolated compound(s) and together with the UV feedback and the HPLC analyses identifies any steps that may require further optimization. The final stage of the process removes remaining protecting groups from the oligosaccharides. MS analysis is then used to confirm the removal of these groups and NMR analysis is used to confirm the structure of the conjugation-ready oligosaccharide. For a more sensitive quality control, IM-MS is used to differentiate between oligosaccharide stereoisomers down to a level of 0.2% and to sequence the oligosaccharide (SI Appendix, Figs. S10–S13) (18). The overall success of the synthesis provides feedback on the optimization process such that an AGA strategy based on approved building blocks, improved automated protocols, and standardized purification steps evolved.
Fig. 3.
Flowchart of the standardized AGA process including purification and process feedback. Full forward arrows reflect the AGA process, synthesis (black) and analysis (blue), while thin backward arrows represent the various feedback levels. (I) Preautomated step is designed with standard building blocks, linker, and module system cycles (red box). (II) Automated synthesis with UV analysis provides the first-level feedback that reflects mostly on the success of glycosylation conversions (green box). (III) Following cleavage and partial deprotection, HPLC, MS, and NMR analysis of the crude products provides a second-level feedback reflecting on the conversion success and on the stereo- and regioselectivity (blue box). (IV) After global deprotection HPLC, IM-MS, and NMR analysis provides third-level feedback that reflects on the overall process including isolated yields (black box). Using the combination of the three levels of feedback, an optimized protocol is produced in minimum effort to allow for the reproducible synthesis of a single-entity oligosaccharide (magenta boxes).
Even though the AGA method has advanced to the point of commercialization, further improvements will accelerate and improve the process to procure glycans even quicker and cheaper. The insertion of a capping step that is common during oligonucleotide assembly will ensure that deletion sequences cannot accumulate and simplifies purification. Further streamlining and optimization of all washing steps, the cooling of the reaction vessel, and all other manipulations will accelerate the syntheses and reduce the amount of building blocks and solvents used. The application of the instrument to procure ever more diverse glycans will be aided by the number of building blocks that are now becoming commercially available. Importantly, increasing adaptation of the technology will generate the demand for diverse structures and more building blocks. Increasing demand is expected to lower the cost per molecule due to the economy of scale, as was the case for oligonucleotide and peptide syntheses.
Conclusion
The streamlined automated glycan assembly using a commercially available instrument (Glyconeer 2.1), resin with linkers, and a set of approved commercially available building blocks enables the synthesis of variety of oligosaccharides. The analysis and feedback process including quality control using NMR and IM-MS guarantee the purity of the ready-to-use oligosaccharides. The process described here enables access to large collections of tailor-made glycans for biological, biochemical, vaccinology, and materials research as well as many other applications.
Materials and Methods
All building blocks were synthesized according to previously described procedures or as described in the SI Appendix. The building blocks were dried before use and transferred to the designated vials and either kept dry or predissolved depending on their solubility. Building-block vials were placed on the synthesizer according to the order of addition. The solvents and reagents were refilled or freshly prepared according to need and placed in the designated position on the synthesizer. The automated synthesis modules and commands were organized according to the oligosaccharide structure. The automated synthesis run was monitored by measuring the UV transmittance after each deprotection step. After the synthesis, the protected oligosaccharides were cleaved from the solid support by irradiation and the crude oligosaccharides were purified by NP-HPLC and analyzed by NMR, MS, and RP-HPLC to confirm their structure and purity. The permanent protecting groups were removed as a last step to provide a conjugation-ready oligosaccharides that were analyzed using NMR and IM-MS.
Detailed information on the methods is provided in the SI Appendix.
Supplementary Material
Acknowledgments
We are grateful to the Max Planck Society and the European Research Council (ERC Advanced Grant AUTOHEPARIN to P.H.S.) for generous financial support. M.H. thanks the Max Planck Society for the Minerva Postdoctoral Fellowship. J.H. and K.P. thank the Free University Berlin for generous financial support.
Footnotes
Conflict of interest statement: P.H.S. declares a significant financial interest in GlycoUniverse GmbH & Co. KGaA, the company that commercializes the synthesis instrument, building blocks, and other reagents.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700141114/-/DCSupplemental.
References
- 1.Lashkari DA, Hunicke-Smith SP, Norgren RM, Davis RW, Brennan T. An automated multiplex oligonucleotide synthesizer: Development of high-throughput, low-cost DNA synthesis. Proc Natl Acad Sci USA. 1995;92:7912–7915. doi: 10.1073/pnas.92.17.7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mäde V, Els-Heindl S, Beck-Sickinger AG. Automated solid-phase peptide synthesis to obtain therapeutic peptides. Beilstein J Org Chem. 2014;10:1197–1212. doi: 10.3762/bjoc.10.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Domon B, Aebersold R. Mass spectrometry and protein analysis. Science. 2006;312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- 4.Venter JC, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 5.Hsu CH, Hung SC, Wu CY, Wong CH. Toward automated oligosaccharide synthesis. Angew Chem Int Ed Engl. 2011;50:11872–11923. doi: 10.1002/anie.201100125. [DOI] [PubMed] [Google Scholar]
- 6.Seeberger PH. The logic of automated glycan assembly. Acc Chem Res. 2015;48:1450–1463. doi: 10.1021/ar5004362. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Ye XS, Zhang LH. Oligosaccharide assembly by one-pot multi-step strategy. Org Biomol Chem. 2007;5:2189–2200. doi: 10.1039/b704586g. [DOI] [PubMed] [Google Scholar]
- 8.Merrifield RB. Solid phase peptide synthesis. I. The synthesis of a tetrapeptide. J Am Chem Soc. 1963;85:2149–2154. [Google Scholar]
- 9.Matteucci MD, Caruthers MH. Nucleotide chemistry. 4. Synthesis of deoxyoligonucleotides on a polymer support. J Am Chem Soc. 1981;103:3185–3191. [PubMed] [Google Scholar]
- 10.Plante OJ, Palmacci ER, Seeberger PH. Automated solid-phase synthesis of oligosaccharides. Science. 2001;291:1523–1527. doi: 10.1126/science.1057324. [DOI] [PubMed] [Google Scholar]
- 11.Calin O, Eller S, Seeberger PH. Automated polysaccharide synthesis: Assembly of a 30mer mannoside. Angew Chem Int Ed Engl. 2013;52:5862–5865. doi: 10.1002/anie.201210176. [DOI] [PubMed] [Google Scholar]
- 12.Eller S, Collot M, Yin J, Hahm HS, Seeberger PH. Automated solid-phase synthesis of chondroitin sulfate glycosaminoglycans. Angew Chem Int Ed Engl. 2013;52:5858–5861. doi: 10.1002/anie.201210132. [DOI] [PubMed] [Google Scholar]
- 13.Hahm HS, Hurevich M, Seeberger PH. Automated assembly of oligosaccharides containing multiple cis-glycosidic linkages. Nat Commun. 2016;7:12482. doi: 10.1038/ncomms12482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahm HS, et al. Automated glycan assembly of complex oligosaccharides related to blood group determinants. J Org Chem. 2016;81:5866–5877. doi: 10.1021/acs.joc.6b00554. [DOI] [PubMed] [Google Scholar]
- 15.Kandasamy J, Hurevich M, Seeberger PH. Automated solid phase synthesis of oligoarabinofuranosides. Chem Commun (Camb) 2013;49:4453–4455. doi: 10.1039/c3cc00042g. [DOI] [PubMed] [Google Scholar]
- 16.Kröck L, et al. Streamlined access to conjugation-ready glycans by automated synthesis. Chem Sci (Camb) 2012;3:1617–1622. [Google Scholar]
- 17.Walvoort MTC, et al. Automated solid-phase synthesis of β-mannuronic acid alginates. Angew Chem Int Ed Engl. 2012;51:4393–4396. doi: 10.1002/anie.201108744. [DOI] [PubMed] [Google Scholar]
- 18.Hofmann J, Hahm HS, Seeberger PH, Pagel K. Identification of carbohydrate anomers using ion mobility-mass spectrometry. Nature. 2015;526:241–244. doi: 10.1038/nature15388. [DOI] [PubMed] [Google Scholar]
- 19.Brennan PJ, Nikaido H. The envelope of mycobacteria. Annu Rev Biochem. 1995;64:29–63. doi: 10.1146/annurev.bi.64.070195.000333. [DOI] [PubMed] [Google Scholar]
- 20.Galili U, Chen ZC, DeGeest K. Expression of alpha-gal epitopes on ovarian carcinoma membranes to be used as a novel autologous tumor vaccine. Gynecol Oncol. 2003;90:100–108. doi: 10.1016/s0090-8258(03)00148-3. [DOI] [PubMed] [Google Scholar]
- 21.Hurevich M, et al. Continuous photochemical cleavage of linkers for solid-phase synthesis. Org Lett. 2014;16:1794–1797. doi: 10.1021/ol500530q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.