Significance
New technologies have fostered renewed interest in bacterial epigenetics, with DNA modifications defending against other microbes and controlling gene expression. Here, we describe how two diverse modifications, adenine methylation (6mA) and phosphorothioation (PT), have evolved to occupy the same genomic sites in bacteria. Using mass spectrometry, single-molecule real-time sequencing, and genetic engineering, we discovered coincident PT and 6mA in GPS6mAAC and GPS6mATC, representing PT consensus sequences, a GA-containing consensus for an unknown methyltransferase, and the GATC consensus for Dam methyltransferase. 6mA was found in all GPSATC sites in vivo, with 6mA substituting for PT in preventing DNA cleavage by PT restriction enzymes. These findings show how different DNA modification systems have evolved to avoid conflict.
Keywords: DNA phosphorothioation, DNA methylation, epigenetics, restriction–modification, bioanalytical chemistry
Abstract
Explosive growth in the study of microbial epigenetics has revealed a diversity of chemical structures and biological functions of DNA modifications in restriction–modification (R-M) and basic genetic processes. Here, we describe the discovery of shared consensus sequences for two seemingly unrelated DNA modification systems, 6mA methylation and phosphorothioation (PT), in which sulfur replaces a nonbridging oxygen in the DNA backbone. Mass spectrometric analysis of DNA from Escherichia coli B7A and Salmonella enterica serovar Cerro 87, strains possessing PT-based R-M genes, revealed d(GPS6mA) dinucleotides in the GPS6mAAC consensus representing ∼5% of the 1,100 to 1,300 PT-modified d(GPSA) motifs per genome, with 6mA arising from a yet-to-be-identified methyltransferase. To further explore PT and 6mA in another consensus sequence, GPS6mATC, we engineered a strain of E. coli HST04 to express Dnd genes from Hahella chejuensis KCTC2396 (PT in GPSATC) and Dam methyltransferase from E. coli DH10B (6mA in G6mATC). Based on this model, in vitro studies revealed reduced Dam activity in GPSATC-containing oligonucleotides whereas single-molecule real-time sequencing of HST04 DNA revealed 6mA in all 2,058 GPSATC sites (5% of 37,698 total GATC sites). This model system also revealed temperature-sensitive restriction by DndFGH in KCTC2396 and B7A, which was exploited to discover that 6mA can substitute for PT to confer resistance to restriction by the DndFGH system. These results point to complex but unappreciated interactions between DNA modification systems and raise the possibility of coevolution of interacting systems to facilitate the function of each.
The emergence of convergent technologies has led to a growing appreciation for the diversity of DNA modifications in microbial epigenetics and restriction–modification (R-M) systems (1–3). DNA methylation, the most extensively studied genetic modification, was originally discovered in bacteria in the context of R-M systems involving a methyltransferase (MTase) that modifies “self” DNA at specific target sites and a cognate restriction endonuclease (REase) that discriminates and destroys unmodified invading DNA (3–5). R-M systems are ubiquitous in prokaryotes and are classified into four types (I, II, III, and IV) based on their molecular structure, sequence recognition, cleavage position, and cofactor requirements (3, 6, 7). However, some MTases exist alone, without an apparent cognate REase partner. These so-called “orphan” MTases include DNA adenine methylase (Dam), which modifies the adenine N-6 in the GATC motif, DNA cytosine methylase (Dcm), which methylates C-5 of the second cytosine in CC(A/T)GG sequences, and cell cycle-regulated methylase (CcrM), which methylates the N-6 of adenine in GANTC (N = A, T, C, or G) (8). Despite the absence of cognate REases, orphan MTases still confer immunity against the virulence of a parasitic R-M complex with the same target sites (9). In addition to defense against bacteriophages and transposons, DNA methylation is involved in various cellular functions, including chromosome replication, DNA mismatch repair, transcriptional regulation, transposition, and pathogenesis (1, 10, 11).
The chemical diversity of DNA modifications expanded with our discovery of phosphorothioates (PTs) as physiological modifications of the DNA sugar-phosphate backbone in which a nonbridging oxygen is replaced by sulfur (12). The PT system is more complicated than methylation and entails sequence-selective and RP-stereo–specific incorporation of sulfur by five Dnd proteins (12, 13). Since the original report in Streptomyces lividans, the five-member dndABCDE gene cluster has been found in diverse, taxonomically unrelated bacteria and archaea (14, 15). dndA can be located either adjacent to clustered dndBCDE or elsewhere in the genome, consistent with the function of DndA as a cysteine desulfurase; DndA can also be functionally replaced by an iron–sulfur cluster assembly protein (IscS) homolog in some bacterial strains (16). DndB binds to the promoter of the dnd operon to regulate dndBCDE transcription and affect PT frequency (17, 18). DndC possesses ATP pyrophosphatase activity and shows significant homology to PAPS reductase family proteins (13, 16). By comparison, DndD has ATPase activity that is possibly related to DNA nicking during sulfur incorporation, and structural analysis suggests that DndE binds to nicked double-stranded DNA (13, 19, 20). In some strains, a divergently transcribed cluster, dndFGH, occurs in the vicinity of the dndBCDE operon. Without PT protection, unrestrained DndFGH restriction activity leads to double-stranded DNA damage that triggers the SOS response, cell filamentation, and prophage induction (21, 22). Dnd is thus regarded as an unusual bacterial R-M system.
The complexities of different DNA modification systems have begun to come to light with the application of new technologies. For example, the application of liquid chromatography-coupled tandem quadrupole mass spectrometry (LC-MS/MS) to quantify nuclease-resistant PT-modified dinucleotides has revealed significant diversity in the quantity, sequence setting, and bacterial distribution of PT modifications, such as the d(GPSA) and d(GPST) motifs in 1:1 proportions in the enteric pathogen Escherichia coli B7A (15). Subsequent application of single-molecule, real-time (SMRT) sequencing technology to map genomic PT sites in E. coli B7A revealed that these d(GPSA) and d(GPST) motifs are located within conserved 4-bp complementary GPSAAC/GPSTTC contexts, with no apparent wider consensus sequence (23). However, only 12% of the 40,701 possible GAAC/GTTC sites on the chromosome were found to be PT-modified, even in the presence of active DndFGH (23). These unique features differentiate dndBCDE-dndFGH from typical methylation-based R-M systems and suggest that it represents a type of bacterial defense system with highly unusual target selection by Dnd proteins.
Despite having similar functional roles, DNA methylation and PT modification are considered to be two R-M defense systems that have evolved independently in prokaryotes. Here, we report the discovery of an intriguing collision of the two modification systems, which were found to share DNA substrates to generate a hybrid d(GPS6mA) modification motif. This commonality is associated with significant rules of interaction and interference: sulfur replacement on the DNA backbone remarkably inhibits methylation, and methylation can substitute for PT to confer resistance to restriction by DndFGH. This interaction between two distinct endogenous DNA modification systems suggests that such “conflict” drives the evolution of different consensus sequences to avoid interference and ensure the protective function of each.
Results
Discovery of the Doubly Modified d(GPS6mA) Consensus in Bacteria.
PT modifications occur sequence-selectively in DNA, with all 16 possible dinucleotide contexts represented in archaebacterial and eubacterial organisms (15). The PT context of d(GPSA), which occurs in E. coli B7A and Salmonella enterica serovar Cerro 87 (15), drew our attention because adenine is the substrate of N6-adenine–specific methyltransferases (MTases). This enzyme class catalyzes the 6mA modification widespread in R-M systems, such as M.TaqI, M.ClaI, M.EcoBI, and M.EcoRI, as well as solitary Dam and CcrM MTases. Adenine MTases modify a variety of sequence contexts such as TCGA, ATCGAT, TGA(N)8TGCT, GAATTC, GATC and GANTC (underlined bases indicate methylation). What was notable here is the coincidence of recognition sequences of some adenine MTases with those for PT modifications, such as TGA(N)8TGCT for M.EcoBI and others (24) and GAAC/GTTC for Dnd enzymes.
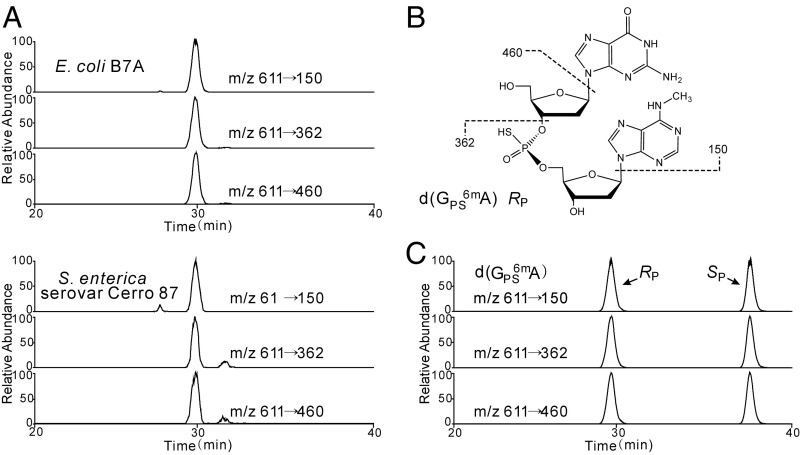
These coincidences prompted us to search for PT and 6mA in the same sequence motifs. Here, we searched first for d(GPS6mA) dinucleotides in E. coli B7A and S. enterica serovar Cerro 87 possessing d(GPSA) motifs, using an LC-MS/MS method that exploits the nuclease resistance of PT-containing dinucleotides. Genomic DNA is hydrolyzed and persistent PT-containing dinucleotides are resolved from the bulk 2′-deoxyribonucleosides by HPLC, which allows sensitive MS identification and detection of the dinucleotides. Systematic MS scanning of the HPLC eluent of hydrolyzed genomic DNA from these bacteria revealed a molecule with m/z 611 (Fig. 1A). Collision-induced dissociation (CID) analysis yielded fragment ions with m/z values of 150, 362, and 460 (Fig. 1A), which are consistent with the structure d(GPS6mA) shown in Fig. 1B. This structure was validated in several ways. First, the m/z 611 species disappeared in DNA from PT-deficient mutant strains CX3 and XTG103 of E. coli B7A and S. enterica serovar Cerro 87, respectively, which points to the presence of a PT structure in the dinucleotide. Second, the CID fragmentation properties parallel those of d(GPSA), which is 14 atomic mass units (amu) lighter due to the absence of the N6-methyl group: a protonated molecular ion [M+H]+ m/z of 597 (vs. 611), a protonated adenine base (m/z 136 vs. 150), and other fragments (m/z 348, 446) lighter by 14 amu (12). Finally, the structure in Fig. 1B was confirmed by LC-MS/MS analysis of a synthetic d(GPS6mA) standard that possessed PT as a mixture of RP and SP stereochemistries (Fig. 1C), with RP as the naturally occurring configuration (Fig. 1A). These results established the cooccurrence of a PT and a methyl group in the same d(GA) context to yield a hybrid d(GPS6mA) in RP configuration.
Fig. 1.
LC-MS/MS analysis of isolated and synthetic d(GPS6mA). (A) Naturally occurring d(GPS6mA) RP in E. coli B7A (Upper) and S. enterica serovar Cerro 87 (Lower) was eluted at 29.5 min, identical to the retention time of synthetic d(GPS6mA) (C) in the RP configuration. (B) The major fragment ions of d(GPS6mA) observed in LC-MS/MS analysis are shown in the structural insert, with the protonated molecular ion [M+H] appearing at m/z 611.
We next performed quantitative analysis to establish the frequency of d(GPS6mA) in the bacterial genomes. LC-MS/MS analysis using calibration curves based on chemical standards revealed that, compared with the frequency of d(GPSA) at 301 ± 2 and 258 ± 17 per 106 nt, the methylated d(GPS6mA) occurred at much lower levels of 15 ± 1 and 2 ± 0 per 106 nt in E. coli B7A and S. enterica serovar Cerro 87, respectively. Interestingly, we are unaware of adenine MTases with GAAC/GTTC as the precise recognition sequence, so it is possible that the frequency of d(GPS6mA), low as it is, resulted from the overlap of GA in the consensus sequences of a MTase and the PT-inserting Dnd proteins in a subset of GAAC/GTTC sites. To further analyze the intersecting DNA modifications, we engineered bacteria with PT and methylation systems, as discussed next.
Interaction Between PT Modification and Methylation in Bacterial Genomes.
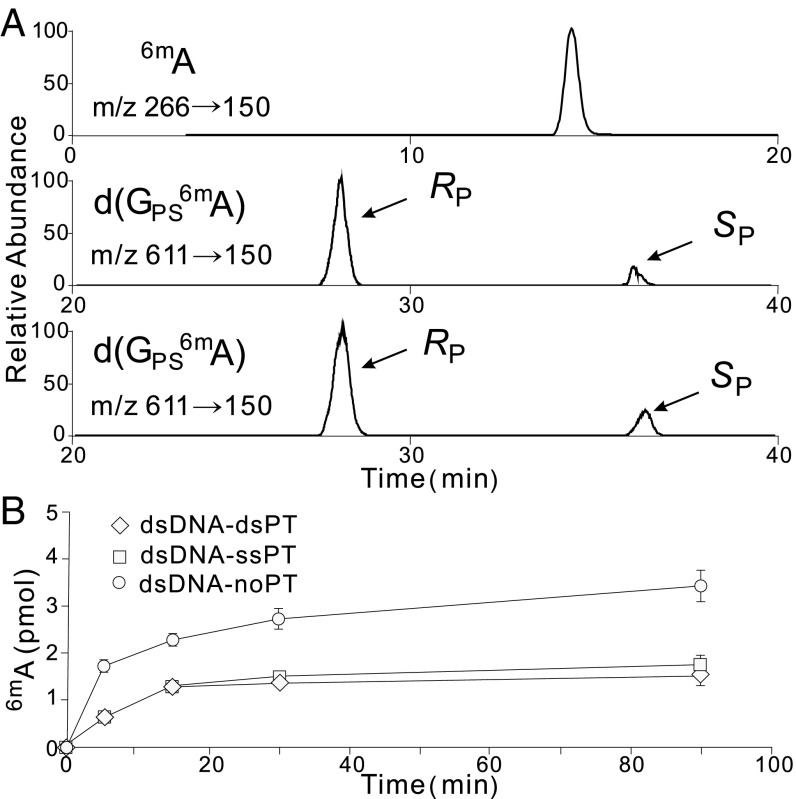
This coincidence of PT and 6mA modifications at the same sites in bacterial genomes raised questions about both the target selection mechanisms underlying the modification proximity and the implications of proximity for modification function. Here, we explored this issue with PTs and Dam-mediated methylation, both of which are considered to be postreplicative modifications, with hemimodified sites created by passage of the replication fork later fully modified (25, 26). We used the PT modification system from Hahella chejuensis KCTC2396 and dam from E. coli DH10B, both of which recognize GATC as a consensus sequence and generate GPSATC and G6mATC, respectively. To assess how Dam reacts with PT-modified DNA, oligonucleotide substrates harboring GATC or GPSATC (Table S1) were reacted with Dam and S-adenosylmethionine (SAM) in vitro. As shown in Fig. 2A, Dam is able to use GPSATC with both RP and SP stereoisomers of PT as a substrate to yield d(GPS6mA). However, a time course of the Dam methylation reaction measured by LC-MS/MS (Fig. 2B) revealed that both the rate and extent of Dam activity were significantly reduced in the presence of PTs, regardless of whether the substrate was single- or double-stranded. The sulfur substitution of GPSATC thus interfered with either recognition or catalysis by Dam in an in vitro setting.
Table S1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Characteristics | Source |
| Strains | ||
| Trans1-T1 | F− φ80(lacZ)ΔM15ΔlacX74hsdR(rk−mk+)ΔrecA 1398endA1 tonA | TransGen Biotech |
| E. coli HST04 | F-, ara, Δ(lac-proAB) [Φ80d lacZ ΔM15], rpsL(str), thi, Δ(mrr-hsdRMS-mcrBC), ΔmcrA, dam, dcm | TAKARA |
| E. coli B7A | WT, d(GPSA), d(GPST), d(GPS6mA) | (40) |
| CX2 | B7A derivative, dndFGH in-frame deletion mutant | This work |
| WXY-1 | B7A derivative, dndBCDE in-frame deletion mutant | This work |
| CX3 | B7A derivative, dndBCDEFGH in-frame deletion mutant | This work |
| S. enterica serovar Cerro 87 | WT, d(GPSA), d(GPST), d(GPS6mA) | (41) |
| XTG103 | S. enterica derivative, dndBCDEFGH deletion mutant | (31) |
| H. chejuensis KCTC2396 | d(GPSA) | (42) |
| Plasmids | ||
| pEASY-Blunt zero | Cloning vector, Ampr, Kanr | TransGen Biotech |
| pBluescript II SK(+) | Cloning vector, Ampr | (43) |
| pKOV-Kan | Shuttle vector, Kanr | (44) |
| pACYC184 | Cloning vector, Cmr | (45) |
| pACYCDuet-1 | Cloning vector, Cmr | Novagen |
| pWHU701 | dndBCDE from H. chejuensis KCTC2396, cloned in SK(+) | This work |
| pWHU702 | dam from E. coli DH10B, cloned in pACYC184 | This work |
| pWHU712 | dam from E. coli DH10B, cloned in SK(+) | This work |
| pWHU705 | dndFGH from H. chejuensis KCTC2396, cloned in pACYCDuet-1 | This work |
| pWHU623 | pKOV-Kan derivative for CX2 construction | This work |
| pWHU624 | pKOV-Kan derivative for CX3 construction | This work |
| pWHU1835 | pKOV-Kan derivative for WXY-1 construction | This work |
| Primers | ||
| Mutant construction | ||
| CX2-LL | GTCGACCTCTGGCAATCCTCAAAC | |
| CX2-LR | GTCAATGTTAAATACCGCCAGATAACGCTGTAACTCTTC | |
| CX2-RL | GAAGAGTTACAGCGTTATCTGGCGGTATTTAACATTGAC | |
| CX2-RR | GGATCCATGGTATGCCATCCTTTG | |
| CX3-LL | GTCGACACGGTTGCTCTGGGTTAT | |
| CX3-LR | AATCCCATCCTCAACATGGATGCTGCTCAACGTAAAT | |
| CX3-RL | ATTTACGTTGAGCAGCATCCATGTTGAGGATGGGATT | |
| CX3-RR | GGATCCTATAAAGATAGCTCTGGC | |
| WXY-1-LL | GGATCCAAGTGAAGGCAGACGG | |
| WXY-1-LR | GATTTCTCAATGCCGCTTTCGGGATGATACGC | |
| WXY-1-RL | GCGTATCATCCCGAAAGCGGCATTGAGAAATC | |
| WXY-1-RR | GTCGACGCTCTGGCCTGAACAT | |
| Heterologous expression | ||
| dndBCDE-1 | GGGGTACCAGCCATCACCACTCGTAAC | |
| dndBCDE-2 | CGGGATCCTGATCTGACAGTAACCTCCATC | |
| dam-SK-1 | GCTCTAGAATGGTCAGCCGTGGTATG | |
| dam-SK-2 | GGAATTCCGTCAGATTGGGAACATAG | |
| dam-184-1 | CCCAAGCTTCTCTAACTACGACAACCTGAAC | |
| dam-184-2 | GCTCTAGATCGGAGGCTTCTGGATGA | |
| dndFGH-L-1 | CGAGCTCATGAGTCCGTAATAAAGGG | |
| dndFGH-L-2 | AGAATGTGAGATGTGCGTAG | |
| dndFGH-R-1 | GTCGTTACATTCATGTAAACC | |
| dndFGH-R-2 | ATAAGAATGCGGCCGCATGGAGGTTACTGTCAGATC | |
| Real-time qRT-PCR | ||
| gapA-1 | CACGCTACTACCGCTACTCA | |
| gapA-2 | AGGACGGGATGATGTTCTGG | |
| B7A-dndFG-1 | AGCGTCGTCAACAATTATGCA | |
| B7A-dndFG-2 | TGTTTGCCACATGATTCCAATAA | |
| B7A-dndGH-1 | GCGTGATAAAGCATTCTGGGA | |
| B7A-dndGH-2 | CCTCTGGAAATCGGGTTGGA | |
| HST04-dndF-1 | GCGGCGTACCCAGAAAATAG | |
| HST04-dndF-2 | TAACGCGCCTGATACTCGAT | |
| HST04-dndGH-1 | AGGAAATCTCGAGCGGATGA | |
| HST04-dndGH-2 | CCGCCAACCAAGTTTTCAAG | |
| In vitro assay | ||
| dsDNA-noPT | ATGAGGTCAGTCTGGGATCAAGTCGATGCTGCGCAGCATCGACTTGATCCCAGACTGACCTCAT | |
| dsDNA-ssPT | ATGAGGTCAGTCTGGGPSATCAAGTCGATGCTGCGCAGCATCGACTTGATCCCAGACTGACCTCAT | |
| dsDNA-dsPT | ATGAGGTCAGTCTGGGPSATCAAGTCGATGCTGCGCAGCATCGACTTGPSATCCCAGACTGACCTCAT |
Underlined bases indicate the recognition motif of Dam methyltransferase.
Fig. 2.
Dam enzymatic activity with different oligonucleotide substrates. (A) LC-MS/MS analysis of the Dam-methylated products using duplex oligonucleotides possessing normal GATC (dsDNA-noPT), single-stranded GPSATC (dsDNA-ssPT), and double-stranded GPSATC (dsDNA-dsPT) as substrates. The oligonucleotides are listed in Table S1. Dam recognizes GPSATC in both RP and SP configurations. (B) Time course of the Dam enzymatic reaction with dsDNA-noPT, dsDNA-ssPT, and dsDNA-dsPT; 4 units of Dam were incubated with 20 pmol of a DNA substrate in 10 µL of 1× dam methyltransferase buffer supplemented with 80 µM SAM for 0 to 90 min at 37 °C, followed by inactivation at 65 °C for another 15 min. 6mA was measured by LC-MS/MS at the indicated times (SI Materials and Methods).
We next assessed the interaction of Dam and the PT-modifying enzymes DndBCDE in vivo. Here, we used E. coli HST04, which lacks dam, dcm, and hsd MTases, as well as PT modification genes, as a host for vectors expressing dnd genes B, C, D, and E (dndBCDE) and dam genes (pWHU701 and pWHU702, respectively); E. coli HST04 possesses the iscS gene that substitutes for dndA. LC-MS/MS analysis revealed the formation of d(GPS6mA) in exponentially growing E. coli HST04(dndBCDE, dam) at a frequency of 219 ± 2 per 106 nt, compared with 228 ± 4 d(GPSA) per 106 nt in E. coli HST04(dndBCDE) lacking dam (Table S2). This finding stands in contrast to the observation of PT inhibition of Dam activity in vitro and suggests either that Dam activity in vivo is not affected by the presence of a PT or that 6mA forms first followed by PT insertion, with Dnd proteins unaffected by 6mA. As described next, however, only a fraction of the 37,610 GATC sites containing 6mA also contained PT, which points to different target selection mechanisms by Dnd and Dam proteins.
Table S2.
PT modifications and hybrid modifications of DNA in bacteria
| Strains | Modifications per 106 nt | |
| d(GPSA) | d(GPS6mA) | |
| HST04(dndBCDE, pACYC184) | 228 ± 4 | — |
| HST04(dndBCDE, dam) | <0.4* | 219 ± 2 |
Values represent the mean ± SD for three analyses of 10 μg of bacterial DNA; a dash indicates that the dinucleotide was not detected.
A signal for the dinucleotide was detected but below the limit of quantification of ∼4 PT per 107 nt.
Comparative Genome Maps of PT and 6mA Sites by SMRT Sequencing.
The evidence for coincidence motivated an attempt to apply the SMRT sequencing platform to compare the locations of PT and 6mA across the genome of engineered E. coli HST04. SMRT sequencing of DNA modifications exploits the kinetic signatures, defined as pulse width and interpulse duration (IPD), arising when the DNA polymerase copies past the modification (27). Building on SMRT applications to nucleobase methylation (27), 5-hydroxymethylcytosine (28), and nucleobase damage (29), we recently demonstrated that SMRT sequencing is applicable to PT mapping, with identification of GPSAAC/GPSTTC and CPSCA in E. coli B7A and Vibrio cyclitrophicus FF75, respectively (23). Both 6mA and d(GPSA) modifications of template DNA generate kinetic signatures with regard to the A base. The discovery of the hybrid d(GPS6mA) prompted us to compare the maps of PT and 6mA across the E. coli HST04 genome.
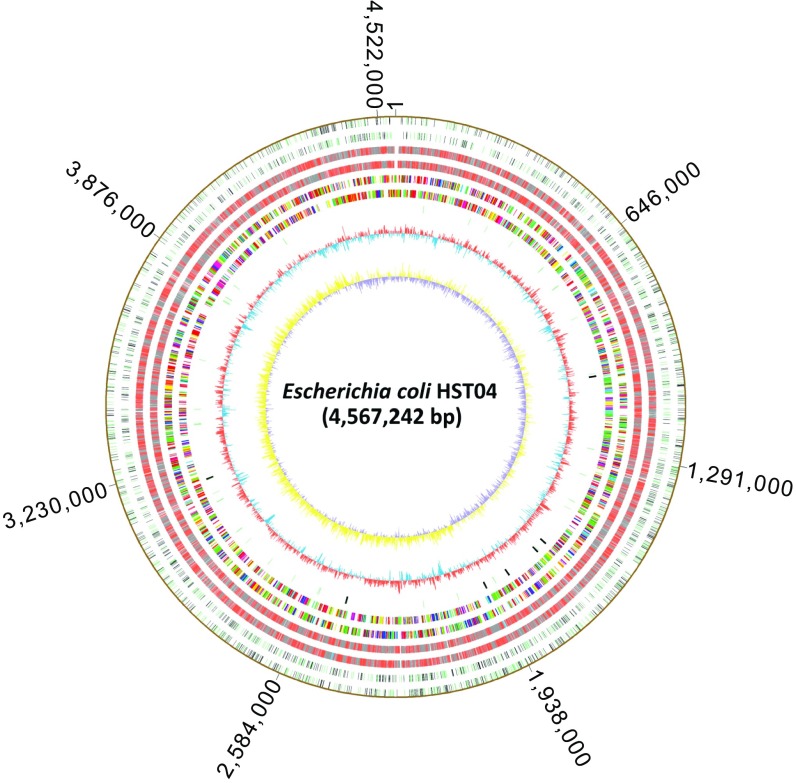
To this end, we used SMRT sequencing to map GPSATC in E. coli HST04(dndBCDE) and G6mATC and hybrid GPS6mATC in E. coli HST04(dndBCDE, dam). In the 4.57 × 106 bp in the E. coli HST04(dndBCDE) genome, 2,058 of the 37,698 GATC sites (5%) were found to be PT-modified (Fig. 3), which is consistent with the d(GPSA) quantitation by LC-MS/MS (228 ± 4 PT per 106 nt) (Table S2). Among the 2,058 GPSATC sites, 1,019 are located on the (+) strand and 1,039 on the (−) strand, with PT occurring on both strands of the same GATC site 517 times. The remaining 1,024 PT modification sites are single-stranded GpsATC sites.
Fig. 3.
PT and methylation site mapping across the E. coli HST04 genome. From outer to inner circles: Circles 1 and 2, (forward, reverse strands) 2,058 GPSATC sites in E. coli HST04 (dndBCDE) detected by SMRT in ORFs (black) and nonencoding regions (green). No site is detected in noncoding RNA. These sites were almost completely converted to methylation-PT double modification sites in E. coli HST04 (dndBCDE, dam). Circles 3 and 4, methylation sites E. coli HST04 (dndBCDE, dam) (excluding sites in circles 1 and 2) in ORFs (gray), noncoding RNA (blue), and nonencoding regions (red). Circles 5 and 6, predicted protein-coding sequences colored according to cluster of orthologous groups of proteins (COG) function categories. Circle 7, tRNA/rRNA operons. Circle 8, GC content. Circle 9, GC skew.
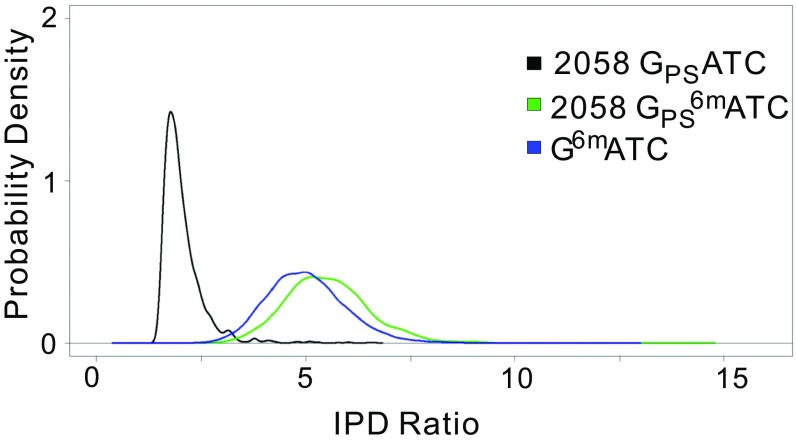
The map of G6mATC and GPS6mATC sites across the E. coli HST04(dndBCDE, dam) genome revealed modification of nearly 100% of all GATC sites (37,610 out of 37,698 GATC sites) (Fig. 3). However, the similarity of the SMRT sequencing IPD ratios for GPS6mATC and G6mATC made it difficult to distinguish between them in the genome modification maps in E. coli HST04(dndBCDE, dam). Knowing that nearly all of the 2,058 PT-modified GATC sites detected in E. coli HST04(dndBCDE) contained 6mA in the dam-expressing E. coli HST04(dndBCDE, dam), we can conclude that all GPSATC sites in E. coli HST04(dndBCDE, dam) also contain 6mA, which leads to the observation that IPD ratios for GPS6mATC sites were 2.8-fold larger than those for GPSATC, with 89% of the GPS6mATC sites having an IPD ratio ≥2× larger than GPSATC. Using a Gaussian distribution as the smoothing kernel, we estimated the probability density functions of the IPD ratio values for 2,058 GPSATC, 2,058 GPS6mATC, and 35,552 G6mATC on chromosomes. As shown in Fig. S1, the immediate observation was that the average IPD ratios of both GPS6mATC and G6mATC were larger than that of GPSATC and that the differences were significant (both P values < 0.0001). The IPD ratios of GPSATC were largely concentrated within a small range of [1.3, 3.5] and formed a sharp peak; in contrast, the GPS6mATC and G6mATC sites exhibited more widely ranging IPD ratios, and the corresponding probability density curves were flatter (Fig. S1). In addition, the GPS6mATC and G6mATC probability density curves were similar in shape: (i) both curves were symmetric about their means; (ii) the highest probability density of each did not differ much between the curves (0.405 for GPS6mATC and 0.437 for G6mATC); and (iii) the ranges covering the middle 90% IPD ratios for the curves did not differ much ([3.98, 7.32] for GPS6mATC and [3.58, 6.67] for G6mATC). Therefore, the similar IPD ratios of GPS6mATC and G6mATC made it difficult to distinguish between them, consistent with the SMRT result that the 2,058 GPS6mATC sites were grouped with the remaining 35,552 methylation G6mATC sites by the SMRT analysis portal (Fig. 3). We nonetheless conclude that PT and 6mA did not interfere with each other in terms of modification location or frequency in vivo, with normal target recognition behavior for proteins in both modification systems.
Fig. S1.
Estimated probability functions of IPD ratios for three types of modifications. GPSATC in E. coli HST04(dndBCDE), GPS6mATC, and G6mATC in E. coli HST04(dndBCDE, dam) are denoted by black, green, and blue, respectively.
Development of a Temperature-Dependent PT-Associated Restriction System.
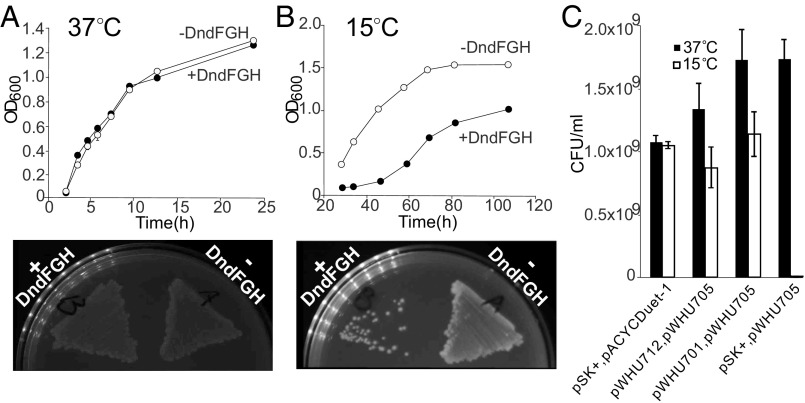
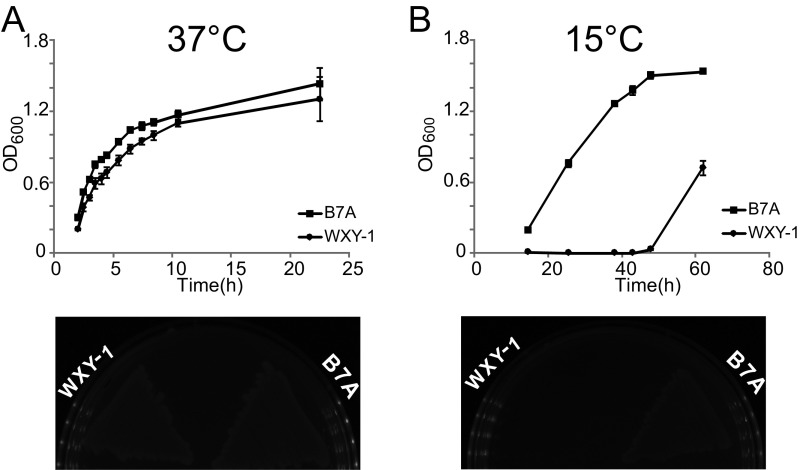
With evidence for proximal PT and 6mA modifications, we next sought to assess the effects of 6mA on PT-dependent restriction. Here, we engineered E. coli HST04 to express the DndFGH restriction proteins from H. chejuensis KCTC2396, which, as noted for H. chejuensis Dnd modification proteins described earlier, lacks Dam MTase and possesses PTs as d(GPS6mA) in the Dam methylation consensus GATC. To this end, we first cloned the dndFGH gene from H. chejuensis KCTC2396 into the low-copy plasmid pACYCDuet-1 to generate pWHU705. Without the protection of PT modifications, the DndFGH restriction endonuclease activity should be lethal or at least toxic to bacteria as a result of DNA damage and the SOS response (5, 21, 22). Surprisingly, we were able to prepare the pWHU705 plasmid, and the restriction proteins DndFGH exerted no apparent toxicity or growth inhibition in E. coli HST04 at 37 °C (Fig. 4A). However, a DndFGH-mediated bactericidal effect emerged when the cells were grown at 15 °C (Fig. 4B). Similar thermoregulated restriction was also observed for WXY-1, a dndBCDE-deficient mutant of E. coli B7A. Without PT protection, WXY-1 demonstrated a slight growth defect in liquid culture at 37 °C, but restriction became more evident when the cells were cultured at 15 °C (Fig. S2). To characterize this effect of growth temperature, we examined dndFGH transcription in E. coli HST04(pWHU705) and WXY-1 at different temperatures using real-time quantitative RT-PCR (qRT-PCR) with GAPDH as an internal control. Levels of dndFGH transcript increased significantly when cells growing at 37 °C were shifted to 15 °C for 1 h (Table S3). These results reveal that DndFGH restriction systems in E. coli B7A and H. chejuensis KCTC2396 are thermoregulated, with DndBCDE-DndFGH possibly functioning as an unusual temperature-dependent defense system. The thermoregulated behavior of DndFGH proved useful for studying the effects of 6mA on PT restriction activity.
Fig. 4.
Growth and cell viability analysis of bacterial strains. (A and B) Effect of pWHU705, expressing DndFGH of H. chejuensis KCTC2396, on the growth of E. coli HST04 at 37 °C (A) and 15 °C (B) in liquid LB broth (Upper) and on LB-agar plates (Lower). E. coli HST04 harboring empty pBluescript SK+ or pACYCDuet-1 plasmid was used as the control. (C) Cell viability assessment to compare protection mediated by DNA methylation and PT modification against DndFGH restriction. Exponentially growing cells at 37 °C were appropriately diluted and plated onto fresh LB-agar plates, which were incubated at 37 °C or 15 °C for cfu counting. Error bars indicate the SDs of three independent experiments.
Fig. S2.
Growth analysis of WT B7A and the in-frame deletion mutant. Growth was assessed at 37 °C (A) and 15 °C (B) both in LB broth (Upper) and on LB-agar plates (Lower).
Table S3.
Quantification of dndFGH transcription by real-time qRT-PCR
| Average CT ± SD | |||||
| Strains | dndGH | gapA | ΔCT ± SD | ΔΔCT ± SD | dndGH fold-change |
| HST04(pWHU705) | |||||
| 37 °C | 21.72 ± 0.08 | 19.07 ± 0.05 | 2.65 ± 0.04 | N/A | N/A |
| 15 °C | 22.89 ± 0.04 | 23.03 ± 0.00 | −0.15 ± 0.04 | −2.79 ± 0.05 | 6.7–7.2 |
| WXY-1 | |||||
| 37 °C | 25.36 ± 0.04 | 24.73 ± 0.05 | 3.77 ± 0.09 | N/A | N/A |
| 15 °C | 24.92 ± 0.01 | 24.21 ± 0.05 | 2.72 ± 0.03 | −1.05 ± 0.09 | 1.9–2.2 |
N/A, not applicable.
DNA Methylation Protects Against PT-Associated Restriction.
The temperature-sensitive DndFGH system in E. coli HST04 provided an opportunity to test the effects of 6mA on PT-dependent restriction. Here, we compared the bactericidal activity of DndFGH in the presence of PT or 6mA by quantifying viability [colony-forming units (cfu)] after exponentially growing cells were plated and cultured at 37 °C or 15 °C. In the absence of either PT or 6mA, the viability of E. coli HST04 cells at 15 °C was significantly decreased in the presence of DndFGH (Fig. 4C). Cotransformation of E. coli HST04 with pWHU705 and pWHU701, which express H. chejuensis KCTC2396 DndFGH and DndBCDE, respectively, restored viability at 15 °C to 66% of the level at 37 °C (Fig. 4C). This partial recovery of cell viability could result from the amount of DndBCDE expressed from pWHU701 under these growth conditions, which may not be fully sufficient to counteract the temperature-induced DndFGH. Remarkably, methylation of GATC sites conferred protection against DndFGH restriction activity to almost the same extent as PT modification (64%) (Fig. 4C), which suggests that the A methylation of GATC sites hampered DNA cleavage activity or interfered with the ability of DndFGH to bind to target sites. The experiment to assess the mechanism of A methylation on DndFGH is not yet possible due to the lack of a system for in vitro reconstitution of DndFGH activity.
Discussion
The advent of SMRT sequencing has accelerated the pace of discovery and characterization of prokaryotic epigenetics. In particular, methylation-based R-M systems and epigenetic marks have been detected in the vast majority of bacterial and archaeal genomes (3, 30). The chemical diversity of DNA modifications in bacteria has recently grown beyond methylation with the discovery of 7-deazaguanine modifications (2) and PT modifications of the DNA backbone (12, 15). We have shown that PTs function in R-M systems with DndFGH (22, 23, 31) but that PTs are also present in bacteria that lack restriction genes. Although it is well-established that many bacteria possess multiple R-M systems and MTases, each with unique methylation consensus sequences (3, 32), we have discovered a proximity of two diverse DNA modifications in the same core consensus sequence, with interactions that point to a functional cooperation of the two modification systems.
Although SMRT sequencing is not able to resolve closely spaced DNA modifications, including PT and 6mA in GPS6mATC consensus sequences as observed here, we were able to detect d(GPS6mA) dinucleotides by LC-MS/MS in two unrelated bacteria, E. coli B7A and S. enterica serovar Cerro 87, by virtue of the PT-induced nuclease inhibition that produces a limit digest of PT-containing dinucleotides. The fact that d(GPS6mA) occurred at much lower frequencies than the d(GPSA) dinucleotide (2 to 15 per 106 nt versus 250 to 300 per 106 nt) supports the idea that d(GPS6mA) is occurring in the larger consensus sequence of a DNA methyltransferase. Examples of such sequences in other E. coli strains include the GAACC consensus of many E. coli R-M systems (33) and the larger recognition sequences for EcoBI/Eco1158I (TGAN8TGCT), Eco377I (GGAN8ATGC), and Eco777I (GGAN6TATC) (24). However, E. coli B7A does not possess genes for these enzymes, so the source of d(GPS6mA) in B7A must be another MTase. Similarly, d(GPS6mA) dinucleotides must occur in the GAAC consensus sequence for Dnd proteins in S. enterica serovar Cerro 87, so DNA N6-adenine-methyltransferase from phage origin (M2.SenCer87ORF144P) and methyl-directed repair DNA adenine methylase (M.SenCer87DamP) (33), which target GATC, cannot account for the d(GPS6mA) modification. Interestingly, the GPSATC sites may also contain 5mC due to the activity of C5-cytosine–specific DNA methylase (M1.SenCer87ORF144P) (33), which raises the possibility of sites containing all three modifications, GPS6mAT5mC, further complicating the interplay among DNA modification systems.
The discovery of proximate PT and 6mA motivated the development of a model system to further characterize coincident DNA modification consensus sequences. Here, we focused on the GATC recognition sequence for DndBCDE from H. chejuensis KCTC2396 and Dam MTase. Although the in vitro activity of Dam was partially inhibited by a preexisting PT modification, the SMRT sequencing map of 6mA in GATC sites in E. coli HST04(dndBCDE, dam) showed complete modification of all GATC sites (37,610 of 37,698 sites), which suggests either that the Dam inhibition is an artifact of the in vitro situation, with in vivo factors overcoming any inhibitory effect of PT, or that Dam methylation occurs in synchrony with or before PT modification. As a double-stranded DNA modification at GATC, 6mA methylation by Dam does indeed occur quickly after DNA replication (34) so it is possible that PT and 6mA are inserted in tandem at hemimodified sites postreplication.
The E. coli HST04 model system provided an opportunity to test these in vitro observations in vivo. The observation of PT modifications in 2,058 of the 37,698 GATC sites (5%) in E. coli HST04(dndBCDE) is similar to previous SMRT analyses in E. coli B7A (12% of GAAC/GTTC) and V. cyclitrophicus FF75 (14% of CCA) (23), with the 228 ± 4 PT per 106 nt measured by LC-MS/MS analysis corroborating the quantitative nature of SMRT mapping. However, the map of the GPSATC sites across the genome (GenBank accession no. CP013952) showed PTs in 939 ORFs, 1,119 in noncoding regions, and none in tRNAs or rRNA genes (Fig. 3), which stands in contrast to the slight enrichment of PTs in tRNA genes in E. coli B7A (23), in which dnd genes are native.
We also made the surprising observation of the temperature dependence of the DndFGH restriction system in two bacteria. Thermoregulated R-M systems are relatively rare, with two recent studies describing this phenotype for the restriction endonucleases LlaJI and LmoH7 (35, 36). The first is a plasmid-associated R-M system that mediates temperature-dependent resistance to phages in Lactococcus lactis, with LlaJI activity most pronounced when plaque assays are performed at 19 °C and completely abolished at 37 °C (35). The other reported temperature-sensitive restriction phenomenon involves the endonuclease LmoH7, which shares no sequence similarity, genetic components, or recognition sites with LlaJ1, but it can confer phage resistance in Listeria monocytogenes ECII strains at low temperature (36). Similar to LlaJI and LmoH7, the mechanism responsible for dndFGH induction may be due to the instability of mRNA at high temperature or the involvement of an unknown temperature-sensitive transcriptional regulator. At this point, we do not know whether the temperature sensitivity occurs at the level of biochemical activity or protein levels, but the phenomenon points to the potential for an intriguing fitness advantage for bacteria in terms of resisting phage infection, for example, in moderate-temperature environments outside the core temperature of homeothermic animals (e.g., soil, water).
Although the coincidence of PT and 6mA in the same DNA sequence allows an organism to repurpose consensus sequences, the phenomenon has implications for the effectiveness of R-M systems and the real and potential epigenetic functions of both PT and 6mA. The artificial GPS6mATC system used here ignores the potential disruptive effects of PT on the binding of 6mA-sensitive protein binding in Dam-regulated epigenetic functions so it will be important to explore the coincidence of PT and DNA methylation in bacteria with both native modification systems. There are also interesting implications for R-M activity against phage infection. Many phages possess orphan MTases that modify and protect the phage genome against host restriction enzymes. For example, several phages have been found to possess GATC-specific Dam MTase activities, which not only modulate phage propagation but also protect against hundreds of R-M systems (37). Based on our observation of the interchangeability of 6mA and PT against PT-dependent restriction, this protection would also be extended against GATC-specific PT R-M systems in diverse bacteria. Interestingly, phages such as Staphylococcus phage K have evolved to lack 5′-GATC-3′ motifs in their DNA genome (38).
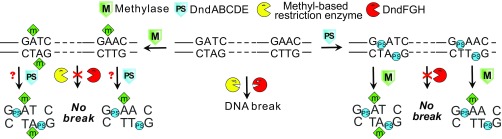
In summary, the discovery of proximate PT and 6mA DNA modifications in several bacteria point to the potential for widespread coincidence of epigenetics systems (Fig. S3). The results reveal complex but unappreciated interactions between DNA modification systems and raise the possibility of coevolution of interacting systems to facilitate the function of each.
Fig. S3.
Summary of the interaction and interference between DNA methylation and PT modification. The diagram illustrates the idea of a DNA methyltransferase and the DndABCDE proteins sharing the same site in DNA to generate a hybrid modification motif, d(GPS6mA). In a bacterium possessing Dam MTase and the DndABCDE proteins similar to those from H. chejuensis KCTC 2396, both sets of proteins recognize the same GATC motif to generate hybrid GPS6mATC structures, with GPSATC and G6mATC able to replace each other to protect against restriction by DndFGH, respectively. A similar situation may exist for the d(GPS6mA) hybrid modification in the GAAC motif in E. coli B7A and S. enterica serovar Cerro 87.
Materials and Methods
Detailed descriptions of materials and methods are provided in SI Materials and Methods.
Bacterial Strains, Plasmids, and Growth Conditions.
All strains, plasmids and primers used in this study are listed in Table S1. In-frame deletion mutants of E. coli B7A (WXY-1, CX2 and CX3, dndBCDE, dndFGH, and dndBCDEFGH) were constructed by homologous recombination using the thermo- and sucrose-sensitive plasmid pKOV-Kan, as previously described (31). Cell viability was determined by colony counting on LB-agar plates, plates placed at 15 °C for 24 h and shifted to 37 °C for colony counting for assessing low-temperature viability. qRT-PCR analysis of gene transcription was performed as described in detail in SI Materials and Methods.
DNA Modification Analysis.
For in vitro DNA methylation, duplex oligonucleotides containing PT modifications were reacted with Dam MTase, and modifications were analyzed by LC-MS/MS as described in SI Materials and Methods. PT modifications were detected by LC-MS/MS analysis of enzymatically hydrolyzed DNA as previously described (39); SI Materials and Methods for details. SMRT sequencing to map PT and methylation modifications was performed as described previously (23) and in SI Materials and Methods.
SI Materials and Methods
Dam Methylation in Vitro.
For double-stranded oligonucleotide preparation, both complementary single-stranded oligonucleotides were mixed at the same molar concentration. The oligonucleotide mixture was heated to 95 °C for 5 min and gradually cooled to ambient temperature. For dsDNA methylation in vitro, 20 pmol of dsDNA was mixed in a 10-μL volume with 80 mM SAM, 1× dam methyltransferase buffer, and 4 units (U) of Dam (New England Biolabs) at 37 °C for an appropriate time, followed by 65 °C for 15 min.
In-Frame Deletion of dnd Genes in E. coli B7A.
WXY-1, CX2 and CX3, dndBCDE, dndFGH, and dndBCDEFGH in-frame deletion mutants of E. coli B7A were constructed by homologous recombination using the thermo- and sucrose-sensitive plasmid pKOV-Kan, as previously described (31). In general, upstream and downstream fragments of dndBCDE, dndFGH, and dndBCDEFGH were first generated by PCR using the primer pairs WXY-1-LL/WXY-1-LR and WXY-1-RL/WXY-1-RR, CX2-LL/CX2-LR and CX2-RL/CX2-RR, and CX3-LL/CX3-LR and CX3-RL/CX3-RR (Table S1). The recombinant fragments were next PCR-amplified using the mixture of upstream and downstream arms as templates, overlapping by ∼40 bp, and cloned into pEASYTM-T5 zero for sequencing. The correct recombinant fragments were released from pEASYTM-T5 zero and cloned into pKOV-Kan to generate pWHU1835, pWHU623, and pWHU624 for in-frame deletions of dndBCDE, dndFGH, and dndBCDEFGH, respectively. For instance, recombinant plasmid pWHU623 was introduced into WT E. coli B7A for the construction of CX2. A single-crossover intermediate was obtained at 43 °C, and double crossover was obtained on a plate containing 15% sucrose at 43 °C to generate CX2. CX3 was constructed from CX2, and WXY-1 was obtained directly from E. coli B7A.
Enzymatic Digestion of DNA.
For PT quantitation, DNA samples were prepared, hydrolyzed, and dephosphorylated as described previously (39). To completely release 6mA from d(GPS6mA), DNA samples (20 pmol in 100 μL) were hydrolyzed with 2 U of nuclease P1 (USBiological) in 30 mM sodium acetate, pH 5.3, and 0.5 mM ZnCl2 at 50 °C for 12 h. After pH adjustment (15 μL of 1 M Tris-Cl, pH 8.0), 1.25 U of Bezonase (Sigma), 16 U of snake venom phosphodiesterase (Worthington), 1 U of alkaline phosphatase (Sigma), and 3 μL of 5 mg/mL BSA were added, and the 150-μL mixture was incubated at 37 °C for 48 h. The enzymes were subsequently removed by ultrafiltration (Nanosep 10K-column; PALL) before LC-MS/MS analysis.
LC-MS/MS Analysis.
For LC-MS/MS detection of 6mA and d(GPS6mA), the HPLC retention time was determined using a Thermo Hypersil Gold aQ column (150 × 2.1 mm, 3 µm). Elution was performed at a flow rate of 0.2 mL/min and a gradient of buffer A (water with 0.1% acetic acid) and buffer B (acetonitrile with 0.1% acetic acid) according to the following profile: 97% buffer A and 3% buffer B for 5 min, followed by 3 to 6% buffer B over a period of 42 min and 6 to 98% buffer B over a period of 1 min. The column was coupled to a Thermo TSQ Quantum Access MAX mass spectrometer with an electrospray ionization source in positive mode. The selected reaction monitoring (SRM) scan mode in MS/MS was used for the detection of product ions derived from the precursor ions. All instrument parameters were optimized for maximal sensitivity as follows (precursor ion m/z, product ion m/z, collision energy, tube lens): 6mA, 266, 150, 22 V, 32 V; d(GPS6mA), 611, 150, 36 V, 96 V. The following optimized parameters were used: sheath gas pressure, 40 psi; vaporizer temperature, 30 °C; auxiliary gas pressure, 8 psi; capillary temperature, 320 °C; source spray voltage, 3.5 kV.
Real-Time qRT-PCR.
Extraction of total RNA, DNase treatment, and reverse transcription were described in our previous study (39). To measure dndFGH cluster transcription, primers were designed within dndGH; the housekeeping gene gapA, encoding GAPDH, was used for normalization. The real-time qRT-PCR data analysis was performed according to the comparative threshold cycle method (also known as 2–ΔΔCT). The fold change in expression of dndFGH according to temperature was expressed relative to that at 37 °C.
SMRT Sequencing.
For DNA isolation, an overnight bacterial culture was diluted and regrown to an OD600 of ∼0.8 to achieve logarithmic growth. DNA was isolated from the bacteria using a QIAGEN Genomic-tip 500/G kit with a standard protocol. P6/C4 chemistry was applied for library preparation and SMRT sequencing steps. The genomes were assembled using HGAP2.3.3. Base modification analysis was performed using the base modification detection workflow of SMRT Analysis version 1.3.
Cell Viability Assay.
Cell viability was determined by plating exponentially growing E. coli cells (OD600 = 0.8), serially diluted, onto LB-agar plates and counting the colonies that developed. Colonies formed at 37 °C for 12 h were used as a reference. To assess cell viability at low temperature, the plates were placed at 15 °C for 24 h and then shifted to 37 °C for cfu counting.
Acknowledgments
We thank Bochen Zhu and Yuanjie Ge for research assistance. This work was supported by grants from the National Science Foundation of China (31520103902, 31670086, and 31300038) and the 973 program of the Ministry of Science and Technology (2013CB734003), Hubei Province’s Outstanding Medical Academic Leader Program, the Young One Thousand Talent program of China, the US National Science Foundation (CHE-1019990), the US National Institute of Allergy and Infectious Diseases (AI112711), and the Singapore-MIT Alliance for Research and Technology sponsored by the National Research Foundation of Singapore.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1702450114/-/DCSupplemental.
References
- 1.Casadesús J, Low D. Epigenetic gene regulation in the bacterial world. Microbiol Mol Biol Rev. 2006;70:830–856. doi: 10.1128/MMBR.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thiaville JJ, et al. Novel genomic island modifies DNA with 7-deazaguanine derivatives. Proc Natl Acad Sci USA. 2016;113:E1452–E1459. doi: 10.1073/pnas.1518570113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blow MJ, et al. The epigenomic landscape of prokaryotes. PLoS Genet. 2016;12:e1005854. doi: 10.1371/journal.pgen.1005854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meselson M, Yuan R, Heywood J. Restriction and modification of DNA. Annu Rev Biochem. 1972;41:447–466. doi: 10.1146/annurev.bi.41.070172.002311. [DOI] [PubMed] [Google Scholar]
- 5.Blumenthal RM, Cheng X. Restriction-modification systems. In: Streips UN, Yasbin RE, editors. Modern Microbial Genetics. 2nd Ed. John Wiley & Sons; New York: 2002. pp. 177–225. [Google Scholar]
- 6.Roberts RJ, et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loenen WA, Raleigh EA. The other face of restriction: Modification-dependent enzymes. Nucleic Acids Res. 2014;42:56–69. doi: 10.1093/nar/gkt747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Low DA, Weyand NJ, Mahan MJ. Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect Immun. 2001;69:7197–7204. doi: 10.1128/IAI.69.12.7197-7204.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takahashi N, Naito Y, Handa N, Kobayashi I. A DNA methyltransferase can protect the genome from postdisturbance attack by a restriction-modification gene complex. J Bacteriol. 2002;184:6100–6108. doi: 10.1128/JB.184.22.6100-6108.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heusipp G, Fälker S, Schmidt MA. DNA adenine methylation and bacterial pathogenesis. Int J Med Microbiol. 2007;297:1–7. doi: 10.1016/j.ijmm.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Wion D, Casadesús J. N6-methyl-adenine: An epigenetic signal for DNA-protein interactions. Nat Rev Microbiol. 2006;4:183–192. doi: 10.1038/nrmicro1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L, et al. Phosphorothioation of DNA in bacteria by dnd genes. Nat Chem Biol. 2007;3:709–710. doi: 10.1038/nchembio.2007.39. [DOI] [PubMed] [Google Scholar]
- 13.Zhou X, et al. A novel DNA modification by sulphur. Mol Microbiol. 2005;57:1428–1438. doi: 10.1111/j.1365-2958.2005.04764.x. [DOI] [PubMed] [Google Scholar]
- 14.He X, et al. Analysis of a genomic island housing genes for DNA S-modification system in Streptomyces lividans 66 and its counterparts in other distantly related bacteria. Mol Microbiol. 2007;65:1034–1048. doi: 10.1111/j.1365-2958.2007.05846.x. [DOI] [PubMed] [Google Scholar]
- 15.Wang L, et al. DNA phosphorothioation is widespread and quantized in bacterial genomes. Proc Natl Acad Sci USA. 2011;108:2963–2968. doi: 10.1073/pnas.1017261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.You D, Wang L, Yao F, Zhou X, Deng Z. A novel DNA modification by sulfur: DndA is a NifS-like cysteine desulfurase capable of assembling DndC as an iron-sulfur cluster protein in Streptomyces lividans. Biochemistry. 2007;46:6126–6133. doi: 10.1021/bi602615k. [DOI] [PubMed] [Google Scholar]
- 17.Cheng Q, et al. Regulation of DNA phosphorothioate modifications by the transcriptional regulator DptB in Salmonella. Mol Microbiol. 2015;97:1186–1194. doi: 10.1111/mmi.13096. [DOI] [PubMed] [Google Scholar]
- 18.He W, et al. Regulation of DNA phosphorothioate modification in Salmonella enterica by DndB. Sci Rep. 2015;5:12368. doi: 10.1038/srep12368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu W, et al. Structural insights into DndE from Escherichia coli B7A involved in DNA phosphorothioation modification. Cell Res. 2012;22:1203–1206. doi: 10.1038/cr.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao F, Xu T, Zhou X, Deng Z, You D. Functional analysis of spfD gene involved in DNA phosphorothioation in Pseudomonas fluorescens Pf0-1. FEBS Lett. 2009;583:729–733. doi: 10.1016/j.febslet.2009.01.029. [DOI] [PubMed] [Google Scholar]
- 21.Cao B, et al. Pathological phenotypes and in vivo DNA cleavage by unrestrained activity of a phosphorothioate-based restriction system in Salmonella. Mol Microbiol. 2014;93:776–785. doi: 10.1111/mmi.12692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gan R, et al. DNA phosphorothioate modifications influence the global transcriptional response and protect DNA from double-stranded breaks. Sci Rep. 2014;4:6642. doi: 10.1038/srep06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao B, et al. Genomic mapping of phosphorothioates reveals partial modification of short consensus sequences. Nat Commun. 2014;5:3951. doi: 10.1038/ncomms4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kasarjian JK, Iida M, Ryu J. New restriction enzymes discovered from Escherichia coli clinical strains using a plasmid transformation method. Nucleic Acids Res. 2003;31:e22. doi: 10.1093/nar/gng022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dyson P, Evans M. Novel post-replicative DNA modification in Streptomyces: Analysis of the preferred modification site of plasmid pIJ101. Nucleic Acids Res. 1998;26:1248–1253. doi: 10.1093/nar/26.5.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barras F, Marinus MG. The great GATC: DNA methylation in E. coli. Trends Genet. 1989;5:139–143. doi: 10.1016/0168-9525(89)90054-1. [DOI] [PubMed] [Google Scholar]
- 27.Flusberg BA, et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010;7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song CX, et al. Sensitive and specific single-molecule sequencing of 5-hydroxymethylcytosine. Nat Methods. 2011;9:75–77. doi: 10.1038/nmeth.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark TA, Spittle KE, Turner SW, Korlach J. Direct detection and sequencing of damaged DNA bases. Genome Integr. 2011;2:10. doi: 10.1186/2041-9414-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE—A database for DNA restriction and modification: Enzymes, genes and genomes. Nucleic Acids Res. 2010;38:D234–D236. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu T, Yao F, Zhou X, Deng Z, You D. A novel host-specific restriction system associated with DNA backbone S-modification in Salmonella. Nucleic Acids Res. 2010;38:7133–7141. doi: 10.1093/nar/gkq610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krebes J, et al. The complex methylome of the human gastric pathogen Helicobacter pylori. Nucleic Acids Res. 2014;42:2415–2432. doi: 10.1093/nar/gkt1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE—A database for DNA restriction and modification: Enzymes, genes and genomes. Nucleic Acids Res. 2015;43:D298–D299. doi: 10.1093/nar/gku1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marinus MG, Løbner-Olesen A. DNA methylation. Ecosal Plus. 2014;6(1) doi: 10.1128/ecosalplus.ESP-0003-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Driscoll J, et al. Lactococcal plasmid pNP40 encodes a novel, temperature-sensitive restriction-modification system. Appl Environ Microbiol. 2004;70:5546–5556. doi: 10.1128/AEM.70.9.5546-5556.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim JW, et al. A novel restriction-modification system is responsible for temperature-dependent phage resistance in Listeria monocytogenes ECII. Appl Environ Microbiol. 2012;78:1995–2004. doi: 10.1128/AEM.07086-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murphy J, Mahony J, Ainsworth S, Nauta A, van Sinderen D. Bacteriophage orphan DNA methyltransferases: Insights from their bacterial origin, function, and occurrence. Appl Environ Microbiol. 2013;79:7547–7555. doi: 10.1128/AEM.02229-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krüger DH, Barcak GJ, Smith HO. Abolition of DNA recognition site resistance to the restriction endonuclease EcoRII. Biomed Biochim Acta. 1988;47:K1–K5. [PubMed] [Google Scholar]
- 39.Lai C, et al. In vivo mutational characterization of DndE involved in DNA phosphorothioate modification. PLoS One. 2014;9:e107981. doi: 10.1371/journal.pone.0107981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DuPont HL, et al. Pathogenesis of Escherichia coli diarrhea. N Engl J Med. 1971;285:1–9. doi: 10.1056/NEJM197107012850101. [DOI] [PubMed] [Google Scholar]
- 41.Murase T, Nagato M, Shirota K, Katoh H, Otsuki K. Pulsed-field gel electrophoresis-based subtyping of DNA degradation-sensitive Salmonella enterica subsp. enterica serovar Livingstone and serovar Cerro isolates obtained from a chicken layer farm. Vet Microbiol. 2004;99:139–143. doi: 10.1016/j.vetmic.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 42.Lee HK, et al. Hahella chejuensis gen. nov., sp. nov., an extracellular-polysaccharide-producing marine bacterium. Int J Syst Evol Microbiol. 2001;51:661–666. doi: 10.1099/00207713-51-2-661. [DOI] [PubMed] [Google Scholar]
- 43.Alting-Mees MA, Short JM. pBluescript II: Gene mapping vectors. Nucleic Acids Res. 1989;17:9494. doi: 10.1093/nar/17.22.9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lalioti M, Heath J. A new method for generating point mutations in bacterial artificial chromosomes by homologous recombination in Escherichia coli. Nucleic Acids Res. 2001;29:E14. doi: 10.1093/nar/29.3.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang AC, Cohen SN. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]