Significance
A vaccine against HIV-1 should elicit antibodies that prevent infection (“neutralize”) by binding the viral envelope glycoprotein (Env). The extraordinary genetic diversity of Env means that protective antibodies must recognize one of a few broadly conserved sites (“epitopes”). The trimeric Env protein fluctuates among a range of conformations: Closed conformations expose only the conserved epitopes, whereas various open conformations expose many other, more variable epitopes. The latter dominate immune responses and restrict protection to a narrow range of viral strains. We describe here the properties of Env from an extremely neutralization-resistant HIV-1 variant. We find that it binds only antibodies that recognize conserved epitopes, consistent with a tightly closed conformation, and suggest that it might be a suitable vaccine template.
Keywords: HIV-1 gp160, neutralizing antibodies, vaccine design
Abstract
The extraordinary genetic diversity of the HIV-1 envelope spike [Env; trimeric (gp160)3, cleaved to (gp120/gp41)3] poses challenges for vaccine development. Envs of different clinical isolates exhibit different sensitivities to antibody-mediated neutralization. Envs of difficult-to-neutralize viruses are thought to be more stable and conformationally homogeneous trimers than those of easy-to-neutralize viruses, thereby providing more effective concealment of conserved, functionally critical sites. In this study we have characterized the antigenic properties of an Env derived from one of the most neutralization-resistant HIV-1 isolates, CH120.6. Sequence variation at neutralizing epitopes does not fully account for its exceptional resistance to antibodies. The full-length, membrane-bound CH120.6 Env is indeed stable and conformationally homogeneous. Its antigenicity correlates closely with its neutralization sensitivity, and major changes in antigenicity upon CD4 engagement appear to be restricted to the coreceptor site. The CH120.6 gp140 trimer, the soluble and uncleaved ectodomain of (gp160)3, retains many antigenic properties of the intact Env, consistent with a conformation close to that of Env spikes on a virion, whereas its monomeric gp120 exposes many nonneutralizing or strain-specific epitopes. Thus, trimer organization and stability are important determinants not only for occluding many epitopes but also for conferring resistance to neutralization by all but a small set of antibodies. Env preparations derived from neutralization-resistant viruses may induce irrelevant antibody responses less frequently than do other Envs and may be excellent templates for developing soluble immunogens.
HIV-1 envelope spike (Env), a heavily glycosylated type I membrane protein, induces strong antibody responses in infected patients (1–3). The Env polypeptide chain, produced as a gp160 precursor that trimerizes to (gp160)3, undergoes cleavage by a furin-like protease into two noncovalently associated fragments: the receptor-binding fragment, gp120, and the fusion fragment, gp41 (4). Three copies of each fragment form the mature viral spike (gp120/gp41)3. Sequential binding of gp120 to the primary receptor CD4 and a coreceptor induces large conformational changes and a cascade of refolding events in gp41, possibly accompanied by dissociation of gp120 (5, 6). During the fusion process, Env has at least three distinct conformational states: a prefusion state, a fusion intermediate, and a postfusion state. The prefusion state is an ensemble of closed and open (or partly open) conformations (substates) (7–9), and the extended intermediate probably also populates a dynamically exchanging set of conformations. This conformational plasticity creates substantial difficulties for producing stable recombinant protein that mimics the structure and antigenicity of the native HIV-1 Env on the virion surface.
Most broadly neutralizing antibodies (bnAbs) appear to recognize a closed prefusion conformation of HIV-1 Env (10–12), which therefore is considered advantageous as a candidate for a vaccine immunogen. Many Env preparations, including some on the surface of infectious virions, are unstable and conformationally heterogeneous, confounding antigenicity studies and immunogen design. Indeed, it has been suggested that HIV-1 has evolved to express a substantial amount of nonfunctional forms of Env on the virion surface, in addition to the functional trimer, as decoys to evade host immune responses (13). Moreover, the intrinsic, strain-dependent dynamics of Env also cause exposure of a varied range of epitopes (8). We recently reported that Envs derived from two difficult-to-neutralize HIV-1 primary isolates (92UG037.8 and C97ZA012) are antigenically homogeneous and hence conformationally restricted when expressed on cell surfaces (10). These Env trimers adopt a conformation that exposes only bnAb epitopes and buries nonneutralizing and strain-specific neutralizing epitopes. Because of the conformational homogeneity of these Envs, we could evaluate the contributions of the cytoplasmic tail (CT) to the stability and antigenicity of the Env ectodomain. We found that deletion of the CT has minimal impact on the membrane-fusion function of the Env trimer but has an unexpectedly large influence on the antigenic properties of the ectodomain (10). Moreover, to understand the physical coupling between the CT and the ectodomain, we then determined an atomic structure of the transmembrane domain (TMD) (14). The TMD forms a well-ordered trimer. Mutations that destabilize this trimer resemble the CT deletion in influencing Env stability and altering its antibody sensitivity.
Because stable and homogenous preparations of HIV-1 Env are essential for proper analysis of its structure, function, antigenicity, and likely immunogenicity, we have analyzed the antigenic properties of various forms of an HIV-1 Env derived from an isolate, CH120.6, that is one of the most difficult-to-neutralize viruses characterized so far (15). Consistent with our previous findings, the intact membrane-bound CH120.6 gp160 was structurally stable and antigenically homogeneous, and its antigenicity fully correlated with neutralization. Sequence analysis indicated that escape mutations at various neutralizing epitopes do not explain its exceptional resistance to antibody-mediated neutralization. When we expressed this Env as a soluble trimer of gp140, the uncleaved ectodomain of (gp160)3, many of its antigenic properties were similar to those of the intact, native protein. Its monomeric gp120 bound many nonneutralizing or strain-specific neutralizing antibodies, as expected. These data suggest that Envs derived from difficult-to-neutralize strains adopt a very narrow range of prefusion conformations, with only broadly neutralizing epitopes accessible. When properly produced, they may be less likely than other forms of Env to induce nonneutralizing or strain-specific antibodies and thus may be excellent templates for Env-based immunogen design.
Results
Neutralization of the HIV-1 Isolate CH120.6.
CH120.6 Env is a chronic circulating recombinant form (CRF) 07_BC gp160 clone isolated from transmissions of i.v. drug use in China (16). It is a tier 3 virus with exceptional resistance to antibody-mediated neutralization when tested against pools of diverse HIV+ plasma samples, polyclonal HIV Ig (HIVIG) derived from HIV antibody-positive donors, and several monoclonal antibodies (15). To obtain a complete antigenic picture of this isolate, we analyzed antibody-mediated inhibition of viral infectivity in a pseudovirus-based neutralization assay using a panel of nonneutralizing antibodies, strain-specific neutralizing antibodies, and various bnAbs (see Table S1 for references). As summarized in Table 1, the CH120.6 virus was resistant to all nonneutralizing and strain-specific neutralizing antibodies, regardless of the location of their epitopes. Several bnAbs, including VRC01, NIH45-46, 12A12, PG9, PG16, PGT128, 4E10, and 10E8, did neutralize the virus, but at much higher effective concentrations than required for most other HIV-1 isolates (Table S2). More potent neutralization was observed for two other bnAbs, PGT145 (IC50: 0.75 μg/mL for Fab and 0.24 for IgG) and 10-1074 (IC50: 0.27 for Fab and 0.50 for IgG). These results indicate that CH120.6 Env imparts resistance not only to nonneutralizing and strain-specific neutralizing antibodies but also, to some extent, to many bnAbs with great potency against other HIV-1 strains.
Table 1.
Neutralization of HIV-1 isolate CH120.6 by known monoclonal antibodies
| Antibody | Neutralization titer | |||
| Epitope | Name | MPI, % | IC50, μg/mL | IC80, μg/mL |
| CD4bs | VRC01 | 95 | 11 | 25 |
| NIH45-46 | 92 | 8.8 | 35 | |
| 12A12 | 98 | 7.3 | 24 | |
| DH583 | 25 | >50 | >50 | |
| CD4i | 17b | 0 | >50 | >50 |
| 412d | 2 | >50 | >50 | |
| V1V2-glycan | PG9 | 64 | 18 | >50 |
| PG16 | 58 | 27 | >50 | |
| PGT145 | 96 | 0.75 | 3.8 | |
| V2 | 2158 | 0 | >50 | >50 |
| V3 | 3791 | 0 | >50 | >50 |
| 447-52D | 22 | >50 | >50 | |
| V3-glycan | PGT128 | 77 | 12 | >50 |
| 10-1074 | 100 | 0.27 | 0.70 | |
| Glycan | 2G12 | 14 | >50 | >50 |
| MPER | 4E10 | 54 | 47 | >50 |
| 10E8 | 99 | 4.4 | 15 | |
| Cluster I | 240-D | 7 | >50 | >50 |
| 246-D | 8 | >50 | >50 | |
| Cluster II | 1281 | 16 | >50 | >50 |
| PGT151 (IgG) | 45 | >50 | >50 | |
To address whether the extraordinary resistance of CH120.6 Env has resulted from escape mutations at various neutralizing epitopes, we compared its sequence with those of other HIV-1 isolates. Based on an analysis of 50 bnAbs, we first defined mutational patterns that are statistically associated with resistance and sensitivity, using phylogenetically corrected methods (17), for four classes of bnAbs: the CD4 binding site; a trimer-specific, glycan-dependent V1V2 region; an N332-glycan–dependent V3 region; and the membrane proximal external region (MPER). The CH120.6 sequence retained key epitope-defining patterns of specific amino acid residues and N-linked glycosylation sites in these bnAb epitopes (18–20). We then used these signature patterns to predict the relative sensitivity of CH120.6 to the different antibodies shown in Table S2 using the machine-learning strategy Random Forests (21) implemented in the Python package scikit-learn (22). The measured values were 2.6- to 124-fold higher (i.e., less potent neutralization) than the predicted ones, except for antibodies NIH45-46, PG9, and PGT145, for which the measurements gave essentially the predicted values. These data suggest that the antibody resistance of CH120.6 cannot be fully explained by local escape mutations at these neutralizing epitopes. The glycan shield of the CH120.6 Env appeared to be no different from those of most the other M-group Envs (Fig. S1). Thus, CH120.6 Env is exceptionally resistant to antibody-mediated neutralization, but sequence variations or differences in glycosylation at or near the neutralizing epitopes do not appear to explain the observed antibody resistance.
Antigenic Properties of the Native CH120.6 Env Expressed on Cell Surfaces.
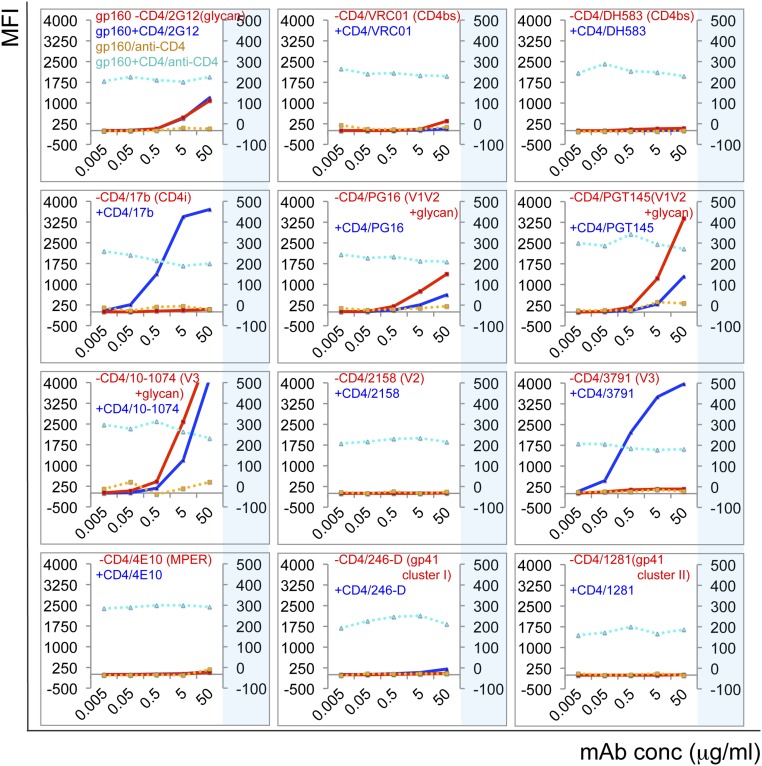
To test whether the neutralization profile of the CH120.6 virus correlates with the antigenic properties of CH120.6 Env, we generated 293T cell lines stably transfected with the CH120.6 gp160 (Fig. S2A). These cells expressed functional CH120.6 Env on their surface, as judged by efficient fusion with TZM.bl cells (23). The observed cell–cell fusion was sensitive to the known fusion inhibitor T20 (24), as expected. Approximately 50% of gp160s were cleaved into gp120 and gp41 both inside the cells and on the cell surface (Fig. S2B). We analyzed by flow cytometry the antibody-binding properties of the CH120.6 Env on the cell surface in the presence or absence of soluble CD4 (10). As shown in Fig. 1, soluble CD4 had no effect on binding by antibody 2G12, which targets a glycan epitope on the well-exposed surface of gp120, showing that CD4 binding had not induced gp120 dissociation. Three CD4 binding site (CD4bs) bnAbs, VRC01, 12A12, and NIH45-46, bound at only low levels to the cell-surface CH120.6 Env when CD4 was absent, as is consistent with their weak neutralizing potency for this virus (Fig. 1, Fig. S3, and Table 1). The nonneutralizing CD4bs antibody DH583 did not bind at all to the Env, suggesting that this epitope is occluded on both the cleaved, functional trimer (gp120/gp41)3 and the uncleaved, nonfusogenic trimer (gp160)3. The binding of nonneutralizing CD4-induced (CD4i) antibodies 17b and 412d depended strictly on CD4 engagement (Fig. 1 and Fig. S3). The trimer-specific bnAbs PG9, PG16, and PGT145 bound the CH120.6 Env trimer when CD4 was absent, but they also showed detectable binding when CD4 was present (Fig. 1 and Fig. S3), differing in that respect from what we have found for the 92UG037.8 and C97ZA012 Envs (compare, for example, the right-most panel in row 2 of Fig. 1, for PGT145, with the panel for PGT145 in figure S5A of ref. 10). Another two bnAbs, 10-1074 and PGT128, which recognize the N332 glycan-dependent epitopes in the V3 stem, also bound almost equally well to both the native, unliganded Env and the CD4-bound form (compare the 10-1074 and PGT128 panels in Fig. 1 and Fig. S3 with those in figure S5 A and B of ref. 10). Thus, CD4 binding appears to have a much smaller impact on these bnAb epitopes of the CH120.6 Env than it does on those of the 92UG037.8 and C97ZA012 Envs (10).
Fig. 1.
Antigenic characteristics of the CH120.6 Env trimer expressed on cell surfaces. Plots of antibody binding to the Env trimer on the CH120.6 gp160 cell surfaces in the absence (red) or presence (blue) of soluble CD4. The fluorescent signal for bound CD4 is shown in the presence (cyan) or absence (orange) of CD4. Antibody binding to Env was detected by R-phycoerythrin and plotted against the y axis on the left; CD4 binding was detected by FITC and plotted against the y axis on the right. Antibodies and their epitopes are indicated. Unless specified, all antibodies used are Fab fragments. Extensive control experiments were carried out to ensure the binding specificity and the experiments were repeated at least twice with almost identical results. MFI, mean fluorescence intensity.
Other nonneutralizing epitopes, including V3 loop (3791 and 447-52D), V2 (2158), gp41 cluster I epitopes 246-D and 240-D, and the gp41 cluster II epitope 1281, were not exposed on the native CH120.6 Env trimer, as expected (Fig. 1 and Fig. S3). As we have found for the 92UG037.8 and C97ZA012 Envs, occlusion of these nonneutralizing epitopes is independent of the cleavage between gp120 and gp41, because the Env on the cell surfaces is a 50:50 mixture of uncleaved and cleaved Env (Fig. S2). Upon CD4 binding, the nonneutralizing epitopes in V3 became fully accessible, but the cluster I epitopes and those in the V2 loop did not. The MPER-directed bnAbs 10E8 and 4E10 did not bind the native Env trimer, because they target a fusion-intermediate conformation of gp41 (25, 26). Another bnAb, PGT151, which binds at the gp120–gp41 interface, neutralized only very weakly with a maximum percent inhibition (MPI) of 45%; it bound at detectable levels to the CH120.6 Env trimer with or without CD4 (Fig. S3). Although high temperatures (37 or 42 °C) reduced the level of bound CD4 and diminished CD4i changes in antigenicity (except for increased exposure of the cluster I epitopes upon CD4 binding), they had little impact on the antigenicity of the unliganded Env (Fig. S4). Overall, the antigenicity of the native CH120.6 Env expressed on cell surfaces correlates closely with antibody neutralization, but CD4 engagement appears to induce localized antigenic changes near the V3 tip and the CD4i epitopes, which form the coreceptor binding site, rather than a global “opening-up” of the trimer, as observed for the two tier 2 Envs (92UG037.8 and C97ZA012). These comparisons suggest that the CH120.6 Env adopts an even narrower range of prefusion conformations than do many other difficult-to-neutralize viruses (10).
Antigenic Properties of Soluble Forms of the CH120.6 Env.
We further asked whether any of the soluble forms of the CH120.6 Env can mimic the antigenic properties of the membrane-bound native trimer. We expressed and purified from Expi293F cells the trimeric gp140 of CH120.6 fused with a foldon trimerization tag (27) as well as its monomeric gp120 (Fig. S5). These proteins eluted as single sharp peaks from a size-exclusion column, and Coomassie blue-stained SDS/PAGE showed little if any contamination with other proteins (Fig. S6, Insets). The gp140 protein migrated as a trimer under nonreducing and nonboiling conditions. As expected, surface plasmon resonance (SPR) binding experiments showed that the gp140 trimer and the gp120 monomer both bound soluble CD4 very tightly (Figs. S7 and S8).
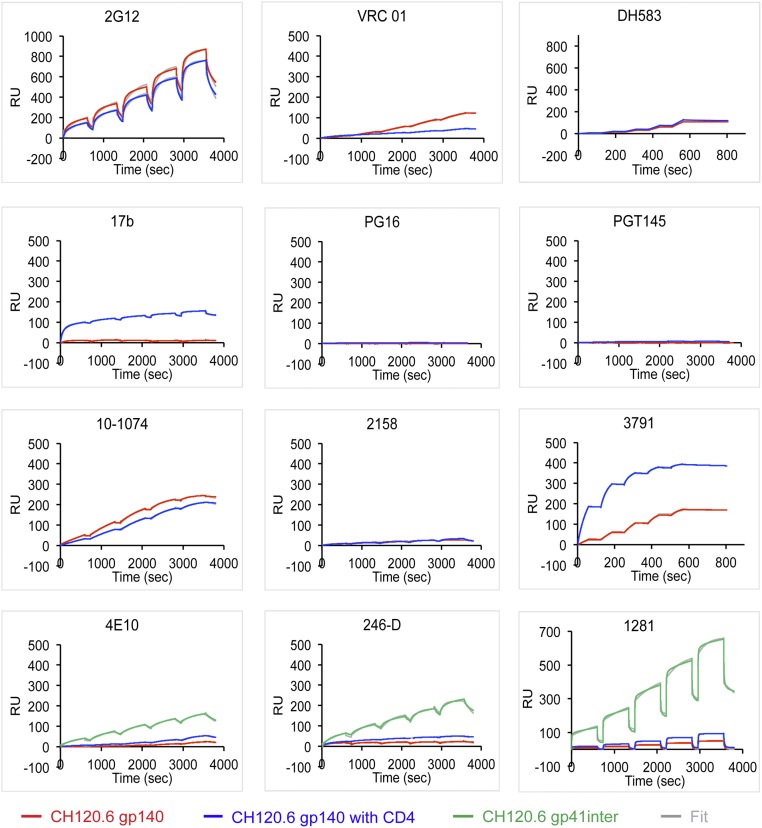
We then used SPR to assess the reactivity of various antibodies to both gp140 and gp120 from CH120.6 in either the absence or presence of soluble CD4. The results in Fig. 2 show that the antigenic properties of the CH120.6 gp140 trimer resemble those of cell surface-expressed Env. The unliganded gp140 bound bnAbs, such as VRC01, NIH45-46, 12A12, 10-1074, PGT128, and PGT151, but failed to bind nonneutralizing antibodies, such as DH583, 17b, 412d, 2158, 240-D, 246-D, and 1281. CD4 binding exposed the epitopes for the CD4i antibodies 17b and 412d, indicating that CH120.6 gp140 undergoes the expected conformational changes. We used CH120.6 gp41-inter, a construct designed to capture the prehairpin-intermediate conformation of gp41 (26), as a positive control for all anti-gp41 antibodies. Our uncleaved CH120.6 gp140 trimer bound strongly with PGT151 (Fig. S7), which has been previously reported to be specific for cleaved Env trimers (28).
Fig. 2.
Interactions of the CH120.6 gp140 with monoclonal antibodies. Envelope proteins or their purified complexes with two-domain CD4 were captured on the surface of a sensor chip coated with an anti-histidine antibody to avoid potential artifacts introduced by protein immobilization. Fab fragments of each antibody at various concentrations were passed over the envelope trimer surface individually without regeneration for single-cycle kinetic (SCK) analysis. The recorded sensorgram for gp140 is in red, for the gp140-CD4 complex in blue, and for gp41-inter in green; the fitted curves are in gray. Sensorgrams were fit using a 1:1 binding model; binding constructs are summarized in Tables S3 and S4.
There were two sets of differences between the antigenic properties of the CH120.6 gp140 and those of the intact, cell surface-borne molecule. First, all three trimer-specific bnAbs, PG9, PG16, and PGT145, failed to bind the gp140 (Fig. 2 and Fig. S6), suggesting that the V1V2 loop in this gp140 construct has shifted away from its position and conformation on intact cell-surface Env. Second, the nonneutralizing V3 epitope recognized by 3791 was partially exposed in the unliganded gp140 but was fully exposed when CD4 was bound. These results suggest that the CH120.6 gp140 trimer stabilized by an artificial trimerization tag foldon has an antigenic profile different from that of the corresponding intact Env on the cell surface but similar to that of the cell-surface Env with a truncated or deleted cytoplasmic tail, just as we reported for the 92UG037.8 and C97ZA012 Envs (10).
The unliganded, monomeric CH120.6 gp120, which lacks structural constraints of the Env trimer organization, bound all the anti-gp120 antibodies, including nonneutralizing ones such as DH583 (CD4bs), 17b (CD4i), 2158 (V2), and 3791 (V3). As expected, it failed to bind the trimer-dependent bnAbs PG9, PG16, and PGT145 (Fig. S8). We also found that the epitope of 3791 near the tip of V3 loop was not fully exposed until CD4 bound, suggesting that the CH120.6 gp120 is intrinsically less dynamic than the 92UG037.8 gp120, which binds 3791 as well when CD4 is bound as when it is not (29).
Discussion
An effective HIV-1 vaccine must protect against viruses with an extraordinary genetic diversity in their envelope spike. Sequence differences in HIV-1 Env lead to large variations in their sensitivity to antibody-mediated neutralization among different viral isolates. According to the conventional classification, the viruses designated as “tier 1” (including laboratory-adapted strains) are particularly neutralization sensitive; tier 2 viruses are substantially more resistant; and tier 3 viruses are even harder to neutralize (30).
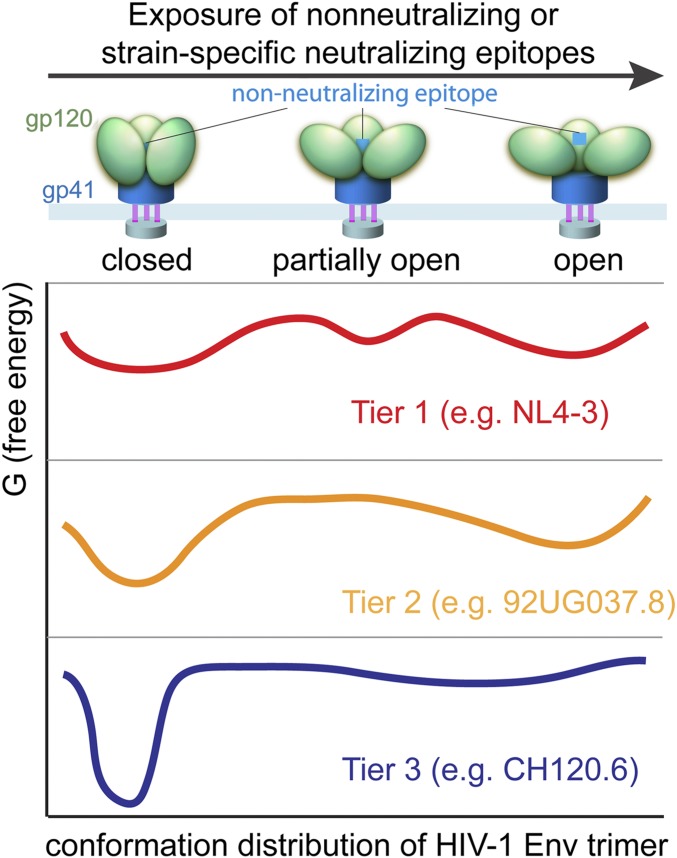
Our characterization of the tier 3 Env from the HIV-1 isolate CH120.6, which exhibits exceptional resistance to antibody-mediated neutralization (15), our earlier studies of tier 2 Envs, and work from other laboratories on the dynamics of Env trimers (8) and on structures of different Env conformers (9) together suggest the schematic representation in Fig. 3. Tier 3 Envs, such as CH120.6 described here, spend nearly all the time in a relatively narrow range of closed conformations; the tier 1 Envs of many laboratory-adapted strains spend a much larger fraction of the time in open conformations, with a relatively low barrier to complete dissociation of gp120 if the processing cleavage has occurred. Tier 2 Envs represent an intermediate case; they visit open conformations but with much lower frequency than tier 1 Envs and with no detectable tendency to shed gp120. Indeed, the Env of the HIV-1NL4-3 laboratory-adapted strain has been shown to occupy the CD4-triggered conformation more frequently than does the Env of the tier 2 primary isolate, HIV-1JR-FL, explaining the increased sensitivity of the former to 17b neutralization (8).
Fig. 3.
Proposed conformation distribution of the HIV-1 Env trimer. A free-energy (G) landscape is proposed for unliganded HIV-1 Env that samples closed, partially open, and open conformations, including tier 1 viruses (red) such as NL4-3 studied in ref. 8, tier 2 viruses (orange) such as 92UG037.8 reported in ref. 10, and tier 3 viruses (blue) such as CH120.6 (this study).
As Fig. 3 illustrates, mutations that affect the strength of intersubunit contacts in an Env trimer may change its sensitivity to particular classes of antibodies. Indeed, primary (tier 2 and tier 3) and laboratory-adapted (tier 1) simian/human immunodeficiency virus (SHIV) or HIV-1 isolates can interconvert during in vitro and in vivo passages (30, 31), suggesting that all viruses, regardless of their genetic background, can become neutralization resistant under immune pressure in infected individuals. Because HIV-1 transmission has no preference for easily neutralized viruses (31, 32), any successful vaccine must be effective against even very difficult-to-neutralize variants. In this context, our antigenic analysis of the exceptionally neutralization-resistant CH120.6 Env leads to three broad conclusions relevant to Env-based immunogen design.
First, the antigenicity of full-length, native CH120.6 Env correlates closely with its neutralization profile. Flow cytometry did not show any irrelevant cell-surface forms of Env, which would have shown high-affinity binding to various nonneutralizing antibodies. The absence of binding of the CH120.6 Env to anti-gp41 cluster I and II antibodies and equivalent binding to 2G12 with and without CD4 show that there is no significant spontaneous or CD4-induced gp120 shedding, unlike Envs from many other HIV-1 isolates (33). Stability of this kind is an important criterion for a vaccine candidate. These properties are independent of the cleavage between gp120 and gp41, because the surface-expressed CH120.6 Env is a roughly 50:50 mixture of cleaved and uncleaved Env. Our data thus indicate that the CH120.6 Env, whether cleaved or uncleaved, samples a narrow range of conformations that expose only the bnAb epitopes, but none of the nonneutralizing or strain-specific neutralizing epitopes, even at physiological temperature or higher. It appears to be an excellent starting point for producing Env immunogens that mimic the native prefusion conformation.
Second, we find that the extraordinary resistance of CH120.6 Env cannot be explained simply by escape mutations at various neutralizing epitopes, because measured IC50 values for most neutralizing antibodies are much greater than those predicted from sequence analysis (Table S2). These data suggest that the stability of the trimer is an important determinant of antibody resistance. The effect of CD4 binding on the antigenicity of CH120.6 Env is confined to the coreceptor binding site, including the CD4i epitopes and the V3 loop. CD4 binding to tier 2 Envs such as 92UG037.8 disrupts the trimer-specific quaternary epitopes recognized by PG16 or PG145 (34), but CD4 binding to CH120.6 Env led only to reduced, but clearly detectable, binding to those antibodies. Moreover, CD4 binding did not induce exposure of the gp41 cluster I epitopes or of nonneutralizing V2 epitopes on CH120.6 Env, unlike its effect on 92UG037.8 Env (10). These observations suggest that CD4 binding does not, in the case of CH120.6, produce a fully open trimer, which would not bind PG9 or PG145 at all and which would expose cluster I epitopes and nonneutralizing sites on V2.
Third, vaccine development requires the production of a recombinant, prefusion Env in quantities suitable for clinical trials. Two such preparations—a soluble Env trimer, known as “SOSIP.664,” stabilized by several modifications (12, 35), and a detergent-solubilized clade B Env with the CT deleted (JR-FL EnvΔCT) (36)—appear to present substantial technical barriers to large-scale, clinical-grade production (37, 38). Purification of full-length, membrane-bound CH120.6 Env in large quantities will be difficult also. We therefore have expressed and characterized its soluble forms, including a foldon-stabilized gp140 trimer and monomeric gp120. Although gp120 exposes many nonneutralizing epitopes, as expected, the CH120.6 gp140 trimer is stable and conformationally homogenous, but it does not bind trimer-specific bnAbs such as PG16 or PGT145 and has detectable affinity for the nonneutralizing, V3-directed antibody 3791, even in the absence of CD4. Thus, like many other soluble Env preparations (12, 29), it does not fully recapitulate the antigenic structure of virion-borne Env. Our recent observation that the TMD of HIV-1 Env is a structural component critical for the stability of the functional Env spike provides a plausible explanation for why the antigenic properties of most recombinant, soluble Env preparations with the TMD deleted deviate from those of the native protein (14, 39). We therefore must take the structural constraints imposed by the TMD on the ectodomain into consideration when designing a soluble immunogen, e.g., by mutating surface-exposed, lipid-interacting hydrophobic residues on the TMD while maintaining the trimeric structure seen in the high-resolution structure (14, 40).
Materials and Methods
HIV-1 envelope proteins and soluble CD4 were produced in 293T cells following protocols described previously (29). Monoclonal antibody IgGs or Fab fragments were expressed in 293T cells by transient transfection, by using selected stably transfected clones, or from hybridomas. The antibodies were purified by affinity chromatography using GammaBind-plus resin (GE Healthcare), followed by gel-filtration chromatography. Fab preparations by papain digestion were carried out as described (41). SPR binding assays, immunoprecipitation, and Western blot, flow cytometry, and antibody neutralization assays were carried out following the detailed protocols published previously (10) and as described in SI Materials and Methods. We used a trimeric Env crystal structure of a subtype G Env, X1193 [Protein Data Bank (PDB) ID code 5FYJ] (42), to visualize Env glycan shields. Potential N-linked glycosylation sites (PNGs) were mapped on the crystal structure, assuming that the protein surface within a radius of 10 Å from Cα of the PNG asparagine was shielded by the glycan. Additional details are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Wei Ting Chang and Hanqin Peng for technical assistance, Daniela Fera for antibody preparation, and the NIH AIDS Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, NIH for reagents. This work was supported by NIH Grants AI106488, AI127193, and AI112489; Collaboration for AIDS Vaccine Discovery Grant OPP1040741 from the Bill and Melinda Gates Foundation; and the Center for HIV/AIDS Vaccine Immunology – Immunogen Design Grant AI-100645. S.C.H. is an Investigator in the Howard Hughes Medical Institute.
Footnotes
Conflict of interest statement: S.K-.G., S.T., C.E.L., and A.C. were employees of Novartis Vaccines and Diagnostics at the time of the study. Following the acquisition of Novartis Vaccines and Diagnostics by the GlaxoSmithKline (GSK) group of companies in March 2015, S.K.-G., S.T., C.E.L., and A.C. became employees of the GSK group of companies. A.C. reports ownership of GSK shares. M.S. and W. Weissenhorn (reviewer) are coauthors of several papers (the last in 2014). The role of M.S. in the work in those papers was to provide neutralization analyses, and he does not have an active research collaboration with W. Weissenhorn.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700634114/-/DCSupplemental.
References
- 1.Decker JM, et al. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J Exp Med. 2005;201:1407–1419. doi: 10.1084/jem.20042510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci USA. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 4.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 6.Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 7.Julien JP, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munro JB, et al. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science. 2014;346:759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scharf L, et al. Broadly neutralizing antibody 8ANC195 recognizes closed and open states of HIV-1 Env. Cell. 2015;162:1379–1390. doi: 10.1016/j.cell.2015.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J, et al. HIV-1 ENVELOPE. Effect of the cytoplasmic domain on antigenic characteristics of HIV-1 envelope glycoprotein. Science. 2015;349:191–195. doi: 10.1126/science.aaa9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kovacs JM, et al. HIV-1 envelope trimer elicits more potent neutralizing antibody responses than monomeric gp120. Proc Natl Acad Sci USA. 2012;109:12111–12116. doi: 10.1073/pnas.1204533109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanders RW, et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore PL, et al. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J Virol. 2006;80:2515–2528. doi: 10.1128/JVI.80.5.2515-2528.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dev J, et al. Structural basis for membrane anchoring of HIV-1 envelope spike. Science. 2016;353:172–175. doi: 10.1126/science.aaf7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seaman MS, et al. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol. 2010;84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang W, et al. Identification of subtype B, multiple circulating recombinant forms and unique recombinants of HIV type 1 in an MSM cohort in China. AIDS Res Hum Retroviruses. 2008;24:1245–1254. doi: 10.1089/aid.2008.0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gnanakaran S, et al. Genetic signatures in the envelope glycoproteins of HIV-1 that associate with broadly neutralizing antibodies. PLOS Comput Biol. 2010;6:e1000955. doi: 10.1371/journal.pcbi.1000955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pejchal R, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Julien JP, et al. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog. 2013;9:e1003342. doi: 10.1371/journal.ppat.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doria-Rose NA, et al. A short segment of the HIV-1 gp120 V1/V2 region is a major determinant of resistance to V1/V2 neutralizing antibodies. J Virol. 2012;86:8319–8323. doi: 10.1128/JVI.00696-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 22.Pedregosa F, et al. Scikit-learn: Machine learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 23.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kilby JM, et al. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- 25.Chen J, et al. Mechanism of HIV-1 neutralization by antibodies targeting a membrane-proximal region of gp41. J Virol. 2014;88:1249–1258. doi: 10.1128/JVI.02664-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frey G, et al. A fusion-intermediate state of HIV-1 gp41 targeted by broadly neutralizing antibodies. Proc Natl Acad Sci USA. 2008;105:3739–3744. doi: 10.1073/pnas.0800255105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Letarov AV, Londer YY, Boudko SP, Mesyanzhinov VV. The carboxy-terminal domain initiates trimerization of bacteriophage T4 fibritin. Biochemistry (Mosc) 1999;64:817–823. [PubMed] [Google Scholar]
- 28.Blattner C, et al. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity. 2014;40:669–680. doi: 10.1016/j.immuni.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kovacs JM, et al. Stable, uncleaved HIV-1 envelope glycoprotein gp140 forms a tightly folded trimer with a native-like structure. Proc Natl Acad Sci USA. 2014;111:18542–18547. doi: 10.1073/pnas.1422269112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seaman MS, et al. Standardized assessment of NAb responses elicited in rhesus monkeys immunized with single- or multi-clade HIV-1 envelope immunogens. Virology. 2007;367:175–186. doi: 10.1016/j.virol.2007.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parrish NF, et al. Phenotypic properties of transmitted founder HIV-1. Proc Natl Acad Sci USA. 2013;110:6626–6633. doi: 10.1073/pnas.1304288110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parker ZF, et al. Transmitted/founder and chronic HIV-1 envelope proteins are distinguished by differential utilization of CCR5. J Virol. 2013;87:2401–2411. doi: 10.1128/JVI.02964-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore JP, McKeating JA, Weiss RA, Sattentau QJ. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science. 1990;250:1139–1142. doi: 10.1126/science.2251501. [DOI] [PubMed] [Google Scholar]
- 34.Tran EE, et al. Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog. 2012;8:e1002797. doi: 10.1371/journal.ppat.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pugach P, et al. A native-like SOSIP.664 trimer based on an HIV-1 subtype B env gene. J Virol. 2015;89:3380–3395. doi: 10.1128/JVI.03473-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee JH, Ozorowski G, Ward AB. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science. 2016;351:1043–1048. doi: 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chung NP, et al. Stable 293 T and CHO cell lines expressing cleaved, stable HIV-1 envelope glycoprotein trimers for structural and vaccine studies. Retrovirology. 2014;11:33. doi: 10.1186/1742-4690-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kong L, et al. Uncleaved prefusion-optimized gp140 trimers derived from analysis of HIV-1 envelope metastability. Nat Commun. 2016;7:12040. doi: 10.1038/ncomms12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeffs SA, et al. Expression and characterisation of recombinant oligomeric envelope glycoproteins derived from primary isolates of HIV-1. Vaccine. 2004;22:1032–1046. doi: 10.1016/j.vaccine.2003.08.042. [DOI] [PubMed] [Google Scholar]
- 40.Chen B, Chou JJ. Structure of the transmembrane domain of HIV-1 envelope glycoprotein. FEBS J. November 20, 2016 doi: 10.1111/febs.13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Freeman MM, et al. Crystal structure of HIV-1 primary receptor CD4 in complex with a potent antiviral antibody. Structure. 2010;18:1632–1641. doi: 10.1016/j.str.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stewart-Jones GB, et al. Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B, and G. Cell. 2016;165:813–826. doi: 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.