Significance
The Integrator complex (INT) has important functions in the 3′-end processing of noncoding RNAs and RNA polymerase II transcription. The INT contains at least 14 subunits, but its molecular mechanism of action is still poorly understood. The endonuclease activity of INT is mediated by its subunit 11 (IntS11), which forms a stable complex with Integrator complex subunit 9 (IntS9) through their C-terminal domains (CTDs). Here, we report the crystal structure of the IntS9–IntS11 CTD complex at 2.1-Å resolution and detailed, structure-based biochemical and functional studies. Highly conserved residues are located in the extensive interface between the two CTDs. Yeast two-hybrid assays and coimmunoprecipitation experiments confirm the structural observations. Functional studies demonstrate that the IntS9–IntS11 interaction is crucial for INT in snRNA 3′-end processing.
Keywords: snRNA processing, protein complex, integrator complex
Abstract
The metazoan Integrator complex (INT) has important functions in the 3′-end processing of noncoding RNAs, including the uridine-rich small nuclear RNA (UsnRNA) and enhancer RNA (eRNA), and in the transcription of coding genes by RNA polymerase II. The INT contains at least 14 subunits, but its molecular mechanism of action is poorly understood, because currently there is little structural information about its subunits. The endonuclease activity of INT is mediated by its subunit 11 (IntS11), which belongs to the metallo-β-lactamase superfamily and is a paralog of CPSF-73, the endonuclease for pre-mRNA 3′-end processing. IntS11 forms a stable complex with Integrator complex subunit 9 (IntS9) through their C-terminal domains (CTDs). Here, we report the crystal structure of the IntS9–IntS11 CTD complex at 2.1-Å resolution and detailed, structure-based biochemical and functional studies. The complex is composed of a continuous nine-stranded β-sheet with four strands from IntS9 and five from IntS11. Highly conserved residues are located in the extensive interface between the two CTDs. Yeast two-hybrid assays and coimmunoprecipitation experiments confirm the structural observations on the complex. Functional studies demonstrate that the IntS9–IntS11 interaction is crucial for the role of INT in snRNA 3′-end processing.
The Integrator complex (INT) was originally characterized as an important factor for U-rich small nuclear RNA (UsnRNA) 3′-end processing and as a binding partner for phosphorylated RNA polymerase II (Pol II) (1–3). Since then it also has been found to participate in Pol II transcription initiation, pause release, elongation, and termination at protein-coding genes (4–6), thereby broadening the scope of INT function. Most recently INT was shown to be important for enhancer RNA (eRNA) 3′-end processing as well (7). The physiological importance of INT is reflected by its involvement in other cellular processes, such as embryogenesis (8), ciliogenesis (9), adipose differentiation (10), human lung function (11), and maturation of herpesvirus microRNA 3′ ends (12).
The INT contains at least 14 subunits (IntS1 through IntS14), ranging from 49 to 244 kDa, and the mass of the full complex is more than 1 million daltons. Despite its functional importance, INT is poorly understood at the molecular level, and currently there is very little structural information for any of its subunits. Moreover, the majority of the INT subunits share little sequence homology with other proteins and have few recognizable domains, making it difficult to predict the function and the mechanism of the subunits (2).
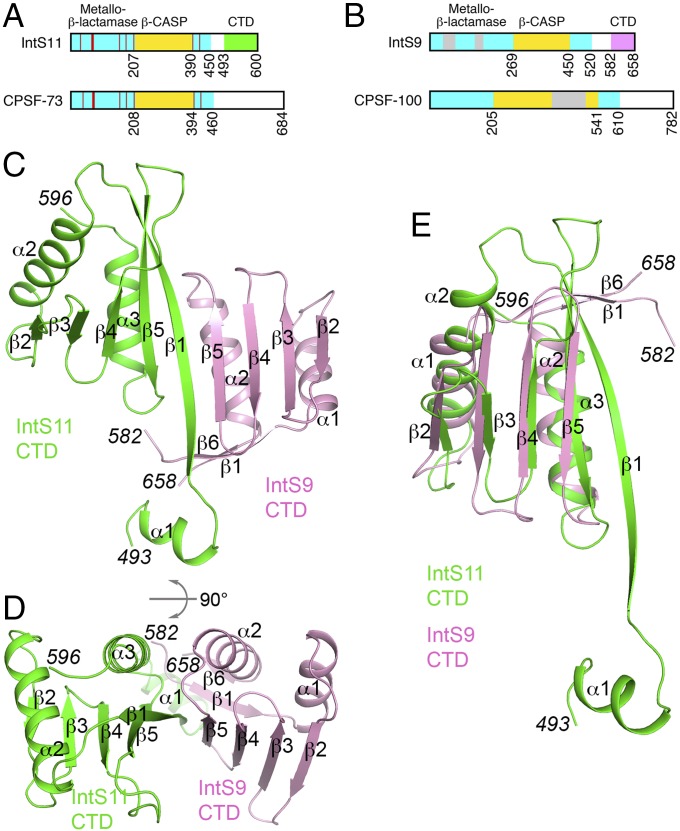
Two INT subunits that do share significant sequence conservation with other proteins are IntS9 and IntS11 (1, 2). Most importantly, IntS11 is a close homolog of CPSF-73 (cleavage and polyadenylation specificity factor, 73 kDa), the endonuclease for the pre-mRNA 3′-end processing machinery (13–16). Similarly, IntS11 is the endonuclease subunit for INT, responsible for the cleavage reaction at the 3′ end of target RNAs (1, 2). CPSF-73 and IntS11 belong to the metallo-β-lactamase superfamily of proteins (17). The metallo-β-lactamase domain of each protein contains conserved amino acids that coordinate metal ions for catalysis (Fig. 1A and Fig. S1) (18). CPSF-73, IntS11, and their close homologs also have a β-CASP domain (named after its founding members: CPSF73, Artemis, SNM, and PSO), which is an insert in the metallo-β-lactamase domain (Fig. 1A). The active site is located at the interface between the two domains, and therefore the β-CASP domain likely regulates substrate access to the active site. The metallo-β-lactamase and the β-CASP domains of human IntS11 (residues 1–450) (Fig. S1) and CPSF-73 share 40% amino acid sequence identity, indicating their high degree of sequence conservation and underscoring the importance of these two domains.
Fig. 1.
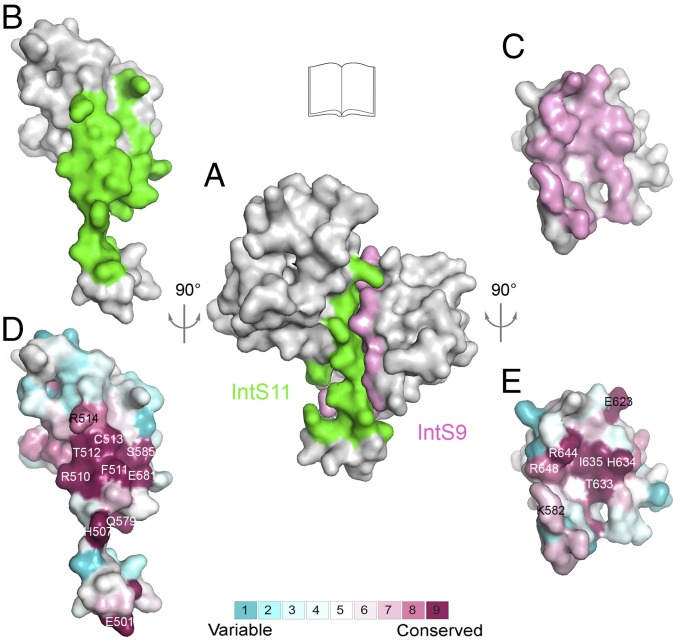
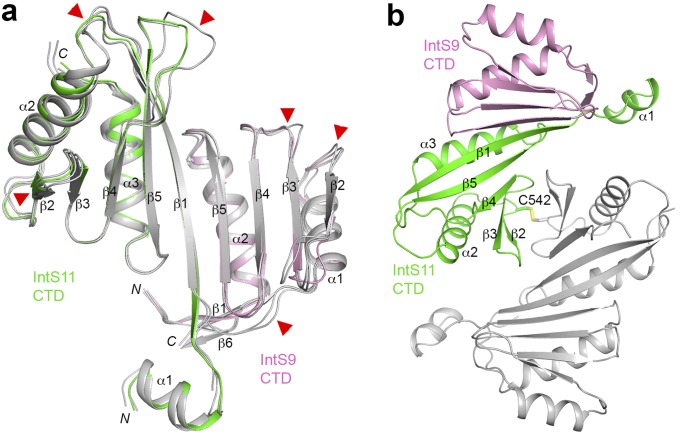
Crystal structure of the human IntS9–IntS11 CTD complex. (A) Domain organizations of human IntS11 and CPSF-73. The metallo-β-lactamase and β-CASP domains are shown in cyan and yellow, respectively. The conserved residues in the active site are indicated by red lines. The CTD of IntS11 is shown in green. CPSF-73 also has a CTD, but its sequence is highly divergent from that of IntS11, and its exact boundary is not known. (B) Domain organizations of human IntS9 and CPSF-100. The CTD of IntS9 is shown in pink. An insert in the β-CASP domain of CPSF-100 and two inserts in the metallo-β-lactamase domain of IntS9 are shown in gray. (C) Structure of the human IntS9–IntS11 CTD complex. The IntS9 CTD is in pink, and the IntS11 CTD is in green. (D) Structure of the human IntS9–IntS11 CTD complex, viewed after 90° rotation around the horizontal axis. (E) Overlay of the structure of the IntS9 CTD (pink) with the structure of the IntS11 CTD (green). The structure figures were produced with PyMOL (www.pymol.org).
Fig. S1.
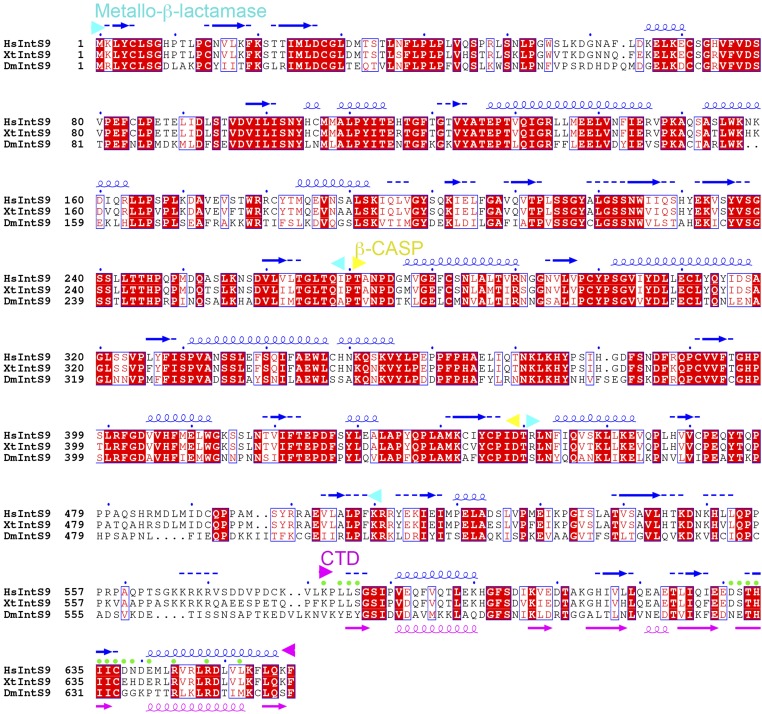
Sequence alignment of IntS11 subunits. The predicted secondary structure elements in human IntS11 (HsIntS11) are shown in blue above the sequence, and the secondary structure elements in the structure of HsIntS11 CTD are shown in green below the sequence. Residues in contact with IntS9 are indicated by pink dots. Conserved residues in the active site are indicated by red dots. The domain boundaries are indicated by arrowheads and are labeled. Dm, Drosophila melanogaster; Hs, Homo sapiens; Xt, Xenopus tropicalis. Modified from an output from ESPript (41).
The CPSF complex also contains an inactive homolog of CPSF-73, CPSF-100, in which several of the active-site residues have been changed during evolution. Likewise, IntS9 is the inactive homolog of IntS11 in INT (Fig. 1B and Fig. S2). The sequence conservation between IntS9 and CPSF-100 is weaker, and IntS9 has two inserted segments in the metallo-β-lactamase domain (Fig. 1B). The functional roles of IntS9 and CPSF-100 in their respective complexes remain to be clarified.
Fig. S2.
Sequence alignment of IntS9 subunits. The predicted secondary structure elements in human IntS9 (HsIntS9) are shown in blue above the sequence, and the secondary structure elements in the structure of HsIntS9 CTD are shown in pink below the sequence. Residues in contact with IntS11 are indicated by green dots. The sequence conservation between IntS9 and CPSF-100/Ydh1 is weak, and the boundaries for the metallo-β-lactamase and β-CASP domains can be defined only tentatively.
IntS9, IntS11, CPSF-73, and CPSF-100 all contain a C-terminal domain (CTD) beyond their metallo-β-lactamase and β-CASP domains (Fig. 1). However, the sequence conservation for these domains is much poorer. We showed previously that the CTDs of IntS9 and IntS11 are required and sufficient to mediate their association, suggesting that these domains ensure specific heterodimerization of the two subunits, thereby compartmentalizing INT from CPSF (19). However, the molecular basis for this association is not known. Currently available dimer structures of other β-CASP homologs (20–23) do not provide any insights into the IntS9–IntS11 heterodimer, nor are structures available for the CPSF-73/CPSF-100 CTDs.
We have determined the crystal structure of the IntS9–IntS11 CTD complex at 2.1-Å resolution. The structure is composed of a continuous nine-stranded β-sheet, with four strands from IntS9 and five from IntS11. Four helices cover one face of this β-sheet, and the other face is exposed to solvent. Highly conserved residues are located in the extensive interface between the two CTDs formed by the two neighboring strands and two helices. We designed truncation and site-specific mutants based on the structure, and both our yeast two-hybrid assays with the CTDs and coimmunoprecipitation experiments with the full-length proteins confirm the structural observations on the complex. Finally, we demonstrated that mutations that disrupt the IntS9–IntS11 interaction also abolish U7 snRNA 3′-end processing, indicating that this interaction is crucial for the function of the Integrator complex.
Results
Structures of the IntS9 and IntS11 CTDs.
The crystal structure of the complex of IntS9 and IntS11 CTDs has been determined at 2.1-Å resolution. The atomic model has good agreement with the X-ray diffraction data and the expected geometric parameters (Table S1). Of the residues, 98.1% are in the favored region of the Ramachandran plot, and no residues are in the disallowed region.
Table S1.
Summary of crystallographic information
| Structure | IntS9–IntS11 CTD complex |
| Data collection | |
| Space group | P21 |
| Cell dimensions | |
| a, b, c, Å | 63.0, 67.8, 98.6 |
| α, β, γ, degrees | 90, 100.6, 90 |
| Resolution, Å | 25–2.1 (2.2–2.1) |
| Rmerge, % | 7.8 (42.4) |
| CC1/2 | (0.898) |
| I/σI | 20.5 (3.5) |
| No. reflections | 51,362 (5,038) |
| Completeness | 99.6 (96.8) |
| Redundancy | 6.8 (5.4) |
| Refinement | |
| Resolution, Å | 25–2.1 (2.15–2.1) |
| No. reflections | 47,760 (3,362) |
| Rwork/Rfree, % | 16.7 (21.5)/22.0 (29.7) |
| No. atoms | |
| Protein | 5,636 |
| Water | 419 |
| B-factors | |
| Protein | 38.3 |
| Water | 39.2 |
| Rmsd | |
| Bond lengths, Å | 0.012 |
| Bond angles, degrees | 1.3 |
The numbers in parentheses are for the highest resolution shell.
The structure of the IntS9 CTD (covering residues 582–658) (Fig. 1B) contains a four-stranded, antiparallel β-sheet (β2–β5) (Fig. 1C). Two helices (α1–α2), formed by residues just before and after the β-sheet, cover one of its faces (Fig. 1D). A two-stranded antiparallel β-sheet (β1, β6) formed by residues near the beginning and end of the domain likely provides further stability to this domain.
The structure of the IntS11 CTD (covering residues 493–596) (Fig. 1A) contains a five-stranded antiparallel β-sheet (β1–β5) with two helices on one of its faces (α2-α3) (Fig. 1C). A short helix (α1) precedes the first β-strand and is partly stabilized by interactions with IntS9 (see next section).
The up–down organization of the last four strands (β2–β5) of the IntS11 CTD is similar to that for the β-sheet in IntS9 CTD. In fact, the two structures can be superposed with an rmsd of 2.4 Å for 64 equivalent Cα atoms (Fig. 1E), although the sequence identity between the two proteins in this region is only 16%. The two helices covering the β-sheet are located at similar positions in the two structures as well. A unique feature of IntS11 is strand β1, which is the longest strand in the structure and is located in the center of the interface with IntS9.
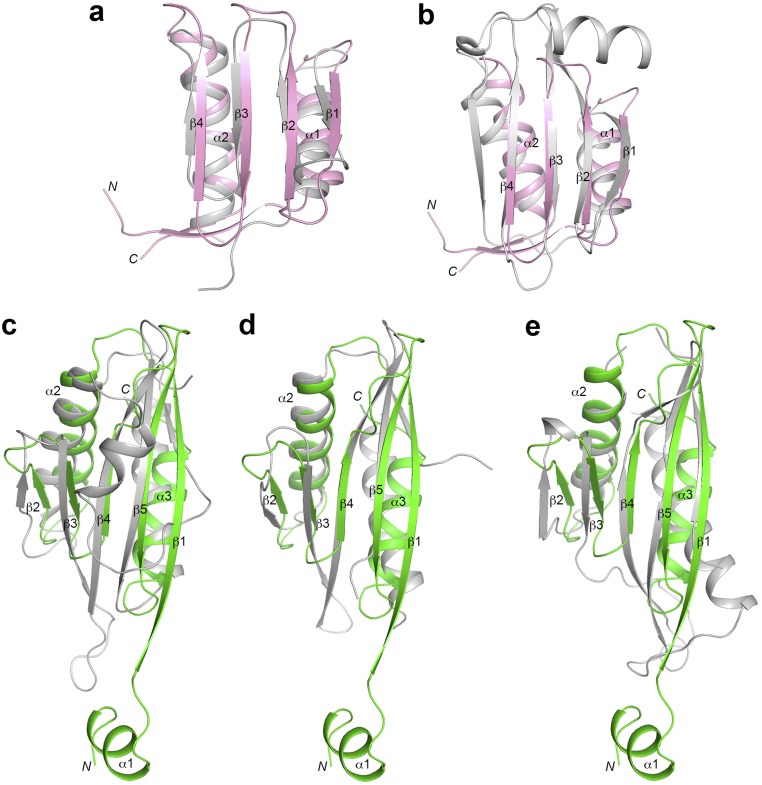
Close structural homologs for IntS9 CTD include the CTD of an atypical Sm-like archaeal protein (24) and the platform subdomain of the AP-2 complex β subunit (Fig. S3) (25), based on a DaliLite search (26). Close structural homologs for IntS11 CTD include the kinase associated-1 domain (KA1 domain) at the C terminus of yeast septin-associated kinases and human MARK/PAR1 kinases (27), the C-terminal domain of the catalytic subunit of AMP-activated protein kinase (AMPK, SNF1 in yeast) that mediates heterotrimer formation (28–30), and the N-terminal domain of BamC, part of the β-barrel assembly machinery (Fig. S3) (31). These structural homologs do not offer much insight into the functions of the two CTDs.
Fig. S3.
Structural homologs of IntS9 and IntS11 CTDs. (A and B) Overlay of the IntS9 CTD (pink) with the C-terminal domain of an atypical Sm-like archaeal protein [Protein Data Bank (PDB) ID code 1M5Q; Z-score 7.3; 17% identity] (gray) (A) and the platform subdomain of the AP-2 complex β subunit (PDB ID code 2IV9, Z-score 6.8; 4% identity) (B). (C–E) Overlay of the IntS11 CTD (green) with the KA1 domain of yeast Kcc4 (PDB ID code 3OSM; Z-score 7.9, 8% identity) (C), the C-terminal domain of the catalytic subunit of AMPK (PDB ID code 4EAK, Z-score 6.9, 16% identity) (D), and the N-terminal domain of BamC (PDB ID code 2YH6, Z-score 6.3, 10% identity) (E).
Crystal Structure of the IntS9–IntS11 CTD Complex.
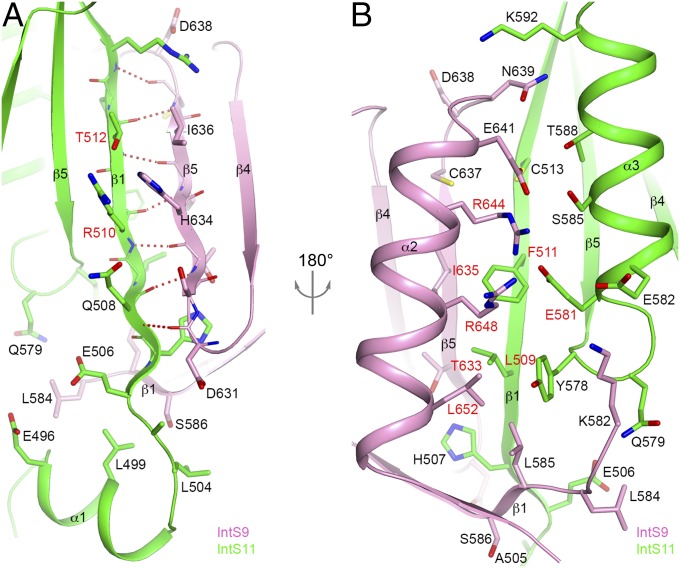
The complex of IntS9–IntS11 CTDs is formed by juxtaposing the β-sheets of the two domains, such that strand β5 of IntS9 forms a parallel β-sheet with strand β1 of IntS11 (Fig. 1C). This juxtaposition creates a nine-stranded, mostly antiparallel β-sheet in the IntS9–IntS11 CTD heterodimer, with only the two strands at the subunit interface being in parallel. The four flanking helices cover the same face of this β-sheet, and the other face of the β-sheet is open to the solvent (Fig. 1D). Seven hydrogen bonds are formed between the two β-strands at the center of the interface (Fig. 2A). In addition to these interactions, many side chains mediate the formation of this heterodimer as well, and ∼1,200 Å2 of the surface area of each subunit is buried in this interface (Fig. 3 A–C). The neighboring side chains of the two strands on the exposed face of the β-sheet are in contact with each other. In addition, the N-terminal helix (α1) of IntS11 contacts the N-terminal segment of IntS9, likely stabilizing both proteins in this region of the interface (Fig. 2A).
Fig. 2.
Detailed interactions at the interface of the IntS9–IntS11 CTD complex. (A) Hydrogen-bonding interactions between strand β1 of IntS11 (green) and strand β5 of IntS9 (pink) are indicated by the dashed lines in red. The side chains of the two β-strands are placed next to each other. Interactions between helix α1 of IntS11 and residues in IntS9 are also shown. Residues selected for mutagenesis studies are labeled in red. (B) Interactions between residues in helix α3 of IntS11 (green) and residues in helix α2 of IntS9 (pink). Residues in strand β1 of IntS11 and strand β5 of IntS9 also contribute to this part of the interface.
Fig. 3.
(A) Molecular surface of the IntS9–IntS11 CTD complex. Residues in IntS9 that contribute to the interface with IntS11 are colored in pink, and those in IntS11 that contact IntS9 are in green. The other residues are in gray. (B) An “open-book” view of the IntS9–IntS11 interface showing the surface area of IntS11 in contact with IntS9 after 90° rotation around the vertical axis. (C) An open-book view of the IntS9–IntS11 interface showing the surface area of IntS9 in contact with IntS11 after 90° rotation around the vertical axis. (D) Molecular surface of IntS11 colored by sequence conservation, produced by ConSurf (40). Highly conserved residues are labeled. The color scheme runs from dark red (highly conserved) to cyan (poorly conserved) (color bar at bottom). The view is the same as in B. (E) Molecular surface of IntS9 colored by sequence conservation.
On the other face of the β-sheet, helix α2 of IntS9 and helix α3 of IntS11 are positioned next to each other, allowing favorable interactions among some of their side chains as well as the side chains of the two β-strands in the center of the interface (Fig. 2B). Residues from the two β-strands are mostly hydrophobic in this part of the interface, whereas those from the two helices are mostly hydrophilic or charged. Most of the residues at this interface are highly conserved among IntS11 (Fig. 3D and Fig. S1) and IntS9 (Fig. 3E and Fig. S2) homologs, especially near the center of the interface.
There are four copies of the IntS9–IntS11 CTD complex in the crystallographic asymmetric unit. The overall structures of the two subunits in the four complexes are similar, with rmsds of ∼0.5 Å for equivalent Cα atoms between any pair of them. The overall structures of the four complexes are similar as well, especially for the β-sheet and the four flanking helices (Fig. S4). However, there are large differences in the conformations of several of the loops, suggesting that these regions are somewhat flexible. In addition, the Cys542 residues from two IntS11 subunits in neighboring complexes form a disulfide bond covalently linking two complexes (Fig. S4), and this disulfide linkage is likely a crystallization artifact. The other cysteine residues in the structure are in the fully reduced state. Cys542 is located just before strand β2 in IntS11, and some conformational differences in this strand are observed among the four complexes (Fig. S4). It is unlikely that this disulfide bond affects the overall structure of the complex, because it creates only a relatively small region of contact between two complexes (Fig. S4).
Fig. S4.
Structural comparisons of the four copies of IntS9–IntS11 CTD complexes. (A) Overlay of the structures of the four IntS9–IntS11 CTD complexes. One complex is shown in color, and the other three are shown in gray. Regions of large conformational differences among the four complexes are indicated by red arrowheads. The α1 helix of IntS11 is missing in one of the subunits. (B) The two Cys542 residues from neighboring IntS11 subunits in the crystal are involved in a disulfide bond, and the two complexes are related by a noncrystallographic twofold axis. One complex is shown in color, and the other is shown in gray.
Biochemical Studies Confirm the Structural Observations.
To assess the structural observations on the IntS9–IntS11 CTD complex, we carried out yeast two-hybrid assays to evaluate interactions between different variants of the two CTDs as well as coimmunoprecipitation experiments with the full-length proteins. We previously demonstrated that using yeast two-hybrid assay to characterize the interaction between IntS9 and IntS11 is remarkably robust, because binding could be detected even when 3-amino-1,2,4-triazole is present at a 100-mM concentration (32). We carefully mapped the regions within the CTDs of IntS9 and IntS11 that are both required and sufficient to mediate their interaction. We created a series of truncation mutants, removing 10 amino acid residues at a time, and found that residues 500–600 of IntS11 interacted strongly with IntS9 CTD, whereas residues 510–600 showed no interaction (Fig. 4A). Residue 500 is located near the end of helix α1, and residue 510 is in the middle of strand β1 (Fig. 1C), indicating the importance of β1. This assay also determined that helix α1 of IntS11 is not required for the interaction, as is consistent with its being located at the periphery of the interface. On the other hand, deleting only 10 residues from the C terminus of IntS11 (resulting in a variant with residues 500–590) abolished the interaction (Fig. 4A). Residue 590 is located in the last turn of helix α3, confirming its importance for the IntS9–IntS11 interaction.
Fig. 4.
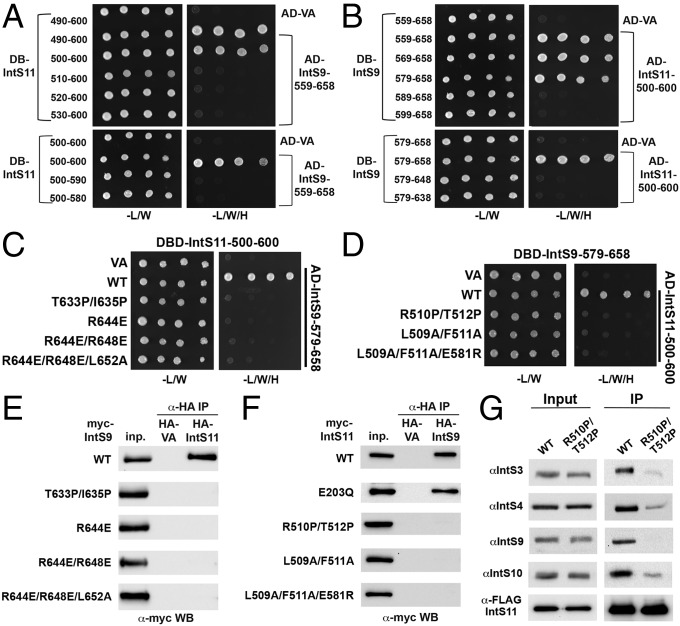
Biochemical studies confirm the structural observations on the IntS9–IntS11 CTD complex. (A) Yeast two-hybrid assay to define the minimal region of IntS11 sufficient to bind IntS9. (Upper) Ten amino acid deletions starting from residue 490. (Lower) Ten amino acid deletions starting from the C terminus of IntS11. AD, activation domain; BD, DNA-binding domain; VA, vector alone control. (B) Yeast two-hybrid assay to define the minimal region of IntS9 sufficient to bind IntS11. (C) Yeast two-hybrid assay using the minimal regions of IntS9 and IntS11 sufficient for their interaction, with structure-based mutations in IntS11 CTD. (D) Yeast two-hybrid assay using the minimal regions of IntS9 and IntS11 sufficient for their interaction, with structure-based mutations in IntS9 CTD. (E) Coimmunoprecipitation of full-length myc-tagged wild-type and mutant IntS9 with full-length wild-type HA-tagged IntS11. Proteins bound to HA affinity resin were probed with anti-myc antibody by Western blot (WB). (F) Coimmunoprecipitation of full-length myc-tagged wild-type and mutant IntS11 with full-length wild-type HA-tagged IntS9. (G) Purification of endogenous INT from stable 293T cells expressing either wild-type FLAG-IntS11 or the heterodimeric mutant (R510P, T512P) using FLAG affinity resin.
Similarly, we found that residues 579–658 of IntS9 interacted strongly with the IntS11 CTD, whereas residues 589–658 showed no interactions (Fig. 4B). Residue 579 is before the first residue observed in the current structure, and residue 589 is just after strand β1 (Fig. 1C). Deleting 10 residues from the C terminus of IntS9 (579–648) also abolished the interaction (Fig. 4B). Residue 648 is located in the middle of helix α2. Overall, the results from the truncation mutants define the minimal regions of IntS9 and IntS11 that are important for their interactions; these results are fully consistent with the structural observations.
We next designed a series of point mutations that are expected to perturb the IntS9–IntS11 interaction based on the structural observations. To perturb the hydrogen-bonding interactions between the two β-strands in the dimer interface, we mutated two residues in the middle of each strand to proline, i.e., the T633P/I635P double mutant for IntS9 (Fig. 2B) and the R510P/T512P double mutant for IntS11 (Fig. 2A). We also designed mutations to disrupt interactions among the side chains, including the R644E single mutant, the R644E/R648E double mutant, and the R644E/R648E/L652A triple mutant in helix α2 of IntS9, the L509A/F511A double mutant in strand β1 of IntS11, and the L509A/F511A/E581R triple mutant in strand β1 and helix α3 of IntS11 (Fig. 2B). Most of these residues are strictly conserved among the homologs, whereas Leu652 of IntS9 and Leu509 of IntS11 show conservative variations to other hydrophobic residues (Figs. S1 and S2). We introduced these mutations into the minimal CTDs of IntS9 or IntS11 and observed a complete loss of interaction based on the yeast two-hybrid assays (Fig. 4 C and D).
To extend upon these data, we also introduced the point mutations into the full-length cDNAs encoding IntS9 and IntS11 and tested their impact on the interaction using a coimmunoprecipitation assay. We had established previously that the IntS9–IntS11 heterodimer could withstand rigorous washing with detergent and high salt (19) and therefore tested these same conditions here. We transfected various myc-tagged IntS9 cDNAs with HA-tagged wild-type IntS11 into 293T cells and subjected the lysates to anti-HA immunoaffinity matrix followed by probing with anti-myc antibodies using Western blot analysis. Only the wild-type IntS9 was able to coimmunoprecipitate with IntS11; none of the mutants was detected in the immunoprecipitate (Fig. 4E). Importantly, all four mutants tested were expressed at levels similar to the wild type, suggesting that these proteins are folded properly and that the lack of coimmunoprecipitation is caused by the disruption of the interaction by the mutations.
We then performed the reciprocal coimmunoprecipitation in which we transfected HA-tagged wild-type IntS9 with several mutants of IntS11. We also included a catalytic mutant (E203Q, mutating one of the conserved residues in the active site of the metallo-β-lactamase domain) as a control; this mutation is not expected to disrupt interaction with IntS9. Both the wild type and the E203Q mutant of IntS11 were able to interact with IntS9, but the interface mutants did not show interaction (Fig. 4F). Finally, we extended these analyses to ask whether disruption of the IntS9–IntS11 interface inhibits the ability of the heterodimer to be incorporated into the endogenous Integrator complex. Previously, we established that purification of the intact INT could be achieved by pulling down a FLAG-tagged IntS11 from nuclear extracts derived from stable cells lines expressing FLAG-IntS11 (33). Therefore, we created cells stably expressing either wild-type IntS11 or a mutant IntS11 and purified the INT from nuclear extracts made from these cells lines. Almost no IntS9 was associated with the mutant IntS11, as expected, but we also observed a significant reduction in the levels of copurifying IntS3, IntS4, and IntS10 relative to wild-type IntS11 (Fig. 4G). Taken together, these results strongly validate the structure of the IntS9–IntS11 CTD complex and demonstrate that this region is essential for the two Integrator subunits to interact and may play a significant role in the incorporation of the IntS11 endonuclease into the endogenous INT.
Functional Importance of the IntS9–IntS11 Complex.
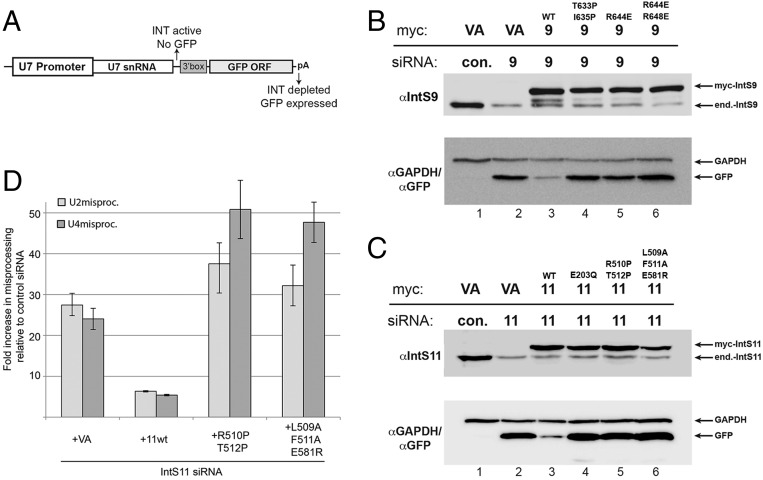
Currently an in vitro system is not available to assess INT function. Therefore, to address the functional relevance of the IntS9–IntS11 interactions observed in the crystal structure, we used a cell-based fluorescence reporter that assays for UsnRNA 3′-end formation (34). The U7-GFP reporter consists of the human U7 snRNA promoter, the U7 snRNA gene body, a 3′ box sequence for snRNA 3′-end processing, and then the coding sequence for GFP followed by a strong polyadenylation signal (Fig. 5A). When transfected into untreated cells, the reporter gives rise to no GFP expression because of the 3′-end processing activity of endogenous INT. If cells are transfected with siRNA targeting INT subunits, the efficiency of snRNA 3′-end formation is reduced, resulting in transcriptional read-through and the production of a GFP mRNA containing the U7 snRNA as its 5′ UTR using the native start codon of GFP because U7 lacks an AUG sequence.
Fig. 5.
Functional importance of the IntS9–IntS11 interactions for snRNA 3′-end processing. (A) Schematic of the U7-GFP reporter that is transfected into human cells. (B) Western blot analysis of lysates from HeLa cells transfected with either control siRNA or IntS9 siRNA that then were transfected with either empty vector or myc-tagged RNAi-resistant IntS9. All cells were also transfected with the U7-GFP reporter. (C) The same analysis as in B, except that cells were treated with siRNA targeting IntS11 rather than IntS9. (D) Quantitative RT-PCR analysis of misprocessed U2 or U4 snRNA that are endogenously expressed. The bar graph represents the fold increase in the levels of misprocessed snRNA; data show the results of biological triplicates; error bars represent the SD from the mean.
We transfected cDNA constructs containing silent point mutations rendering them RNAi-resistant into IntS9- or IntS11-depleted cells to determine how effective these constructs are at restoring INT activity through the reduction of GFP expression. In control siRNA-treated cells, we detected little to no expression of GFP after transfection with the U7-GFP reporter (Fig. 5 B and C). In contrast, we could detect robust GFP expression in cells treated with siRNA targeting either IntS11 or IntS9. Expression of RNAi-resistant myc-tagged wild-type IntS11 or IntS9 could reduce GFP expression nearly back to the levels observed in control siRNA-transfected cells. We observed that, as expected, the E203Q catalytic mutant of IntS11 was unable to rescue U7 snRNA processing (Fig. 5B). Most importantly, mutations in IntS11 or IntS9 that disrupt the interactions between their CTDs also failed to rescue U7 snRNA processing, despite being expressed at levels similar to the wild-type protein (Fig. 5 B and C). Finally, we created stable cell lines expressing either RNAi-resistant wild-type IntS11 or a subset of the heterodimerization IntS11 mutants and assessed the levels of misprocessed, endogenously expressed U2 or U4 snRNA after knockdown. Upon depletion of IntS11 we observed an ∼25-fold increase in the levels of misprocessed snRNA relative to the levels observed in control siRNA-treated cells (Fig. 5D). This level of misprocessing was similar to what we observed in Drosophila S2 cells upon depletion of INT subunits (35). The levels of misprocessed snRNA present in IntS11 knockdown cells could be rescued upon stable expression of the wild-type IntS11, but, as in the U7-GFP reporter experiments, we did not observe any rescue of snRNA misprocessing in cells expressing RNAi-resistant IntS11 heterodimerization mutants (Fig. 5D). Collectively, these results demonstrate that interactions observed in the structure of the IntS9–IntS11 CTD complex are as critical to INT activity in snRNA processing as the residues within the active site of IntS11.
Discussion
Although a role for IntS11 in the cleavage of UsnRNA and eRNA has been established using mutations of the metal-coordinating residues within the active site, the molecular mechanism by which IntS11 is recruited to these substrates and carries out specific endonucleolytic processing is not known. The structure of the IntS9–IntS11 CTD heterodimer reveals an extensive molecular interface mediated by numerous interactions and explains the high binding affinity that has been reported for the two proteins. The structure also indicates a role for the catalytically inactive IntS9, which provides a distinct structural surface established only through heterodimerization with IntS11. This specific interface could allow recognition of only the active cleavage factor by the other members of the INT complex. Such a mechanism also might be operative for CPSF-73 and CPSF-100 in the pre-mRNA 3′-end processing machinery.
The CTDs of IntS9 and IntS11 are substantially larger than that of the homologous enzyme RNase J, which is comprised of a three-stranded β-sheet and two facing α-helices (23). Deletion of the RNase J CTD makes the enzyme become monomeric in solution and also abrogates all catalytic activity in vitro, even though the ΔCTD RNase J retains structurally intact metallo-β-lactamase and β-CASP domains. Based on results from our studies using the U7-GFP reporter, it is clear the mutations that specifically disrupt the formation of the IntS9–IntS11 CTD heterodimer have effects equivalent to those of the mutation (E203Q) that disrupts the active site of IntS11. This finding demonstrates that the binding to IntS9 is essential for IntS11 function in cells and suggests that homo- or heterodimerization of β-CASP RNA endonucleases either plays an important role in the recruitment to RNA substrates or somehow impacts the activity of the catalytic domain. One potential explanation is that formation of the IntS9–IntS11 CTD complex induces obligatory conformational changes in IntS11, for example in the interface between IntS11 metallo-β-lactamase and β-CASP domains, to allow access to and cleavage of the RNA substrates. This structural requirement would ensure that any IntS11 not associated with IntS9 would be inactive, and, by analogy, the same would hold true for CPSF-73 and CPSF-100.
CTD heterodimerization may provide another important function in addition to modulating the catalytic activity of IntS11. In this model, the IntS9–IntS11 CTD complex produces an essential surface that is recognized by a different member of the Integrator complex to recruit the dimerized cleavage factor into the complex. This mechanism would provide an elegant way of ensuring that only the authentic IntS11–IntS9 heterodimer is incorporated into INT and could represent an additional layer of regulation to prevent spurious cleavage events. This model is supported by our experiments demonstrating that a heterodimer-deficient IntS11 failed to associate with other members of INT in addition to IntS9 (Fig. 4G). Indeed, a large scaffolding protein, symplekin, interacts with CPSF-73 and CPSF-100 and likely plays a critical role in mediating cleavage of pre-mRNA substrates (13–16). Such a protein is likely to exist within the Integrator complex, but currently there is no candidate based upon sequence comparison with symplekin.
The linkers between the metallo-β-lactamase domain and the CTD in IntS9 and IntS11 are expected to contain secondary structure elements (Figs. S1 and S2) and hence are likely to be organized structurally. In the structure of RNase J, the linker also contains secondary structure elements and has interactions with both domains (23). Exactly how the linkers in IntS9 and IntS11 connect the two parts of these proteins remains to be determined. It is possible that this region of the proteins functions to communicate heterodimerization to the active site to allow cleavage to take place.
Methods
Protein Expression, Purification, and Crystallization.
The C-terminal domain of human IntS11 (residues 491–600) was subcloned into the pET28a vector (Novagen), which introduced an N-terminal His-tag. The C-terminal domain of human IntS9 (residues 582–658) was subcloned into pCDFDuet vector (Novagen) without any affinity tag. The two proteins were coexpressed in Escherichia coli BL21Star (DE3) cells at 23 °C for 16–20 h. The cells were lysed by sonication in a buffer containing 20 mM Tris (pH 8.5), 200 mM NaCl, and 5% (vol/vol) glycerol. The IntS9–IntS11 heterodimer was purified by Ni-NTA (Qiagen) chromatography. The eluted protein was treated overnight with thrombin at 4 °C to remove the His-tag and was further purified by gel filtration chromatography (Sephacryl S-300; GE Healthcare). The purified protein was concentrated to 30 mg/mL in a solution containing 20 mM Tris (pH 8.5), 200 mM NaCl, and 10 mM DTT before being flash-frozen in liquid nitrogen and stored at −80 °C.
Crystals of the IntS9–IntS11 complex were obtained at 20 °C using the sitting-drop vapor-diffusion method. The reservoir solution contained 0.1 M Bis-Tris (pH 6.5) and 21–24% (wt/vol) PEG 3350. The protein concentration was 10 mg/mL. Crystals took 2 wk to grow to full size. A heavy-atom derivative was prepared by soaking native crystals in the mother liquor with 1 mM HgCl for 3 h. All crystals were cryo-protected by the reservoir solution supplemented with 5% (vol/vol) ethylene glycol and were flash-frozen in liquid nitrogen for data collection at 100 K.
Data Collection and Structure Determination.
X-ray diffraction data of native and heavy-atom–derivative crystals were collected at a wavelength of 0.979 Å on an ADSC Q315R CCD at the 5.0.1 beamline of Advanced Light Source (ALS). The diffraction images were processed with the HKL program (36). The crystals belonged to space group P21 with cell dimensions of a = 63.0 Å, b = 67.8 Å, c = 98.6 Å, and β = 100.6°. There are four copies of the IntS9–IntS11 complex in the crystallographic asymmetric unit.
A native dataset was collected to 2.1-Å resolution, and the derivative dataset was collected to 2.3-Å resolution. Four Hg atoms were located and used for phasing by the AutoSol routine in PHENIX (37), using the single isomorphous replacement (SIR) method. Most of the protein residues were automatically built by the AutoBuild routine in PHENIX, and further manual building was carried out with the program Coot (38). The structure was refined using PHENIX. The crystallographic information is summarized in Table S1.
Yeast Two-Hybrid Assays.
Yeast two-hybrid assays were carried out in PJ69-4a and PJ49-4alpha. Human IntS11 or IntS9 CTD fragments were cloned into either pOBD or pOAD vectors using conventional cloning. Clones were sequenced to verify identity; PCR primers are available upon request. pOBD plasmids were transformed into PJ69-4a yeast and were selected on tryptophan-dropout medium; pOAD plasmids were transformed into PJ49-4alpha yeast and were selected on leucine-dropout medium. Double transformants were created by mating the yeast strains followed by selection on medium lacking both tryptophan and leucine. Interactions were tested through serial dilution of diploid yeast followed by plating on medium lacking tryptophan and leucine or on medium lacking tryptophan, leucine, and histidine that also was supplemented with 1 mM 3-amino-1,2,4-triazole.
Coimmunoprecipitation.
IntS11 and IntS9 cDNAs were cloned into pcDNA3 expression plasmids and were subjected to site-directed mutagenesis as described previously (10). All clones were sequenced to confirm identity. Approximately 5 × 105 293T cells (in one well of a six-well plate) were transfected with 1 μg of each plasmid encoding either HA-tagged or myc-tagged IntS9 or IntS11 using Lipofectamine 2000 according to the manufacturer’s instructions (Thermo Fisher). Forty-eight hours after transfection, cells were lysed in 500 μL of denaturing lysis buffer (19), and 50 μL was removed for input lanes. To the remaining lysate, 20 μL of anti-HA affinity resin (Sigma) was added and incubated at 4 °C for 1 h with rotation. Following immunoprecipitation, beads were washed three times in lysis buffer and eluted with SDS loading buffer. Western blots were performed using SDS/PAGE as described previously (19). Affinity purification of FLAG-IntS11 was conducted essentially as described previously (33). Western blots were conducted using antibodies raised to IntS3 (PTGlab), IntS4 (Bethyl), IntS9 (Bethyl), IntS10 (PTGlab), and FLAG epitope (Sigma).
Cell Culture and RNAi Assays.
RNAi-rescue experiments were performed using HeLa cells, which were grown under standard conditions using DMEM and 10% FBS. Cells were plated initially at 8.5 × 104 cells per well in a 24-well plate. Cells were transfected with control siRNA (GGUCCGGCUCCCCCAAAUGdTdT), IntS9 siRNA (GAAAUGCUUUCUUGGACAAdTdT), or IntS11 siRNA (CAGACUUCCUGGACUGUGUdTdT) using a two-hit protocol (39). Twenty-four hours after the second siRNA transfection, cells were transfected a third time with 500 ng of the U7-GFP reporter (19) and with 200 ng of empty pcDNA-myc vector or with pcDNA-myc where either RNAi-resistant wild-type IntS9/IntS11 or mutant versions were cloned. Two days after the transfection, cells were lysed in denaturing buffer and probed using Western blot analysis with antibodies raised against GFP (Clontech), IntS11 (Bethyl), or GAPDH (Thermo). To monitor endogenous snRNA misprocessing, RNA was isolated from cells using TRIzol (Thermo Scientific) and was subjected to MMLV reverse transcription according to the manufacturer’s instructions (Life Sciences). Real-time PCR was conducted using SYBR Green PCR mix on a CFX quantitative PCR machine (Bio-Rad), and fold calculation was done as described previously (35). Primers to measure U2snRNA misprocessing are 5′-CTTCGGGGAGAGAACAACC-3′ and 5′-GACACTCAAACACGCGTCA-3′. Primers to measure U4snRNA misprocessing are 5′-GCATTGGCAATTTTTGACAG-3′ and 5′-GAACCCCGGACATTCAATC-3′.
Acknowledgments
We thank Marc Allaire and Nathan Smith for access to beamline 5.0.1 at the Advanced Light Source. This research was supported by NIH Grants R35GM118093 and S10OD012018 (to L.T.) and by Grants H1880 from the Welch Foundation and Cancer Prevention and Research Institute of Texas Grant RP140800 (to E.J.W.). The Berkeley Center for Structural Biology is supported in part by the NIH, the National Institute of General Medical Sciences, and the Howard Hughes Medical Institute. The Advanced Light Source is supported by the US Department of Energy under Contract DE-AC02-05CH11231.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.S. is a Guest Editor invited by the Editorial Board.
Data deposition: Crystallography, atomic coordinates, and structure factors reported in this paper have been deposited in the Protein Data Bank (accession code 5V8W).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1616605114/-/DCSupplemental.
References
- 1.Baillat D, et al. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–276. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 2.Baillat D, Wagner EJ. Integrator: Surprisingly diverse functions in gene expression. Trends Biochem Sci. 2015;40:257–264. doi: 10.1016/j.tibs.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamamoto J, et al. DSIF and NELF interact with Integrator to specify the correct post-transcriptional fate of snRNA genes. Nat Commun. 2014;5:4263. doi: 10.1038/ncomms5263. [DOI] [PubMed] [Google Scholar]
- 4.Gardini A, et al. Integrator regulates transcriptional initiation and pause release following activation. Mol Cell. 2014;56:128–139. doi: 10.1016/j.molcel.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skaar JR, et al. The Integrator complex controls the termination of transcription at diverse classes of gene targets. Cell Res. 2015;25:288–305. doi: 10.1038/cr.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stadelmayer B, et al. Integrator complex regulates NELF-mediated RNA polymerase II pause/release and processivity at coding genes. Nat Commun. 2014;5:5531. doi: 10.1038/ncomms6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai F, Gardini A, Zhang A, Shiekhattar R. Integrator mediates the biogenesis of enhancer RNAs. Nature. 2015;525:399–403. doi: 10.1038/nature14906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kapp LD, Abrams EW, Marlow FL, Mullins MC. The integrator complex subunit 6 (Ints6) confines the dorsal organizer in vertebrate embryogenesis. PLoS Genet. 2013;9:e1003822. doi: 10.1371/journal.pgen.1003822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jodoin JN, et al. The snRNA-processing complex, Integrator, is required for ciliogenesis and dynein recruitment to the nuclear envelope via distinct mechanisms. Biol Open. 2013;2:1390–1396. doi: 10.1242/bio.20136981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otani Y, et al. Integrator complex plays an essential role in adipose differentiation. Biochem Biophys Res Commun. 2013;434:197–202. doi: 10.1016/j.bbrc.2013.03.029. [DOI] [PubMed] [Google Scholar]
- 11.Obeidat M, et al. GSTCD and INTS12 regulation and expression in the human lung. PLoS One. 2013;8:e74630. doi: 10.1371/journal.pone.0074630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie M, et al. The host Integrator complex acts in transcription-independent maturation of herpesvirus microRNA 3′ ends. Genes Dev. 2015;29:1552–1564. doi: 10.1101/gad.266973.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 2010;38:2757–2774. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Q, Doublié S. Structural biology of poly(A) site definition. Wiley Interdiscip Rev RNA. 2011;2:732–747. doi: 10.1002/wrna.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jurado AR, Tan D, Jiao X, Kiledjian M, Tong L. Structure and function of pre-mRNA 5′-end capping quality control and 3′-end processing. Biochemistry. 2014;53:1882–1898. doi: 10.1021/bi401715v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romeo V, Schümperli D. Cycling in the nucleus: Regulation of RNA 3′ processing and nuclear organization of replication-dependent histone genes. Curr Opin Cell Biol. 2016;40:23–31. doi: 10.1016/j.ceb.2016.01.015. [DOI] [PubMed] [Google Scholar]
- 17.Pettinati I, Brem J, Lee SY, McHugh PJ, Schofield CJ. The chemical biology of human metallo-b-lactamase fold proteins. Trends Biochem Sci. 2016;41:338–355. doi: 10.1016/j.tibs.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Callebaut I, Moshous D, Mornon J-P, de Villartay J-P. Metallo-beta-lactamase fold within nucleic acids processing enzymes: The beta-CASP family. Nucleic Acids Res. 2002;30:3592–3601. doi: 10.1093/nar/gkf470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Albrecht TR, Wagner EJ. snRNA 3′ end formation requires heterodimeric association of integrator subunits. Mol Cell Biol. 2012;32:1112–1123. doi: 10.1128/MCB.06511-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li de la Sierra-Gallay I, Pellegrini O, Condon C. Structural basis for substrate binding, cleavage and allostery in the tRNA maturase RNase Z. Nature. 2005;433:657–661. doi: 10.1038/nature03284. [DOI] [PubMed] [Google Scholar]
- 21.Mir-Montazeri B, Ammelburg M, Forouzan D, Lupas AN, Hartmann MD. Crystal structure of a dimeric archaeal cleavage and polyadenylation specificity factor. J Struct Biol. 2011;173:191–195. doi: 10.1016/j.jsb.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 22.Silva AP, et al. Structure and activity of a novel archaeal β-CASP protein with N-terminal KH domains. Structure. 2011;19:622–632. doi: 10.1016/j.str.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li de la Sierra-Gallay I, Zig L, Jamalli A, Putzer H. Structural insights into the dual activity of RNase J. Nat Struct Mol Biol. 2008;15:206–212. doi: 10.1038/nsmb.1376. [DOI] [PubMed] [Google Scholar]
- 24.Mura C, Phillips M, Kozhukhovsky A, Eisenberg D. Structure and assembly of an augmented Sm-like archaeal protein 14-mer. Proc Natl Acad Sci USA. 2003;100:4539–4544. doi: 10.1073/pnas.0538042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmid EM, et al. Role of the AP2 beta-appendage hub in recruiting partners for clathrin-coated vesicle assembly. PLoS Biol. 2006;4:e262. doi: 10.1371/journal.pbio.0040262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holm L, Kääriäinen S, Rosenström P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moravcevic K, et al. Kinase associated-1 domains drive MARK/PAR1 kinases to membrane targets by binding acidic phospholipids. Cell. 2010;143:966–977. doi: 10.1016/j.cell.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Townley R, Shapiro L. Crystal structures of the adenylate sensor from fission yeast AMP-activated protein kinase. Science. 2007;315:1726–1729. doi: 10.1126/science.1137503. [DOI] [PubMed] [Google Scholar]
- 29.Amodeo GA, Rudolph MJ, Tong L. Crystal structure of the heterotrimer core of Saccharomyces cerevisiae AMPK homologue SNF1. Nature. 2007;449:492–495. doi: 10.1038/nature06127. [DOI] [PubMed] [Google Scholar]
- 30.Xiao B, et al. Structural basis for AMP binding to mammalian AMP-activated protein kinase. Nature. 2007;449:496–500. doi: 10.1038/nature06161. [DOI] [PubMed] [Google Scholar]
- 31.Albrecht R, Zeth K. Structural basis of outer membrane protein biogenesis in bacteria. J Biol Chem. 2011;286:27792–27803. doi: 10.1074/jbc.M111.238931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dominski Z, Yang X-C, Purdy M, Wagner EJ, Marzluff WF. A CPSF-73 homologue is required for cell cycle progression but not cell growth and interacts with a protein having features of CPSF-100. Mol Cell Biol. 2005;25:1489–1500. doi: 10.1128/MCB.25.4.1489-1500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baillat D, Russell WK, Wagner EJ. CRISPR-Cas9 mediated genetic engineering for the purification of the endogenous integrator complex from mammalian cells. Protein Expr Purif. 2016;128:101–108. doi: 10.1016/j.pep.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 34.Peart N, Wagner EJ. Gain-of-function reporters for analysis of mRNA 3′-end formation: Design and optimization. Biotechniques. 2016;60:137–140. doi: 10.2144/000114390. [DOI] [PubMed] [Google Scholar]
- 35.Ezzeddine N, et al. A subset of Drosophila integrator proteins is essential for efficient U7 snRNA and spliceosomal snRNA 3′-end formation. Mol Cell Biol. 2011;31:328–341. doi: 10.1128/MCB.00943-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 37.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 38.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 39.Wagner EJ, Garcia-Blanco MA. RNAi-mediated PTB depletion leads to enhanced exon definition. Mol Cell. 2002;10:943–949. doi: 10.1016/s1097-2765(02)00645-7. [DOI] [PubMed] [Google Scholar]
- 40.Armon A, Graur D, Ben-Tal N. ConSurf: An algorithmic tool for the identification of functional regions in proteins by surface mapping of phylogenetic information. J Mol Biol. 2001;307:447–463. doi: 10.1006/jmbi.2000.4474. [DOI] [PubMed] [Google Scholar]
- 41.Gouet P, Courcelle E, Stuart DI, Métoz F. ESPript: Analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]