Abstract
Genetic studies of complex traits have mainly identified associations with non-coding variants. To further determine the contribution of regulatory variation, we combined whole genome and transcriptome data for 624 individuals from Sardinia in order to identify common and rare variants that influence gene expression and splicing. We identified 21,183 expression quantitative trait loci (eQTLs) and 6,768 splicing quantitative trait loci (sQTLs), including 619 novel QTLs. We identified high-frequency QTLs and evidence of selection near genes involved in malarial resistance and increased multiple sclerosis risk, reflecting the epidemiological history of Sardinia. Using family relationships, we identified 809 segregating expression outliers (median z-score of 2.97), averaging 13.3 genes per individual. Outlier genes were enriched for proximal rare variants, providing a new approach to study large-effect regulatory variants and their relevance to traits. Our results provide insight into the effects of regulatory variants and their relationship to population history and individual genetic risk.
INTRODUCTION
Human migration and rapid population expansion have led to an abundance of population and individual-specific genetic variation1–5. Within protein-coding regions of the genome, multiple studies have identified numerous rare loss-of-function alleles6–11 that affect monogenic disorders and, to a lesser extent and especially in founder populations, common diseases and complex traits12–14. Most of the variants associated with complex traits are found outside protein-coding regions, however, and their functional consequences remain elusive. Large studies of gene expression have greatly advanced our ability to identify functional variation in non-coding regions of the genome15–17, and many of these variants have been connected to common genetic diseases18,19. However, few studies to date have had access to whole genome sequencing data, family relationships, and auxiliary complex trait data from research participants. Such data has the potential to empower the assessment of population and individual-specific consequences of regulatory variants.
To overcome this, we sequenced RNA isolated from the white blood cells of 624 individuals from the founder population of Sardinia. The Sardinian population has several advantages: their DNA includes the bulk of mainland European DNA variation, but due to a period of relative isolation for >10,000 years, many alleles have been added, and many old and novel variants have reached dramatically higher frequencies which should improve power to detect associations between those variants and traits such as gene expression20–22. In addition, the SardiNIA study cohort has been extensively genotyped and phenotyped and consists of both unrelated and related individuals23. By combining RNA-seq data with whole genome sequencing data, we discovered expression and splicing quantitative trait loci (e/sQTLs) that are specific to the isolated Sardinian population. As this is the first e/sQTL study to integrate both whole genomes and transcriptomes from multiple families, we developed a framework that leverages these family relationships in order to identify large-effect rare regulatory variants. We identified extreme gene expression outliers that segregate within these families and investigate the distribution and associated functional annotations of putatively causal rare variants as well as their influence on individual disease risk. This approach enhances ongoing studies of loss-of-function variants by demonstrating a new approach to identifying and studying large-effect alleles.
RESULTS
Expression and splicing quantitative trait discovery in Sardinia
The 624 participants, all from four towns in the Lanusei Valley in the Ogliastra region of Sardinia, were enrolled from a cohort of 6,921 in the SardiNIA longitudinal study of aging24. The entire SardiNIA cohort was genotyped using the Cardio-MetaboChip, ImmunoChip, exomeChip, and OmniExpress arrays. A subset of 2,120 Sardinians were additionally whole-genome sequenced at low coverage (average four-fold), producing an integrated map of ~15 million SNPs after imputation. This cohort and imputation pipeline has been previously described20,23,25. For RNA, we sequenced a median of ~59 million 51 bp paired-end reads per participant (over 36 billion reads in total). After quantification and quality control, 15,243 and 12,603 genes were sufficiently expressed for eQTL and sQTL analyses, respectively (Table 1). To account for confounding factors that can reduce power to discover cis-QTLs, we performed hidden factor correction with PEER26. We were able to identify and remove factors correlated with gender, age, various blood cell counts, and sequencing (Supplementary Figure 1, Supplementary Table 1).
Table 1. Expression traits with at least one eQTL.
We report the number of tests performed and the number of significant QTL associations for different expression traits at a false discovery rate of 5%. Associations that are significant by BH are significant after Bonferroni correction and Benjamini-Hochberg adjustment (see Methods)
| Measurement | Trait type | # of tested traits | # of traits with at least one QTL (FDR 5%) | |
|---|---|---|---|---|
|
| ||||
| BH | By permutation | |||
| Gene-level | Protein coding | 11,477 | 8,381 (73%) | 9,019 (79%) |
| lncRNA | 1,694 | 991 (69%) | 1,258 (74%) | |
| miRNA precursors | 172 | 48 (27%) | 55 (32%) | |
| Other | 1,900 | 935 (39%) | 835 (44%) | |
|
| ||||
| Total | 15,243 | 10,329 (68%) | 11,167 (73%) | |
|
| ||||
| Isoform-proportion* | Protein coding | 11,116 | 3,865 (35%) | 4,515 (41%) |
| lncRNA | 826 | 335 (41%) | 373 (45%) | |
| Other | 661 | 213 (32%) | 323 (49%) | |
|
| ||||
| Total | 12,603 | 4,413 (35%) | 5,120 (41%) | |
Isoforms results are reported at gene-level (only one sQTL per gene is reported)
To discover eQTLs, we tested the association of genotype with expression level for all variants within ±1 Mb of a target gene’s transcription start site (TSS) for all individuals with genetic data in the integrated map (n = 606). At a false discovery rate (FDR) of 5%, we identified eQTLs for the majority of tested genes (Table 1). We then used a forward-stepwise regression approach to characterize the number of independent eQTLs per gene (see Methods). We found that approximately half of all protein-coding and lncRNA transcripts were influenced by at least two independent eQTLs; miRNAs, however, were mostly associated with a single eQTL (Table 2). At the extreme, we found a single protein-coding gene, ITGB1BP1, affected by 14 independent eQTLs. ITGB1BP1 encodes an integrin binding protein that is implicated in upstream regulation of immune-critical TNF/NF-kB transcriptional regulation. We also identified a lncRNA of unknown function, NBPF1, that was affected by 11 independent eQTLs (Supplementary Table 2). In total, we mapped at least one eQTL for 73% of tested genes, corresponding to 11,167 primary eQTLs. Our forward-stepwise regression analysis identified an additional 10,016 secondary eQTLs for a total of 21,183 eQTLs (Table 2). We observed that both primary and secondary QTLs were enriched in diverse functional annotations (Supplementary Figure 2; Supplementary Tables 3–4).
Table 2. Independent QTLs segmented by gene type.
We report the number of independent QTLs for gene-level and isoform-level analyses. Isoform results are grouped by their respective gene.
| Max # of independent QTLs | Gene-level (# of genes) | Isoform-proportion (# of genes) | |||
|---|---|---|---|---|---|
|
| |||||
| Protein coding | lncRNA | miRNA precursors | Protein coding | lncRNA | |
| 1 | 4,215 | 598 | 44 | 3,489 | 281 |
| 2 | 2,833 | 386 | 8 | 799 | 60 |
| 3 | 1,235 | 170 | 2 | 165 | 22 |
| 4 | 428 | 66 | 0 | 36 | 8 |
| 5 | 175 | 18 | 1 | 18 | 1 |
| 6 | 82 | 6 | 0 | 3 | 1 |
| 7 | 27 | 5 | 0 | 5 | 0 |
| 8 | 14 | 3 | 0 | 0 | 0 |
| ≥9 | 10 | 6 | 0 | 0 | 0 |
|
| |||||
| Total | 9,019 | 1,258 | 55 | 4,515 | 373 |
To discover sQTLs, we tested the association of genotype with the ratio of known transcript abundances calculated using Cufflinks27. At an FDR of 5%, we observed significant sQTLs for nearly half of the protein-coding genes and lncRNAs we tested. In total, this is over a thousand more sQTLs than previously reported15,17. In comparison to eQTLs, we found that protein-coding genes and lncRNAs were less likely to have multiple independent sQTLs (Table 2); however, we found five protein-coding genes that were influenced by as many as seven independent sQTLs (Supplementary Table 5). Notably, two of these genes affect transcription and splicing itself, and are expected to impact the immune system. These genes include: POLR2J2, which encodes one of two nearly identical polymerase II subunit genes known to produce alternative transcripts; and SMN1, whose product functions in the assembly of the spliceosome. The other three sQTL genes are directly related to immune function: the non-classical class I heavy chain paralog HLA-G; the class I heavy chain receptor HLA-C; and ITGB1BP1. The ITGB1BP1 gene, which has 8 exons that extend over 16 kb and are spliced into 21 isoforms, had extreme numbers of both independent eQTLs and sQTLs, suggesting that it is a large mutational target for modulators of expression. While less pervasive than eQTLs, we mapped at least one sQTL for 41% of tested genes, corresponding to 5,120 primary sQTLs. Our forward-stepwise regression analysis identified an additional 1,648 secondary sQTLs for a total of 6,768 sQTLs (Table 2).
We compared our forward-stepwise regression approach to an alternative method implemented in the MLMM software package28 that uses a stepwise mixed-model regression with forward inclusion and backward elimination in order to identify independent associations. Both approaches resulted in a similar number of independent eQTL associations (Supplementary Table 6) and sQTL associations (Supplementary Table 7) that were largely consistent with our original findings (see Supplementary Note). Furthermore, we performed simulations to assess the impact of statistical noise and missing SNPs on our independent association analyses. We ran our pipeline on simulated expression traits where a single, randomly selected SNP in the ±1 Mb region of each gene explained 25% of the trait’s variance (we did this for each gene where at least one eQTL was found in the original analysis). Only a small fraction of these simulations resulted in multiple independent associations compared to the actual analysis (Supplementary Table 8). We repeated this simulation a second time but excluded the randomly chosen causal SNP from the association-mapping phase. While we observed more independent associations relative to the first analysis (Supplementary Table 9), the similarity between the results of these simulations and the consistency observed between our pipeline and MLMM suggest our approach is identifying independent associations and is robust to statistical noise and residual LD blocks near these genes.
Comparison of Sardinia and European eQTLs identifies novel functional and trait-associated variants
We next measured the replication of Sardinian QTLs with European QTLs found in LCLs (GEUVADIS15) and whole blood (Depression Genes and Networks17; DGN). For Sardinian eQTLs that were tested in each study, the replication rate was 92% in DGN and 72% in GEUVADIS, reflecting the high-degree of sharing of common European alleles within Sardinia (Supplementary Table 10). For sQTLs, the replication rate was 72% in DGN and 76% in GEUVADIS. Additionally, we tested eQTLs and sQTLs found in either DGN or GEUVADIS for replication in the Sardinia cohort and found that between 89–92% of eQTLs and 70–97% of sQTLs replicated (Supplementary Table 11). Replication could not be tested for 2,568 eQTLs and 1,152 sQTLs found in Sardinia because the SNPs were either absent in Europe or only present at a minor allele frequency below 1%. Of these QTLs, 437 eQTLs and 182 sQTLs were novel in Sardinia when compared to the 1000 Genomes, dbSNP, UK10K, and ExAC databases, representing new and/or previously uncaptured functional variation.
We first observed that novel eQTLs were depleted from known disease genes (Supplementary Figure 3). To determine if these novel eQTLs were associated with traits measured in Sardinia, we tested all 437 novel eQTL variants for associations with 15 blood cell measurements in the whole SardiNIA cohort (N ≈ 6,000; Supplementary Table 12). We identified 5 associations (5 traits and 4 variants, in total) that were significant after correcting for multiple testing (p-value < 8.8 × 10−6). For each association, we then retested the trait association for all variants within ±4 Mb of the target gene to identify the subset of loci where both the Sardinia-specific eQTL variant and the top trait-associated variant within this region were in high LD (r2 > 0.8). We identified a Sardinia-specific eQTL for ARHGDIB that was also linked to the top trait-associated variant for neutrophil percentage, which is also Sardinia-specific (Supplementary Figure 4; top neutrophil percentage variant chr12:g.14190223T>C, p-value = 3.8 × 10−6; top eQTL, chr12:g.15553026G>T p-value = 7.69 × 10−6, r2 = 0.86). We further performed an eQTL/trait colocalization analysis with eCAVIAR29 and observed strong local colocalization between the ARHGDIB eQTL and both neutrophil and lymphocyte percentages (Supplementary Figure 5). Within this locus, only 3 of 14 variants that passed our LD filter have been previously reported outside of Sardinia (allele frequencies in Europe below 0.002). Of note, one of these variants (chr12:g.15095546G>C, p-value = 3.85 × 10−6, r2 with top neutrophil signal = 0.84) is a nonsense mutation that had been observed only once in the ExAC database but has a frequency >1% in Sardinia, with the direction of effect on expression consistent with nonsense-mediated decay. ARHGDIB presents a biologically plausible target for this association as it is a multi-function protein with a central role in inhibition of cell migration, and ARHGDIB−/− mice show changes in lymphocyte expansion and survival in culture30.
Sardinia eQTLs exhibit founder population effects and evidence of selection
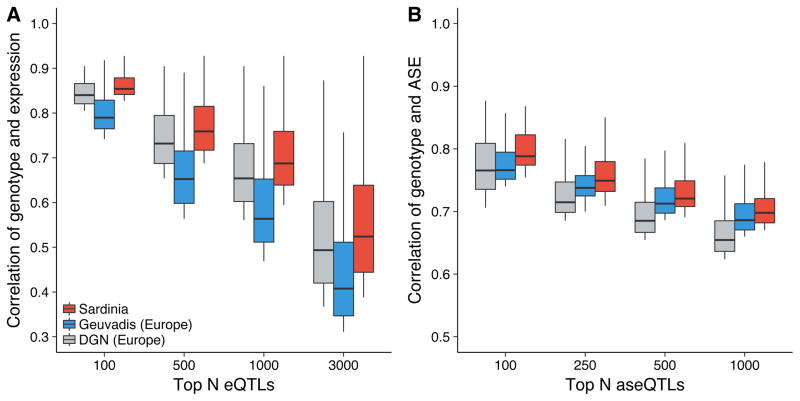
As genetic analyses in founder populations like Sardinia are expected to have increased statistical power based on relatively low genetic heterogeneity and shared environment21, we compared the observed impact of Sardinian eQTLs to European eQTLs. Using an identical pipeline and controlling for various differences in study parameters, we regenerated European eQTLs from the DGN and GEUVADIS studies (see Methods). When comparing eQTLs between these studies and Sardinia, we observed increased correlation between expression and genotype for Sardinian eQTLs (Figure 1A). This could reflect founder population effects or reduced technical noise in our study. As allele-specific expression signals have been demonstrated to be more robust to technical noise, we also compared Sardinian allele-specific expression QTLs (aseQTLs) to European aseQTLs31. We observed an increased correlation of genotype and allelic expression for Sardinia aseQTLs, similar to the trend we observed for eQTLs and consistent with a founder population effect (Figure 1B).
Figure 1. QTLs show larger effect sizes in Sardinia compared to Europe.
The distribution of Spearman correlation coefficients (absolute value) is shown for (a) top expression QTLs (eQTLs) and (b) top allele-specific expression QTLs (aseQTLs) in Sardinia, Geuvadis, and DGN. Top eQTLs and aseQTLs in Sardinia show increased correlations relative to Geuvadis and DGN. To make analyses comparable across studies, 188 unrelated individuals from each study were uniformly processed and analyses were performed on a subset of genes that were quantifiable in all three studies.
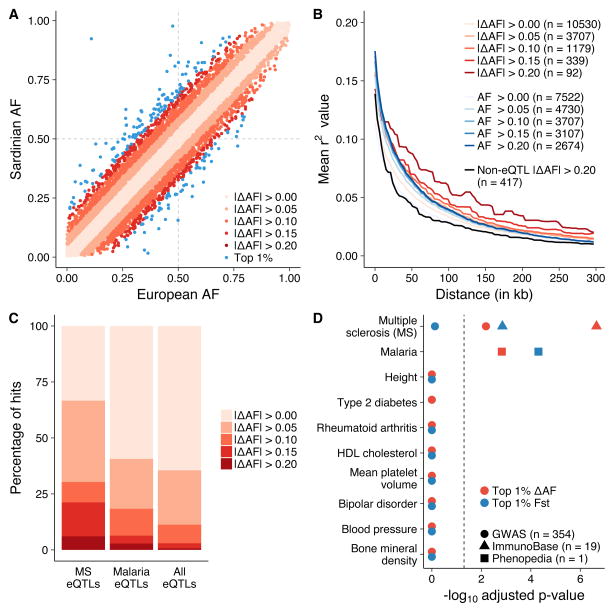
To identify eQTLs where founder effects, genetic drift, or selective pressures have significantly influenced the prevalence of these alleles in Sardinia, we first compared the Sardinian allele frequencies of eQTLs and sQTLs with the corresponding European allele frequency reported by the 1000 Genomes Project5. We found that 11% of significant eQTLs were differentiated at an allele frequency greater than 10% (Figure 2A). In addition, we observed longer tracts of linkage disequilibrium (LD) decay in Sardinians conditioned on the extent of allelic differentiation for eQTLs versus non-eQTLs (Figure 2B). Furthermore, ten of the top 1% of differentiated eQTLs showed evidence of hard selective sweeps (integrated haplotype scores |iHS| > 2.5), consistent with a proportion of these eQTLs having undergone recent positive selection32,33 (Supplementary Figure 6, Supplementary Table 13).
Figure 2. Differentiated eQTLs in Sardinia.
(a) Sardinian eQTLs are plotted based on their allele frequency in Europe (measured in the 1000 Genomes Project) and Sardinia. Blue points represent eQTLs in the top 1% of the |ΔAF| distribution. Sample sizes: |ΔAF| > 0.00 (n = 19,108 eQTLs), > 0.05 (n = 6,793), > 0.10 (n = 2,151), > 0.15 (n = 567), > 0.20 (n = 134), and Top 1% (n = 192). (b) eQTLs with larger allele frequency differences compared to Europe have longer tracts of LD decay as potential evidence for recent positive selection. These are compared to eQTLs that have comparable allele frequencies in Sardinia and Europe (allele frequencies within ±2.5%; blue lines) as well as randomly selected, distance to TSS-matched, non-eQTL variants with large allele frequency changes (black line). (c) eQTLs linked to multiple sclerosis variants and malaria-associated genes are both enriched in allele frequency difference changes between Sardinia and Europe. (d) The significance of the top ten trait enrichments for differentiated eQTLs (red = ΔAF, blue = FST) after Bonferroni correction for all possible tests. Traits with less than 10 eQTLs in LD were filtered out.
Highly differentiated eQTLs are enriched for malaria and multiple sclerosis genes
We next tested whether two epidemiological factors present in Sardinia were reflected among highly differentiated eQTLs. Until the mid-twentieth century, the Sardinian population suffered high mortality rates due to malaria34,35, and continues to have a higher prevalence of multiple sclerosis (MS) relative to other Caucasian populations in the Mediterranean basin36,37. Indeed, we identified a significant enrichment for known malarial resistance genes (p-value = 0.0015) and genes associated with MS (EBI/NHGRI GWAS Catalog nominal p-value = 1.84 × 10−5 and ImmunoBase nominal p-value = 1.17 × 10−8) among the top 1% of differentiated eQTLs (mean allele frequency difference of ~17%) (Figure 2C–D, Supplementary Table 14). MS had the highest enrichment among 354 traits tested from the EBI/NHGRI GWAS catalog and among 19 traits tested from the ImmunoBase catalog (Figure 2D, Supplementary Figure 7). Furthermore, GWAS hits for MS show evidence for co-localization with eQTLs identified in Sardinia, suggesting that regulation of these genes mediates the association signals at these loci (Supplementary Table 15).
One of the most differentiated eQTLs was associated with expression levels of the BAFF gene (p-value = 8.051 × 10−12, ΔAFSRD-EUR = 0.25), which is known to be involved in the response and survival to malaria infection38–40 and has unique evolutionary history in Sardinia (Steri et al, submitted). We also identified several regulatory variants for the CR1 gene whose product is involved in complement activation and immune complex formation during malaria infection41,42. CR1 has two eQTLs (chr1:g.207275799G>A and chr1:g.207667190G>C) and 9 sQTLs. The eQTL at chr1:g.207667190G>C is highly differentiated between Sardinia and Europe (ΔAFSRD-EUR = −0.25) (Supplementary Figure 8). Among the 9 sQTLs associated with CR1, two of them are highly differentiated: chr1:g.207681501C>G (ΔAFSRD-EUR = 0.42) influences the abundance of ENST00000367051 and chr1:g.207716099A>C (ΔAFSRD-EUR = 0.43) influences the abundance of ENST00000529814. Both sQTLs are tightly linked and in high LD (r2 = 0.99 and 0.95) with a variant at chr1:g.207757515A>G that has been previously associated with erythrocyte sedimentation rate in the SardiNIA cohort43.
Finally, as ΔAF itself does not account for background selection near genes, we used an alternative method to define differentiated Sardinia eQTLs based on FST values (see Supplementary Note). Differentiated eQTLs identified with this method were similarly enriched near genes associated with malaria (p-value = 4.91 × 10−5, Supplementary Table 16) and near MS loci (Figure 2D), with MS being the most significantly enriched trait in both the EBI/NHGRI GWAS catalog (nominal p-value = 2.11 × 10−3, Supplementary Table 17) and ImmunoBase (nominal p-value = 7.41 × 10−5, Supplementary Table 18).
Heritable patterns of extreme gene expression in families
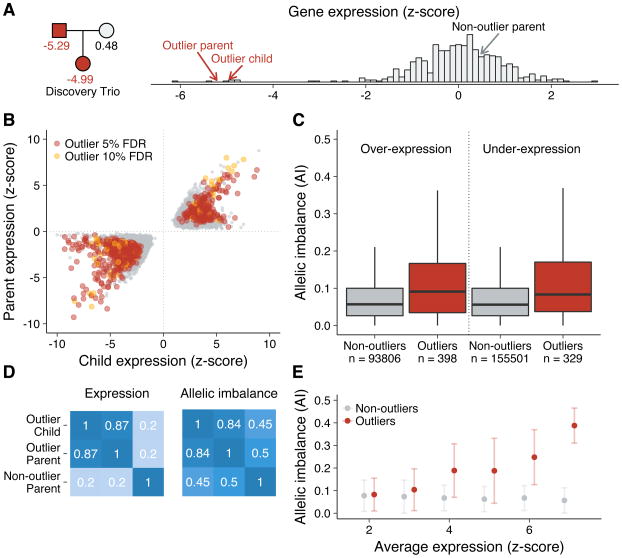
Beyond the unique history of the Sardinia population, the availability of family relationship data in the SardiNIA cohort provided an opportunity to identify the impact of rare, large-effect regulatory variation. Specifically, we developed a likelihood ratio test to identify patterns of extreme gene expression that segregated in families (Figure 3A; see Methods). We tested 61 Sardinian trios for the 15,243 genes included in our eQTL analyses and identified 809 genes where a parent and child are both expression outliers (median z-score = 2.97) at an FDR of 10% (Figure 3B). On average we found 13.3 shared gene expression outliers per child.
Figure 3. Outlier gene expression in Sardinian trios.
(a) An example of a significant gene expression outlier effect that segregates in a single Sardinia trio. The father and daughter both under express the RINL gene and share a rare splicing variant. (b) A scatterplot showing the sharing of extreme gene expression patterns between parents and children in 61 Sardinian trios, with significant outliers highlighted (orange = 5% FDR and yellow = 10% FDR). (c) Heterozygous sites in outlier genes show elevated levels of allelic imbalance (AI) in outlier individuals (red) versus the rest of the population (gray). Allelic imbalance (AI) measures the absolute deviance of the reference allele ratio from 0.5 at heterozygous sites. (d) Correlation matrices for gene expression and allele-specific expression within outlier trios suggest that the extreme regulatory effects are restricted to the affected individuals and not primarily a family-specific event due to a shared environment. (e) The relationship between outlier gene expression and allelic imbalance (AI) in outlier (red) and non-outlier (gray) individuals. The mean ± one s.d. is shown for each bin.
Several lines of evidence suggest shared expression outliers are not due simply to parent-offspring shared environment. There was little correlation of gene expression between the outlier parent and the non-outlier partner (Pearson r = 0.20) (Figure 3D). Additionally, mothers and fathers were equally likely to be the outlier parent (p = 0.20), regardless of the sex of the child (p = 0.83) (Supplementary Figure 9). In addition, we used a separate method to identify outliers based on z-scores alone and found that approximately 10% of the average child’s extreme expression outliers were shared with one parent alone and the remaining 90% are likely not caused by genetics44 (Supplementary Figure 10). These results are concordant with Tabassum et al44 who found ~100 expression outliers per individual that could be largely explained by extrinsic factors, e.g. cell type proportions.
We found almost twice as many shared under-expression outliers (529 outliers, 65%) as over-expression outliers (280 outliers, 35%), consistent with observations of the effects of random substitutions in promoters and enhancers in massively parallel reporter assays45–47. Furthermore, since rare variants tend to be heterozygotic and thus only influence one allele, we hypothesized that outlier parents and children would be enriched for allele-specific expression compared to non-outlier controls. We found that allele-specific expression was significantly enriched in outlier individuals for both under- and over-expression outliers (adjusted Wilcoxon rank-sum p-value = 6.0 × 10−6) (Figure 3C). This is likely a conservative estimate of the true enrichment, given the inherently low levels of read depth in under-expression outliers that limits the ability to measure allelic effects in outlier genes. These allelic effects were consistent between outlier parents and children (Pearson r = 0.84) but not with the other, non-outlier parent (Figure 3D). The strength of the outlier effect was also significantly associated with the enrichment of allele-specific expression (Spearman ρ = 0.338, p-value < 1 × 10−6), reflecting the capacity of allele-specific effects to impact total expression (Figure 3E).
Rare variants can underlie extreme gene expression in families
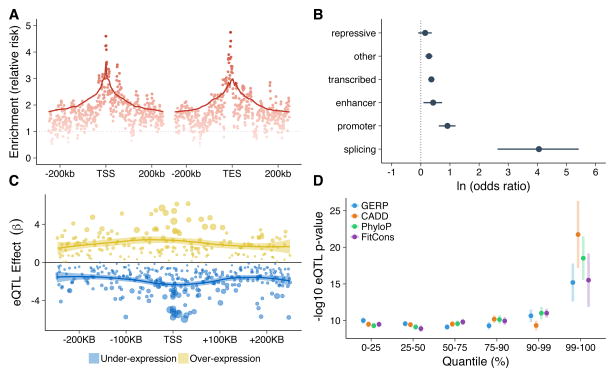
Using the combination of whole genome data and family relationships, we were able to characterize potential causal variants underlying expression outliers. We first identified 3,464 rare variants (Sardinia MAF < 1%) that were located in 250 kb windows adjacent to the transcription start site (TSS) and end site (TES) of outlier genes and were unambiguously transmitted from the outlier parent to the outlier child (i.e. the variant was heterozygous in both the outlier parent and child and the other parent was homozygous for the reference allele). We also identified an equivalent set of 245,165 rare variants in the same genomic loci that were unambiguously transmitted between non-outlier parents and their children. We found at least one shared rare variant for 509 of the outlier genes (63%), with an average of 6.8 variants shared by outliers versus 4.0 shared by non-outliers (enrichment = 1.71, 95% confidence interval 1.65 – 1.77). Of interest, rare variants shared by outlier individuals were concentrated within 5 kb of the TSS (enrichment = 3.61, 95% confidence interval 2.96 – 4.24) and TES (enrichment = 3.00, 95% confidence interval 2.44 – 3.54) (Figure 4A) of outlier genes, similar to what has been observed for common regulatory variation48. Furthermore, rare variants shared by outliers were enriched in multiple functional annotations49 (Figure 4B, Supplementary Figure 11). For variants in the 50 kb window adjacent to the TSS, this enrichment was most notable in splice donor/acceptor sites (log odds = 4.05, p-value = 2.52 × 10−7) and regions associated with active transcription, including promoters (log odds = 0.91, p-value = 8.8 × 10−9) and enhancers (log odds = 0.42, p-value = 0.0094) (enrichment data for different genomic window sizes is provided in Supplementary Tables 19–20). We further investigated whether other carriers of these variants had the same outlier expression profile as the parent-child pairs. We analyzed 2,912 variants (84% of the 3,464 outlier variants) that were heterozygous in at least four individuals in the cohort, regressing outlier gene expression on genotype at the rare variant position. The largest and most significant of these genotype-expression associations for both over- and under-expression outliers were concentrated at the TSS of outlier genes (Figure 4C). Additionally, we found that metrics of conservation (GERP, PhyloP) and predicted functional relevance (FitCons, CADD) all discriminated the most significant associations (Figure 4D)50–53.
Figure 4. Properties of rare, shared variants near outlier genes.
(a) Relative enrichment in the number of rare variants transmitted between outlier parents and children versus non-outlier parents and children. Relative enrichments were calculated in overlapping windows of 5 kb for the 250 kb regions adjacent to the TSS and TES of outlier genes. Enrichment is measured as the relative risk of finding rare shared variants in outlier versus non-outlier lineages in each window. (b) Shared rare variants in outlier lineages are enriched for functional regions of chromatin in peripheral blood and splice donor/acceptor regions. Enrichments are shown as the log odds ratio derived from Fisher’s exact tests with 95% confidence intervals. (c) The position of shared rare variants is plotted relative to the TSS against the regression coefficient derived from the rare eQTL analysis. The color represents under-expression (blue) and over-expression (yellow) rare eQTLs, and the size indicates relative significance. (d) Metrics of conservation, evolutionary constraint, fitness, and deleteriousness can identify the most significant rare eQTLs. The mean ± one s.d. is shown for each bin.
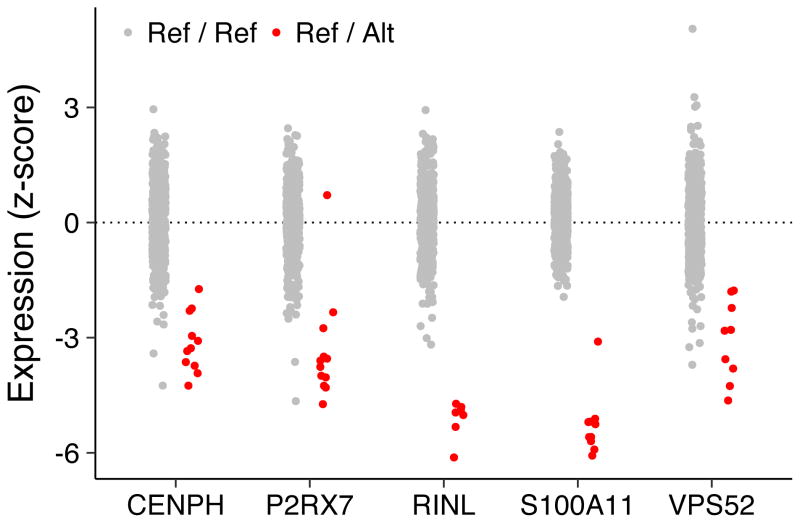
Based on these observations, we developed a strict set of rules to distinguish putatively causal rare variants by prioritizing variants that were close to the TSS or likely involved in splicing, highly conserved, and replicated their effects in the larger population (see Methods). We identified candidate causal variants for 30 outlier genes (Supplementary Table 21), including five rare splicing variants. One of these splicing variants, chr12:g.121570899G>T, is found at the first exon-intron boundary of the P2RX7 gene, which codes for a ligand-gated ion channel responsible for ATP-dependent lysis of macrophages. While chr12:g.121570899G>T is rare in all European populations including Sardinia, where it is most frequent with a MAF = 0.009% (Supplementary Table 22), it has been previously shown to disrupt proper splicing of P2RX7, leading to an elongated transcript that is subsequently degraded by nonsense-mediated decay and results in mono-allelic expression54. As expected, all carriers of chr12:g.121570899G>T (n = 12) in the Sardinia cohort under-expressed P2RX7 and all reads showed the same allele. While the other splicing variants have not been characterized, we saw similar trends for all five splicing variants suggesting that all of these putative splicing variants are effectively null alleles (Figure 5).
Figure 5. Gene expression patterns in carriers of rare splicing variants.
We identified five splicing variants in under-expression outliers - for each variant, the expression level of the affected gene is shown in red for heterozygous carriers in the Sardinia cohort and gray for individuals homozygous for the reference allele. The rare splicing variants for each gene are given here: chr12:g.121570899G>T for P2RX7; chr5:g.68490523G>A for CENPH; chr1:g.152009388C>T for S100A11; chr19:g.39368871C>T for RINL; and chr6:g.33237597C>G for VPS52.
Because the SardiNIA cohort has been extensively phenotyped, we were able to test for the association of rare variants with measured traits. Of the 30 putatively causal variants, 11 were associated with the expression of genes near significant GWAS loci. Of these, five genes (SPECC1, GLB1, CADM1, BRI3BP, and ANXA5) were associated with traits measured in the Sardinia cohort. However, we found no significant association between the five candidate variants for these genes and their matched GWAS traits (Supplementary Table 23). Furthermore, we found no significant relationship between expression levels of these genes and their matched GWAS trait (Supplementary Table 23), suggesting that either the gene is not involved in the trait or that dosage is not a critical factor. We next searched for outlier genes that have established roles in the manifestation of rare clinical traits. We were able to identify three outlier genes associated with clinical traits in our database: VPS13D is known to repress interleukin-6 (IL6) production; TSSC1 suppresses osteolysis; and mutations in POMGNT1 disrupt dystroglycan and can interfere with skeletal muscle function. For each gene, we tested the genotype of the candidate rare variant with levels of the appropriate trait and then for the overall association between gene expression and the trait. We were, however, unable to find any significant evidence for association (Supplementary Table 24), consistent with recent observations in British-Pakistani cohorts for association testing of rare protein-coding variants in trait-associated genes6,55. While we were unable to identify any direct association between rare variants and clinical traits, we did observe a modest enrichment of outliers in potential disease genes and a marked enrichment of outlier genes in loss-of-function intolerant genes relative to common eQTLs (Supplementary Figure 12).
DISCUSSION
Our study focused on identifying the effect of population and individual-specific regulatory variants in Sardinia. We identified hundreds of novel or highly differentiated regulatory alleles and observed that these alleles reveal novel trait associations and reflect the island’s epidemiological history of multiple sclerosis and malaria. By combining whole genome sequencing data with transcriptomes from many families, we were able to identify patterns of outlier gene expression and implicate the functional role of rare regulatory variants56–59. While such observations have previously been limited to unrelated individuals58,59, we were able to identify hundreds of genes with large heritable effects and candidate rare regulatory variants. Relating the effects of candidate rare regulatory variants to phenotypes remained a significant challenge, comparable to systematic efforts to identify the phenotypic consequences of rare, protein-coding loss-of-function alleles. However, we observed that outlier expression effects were more prevalent in genes intolerant of loss-of-function variation, consistent with their increased potential for important individual consequences.
As gene expression assays complement whole-genome sequencing, discovery of population-specific and rare, large-effect regulatory variants will enable the generation of new hypotheses to understand the molecular etiology of diverse disorders44 and increase our understanding of the utility of different genes as potential therapeutic targets. In particular, identifying extreme patterns of gene expression can be used to provide a more nuanced view of genic dosage tolerance than revealed by naturally occurring knockouts. We anticipate that large catalogs of rare, large-effect regulatory variants, found in either isolated populations or families, will yield new opportunities for clinical interpretation of the non-coding genome, precision health, and our understanding of genome biology.
Data availability statement
The RNA sequencing data that supports the findings of this study has been deposited in the European Genome-phenome Archive (EGA) under accession number EGAS00001002105 (https://www.ebi.ac.uk/ega/datasets/EGAD00001003102). The whole-genome sequencing data used in this study has been deposited in dbGaP under accession number phs000313.v3.p2 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000313.v3.p2).
Code availability
All code used to generate figures and identify outliers is available at http://montgomerylab.stanford.edu/resources/ under the heading “Supplemental pages for manuscripts.”
URLs
eQTL, sQTL, and ASE data, http://eqtlsdownload.irgb.cnr.it and http://montgomerylab.stanford.edu/resources/sardinia.html; Kinship R package, http://www.inside-r.org/packages/kinship; European Genome-phenome Archive, https://www.ebi.ac.uk/ega/home; MERLIN, https://csg.sph.umich.edu/abecasis/Merlin/; Ever-seq, https://code.google.com/archive/p/ever-seq; BWA, http://bio-bwa.sourceforge.net/; Picard, http://broadinstitute.github.io/picard/; STAR, https://github.com/alexdobin/STAR; RSeQC, http://dldcc-web.brc.bcm.edu/lilab/liguow/CGI/rseqc/_build/html/; EPACTS, http://genome.sph.umich.edu/wiki/EPACTS; MLMM, https://github.com/Gregor-Mendel-Institute/mlmm; vcftools, https://vcftools.github.io/index.html; selscan, https://github.com/szpiech/selscan; eCAVIAR, http://genetics.cs.ucla.edu/caviar/; PEER, https://github.com/PMBio/peer; OrphaNet, http://www.orphadata.org/; OMIM, http://www.omim.org; ExAC, http://exac.broadinstitute.org/
ONLINE METHODS
Study population and sample acquisition
Our study was performed on a subset of 624 participants from the larger SardiNIA cohort. All 624 participants live in the Lanusei Valley in the Ogliastra region of Sardinia. Participants represented a mixture of related individuals, including 61 complete trios, and unrelated individuals (n = 188; Supplementary Figure 13). Whole genomes for 606 of these samples were available from a previous published study20. For each participant, leukocytes were isolated from whole blood using the LeukoLOCK™ fractionation kit and RNA was extracted using TRI Reagent® (Ambion #AM9738) and isolated using the PureLink® RNA Mini Kit (Ambion #12183018A). The quantity and the integrity of isolated RNA samples was evaluated using the Agilent Technologies 2100 Bioanalyzer platform with the RNA 6000 LabChip® kit (Agilent #5067-1511) - samples with an RNA integrity number (RIN) less than 7.5 were discarded. Poly-A+ RNA was isolated from 4μg of high-quality total RNA samples through two rounds of positive selection and purification using magnetic beads following the TruSeq RNA Sample Preparation manual (Illumina #15015050).
Sequencing library preparation, alignment, and quality control
Prior to library preparation, we added one of two ERCC RNA Spike-in Control Mixes (Ambion #4456740) to 288 samples at a 1:625 final dilution in order to assess the uniformity of library preparation across samples. Purified RNA samples were then processed into indexed, paired-end cDNA libraries using the TruSeq RNA-Seq Library Preparation Kit. Following purification, amplification, and cleanup, cDNA libraries were quantified using the Agilent Technologies 2100 Bioanalyzer with the Agilent DNA 1000 assay (Agilent #5067-1504). Sample-specific cDNA libraries were then pooled to obtain equimolar concentrations and loaded on to a paired-end flow cell using the Illumina cBot System and the TruSeq PE Cluster Generation kit v3 (Illumina #PE-401-3001). 51 bp paired-end reads were generated on an Illumina HiSeq 2000 using TruSeq SBS v3 reagents (Illumina #FC-401-3001). De-multiplexed FASTQ files were generated and aligned to the hs37d5 reference genome supplemented with ERCC spike-in sequences using STAR (version 2.2.0c)60. Three of the 627 samples were discarded due to extreme GC-content biases, and we observed several other well-known technical biases that we ultimately correct for (Supplementary Figure 14). A full description of library preparation and quality control procedures is available in the Supplementary Note.
Quantification and normalization of gene, isoform, and allele-specific expression
Gene levels were quantified using HTSeq61 (0.5.4p5) over the GENCODE v14 annotation; counts were converted to FPKMs27,62 and variance stabilized using DESeq63 (1.10.1). We then ran PEER26 (v1.3) in order to identify and remove confounding factors. The number of hidden factors to remove was decided by empirically optimizing our ability to discover eQTLs on a random subset of 1,500 genes. eQTLs were mapped using Merlin64 on PEER residuals after removing k hidden factors (we tested various k in the range of 0 to 100, see Supplementary Note) – we found that removing 30 hidden factors maximized our power to discover eQTLs (Supplementary Figure 1). We attempted to identify the biological or technical sources of these hidden factors; many corresponded to known technical biases like GC-content, 3′ and 5′ biases, etc. (Supplementary Table 1). We additionally filtered out non-autosomal genes, genes with a mean FPKM less than 0.3 across all 624 samples, and genes with an FPKM of 0 in 50% or more of the 624 samples. Ultimately we mapped eQTLs for 15,243 genes that passed these filters. Isoform quantification was performed for these 15,243 genes using Cufflinks27 (v2.1.1). Isoform proportions were computed as the ratio of the isoform FPKM relative to the sum of FPKMs for all the isoforms for each gene. We then filtered out genes with only one expressed isoform and where the isoform ratios did not follow a normal distribution (see Supplementary Note). For the 606 samples where whole genome data was available, allele-specific expression (ASE) data was generated using samtools65 (v1.2) mpileup and quantified as the deviation of the reference allele ratio from 0.5. We only considered heterozygous sites with at least 30 reads and where both the reference and alternate allele comprised at least 2% of all supporting reads. We additionally restricted our analyses to sites with an ENCODE mappability score equal to one. Finally, we excluded ASE data for 49 genes that showed significantly biased trends in allelic effects across individuals in our study, the DGN cohort, or the GEUVADIS cohort (Supplementary Table 25; Supplementary Figure 15).
Quantitative trait loci (QTL) mapping
We used an integrated map of ~15 million SNPs for the 606 genotyped samples to map eQTLs and sQTLs using Merlin64 (v1.1.2). We excluded variants that were not in Hardy-Weinberg equilibrium (HWE p-value < 1 x 10−6), had a MAF < 1% in the 606 samples, or had an imputation R2 less than 0.3. Expression values (either expression residuals or isoform ratios) were standardized using Merlin’s inverse normal option. For each gene and isoform, we tested the association of the trait with all cis variants within 1 Mb from the transcription start site (TSS) of the gene. We estimated the overall false discovery rate (FDR) by permutation (see Supplementary Note). We additionally calculated adjusted p-values by selecting the top association for each gene/isoform, applying a gene-level Bonferroni correction, and applying the Benjamini-Hochberg procedure66 to the collection of top associations. Independent gene/isoform QTLs were identified by forward step-wise regression, in which significant QTLs were iteratively regressed out until the next best QTL was no longer significant at an FDR of 5% (Supplementary Figure 16). We also identified independent QTLs using MLMM28, a stepwise linear mixed model approach, and found similar results to our Merlin-based pipeline (Supplementary Tables 6–7). Additionally, we performed simulations in order to show that our independent QTL results were not a result of statistical noise, residual LD, and genotyping errors. Specifically, we repeated the following simulation ten times. For each gene with at least one eQTL, we chose a common SNP (MAF > 5%) within 1 Mb of the TSS to explain 25% of the gene expression variance in the simulated trait. We then ran our Merlin-based pipeline to detect independent eQTLs on the simulated expression traits, iteratively regressing out significant SNPs. We repeated these simulations a second time, excluding the randomly selected causal SNP from the association stage. We then compared the number of independent eQTLs identified in the real data versus the simulated datasets (Supplementary Tables 8–9).
We mapped aseQTLs by computing the Spearman correlation of allelic imbalance in the 15,243 expressed genes with the genotype of nearby cis variants (within 1 Mb of the heterozygous site). Genotypes at cis variants were encoded as 0 (samples homozygous for the reference or non-reference allele) or 1 (heterozygous samples). In order to compare effect sizes across studies, we identified eQTLs and aseQTLs in 188 unrelated Sardinians and compared them with a randomly chosen subset of 188 unrelated individuals in DGN17 and GEUVADIS15. eQTLs in the unrelated 188 individuals for each cohort were recalculated using Matrix eQTL67. We estimated the reproducibility of Sardinian eQTLs using the π1 statistic68 after re-processing each dataset with our pipeline (Supplementary Tables 10–11, 26). A full description of how we controlled for power differences across studies is available in the Supplementary Note.
Co-localization of GWAS and eQTL signals
Co-localization analyses were performed with eCAVIAR29 using default parameters. eCAVIAR calculates a posterior probability that two association signals overlap (CLPP score), accounting for linkage disequilibrium in the study population where the two signals are measured. The supplied LD was computed with vcftools (for the GWAS signals outside of Sardinia, we used LD calculated for European genotypes in the 1000 Genomes Project). For the ARHGDIB co-localization analyses, associations with neutrophil and lymphocyte percentages were calculated within SardiNIA. For the co-localization analysis between multiple sclerosis GWAS and eQTL associations, we used the GWAS data provided by the International Multiple Sclerosis Genetics Consortium69 and the primary eQTL association data from Sardinia (i.e. the association for each SNP without adjusting for conditionally independent eQTLs). We calculated the CLPP score for the identified MS gene as well as nearby genes (±1 Mb of the GWAS SNP) and report the rank of the identified gene in the list of all genes tested for that GWAS locus (Supplementary Table 15). For the 21 genes we tested, 14 of the target genes had the highest evidence of co-localization versus background and 3 had the second highest evidence of co-localization. Only two genes showed very little evidence of co-localization (CD6 and CTD-2006C1.2).
Allelic differentiation, selection, and disease association
Analysis of allelic differentiation and selection was carried out on a subsample (n = 691) of the SardiNIA cohort for which phased genotyped data was already available20 and on data from the 1000 Genomes Phase 35. Integrated haplotype scores (iHS) were computed using the selscan70 software on common variants (MAF ≥ 1%) that passed QC filters (see Supplementary Note). The delta allele frequency for Sardinian QTLs, ΔAFSRD–EUR, was computed as the deviation between the Sardinian MAF and the European MAF (as computed by the 1000 Genomes project). We then tested for the enrichment of different eQTLs near significant GWAS loci. Briefly, we identified significant eQTL in high LD with significant GWAS SNPs (r2 greater than 0.8). For each GWAS trait, we then built a 2x2 count table where the rows separated differentiated eQTLs from non-differentiated eQTLs and the columns separated eQTLs in LD with a GWAS SNP and eQTLs not in LD with a GWAS SNP. We then performed a Fisher’s exact test on each GWAS contingency table, where a significant p-value after Bonferroni correction for the number of traits tested implicated an enrichment of differentiated eQTLs for the GWAS trait relative to all significant eQTLs in Sardinia (Supplementary Table 14). We repeated these analyses using different thresholds for differentiation (ΔAFSRD–EUR greater than 0.05, 0.10, 0.15, 0.20, and 0.25) (Supplementary Figure 7). We identified novel eQTLs in Sardinia by excluding SNPs recorded in other SNP databases (1000 Genomes Phase 35, dbSNP71, ExAC11, and the UK10K project4.
Identifying heritable patterns of outlier gene expression
For the 61 trios in our study, we developed a generalized likelihood ratio test that identifies extreme gene expression signatures that are shared between one parent and their child (a full derivation of the test is given in the Supplementary Note). In practice, we ran our outlier pipeline on the same PEER normalized data as we did for the eQTL analyses; we tested another normalization pipeline to see if PEER was over-correcting outlier signals but found less results overall (instead of using PEER, we regressed out covariates that were highly correlated with PEER factors as described in Supplementary Table 27). We tested each trio for all 15,243 expressed genes used in the eQTL analyses and evaluated significance via permutation, selecting the most significant trio for each gene and applying the Benjamini-Hochberg adjustment66. For all genes with an outlier trio at a 10% FDR we compared ASE in the outlier individuals with ASE in the rest of the participants (non-outliers). We next identified rare variants shared between outlier parents and children in the 250 kb window of the outlier gene and measured the relative enrichment of these variants with similarly identified variants in non-outlier individuals; confidence intervals were calculated via bootstrap resampling (B = 1000) of all observed shared rare variants. Shared rare variants were annotated with chromatin state annotations from peripheral blood mononuclear cells (E062) from the Roadmap Epigenomics Consortium49 (Supplementary Table 28). Log odds scores and confidence intervals were calculated using Fisher’s exact tests for all functional annotations (Supplementary Tables 19–20). We then tested whether the effect of these shared rare variants on expression replicated in the larger study cohort (i.e. where there were at least 4 carriers in the population).
Clinical relevance of candidate causal rare variants
We prioritized 30 of these shared rare variants as candidate causal variants based on several annotations (e.g. proximity to the TSS, were either highly conserved/deleterious, or were potential splicing variants) (Supplementary Table 21). Five of these were associated with genes near significant GWAS loci and 3 were associated with genes previously implicated in the manifestation of clinical traits available to us for study. We tested these rare variants for association to the complex traits or disease they were predicted to impact. For categorical traits (e.g. Celiac disease and bipolar disorder), we performed a likelihood ratio test comparing two nested logistic regression models with the full model (genotype at the rare variant locus, sex, age, and age2) and the reduced null model (without the above covariates). Empirical p-values were computed by permuting sample genotypes 1000 times. To test rare variants for continuous traits (e.g. BMI), we ran the lmekin function from the kinship R package to perform a likelihood ratio test comparing two nested linear mixed models with the full model (genotype at the rare variant locus, sex, age, and age2) and the reduced null model (without the above covariates). We then calculated the Pearson correlation between outlier gene expression and the adjusted trait data and calculated the correlation of gene expression with each clinical trait for each outlier gene-trait association; significance was assessed as the percentile of the empirical distribution obtained from the p-values for all tested genes (Supplementary Tables 23–24).
Supplementary Material
Acknowledgments
All participants gave informed consent, with protocols approved by institutional review boards for ASL4 in Sardinia and by the University of Michigan. IRB exemption (OHSRP #11916) applied to analyses on coded data at collaborating institutions. Z.Z. is supported by the National Science Foundation (NSF) GRFP (DGE-114747) and by the Stanford Center for Computational, Evolutionary, and Human Genomics (CEHG). Z.Z., J.R.D., and G.T.H. also acknowledge support from the Stanford Genome Training Program (SGTP; NIH/NHGRI T32HG000044). J.R.D. is supported by the Stanford Graduate Fellowship. K.R.K. is supported by DoD, Air Force Office of Scientific Research, National Defense Science and Engineering Graduate (NDSEQ) Fellowship, 32 CFR 168a. S.B.M. is supported by the National Institutes of Health through R01HG008150, R01MH101814, U01HG007436, and U01HG00908001. The SardiNIA project is supported in part by the intramural program of the National Institute on Aging through contract HHSN271201100005C to the Consiglio Nazionale delle Ricerche of Italy. All of the authors would like to thank the CRS4 and the SCGPM for the computational infrastructure supporting this project.
Footnotes
Author contributions
M.P., Z.Z., Ma.M., G.R.A., D.S., F.Cu., and S.B.M. conceived and designed the experiments. Ma.M., R.C., F.Cr., M.G.P., A.Mu., M.Z., F.B., A.Ma., E.F., and A.A. performed the experiments. M.P., Z.Z., X.L., J.R.D., M.J.G., G.R.A., F.Cu., and S.B.M performed statistical analysis. M.P., Z.Z., X.L., J.R.D., K.R.K., M.J.G., F.R., R.B., Mi.M., M.S., C.S., Se.S., A.J.B., J.N., G.R.A., D.S., F.Cu., and S.B.M. analyzed the data. M.P., Z.Z., M.C.B., A.J.B., J.N., C.J., St.S., G.R.A., D.S., F.Cu., G.T.H., E.S., K.S.S., and S.B.M. contributed reagents/materials/analysis tools. M.P., Z.Z., J.N., G.R.A., D.S., F.Cu., and S.B.M. wrote the paper. M.P. and Z.Z. contributed equally. F.Cu. and S.B.M. jointly supervised research. All authors read and approved the final version of the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Tennessen JA, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nelson MR, et al. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science. 2012;337:100–104. doi: 10.1126/science.1217876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coventry A, et al. Deep resequencing reveals excess rare recent variants consistent with explosive population growth. Nat Commun. 2010;1:131. doi: 10.1038/ncomms1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.UK10K Consortium et al. The UK10K project identifies rare variants in health and disease. Nature. 2015;526:82–90. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Narasimhan VM, et al. Health and population effects of rare gene knockouts in adult humans with related parents. Science. 2016;352:474–477. doi: 10.1126/science.aac8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacArthur DG, et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li AH, et al. Analysis of loss-of-function variants and 20 risk factor phenotypes in 8,554 individuals identifies loci influencing chronic disease. Nat Genet. 2015;47:640–642. doi: 10.1038/ng.3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sulem P, et al. Identification of a large set of rare complete human knockouts. Nat Genet. 2015;47:448–452. doi: 10.1038/ng.3243. [DOI] [PubMed] [Google Scholar]
- 10.Flannick J, et al. Loss-of-function mutations in SLC30A8 protect against type 2 diabetes. Nat Genet. 2014;46:357–363. doi: 10.1038/ng.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Exome Aggregation Consortium et al. Analysis of protein-coding genetic variation in 60,706 humans. bioRxiv. 2015 doi: 10.1101/030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moltke I, et al. A common Greenlandic TBC1D4 variant confers muscle insulin resistance and type 2 diabetes. Nature. 2014;512:190–193. doi: 10.1038/nature13425. [DOI] [PubMed] [Google Scholar]
- 13.Zoledziewska M, et al. Height-reducing variants and selection for short stature in Sardinia. Nat Genet. 2015;47:1352–1356. doi: 10.1038/ng.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bottini N, et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet. 2004;36:337–338. doi: 10.1038/ng1323. [DOI] [PubMed] [Google Scholar]
- 15.Lappalainen T, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Battle A, et al. Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals. Genome Res. 2014;24:14–24. doi: 10.1101/gr.155192.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maurano MT, et al. Systematic Localization of Common Disease-Associated Variation in Regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicolae DL, et al. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sidore C, et al. Genome sequencing elucidates Sardinian genetic architecture and augments association analyses for lipid and blood inflammatory markers. Nat Genet. 2015;47:1272–1281. doi: 10.1038/ng.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peltonen L, Palotie A, Lange K. Use of population isolates for mapping complex traits. Nat Rev Genet. 2000;1:182–190. doi: 10.1038/35042049. [DOI] [PubMed] [Google Scholar]
- 22.Lim ET, et al. Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet. 2014;10:e1004494. doi: 10.1371/journal.pgen.1004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orrù V, et al. Genetic variants regulating immune cell levels in health and disease. Cell. 2013;155:242–256. doi: 10.1016/j.cell.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pilia G, et al. Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genet. 2006;2:e132. doi: 10.1371/journal.pgen.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pistis G, et al. Rare variant genotype imputation with thousands of study-specific whole-genome sequences: implications for cost-effective study designs. European Journal of Human Genetics. 2015;23:975–983. doi: 10.1038/ejhg.2014.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stegle O, Parts L, Piipari M, Winn J, Durbin R. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat Protoc. 2012;7:500–507. doi: 10.1038/nprot.2011.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Segura V, et al. An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat Genet. 2012;44:825–830. doi: 10.1038/ng.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hormozdiari F, et al. Joint Fine Mapping of GWAS and eQTL Detects Target Gene and Relevant Tissue. bioRxiv. 2016:065037. doi: 10.1101/065037. [DOI] [Google Scholar]
- 30.Dovas A, Couchman JR. RhoGDI: multiple functions in the regulation of Rho family GTPase activities. Biochem J. 2005;390:1–9. doi: 10.1042/BJ20050104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castel SE, Levy Moonshine A, Mohammadi P, Banks E, Lappalainen T. Tools and best practices for data processing in allelic expression analysis. Genome Biol. 2015;16:195. doi: 10.1186/s13059-015-0762-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kudaravalli S, Veyrieras JB, Stranger BE, Dermitzakis ET, Pritchard JK. Gene expression levels are a target of recent natural selection in the human genome. Molecular Biology and Evolution. 2009;26:649–658. doi: 10.1093/molbev/msn289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaneko A, et al. Malaria eradication on islands. Lancet. 2000;356:1560–1564. doi: 10.1016/S0140-6736(00)03127-5. [DOI] [PubMed] [Google Scholar]
- 35.Tognotti E. Program to eradicate malaria in Sardinia, 1946–1950. Emerging Infect Dis. 2009;15:1460–1466. doi: 10.3201/eid1509.081317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pugliatti M, Sotgiu S, Rosati G. The worldwide prevalence of multiple sclerosis. Clin Neurol Neurosurg. 2002;104:182–191. doi: 10.1016/s0303-8467(02)00036-7. [DOI] [PubMed] [Google Scholar]
- 37.Pugliatti M, et al. The epidemiology of multiple sclerosis in Europe. Eur J Neurol. 2006;13:700–722. doi: 10.1111/j.1468-1331.2006.01342.x. [DOI] [PubMed] [Google Scholar]
- 38.Liu XQ, et al. Malaria infection alters the expression of B-cell activating factor resulting in diminished memory antibody responses and survival. Eur J Immunol. 2012;42:3291–3301. doi: 10.1002/eji.201242689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scholzen A, Sauerwein RW. How malaria modulates memory: activation and dysregulation of B cells in Plasmodium infection. Trends Parasitol. 2013;29:252–262. doi: 10.1016/j.pt.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 40.Scholzen A, et al. BAFF and BAFF receptor levels correlate with B cell subset activation and redistribution in controlled human malaria infection. J Immunol. 2014;192:3719–3729. doi: 10.4049/jimmunol.1302960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kosoy R, et al. Evidence for malaria selection of a CR1 haplotype in Sardinia. Genes Immun. 2011;12:582–588. doi: 10.1038/gene.2011.33. [DOI] [PubMed] [Google Scholar]
- 42.Stoute JA. Complement receptor 1 and malaria. Cell Microbiol. 2011;13:1441–1450. doi: 10.1111/j.1462-5822.2011.01648.x. [DOI] [PubMed] [Google Scholar]
- 43.Naitza S, et al. A genome-wide association scan on the levels of markers of inflammation in Sardinians reveals associations that underpin its complex regulation. PLoS Genet. 2012;8:e1002480. doi: 10.1371/journal.pgen.1002480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tabassum R, et al. Omic personality: implications of stable transcript and methylation profiles for personalized medicine. Genome Medicine. 2015;7:88. doi: 10.1186/s13073-015-0209-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Melnikov A, et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat Biotechnol. 2012;30:271–277. doi: 10.1038/nbt.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patwardhan RP, et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nat Biotechnol. 2012;30:265–270. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kwasnieski JC, Mogno I, Myers CA, Corbo JC, Cohen BA. Complex effects of nucleotide variants in a mammalian cis-regulatory element. Proc Natl Acad Sci USA. 2012;109:19498–19503. doi: 10.1073/pnas.1210678109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Veyrieras JB, et al. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4:e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cooper GM, et al. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gulko B, Hubisz MJ, Gronau I, Siepel A. A method for calculating probabilities of fitness consequences for point mutations across the human genome. Nat Genet. 2015;47:276–283. doi: 10.1038/ng.3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skarratt KK, et al. A 5′ intronic splice site polymorphism leads to a null allele of the P2X7 gene in 1–2% of the Caucasian population. FEBS Lett. 2005;579:2675–2678. doi: 10.1016/j.febslet.2005.03.091. [DOI] [PubMed] [Google Scholar]
- 55.Johnston JJ, et al. Individualized iterative phenotyping for genome-wide analysis of loss-of-function mutations. Am J Hum Genet. 2015;96:913–925. doi: 10.1016/j.ajhg.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Montgomery SB, Lappalainen T, Gutierrez-Arcelus M, Dermitzakis ET. Rare and common regulatory variation in population-scale sequenced human genomes. PLoS Genet. 2011;7:e1002144. doi: 10.1371/journal.pgen.1002144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li X, et al. Transcriptome sequencing of a large human family identifies the impact of rare noncoding variants. Am J Hum Genet. 2014;95:245–256. doi: 10.1016/j.ajhg.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zeng Y, et al. Aberrant gene expression in humans. PLoS Genet. 2015;11:e1004942. doi: 10.1371/journal.pgen.1004942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao J, et al. A Burden of Rare Variants Associated with Extremes of Gene Expression in Human Peripheral Blood. Am J Hum Genet. 2016;98:299–309. doi: 10.1016/j.ajhg.2015.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2012;29 doi: 10.1093/bioinformatics/bts635. bts635–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Garber M, Grabherr MG, Guttman M, Trapnell C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat Methods. 2011;8:469–477. doi: 10.1038/nmeth.1613. [DOI] [PubMed] [Google Scholar]
- 63.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 65.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B Methodological. 1995;57:289–300. [Google Scholar]
- 67.Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sawcer S, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. - PubMed - NCBI. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen W, Abecasis GR. Family-based association tests for genomewide association scans. Am J Hum Genet. 2007;81:913–926. doi: 10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucl Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing data that supports the findings of this study has been deposited in the European Genome-phenome Archive (EGA) under accession number EGAS00001002105 (https://www.ebi.ac.uk/ega/datasets/EGAD00001003102). The whole-genome sequencing data used in this study has been deposited in dbGaP under accession number phs000313.v3.p2 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000313.v3.p2).