Abstract
Fluorescent proteins and epitope tags are often used as protein fusion tags to study target proteins. One prevailing technique in the budding yeast Saccharomyces cerevisiae is to fuse these tags to a target gene at the precise chromosomal location via homologous recombination. However, several limitations hamper the application of this technique, such as the selectable markers not being reusable, tagging of only the C-terminal being possible, and a “scar” sequence being left in the genome. Here, we describe a strategy to solve these problems by tagging target genes based on a pop-in/pop-out and counter-selection system. Three fluorescent protein tag (mCherry, sfGFP, and mKikGR) and two epitope tag (HA and 3×FLAG) constructs were developed and utilized to tag HHT1, UBC13 or RAD5 at the chromosomal locus as proof-of-concept.
Introduction
The budding yeast Saccharomyces cerevisiae has been widely used in basic research due to the ease with which it can be genetically manipulated. Considerable progress has been made in studies on budding yeast via the use of epitope and fluorescent protein (FP) tags. Epitope tags, such as Flag, HA, and Myc, are useful for western blotting analysis, immunoprecipitation, affinity purification, and immunochemistry [1–3]. FP tags, such as green fluorescent protein (GFP) and its variants, are extensively used in molecular and cellular biology studies. In many experiments, an FP has been fused to a target protein as a reporter to track its cellular dynamics. For instance, the global protein subcellular localization was studied by plasmid-based epitope-tagging and immunofluorescence analyses [4], or by fusing a GFP tag at the C-terminal of the coding region of a gene of interest, followed by microscopic imaging [5].
Integration of an epitope or FP tag into the desired chromosomal locus is particularly easy to carry out in Saccharomyces cerevisiae due to the ease of introducing a tag sequence into the genome by homologous recombination (HR). Compared with the plasmid-based protein-tagging method, chromosomal gene-tagging has higher genetic stability and ensures that there is one copy of the fusion gene in a haploid yeast cell. Moreover, chromosomal gene-tagging can avoid numerous artifacts since the fusion gene is in its native chromosomal context and expressed under the control of its native promoter. In recent decades, techniques have been developed that enable directed insertion of desired genetic elements into a precise location in a budding yeast chromosome. The general process in these techniques involves integrating a given element along with a selectable marker gene into a chromosome via HR, obtaining transformants on corresponding selective medium, and screening for the desired recombinant product by PCR of genomic DNA. Two types of marker gene have been used for selection. One is an auxotrophic marker gene, including TRP1, LEU2, ADE2, MET15 [6], HIS3 [7], LYS2 [8], and URA3 [9]. The other is a dominant marker gene, such as KanMX [10], natMX [11], and amdSYM [12].
For auxotrophic markers, URA3 is the most widely used recyclable marker, although LYS2 and MET15 can be counter-selected by alpha-aminoadipate and methyl-mercury, respectively [8, 9, 12, 13]. Alani et al. successfully developed a pop-in/pop-out system for recycling the URA3 marker as follows. A 3.8 kb DNA fragment consisting of URA3 flanked by direct repeats of 1.1 kb hisG from Salmonella (hisG-URA3-hisG) was integrated into the yeast chromosome. The Ura− transformants resulting from popping out of the URA3 gene along with one copy of hisG were counter-selected in the presence of 5-fluoroorotic acid (5-FOA) [9, 14, 15]. However, in this system, one unwanted copy of hisG still remains in the yeast genome after the URA3 gene has been popped out, which produces “scar” sequences around the target gene.
Using the dominant markers, it is easy to add a tag at the C-terminal of a gene of interest at its native chromosomal location. However, almost none of these markers are counter-selectable. A Cre/loxP recombination system was thus developed to make dominant markers reusable by flanking the markers with loxP [16, 17]. However, similar to the problem in the URA3 pop-in/pop-out system, the Cre/loxP recombination system leaves one copy of loxP in the chromosome. Subsequently, the amdSYM cassette was reported as the first recyclable dominant marker through counter-selection with the acetamide homologue fluoroacetamide[18]. Actually, this new dominant recyclable marker has not been widely used as URA3 marker.
PCR-based epitope tagging using plasmids containing tag-URA-tag as templates in budding yeast have been introduced firstly in 1995[19]. YFGs were tagged with 3×HA and 3×MYC, and the tagged proteins were verfied by western blotting. Then four plasmids containing triple-epitope tags (FLAG, HSV, V5 and VSV-G) were constructed in the same strategy[20]. General primer sequences were provided in the two papers to allow tagging the YFGs with all these epitopes using a single pair of primers, while a short nucleotide fragment will be left in the genome.
In our previous study, we reported a method to integrate a designed promoter into a target locus on a chromosome without introducing any junk sequence [21]. Here, using the same strategy, we fused FP tags (mCherry, sfGFP, and mKikGR) to a yeast gene (UBC13, RAD5 or HHT1) and monitored FP-fused protein expression. We tagged UBC13 with epitope tags (HA and 3×FLAG) and analyzed the results using western blotting. This study provides a reliable protein-tagging method, which is characterized by the recycling of selectable markers, ease of tagging the N-terminal of nonessential genes, and a minimal disturbance of native gene expression level.
Materials and methods
Plasmid construction
To obtain the three fluorescent protein tag constructs, pCUC, psfGUG, and pKUK, pBS-URA3 was constructed by first inserting URA3 into pBluescriptII SK between the HindIII and EcoRI sites. Then, 3′-truncated and 5′-truncated mCherry, sfGFP, and mKikGR were cloned into the XhoI-HindIII site and the EcoRI-BamHI site in pBS-URA3, respectively. For the two peptide tag constructs, pHUH and pFUF, URA3 flanked by HA and that flanked by 3×FLAG were integrated into the HindIII-EcoRI and XhoI-BamHI sites of pBluescriptII SK, respectively.
Yeast strains
In this study, all strains were isogenic derivatives of strains BY4741 (Invitrogen), HK578-10A (H. Klein), HK578-10D (H. Klein), and yRH182[2]. HHT1, UBC13 and RAD5, shown as your favorite genes (YFGs), were targeted for tagging through HR. To increase the efficiency of integration into the yeast genome, ~300-bp homologous sequences from both up- and downstream of each open reading frame (ORF) were added to both ends of FP-URA3-FP by overlapping PCR. The 5’ YFG-FP-URA3-FP-YFG 3’ fragments were amplified and transformed into yeast strains via a LiAc/SS carrier DNA/PEG method, as described previously [22]. The colonies that could grow on plates with SD-Ura medium (synthetic dextrose medium without uracil) were selected and then patched onto 5-FOA plates [14] to obtain marker-free single colonies, as described previously [9]. Genomic PCR and sequencing were used to confirm the correct tagging of YFG of the resulting strains. All of the primers used for plasmid construction, protein tagging, and strain verification are listed in S1–S3 Tables.
Fluorescence microscopy
The yeast strains were cultured overnight at 30°C in synthetic complete medium, and were subcultured in fresh medium for about 4 h until the phase of exponential growth was reached. One milliliter of the culture was then collected by centrifugation at 3,000 g for 1 min, and the pellet was resuspended in 20 μL of phosphate-buffered saline. A total of 1 μg/mL DAPI (D9542, Sigma-Aldrich) was used to stain the nucleus for 10 min at room temperature. Fluorescence was detected by laser scanning confocal microscope (Zeiss LSM780), using excitation at 405 nm for DAPI, 488 nm for sfGFP and green mKikGR, 561 nm for mCherry, and 594 nm for red mKikGR. The images were processed by ZEN software.
Western blot analysis
Protein extracts were prepared using the YeastBusterTM Protein Extraction Reagent (71186, Merck Millipore). The antibodies used in this study were HA-probe antibody (F-7) (sc-7392, Santa Cruz Biotechnology), monoclonal anti-Flag M2 antibody (F1804, Sigma-Aldrich), and anti-Pgk1 (a kind gift from Wei Li, Institute of Zoology, Chinese Academy of Sciences).
Results
Construction of FP- and epitope-tag-integrating cassettes for tagging at the native genomic locus
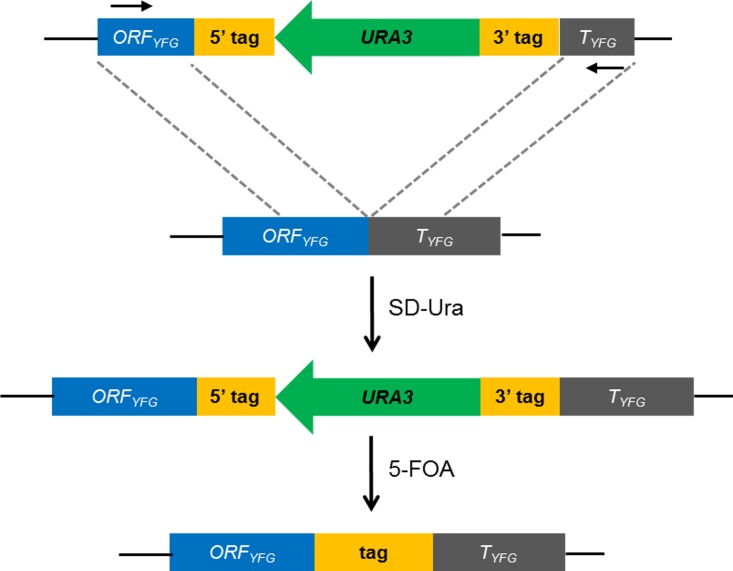
Labeling a gene of interest with a tag at its native locus in the chromosome is a commonly used technique in studies on budding yeast. To solve the two main problems of an inability to recycle the selectable marker and the presence of “scar” sequences after tagging, we proposed that the URA3 flanking sequences hisG in the traditional hisG-URA3-hisG system [9] could be replaced by the desired genetic element in tandem repeats. Thus, after site-specific integration and pop-out, only the designed element will remain at the predetermined locus, as illustrated in Fig 1.
Fig 1. Schematic representation of the protein-tagging via the URA3 gene pop-in/pop-out system.
Here, we use tagging of YFG C-terminal as an example, and the N-terminal tagging of nonessential genes could be done following the same strategy. First, the ORFYFG-5′tag-URA3-3′tag-TYFG fragment is integrated into yeast chromosomes through HR using the homologous sequences of the ORF and terminator of YFG. Positive selection on an SD-Ura plate yields pop-in clones in which the designed sequence has been successfully integrated into the chromosomal locus of the YFG gene. Second, the URA3 gene would be eliminated via homologous recombination due to the duplicated sequences of 5′ and 3′ tags, leaving only one copy of the intact tag. The strains in which URA3 has been popped out are counter-selected on 5-fluoroorotic acid plates.
Here, we designed a system to use FP sequences to replace hisG to form FP-URA3-FP cassettes. After HR between flanking homologous FP sequences, the URA3 gene along with one copy of the repeat FP sequence was removed and formed an intact FP sequence at the target locus. To validate this concept, we chose three typical FPs: mCherry, sfGPF (superfolder GFP), and mKikGR, as tags. mCherry is a red monomer that is widely used due to it having the best photostability among the red FPs [23]. sfGFP is an engineered GFP with increased resistance to denaturation and improved folding kinetics [24]. Finally, mKikGR is a photoswitchable fluorescent protein that emits green fluorescence in its initial state, which is converted to red fluorescence after illumination with a 405-nm laser [25].
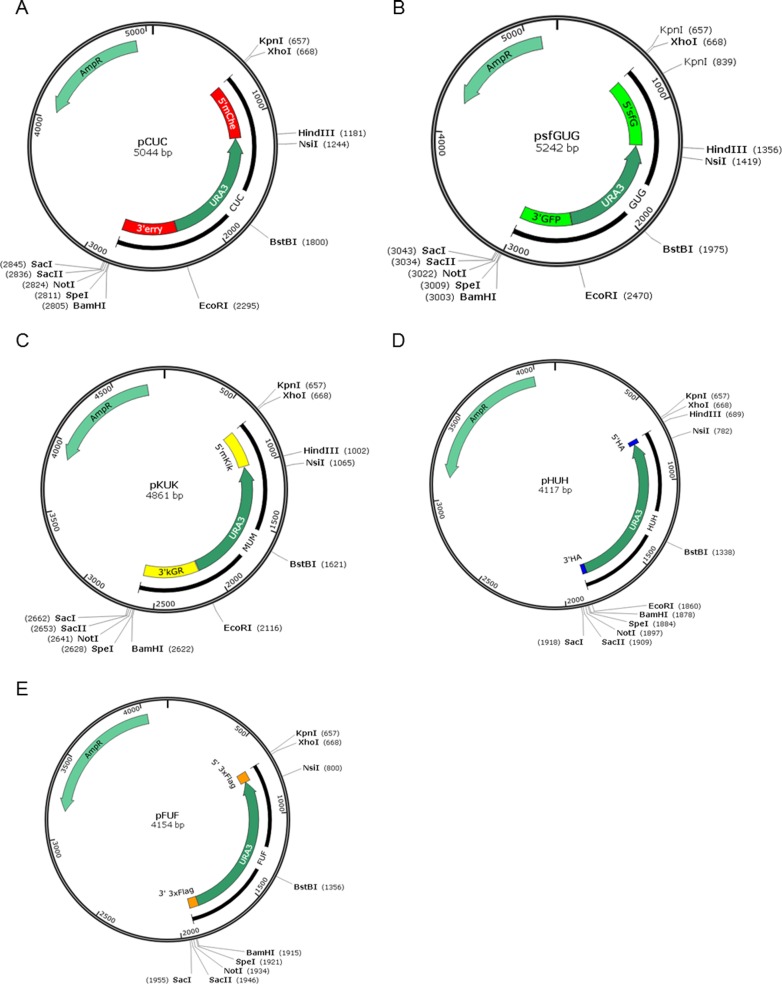
To construct these three FP-URA3-FP cassettes, the URA3 marker gene was first introduced into pBluescriptII SK using HindIII and EcoRI. Based on this new construct, a 5′ section containing C-terminally truncated FP and a 3′ section lacking N-terminal sequences of FP were cloned into XhoI-HindIII sites and EcoRI-BamHI sites, respectively, flanking the URA3 to generate three plasmids: pCUC, psfGUG, and pKUK (Fig 2). In each case, both FP coding sequences were truncated at either the 5′ or the 3′ end (Table 1), instead of using the full-length sequences, for three main reasons. First, compared with an epitope or affinity tag, FP coding sequences are much longer, while only about 50-bp homologous sequences are considered necessary for efficient popping out through HR. Second, differential truncation of the tandem repeats facilitates the selection of unique sequences on each copy to amplify the cassette for transformation by PCR. Third, since neither copy of the tandem repeats is intact, the initial integration product is negative for fluorescence, and only correct pop-out strains can display fluorescence. Accordingly, it is convenient to screen and confirm the desired final product.
Fig 2. Plasmid constructs for protein-tagging.
The original plasmid is pBluescriptII SK, and the maps display the restriction enzyme sites used for construction. (A) The pCUC map contains the 5’ mChe-URA3-3’ erry cassette. (B) The psfGUG map contains the 5’ sfG-URA3-3’ GFP cassette. (C) The pKUK map contains the 5’ mKik-URA3-3’ kGR cassette. For pCUC, psfGUG and pKUK, each pair of sectional tag sequences has a partially duplicated sequence for homologous recombination. (D) The pHUH map contains the 5’HA-URA3-3’HA cassette. (E) The pFUF map contains the 5’3×FLAG-URA3-3’3×FLAG cassette. For these two plasmids, the HA and FLAG tags are too short to be split, so the full-length sequences are used for homologous recombination.
Table 1. Truncation sequences of fluorescent proteins tags flanked by the URA3 gene.
| Fluorescent protein | Full length (bp) |
5’ section (bp) |
3’ section (bp) |
Overlap section (bp) |
|---|---|---|---|---|
| mCherry | 711 | 1–507 | 205–708 | 303 |
| sfGFP | 720 | 1–676 | 200–717 | 477 |
| mKikGR | 699 | 1–322 | 203–696 | 120 |
| HA | 30 | 1–30 | 1–30 | 30 |
| 3×FLAG | 69 | 1–69 | 1–69 | 69 |
In addition to the FP tags, two plasmids containing URA3 flanking with full length HA and 3×FLAG tags were constructed, and named as pHUH and pFUF, respectively (Table 1, Fig 2).
Verification of FP-tagging at the native genomic locus using microscopic imaging
As the proof-of-concept, we aimed to tag chromosomal HHT1, UBC13 and RAD5 genes with three FPs. sfGFP was fused to the C-terminal of HHT1, which encodes one of two identical histone H3 proteins[26]. Based on the fact that the N-terminal fusion may affect the physical interaction of Rad5 with Rev1 [27, 28], the mKikGR tags were tethered to the C-terminal of Rad5, while mCherry was fused to the N-terminal of Ubc13. About 0.3~0.4 kb homologous sequences upstream and downstream of the locus of insertion were introduced into the FP-URA3-FP cassettes to generate 5′YFG-FP-URA3-FP-YFG3′ products by overlap PCR (S1 Fig). Table 2 lists the primers used to amplify each cassette sequence followed by gene-specific sequences, and their relative reading frame. These 5′YFG-FP-URA3-FP-YFG3′ products were used to transform yeast strains. After selection by SD-Ura, followed by counter-selection with 5-FOA, the resulting strains (Table 3) containing the desired FP fusion proteins were generated. Fig 3A shows the results of PCR analysis of the resulting HHT1-sfGFP strain and intermediates, while Fig 4 shows that of RAD5-sfGFP and RAD5-mKikGR strains.
Table 2. Proteins tagging primers used in this studyɑ.
| Primers | Sequences (5’→3’) |
|---|---|
| HA-Ubc13 | |
| pNUbc13-HUH F |
TTAGCAAATAAGGTCAGGTTCATTGTAACATAGTTAGAA ATGTACCCATACGATGTTCC |
| pNHUH-Ubc13 R |
TGAACATACCTTGATTATTCTCTTGGGTAATGATGCCAT AGCGTAATCTGGAACATCGT |
| UBC13-HA | |
| pCUbc13-HUH F |
CGCGAATGGACGAAATTGTATGCAAAGAAGAAACCCGAG ATGTACCCATACGATGTTCC |
| pCHUH-Ubc13 R |
TTATATATTCAGTTGAGAAAACTTATACAGAAATGATCA AGCGTAATCTGGAACATCGT |
| 3xFLAG-Ubc13 | |
| pUbc13-FUF F |
TTAGCAAATAAGGTCAGGTTCATTGTAACATAGTTAGAA ATGGACTACAAAGACCATGA |
| pFUF-Ubc13 R |
TGAACATACCTTGATTATTCTCTTGGGTAATGATGCCAT CTTGTCATCGTCATCCTTGT |
| mCherry-Ubc13 | |
| pUbc13 F pUbc13-mCherry R pUbc13-mCherry F |
AGCTTAATGATTAGATTCTG TCCTCGCCCTTGCTCACCATTTCTAACTATGTTACAATGA TCATTGTAACATAGTTAGAAATGGTGAGCAAGGGCGAGGA |
| pmCherry-Ubc13 R pmCherry-Ubc13 F |
CTCTTGGGTAATGATGCCATCTTGTACAGCTCGTCCATGC GCATGGACGAGCTGTACAAGATGGCATCATTACCCAAGAG |
| pUbc13 R Rad5-sfGFP pRad5 F pRad5-sfGFP R |
AAGTAACGTAAGTTGTCGTC GCCGTTAAAGACTATAGCAG AGCTCTTCGCCTTTACGCATTTCAAACAGCATCTGGATTTC |
| pRad5-sfGFP F psfGFP-Rad5 R psfGFP-Rad5 F |
GAAATCCAGATGCTGTTTGAAATGCGTAAAGGCGAAGAGCT TATGAGTATGTGGTATGACTA TCATCATTTGTACAGTTCATCCATA TATGGATGAACTGTACAAATGATGATAGTCATACCACATACTCATA |
| pRad5 R |
CCACCTACACTTTCTGTCAT |
| Rad5-mKikGR | |
| pRad5-mKikGR F pRad5-mKikGR R pmKikGR-Rad5 F |
GAAATCCAGATGCTGTTTGAA ATGAGTGTGATTACATCAGA TCTGATGTAATCACACTCATTTCAAACAGCATCTGGATTTC GGGCGCCAAGTATGAATTTGAAGCCTAGTCATACCACATACTCATA |
| pmKikGR-Rad5 R |
TATGAGTATGTGGTATGACTA GGCTTCAAATTCATACTTGGCGCCC |
| HHT1-sfGFP | |
| pHHT1-sfGFP F | GATATCAAGTTGGCTAGAAGATTAAGAGGTGAAAGATCA ATGCGTAAAGGCGAAGAGCT |
| pHHT1-sfGFP R |
GTTCGTTTTTTACTAAAACTGATGACAATCAACAAACTA TCATCATTTGTACAGTTCATCCATA |
ɑ Primers used to amplify each cassette are listed here. Each primer consists of two parts. One part is the sequence corresponding to each cassette element. Another part underlined in the list is the sequence complementary to the gene-specific sequence.
Table 3. Yeast strains used in this study.
| Strains | Genotype |
|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 |
| WXY3812 | BY4741 UBC13-HA |
| WXY3814 | BY4741 HA-UBC13 |
| HK578-10A | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 |
| WXY3816 | 10A mCherry-UBC13 |
| WXY3818 | 10A PPGK1-mCherry-UBC13 |
| HK578-10D | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 |
| WXY3822 | 10D RAD5-sfGFP |
| WXY3824 | 10D PPGK1-RAD5-sfGFP |
| WXY3826 | 10D RAD5-mKikGR |
| WXY3828 | 10D PPGK1-RAD5-mKikGR |
| FYV1ɑ | yRH182 HHT1-sfGFP |
ɑThe detail information about strain yRH182 please see[2].
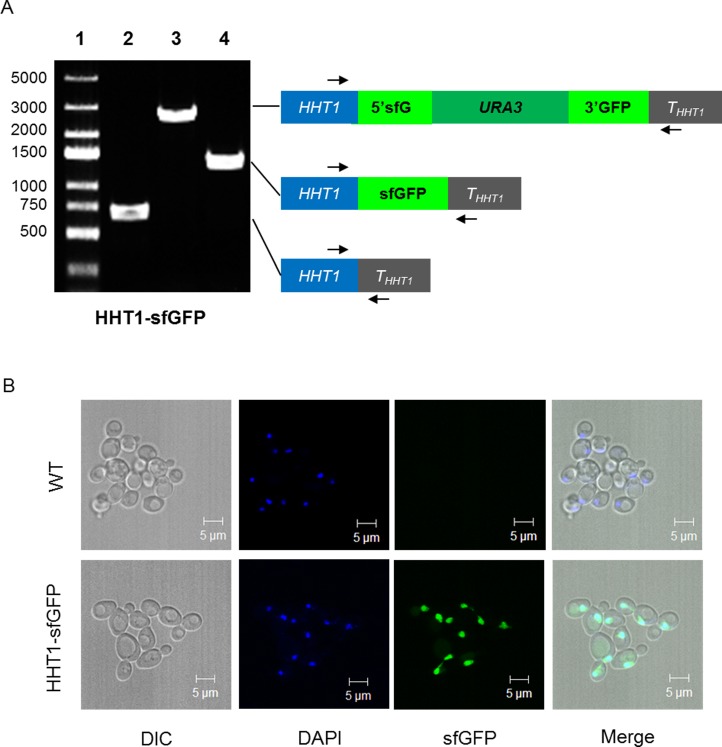
Fig 3. PCR analysis of tag-HHT1 yeast strains and their subcellular localizations.
(A) HHT1-sfGFP strain. The genomic DNA PCR data shows in the left panel. Lane 1: DNA ladder marker. Lane 2: the original strain yRH182. Lane 3 and 4: the pop-in and pop-out strain. (B) The subcellular localizations of HHT1-sfGFP. The yRH182 (upper) is the wild-type yeast strains without modification; the tagged strains HHT1-sfGFP are shown at the bottom. The images were obtained under Plan-Apochromat 63×/1.40 oil (Zeiss 5 Live).
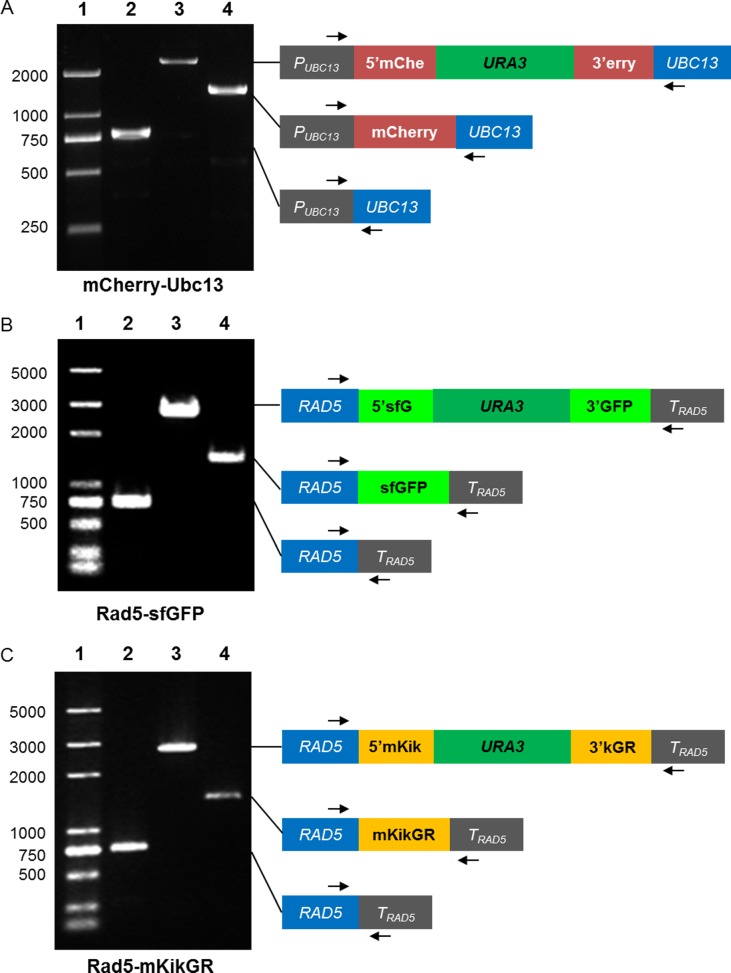
Fig 4. PCR analysis of tag-UBC13/RAD5 yeast strains.
(A) mCherry-Ubc13 strain. The left panel displays the genomic DNA PCR data. Lane 1: DNA ladder marker. Lane 2: the original strain HK578-10A. Lane 3: the pop-in strain CUC-Ubc13. Lane 4: the pop-out strain mCherry-Ubc13. The right panel represents the corresponding genomic organizations at the locus of UBC13 in these three different strains. (B) Rad5-sfGFP strain. The original strain is HK578-10D. (C) Rad5-mKikGR strain. The original strain is HK578-10D. Arrows indicate the locations of primers. The detail information of primers could be found in the part of yeast strains check primers in S3 Table.
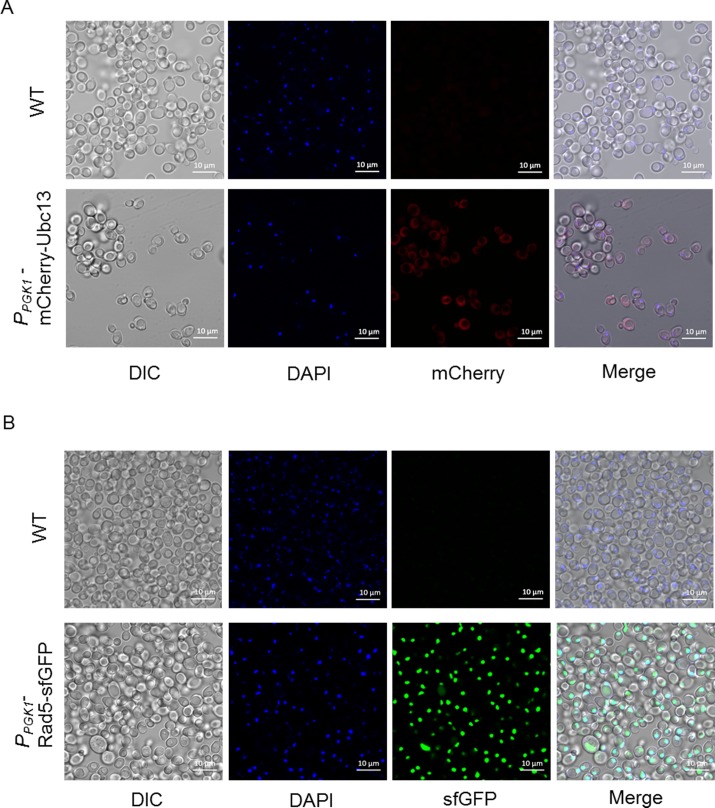
Next, we detected the expression of FP-tagged proteins by laser scanning confocal microscopy. The HHT1-sfGFP was controlled under its native promoter, and sfGFP fusion proteins were detected under the 488nm laser and located in the nucleus. All of the mCherry-Ubc13, Rad5-sfGFP, and Rad5-mKikGR fusion proteins only produced weak fluorescent signals. To enhance these signals, and further confirm the reuse of the URA3 marker, we employed the plasmid pPUP, which contains a tandem PGK1 promoter flanking the URA3 gene [21], and used it to replace the native promoters. The procedure and results confirming the successful PGK1 promoter replacement of the resulting strains are shown in S2 Fig.
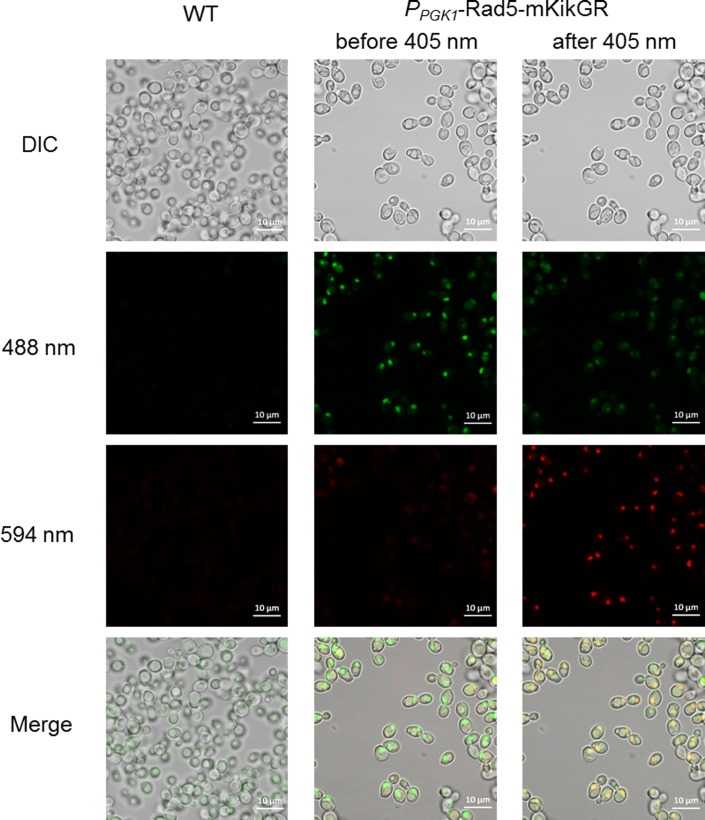
Under control of a constitutive PGK1 promoter, we observed that the mCherry-tagged UBC13 was located in both the cytoplasm and the nucleus (Fig 5A), which is accordance with controlling under its native promoter, as previously reported[5]. Similar to mCherry-Ubc13, the expression of the Rad5-sfGFP fusion protein was enhanced under the regulation of the PPGK1 promoter. Upon illumination with a 488-nm laser, the green fluorescence colocalized with the DAPI staining, indicating the nuclear localization of Rad5 (Fig 5B). This observation is consistent with the predicted function of RAD5 in error-free DNA post-replication repair [29] and the early localization experiment[5]. The expression level of Rad5- mKikGR was also improved when driven by the PGK1 promoter, since we detected a strong fluorescent signal in the resulting strains (Fig 6). To validate the photoswitching of Rad5-mKikGR, yeast cells were first observed under a 488-nm laser, which showed that the fluorescent green of Rad5-mKikGR was localized in the nucleus. After illumination with a pulse of 405-nm light, most of the signals of green fluorescence were converted into red fluorescence upon excitation with 594-nm light (Fig 6). Thus, our data demonstrate that all of these FP-labeled proteins of interest function normally in the resulting strains.
Fig 5. The subcellular localizations of mCherry-Ubc13 and Rad5-sfGFP.
HK578-10A (upper A) and HK578-10D (upper B) are the wild-type yeast strains without modification; the tagged strains PPGK1-Ubc13-mCherry and PPGK1-Rad5-sfGFP are shown at the bottom of A and bottom of B, respectively. The nucleus was stained by DAPI. The images were obtained under Plan-Apochromat 63×/1.40 oil (Zeiss LSM780).
Fig 6. The photoswitching of mKikGR in PPGK1-Rad5-mKikGR strains.
HK578-10D (left panel) is the wild-type strain without modification. The cells containing Rad5-mKikGR were imaged with 488-nm light and 594-nm light before (middle) and after (right) 405-nm laser photoswitching.
Verification of epitope-tagging at the UBC13 native genomic locus using biochemical analysis
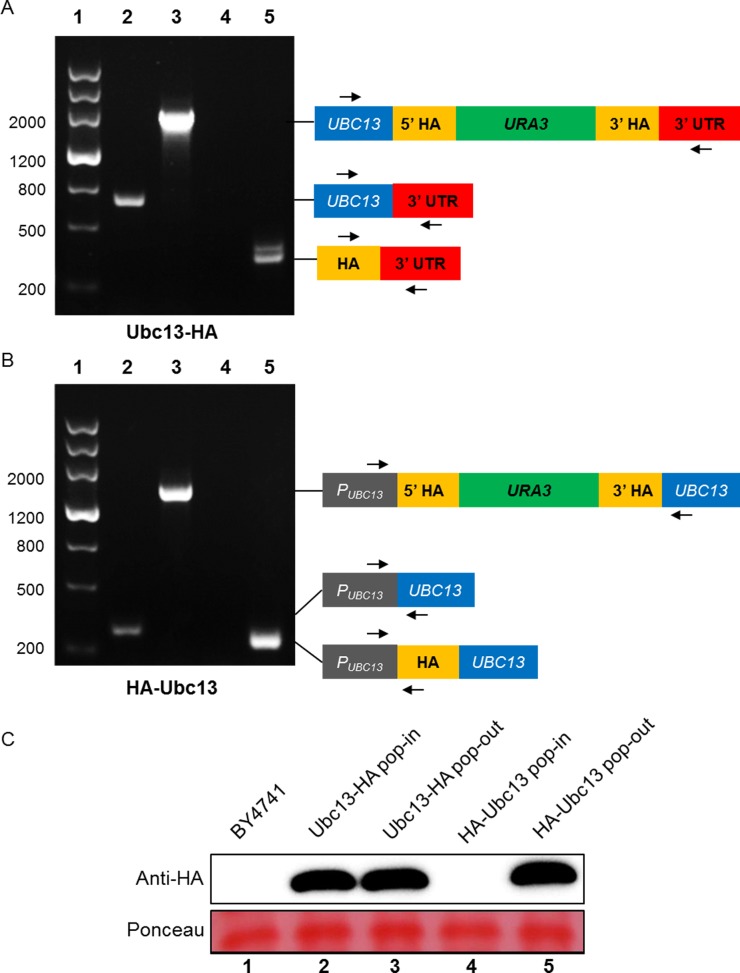
Using the same technique as described above, the N- or C-terminals of UBC13 tagged with HA stains were obtained separately (Table 3). As shown in Fig 7A and 7B, PCR analysis indicated that successful chromosomal integration had occurred as intended. The tagged proteins were examined by western blot analysis. The anticipated Ubc13-HA C-terminal fusion protein was detected in both pop-in and pop-out strains (Fig 7C, cf. lanes 2 and 3). In contrast, integration of the HA-URA3-HA cassette at the 5′ end of the UBC13 ORF interfered with its expression (Fig 7B). Only if the cassette was popped-out were we able to detect the HA-Ubc13 N-terminal fusion protein by western blot analysis (Fig 7C, cf. lanes 4 and 5). Using a different strain, a 3×FLAG-Ubc13 N-terminal fusion cell line was created and the expression was verified by western blot (S3 Fig).
Fig 7. PCR and western blot analysis of HA-tagged Ubc13 yeast strains.
(A) Ubc13-HA strain. (B) HA-Ubc13 strain. Lane 1: DNA ladder marker. Lanes 2 and 4: the original strain BY4741. Lane 3: the pop-in strain. Lane 5: the pop-out strain. Arrows indicate the locations of primers. The detail information of primers could be found in the part of yeast strains check primers in S1 Table. And the strains in each Lane 4 and 5 share the same pair of primers for analysis. (C) (Upper) Anti-HA antibody is used for detecting the HA tag. (Bottom) Ponceau S staining is shown as the total protein loading control.
Compared with current in vivo protein-tagging techniques used in budding yeast, this strategy has three advantages. First, it allows tagging of N -terminals of nonessential genes at the native genomic locus, while the traditional methods only allow tagging of the C-terminal. Second, only the desired tag remains in the yeast genome after tagging, and the selection marker is recyclable. Third, the method described in this report does not alter the sequence of the 3′ noncoding region when C-terminal tagging is performed, meaning that alteration of the expression of the target gene is unlikely.
Discussion
In this report, we describe a strategy to tag genes based on the URA3 pop-in/pop-out system, which replaces “junk” tandem repeats with designed DNA elements for pop-out. As a proof-of-concept, plasmids containing three FPs and two epitope tags were constructed, each of which was demonstrated to tag Ubc13, Histone 3, or Rad5 at the N- or C-terminal successfully. We found that, consistent with the recent published study of tagging in yeast[30], 0.3~0.4 kb homologous sequences of the target intergration site is more efficient than 50 bp mentioned in the the previous epitope tagging studies[19, 31].
Yeast is a powerful eukaryotic model organism for biological and medical science. The development of many valuable tools in recent decades has made the genetic manipulation of budding yeast much easier. However, some limitations remain. For example, tagging of the N-terminal is one of the long-standing challenges in yeast genetics. In early studies, a tag could be introduced at the N-terminal of a gene of interest by promoter fusion; however, its native promoter had to be replaced because of the way in which the tag was integrated into the chromosome [32, 33]. The method we report here is suitable for N-terminal tagging of nonessential genes without any modification in the promoter region after integration. The limited number of selectable markers is another problem in yeast genetic engineering, especially when multiple genes are to be modified. Our URA3 pop-in/pop-out system makes it possible to recycle a selectable marker; thus, one selectable marker can function repeatedly for multiple protein tagging, which is important for complex studies involving many proteins. For recycling of a selectable marker, pop-in/pop-out is the only strategy that has thus far been developed. However, in the pop-in/pop-out system, there is a problem that “junk” sequences such as hisG or loxP remain at the modification locus after popping out the selectable marker via HR. The efficiency of HR using limited homologous sequences, like PCR-mediated gene targeting, is clearly reduced when hisG-URA3-hisG is used more than once in the same strain because the hisG left in the genome causes significant internal homology [34]. Moreover, the unwanted “junk” sequence left in the chromosome might affect the expression of the target gene. For instance, a technique called DAmP (decreased abundance by mRNA perturbation) affects the expression of target gene through the disruption of natural 3′ untranslated region with an antibiotic-resistance marker insertion [35]. Thus, we here designed the DNA sequence from the tag itself for HR pop-out, which could avoid the decreased efficiency of HR and minimize the disturbance of the target gene’s expression level.
Labeling a protein of interest with an FP or epitope tag to study its properties, such as expression, localization, and dynamics, is widely used in biochemistry and cell biology. In addition, fluorescent probes are playing an increasingly important role in the newly developing techniques of single-molecule fluorescence detection and super-resolution microscopy [36, 37]. There are three main kinds of fluorescent probe: organic dyes, FPs, and quantum dots (QDs). Tagging of a protein with a fluorescent probe makes it possible to study the properties of the protein and its related biological processes at the single-molecule level [38, 39]. Using the photoconvertible protein mKikGR, a new powerful imaging technique, PhADE (photoactivation, diffusion, and excitation), was established and proposed to allow single-molecule study at physiological concentrations [37]. With the aid of anti-Flag-antibody-coated QDs, the mechanism by which CRISPR RNA-guided Cas9 interrogates DNA to recognize specific cleavage sites was clarified [40]. While the antibody for a protein of interest could be used for imaging at the single-molecule level [41], an epitope tag antibody would be more widely available from commercial sources. Taking these factors together, the convenient FP- and epitope-tagging techniques are helpful for solving scientific problems at the single-molecule level.
In summary, this protein-tagging method is an excellent tool for fusing an FP or epitope tag scarless to a target protein at its native locus in the yeast S. cerevisiae. Recently, Landgraf et al reported the same technique to tag the yeast genes with mNeonGreen and mCherry[30]. Here five tag constructs were presented and demonstrated to function in an easy-to-use and efficient way with homologous sequences. We believe that this strategy and the plasmids containing with newly developing fluorescent proteins will provide benefits in both bulk assays and single-molecule imaging in budding yeast research.
Supporting information
It details the steps for the 5’YFG-FP-URA3-FP-YFG3’products. The 5’YFG(PYFG) and YFG3’(ORFYFG) are PCR amplified from the yeast genome. The FP-URA3-FP cassettes are obtained by PCR from each plasmid. These three PCR products have a 25–30 bp duplication sequence (marked with wavy line) in the junction part for annealing and matching. With the mixture of these products as template, amplify to generate the final 5’YFG-FP-URA3-FP-YFG3’ products.
(PDF)
(A) PPGK1-mCherry-Ubc13 strain. The PCR products are 1.5 kb (lane 2), 3.9 kb (lane 3) and 2.2 kb (lane 4). The same pair of primers is used for PCR. (B) PPGK1-Rad5-sfGFP strain and (C) PPGK1-Rad5-mKikGR strain. The PCR products are 0.7 kb (lane 2), 3.0 kb (lane 3) and 1.4 kb (lane 4).
(PDF)
(A) 3xFLAG-Ubc13 strain. Lane 1: DNA ladder marker. Lane 2: the original strains-HK578-10A. Lane 3: the pop-in strain. Lane 4: the pop-out strain. Arrows indicate the locations of primers. (B) (Upper) Anti-FLAG antibody is used for detecting the 3xFLAG tag (Bottom). Pgk1 was used as an internal control.
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
The authors wish to thank Wei Li (Institute of Zoology, Chinese Academy of Sciences) for providing anti-Pgk1 antibody and Huiping Guo for technical assistance.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by the National Natural Science Foundation of China grant 31371264 for (YVF), the National Natural Science Foundation of China, 31670068 for (XW), the the Newton Advanced Fellowship (NA140085) for (YVF) from the Royal Society and CAS Interdisciplinary Innovation Team for (YVF). It was partially funded by Student's Platform for Innovation and Entrepreneurship Training Program for (QW). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Chang VK, Fitch MJ, Donato JJ, Christensen TW, Merchant AM, Tye BK. Mcm1 binds replication origins. The Journal of biological chemistry. 2003;278(8): 6093–100. doi: 10.1074/jbc.M209827200 [DOI] [PubMed] [Google Scholar]
- 2.Heller RC, Kang S, Lam WM, Chen S, Chan CS, Bell SP. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell. 2011;146(1): 80–91. doi: 10.1016/j.cell.2011.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yeeles JT, Deegan TD, Janska A, Early A, Diffley JF. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature. 2015;519: 431–5. doi: 10.1038/nature14285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar A, Agarwal S, Heyman JA, Matson S, Heidtman M, Piccirillo S, et al. Subcellular localization of the yeast proteome. Gene and Development. 2002;12(1): 47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huh W, Falvo JV, Gerke LC, Carroll SA, Howson RW, J.S. W, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425: 686–91. doi: 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- 6.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14(2): 115–32. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2 [DOI] [PubMed] [Google Scholar]
- 7.Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13(11): 1065–75. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K [DOI] [PubMed] [Google Scholar]
- 8.Chattoo BB, Sherman F, Azubalis DA, Fjellstedt TA, Mehnert D, Ogur M. Selection of lys2 Mutants of the Yeast SACCHAROMYCES CEREVISIAE by the Utilization of alpha-AMINOADIPATE. Genetics. 1979;93(1): 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alani E, Cao L, Kleckner N. A Method for Gene Disruption That Allows Repeated Use of Ura3 Selection in the Construction of Multiply Disrupted Yeast Strains. Genetics. 1987;116(4): 541–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10(13): 1793–808. [DOI] [PubMed] [Google Scholar]
- 11.Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15(14): 1541–53. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K [DOI] [PubMed] [Google Scholar]
- 12.Solis-Escalante D, Kuijpers NG, Bongaerts N, Bolat I, Bosman L, Pronk JT, et al. amdSYM, a new dominant recyclable marker cassette for Saccharomyces cerevisiae. Fems Yeast Research. 2013;13(1): 126–39. doi: 10.1111/1567-1364.12024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh A, Sherman F. Association of methionine requirement with methyl mercury resistant mutants of yeast. Nature. 1974;247(5438): 227–9. [DOI] [PubMed] [Google Scholar]
- 14.Boeke JD, Lacroute F, Fink GR. A Positive Selection for Mutants Lacking Orotidine-5'-Phosphate Decarboxylase Activity in Yeast—5-Fluoro-Orotic Acid Resistance. Molecular & General Genetics. 1984;197(2): 345–6. [DOI] [PubMed] [Google Scholar]
- 15.Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154: 164–75. [DOI] [PubMed] [Google Scholar]
- 16.Guldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic acids research. 1996;24(13): 2519–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic acids research. 2002;30(6): e23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solis-Escalante D, Kuijpers NG, Bongaerts N, Bolat I, Bosman L, Pronk JT, et al. amdSYM, a new dominant recyclable marker cassette for Saccharomyces cerevisiae. FEMS yeast research. 2013;13(1): 126–39. doi: 10.1111/1567-1364.12024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider B, Seufert W, Steiner B, Yang Q, Futcher A. Use of Polymerase Chain Reaction Epitope Tagging for Protein Tagging in Saccharomyces cerevisiae. Yeast. 1995;11: 1265–74. doi: 10.1002/yea.320111306 [DOI] [PubMed] [Google Scholar]
- 20.Moqtaderi Z, Struhl K. Expanding the repertoire of plasmids for PCR-mediated epitope tagging in yeast. Yeast. 2008;25(4): 287–92. doi: 10.1002/yea.1581 [DOI] [PubMed] [Google Scholar]
- 21.Tian XL, Xu X, Xiao W. Novel Method for Genomic Promoter Shuffling by Using Recyclable Cassettes. Applied and Environmental Microbiology. 2013;79(22): 7042–7. doi: 10.1128/AEM.02159-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gietz RD, Schiestl RH. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2(1): 31–4. doi: 10.1038/nprot.2007.13 [DOI] [PubMed] [Google Scholar]
- 23.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nature Methods. 2005;2(12): 905–9. doi: 10.1038/nmeth819 [DOI] [PubMed] [Google Scholar]
- 24.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nature Biotechnology. 2006;24(1): 79–88. doi: 10.1038/nbt1172 [DOI] [PubMed] [Google Scholar]
- 25.Habuchi S, Tsutsui H, Kochaniak AB, Miyawaki A, van Oijen AM. mKikGR, a monomeric photoswitchable fluorescent protein. PloS one. 2008;3(12): e3944 doi: 10.1371/journal.pone.0003944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dollard C, Ricupero-hovasse SL, Natsoulis G, BOEKE JD, Winston F. SPT1O and SPT21 are required for transcription of particular histone genes in Saccharomyces cerevisiae. Molecular and Celluar biology. 1994;14: 5223–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuang L, Kou H, Xie Z, Zhou Y, Feng X, Wang L, et al. A non-catalytic function of Rev1 in translesion DNA synthesis and mutagenesis is mediated by its stable interaction with Rad5. DNA Repair (Amst). 2013;12(1): 27–37. [DOI] [PubMed] [Google Scholar]
- 28.Xu X, Lin A, Zhou C, Blackwell SR, Zhang Y, Wang Z, et al. Involvement of budding yeast Rad5 in translesion DNA synthesis through physical interaction with Rev1. Nucleic Acids Res. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ball LG, Xu X, Blackwell S, Hanna MD, Lambrecht AD, Xiao W. The Rad5 helicase activity is dispensable for error-free DNA post-replication repair. DNA Repair. 2014;16: 74–83. doi: 10.1016/j.dnarep.2014.02.016 [DOI] [PubMed] [Google Scholar]
- 30.Landgraf D, Huh D, Hallacli E, Lindquist S. Scarless Gene Tagging with One-Step Transformation and Two-Step Selection in Saccharomyces cerevisiae and Schizosaccharomyces pombe. PloS one. 2016;11(10): e0163950 doi: 10.1371/journal.pone.0163950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moqtaderi Z, Struhl K. Expanding the repertoire of plasmids for wow PCR-mediated epitope tagging in yeast. Yeast. 2008;25(4): 287–92. doi: 10.1002/yea.1581 [DOI] [PubMed] [Google Scholar]
- 32.Longtine MS, Mckenzie A III, Douglas JD, G.S. N, Wach A, Brachat A, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14: 953–61. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U [DOI] [PubMed] [Google Scholar]
- 33.Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21(11): 947–62. doi: 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- 34.Davidson JF, Schiestl RH. Mis-targeting of multiple gene disruption constructs containing hisG. Current Genetics 2000;38: 188–90. [DOI] [PubMed] [Google Scholar]
- 35.Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123(3): 507–19. doi: 10.1016/j.cell.2005.08.031 [DOI] [PubMed] [Google Scholar]
- 36.Shaner NC, Patterson GH, Davidson MW. Advances in fluorescent protein technology. Journal of cell science. 2007;120(Pt 24): 4247–60. doi: 10.1242/jcs.005801 [DOI] [PubMed] [Google Scholar]
- 37.Loveland AB, Habuchi S, Walter JC, van Oijen AM. A general approach to break the concentration barrier in single-molecule imaging. Nat Methods. 2012;9(10): 987–92. doi: 10.1038/nmeth.2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finkelstein IJ, Visnapuu ML, Greene EC. Single-molecule imaging reveals mechanisms of protein disruption by a DNA translocase. Nature. 2010;468(7326): 983–7. doi: 10.1038/nature09561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie XS, Choi PJ, Li GW, Lee NK, Lia G. Single-molecule approach to molecular biology in living bacterial cells. Annual review of biophysics. 2008;37: 417–44. doi: 10.1146/annurev.biophys.37.092607.174640 [DOI] [PubMed] [Google Scholar]
- 40.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507(7490): 62–7. doi: 10.1038/nature13011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yardimci H, Loveland AB, van Oijen AM, Walter JC. Single-molecule analysis of DNA replication in Xenopus egg extracts. Methods. 2012;57(2): 179–86. doi: 10.1016/j.ymeth.2012.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
It details the steps for the 5’YFG-FP-URA3-FP-YFG3’products. The 5’YFG(PYFG) and YFG3’(ORFYFG) are PCR amplified from the yeast genome. The FP-URA3-FP cassettes are obtained by PCR from each plasmid. These three PCR products have a 25–30 bp duplication sequence (marked with wavy line) in the junction part for annealing and matching. With the mixture of these products as template, amplify to generate the final 5’YFG-FP-URA3-FP-YFG3’ products.
(PDF)
(A) PPGK1-mCherry-Ubc13 strain. The PCR products are 1.5 kb (lane 2), 3.9 kb (lane 3) and 2.2 kb (lane 4). The same pair of primers is used for PCR. (B) PPGK1-Rad5-sfGFP strain and (C) PPGK1-Rad5-mKikGR strain. The PCR products are 0.7 kb (lane 2), 3.0 kb (lane 3) and 1.4 kb (lane 4).
(PDF)
(A) 3xFLAG-Ubc13 strain. Lane 1: DNA ladder marker. Lane 2: the original strains-HK578-10A. Lane 3: the pop-in strain. Lane 4: the pop-out strain. Arrows indicate the locations of primers. (B) (Upper) Anti-FLAG antibody is used for detecting the 3xFLAG tag (Bottom). Pgk1 was used as an internal control.
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.