Abstract
MicroRNAs (miRNAs) are small regulators of gene expression that act on many different molecular and biochemical processes in eukaryotes. To date, miRNAs have not been considered in the current evaluation system for GM crops. In this study, small RNAs from the dry seeds of a GM wheat line overexpressing GmDREB1 and non-GM wheat cultivars were investigated using deep sequencing technology and bioinformatic approaches. As a result, 23 differentially expressed miRNAs in dry seeds were identified and confirmed between GM wheat and a non-GM acceptor. Notably, more differentially expressed tae-miRNAs between non-GM wheat varieties were found, indicating that the degree of variance between non-GM cultivars was considerably higher than that induced by the transgenic event. Most of the target genes of these differentially expressed miRNAs between GM wheat and a non-GM acceptor were associated with abiotic stress, in accordance with the product concept of GM wheat in improving drought and salt tolerance. Our data provided useful information and insights into the evaluation of miRNA expression in edible GM crops.
Introduction
The global hectarage of genetically modified (GM) crops has increased 100-fold, from 1.7 million hectares in 1996 to 179.7 million hectares in 2015, for 17 to 18 million farmers in 28 countries. The latest data for 1996 to 2014 shows that GM crops contribute to food security, sustainability and climate change [1]. However, in many areas, cultivation of the GM crops was prevented because of the dominant ideological voices of the opponents, debates on their potential negative environmental impact, and their health risks to consumers. Although there is no sufficient scientific experimental evidence [2], the adverse effects of several GM crops that have been published in scientific journals still generate suspicion and controversy in the opinion of both the public and the scientific community [3–5]. Therefore, research assessing the potential risks associated with GM crops is essential for fast scientific adoption and general public acceptance.
The current evaluation system for GM crops is focused on proteins, fats, carbohydrates, toxins, and nutritional ingredients. However, miRNAs have not yet been taken into account in this system of evaluation. It was recently found that several gma-miRNAs are differentially expressed in GM soybean seeds compared with the non-GM control [6]. miRNAs have been widely demonstrated to play fundamental roles in gene regulation in most eukaryotes [7–10]. The first cross-kingdom regulation of gene expression between humans and edible plants by miRNAs was reported in 2012 [11]. Moreover, it was recently demonstrated that Brassica miRNAs can regulate the expression of human genes and proteins in vitro [12], which has raised new concerns regarding the miRNA components of GM crops.
miRNAs regulate gene expression by guiding target mRNA cleavage or by translational inhibition [7–10]. The targets of miRNAs include a large set of transcription factors (TFs) [13], such as Auxin Response Factors (ARFs) and QUAMOSA Promoter Binding Protein-Like (SPLs), which are involved in the regulation of miRNA expression in a negative feedback loop in Arabidopsis [14]. On the other hand, miRNAs are processed from hairpin precursors (pre-miRNAs), and pre-miRNAs are derived from primary transcripts (pri-miRNAs) transcribed from genomic DNA which is most likely regulated by TFs. An over-expressed transcription factor (TaDREB3) in barley affects the expression of miRNAs and other small non-coding RNAs in its leaf [15].
Due to their involvement in the regulation of many stress-related genes, many TFs were utilized in GM crops to improve crop tolerance to abiotic stresses. GM drought tolerance is an extremely important goal given that droughts will likely become more severe and more frequent as climate change impacts crop productivity, agriculture and society. So far, only 3 GM drought tolerant plants have been recommended for commercialization approval including GM maize in America, soybean in Argentina, and sugarcane in Indonesia. GM DroughtGard™ tolerant maize, first planted in the US in 2013, increased more than 15-fold from 50,000 hectares in 2013 to 275,000 hectares in 2014 and 810,000 hectares in 2015, reflecting a high farmer acceptance, at a 3-fold, year-to-year increase between 2014 and 2015 [1]. Wheat (Triticum aestivum L.) is one of the most important cereal crops worldwide, and it is used as a stable food grain and as a primary source of straw for animal feeding. Wheat production is severely affected by various abiotic stresses such as drought, salinity, low temperature, and heat, which results in an estimated 50–60% loss in grain yield annually [16]. Thus far, except for the glyphosate herbicide tolerance wheat (Mon71800), no other GM drought tolerant wheat has been approved for commercialization. Transgenic wheat transformed with a DREB TF (GmDREB1) from soybean (Glycine max L.), which is driven by an ubiquitin promoter showed strong tolerance to drought and salt [17, 18].
In this study, we investigated small RNAs from the dry seeds of a GM wheat line overexpressing GmDREB1 and the non-transgenic control varieties using deep sequencing technology and bioinformatic approaches. Comparative analysis showed that 7 known tae-miRNAs were differentially expressed in the dry seeds between GM wheat and the non-GM acceptor. Combined with miRcheck prediction and experimental verification, 16 conserved and novel tae-miRNA candidates were also identified. This indicates that overexpression of the transcript factor GmDREB1 affects the expression of miRNAs in wheat seeds. Most of the target genes of the differentially expressed miRNAs between GM wheat and the non-GM acceptor were associated with abiotic stress, in accordance with the product concept of the GM wheat line T349 [17, 18]. These results provided useful data for a bio-safety assessment of GM crops and valuable information for wheat miRNA research.
1. Materials and Methods
1.1 GM wheat samples and RNA isolation
The transgenic wheat line T349, non-transgenic acceptor Jimai 19 (J19) and comparable non-transgenic varieties Jimai 22 (J22) and Lumai 21 (L21) were provided by the Institute of Crop Science, Chinese Academy of Agricultural Sciences. The transgenic wheat line T349, transformed with GmDREB1 from soybean, showed strong tolerance to drought and salt stresses [17, 18]. The transgenic and non-transgenic wheat varieties were planted in a field with water-saving measures in the Jinan province of China. The mature wheat seeds were harvested for RNA isolation. The main wheat cultivars, J22 and L21, were cultivated in the same climatic and geographic ecological region as J19. The total RNA was isolated from wheat seeds that were pooled from 3 individuals of the same transgenic or nontransgenic line using the Plant RNA Extraction kit (TaKaRa MineBEST) according to the manufacturer’s instructions. The quantity and quality of total RNA were evaluated using a NanoDrop 2000 Spectrophotometer and an Agilent 2100 RNA 6000 Nano kit.
1.2 Small RNA library construction and sequencing
The sRNAs were isolated from the total RNA using the TruSeq Small RNA Sample Preparation kit. The sRNA was ligated with a 3’-linker and electrophoresed on a 15% polyacrylamide gel. The 15–30 nt sRNAs were excised from the gel and recovered in 0.3 M NaCl. The recovered sRNAs were then ligated with a 5’-linker and subject to reverse transcription PCR. The RT-PCR products were separated on a 3.5% agarose gel. The 140–160 bp products were selected for library construction and submitted for sequencing on a Hi-Seq 2500 analyzer. The raw data from the small RNA libraries were deposited in the NCBI Sequence Read Archive (SRA) under accession No. SRP091415.
1.3 Bioinformatic analysis of high-throughput data
For deep sequencing data analysis, similar methods reported in a previous work were used in this section [6]. In brief, raw reads were cleaned by quality and adaptor trimming. Identical reads were collapsed and recorded for abundance. The SOAP programme was used for genome mapping. Only perfectly matched sRNAs with a length between 15–30 nt were subjected to further analysis [19]. The wheat (Triticum aestivum L.) genome was downloaded from the cerealsDB database (http://www.cerealsdb.uk.net/cerealgenomics/CerealsDB/copyright.php).
1.4 Differential expression analysis of known tae-miRNAs
Known wheat miRNA sequences were downloaded from miRBase (V21.0) [20]. The DESeq2 package [21] was used to determine the differentially expressed miRNAs, which were required to meet 3 prerequisites: raw reads abundance > 100, |log2 value| > 1 and FDR value < 0.001.
1.5 Novel miRNA prediction
Before novel miRNA prediction, small RNA reads were searched against the Rfam database (V11.0) to remove the known non-coding RNAs [22]. Customized Perl scripts, RNAfold and miRCheck programmes were used for context extraction, secondary structure determination and novel miRNA prediction [23]. Novel miRNA candidates were clustered with known miRNAs, including mature and precursor sequences, to find novel members in conserved families. Details of this process are described in a previous study [24].
1.6 Target prediction and function analysis
Target genes were predicted using psRNATarget [25], a plant sRNA target analysis server (http://plantgrn.noble.org/psRNATarget). Eight degradome libraries (GSM1606478, GSM1606479, GSM911923, GSM911924, SRR1197125, SRR1197126, SRR1197127, SRR1197128) were downloaded and analyzed for validating the cleavage sites in target genes using CleaveLand4 [26]. To further understand the function and classification of the predicted miRNA targets, GO classification of the target genes was conducted with WEGO web service (http://www.geneontology.org/), GO terms assigned to the query sequences and catalogued groups were produced based on their biological process, molecular functions, and cellular components.
1.7 RT-PCR assays
Differential expression of the tae-miRNAs was verified by qRT-PCR experiments. One microgram of total RNA was reverse-transcribed and amplified using miRcute miRNA First-Strand cDNA synthesis kit (Tiangen, KR201) and the miRcute miRNA qPCR detection kit (Tiangen, FP401). The comparative ΔΔCT method was used for the relative quantitation of these tae-miRNAs [27]. miR159 was selected as the endogenous reference gene for normalization [28]. For the validation of novel tae-miRNA candidates, a diluted cDNA template was used for RT-PCR assays with specific forward primers and a universal reverse primer. The amplified products were detected by electrophoresis on a 2% (w/v) agarose gel and validated by Sanger sequencing.
The expression profiles of some target genes of the differentially expressed miRNAs were analyzed by qRT-PCR analysis of a randomly selected set of genes. DNaseI-treated RNA was used for first-strand cDNA synthesis using Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA) and oligo (dT)18 primers according to the manufacturer’s protocol. The comparative ΔΔCT method was used for the relative quantitation of genes [27]. The Tubulin gene was amplified as an internal control. The primers used in this section are listed in S1 Table.
2. Results
2.1 Deep sequencing of small RNAs in wheat seeds
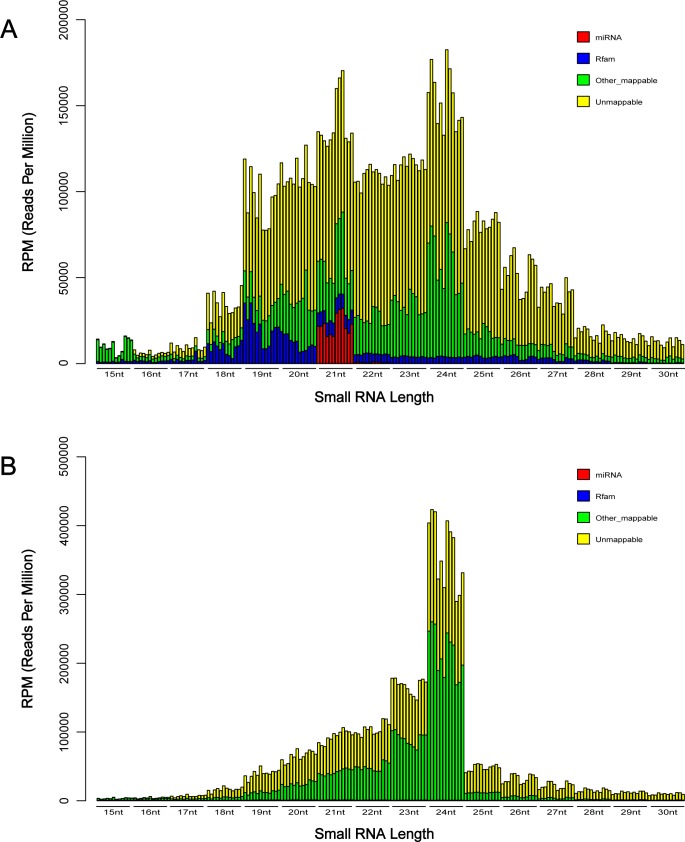
Twelve small RNA libraries from T349, J19, J22 and L21 wheat cultivars, each with 3 replicates, generated 187,919,197 raw reads via the Illumina/Solexa deep sequencing platform. After the removal of low-quality reads and 3’ adaptor trimming, 159,723,105 (100%) clean reads, corresponding to 25,123,347 (100%) unique reads, were obtained for subsequent analysis. Genomic mapping results showed that 53,820,203 (33.7%) clean reads and 11,937,579 (47.5%) unique reads could find at least one perfectly matching locus in the wheat genome. We regarded 40,599,438 (25.4%) clean reads and 11,669,683 (46.4%) unique reads as unknown reads after searching against the miRBase and Rfam databases, which were used for novel miRNA prediction (Table 1). The length distribution of the clean reads showed that both 21-nt and 24-nt classes were dominant in the total reads (Fig 1A) and only the 24-nt class occupied the majority in the unique reads with obvious superiority (Fig 1B).
Table 1. Composition of small RNA libraries from wheat seeds of GM and non-GM plants.
| T349 | J19 | J22 | L21 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | A | B | C | A | B | C | A | B | C | ||
| Raw reads | Total | 19,203,574 | 14,806,199 | 17,440,475 | 17,302,501 | 17,735,423 | 17,113,284 | 10,471,928 | 11,234,870 | 10,844,006 | 16,663,383 | 18,475,545 | 16,628,009 |
| High quality reads | Total | 18,579,060 | 14,317,885 | 16,893,403 | 16,755,380 | 17,163,457 | 16,549,277 | 10,026,859 | 10,675,060 | 10,385,552 | 16,408,397 | 18,189,376 | 16,376,319 |
| Clean reads(15–30 nt) | Unique | 2,721,909 | 2,533,728 | 2,615,689 | 2,091,765 | 2,230,138 | 1,886,868 | 1,769,211 | 1,783,405 | 1,552,281 | 1,852,931 | 2,049,892 | 2,035,530 |
| Total | 16,279,286 | 12,587,688 | 14,854,978 | 14,783,399 | 15,154,921 | 14,235,466 | 9,026,382 | 9,512,116 | 9,184,495 | 13,954,104 | 15,895,052 | 14,255,218 | |
| Not matching genome | Unique | 1,340,349 | 1,237,502 | 1,301,946 | 1,149,935 | 1,214,177 | 1,069,609 | 921,192 | 954,777 | 837,046 | 993,653 | 1,108,576 | 1,057,006 |
| Total | 10,006,079 | 7,872,719 | 9,104,624 | 10,217,273 | 10,499,254 | 9,987,590 | 5,653,864 | 5,993,448 | 5,670,558 | 9,816,304 | 11,266,819 | 9,814,370 | |
| Matching genome | Unique | 1,381,560 | 1,296,226 | 1,313,743 | 941,830 | 1,015,961 | 817,259 | 848,019 | 828,628 | 715,235 | 859,278 | 941,316 | 978,524 |
| Total | 6,273,207 | 4,714,969 | 5,750,354 | 4,566,126 | 4,655,667 | 4,247,876 | 3,372,518 | 3,518,668 | 3,513,937 | 4,137,800 | 4,628,233 | 4,440,848 | |
| Known tae-miRNAs | Unique | 50 | 51 | 54 | 49 | 53 | 49 | 48 | 55 | 55 | 51 | 48 | 52 |
| Total | 362,683 | 283,931 | 356,260 | 239,879 | 262,491 | 230,794 | 273,018 | 308,941 | 303,733 | 290,011 | 285,688 | 334,083 | |
| Matching Rfam database (exclude miRNAs) | Unique | 23,704 | 21,117 | 22,795 | 25,326 | 25,151 | 24,991 | 17,178 | 18,022 | 16,883 | 23,418 | 25,084 | 24,227 |
| Total | 1,673,743 | 1,112,800 | 1,534,831 | 1,316,541 | 1,232,379 | 1,247,230 | 479,033 | 511,618 | 551,157 | 1,083,226 | 1,301,795 | 1,176,412 | |
| Unknown reads | Unique | 1,357,856 | 1,275,109 | 1,290,948 | 916,504 | 990,810 | 792,268 | 830,841 | 810,606 | 698,352 | 835,860 | 916,232 | 954,297 |
| Total | 4,599,464 | 3,602,169 | 4,215,523 | 3,249,585 | 3,423,288 | 3,000,646 | 2,893,485 | 3,007,050 | 2,962,780 | 3,054,574 | 3,326,438 | 3,264,436 | |
Fig 1. Length distribution of small RNA libraries.
A: Length distribution of the total reads. B: Length distribution of the unique reads. For each class, 12 RPM values were arranged in order: T349_rep1, T349_rep2, T349_rep3, J19_rep1, J19_rep2, J19_rep3, J22_rep1, J22_rep2, J22_rep3, L21_rep1, L21_rep2, and L21_rep3.
2.2 Expression profile of known tae-miRNAs in transgenic and non-transgenic wheat
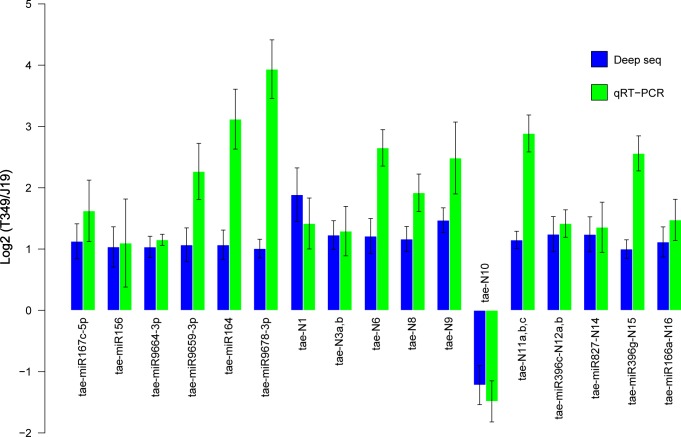
To detect the known tae-miRNAs, the clean reads were aligned against the known tae-miRNAs represented in miRBase 21. As a result, 7 tae-miRNAs were up-regulated in the seeds of GM wheat T349 compared with the non-GM acceptor J19. Moreover, no down-regulated miRNAs were detected (Table 2). The log2 ratios varied from 1.01 to 1.20 and the maximum value was 1.20 for tae-miR319. Among these 7 known tae-miRNAs, 3 miRNAs, tae-miR167c-5p, tae-miR156 and tae-miR9661-3p were up-regulated only in GM wheat seeds compared to a non-GM acceptor, while there were no significant differences between the different non-GM wheat varieties tested in this study (L21/J19 and J22/J19). Four other tae-miRNAs were also differentially expressed between the non-transgenic wheat seeds. Compared with J19, tae-miR319 was up-regulated both in T349 and L21; tae-miR164 and tae-miR9678-3p were up-regulated both in T349 and J22 and tae-miR9659-3p was up-regulated in T349, L21 and J22 (Table 2). Meanwhile, the expressions of these known tae-miRNAs were confirmed by qRT-PCR (Fig 2). The differentially expressed miRNAs between the non-transgenic wheat varieties were not listed here, if they were not differentially expressed between T349 and J19.
Table 2. Differential expression of conserved tae-miRNAs in wheat seeds.
| NO. | Mature miRNA | Sequence(5'-3') | Length (nt) |
Log2 (T349/J19) |
Log2 (L21/J19) |
Log2 (J22/J19) |
RPM in each bank | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T349 | J19 | J22 | L21 | ||||||||||||||||||
| 1 | tae-miR167c-5p | TGAAGCTGCCAGCATGATCTGC | 22 | 1.13 | ↑ | 0.51 | — | 0.02 | — | 5.77 | 7.15 | 10.10 | 3.18 | 4.36 | 4.43 | 3.55 | 5.05 | 3.48 | 3.08 | 5.10 | 5.05 |
| 2 | tae-miR156 | TGACAGAAGAGAGTGAGCACA | 21 | 1.04 | ↑ | -0.36 | — | 0.51 | — | 3.99 | 2.94 | 3.77 | 2.30 | 2.05 | 1.62 | 3.77 | 2.00 | 2.72 | 1.15 | 0.82 | 1.61 |
| 3 | tae-miR9664-3p | TTGCAGTCCTCGATGTCGTAG | 21 | 1.03 | ↑ | 0.45 | — | 0.87 | — | 161.55 | 174.62 | 150.12 | 106.94 | 90.99 | 74.88 | 170.17 | 175.15 | 151.34 | 103.84 | 78.89 | 104.94 |
| 4 | tae-miR319 | TTGGACTGAAGGGAGCTCCCT | 21 | 1.20 | ↑ | 1.02 | ↑ | 0.70 | — | 8.48 | 6.59 | 7.61 | 3.65 | 3.76 | 3.86 | 6.54 | 5.57 | 6.21 | 4.44 | 5.91 | 7.44 |
| 5 | tae-miR9659-3p | TCCAATGGTTGTTCACGGCATC | 22 | 1.07 | ↑ | 1.09 | ↑ | 1.23 | ↑ | 3.93 | 4.45 | 3.90 | 1.96 | 2.38 | 2.32 | 5.54 | 5.68 | 4.46 | 3.22 | 3.15 | 4.70 |
| 6 | tae-miR164 | TGGAGAAGCAGGGCACGTGCA | 21 | 1.07 | ↑ | 0.81 | — | 1.62 | ↑ | 9.28 | 7.31 | 8.55 | 4.87 | 3.96 | 4.92 | 14.73 | 13.56 | 14.05 | 4.94 | 5.54 | 8.28 |
| 7 | tae-miR9678-3p | TCTGGCGAGGGACATACACTGT | 22 | 1.01 | ↑ | 0.17 | — | 1.42 | ↑ | 65.60 | 61.57 | 52.17 | 35.24 | 33.65 | 33.65 | 92.17 | 89.57 | 90.70 | 29.96 | 28.06 | 30.94 |
RPM, reads per million; ↑, up-regulated;—, no significant difference.
Fig 2. Verification of differentially expressed tae-miRNAs by qRT-PCR.
The comparative ΔΔCT method was used for the qRT-PCR experiments and miR159 was selected as the reference.
2.3 Predicted novel miRNAs in GM and non-GM wheat
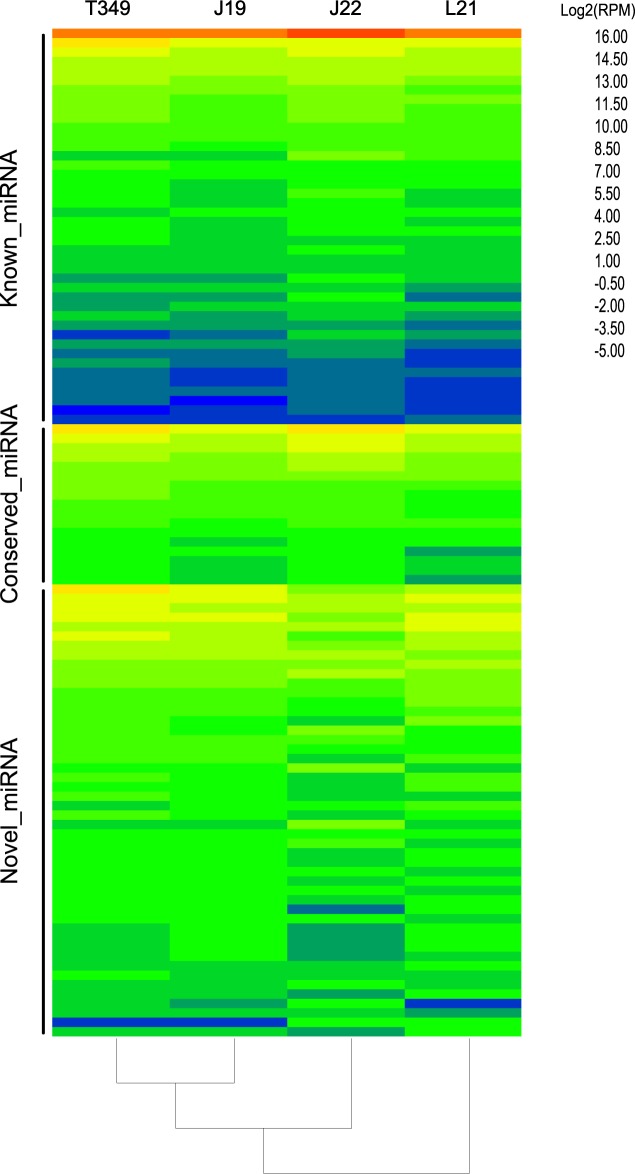
In the 12 libraries, the sequenced miRNAs that were not reported in Triticum aestivum but were in Aegilops tauschii, Brachypodium distachyon, Oryza sativan, Sorghum bicolor or Zea mays according to miRBase 21, were defined as conserved miRNAs. The sequenced miRNAs that were not reported in Triticum aestivum or the other five monocotyledons referenced above were defined as novel miRNAs. Sixteen conserved miRNAs and 49 novel miRNAs were found in wheat seeds after sequence alignment (Fig 3, S2 Table). All 49 candidate novel miRNAs and 16 conserved miRNAs possessed a perfect secondary stem loop structure (S1 File, S2 File). Because the miRNA* strand is degraded during typical miRNA biogenesis, we found both mature miRNA and miRNA* of the 8 miRNAs in 49 novel miRNAs and 6 conserved miRNAs (S1 File).
Fig 3. Heatmap of all miRNAs found in this study.
miRNAs were classified into three categories: known, conserved and novel. For each panel, miRNAs were listed according to their abundance, which is Log2 (mean value of RPM).
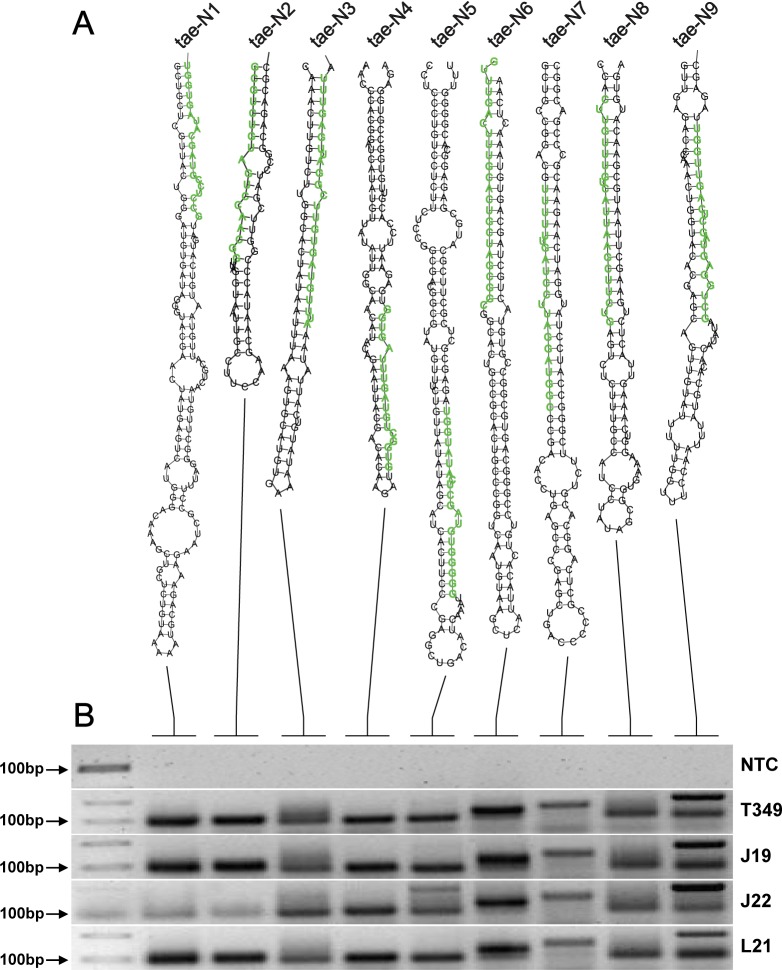
Compared with the non-GM acceptor J19, 5 conserved miRNAs and 11 novel miRNAs were differentially expressed in the seeds of GM wheat T349 (Table 3). One novel miRNA was down-regulated and another 15 miRNAs were up-regulated in T349. In addition to T349, the conserved miRNAs ata-miR396c-5p, osa-miR164c and bdi-miR827-3p were also up-regulated in J22. The miRNAs ata-miR396c-5p and ata-miR166a-3p were also up-regulated in L21 and J22 compared with J19 (Table 3). Among the 11 novel miRNAs families, 2 miRNAs were differentially expressed only in GM wheat seeds, while no significant differences were observed between the different non-transgenic wheat varieties tested in this study (L21/J19 and J22/J19). We defined these two novel miRNAs as tae-N1 (5’GCCTCCGTAGCATAGTGGT3’) and tae-N2 (5’GCGTCTGTAGTCCAACGGT3’). The other 9 miRNA families were differentially expressed in both GM wheat T349 and the non-GM varieties J22 and/or L21 compared with J19. The expressions of some novel and conserved miRNAs were tested by qRT-PCR (Fig 2). The 11 novel miRNAs and the 5 conserved tae-miRNA candidates were subjected to experimental verification by RT-PCR and Sanger sequencing, and they were all positively identified in at least one library (Fig 4, S3 File).
Table 3. Differential expression of novel and conserved tae-miRNAs in wheat seeds.
| NO. | Mature miRNA |
Sequence(5'-3') | Length (nt) |
Log2 (T349/J19) |
Log2 (L21/J19) |
Log2 (J22/J19) |
RPM in each bank | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T349 | J19 | J22 | L21 | ||||||||||||||||||
| 1 | tae-N1 | GCCTCCGTAGCATAGTGGT | 19 | 1.89 | ↑ | -0.12 | — | -0.10 | — | 10.197 | 4.846 | 16.762 | 3.99 | 2.441 | 3.02 | 4.985 | 3.048 | 0.762 | 1.648 | 1.95 | 3.086 |
| 2 | tae-N2 | GCGTCTGTAGTCCAACGGT | 19 | 1.32 | ↑ | -0.01 | — | -0.38 | — | 838.611 | 575.006 | 801.28 | 379.006 | 285.187 | 354.115 | 315.519 | 270.286 | 192.824 | 241.649 | 242.087 | 298.066 |
| 3 | tae-N3a, b | ATTTGTAGTGTTCGGATTGAGTTT | 24 | 1.23 | ↑ | 0.92 | — | 1.18 | ↑ | 10.872 | 14.935 | 12.319 | 7.102 | 6.73 | 4.706 | 15.731 | 14.718 | 11.65 | 8.456 | 7.36 | 11.364 |
| 4 | tae-N4a, b, c, d, e, f, g | GTGGCTGTAGTTTAGTGG | 18 | 1.29 | ↑ | 2.50 | ↑ | -2.00 | ↓ | 11.302 | 22.164 | 9.895 | 14.881 | 9.237 | 15.665 | 9.306 | 9.566 | 6.423 | 6.736 | 5.095 | 4.7 |
| 5 | tae-N5 | GGGGGTGTAGCTCATATGGT | 20 | 1.65 | ↑ | -0.36 | — | -1.80 | ↓ | 18.489 | 23.038 | 19.185 | 23.742 | 27.713 | 19.177 | 37.224 | 38.687 | 33.099 | 16.052 | 15.728 | 17.256 |
| 6 | tae-N6 | GTTTGACTTTGCACTGCTAGCGGC | 24 | 1.21 | ↑ | -2.27 | ↓ | 3.15 | ↑ | 2.088 | 1.668 | 3.433 | 6.29 | 5.74 | 7.656 | 0.221 | 1.471 | 1.85 | 10.534 | 9.499 | 10.031 |
| 7 | tae-N7 | TTTTTTGATCCTTAGGATGGC | 21 | 1.15 | ↑ | -0.06 | — | 1.26 | ↑ | 3.378 | 3.098 | 3.567 | 1.623 | 1.979 | 1.334 | 15.066 | 16.189 | 13.392 | 0.286 | 0.314 | 0.07 |
| 8 | tae-N8 | CTTGTTTGTCATTAAGCTTCTG | 22 | 1.16 | ↑ | -1.34 | ↓ | -1.38 | ↓ | 6.204 | 6.673 | 7.135 | 7.778 | 12.141 | 9.202 | 4.431 | 5.992 | 3.919 | 4.944 | 6.291 | 5.471 |
| 9 | tae-N9 | GCTGGAGTAGCTCAGTTGGT | 20 | 1.47 | ↑ | -0.13 | — | -2.13 | ↓ | 14.435 | 13.902 | 10.77 | 6.29 | 6.928 | 7.024 | 15.731 | 16.189 | 16.549 | 4.156 | 5.284 | 5.471 |
| 10 | tae-N10 | GCGGGAGGCACGGGGTTCG | 19 | -1.22 | ↓ | 0.97 | — | -2.37 | ↓ | 24.878 | 34.16 | 27.061 | 14.949 | 16.298 | 12.714 | 7.311 | 4.625 | 4.681 | 4.228 | 4.718 | 4.279 |
| 11 | tae-N11a, b, c | ATCAGAGTGGCGCAGCGGA | 19 | 1.15 | ↑ | 0.02 | — | -2.36 | ↓ | 26.291 | 24.865 | 27.802 | 11.972 | 9.831 | 10.818 | 3.434 | 1.366 | 2.395 | 7.381 | 8.052 | 7.506 |
| 12 | tae-miR396c-N12a, b | TCCACAGCTTTCTTGAACTG | 20 | 1.24 | ↑ | 1.03 | ↑ | 1.25 | ↑ | 6.941 | 10.565 | 5.789 | 3.652 | 2.837 | 4.636 | 9.084 | 9.777 | 7.839 | 4.371 | 5.85 | 7.576 |
| 13 | tae-miR164c-N13 | TGGAGAAGCAGGGCACGTGCT | 21 | 1.38 | ↑ | 0.32 | — | 1.62 | ↑ | 7.739 | 6.355 | 8.078 | 3.314 | 2.969 | 3.442 | 9.084 | 9.251 | 11.541 | 2.938 | 3.271 | 3.156 |
| 14 | tae-miR827-N14 | TTAGATGACCATCAGCAAACA | 21 | 1.24 | ↑ | 0.00 | — | 1.39 | ↑ | 6.941 | 7.547 | 7.404 | 4.193 | 2.969 | 3.371 | 9.416 | 5.992 | 12.085 | 3.368 | 2.327 | 2.455 |
| 15 | tae-miR396g-N15 | TCCACAGGCTTTCTTGAACTG | 21 | 1.00 | ↑ | 0.54 | — | 1.13 | ↑ | 101.97 | 101.845 | 90.676 | 58.308 | 56.681 | 54.23 | 129.62 | 131.936 | 108.552 | 56.9 | 60.396 | 72.254 |
| 16 | tae-miR166a-N16 | TCGGACCAGGCTTCATTCC | 19 | 1.12 | ↑ | 1.38 | ↑ | 1.20 | ↑ | 48.527 | 57.357 | 46.314 | 29.627 | 35.236 | 30.416 | 88.739 | 87.046 | 63.585 | 39.056 | 36.363 | 54.786 |
↑, up-regulated; ↓, down-regulated;—, no significant difference. No.1-11, novel miRNAs; No. 12–16, conserved miRNAs.
Fig 4. Novel tae-miRNA candidates found in this study.
A: Hairpin structures of the novel tae-miRNA precursors predicted in GM wheat seeds. Mature miRNAs are indicated with green lowercase letters. B: Expression analysis of the novel tae-miRNA candidates by RT-PCR. NTC, no template control; T349, T349; J19, Jimai 19; L21, Lumai 21; J22, Jimai 22
2.4 Target genes of up- and down-regulated miRNAs in GM and non-GM wheat
miRNAs regulate target genes involved in plant development and stress response. miR167 target auxin response factor mediated plant root development [29, 30]. miR156 targets SQUAMOSA promoter binding protein-like (SPL) TFs related to flowering time, phase change and leaf initiation rate [31, 32]. miR319 targets TCP transcription factors and miR164 targets NAC domain gene, which contributes to leaf development [33]. Moreover, these target genes are all involved in stress response [34–38] (Table 4). To reveal the function of the predicted novel miRNAs and the conserved miRNAs, we predicted their targets using psRNATarget (http://plantgrn.noble.org/psRNATarget/). As is common for wheat, some predicted targets were functionally unknown and most miRNAs were found to target more than one gene (S3 Table). Part of these cleave sites were validated by wheat degradome data (S4 Table). T-plots of the identified targets were shown in S1 Fig. The expression profiles of some target genes of differentially expressed miRNAs were analyzed by qRT-PCR, and some target genes were down-regulated in T349 compared with J19 (S2 Fig). GO analysis results of all the target genes of the differentially expressed miRNAs in GM and non-GM wheat showed that many targets were related to water transport and responses to oxygen-containing compounds (S3 Fig), suggesting that the corresponding miRNAs may be involved in drought/salt stress and oxidative stress.
Table 4. Target prediction of known tae-miRNAs in wheat seeds.
| miRNA | Target accession | Known annotation | New annotation | |||
|---|---|---|---|---|---|---|
| tae-miR167c-5p | TC402576 | CV773862 | TC422705 | Auxin Response Factor | No results. | |
| tae-miR156 | Squamosa Promoter-binding-Like protein (SPL) | Teosinte glume architecture 1, | ||||
| TC453361 | CK196549 | AL810223 | TC409846 | Glycosyl/glycerophosphate transferase, | ||
| DR739383 | TC384445 | TC420438 | TC412204 | Predicted permease, | ||
| TC441570 | TC373290 | TC398965 | TC460639 | Cob(I)alamin adenolsyltransferas, | ||
| TC372857 | TC390294 | CA741955 | TC370322 | Cytochrome P450, | ||
| TC413555 | CA612886 | Telomere binding protein, | ||||
| Initiator binding protein. | ||||||
| tae-miR9664-3p | Not available | NB-ARC domain containing protein, | ||||
| CN008390 | CN007906 | CJ899791 | DR734475 | Nodulin-like protein, | ||
| CK202829 | CK203152 | BE425456 | BE604149 | NB-ARC domain containing protein, | ||
| CD876018 | CA499346 | CV779224 | TC436817 | NBS-LRR resistance-like protein, | ||
| CK193467 | TC451016 | BJ245571 | BE585521 | Resistance protein RGA1R, | ||
| CV772438 | TC401974 | DR738720 | TC379417 | LRR19, | ||
| Alternative splicing regulator. | ||||||
| tae-miR319 | TCP and MYB transcription factor | Glycosyl transferase, | ||||
| TC407332 | CK212140 | TC421314 | TC368630 | Histone H2B, | ||
| BF485310 | CA484819 | TC455115 | BE517710 | Acyl-CoA synthetase, | ||
| GH728978 | TC438746 | CA630893 | TC398226 | Trans-cinnamate 4-monooxygenase, | ||
| TC432120 | CK215833 | CJ863381 | Type 1 non specific lipid transfer protein precursor, | |||
| Transcription factor PCF8, | ||||||
| Ribosomal protein L3-A2-II. | ||||||
| tae-miR9659-3p | TC438538 | CA606192 | CV779210 | TC382784 | Not available | Anaeromyxobacter dehalogenans, |
| TC454472 | TC371921 | TC438010 | BQ904504 | Glutathione transferase, | ||
| BE401763 | CA639001 | TC393061 | TC371776 | HvPIP1;5 protein, | ||
| TC370293 | CD875198 | TC383120 | Aquaporin PIP1-2, | |||
| Inositol-1,4,5-triphosphate-5-phosphatase. | ||||||
| tae-miR164 | TC416811 | CA642340 | TC390810 | TC376198 | NAC domain-containing protein | |
| TC394945 | TC373635 | TC371535 | TC410195 | |||
| TC429623 | CA704421 | TC393137 | TC421735 | Efflux ABC transporter permease protein, | ||
| TC369110 | CA681504 | TC398164 | TC406273 | Harpin-induced protein 1 containing protein, | ||
| TC394481 | TC371551 | TC370694 | TC382290 | Mitogen-activated protein kinase homolog 1, | ||
| TC375350 | DR741669 | TC368951 | DR741517 | ZmRR2 protein, | ||
| TC378810 | BE515854 | TC435250 | TC430604 | Phytosulfokine-alpha 1 precursor, | ||
| CK211831 | TC445043 | TC458082 | CA648036 | OTU-like cysteine protease family protein, | ||
| TC408311 | TC407532 | TC430881 | TC407133 | Sugar transport protein. | ||
| TC404361 | TC411029 | TC384650 | CK198447 | |||
| CK216067 | BJ266172 | |||||
| tae-miR9678-3p | TC427317 | DR736808 | TC381152 | CJ648199 | Not available | F-box protein-like. |
3. Discussion
3.1 Over-expressed transcription factor (GmDREB1) in wheat affects the expression of miRNA in seeds
The global area for the cultivation of transgenic plants has grown continuously during the past two decades, despite constant controversy concerning their unforeseeable biosafety issues [1]. The application of transgenic technology has led to significant changes in modern agriculture and this trend seems irreversible [1]. Because of the dynamic property of RNA molecules, it is reasonable to propose that some endogenous RNAs might be diverse in transgenic plants. To date, very few reports have discussed the RNA ingredient content difference between GM and non-GM plants [6, 15]. Herein, we investigated the differential expression of miRNAs in wheat seeds from a GM line (T349), its non-GM receptor (J19) and local major wheat cultivars (J22 and L21) using high throughput sequencing technology. As a result, GmDREB1over-expression in wheat affects the expression of miRNAs in seeds. Other reports also mentioned the relationship between miRNAs and DREB. The expression levels of DREB1A and DREB2A were relatively induced in miR408 overexpressed plants compared to the vector control upon drought stress [39]. An over-expressed transcription factor (TaDREB3) in barley affects the expression of miRNAs and other small non-coding RNAs in its leaf [15].
In this study, the degree of variance, including miRNAs (Fig 2) and sRNA variance (S4 Fig), between cultivars was much higher than that induced by the transgenic event. Twenty-three differentially expressed miRNAs were found and confirmed between the T349 and J19 lines. Moreover, some of the miRNAs affected by DNA manipulation overlapped with the differentially expressed miRNAs related to varietal influence. Further analysis showed that most target genes of these differentially expressed miRNAs were associated with abiotic stress. This is understandable because a GmDREB1 transcription factor was introduced in the transgenic line T349 to elevate its drought and salt tolerance ability [17, 18].
3.2 Target genes of the differentially expressed miRNAs were associated with abiotic stress
Differentially expressed miRNAs were observed between the GM wheat T349 and the non-GM acceptor J19. Several known miRNAs (miR319, miR164, miR167, and miR156) are involved in drought response. miR319 regulates TCP transcription factors [33] and the NAC domain gene, which contributes to leaf development and drought stress response [38]. Moreover, the NAC domain gene is known as the target gene of miR164 in rice [40], and TCP genes have been shown to regulate miR164 in Arabidopsis [34]. It was reported that miR319 also target MYB transcription factors [41] that play significant roles in stress responses. miR167 regulates its target gene, ARF, which mediates plant root development [30, 37] and responds to drought stress [35]. Another identified target of miR167, IAA-Ala Resistant3 (IAR3) has the same roles as ARF in drought stress and root development [29]. miR156 targets SQUAMOSA promoter binding protein-like (SPL) TFs, which control flowering time, phase change and leaf initiation rate [31, 32, 42]. The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Blocking the miR156 signalling pathway with 35S::MIM156 (via target mimicry) increased the sensitivity of the plant to stress treatment, whereas the overexpression of miR156 increased stress tolerance [36]. In transgenic barley overexpressing the drought tolerant gene TaDREB3, miR156 was over two-fold up-regulated when comparing with the non-transgenic control. Thus these known miRNAs up-regulated in GM wheat may enhance the stress tolerance of transgenic wheat. In another study, miRNA microarray analysis showed that miR156, miR167, miR164, miR319, miR396 and miR166 were up-regulated in leaf or root of bread wheat under drought stress [43]. GO analysis of all the target genes of differentially expressed miRNAs in the GM and non-GM wheat lines show that many targets are related to water transport and response to oxygen-containing compounds (S3 Fig), suggesting that the corresponding miRNAs may be involved in drought/salt stress and oxidative stress.
3.3 Possible environmental risks of differentially expressed miRNAs in GM wheat
A controversial and attention-drawing issue is whether the exogenous miRNAs of GM crops could regulate human gene expression by cross-kingdom RNAi, which has been observed in both animal and plant systems. In animal systems, there are a few instances of cross-kingdom gene silencing. One example is that RNAi can be induced by soaking Caenorhabditis elegans worms in RNA solutions or by feeding them antisense RNA-expressing bacteria, such as Escherichia coli [44]. Plants transfer RNAi signals into interacting organisms, such as filamentous fungi, oomycetes, nematodes, parasitic plants, and pests [45–48] to silence their genes; however, it is not confirmed whether miRNA mediates human-plant interactions. Although it was reported that exogenous rice MIR168a is present in the human and mouse sera and the exogenous MIR168a inhibits human/mouse LDLRAP1 gene expression in liver [11], many subsequent reports have also questioned this controversial result [49–52]. It was recently demonstrated that Brassica miRNAs could regulate the expression of human genes and proteins, but only in vitro [12].
In this study, the targets of 23 differentially expressed miRNAs in GM wheat seeds were predicted in human (S5 Table) and chicken (S6 Table), both of which may eat GM wheat seeds. The top 50 predicted targets with the highest scores of 23 differentially expressed miRNAs in GM wheat seeds were mainly involved in the regulation of metabolic process, including primary, macromolecule and RNA metabolic processes in humans (S5A Fig) Moreover, they were primarily involved in epithelial cell differentiation in chicken (S5B Fig). However, our study did not conclude whether there are possible environmental risks of eating GM wheat seeds exist until it was the cross-kingdom RNAi was proven to be induced between animal/human-plant in vivo, and the differentially expressed miRNAs target these genes as predicted.
In addition to miRNA, sRNA includes other classes, such as small interfering RNAs, a second class of small regulatory RNAs that can direct DNA methylation and regulate salt tolerance and disease resistance [53, 54]. Moreover, long noncoding RNAs that emerged as important regulators of many biological processes in animals were identified in higher plants [55]. In this study, besides the 23 miRNAs discussed above, other sRNAs may be taken into account during GM crop evaluations in the future. The function of these different types of sRNA will gradually be identified in future studies. Moreover, the targets of some miRNAs were not predicted. For example, tae-N1 and tae-N2 from this study may function by interacting with other mRNA or lincRNA. The complications of sRNA justify taking sRNA into account in the evaluation system for GM crops. Our findings provide useful data for assessing the potential risks associated with GM crops and provide valuable information for wheat miRNA research.
Supporting information
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(TXT)
(PDF)
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
This study was supported by the National Transgenic Key Project from the Ministry of Agriculture of China (2016ZX08011-003), National Natural Science Foundation of China (No. 31671964) and the Agricultural Science and Technology Program for Innovation Team on Identification and Excavation of Elite Crop Germplasm, CAAS.
Data Availability
The raw data from the small RNA libraries were deposited at the NCBI Sequence Read Archive (SRA) under accession No. SRP091415.
Funding Statement
This study was supported by the National Transgenic Key Project from the Ministry of Agriculture of China (2016ZX08011-003), General Project, National Natural Science Foundation of China (No. 31671964) and the Agricultural Science and Technology Program for Innovation Team on Identification and Excavation of Elite Crop Germplasm, CAAS.
References
- 1.James C. Global status of commercialized Biotech/GM crops: 2015. ISAAA briefs. 2015;(51). [Google Scholar]
- 2.Domingo JL. Safety assessment of GM plants: An updated review of the scientific literature. Food and chemical toxicology: an international journal published for the British Industrial Biological Research Association. 2016;95:12–8. [DOI] [PubMed] [Google Scholar]
- 3.Ewen SW, Pusztai A. Effect of diets containing genetically modified potatoes expressing Galanthus nivalis lectin on rat small intestine. Lancet. 1999;354(9187):1353–4. doi: 10.1016/S0140-6736(98)05860-7 [DOI] [PubMed] [Google Scholar]
- 4.Losey JE, Rayor LS, Carter ME. Transgenic pollen harms monarch larvae. Nature. 1999;399(6733):214. [DOI] [PubMed] [Google Scholar]
- 5.Quist D, Chapela IH. Transgenic DNA introgressed into traditional maize landraces in Oaxaca, Mexico. Nature. 2001;414(6863):541–3. doi: 10.1038/35107068 [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Lan Q, Zhao X, Xu W, Li F, Wang Q, et al. Comparative profiling of microRNA expression in soybean seeds from genetically modified plants and their near-isogenic parental lines. PloS one. 2016;11(5):e0155896 doi: 10.1371/journal.pone.0155896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu HH, Tian X, Li YJ, Wu CA, Zheng CC. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA. 2008;14(5):836–43. doi: 10.1261/rna.895308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Unver T, Namuth-Covert DM, Budak H. Review of current methodological approaches for characterizing microRNAs in plants. International journal of plant genomics. 2009;2009:262463 doi: 10.1155/2009/262463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eldem V, Okay S, ÜNVER T. Plant microRNAs: new players in functional genomics. Turkish Journal of Agriculture and Forestry. 2013;37(1):1–21. [Google Scholar]
- 10.Zhang B. MicroRNA: a new target for improving plant tolerance to abiotic stress. Journal of experimental botany. 2015;66(7):1749–61. doi: 10.1093/jxb/erv013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L, Hou D, Chen X, Li D, Zhu L, Zhang Y, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell research. 2012;22(1):107–26. doi: 10.1038/cr.2011.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pastrello C, Tsay M, McQuaid R, Abovsky M, Pasini E, Shirdel E, et al. Circulating plant miRNAs can regulate human gene expression in vitro. Scientific Reports. 2016;6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218 [DOI] [PubMed] [Google Scholar]
- 14.Xie Z, Khanna K, Ruan S, editors. Expression of microRNAs and its regulation in plants. Seminars in cell & developmental biology; 2010: Elsevier. [DOI] [PMC free article] [PubMed]
- 15.Hackenberg M, Shi B-J, Gustafson P, Langridge P. A transgenic transcription factor (TaDREB3) in barley affects the expression of microRNAs and other small non-coding RNAs. PloS one. 2012;7(8):e42030 doi: 10.1371/journal.pone.0042030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alvarez S, Roy Choudhury S, Pandey S. Comparative quantitative proteomics analysis of the ABA response of roots of drought-sensitive and drought-tolerant wheat varieties identifies proteomic signatures of drought adaptability. Journal of proteome research. 2014;13(3):1688–701. doi: 10.1021/pr401165b [DOI] [PubMed] [Google Scholar]
- 17.Shiqing G, Huijun X, Xianguo C, Ming C, Zhaoshi X, Liancheng L, et al. Improvement of wheat drought and salt tolerance by expression of a stress-inducible transcription factor Gm DREB of soybean (Glycine max). Chinese Science Bulletin. 2005;50(23):2714–23. [Google Scholar]
- 18.Jiang Q, Hu Z, Zhang H, Ma Y. Overexpression of GmDREB1 improves salt tolerance in transgenic wheat and leaf protein response to high salinity. The Crop Journal. 2014;2(2):120–31. [Google Scholar]
- 19.Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24(5):713–4. doi: 10.1093/bioinformatics/btn025 [DOI] [PubMed] [Google Scholar]
- 20.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic acids research. 2014;42(D1):D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15(12):550 doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burge SW, Daub J, Eberhardt R, Tate J, Barquist L, Nawrocki EP, et al. Rfam 11.0: 10 years of RNA families. Nucleic acids research. 2012:gks1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Molecular cell. 2004;14(6):787–99. doi: 10.1016/j.molcel.2004.05.027 [DOI] [PubMed] [Google Scholar]
- 24.Chen R, Jiang N, Jiang Q, Sun X, Wang Y, Zhang H, et al. Exploring microRNA-like small RNAs in the filamentous fungus Fusarium oxysporum. PloS one. 2014;9(8):e104956 doi: 10.1371/journal.pone.0104956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai X, Zhao PX. psRNATarget: a plant small RNA target analysis server. Nucleic acids research. 2011;39(suppl 2):W155–W9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Addo-Quaye C, Miller W, Axtell MJ. CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics. 2009;25(1):130–1. doi: 10.1093/bioinformatics/btn604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nature protocols. 2008;3(6):1101–8. [DOI] [PubMed] [Google Scholar]
- 28.Feng H, Huang X, Zhang Q, Wei G, Wang X, Kang Z. Selection of suitable inner reference genes for relative quantification expression of microRNA in wheat. Plant Physiology and Biochemistry. 2012;51:116–22. doi: 10.1016/j.plaphy.2011.10.010 [DOI] [PubMed] [Google Scholar]
- 29.Kinoshita N, Wang H, Kasahara H, Liu J, MacPherson C, Machida Y, et al. IAA-Ala Resistant3, an evolutionarily conserved target of miR167, mediates Arabidopsis root architecture changes during high osmotic stress. The Plant Cell. 2012;24(9):3590–602. doi: 10.1105/tpc.112.097006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Li K, Chen L, Zou Y, Liu H, Tian Y, et al. microRNA167-directed regulation of the auxin response factors, GmARF8a and GmARF8b, is required for soybean (Glycine max L.) nodulation and lateral root development. Plant physiology. 2015:pp. 00265.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J- W, Czech B, Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;138(4):738–49. doi: 10.1016/j.cell.2009.06.014 [DOI] [PubMed] [Google Scholar]
- 32.Cao D, Li Y, Wang J, Nan H, Wang Y, Lu S, et al. GmmiR156b overexpression delays flowering time in soybean. Plant molecular biology. 2015;89(4–5):353–63. doi: 10.1007/s11103-015-0371-5 [DOI] [PubMed] [Google Scholar]
- 33.Nag A, King S, Jack T. miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proceedings of the National Academy of Sciences. 2009;106(52):22534–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koyama T, Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. The Plant Cell. 2010;22(11):3574–88. doi: 10.1105/tpc.110.075598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frazier TP, Sun G, Burklew CE, Zhang B. Salt and drought stresses induce the aberrant expression of microRNA genes in tobacco. Molecular biotechnology. 2011;49(2):159–65. doi: 10.1007/s12033-011-9387-5 [DOI] [PubMed] [Google Scholar]
- 36.Cui LG, Shan JX, Shi M, Gao JP, Lin HX. The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. The Plant Journal. 2014;80(6):1108–17. doi: 10.1111/tpj.12712 [DOI] [PubMed] [Google Scholar]
- 37.Phookaew P, Netrphan S, Sojikul P, Narangajavana J. Involvement of miR164-and miR167-mediated target gene expressions in responses to water deficit in cassava. Biologia plantarum. 2014;58(3):469–78. [Google Scholar]
- 38.Zhou M, Luo H. Role of microRNA319 in creeping bentgrass salinity and drought stress response. Plant signaling & behavior. 2014;9(4):1375–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hajyzadeh M, Turktas M, Khawar K M, Unver T. miR408 overexpression causes increased drought tolerance in chickpea. Gene, 2015; 555(2):186–93. doi: 10.1016/j.gene.2014.11.002 [DOI] [PubMed] [Google Scholar]
- 40.Wu L, Zhang Q, Zhou H, Ni F, Wu X, Qi Y. Rice microRNA effector complexes and targets. The Plant Cell. 2009;21(11):3421–35. doi: 10.1105/tpc.109.070938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li C, Lu S. Genome-wide characterization and comparative analysis of R2R3-MYB transcription factors shows the complexity of MYB-associated regulatory networks in Salvia miltiorrhiza. BMC genomics. 2014;15(1):277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009;138(4):750–9. doi: 10.1016/j.cell.2009.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Akdogan G, Tufekci E D, Uranbey S, Unver T. miRNA-based drought regulation in wheat. Functional & Integrative Genomics, 2016; 16(3):221–233. [DOI] [PubMed] [Google Scholar]
- 44.Knip M, Constantin ME, Thordal-Christensen H. Trans-kingdom cross-talk: small RNAs on the move. PLoS Genet. 2014;10(9):e1004602 doi: 10.1371/journal.pgen.1004602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nowara D, Gay A, Lacomme C, Shaw J, Ridout C, Douchkov D, et al. HIGS: host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. The Plant Cell. 2010;22(9):3130–41. doi: 10.1105/tpc.110.077040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nunes CC, Dean RA. Host-induced gene silencing: a tool for understanding fungal host interaction and for developing novel disease control strategies. Molecular plant pathology. 2012;13(5):519–29. doi: 10.1111/j.1364-3703.2011.00766.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koch A, Kumar N, Weber L, Keller H, Imani J, Kogel K-H. Host-induced gene silencing of cytochrome P450 lanosterol C14α-demethylase–encoding genes confers strong resistance to Fusarium species. Proceedings of the National Academy of Sciences. 2013;110(48):19324–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiberg A, Wang M, Lin F-M, Zhao H, Zhang Z, Kaloshian I, et al. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science. 2013;342(6154):118–23. doi: 10.1126/science.1239705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang M, Sang X, Hong Z. Beyond nutrients: Food‐derived microRNAs provide cross‐kingdom regulation. Bioessays. 2012;34(4):280–4. doi: 10.1002/bies.201100181 [DOI] [PubMed] [Google Scholar]
- 50.Wang K, Li H, Yuan Y, Etheridge A, Zhou Y, Huang D, et al. The complex exogenous RNA spectra in human plasma: an interface with human gut biota? PloS one. 2012;7(12):e51009 doi: 10.1371/journal.pone.0051009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Wiggins BE, Lawrence C, Petrick J, Ivashuta S, Heck G. Analysis of plant-derived miRNAs in animal small RNA datasets. BMC genomics. 2012;13(1):381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Witwer KW, McAlexander MA, Queen SE, Adams RJ. Real-time quantitative PCR and droplet digital PCR for plant miRNAs in mammalian blood provide little evidence for general uptake of dietary miRNAs: limited evidence for general uptake of dietary plant xenomiRs. RNA biology. 2013;10(7):1080–6. doi: 10.4161/rna.25246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu J-K. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123(7):1279–91. doi: 10.1016/j.cell.2005.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas A, Zhu J-K, et al. A pathogen-inducible endogenous siRNA in plant immunity. Proceedings of the National Academy of Sciences. 2006;103(47):18002–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu J, Wang H, Chua NH. Long noncoding RNA transcriptome of plants. Plant biotechnology journal. 2015;13(3):319–28. doi: 10.1111/pbi.12336 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(TXT)
(PDF)
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
The raw data from the small RNA libraries were deposited at the NCBI Sequence Read Archive (SRA) under accession No. SRP091415.