Abstract
Nrf2/skn-1, a transcription factor known to mediate adaptive responses of cells to stress, also regulates energy metabolism in response to changes in nutrient availability. The ability to locate food sources depends upon chemosensation. Here we show that Nrf2/skn-1 is expressed in olfactory interneurons, and is required for proper integration of multiple food-related sensory cues in Caenorhabditis elegans. Compared to wild type worms, skn-1 mutants fail to perceive that food density is limiting, and display altered chemo- and thermotactic responses. These behavioral deficits are associated with aberrant AIY interneuron morphology and migration in skn-1 mutants. Both skn-1-dependent AIY autonomous and non-autonomous mechanisms regulate the neural circuitry underlying multisensory integration of environmental cues related to energy acquisition.
Introduction
The nuclear factor erythroid 2-related factor 2 (Nrf2) is a member of the Cap'n'Collar basic leucine zipper family known to play a key role in the cellular antioxidant and detoxification responses to stress [1]. In mammals under homeostatic conditions Nrf2 is maintained inactive via interaction with kelch-like ECH associated protein 1 (Keap1) and Cullin3. Cullin3 ubiquitinates and targets Nrf2 for continuous proteasomal degradation. In response to oxidative or electrophilic agents the interaction of Nrf2 with Keap1 is disrupted preventing Cullin 3-dependent degradation. The stabilized Nrf2 protein translocates into the nucleus where it dimerizes with small musculoaponeurotic fibrosarcoma (Maf) proteins to drive the transcription of cytoprotective genes [1]. Nrf2 and its invertebrate orthologs skn-1 (Caenorhabditis elegans) and CncC (Drosophila melanogaster) have also been shown to promote health span and longevity. In C. elegans and D. melanogaster the genetic activation of the Nrf2 signaling pathway leads to enhanced longevity [2,3]. The comparison of Nrf2 activity in rodents with different maximum lifespan potential revealed a positive correlation between Nrf2 activation status and longevity [4]. Interestingly, despite showing high constitutive levels of active Nrf2 [4], the naturally long lived rodent naked mole rat has very low antioxidant enzyme expression [5]. This suggests that Nrf2 may exert pro-longevity benefits beyond its well characterize role as an antioxidant modulator. One of the best studied examples of biological conditions able to promote longevity in a wide range of experimental models is decreased nutrient availability [6, 7]. Caloric restriction and signaling pathways involved in metabolic homeostasis are recognized as universal effectors of longevity phenotypes [6–8]. Optimal metabolic homeostasis depends upon proper integration of intrinsic and extrinsic environmental signals by the nervous system. The impact of sensory integration (i.e. gustatory, olfactory, thermosensory) in health span and aging phenotypes has been extensively studied in invertebrates models [9]. For example, experimental impairment of olfaction extends lifespan in flies [10]. In C. elegans, mutations or manipulations impairing sensory neuronal function extend lifespan [11,12], and the longevity phenotype induced by caloric restriction depends upon the activity of Nrf2/skn-1 in the ASI chemosensory neurons [13].
Activation of Nrf2 protects neurons in experimental models of stroke [14–16], Parkinson’s disease [17,18], and Huntington’s disease [19]. However, the contribution of the Nrf2 pathway to neuronal functions under physiological conditions is still largely unknown. In the present study we provide evidence that Nrf2 ortholog skn-1 plays a fundamental role in foraging and food-related sensory integration in C. elegans.
Material and methods
Mice
C57BL/6 mice were breed and housed at the National Institute on Aging facility. Male (3 months old) were euthanized by carbon dioxide inhalation, and brains were removed and either dissected for immunoblot analysis, or processed for immunohistochemistry. All procedures were approved by the Institutional Care and Use Committee (IACUC) of the National institute on Aging (ASP 290-LNS-2019), and complied with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Western blot
Tissue extracts and western blot analysis were performed as described previously [14]. The primary antibodies used in this study were: Nrf2 (Cat: EP1808Y, Epitomics, Burlingame, CA), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Cat: 2-RGM2, Advanced Immunochemical Inc, Long Beach, CA), glial-fibrillary acidic protein (GFAP) (GA5) (Cat: MAB360, EMD Millipore).
Immunohistochemistry
Brains were fixed in 4% paraformaldehyde in PBS for 24 hours, then transferred to 30% sucrose in PBS at 4°C. Following antigen retrieval, sagittal sections were incubated for 1 h in blocking solution and then incubated overnight at 4°C with Nrf2 and glial fibrillary acidic protein (GFAP) antibodies. After extensive washing, the primary antibodies were detected using Alexa Flour 488- and 633-conjugated secondary antibody. Nonspecific labeling was determined by omission of the primary antibody. The cell nuclei were counterstained with DAPI (4',6'-diamidino-2-phenylindole) dye. Confocal images were acquired using a Zeiss 510 LSM microscope with 10X and 40X objective lenses.
Caenorhabditis elegans strains
Worms were maintained on NGM agar plates with E. coli OP50 as food at room temperature (23°C) according to standard protocols [20]. Strains N2 Bristol wild-type strain, EU31 skn-1(zu135)/DnT1) putative skn-1 null, EU1 skn-1(zu67)/DnT1) skn-1a/c null, FK134 ttx-3(ks5) putative ttx-3 null, OH1098 otIS133[ttx-3::RFP + unc-4(+)], OH2246 otIS107[ser-2::GFP] and OP342 unc-119(ed3);wgIS342[skn-1::TY1::EGFP::3xFLAG + unc-119(+)] were obtained from the Caenorhabditis Genetics Center at the University of Minnesota. OH1098 spontaneous males were crossed to OP342, and RFP/GFP double positive progeny bred to homozygosity to produce wgIS342;otIS133 double transgenic worms. N2 males were crossed to OH2246, and F1 GFP+ males were then crossed to EU1. GFP+ F1 unc/het progeny were then crossed to zu135/+ males, and resultant unc/het GFP+ progeny singled and selected for presence of the zu135 allele, then bred to homozygosity for otIS107 to introduce ser2promoter1::GFP into the skn-1(zu135) background.
Transgenics
Transgenic strains were generated by microinjection. For all lines, 50 ng/μl of each expression construct was injected into N2 worms. Transgenes were crossed into the skn-1 background as described above. skn-1b coding sequence was cloned from N2 young adult total RNA by rtPCR. 5kb ttx-3 and 1kb ric-19 promoter fragments were cloned from N2 DNA by PCR. Promoter and skn-1b insert fragments were cloned into pPD95.75 GFP expression vector [21]. The mCherry::unc-54utr fragment of pCFJ104 [22] was used to replace gfp::unc54utr of pPD95.75, then the 5kb ttx-3 promoter fragment was inserted to create pttx-3::mCherry AIY specific marker CY691(zu135/dnT1;bvEx177(pttx-3::mcherry;pric19::skn-1b::gfp)). CY696(zu135/dnT1;bvEx181 (pttx-3::mcherry;pttx-3::skn-1b::gfp)). Primers are listed in S1 Table. PCR fragments were sequence verified.
Behavioral assays
Chemotaxis assays were performed on 10 cm plastic petri dishes containing 10 ml assay medium (1.6% agar, 1mM CaCl2, 1mM MgSO4, 4mM NH4Cl, 25mM KH2PO4 pH 6.0), and analyzed as previously described [23]. Trials were performed in triplicate, with 25 animals per plate, unless otherwise indicated. Briefly, worms were transferred from culture plates to an empty assay plate and allowed to crawl freely for 10 minutes to remove residual bacteria. Worms were transferred to the center 1 cm of the assay plate (origin) and incubated at room temperature for 1 hour, at which time their positions were scored immediately. For sodium chloride assays, 5 μl of 5M NaCl was spotted onto one edge of the assay plate and left for 16 hours at room temperature to form a gradient prior to use [24]. Chemotaxis index was calculated as (O-S)/(T) where O was the number of animals at the odorant location and S the number of animals at the solvent control location after 1 hour and T the total number of worms. Thermotaxis assays were performed on radial thermal gradients on chemotaxis assay plates [25]. For dwelling assays, a 10 μl spot of diluted OP50 was spotted onto the center of the assay plate and allowed to dry for 1 hour. OP50 bacteria were grown overnight at 37°C with shaking, then washed twice with M9 buffer and diluted to an absorbance at A600 nm of 1.5 (1x concentration).
C. elegans microscopy
Z-stack photomicrographs were acquired and analyzed on a Zeiss LSM510 inverted confocal microscope with LSM5 software. All images were taken using a 63x oil immersion objective with differential interference contrast. For quantification, identical areas of the worms were selected, and the total fluorescence was calculated as the mean fluorescence per μm2.
Gene expression
Semi-quantitative rtPCR was performed on a Chromo4 system with Opticon 3 software (BioRad). Fold change was calculated using the ΔΔCt model [26], normalized to actin (act-1). Primers are listed in S1 Table.
Results and discussion
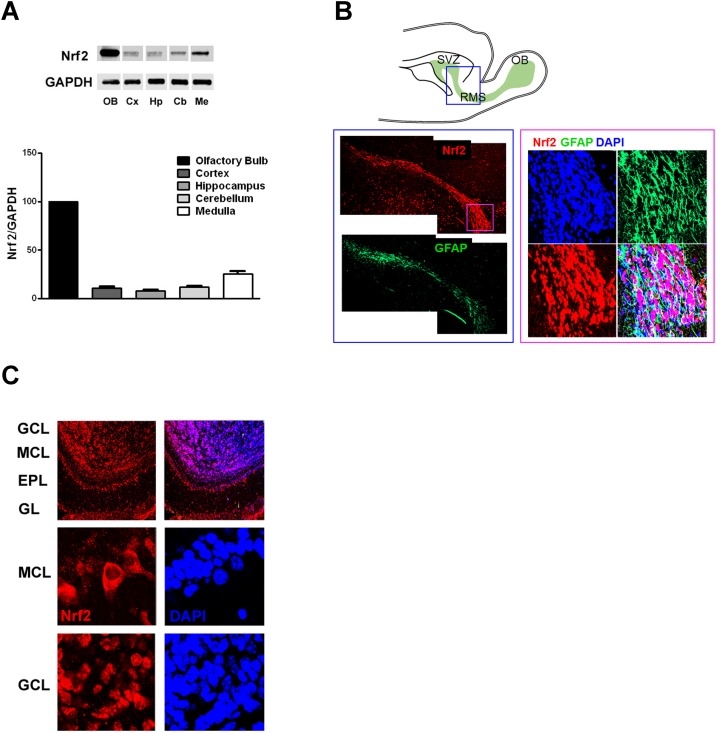
In order to gain insights into the potential roles for Nrf2 in brain physiology, we first analyzed the levels of Nrf2 in five different regions of the mouse brain (olfactory bulb, cerebral cortex, hippocampus, cerebellum, and medulla). Immunoblot analysis revealed that Nrf2 protein levels in the olfactory bulb were four- to ten-fold greater than in any other brain regions evaluated (Fig 1A). Histological analysis of sagittal brain sections revealed high levels of Nrf2 expression in the rostral migratory stream (RSM), as well as in the mitral cell layer (MCL), and granule cell layer (GCL) of the olfactory bulb (Fig 1B and 1C). The RMS is a specialized pathway through which neuronal precursor cells originating in the subventricular zone migrate into the olfactory bulb (Fig 1B). Our findings of high Nrf2 expression in RMS cells substantiates recent evidence showing that in mice Nrf2 ablation and age-dependent alterations of the Nrf2 pathway, impair subventricular zone neuronal progenitor cell proliferation and survival [27–29]. Once the newly generated cells reach the olfactory bulb they differentiate into γ-aminobutyric acid (GABA) interneurons that integrate into the granule cell layer (GCL), or replace periglomerular cells in the glomerular layer (GL). In the olfactory bulb in addition to specific regional distribution we also observed distinct subcellular patterns of expression. In mitral cells Nrf2 is mainly located in the cytoplasm (Fig 1C), but in granule cell interneurons it is mostly concentrated in the nucleus, suggesting a constitutively active status in these cells (Fig 1C).
Fig 1. Nrf2 is highly expressed in olfactory bulb interneurons.
(A) Representative immunoblots and quantification of Nrf2 protein levels in adult murine brain (n = 4). Values were normalized by GAPDH and expressed as mean percentage (and S.E.M.) compared to olfactory bulb (OB). Cx, cortex; Hp, hippocampus; Cb, cerebellum; Me, medulla. (B) Schematic showing the rostral migratory stream (RMS), the route followed by neuroblasts originating in the sub ventricular zone (SVZ) to reach the olfactory bulb (OB). Immunohistochemistry showing the expression of Nrf2 in the RMS at low magnification (left) and high magnification (right). The boxed area indicates the regions shown in the immunostaining. (C) Immunohistochemistry showing the distribution of Nrf2 in the various regions of the olfactory bulb (upper panel). GCL: granule cell layer; MCL: mitral cell layer; EPL: external plexiform layer; GL: glomerular layer. The higher magnification panels show Nrf2 subcellular localization in mitral cells (MCL) and granule cells (GCL).
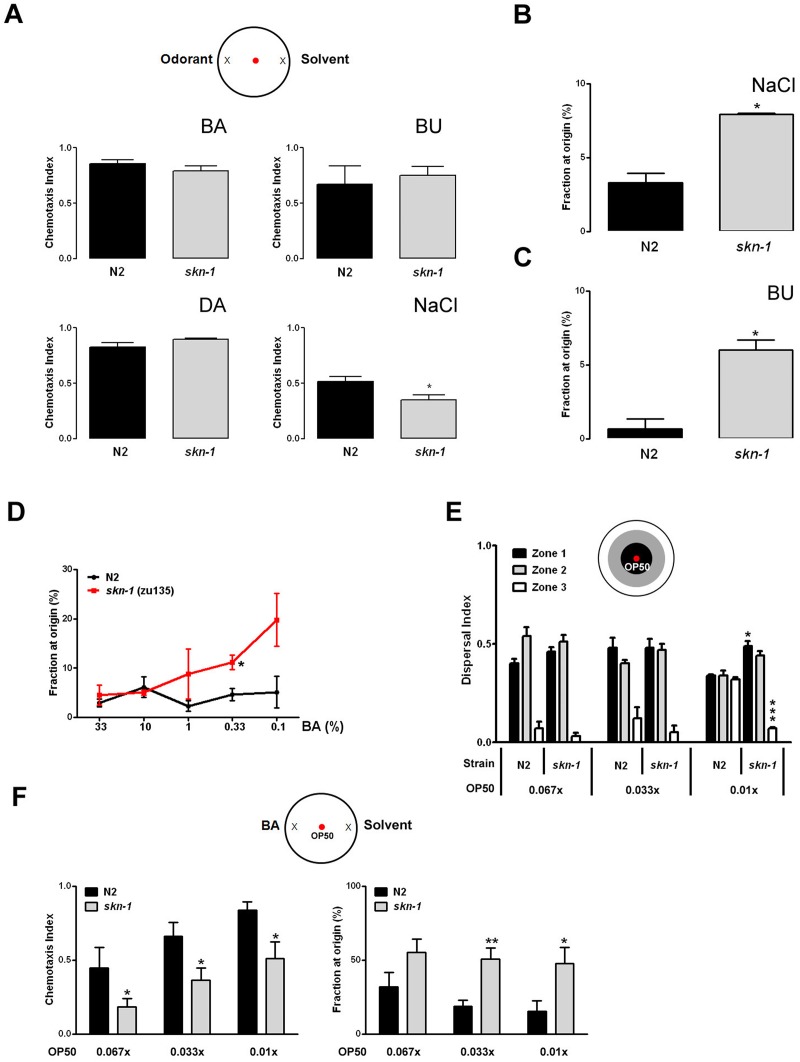
Because of its prominence in olfactory neurons we asked whether Nrf2 plays roles in chemosensation-related behaviors that enable animals to locate food sources and mates, as well as to avoid potentially harmful substances and predators. As the predominant sensory input, chemosensation is particularly important in the nematode C. elegans, where it regulates several metabolic and behavioral responses including pharyngeal pumping, locomotion, egg laying, life span and dauer formation [30]. In C. elegans the ortholog of Nrf2, skn-1, encodes three major isoforms (a, b and c) which each play distinct functional roles. skn-1a has been recently shown to localize to the mitochondrial membrane and respond to starvation [31]. skn-1b is expressed in neurons and mediates dietary restriction-induced longevity [13]. skn-1c on the other hand is intestinal and has been linked to oxidative stress resistance and longevity [2, 32]. To assess the role of skn-1/Nrf2 in chemosensory perception we took advantage of the fact that the C. elegans nervous system and behavioral responses to odorants are well-characterized. We performed standard chemotaxis experiments comparing the ability of wild type N2 and skn-1(zu135) null worms to respond to known chemoattractants. There were no significant differences in the chemotaxis index for volatile compounds such as benzaldehyde (BA) (Fig 2A and S2 Table), butanone (BU) and diacetyl (DA) which are recognized by AWC and AWA sensory neurons. However, we did find a small significant decrease in ASE-dependent chemotaxis index toward sodium chloride in skn-1 worms (Fig 2A). During the course of our chemotaxis experiments we noted a tendency of skn-1 worms to remain in close proximity to the origin of the assay plate. When we analyzed this behavior in more detail, we found that a significant fraction of skn-1 worms preferred to remain in the center area of the plate regardless of the type of the compound, soluble or volatile, used as chemoattractant (Fig 2B and 2C). This behavior was more pronounced when suboptimal concentrations of chemoactractant were used (Fig 2D and S3 Table), did not depend upon the skn-1 background as the skn-1(zu67) strain showed a similar phenotype (S1A Fig and S3 Table), and was not related to impaired motility as demonstrated by identical dispersal indexes (S1B Fig).
Fig 2. skn-1 regulates sensory integration in C. elegans.
(A) skn-1(zu135) worms showed wild type chemotaxis to optimal doses of volatile compounds and reduced migration to sodium chloride. Chemotaxis indexes were determined at the following odorant concentrations: 1% benzaldehyde (BA); 0.1% butanone (BU); 1% diacetyl (DA). Sodium chloride chemotaxis was tested on a 5 mM gradient. The percentages of animals remaining at the origin in presence of NaCl (B), butanone (C), or the indicated concentrations of benzaldehyde (D), are shown. (E) skn-1 food-leaving behavior is impaired in the presence of very low local concentrations of food (OP50 E. coli). (F) skn-1 chemotaxis toward BA is significantly decreased and dwelling behavior enhanced in the presence of known concentrations of bacterial lawn. Data are mean and S.E.M. of 3–6 trials performed in triplicate. *p<0.05; **p<0.01; ***p<0.001 versus N2 (Student’s t-test).
In addition to salt chemoattraction, ASE neurons have recently been shown to promote adaptive food-leaving behavior as food becomes limiting [33]. Chemotaxis enables nematodes to locate new food sources, and many chemoattractants are products of bacterial decay. One explanation for the behavioral phenotype we observed is that skn-1 worms can detect very small amounts of carried over bacteria and prefer to dwell around the origin area. To test this possibility we performed ‘leaving behavior’ assays in which known dilutions of OP50 were placed at the origin. As shown in Fig 2E at concentrations that N2 worms considered indicative of no or negligible levels of food, and led them to roam throughout the plate, skn-1 worms preferred to remain in the area close to the bacterial lawn. Because of this enhanced sensitivity to very small quantities of bacteria, it is possible that for skn-1 worms the chemotaxis experiments performed under standard conditions were actually multi-sensory integration experiments. We thus decided to compare the behavior of N2 and skn-1 worms in conditions where an attractant and a specific concentration of OP50 E. coli placed at the origin were provided as sensory cues. We found that in such experimental settings skn-1 worms preferred to dwell on the OP50 lawn rather than move toward the chemoattractant (Fig 2F and S4 and S5 Tables). These results suggest that skn-1 worm behavior under conditions requiring integration of multiple food-related sensory cues is impaired compared to wild type animals.
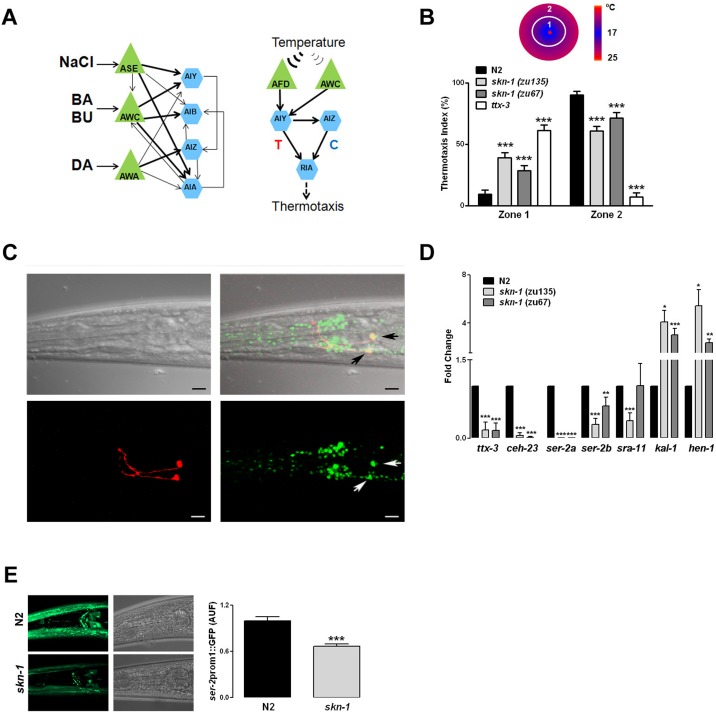
Interneurons play a central role in the integration of different sensory modalities. Based on the distinct distribution of Nrf2 in the mammalian olfactory interneurons, the perturbed chemotaxis behavior in skn-1 mutant C. elegans and the known C. elegans neuronal wiring (Fig 3A), we focused on AIY and AIA as interneurons potentially responsible for the observed skn-1 behavioral phenotype. Previous findings suggest that AIY interneurons are critical for chemotaxis responses to near-threshold levels of odors [34], food choice behavior [35] and dwelling [34–36]. We took advantage of the fact that AIY interneurons are essential for C. elegans thermotaxis behavior through AFD sensory neurons (Fig 3A) [37]. We hypothesized that if AIY functions are altered in skn-1 worms, they will likely display abnormal thermotaxis behavior. The ability of N2 and skn-1 worms to migrate toward their cultivation temperature (23°C) was assessed in population thermotaxis assays using ttx-3 mutant worms in which AIY neurons are dysfunctional [38–40]. While N2 animals moved toward the region of the plate closer to their previous cultivation temperature, both skn-1 loss-of-function strains zu135 and zu67 showed a cryophilic phenotype similar to, although not as pronounced as ttx-3 null worms (Fig 3B and S6 Table). Overall our behavioral results indicate that skn-1 is required for proper integratory functions of sensory interneurons. Notably, altered olfactory integration with impaired fine discrimination has also recently been shown in Nrf2 knock out mice [29].
Fig 3. skn-1 is expressed in AIY neurons and regulates their functions.
(A) Schematic of the neuronal circuits of C. elegans chemoattractive and thermotactic behaviors. Green triangles represent sensory neurons and blue hexagons command interneurons. Arrows indicate direct interactions, and their thickness is proportional to the frequency of synaptic contacts between the neurons (Adapted from [36]). T, thermophilic; C, cryophilic.(Adapted from [41]). (B) Population thermotaxis experiments were performed using a radial thermal gradient. After 60 minutes the position of the worms was scored and the percentages of worms in the colder (Zone 1) and warmer (Zone 2) regions determined. N2 animals moved toward the region of the plate closer to their previous cultivation temperature (23°C), while both skn-1 null strains zu135 and zu67 showed a cryophilic phenotype similar to ttx-3 (AIY) null worms. (C) Representative 3D reconstruction of confocal sections through the head region of double transgenic wglS342;otlS133 worms is shown. The skn-1:EGFP reporter (green), containing the entire skn-1 gene, is highly expressed in neurons in the head ganglia, and colocalizes with the AIY-specific marker ttx3:RFP (Red). Arrows point at AIY. Scale bar 10 μm. (D) Relative mRNA levels of the AIY cell fate specification homeobox genes ttx-3 and ceh-23, and their targets ser-2a, ser-2b, sra-11, kal-1 and hen-1 in N2 and skn-1 null worms. (E) Representative 3D reconstructions showing the expression of the transcriptional reporter ser2:prom1:EGFP in the head region of N2 and skn-1(zu135) null worms. Data are mean and S.E.M. of 3–5 experiments. *p<0.05; **p<0.01; ***p<0.001 versus N2 (Student’s t-test).
With regard to the C. elegans nervous system, skn-1 has been shown to be expressed in ASI [13] and dopaminergic neurons [42]. Because our data suggest that skn-1 is required for normal AIY function, we analyzed wgIS342;otIS133 double transgenic animals using confocal microscopy. Because of the large genomic DNA sequence used to create the construct, wglS342 drives the expression of all skn-1 isoforms under endogenous coding and regulatory sequences, thus providing a more comprehensive and faithful expression pattern. As shown in Fig 3C, skn-1::EGFP is highly expressed in neurons in the lateral, ventral and dorsal ganglia, and to a lesser extent in the anterior ganglia. Furthermore, skn-1::EGFP clearly colocalizes with the specific AIY marker ttx-3::RFP, confirming that skn-1 is indeed expressed in AIY interneurons.
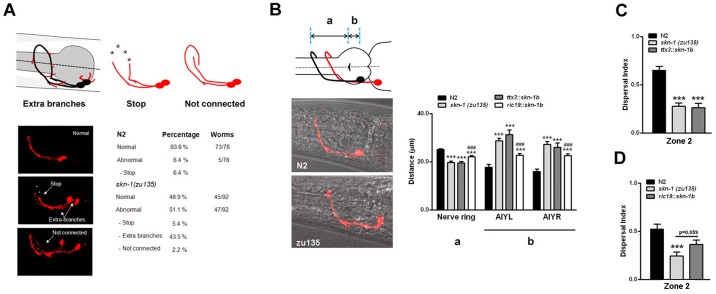
The homeodomain transcription factors ceh-10, ttx-3 and ceh-23 coordinate the AIY cell fate specification [43]. We used qRT-PCR to quantify the expression of these transcription factors and some of their known targets in N2 and skn-1 worms. Levels of ceh-10 were comparable amongst the various strains (data not shown), however both ttx-3 and ceh-23 were significantly reduced in skn-1(zu135) and (zu67) worms (Fig 3D). ser-2a and ser-2b, which encode a tyramine receptor, were also significantly reduced in skn-1 worms (Fig 3D). skn-1(zu135), but not skn-1(zu67), showed decreased levels of sra-11 (Fig 3D), suggesting a specific effect of the skn-1b isoform in the regulation of this G protein-coupled receptor implicated in olfactory imprinting [44]. We verified the changes in ser-2 levels by analyzing the expression pattern of GFP driven by ser2::prom1 in N2 and skn-1(zu135) transgenic animals. As shown in Fig 3E a significant decrease for ser-2:: GFP expression was observed in neurons as well as pharyngeal cells and head muscles in skn-1 worms. Surprisingly, levels of hen-1 and kal-1 were increased in both skn-1 strains (Fig 3D). In humans Kallmann syndrome is characterized by hypogonadism, and hyposmia or anosmia. In C. elegans misregulation of the Kallmann syndrome gene ortholog kal-1 has been shown to alter neurite branching and the formation of axon collaterals, as well as axon pathfinding, and establishment of target connections [45,46]. The analysis of AIY morphology in skn-1 worms showed a significant increase of aberrant branching from the axon, as well as of short neurite-like projections from the cell body (Fig 4A). We also observed a less penetrant phenotype of axonal misrouting characterized by the axon of one of the AIY pair running over and around the dorsal midline, and failing to interact with the contralateral axon (Fig 4A). Such defects depended upon levels of skn-1 expression, as heterozygous skn-1 animals displayed an intermediate percentage of abnormalities (normal = 73.7%; abnormal = 26.3%). Furthermore, regardless of the presence of morphological defects we observed that, relative to the pharynx grinder, the position of the AIY cell bodies in skn-1 worms was far more caudal compared to N2 worms (Fig 4B), while the distance of the AIY axon fasciculation at the nerve ring was shorter (Fig 4B). Similar neuroanatomical defects have been described in ttx-3 mutant animals [42,46]. We assessed the possibility that our behavioral and morphological findings result from skn-1 dependent misregulation of AIY by generating transgenic animals in which skn-1b cDNA is specifically and exclusively driven in AIY by a ttx-3 promoter (CY696). Compared to skn-1 mutants, CY696 animals displayed about 54% less AIY morphological defects (normal = 76.3%; abnormal = 23.7%), but identical AIY migratory (Fig 4B), and dwelling behavioral defects (Fig 4C and S7 Table). These findings suggest that skn-1b acts at least partially in a cell autonomous fashion to control AIY morphogenesis, whereas the AIY migratory and behavioral impairments are non-AIY autonomous defects.
Fig 4. skn-1 regulates AIY neuron morphology and food-seeking behavior by cell-autonomous and non-autonomous mechanisms.
(A) Schematic depiction, representative images, and quantification of the neuroanatomical defects observed in skn-1(zu135) worms. Wild type morphology is shown in black, while aberrant branches from the axon or cell body are depicted in red. Also shown are examples of premature stops, and axon misrouting. (B) Schematic depiction and representative images of the AIY position relative to the pharynx grinder in N2 (black line) and skn-1(zu135) worms (red line). a, distance between grinder and the AIY axon fasciculation at the nerve ring; b, distance between the grinder and AIY cell body. Quantification of the AIY migratory defects in the indicated worm strains. AIY-specific expression of skn-1b has no effect on migration (B), and food-leaving behavior (C) (OP50 = 0.033x). Pan-neuronal expression of skn-1b significantly decreases AIY migratory defects (B) and partially rescues the food-leaving behavior (D). Data are mean and S.E.M. of 3–7 experiments. ***p<0.001 versus N2; ###p<0.001 versus skn-1 (zu135) (Student’s t-test).
Because ttx-3 is normally expressed in other head neurons (AIA, ADL, ASI, ADF) [34,43], and the AIY specific expression of skn-1b is not sufficient to rescue the behavioral abnormalities, we tested a ric-19 promoter to drive the pan-neuronal expression of skn-1b (CY691). CY691 transgenic animals had about 41% less AIY anatomical defects (normal = 69.9%; abnormal = 30.1%), and an intermediate phenotype between N2 and skn-1 null worms in terms of AIY migratory defects (Fig 4B), and food-leaving behavior (Fig 4D and S8 Table). This result, together with the behavioral similarities we observed between the skn-1 strains lacking all three skn-1 isoforms (zu135) and skn-1a/c isoforms (zu67), suggest that expression of skn-1b in non-neuronal cell types is required for a full functional recovery.
Nrf2/skn-1 is widely known for its role in adaptive responses to many different types of stress, particularly oxidative stress. Accordingly, much attention has been given to Nrf2/skn-1 target genes that encode antioxidant proteins [47]. Studies of cell culture and animal models have shown that Nrf2/skn-1 plays a major role in the process of hormesis, in which exposure of a cell or organism to a low level of stress (e.g., oxidative, metabolic, xenobiotic) results in resistance to greater levels of the same or different stressor [48]. Moreover, recent findings suggest that Nrf2/skn-1 mediates, in part, beneficial effects of bioenergetics challenges such as food restriction on organ system function, disease resistance, and longevity [49–52]. Previous studies of C. elegans have shown that phytochemicals that activate Nrf2/skn-1 can significantly increase lifespan [53], and that skn-1 in ASI neurons is required for the lifespan-extending effect of food deprivation [13]. Our data reveal a previously unknown role for Nrf2/skn-1 in regulating the organization and functionality of neuronal networks involved in behavioral responses of worms to food availability, temperature, and specific odorants. We speculate that this ‘decision-making’ role for skn-1 in AIY neurons evolved to optimize adaptive behavioral responses to a wide range of environmental challenges faced by worms in their normal habitat.
Conclusions
Our findings show that Nrf2/skn-1 is expressed in interneurons involved in chemosensation in mice and C. elegans, and that genetic deletion of skn-1 results in impaired sensory integration. The inability to activate skn-1 signaling causes the skn-1 worms to linger in inadequate food patches rather than migrate toward new potential food sources. Lack of skn-1 results in morphological, migratory and functional alterations of AIY suggesting that skn-1 is important for the proper development of a pair of neurons that play a pivotal role in coordinating behavioral responses to food and temperature, two fundamental environmental factors influencing growth, reproduction and survival.
Supporting information
(A) Percentage of N2 and skn-1(zu67) animals remaining at the origin in presence of the indicated concentrations of benzaldehyde. (B) N2 and skn-1(zu135) worms display identical locomotion abilities. Animals were placed on an empty plate and allowed to crawl freely for 60 min. Their position was scored and the percentage of animals in the center (Zone 1) and periphery (Zone 2) determined. Data are mean and S.E.M. * p<0.05 versus N2.
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by the Intramural Research Program of the National Institute on Aging. JVCC was sponsored by the Brazilian Scientific Mobility Program (Ciência sem Fronteiras #7610-11-4). Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
References
- 1.Son TG, Camandola S, Mattson MP. Hormetic dietary phytochemicals. Neuromolecular Med. 2008;10: 236–246. 10.1007/s12017-008-8037-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, Liu S, et al. Direct inhibition of the longevity -promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008; 132:1025–1038. 10.1016/j.cell.2008.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sykiotis GP, Bohmann D. Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev Cell. 2008; 14:76–85. 10.1016/j.devcel.2007.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis KN, Wason E, Edrey YH, Kristan DM, Nevo E, Buffenstein R. Regulation of Nrf2 signaling and longevity in naturally long-lived rodents. Proc. Natl. Acad. Sci. USA 2015; 112:3722–3727. 10.1073/pnas.1417566112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis KN, Andziak B, Yang T, Buffenstein R. The naked mole-rat response to oxidative stress: just deal with it. Antioxid. Redox Signal. 2013; 19: 1388–1399. 10.1089/ars.2012.4911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lopez-Otin C, Galluzzi L, Freije JM, Madeo F, Kroemer G. Metabolic control of longevity. Cell. 2016; 166:802–821. 10.1016/j.cell.2016.07.031 [DOI] [PubMed] [Google Scholar]
- 7.Bishop NA, Guarente L. Genetic Links between diet and lifespan; shared mechanisms from yeast to humans. Nat. Rev. Genet. 2007; 8: 835–844. 10.1038/nrg2188 [DOI] [PubMed] [Google Scholar]
- 8.Fontana L, Partridge L, Longo VD. Extending healthy life span- from yeast to humans. Science 2010: 328:321–326. 10.1126/science.1172539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeong DE, Artan M, Seo K, Lee SJ. Regulation of lifespan by chemosensory and thermosensory systems: findings in invertebrates and their implivcations in mammalian aging. Front. Genet. 2012; 3: 218 10.3389/fgene.2012.00218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Libert S, Zwiener J, Chu X, Vanvoorhies W, Roman G, Pletcher SD. Regulation of Drosophila life span by olfaction and food-derived odors. Science. 2007; 315:1133–1137. 10.1126/science.1136610 [DOI] [PubMed] [Google Scholar]
- 11.Apfeld J, Kenyon C. Regulation of lifespan by sensory perception in Caenorhabditis elegans. nature. 1999; 402:804–809. 10.1038/45544 [DOI] [PubMed] [Google Scholar]
- 12.Alcedo J, Kenyon C. Regulation of C. elegans longevity by specific gustatory and olfactory neurons. Neuron. 2004; 41:45–55. [DOI] [PubMed] [Google Scholar]
- 13.Bishop N.A. & Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature 447, 545–549 (2007) 10.1038/nature05904 [DOI] [PubMed] [Google Scholar]
- 14.Son TG, Camandola S, Arumugam TV, Cutler RG, Telljohann RS, Mughal MR, et al. Plumbagin a novel Nrf2/ARE activator, protects against cerebral ischemia. J Neurochem. 2010; 112: 1316–925. 10.1111/j.1471-4159.2009.06552.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shih AY, Li P, Murphy TH. A small-molecule-inducible Nrf2-mediated antioxidant response provide prophylaxis against cerebral ischemia in vivo. J Neurosci. 2005; 25:10321–10335. 10.1523/JNEUROSCI.4014-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Satoh T, Okamoto SI, Cui J, Watanabe Y, Furuta K, Suzuki M, et al. Activation of Keap1/Nrf2 pathway for neuroprotection by electrophilic phase II inducers. Proc Natl Acad Sci USA. 2006; 103: 768–773. 10.1073/pnas.0505723102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen PC, Vargas MR, Pani AK, Smeyne RJ, Johnson DA, Kan YW, et al. Nrf2-mediated neuroprotection in the MPTP mouse model of Parkinson's disease: Critical role for the astrocyte. Proc Natl Acad Sci USA. 2009; 106: 2933–2938. 10.1073/pnas.0813361106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cook AL, Vitale AM, Ravishankar S, Matigian N, Sutherland GT, Shan GT, et al. NRF2 activation restores disease related metabolic deficiencies in olfactory neurosphere-derived cells from patients with sporadic Parkinson's disease. PLoS One. 2011;6: e21907 10.1371/journal.pone.0021907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stack C, Ho D, Wille E, Calingasan NY, Williams C, Liby K, et al. Triterpenoids CDD)-ethylamide and CCDO-trifluoroethylamide improve the behavioral phenotype and brain pathology in a transgenic mouse model of Hungtington's disease. Free Radic Biol Med. 2010; 49:147–158. 10.1016/j.freeradbiomed.2010.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974; 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fire A, Harrison SW, Dixon D. A modular set of lacZ fusion vectors for studying gene expression in Caenorhabditis elegans. Gene. 1990; 93: 189–198. [DOI] [PubMed] [Google Scholar]
- 22.Frøkjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, et al. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008; 40: 1375–1383. 10.1038/ng.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bargmann CI, Horvitz HR. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron. 1991; 7: 729–742. [DOI] [PubMed] [Google Scholar]
- 24.Uchida O, Nakano H, Koga M, Ohshima Y. The C. elegans che-1 gene encodes a zinc finger transcription factor required for specification of the ASE chemosensory neurons. Development. 2003;130: 1215–1224. [DOI] [PubMed] [Google Scholar]
- 25.Hedgecock EM, Russell RL. Normal and mutant thermotaxis in the nematode Caenorhabditis elegans. Proc Natl Acad Sci USA. 1975;72: 4061–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2005;25: 402–408. [DOI] [PubMed] [Google Scholar]
- 27.L'Episcopo F, Tirolo C, Testa N, Caniglia S, Morale MC, Impagnatiello F, et al. Aging-induced Nrf2-ARE pathway disruption in the subventricular zone drives neurogenic impairment in parkinsonian mice via PI3K-Wnt/β-catenin dysregulation. J Neurosci. 2013; 33:1462–1485. 10.1523/JNEUROSCI.3206-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kärkkäinen V, Pomeshchik Y, Savchenko E, Dhungana H, Kurronen A, et al. Nrf2 regulates neurogenesis and protects neural progenitor cells against Aβ toxicity. Stem Cells. 2014;32:1904–1916. 10.1002/stem.1666 [DOI] [PubMed] [Google Scholar]
- 29.Corenblum MJ, Ray S, Remley QW, Long M, Harder B, Zhang DD, et al. Reduced Nrf2 expression mediates the decline in neural stem cell function during a critical middle-age period. Aging Cell. 2016;15:725–736. 10.1111/acel.12482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bargmann CI. Chemosensation in C. elegans. WormBook; 2006;1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paek J, Lo JY, Narasimhan SD, Nguyen TN, Glover-Cutter K, Robida-Stubbs S, et al. Mitochondrial SKN-1/Nrf mediates a conserved starvation response. Cell Metab. 2012;16:526–537. 10.1016/j.cmet.2012.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.An JH, Blackwell TK. skn-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 2003;17: 1882–1893. 10.1101/gad.1107803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milward K, Busch KE, Murphy RJ, de Bono M, Olofsson B. Neuronal and molecular substrates for optimal foraging in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2011;108: 20672–20677. 10.1073/pnas.1106134109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsalik EL, Hobert O. Functional mapping of neurons that control locomotory behavior in Caenorhabditis elegans. J Neurobiol. 2003;56:178–197. 10.1002/neu.10245 [DOI] [PubMed] [Google Scholar]
- 35.Shtonda BB, Avery L. Dietary choice behavior in Caenorhabitis elegans. J Exp Biol. 2006; 209: 89–102. 10.1242/jeb.01955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gray JM, Hill JJ, Bargmann CI. A circuit for navigation in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2005;102: 3184–3191. 10.1073/pnas.0409009101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mori I, Ohshima Y. Neural regulation of thermotaxis in Caenorhabditis elegans. Nature 1995; 376: 344–348. 10.1038/376344a0 [DOI] [PubMed] [Google Scholar]
- 38.Tsalik EL, Niacaris T, Wenick AS, Pau K Avery L, Hobert O. LIM homeobox gene-dependent expression of biogenic amine receptors in restricted regions of the C.elegans nervous system. Dev Biol. 2003;263:81–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wenick AS, Hobert O. Genomic cis-regulatory architecture and trans-acting regulators of a single interneuron-specific gene battery in C.elegans. Dev Cell. 2004; 6: 757–770. 10.1016/j.devcel.2004.05.004 [DOI] [PubMed] [Google Scholar]
- 40.Faumont S, Boulin T, Hobert O, Lockery SR. Developmental regulation of whole cell capacitance and membrane current in identified interneurons in C.elegans. J Neurophysiol. 2006; 95: 3665–3673. 10.1152/jn.00052.2006 [DOI] [PubMed] [Google Scholar]
- 41.Hobert O, Mori I, Yamashita Y, Honda H, Ohshima Y, Liu Y, et al. Regulation of interneuron function in the C.elegans thermoregulatory pathway by the ttx-3 LIM homeobox gene. Neuron 1997;19: 345–357. [DOI] [PubMed] [Google Scholar]
- 42.VanDuyn N, Settivari R, Wong G, Nass R. SKN-1/Nrf2 inhibits dopamine neuron degeneration in a Caenorhabditis elegans model of methylmercury toxicity. Toxicol Sci. 2010; 118: 613–624. 10.1093/toxsci/kfq285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altun-Gultekin Z, Andachi Y, Tsalik EL, Pilgrim D, Kohara Y, Hobert O. A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans. Development 2011;128: 1951–1969. [DOI] [PubMed] [Google Scholar]
- 44.Remy JJ, Hobert O. An interneuronal chemoreceptor required for olfactory imprinting in C. elegans. Science 2005; 309: 787–790. 10.1126/science.1114209 [DOI] [PubMed] [Google Scholar]
- 45.Bulow HE, Berry KL, Topper LH, Peles E, Hobert O. Heparan sulfate proteoglycan-dependent induction of axon branching and axon misrouting by the Kallmann syndrome gene kal-1. Proc Natl Acad Sci USA. 2002; 99: 6346–6351. 10.1073/pnas.092128099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rugarli EI, Di Sciavi E, Hillard MA, Arbucci S, Ghezzi C, Faccioli A, et al. The Kallmann syndrome gene homolog in C. elegans is involved in epidermal morphogenesis and neurite branching. Development 2002;129: 1283–1294. [DOI] [PubMed] [Google Scholar]
- 47.Kaspar JW, Niture SK, Jaiswal AK. Nrf2:INrf2 (Keap1) signaling in oxidative stress. Free Radic Biol Med 2009; 47:1304–1309. 10.1016/j.freeradbiomed.2009.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mattson MP. Hormesis defined. Ageing Res Rev 2008; 7:1–7. 10.1016/j.arr.2007.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Done AJ, Traustadóttir T. Nrf2 mediates redox adaptations to exercise. Redox Biol 2016; 10:191–199. 10.1016/j.redox.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Satoh T, McKercher SR, Lipton SA. Nrf2/ARE-mediated antioxidant actions of -electrophilic drugs. Free Radic Biol Med 2013; 65:645–57. 10.1016/j.freeradbiomed.2013.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mattson MP. WHAT DOESN'T KILL YOU…. Sci Am. 2015; 313:40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee J, Jo DG, Park D, Chung HY, Mattson MP. Adaptive cellular stress pathways as therapeutic targets of dietary phytochemicals: focus on the nervous system. Pharmacol Rev. 2014; 66:815–868. 10.1124/pr.113.007757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hunt PR, Son TG, Wilson MA, Yu QS, Wood WH, Zhang Y, et al. Extension of lifespan in C. elegans by naphtoquinones that act through stress hormesis mechanisms. PLoS One 2011;6:e21922 10.1371/journal.pone.0021922 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Percentage of N2 and skn-1(zu67) animals remaining at the origin in presence of the indicated concentrations of benzaldehyde. (B) N2 and skn-1(zu135) worms display identical locomotion abilities. Animals were placed on an empty plate and allowed to crawl freely for 60 min. Their position was scored and the percentage of animals in the center (Zone 1) and periphery (Zone 2) determined. Data are mean and S.E.M. * p<0.05 versus N2.
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.