Abstract
Streptococcus gallolyticus subsp. gallolyticus was identified in humans and animals as commensal of the gut and can act as a causative agent of endocarditis and septicemia. A case-control study was performed to identify yet unknown risk factors for the transmission of this facultative pathogen. The prevalence in the gut of 99 healthy volunteers was determined using real-time polymerase chain reaction resulting in 62.5% S. gallolyticus subsp. gallolyticus positive excrements. Subsequent cultivation offered three isolates and epidemiological analysis based on MLST revealed sequence type (ST) 3 and ST 7, previously detected from bovine and endocarditis patients. These results support the hypotheses of the zoonotic potential of this bacterium. Participant questionnaires were evaluated concerning personal characteristics, nutritional habits and animal contact. Specifically, closer contact between participants and animals influenced the colonization of the human gut significantly and was further affected if volunteers used excrement for the fertilization of plants.
Introduction
Streptococcus gallolyticus subsp. gallolyticus, formally known as Streptococcus bovis biotype I, belongs to the Lancefield group D Streptococci [1], is a normal inhabitant of the animal and human gastrointestinal tract, and appears in 2.5 to 15% of healthy humans [2]. On the contrary, its frequency in the digestive tract of animals and the absolute frequencies in various species are not well described. To date, there is only one study, which estimated the percentage of S. gallolyticus subsp. gallolyticus in feces of turkeys. The detection rate in fecal samples of turkeys is 91% [3]. It was also identified in pigeon, bovine and chicken as commensal bacterium [4–6]. However, S. gallolyticus subsp. gallolyticus can also act as a facultative pathogen, causing sepsis, meningitis and infective endocarditis (IE) in humans and animals [6–8]. Human IE is especially associated with colorectal cancer [9–11]. The incidence of group D Streptococcus-associated diseases is increasing in the south of Europe [12]. The detection in humans and animals as a causative agent producing the same clinical symptoms leads to the assumption that S. gallolyticus subsp. gallolyticus may be a zoonotic pathogen [13]. Investigations in France and Spain suggest a correlation between a rural residency and the presence of the facultative pathogen [14,15]. The transmission of the potential zoonotic pathogen may be directly by smear or droplet infections or indirectly from surfaces contaminated with S. gallolyticus subsp. gallolyticus [16]. The transfer of the bacterium through a closer contact with colonized or infected animals is also discussed as a possible mechanism [13,14]. It was described as an important risk factor for the transmission of Streptococcus suis between infected animals and humans [17,18]. Epidemiologic analyses in a laying hen flock in North Rhine Westphalia also contribute to the assumption that a closer occupational contact with colonized laying hens may be a potential risk factor for the colonization of the gastrointestinal tract with S. gallolyticus subsp. gallolyticus, since the bacterium was identified as the causative agent of IE of the farm owner [13]. In addition to the detection of S. gallolyticus subsp. gallolyticus in eukaryotic organisms, it was also identified in milk and raw milk products (especially in dairy cows with mastitis) and red meat [14,19–21]. The detection in food leads to the assumption that the transmission of S. gallolyticus subsp. gallolyticus between animals and humans can be connected to dietary habits. Exemplarily, Streptococcus equi subsp. zooepidemicus was transmitted through the consumption of unpasteurized raw milk in an outbreak setting [22]. There have been no investigations to date which systematically analyze the correlation between dietary habits or the contact with animals and the detection of S. gallolyticus subsp. gallolyticus in the human gut. Therefore, we conducted an epidemiological study which is comprised of two parts: Firstly, the case-control study to determine the prevalence of S. gallolyticus subsp. gallolyticus and the associated risk factors for the colonization of the human gut, and secondly, multilocus sequence typing (MLST) to characterize the S. gallolyticus subsp. gallolyticus population structure. This analysis identified a correlation between lifestyle habits and the human gastrointestinal colonization with S. gallolyticus subsp. gallolyticus.
Material and methods
Sample and data acquisition
A retrospective case-control study was conducted at the Herz- und Diabeteszentrum Nordrhein-Westfalen (Bad Oeynhausen, Germany) from December 2012 to July 2015. The case-control study used word of mouth to recruit people. All data are collected in pseudonymous form. A total of 135 volunteers from the north and west of Germany participated in this study. Written consent was required for the case-control study. Fecal samples were tested and a questionnaire was completed by each volunteer to analyze the correlation between the fecal presences of S. gallolyticus subsp. gallolyticus and potential risk factors. Furthermore, seven SGG-culture-positive tested healthy volunteers were selected and were analyzed two to three times to estimate the gastrointestinal presence of S. gallolyticus subsp. gallolyticus in a follow-up period (follow-up study). Participants were excluded from the study if there was no fecal sample, no completed questionnaire or no written consent. In addition, only healthy volunteers (without gastrointestinal diseases or IE) over 18 years were included for the identification of risk factors. These data were received from the questionnaires. People were also excluded if an antibiotic therapy was indicated six months prior to participation.
The study was approved by the ethics commission of the Ruhr University Bochum Faculty of Medicine.
Stool investigations
DNA extraction
DNA extraction of homogenized fecal samples was performed by using NucliSENS easyMAG (Biomerieux, Nürtingen, Germany). DNA extraction was generally performed according to the manufacturer’s instruction. The fecal samples were pretreated by inoculating material (about 0.1 g) in 1 ml PBS in a tube with Zirconia beads, mixed for 5 min, incubated for 10 min at room temperature and then centrifuged at 12000 × g for 2 min. A quantity of 200 μl of the supernatant was used for the extraction of the whole DNA. After prelysis within the NucliSENS easyMAG, 100 μl magnetic silica particles were added and extraction was performed as described by the manufacturer. DNA was eluted in 55 μl elution buffer.
Real-Time PCR
The detection of an internal fragment of the recN gene was used to screen fecal specimens for the detection of S. gallolyticus subsp. gallolyticus [23]. The PCR amplification for the presence or absence of this gene was carried out within a 50 μl reaction volume containing 5 μl template DNA, 5 μl Platinum-Taq-buffer (ThermoFisherScientific, Darmstadt, Germany), 200 nM of each primer (F-recN SGG/P: 5’-GATTTTCAAGTCCAATTCACCAAAG-3’, R-recN SGG/P: 5’-GGTTYGTTGAAATGTAAAATTCAACAG-3’; LifeTechnologies, Darmstadt, Germany), 100 nm of the Pf-recN/SGG-probe (5’-FAM-TTCAATCGTGATGGCAA-MGB-3’; LifeTechnologies, Darmstadt), 240 μM dNTPs (Fermentas, Leon-Rot, Germany) and 0.25 μl Platinum-Taq-polymerase (ThermoFisherScientific, Darmstadt). The detection of S. gallolyticus subsp. pasteurianus was performed using the Pv-recN/SGP-probe (5’-VIC-TCAACCGTGATGGAAA-MGB-3’) and the same primers denoted above [23]. The internal control used in the reaction mix was CMV-DNA (CMV-TM2-F: TTYTTAGCACGGGCCTTAGC, CMV-TM2-R: AAGGAGCTGCATGATGTGASC; CMV-TM2-S: CY5-TGCAGTGCACCCCCCAACTTGTT-BHQ2; [24]). Diluted DNA extracted from a bacterial overnight culture of S. gallolyticus subsp. gallolyticus (ATCC BAA-2069) or S. gallolyticus subsp. pasteurianus (DSM 15351) was used as positive control and water as negative control to verify the specificity of the PCR reaction. A two-step PCR on the Rotor Gene Q platform (Qiagen, Hilden, Germany) was performed. Amplification of PCR products was carried out as follows: initial denaturation at 95°C (5 min) followed by 50 cycles, and a denaturation 95°C (15 s), annealing and elongation step at 60°C (60 s).
Selective cultivation
Real-time PCR S. gallolyticus subsp. gallolyticus positive-tested fecal samples were further selectively cultivated on modified trypton soya agar (TSA) (0.5% tannic acid pure (AppliChem GmbH, Darmstadt, Germany, 0.25 g/l sodium acetate, Merck, Darmstadt, Germany), as described previously [3]. Briefly, the homogenized fecal sample was streaked out onto selective medium before weighing and suspending in PBS buffer. Then, 1 g of homogenized fecal sample was suspended in 1 ml PBS medium, mixed and streaked out with PBS (duplicate) and 100 μl was plated as triplicate onto sodium acetate tannic acid TSA. It was then incubated at 37°C and 5% CO2 for 48 h [3]. In parallel, an overnight grown culture of S. gallolyticus subsp. gallolyticus was plated onto modified TSA. Single putative S. gallolyticus subsp. gallolyticus colonies were selected and analyzed regarding species and subspecies level by matrix-assisted laser desorption ionization—time of flight mass spectrometry (MALDI-TOF-MS) and sodA sequencing [25].
Multilocus sequence typing
Multilocus sequence typing was performed, as described preciously [16]. In brief, the total DNA of S. gallolyticus subsp. gallolyticus isolates was isolated by using a QIAamp Blood Mini Kit (Qiagen, Hilden, Germany) and 5 μl was used for each fragment amplification [16]. Partial sequences of the housekeeping genes aroE, glgB, nifS, p20, tkt, trpD and uvrA were amplified, sequenced and analyzed [16]. All detailed protocols can also be found on www.pubmlst.org [16]. The determination of sequence types (STs) was undertaken using the pubMLST database and Bionumerics Software 6.6 (Applied Maths, Sint-Martens-Latem, Belgium) [16,26]. For the characterization of the strains a minimum spanning tree was generated and eBURST version 3 (based upon related sequence types; www.mlst.net) was used to calculate clonal complexes [16,27].
Questionnaire
The questionnaire included 25 questions and sought data on the following aspects: personal characteristics (age, gender, gastrointestinal diseases, residence [urban, rural, landscape—near the forest/farm] and antibiosis), contact with animals (living or working on a farm, private or occupational contact) and dietary habits (consumption and handling of minced meat, raw milk or raw milk products). The exposure factors as well as the absolute frequencies can be found in S1 Table.
Statistical analysis
The statistical analysis software SPSS version 21 was used. Binary logistic regression was utilized to establish a model to determine the simultaneous influence of potential risk factors. Six independent variables (age, gender, consumption of raw animal products, close animal contact, usage of animal waste) were tested within the multiple logistic regression model to verify adjusted odds ratios (ORs). Statistical tests were considered to be significant if the p-value was less than 0.05. A confidence interval of 95% was used for both calculations. Forest plots were generated using Microsoft Excel. The age was listed as mean plus/minus standard deviation.
Results
A total of 134 participants (65 men and 69 women with a mean age of 48.4 ± 14.9 years) were included in the case-control study. After the application of exclusion criteria, 99 healthy volunteers were included to identify potential risk factors for the colonization of the human gut with S. gallolyticus subsp. gallolyticus. The fecal samples of healthy volunteers were screened by PCR for the presence of the facultative pathogen. S. gallolyticus subsp. gallolyticus was detected in 62.5% (n = 59) of the 99 fecal specimens of healthy volunteers. The presence of S. gallolyticus subsp. pasteurianus was also estimated using the VIC labeled probe in the PCR reaction mix. This bacterium was detected seven times out of 99 volunteers. Real-time PCR testing also identified three subjects who were colonized with S. gallolyticus subsp. gallolyticus as well as S. gallolyticus subsp. pasteurianus simultaneously. These volunteers are recognized in the S. gallolyticus subsp. gallolyticus-positive group.
S. gallolyticus subsp. gallolyticus PCR-positive specimens were cultured on selective medium to isolate this bacterium for epidemiologic characterization by MLST. Three isolates, namely HDZ 1323, HDZ 1330 and HDZ 1332, were detected by culture and mass spectrometric analyses, and sodA sequencing confirmed S. gallolyticus subsp. gallolyticus. In addition to S. gallolyticus subsp. gallolyticus, three out of seven S. gallolyticus subsp. pasteurianus isolates were identified by MALDI-TOF MS and sequencing of the partial fragment of the sodA gene. In this regard, the real-time PCR demonstrated one inconsistent result: S. gallolyticus subsp. gallolyticus was detected instead of S. gallolyticus subsp. pasteurianus.
The S. gallolyticus subsp. gallolyticus isolates selected were further typed using MLST. It revealed the sequence types ST 3 (HDZ1330), ST 7 (HDZ1323) and the newly defined ST 105 (HDZ1332). Bionumerics Software 6.6 was used to utilize for the construction of a minimum spanning tree (S1 Fig). The minimum spanning tree of the strains revealed no phylogenetic relationship of these detected STs (S1 Fig). The allelic profiles (STs) show no identical allelic numbers within the identified sequence types of the case control study (one exception: the number of the nifS allele from ST 3 and 7). The ST 3 was already identified in human heart valve cultures and from the intestine of a bovine, which was also detected for ST 7 isolates ([16], www.pubmlst.org).
In order to identify the S. gallolyticus subsp. gallolyticus status in the gastrointestinal tract over time, a follow-up investigation of seven culture positive tested healthy volunteers was performed until the end of the study (a total of 2 to 3 samples per person) (Table 1). Initially, six fecal specimens were screened as real-time PCR positive for S. gallolyticus subsp. gallolyticus and one sample was tested as positive for S. gallolyticus subsp. pasteurianus. Selective cultivation offered three S. gallolyticus subsp. gallolyticus and one S. gallolyticus subsp. pasteurianus isolate. Further analyses of specimens revealed S. gallolyticus subsp. gallolyticus in four cases at any tested time point by using real-time PCR (volunteer 4 to 7). As an example, the first (January 2015) and second sample (March 2015) were tested as positive and the third specimen in July 2015 was tested as negative (volunteer 3). The fecal sample of the 7th volunteer was initially tested as positive (April 2013) and the second sample 26 months later was detected as positive for the presence of S. gallolyticus subsp. gallolyticus (Table 1). At least one exception was identified. The PCR results of the second sample from volunteer 1 showed the presence of both subspecies, and S. gallolyticus subsp. pasteurianus was isolated using modified trypton soya agar (Table 1).
Table 1. Follow-up study of healthy volunteers.
| Fecal sample 1 | Fecal sample 2 | Fecal sample 3 | ||||
|---|---|---|---|---|---|---|
| Real-time PCR | Selective cultivation | Real-time PCR | Selective cultivation | Real-time PCR | Selective cultivation | |
| Sample date | Sample date | Sample date | ||||
| Volunteer 1 | SGP | SGP | SGP, SGG | SGP | - | |
| December 2014 | March 2015 | |||||
| Volunteer 2 | SGG | SGG | negative | negative | negative | negative |
| November 2014 | March 2015 | July 2015 | ||||
| Volunteer 3 | SGG | SGG | SGG | negative | negative | negative |
| January 2015 | March 2015 | July 2015 | ||||
| Volunteer 4 | SGG | SGG | SGG | negative | SGG | negative |
| December 2014 | March 2015 | July 2015 | ||||
| Volunteer 5 | SGG | Negative | SGG | Negative | - | |
| November 2014 | March 2015 | |||||
| Volunteer 6 | SGG | negative | SGG | negative | SGG | negative |
| December 2014 | March 2015 | - | ||||
| Volunteer 7 | SGG | negative | SGG | negative | - | |
| April 2013 | Juni 2015 | |||||
SGG: S. gallolyticus subsp. gallolyticus; SGP: S. gallolyticus subsp. pasteurianus
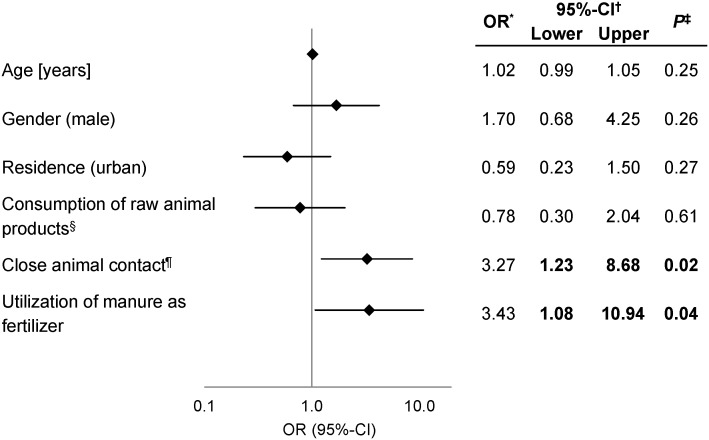
Based on the real-time PCR detection of the bacterium in fecal samples, cases and controls were defined: 59 cases (male/female [m/f] ratio: 25/34) and 40 controls (m/f ratio: 21/19). The cases included an age distribution from 20 to 70 years with a mean value of 44.2 ± 14.6 years and controls from 22 to 82 years with an average of 49.2 ± 15.0 years. The cases and controls were analyzed in terms of their nutrition habits, contact with animals and residence (rural, urban; forest or farm next to their residence). The frequencies of potential risk factors observed in cases and controls are listed in S1 Table. Logistic regression was performed to analyze the simultaneous effect of risk factors and adjusted ORs were calculated and presented as a forest plot (Fig 1).
Fig 1. Binary logistic regression model of exposure factors for the colonization of the human gastrointestinal tract.
Potential risk factors discussed in the literature were included in the logistic regression model and adjusted ORs* were calculated with a confidence interval (CI†) of 95%. The x-axis is displayed logarithmically. P-values were calculated using the chi-square test (‡). Results were significant if a p-values less than 0.05 was detected or the 95%-CI does not include 1 and were indicated in bold. § raw minced meat, raw milk and raw milk products; ¶ Contact with excrement or saliva of animals, striking of animals.
Multiple expositions often characterize the outcome of a disease or different event such as the S. gallolyticus subsp. gallolyticus colonization of the human gastrointestinal tract. Adjusted ORs were calculated to assess the simultaneous effect of the variables. A closer animal contact between volunteers and animals (OR: 3.27, CI: 1.23–8.68; p = 0.02) and the usage of animal waste to fertilize plants (OR: 3.43, CI: 1.08–10.94; p = 0.04) demonstrate significant risk factors for the transmission between animals and humans and to colonize the gastrointestinal tract of healthy people (Fig 1).
In conclusion, simultaneous testing of exposure factors indicate a higher risk of being colonized with S. gallolyticus subsp. gallolyticus of participants who have a direct contact with animals and utilization of manure. Furthermore, two out of three S. gallolyticus subsp gallolyticus isolates reveal the STs 3 and 7. These STs were previously isolated from human blood cultures and bovine ([16], www.pubmlst.org).
Discussion
The knowledge of transmission pathways and the zoonotic potential of the facultative pathogen S. gallolyticus subsp. gallolyticus are quite limited. Thus, a systematic approach was conducted for the first time to determine the latter’s occurrence in the gut of healthy people and to describe the risk factors for the transmission of the bacterium and its colonization of the human gastrointestinal tract. S. gallolyticus subsp. gallolyticus is an opportunist of the gastrointestinal tract in humans and animals with varying prevalence in the healthy human population of 2.5 up to 15% [2], but the S. bovis fecal carriage increased three to five times in patients with colorectal cancer and inflammatory bowel disease [8,10,28,29]. In comparison to the previous studies real-time PCR screenings of fecal samples offer a much higher prevalence in healthy volunteers of 62.5%. Spanish real-time PCR investigations of patients who underwent colonoscopy revealed 11.1% S. gallolyticus subsp. gallolyticus-positive and 13% S. gallolyticus subsp. pasteurianus-positive rectal swabs. It indicates a similar portion of both subspecies in the gastrointestinal tract [23], which cannot be confirmed by real-time PCR screenings of feces from participants of the case-control study. Similar results were detected for the presence of S. gallolyticus subsp. pasteurianus. The detection of both subspecies using real-time PCR may indicate a co-occurrence in the digestive tract, which was also suggested by Lopes et al. [23].
The divergences observed between previous studies may be a resumé of the sample sets analyzed (colonoscopy [30], feces [28], rectal swabs [23]) or the kind of screening methods (cultivation [30,31], molecular techniques [23,32]) to identify or isolate S. gallolyticus subsp. gallolyticus. A higher sensitivity of the molecular screening method was demonstrated by positive real-time PCR results in comparison to selective cultivation and was also confirmed in the follow-up study. The complexity and characteristics of the sample type and difficulties in homogenization as well as the gut microbiota may influence the PCR results (e. g. inhibitors) and the selective cultivation [33]. It explains not only false negative real-time PCR results, but also discrepancies between S. gallolyticus subsp. gallolyticus-positive culture and the S. gallolyticus subsp. pasteurianus-positive real-time PCR screening results. As suggested in a 17-year follow-up study [30], the prospective investigations of seven participants achieved shifts in the composition of the gut microbiome, which was e. g. demonstrated for the participant three. Nutrition and medicines shape the gut microflora (e. g. antibiotics), whereas antibiotics change the gut composition up to one year [34–36]. This may also be transferred to our follow-up study. Although the participants with an antibiotic therapy were excluded it is not known if the composition of the gut microbiome affects the presence of S. gallolyticus subsp. gallolyticus in the gut. However, the sample age may be of more importance than the sample characteristics, microbiota or processing. The fecal samples were sent to the laboratory by mail. Consequently, samples could be in transit for three days before processing in the laboratory. Although a survival of S. gallolyticus subsp. gallolyticus was demonstrated in vitro in S. gallolyticus subsp. gallolyticus-negative tested human stool specimens for 14 d at RT (20°C) (real-time PCR and cultivation; data not shown) it cannot be ruled out, that the growth of other gastrointestinal bacteria may inhibit growth of S. gallolyticus subsp. gallolyticus or a less concentration of S. gallolyticus subsp. gallolyticus in the feces may effect a false negative cultivation. It is assumed that the presence of the same diseases in animals and humans may be a hint that S. gallolyticus subsp. gallolyticus is a zoonotic pathogen [16]. This was supported by MLST, which typed a blood culture isolate of an animal farmer and excrements of the chicken of his laying hen farm with the same ST [13]. Interestingly, the human fecal isolates which were identified in this case-control study were differentiated into the STs 3 and 7 and the new identified ST 105. ST3 and 7 were associated with human blood culture isolates and were also identified in cattle (unknown infective status) [16]. Thus, these results support the potential transmission of S. gallolyticus subsp. gallolyticus between animals and humans and highlight the zoonotic potential of the facultative pathogen. As described previously, it might be the case, as there seems to be some STs (as is the case of ST 7 or ST 3) that seem more associated with humans or animals whereas other isolates are more predominant in animals and humans (e. g. isolates of the clonal complex 45 and 6) [16]. A prevalence of 62.5% in human feces described in this study and a high prevalence in organic turkey flocks give rise to the question if S. gallolyticus subsp. gallolyticus belongs to the common gut microbiome of animals and humans [3]. However, for this conclusion further systematic analyses have to be performed.
Among other diseases, S. gallolyticus subsp. gallolyticus causes IE and is associated with colorectal diseases in humans [10,11,37–40]. Both diseases are generally more often observed in male patients over 50 years old [41,42] and various studies demonstrated the same positive association between the isolation of S. gallolyticus subsp. gallolyticus in men and the elderly population and IE [15,43–47], which was not identified in this study (Fig 1). To identify a relationship between the one-year increasing age and the detection of the bacterium in the digestive tract more people have to be tested.
The facultative pathogen is the most frequently detected agent in cases of infective endocarditis in rural regions (especially in the cattle and milk production area) in the south of Europe [12,14,15], which cannot be observed in this case control study (Fig 1) and should be figured out in further investigations. Joined together with living in the countryside, a close contact with animals was supposed to be a transmission source of the bacterium [8,14], it was demonstrated for the first time that a closer contact with animals is a significant exposure factor to be colonized with S. gallolyticus subsp. gallolyticus. It can be assigned as risk factors for the transmission of S. gallolyticus subsp. gallolyticus between animals and humans and its establishment in the human gut. Another interesting fact is the kind of animal species which come into contact with people. Although dogs and horses are described in the literature as a source of isolation and are common pets [1,5], it is not known whether these animals belonging to the volunteers are colonized with the bacterium. In this context, it is remarkable that fertilization of plants with the excrement of animals increases the risk of carrying S. gallolyticus subsp. gallolyticus significantly. Consequently, it is imaginable that S. gallolyticus subsp. gallolyticus may be transferred directly from animals to humans by smear infections and colonizes the gastrointestinal gut and, thus, can be accounted as a significant risk factor. However, the prevalence of S. gallolyticus subsp. gallolyticus in animals is still unknown [3]. The bacterium has often been identified in, for example, pigeons, chicken and turkeys [3–6,13,48,49]. Therefore, future studies should also include investigations of animal excrement in addition to human specimens to verify the prevalence in animals, too, and to estimate the real risk to human health.
Derived from this hypothesis, not only the animal contact, but also the consumption of or the contact with contaminated food, such as red meat or milk products, may promote the colonization of the human gut and are propagated as exposure factors for the transmission of the bacterium from animals to humans [15]. In total, statistical analyses demonstrate that raw food products play a minor role the colonization process. It was assumed that eating raw minced meat may promote the colonization of the human gut [14]. More interestingly, because of the high frequencies of isolation in turkeys and laying hens [3,13], the consumption and processing of poultry and eggs and its function as a transmission source from animals to humans should be focused on in following research perspectives and participants should be ask about their habits in terms of processing and eating chicken meat and eggs. At the beginning of this case-control study the high prevalence was not known. The transmission from poultry to humans is well-known for Campylobacter. The main significant risk factor for the transmission of this species to humans is particularly bought fresh chicken (OR 5.80; 95%-CI: 2.11–15.93), whereby it was decreased by eating fruit, raw vegetables, high-fiber cereals, vitamins and acidified milk products [50].
In summary, the case-control study conducted demonstrates a very high prevalence of S. gallolyticus subsp. gallolyticus in the gastrointestinal tract of healthy volunteers. In accordance with other researchers [8], it is essential to determine at least the subspecies or the biotype of the S. bovis strains to establish the identification of the frequency of S. gallolyticus in correlation with colorectal cancer, IE and other diseases, as well as its global impact to assess the risk to the human population. The data collected were evaluated with the help of multivariate statistical analyses to identify risk factors for the colonization of the human gut with the facultative pathogen. The simultaneous observation of exposure factors identified the closer contacts with animals and the usage of animals waste as significant risk factors for the detection of S. gallolyticus subsp. gallolyticus in human feces. Further investigations have to be performed to clarify the impact of chicken meat products and protective factors, such as vegetables. In addition, future studies should also include participants with e. g. gastrointestinal disorders associated with S. gallolyticus subsp. gallolyticus to detect the zoonotic pathogenicity of the Gram-positive bacterium to the health status of animals and humans and to determine the rate of S. gallolyticus subsp. gallolyticus fecal colonization. Another approach would be a prospective cohort study of people carrying S. gallolyticus subsp. gallolyticus or modifications of the gut microbiome along with environmental factors to detect the time-dependent influences on human health.
In addition to the detected potential risk factors, the vitality outside the gastrointestinal tract is also important for the direct or indirect transmission of the bacterium between animals and humans or between the environment and animals or humans, and should be pointed out in future studies.
Supporting information
Based on the allelic profile an Minimum spanning tree (MST) was constructed and clonal complexes (CC) were calculated by use of eBURST. CCs are presented as black lines. Each circle represents a ST and the size corresponds with number of bacterial isolates included. The STs of the case-control are presented as red dotted lines.
(TIF)
(DOCX)
Acknowledgments
This work was supported by Ruhr-Universität Bochum Medizinische Fakultät (FoRUM). We thank Philip Saunders (LL.B., B.Ed., FRSA) for his linguistic advice.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Forschungsförderung Ruhr-Universität Medizin (FoRUM). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Schlegel L, Grimont F, Ageron E, Grimont PA, Bouvet A. Reappraisal of the taxonomy of the Streptococcus bovis / Streptococcus equinus complex and related species: description of Streptococcus gallolyticus subsp. gallolyticus subsp. nov., S. gallolyticus subsp. macedonicus subsp. nov. and S. gallolyticus subsp. pasteurianus subsp. nov. International Journal of Systematic and Evolutionary Microbiology. Soc General Microbiol; 2003;53(3):631–45. [DOI] [PubMed] [Google Scholar]
- 2.Sillanpää J, Nallapareddy SR, Qin X, Singh KV, Muzny DM, Kovar CL, et al. A collagen-binding adhesin, Acb, and ten other putative MSCRAMM and pilus family proteins of Streptococcus gallolyticus subsp. gallolyticus (Streptococcus bovis Group, biotype I). Journal of Bacteriology. Am Soc Microbiol; 2009;191(21):6643–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schulz J, Dumke J, Hinse D, Dreier J, Habig C, Kemper N. Organic Turkey Flocks: A Reservoir of Streptococcus gallolyticus subspecies gallolyticus. PloS one. Public Library of Science; 2015;10(12):e0144412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Devriese L, Uyttebroek E, Gevaert D, Vandekerckhove P, Ceyssens K. Streptococcus bovis infections in pigeons. Avian Pathology. Taylor \& Francis; 1990;19(3):429–34. [DOI] [PubMed] [Google Scholar]
- 5.Sasaki E, Osawa R, Nishitani Y, Whiley RA. ARDRA and RAPD analyses of human and animal isolates of Streptococcus gallolyticus. Journal of Veterinary Medical science. Japanese Society of Veterinary Science; 2004;66(11):1467–70. [DOI] [PubMed] [Google Scholar]
- 6.Sekizaki T, Nishiya H, Nakajima S, Nishizono M, Kawano M, Okura M, et al. Endocarditis in chickens caused by subclinical infection of Streptococcus gallolyticus subsp. gallolyticus. Avian Diseases. 2008;52(1):183–6. 10.1637/8048-070307-Case [DOI] [PubMed] [Google Scholar]
- 7.Headings DL, Herrera A, Mazzi E, Bergman MA. Fulminant neonatal septicemia caused by Streptococcus bovis. The Journal of Pediatrics. Mosby; 1978;92(2):282–3. [DOI] [PubMed] [Google Scholar]
- 8.Boleij A, Tjalsma H. The itinerary of Streptococcus gallolyticus infection in patients with colonic malignant disease. The Lancet Infectious Diseases. Elsevier; 2013; [DOI] [PubMed] [Google Scholar]
- 9.Abeni C, Rota L, Ogliosi C, Bertocchi P, Centurini PB, Zaniboni A. Correlation among Streptococcus bovis, endocarditis and septicemia in a patient with advanced colon cancer: a case report. Journal of Medical Case Reports. BioMed Central Ltd; 2013;7(1):185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein RS, Recco RA, Catalano MT, Edberg SC, Casey JI, Steigbigel NH. Association of Streptococcus bovis with carcinoma of the colon. New England Journal of Medicine. Mass Medical Soc; 1977;297(15):800–2. [DOI] [PubMed] [Google Scholar]
- 11.Boleij A, Schaeps RM, Tjalsma H. Association between Streptococcus bovis and colon cancer. Journal of Clinical Microbiology. Am Soc Microbiol; 2009;47(2):516–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoen B, Chirouze C, Cabell CH, Selton-Suty C, Duchene F, Olaison L, et al. Emergence of endocarditis due to group D streptococci: findings derived from the merged database of the International Collaboration on Endocarditis. European Journal of Clinical Microbiology and Infectious Diseases. Springer; 2005;24(1):12–6. [DOI] [PubMed] [Google Scholar]
- 13.Dumke J, Hinse D, Vollmer T, Schulz J, Knabbe C, Dreier J. Potential Transmission Pathways of Streptococcus gallolyticus subsp. gallolyticus. PLoS ONE 10(5): e0126507 2015; 10.1371/journal.pone.0126507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corredoira J, Grau I, Garcia-Rodriguez JF, Alonso-Garcia P, Garcia-Pais M, Rabuñal R, et al. The Clinical Epidemiology and Malignancies Associated with Streptococcus bovis Biotypes in 506 Cases of Bloodstream Infections. Journal of Infection. Elsevier; 2015; [DOI] [PubMed] [Google Scholar]
- 15.Giannitsioti E, Chirouze C, Bouvet A, Béguinot I, Delahaye F, Mainardi J-L, et al. Characteristics and regional variations of group D streptococcal endocarditis in France. Clinical Microbiology and Infection. Wiley Online Library; 2007;13(8):770–6. [DOI] [PubMed] [Google Scholar]
- 16.Dumke J, Hinse D, Vollmer T, Knabbe C, Dreier J. Development and Application of a Multilocus Sequence Typing Scheme for Streptococcus gallolyticus subsp. gallolyticus. Journal of Clinical Microbiology. Am Soc Microbiol; 2014;52 (7):2472–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Francois B, Gissot V, Ploy M, Vignon P. Recurrent septic shock due to Streptococcus suis. Journal of Clinical Microbiology. Am Soc Microbiol; 1998;36(8):2395–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lun Z-R, Wang Q-P, Chen X-G, Li A-X, Zhu X-Q. Streptococcus suis: an emerging zoonotic pathogen. The Lancet Infectious Diseases. Elsevier; 2007;7(3):201–9. [DOI] [PubMed] [Google Scholar]
- 19.Fortin M, Messier S, Paré J, Higgins R. Identification of catalase-negative, non-beta-hemolytic, gram-positive cocci isolated from milk samples. Journal of Clinical Microbiology. Am Soc Microbiol; 2003;41(1):106–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Randazzo CL, Vaughan EE, Caggia C. Artisanal and experimental Pecorino Siciliano cheese: microbial dynamics during manufacture assessed by culturing and PCR-DGGE analyses. International Journal of Food Microbiology. Elsevier; 2006;109(1):1–8. [DOI] [PubMed] [Google Scholar]
- 21.Tsakalidou E, Zoidou E, Pot B, Wassill L, Ludwig W, Devriese L, et al. Identification of streptococci from Greek Kasseri cheese and description of Streptococcus macedonicus sp. nov. International Journal of Systematic Bacteriology. Soc General Microbiol; 1998;48(2):519–27. [DOI] [PubMed] [Google Scholar]
- 22.Bordes-Benitez A, Sánchez-Oñoro M, Suárez-Bordón P, Garcia-Rojas A, Saéz-Nieto J, González-Garcia A, et al. Outbreak of Streptococcus equi subsp. zooepidemicus infections on the island of Gran Canaria associated with the consumption of inadequately pasteurized cheese. European Journal of Clinical Microbiology and Infectious Diseases. Springer; 2006;25(4):242–6. [DOI] [PubMed] [Google Scholar]
- 23.Lopes PGM, Cantarelli VV, Agnes G, Costabeber AM, d’ Azevedo PA. Novel Real-Time PCR Assays Using TaqMan Minor Groove Binder Probes for Identification of Fecal Carriage of Streptococcus bovis / Streptococcus equinus Complex from Rectal Swab Specimens. Journal of clinical microbiology. Am Soc Microbiol; 2014;52(3):974–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yun Z, Lewensohn-Fuchs I, Ljungman P, Vahlne A. Real-Time monitoring of Cytomegalovirus infections after stem cell transplantation using the TaqMan polymerase chain reaction assay1. Transplantation. LWW; 2000;69(8):1733–6. [DOI] [PubMed] [Google Scholar]
- 25.Hinse D, Vollmer T, Erhard M, Welker M, Moore E, Kleesiek K, et al. Differentiation of species of the Streptococcus bovis / equinus-complex by MALDI-TOF Mass Spectrometry in comparison to sodA sequence analyses. Systematic and Applied Microbiology. Elsevier; 2011;34(1):52–7. [DOI] [PubMed] [Google Scholar]
- 26.Jolley KA, Maiden MC. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. BioMed Central Ltd; 2010;11(1):595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. Journal of Bacteriology. Am Soc Microbiol; 2004;186(5):1518–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Al-Jashamy K, Murad A, Zeehaida M, Rohaini M, Hasnan J. Prevalence of colorectal cancer associated with Streptococcus bovis among inflammatory bowel and chronic gastrointestinal tract disease patients. Asian Pac J Cancer Prev. 2010;11(6):1765–8. [PubMed] [Google Scholar]
- 29.Shanan S, Gumaa SA, Sandstrӧm G, Abd H. Significant association of Streptococcus bovis with malignant gastrointestinal diseases. International journal of microbiology. Hindawi Publishing Corporation; 2011;2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boltin D, Goldberg E, Bugaevsky O, Kelner E, Birkenfeld S, Gingold-Belfer R, et al. Colonic carriage of Streptococcus bovis and colorectal neoplasia: a prospective 17-year longitudinal case-control study. European journal of gastroenterology \& hepatology. 2015; [DOI] [PubMed] [Google Scholar]
- 31.Chirouze C, Patry I, Duval X, Baty V, Tattevin P, Aparicio T, et al. Streptococcus bovis / Streptococcus equinus complex fecal carriage, colorectal carcinoma, and infective endocarditis: a new appraisal of a complex connection. European Journal of Clinical Microbiology & Infectious Diseases. Springer; 2013;1–6. [DOI] [PubMed] [Google Scholar]
- 32.Abdulamir AS, Hafidh RR, Bakar FA. Molecular detection, quantification, and isolation of Streptococcus gallolyticus bacteria colonizing colorectal tumors: inflammation-driven potential of carcinogenesis via IL-1, COX-2, and IL-8. Molecular Cancer. BioMed Central Ltd., Middlesex House London W 1 T 4 LB UK; 2010;9:249–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holland J, Louie L, Simor A, Louie M. PCR detection of Escherichia coli O157: H7 directly from stools: evaluation of commercial extraction methods for purifying fecal DNA. Journal of clinical microbiology. Am Soc Microbiol; 2000;38(11):4108–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. Nature Publishing Group; 2006;444(7122):1027–131. [DOI] [PubMed] [Google Scholar]
- 35.Sekirov I, Russell SL, Antunes LCM, Finlay BB. Gut microbiota in health and disease. Physiological reviews. Am Physiological Soc; 2010;90(3):859–904. [DOI] [PubMed] [Google Scholar]
- 36.Rashid M-U, Zaura E, Buijs MJ, Keijser BJ, Crielaard W, Nord CE, et al. Determining the long-term effect of antibiotic administration on the human normal intestinal microbiota using culture and pyrosequencing methods. Clinical Infectious Diseases. Oxford University Press; 2015;60(suppl 2):S77–S84. [DOI] [PubMed] [Google Scholar]
- 37.Vollmer T, Hinse D, Kleesiek K, Dreier J. Interactions between endocarditis-derived Streptococcus gallolyticus subsp. gallolyticus isolates and human endothelial cells. BMC Microbiology. BioMed Central Ltd; 2010;10(1):78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sillanpää J, Nallapareddy SR, Singh KV, Ferraro MJ, Murray BE. Adherence characteristics of endocarditis-derived Streptococcus gallolyticus ssp. gallolyticus (Streptococcus bovis biotype I) isolates to host extracellular matrix proteins. FEMS Microbiology Letters. Wiley Online Library; 2008;289(1):104–9. [DOI] [PubMed] [Google Scholar]
- 39.Boleij A, Muytjens CM, Bukhari SI, Cayet N, Glaser P, Hermans PW, et al. Novel clues on the specific association of Streptococcus gallolyticus subsp gallolyticus with colorectal cancer. Journal of Infectious Diseases. Oxford University Press; 2011;203(8):1101–9. [DOI] [PubMed] [Google Scholar]
- 40.Danne C, Entenza JM, Mallet A, Briandet R, Débarbouillé M, Nato F, et al. Molecular characterization of a Streptococcus gallolyticus genomic island encoding a pilus involved in endocarditis. Journal of Infectious Diseases. Oxford University Press; 2011;204(12):1960–70. [DOI] [PubMed] [Google Scholar]
- 41.Werdan K, Dietz S, Lӧffler B, Niemann S, Bushnaq H, Silber R-E, et al. Mechanisms of infective endocarditis: pathogen-host interaction and risk states. Nature Reviews Cardiology. Nature Publishing Group; 2013; [DOI] [PubMed] [Google Scholar]
- 42.Mager D. Bacteria and cancer: cause, coincidence or cure? A review. Journal of Translational Medicine. BioMed Central Ltd; 2006;4(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kupferwasser I, Darius H, Müller A, Mohr-Kahaly S, Westermeier T, Oelert H, et al. Clinical and morphological characteristics in Streptococcus bovis endocarditis: a comparison with other causative microorganisms in 177 cases. Heart. BMJ Publishing Group Ltd and British Cardiovascular Society; 1998;80(3):276–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoen B, Alla F, Selton-Suty C, Béguinot I, Bouvet A, Briançon S, et al. Changing profile of infective endocarditis: results of a 1-year survey in France. Jama. American Medical Association; 2002;288(1):75–81. [DOI] [PubMed] [Google Scholar]
- 45.Tripodi M-F, Fortunato R, Utili R, Triassi M, Zarrilli R. Molecular epidemiology of Streptococcus bovis causing endocarditis and bacteraemia in Italian patients. Clinical microbiology and infection. Wiley Online Library; 2005;11(10):814–9. [DOI] [PubMed] [Google Scholar]
- 46.Romero B, Morosini M-I, Loza E, Rodiguez-Baños M, Navas E, Cantón R, et al. Reidentification of Streptococcus bovis isolates causing bacteremia according to the new taxonomy criteria: still an issue? Journal of Clinical Microbiology. Am Soc Microbiol; 2011;49(9):3228–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coffey S, Nadarasa K, Pan A, van der Linden A, Chu J, Schultz M. The increasing incidence of Streptococcus bovis endocarditis and bacteraemia: A case series from 1997 to 2010. International journal of cardiology. Elsevier; 2012;161(2):111–3. [DOI] [PubMed] [Google Scholar]
- 48.Droual R, Ghazikhanian GY, Shivaprasad HL, Barr BC, Bland MB. Streptococcus bovis infection in turkey poults. Avian Pathol. 1997;26(2):433–9. 10.1080/03079459708419225 [DOI] [PubMed] [Google Scholar]
- 49.De Herdt P, Haesebrouck F, Ducatelle R, De Groote B, Devriese L. Streptococcus bovis infections in pigeons: virulence of different serotypes. Veterinary Microbiology. Elsevier; 1994;41(4):321–32. [DOI] [PubMed] [Google Scholar]
- 50.Wingstrand A, Neimann J, Engberg J, Nielsen EM, Gerner-Smidt P, Wegener HC, et al. Fresh chicken as main risk factor for campylobacteriosis, Denmark. Emerg Infect Dis. 2006;12(2):280–5. 10.3201/eid1202.050936 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Based on the allelic profile an Minimum spanning tree (MST) was constructed and clonal complexes (CC) were calculated by use of eBURST. CCs are presented as black lines. Each circle represents a ST and the size corresponds with number of bacterial isolates included. The STs of the case-control are presented as red dotted lines.
(TIF)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.